Abstract
The Rudiviridae are a family of rod-shaped archaeal viruses with covalently closed, linear double-stranded DNA (dsDNA) genomes. Their replication mechanisms remain obscure, although parallels have been drawn to the Poxviridae and other large cytoplasmic eukaryotic viruses. Here we report that a protein encoded in the 34-kbp genome of the rudivirus SIRV1 is a member of the replication initiator (Rep) superfamily of proteins, which initiate rolling-circle replication (RCR) of diverse viruses and plasmids. We show that SIRV Rep nicks the viral hairpin terminus, forming a covalent adduct between an active-site tyrosine and the 5′ end of the DNA, releasing a 3′ DNA end as a primer for DNA synthesis. The enzyme can also catalyze the joining reaction that is necessary to reseal the DNA hairpin and terminate replication. The dimeric structure points to a simple mechanism through which two closely positioned active sites, each with a single tyrosine residue, work in tandem to catalyze DNA nicking and joining. We propose a novel mechanism for rudivirus DNA replication, incorporating the first known example of a Rep protein that is not linked to RCR. The implications for Rep protein function and viral replication are discussed.
Archaeal viruses display tremendous diversity of both morphology and genome content. Seven viral families from the Crenarchaeota have been described, all with linear or circular double-stranded DNA (dsDNA) genomes but with few genes in common (28). The Rudiviridae are a family of nonenveloped rod-shaped viruses infecting thermophilic crenarchaea (27). Members include SIRV1 and SIRV2 (24), ARV1 (36), and SRV1 (37), all of which have covalently closed linear dsDNA genomes of 30 to 40 kb encoding up to 50 open reading frames (ORFs). The genome arrangement, including the presence of inverted terminal repeats (ITRs), is reminiscent of those of the large cytoplasmic DNA viruses, such as the Poxviridae (3, 10). Replication of the poxvirus genome, which occurs in cytoplasmic DNA replication factories (29), results in the generation of head-to-head concatemers that are resolved by a Holliday junction-resolving enzyme (7, 12). The Rudiviridae also generate head-to-head intermediates, ruling out a rolling-circle mechanism of replication (24), and encode a resolving enzyme that is assumed to be required for the deconcatamerization of the replicated rudiviral genome (2). A replication model for rudiviruses has been postulated based on the observed similarities with the Poxviridae. This involves replication initiation by an unidentified nicking enzyme at a site that is 11 nucleotides (nt) from the covalently closed terminus, yielding a 3′-hydroxyl suitable for priming DNA synthesis (3).
Small circular DNA viruses such as phiX174 and M13 in bacteria, geminiviruses in plants, and circoviruses in animals replicate by rolling-circle replication (RCR). RCR requires a replication initiator (Rep) protein, which nicks the circular DNA, forming a 5′ DNA adduct and releasing a free 3′ end to act as a primer for DNA synthesis. Rep proteins are virally encoded and typically fused to a helicase domain for DNA unwinding and translocation (14). In RCR, Rep proteins also catalyze closure or ligation of the newly synthesized genomes once replication is complete. A related replication mechanism known as rolling-hairpin replication (RHR) has been demonstrated for small linear single-stranded DNA (ssDNA) viruses such as the parvovirus adeno-associated virus (AAV). This shares many parallels with RCR and requires a Rep protein to initiate replication, although no joining activity is required in this case (reviewed in reference 6).
Here we report that the protein encoded by ORF119 from SIRV1 is a member of the replication initiator (Rep) family of proteins. Key active-site motifs are conserved, and homologs are found in all the rudiviruses. We show that the SIRV Rep protein can introduce a nick at the predicted initiation site at the viral terminus, forming a covalent adduct with the newly created 5′ ends and releasing a 3′-hydroxyl terminus suitable for priming DNA replication. The SIRV Rep protein is a dimer and is capable of joining DNA fragments by a strand transfer (flip-flop) mechanism. This is the first example of a Rep protein that is used to initiate a non-RCR mechanism. The implications for rudiviruses and for the evolution of viral replication strategies in general are discussed.
MATERIALS AND METHODS
Protein purification of wild-type and mutant proteins.
The cloning, expression, purification, crystallization, and structure solution of SIRV1 ORF119 have been reported (23), and the coordinates are available from the Protein Data Bank (PDB) (identifier [ID] 2X3G). The SIRV RepY108F mutant was cloned with a modified site-directed mutagenesis protocol (18) using the following primers: 5′-GATATAAAAAATGTATATAAGTTTATGCTAAAGACAAAAAAAGATATAAAAATGTCATAA and 5′-CATAAACTTATATACATTTTTTATATCGCTAATAGACTTTGGAACTAATTCAATTCTAATGTC (Eurogentec). Recombinant plasmid DNA was sequenced by the University of Dundee Sequencing Service. Recombinant mutant protein was expressed and purified as described for the native protein.
Cleavage activity assays.
The following oligonucleotides were obtained from Operon Biotechnologies: upper strand (US), 5′-ACTCCTATTACCTTCTTACTGCTCTACAA; lower strand (LS), 5′-TTGTAGAGCAGTAAGAAGGTAATAGGAGT; and hairpin (HP), 5′-ACTCCTATTACCTTCTTACTGCTCTACAATTGTAGAGCAGTAAGAAGGTAATAGGAGT.
The oligonucleotide US corresponds to the top strand of the SIRV1 hairpin terminus, as described by Blum et al. (3). Oligonucleotide LS was fully complementary to the US oligonucleotide, representing the other strand of the terminus. The oligonucleotide HP was a concatenation of the US and LS oligonucleotides and matches the covalently closed hairpin terminus of the virus. Versions of these oligonucleotides with fluorescein molecules on the 5′ or 3′ end were used to allow fluorescent detection of the oligonucleotides in nicking assays.
Reaction mixtures containing 20 μM SIRV Rep protein and 10 μM oligonucleotide in reaction buffer (20 mM Tris-HCl, pH 8.5; 20 mM NaCl; 2.5 mM MnCl2) were incubated for 20 min at 60°C. For analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), the reactions were terminated by addition of 2× SDS-PAGE sample buffer. The mixture was then heated at 90°C for 2 min and applied to the 4 to 12% NuPage Bis-Tris gel (Invitrogen), which was run at room temperature. The gels were scanned to visualize fluorescent products using a Fujifilm FLA-5000 imager at a wavelength of 473 nm. To visualize protein moieties, gels were stained with Coomassie blue or silver by following standard protocols. For high-resolution DNA mapping, reactions were terminated with gel loading buffer (95% formamide, 0.25% bromophenol blue, 0.25% xylene cyanol), heated to 90°C for 2 min, applied to a 20% acrylamide-7 M urea Tris-borate-EDTA (TBE) gel, and run at 48°C under a constant current of 95 W for 2.5 h. Following electrophoresis, fluorescent products were detected by imaging, as described above.
DNA joining assay.
Oligonucleotides corresponding to recognition site 2 (see Fig. 2D) were designed as follows. The first strand (5′-6-carboxyfluorescein [FAM]-TTTTTTTTAATAGGA/GTT), with a 5′ fluorescein label, and the second strand (5′-TTTTTTTTAATAGGA/GTTTTT) were purchased from MWG Eurofins. A second set of oligonucleotides corresponding to recognition site 3 were also designed, corresponding to the following sequences: first strand (5′-FAM-TTTTTTTTTAGAGCA/GTA) and second strand (5′-TTTTTTTTTAGAGCA/GTATTT). The forward slashes indicate the nicking sites. Reaction mixtures at total volumes of 20 μl comprised 10 μM SIRV Rep protein and two oligonucleotides (18-mer fluorescein labeled and 21-mer unlabeled) (see Fig. 4 for the amounts) in buffer (20 mM Tris-HCl, pH 8.5; 20 mM NaCl; 2.5 mM MnCl2) were incubated for 30 min at 60°C prior to incubation with 10 μg proteinase K for 30 min at 37°C. All samples were treated as described in the cleavage assay prior to analysis on a 20% acrylamide-7 M urea-TBE gel and imaging.
RESULTS
Structure of SIRV ORF119 resembles that of Rep superfamily proteins.
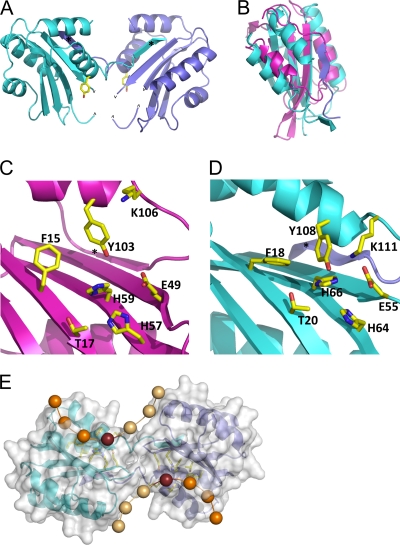
The SIRV Rep protein elutes from a size exclusion column with a retention time consistent with that of a dimeric arrangement. The asymmetric unit of the protein contained one monomer, which consisted of a central antiparallel β-sheet consisting of four strands, β1 (residues 15 to 23), β2 (residues 49 to 55), β3 (residues 63 to 71), and β4 (residues 89 to 94), flanked by four α-helices. Residues 57 to 59 were disordered and could not be modeled into the experimental electron density. The C terminus (residues 112 to 119) has an extended strand structure which exchanges with another monomer (Fig. 1 A), related by the 2-fold rotational symmetry of the crystal, adding a fifth β-strand (β5) to the edge of the central β-sheet. The interface is dominated by the β-sheet hydrogen bonds and by hydrophobic interactions among Val50, Phe51, and Ile53 on the β2-strand and Ile116 and Met118 on the β5-strand of the other monomer, burying 1,950 Å2 of surface area (15).
FIG. 1.
Structure of the SIRV1 Rep protein. (A) The dimer of SIRV 29 Rep protein. The Tyr residue, which forms the covalent link to the DNA, is shown in sticks for each subunit. The carets mark disordered loops (A57 to A59, A64, and B57 to B59). Each subunit exchanges a β-strand (marked with an asterisk) with the other. Subunit A is colored in cyan, and subunit B colored in slate. (B) Superposition of the TLYCV Rep protein (purple) with SIRV Rep monomer A (cyan). (C) The active site of the TYLCV Rep protein (12). One monomer of the NMR ensemble is shown. The key active-site residues conserved in the Rep superfamily are labeled. The asterisk denotes the fifth β-strand. (D) The active site of the SIRV Rep protein. The key residues are shown and correspond well with their equivalents in TLYCV. The asterisk denotes the strand from the other monomer which completes the 5-stranded β-sheet. (E) Potential path of single-stranded DNA prior to recognition and cleavage. A semitransparent surface model overlies the Rep dimer with active-site residues, shown in yellow. Spheres represent the backbone phosphates of the DNA. The phosphate that forms a covalent link with Y108 is shown as a dark purple sphere. Phosphates lighter in color will become the newly exposed 3′ end.
Analysis of the structure using ProFunc (16) suggested significant similarities with the structure of the N-terminal DNA binding domain of the replication initiator protein from tomato yellow leaf curl virus (TYLCV) (PDB ID 1L2M). The TYLCV Rep protein structure comprises a central antiparallel β-sheet surrounded by two α-helices and a β-hairpin (5). The TYLCV Rep protein belongs to the superfamily II group of Rep proteins, which are found in bacterial plasmids and eukaryotic viruses (14, 32), in which rolling-circle replication (RCR) takes place. The match covers the central β-sheet with a root mean square deviation (RMSD) of 3.0 Å over 73 common Cα atoms (Fig. 1B). Interestingly, TYLCV has a five-stranded β-sheet strand, and thus, the domain-swapped strand (β5) of SIRV ORF119 is matched in the superposition. Looking more widely at structurally characterized Rep proteins from eukaryotic viruses, all share a basic fold comprising the central β-sheet, with an α-helix packed against both faces of the β-sheet (32).
Active-site sequence motifs in the Rep superfamily.
Structure-based sequence alignment has revealed that all Rep proteins have three short conserved motifs, denoted I, II, and III (13, 32, 34, 35) (see Fig. S1 in the supplemental material). These motifs contribute to the active sites of RCR Rep proteins (35) (Fig. 1C). Motif I is located on strand β1. The first residue (an invariant phenylalanine) and third residue (typically a threonine or leucine) point up toward the active site of the protein and are involved in specific DNA recognition (5). Motif II resides on β3 and has the consensus sequence HhHh3, where “h” is any hydrophobic amino acid residue. The two histidine residues in motif II are thought to serve as ligands for a divalent metal ion such as Mn2+ or Mg2+, required for efficient DNA cleavage (17, 31, 34, 35). In the TYLCV structure, Glu49 on β2 has been proposed to provide an additional residue for metal ion binding. Motif III is located on α4 and is typically YxxK (14, 32). The first tyrosine in motif III is the putative catalytic residue, which forms a covalent ester linkage to the 5′ end of the DNA following cleavage (17). The lysine in motif III is in a suitable position to help catalyze the phosphotransfer reaction (14, 32).
ORF119 of SIRV1 conforms to the Rep consensus sequences for motifs I, II, and III (see Fig. S1 in the supplemental material). Putative Rep proteins with these motifs can also be identified in the other rudiviruses. When these residues are viewed in a structural context for ORF119 and compared to the equivalent region of the Rep protein of the TYLCV, the conservation of the active site is striking (Fig. 1D). In addition to the three motifs, the metal ligand Glu49 of TYLCV is conserved as Glu55 in ORF119. Thus, the structural data suggest strongly that SIRV1 ORF119 and its homologs in other rudiviruses are Rep superfamily proteins, with a potential role in virus replication. Accordingly, the protein will henceforth be termed “SIRV Rep.” A simple model for the SIRV Rep dimer with ssDNA bound at both active sites (Fig. 1E) suggests that transesterification (joining) could occur when a newly released 3′ DNA end generated at one site attacks the covalently linked 5′ DNA end at the other site.
SIRV Rep cleaves one strand of the SIRV termini, forming covalent adducts.
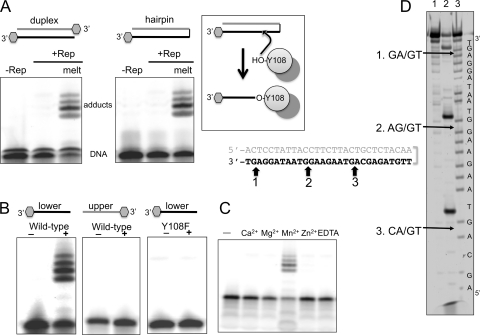
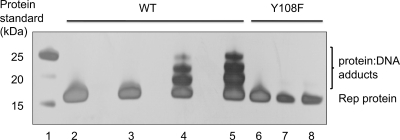
SIRV Rep and a Y108F mutant variant were expressed in Escherichia coli and purified as described previously (23). To test the nicking activity of SIRV Rep, we constructed oligonucleotides with sequences matching the upper and lower strands (US and LS, respectively) of the SIRV1 terminal hairpin. These oligonucleotides have complementary sequences and can be annealed to generate duplex DNA matching the SIRV terminal sequence. A oligonucleotide hairpin (HP) was also synthesized to model the covalently closed hairpin terminus more closely. These substrates were labeled with a 3′ fluorescein tag to allow detection by fluorescence imaging. When SIRV Rep was incubated with the duplex or hairpin substrates in the presence of Mn2+ ions and the reactants were separated by SDS-PAGE, followed by fluorescence imaging, four fluorescent protein-DNA adducts were detected, suggesting that a covalent complex between the protein and DNA had been formed (Fig. 2 A). This activity was observed only when dsDNA was first melted by being heated to 90°C, suggesting that ssDNA is the substrate for SIRV Rep. In vivo, the viral hairpins are likely to adopt transient ssDNA character at the normal growth temperature of the host cells, which is around 80°C. We next tested Rep cleavage of the upper strand (US) and lower strand (LS) of the terminus individually. Incubation of SIRV Rep with the LS oligonucleotide yielded the same four adducts as those observed for the melted hairpin (Fig. 2B), while the US oligonucleotide was not cleaved, suggesting clear sequence dependence for the reaction. The transfer of the 3′-end-labeled fluorescent DNA to form a protein adduct is consistent with the formation of an ester linkage between Tyr108 and the new 5′ DNA end created by the nicking reaction, which was confirmed by the inability of the Y108F mutant to form adducts (Fig. 2B). Adduct formation was dependent on the presence of manganese; no activity was detected with magnesium or any other tested divalent metal (Fig. 2C). Reaction products were also visualized by SDS-PAGE and silver staining to visualize the protein, confirming that the retarded species consisted of DNA-protein adducts (Fig. 3). No adducts were observed with the Y108F mutant, confirming that these are a specific property of the wild-type Rep protein and not due to any contaminating protein.
FIG. 2.
SIRV Rep nicks the viral terminus at specific sites. (A) Fluorescence imaging of the reaction products formed upon incubation of the SIRV1 terminal sequence labeled with a fluorescein at the 3′ ends reveal covalent protein-DNA adduct formation when the duplex is melted by heating it to 90°C for 5 min. (B) The lower strand of the hairpin is the site of nicking and adduct formation, which requires the Tyr108 residue. (C) SIRV Rep activity is dependent on the presence of manganese; a panel of alternative metal ions at 2.5 mM does not support Rep activity (D) The SIRV1 cleavage sites on the lower strand of the SIRV1 terminal hairpin (shown in boldface) were mapped by separating the 5′-fluorescein-labeled reaction products by denaturing gel electrophoresis alongside a sequence ladder. Arrows show the sites of cleavage with respect to the chemically generated sequence ladder, which generates products that run faster than the corresponding enzymatic cleavage products. Lane 1, DNA alone; lane 2, reaction products after incubation of SIRV Rep; lane 3, A+G sequence ladder.
FIG. 3.
DNA-protein adduct formation monitored by SDS-PAGE. SIRV1 Rep was incubated with the LS or US oligonucleotide, and protein-DNA adducts were monitored by SDS-PAGE and silver staining. Three retarded species corresponding to protein adducted to DNA of 3 different lengths were observed in reactions of the wild-type enzyme (WT) with the LS oligonucleotide. Lane 1, protein markers; lane 2, Rep protein control; lane 3, Rep plus US oligonucleotide; lanes 4 and 5, Rep plus LS oligonucleotide; lane 6, Y108F control; lanes 7 and 8, Y108F plus LS oligonucleotide.
Together, these data suggest that SIRV Rep is specific for certain sequences present in one strand of the SIRV terminal hairpin, cleaving the DNA in the presence of manganese when it is in a single-stranded form. The specificity for ssDNA observed for SIRV Rep is consistent with the known activity of that of other Rep family proteins, which cleave terminal sequences that are not base paired (13). In SIRV, the terminal hairpins are likely to adopt spontaneously some ssDNA character at the high-growth temperature of the host species. Many Rep proteins include a helicase functionality that is used to unwind dsDNA, and while this is clearly not present in the 119-amino-acid (aa) SIRV Rep protein, there is a potential helicase in the Rep genome that might function to open and unwind DNA in vivo.
Mapping the SIRV Rep nick sites.
The visualization of four distinct adducted species suggested that four cleavage sites were present in the lower strand. In order to visualize DNA cleavage directly and map the nicking site, we incubated SIRV Rep with oligonucleotide LS, labeled with fluorescein at the 5′ end, and mapped the cleavage sites using a Maxam-Gilbert sequence ladder (Fig. 2D). As the fluorescent label was on the 5′ end, the nonadducted DNA products could be visualized and mapped. As expected from the observation of four adducted species, we observed four distinct cleavage sites. Three of these could be mapped accurately using the sequence ladder, while the one closest to the terminus was too small and ran at the bottom of the gel. The three mapped sites shared a consensus sequence, with cleavage observed in AG-rich regions and always at the 5′ end of a GT dinucleotide (Fig. 2D). The upper strand of the SIRV terminal sequence is notably lacking in guanine residues and has no sequence corresponding to the consensus sequence identified, explaining the nonreactive nature of this region. Cleavage site 3 corresponded to the main nicking site identified by sequencing of SIRV hairpin termini from SIRV-infected cells (3), suggesting that cleavage of the hairpin sequence by SIRV Rep observed in vitro is relevant in vivo.
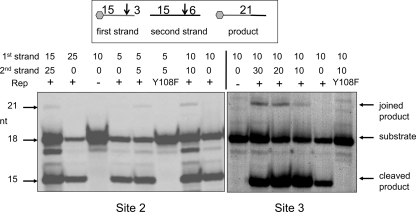
DNA joining by the SIRV Rep protein.
As well as initiating replication by nicking at the origin, Rep proteins involved in RCR can catalyze transesterification, the sealing or joining activity necessary to reform the covalently closed hairpin terminus at the completion of replication (30). To assay for joining activity, we incubated SIRV Rep with a 5′-fluorescein-labeled 18-mer oligonucleotide with a single consensus nicking site (Fig. 2D, site 2 or 3) at position 15, along with an unlabeled 21-mer oligonucleotide containing an identical single nicking site in the same position (Fig. 4). If a transesterification reaction takes place, one expects to see the formation of a fluorescently labeled 21-mer product. This product was observed only when both oligonucleotides were added along with wild-type SIRV Rep. The presence of this species can be explained only by DNA joining via a transesterification reaction mediated by the SIRV Rep protein. The joined product was present at low levels compared to the labeled 18-mer substrate oligonucleotide and the 15-mer cleavage product, as observed for other Rep proteins (22, 30). This may be explained partly, because in this highly simplified assay, the joined product is itself a substrate for the enzyme. In vivo, further cellular and viral replication factors are expected to cooperate to maximize the efficiency of viral replication.
FIG. 4.
DNA joining activity of SIRV Rep. Transesterification (DNA joining) was monitored following incubation of Rep with one fluorescein-labeled (18-mer) and one unlabeled (21-mer) oligonucleotide. The appearance of a fluorescent 21-mer indicated successful DNA joining when both oligonucleotides and wild-type Rep were present. Sequences corresponding to site 2 (left) and site 3 (right), as defined in the legend to Fig. 2D, supported weak formation of ligated products. The inefficiency of this reaction in vitro may reflect the absence of important accessory proteins that are involved in vivo. The first- and second-strand oligonucleotides were present at the amounts indicated above each lane. The presence of a minor band below the 18-mer substrate is explained by the presence of a 17-mer (n-1) oligonucleotide.
DISCUSSION
Implications for the mechanism of Rep proteins.
Rep proteins, often fused to a helicase for DNA unwinding, are encoded by many genetic elements that replicate via RCR and related RHR mechanisms. In all Rep-mediated RCR mechanisms, Rep must catalyze multiple rounds of cleavage and ligation at ori sites by what has been termed a “flip-flop” mechanism. This flip-flop catalysis was first proposed in early studies of Rep proteins, in which two apparently conserved tyrosines separated by three intervening residues in the active site of the phiX174 gene A protein were proposed to carry out the nicking and nicking-joining activities (33). Both Tyr residues were shown to be essential for Rep activity, and it was proposed that they might have different preferences for the nicking or joining reactions (17, 22). The monomeric structure of the adeno-associated virus (AAV) Rep protein domain also revealed two Tyr residues located inside a cleft adjacent to the presumed metal ion nuclease site (13). Studies of the plasmid pC194 Rep protein domain, also a monomer, revealed a Glu residue at the first of the two Tyr positions (20). Those authors suggested that the Glu residue could itself direct a second hydrolytic cleavage of the ori site in the presence of the DNA-Tyr adduct within the same monomer, generating a new 3′ end that could catalyze a transesterification of the DNA-Tyr moiety to release the Rep protein and seal the nick (21). Subsequently, structures of the monomeric catalytic domains of the viral Rep proteins of TYLCV (5), porcine circovirus type 2 (PCV2) (34), and faba bean necrotic yellows virus (FBNYV) (35) were reported. These structures revealed one catalytic Tyr in the nuclease domain, requiring that a flip-flop mechanism could be accommodated only by relying on a non-Tyr residue (e.g., Glu [22]) within the same active site. These monomeric structures were obtained after proteolytic cleavage, preventing characterization of the quaternary structure of the full-length proteins. Recently, the full-length structure of the RepB initiator of plasmid pMV158 was obtained, showing that the oligomerization domain formed a hexamer with six associated mobile Rep nuclease domains (4), the first evidence that a multimeric quaternary structure might be important.
The SIRV Rep protein has a simple organization without any accessory domains and adopts a dimeric structure with the two catalytic Tyr residues (one from each monomer) in close proximity, with the result that a newly released 3′ DNA end formed by a nicking reaction in one active site could flip into the second site to capture a 5′ DNA end in a joining reaction (Fig. 1E). This simple model avoids several important problems implicit in the “single-active-site” models that have predominated: most notably, the difficulty in fitting two DNA strands one after the other into a single active site while preserving the correct geometry for catalysis and recognition. We suggest that the dual-capability single-active-site models are incorrect and, rather, that all Rep proteins function as oligomers, with DNA transferred between monomers in the classic “flip flop” relying on the single absolutely conserved Tyr residue within each monomer. The second Tyr residue, where it exists, must play a subsidiary role in catalysis.
A new model for rudivirus replication.
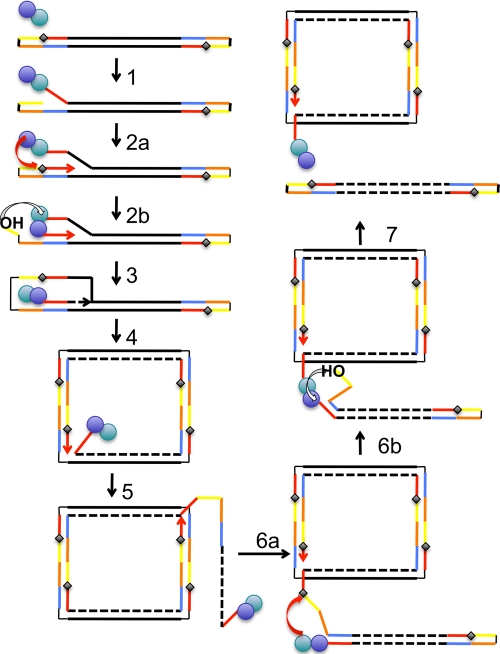
Viruses with double-stranded linear genomes and covalently closed ends are found in all three domains of life. Examples include the bacteriophage N15 that infects E. coli (19), the eukaryal poxviruses (1), and the archaeal rudiviruses (3). Their replication strategies are not fully understood and appear to be quite diverse. The structural and biochemical characterization of SIRV Rep presented here allows a framework for rudivirus replication to be proposed that builds significantly on previous models (Fig. 5). The Rep-mediated initialization of DNA replication could occur at either end of the genome. A series of Rep-mediated transesterification reactions coupled with strand displacement DNA synthesis can account for all the known features of SIRV replication. This includes the observation of head-to-head concatemers and DNA nicks near the hairpins in SIRV DNA studied in infected cells (24). The DNA smearing observed by Peng et al. when they analyzed the dimeric terminal fragments of SIRV1 in infected cells may have been due to covalent adduct formation by Rep at this sequence (24). The model is also consistent with the need for a virally encoded Holliday junction-resolving enzyme (2) to recover from situations in which linear SIRV concatemers are generated by Rep cleavage failure (Fig. 5, step 6). It should be possible to test this model using two-dimensional gel electrophoresis to look for the specific branched replication intermediates predicted to occur near the termini during replication. This technique has been used successfully to monitor the replication of viruses utilizing RCR, such as maize streak virus (11).
FIG. 5.
Model for DNA replication in rudiviruses. We propose that one of the rudivirus Rep protein subunits initiates replication by nicking at one of the target ori sites in the telomeric region (step 1), releasing a new 3′ DNA end which can be used as a primer to initiate DNA replication and forms a covalent adduct with the newly created 5′ end. Replication quickly regenerates the ori site, which is attacked by the second subunit of Rep to form a dual-adducted Rep dimer (step 2a). This species is likely to be transient, as the new 3′ DNA end generated in subunit 2 can flip over to attack the tyrosyl-phosphoester in subunit 1, forming a new contiguous DNA strand (step 2b). Displacement replication is then used to replicate the rest of the genome, which generates a double-stranded DNA circle (step 4). As replication continues around the circle, the previously copied viral DNA is displaced and can fold up into the linear DNA structure found in the virion (step 5). This folding would ensure that Rep was suitably positioned to attack the newly displaced ori site shown in step 6a, generating a transient double-adducted Rep dimer and a new 3′ end that can immediately attack the other Rep subunit (step 6b), releasing a covalently closed linear viral genome and leaving Rep attached to the emerging DNA strand ready for another round of replication (step 7). A missed cleavage at step 6 could result in the generation of DNA concatemers that would require resolution by a Holliday junction-resolving enzyme.
Evolutionary considerations.
Distant homologs of the Rep domain are found in the DNA binding domains of non-RCR initiator proteins, such as the papillomavirus E1 helicase and simian virus 40 (SV40) T antigen (4, 5, 13). These are thought to have retained the origin binding activity of Rep but have lost any catalytic role. The active Rep protein we have identified in the rudiviruses is the first example in a virus that does not utilize rolling-circle or rolling-hairpin replication. The rudiviruses may represent an evolutionary link between the small DNA viruses and plasmids on the one hand and the large cytoplasmic eukaryotic DNA viruses (e.g., poxviruses) on the other. The initiation of poxviral DNA replication is still not fully understood. It may involve a Rep-like nicking enzyme (so far unidentified) that cleaves the covalently closed hairpin termini (8, 25, 26). Alternatively, poxviral replication may utilize the virally encoded D5 primase enzyme, whose activity has recently been confirmed in vitro (9), or a combination of both primase and Rep activities.
Concluding remarks.
The discovery of rudivirus Rep demonstrates that replication initiator proteins are not restricted to RCR and related mechanisms and may thus have wider roles than those currently envisaged, particularly in the replication of large linear DNA viruses. The simple dimeric structure of the protein, bringing two active sites together, provides a neat explanation for the nicking-joining mechanism of Rep proteins in general, with broad implications across biology.
Supplementary Material
Acknowledgments
We thank the St. Andrews Mass Spectrometry service, which is funded by the Wellcome Trust, and Stuart MacNeill for helpful discussions.
This work was funded by the Biotechnology and Biological Sciences Research Council (reference BB/S/B14450). The work done at the Institut Pasteur was supported by the Agence Nationale de la Recherche, France (program blanc).
Footnotes
Published ahead of print on 10 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Beaud, G. 1995. Vaccinia virus DNA replication: a short review. Biochimie 77:774-779. [DOI] [PubMed] [Google Scholar]
- 2.Birkenbihl, R. P., K. Neef, D. Prangishvili, and B. Kemper. 2001. Holliday junction resolving enzymes of archaeal viruses SIRV1 and SIRV2. J. Mol. Biol. 309:1067-1076. [DOI] [PubMed] [Google Scholar]
- 3.Blum, H., W. Zillig, S. Mallok, H. Domdey, and D. Prangishvili. 2001. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology 281:6-9. [DOI] [PubMed] [Google Scholar]
- 4.Boer, D. R., J. A. Ruiz-Maso, J. R. Lopez-Blanco, A. G. Blanco, M. Vives-Llacer, P. Chacon, I. Uson, F. X. Gomis-Ruth, M. Espinosa, O. Llorca, G. del Solar, and M. Coll. 2009. Plasmid replication initiator RepB forms a hexamer reminiscent of ring helicases and has mobile nuclease domains. EMBO J. 28:1666-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campos-Olivas, R., J. M. Louis, D. Clerot, B. Gronenborn, and A. M. Gronenborn. 2002. The structure of a replication initiator unites diverse aspects of nucleic acid metabolism. Proc. Natl. Acad. Sci. U. S. A. 99:10310-10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cotmore, S. F., and P. Tattersall. 1996. Parvovirus DNA replication, p. 799-813. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells, vol. 31. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 7.Culyba, M. J., J. E. Harrison, Y. Hwang, and F. D. Bushman. 2006. DNA cleavage by the A22R resolvase of vaccinia virus. Virology 352:466-476. [DOI] [PubMed] [Google Scholar]
- 8.DeMasi, J., S. Du, D. Lennon, and P. Traktman. 2001. Vaccinia virus telomeres: interaction with the viral I1, I6, and K4 proteins. J. Virol. 75:10090-10105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Silva, F. S., W. Lewis, P. Berglund, E. V. Koonin, and B. Moss. 2007. Poxvirus DNA primase. Proc. Natl. Acad. Sci. U. S. A. 104:18724-18729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Du, S., and P. Traktman. 1996. Vaccinia virus DNA replication: two hundred base pairs of telomeric sequence confer optimal replication efficiency on minichromosome templates. Proc. Natl. Acad. Sci. U. S. A. 93:9693-9698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erdmann, J. B., D. N. Shepherd, D. P. Martin, A. Varsani, E. P. Rybicki, and H. Jeske. 2010. Replicative intermediates of maize streak virus found during leaf development. J. Gen. Virol. 91:1077-1081. [DOI] [PubMed] [Google Scholar]
- 12.Garcia, A. D., L. Aravind, E. Koonin, and B. Moss. 2000. Bacterial-type DNA Holliday junction resolvases in eukaryotic viruses. Proc. Natl. Acad. Sci. U. S. A. 97:8926-8931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hickman, A. B., D. R. Ronning, R. M. Kotin, and F. Dyda. 2002. Structural unity among viral origin binding proteins: crystal structure of the nuclease domain of adeno-associated virus Rep. Mol. Cell 10:327-337. [DOI] [PubMed] [Google Scholar]
- 14.Ilyina, T. V., and E. V. Koonin. 1992. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 20:3279-3285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski, R. A., J. D. Watson, and J. M. Thornton. 2005. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 33:W89-W93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laufs, J., W. Traut, F. Heyraud, V. Matzeit, S. G. Rogers, J. Schell, and B. Gronenborn. 1995. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. U. S. A. 92:3879-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu, H., and J. H. Naismith. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mardanov, A. V., and N. V. Ravin. 2009. Conversion of linear DNA with hairpin telomeres into a circular molecule in the course of phage N15 lytic replication. J. Mol. Biol. 391:261-268. [DOI] [PubMed] [Google Scholar]
- 20.Noirot-Gros, M. F., V. Bidnenko, and S. D. Ehrlich. 1994. Active site of the replication protein of the rolling circle plasmid pC194. EMBO J. 13:4412-4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noirot-Gros, M. F., and S. D. Ehrlich. 1996. Change of a catalytic reaction carried out by a DNA replication protein. Science 274:777-780. [DOI] [PubMed] [Google Scholar]
- 22.Odegrip, R., and E. Haggard-Ljungquist. 2001. The two active-site tyrosine residues of the A protein play non-equivalent roles during initiation of rolling circle replication of bacteriophage p2. J. Mol. Biol. 308:147-163. [DOI] [PubMed] [Google Scholar]
- 23.Oke, M., L. G. Carter, K. A. Johnson, H. Liu, S. A. McMahon, X. Yan, M. Kerou, N. D. Weikart, N. Kadi, M. A. Sheikh, S. Schmelz, M. Dorward, M. Zawadzki, C. Cozens, H. Falconer, H. Powers, I. M. Overton, C. A. van Niekerk, X. Peng, P. Patel, R. A. Garrett, D. Prangishvili, C. H. Botting, P. J. Coote, D. T. Dryden, G. J. Barton, U. Schwarz-Linek, G. L. Challis, G. L. Taylor, M. F. White, and J. H. Naismith. 2010. The Scottish Structural Proteomics Facility: targets, methods and outputs. J. Struct. Funct. Genomics 11:167-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng, X., H. Blum, Q. She, S. Mallok, K. Brugger, R. A. Garrett, W. Zillig, and D. Prangishvili. 2001. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology 291:226-234. [DOI] [PubMed] [Google Scholar]
- 25.Pogo, B. G., E. M. Berkowitz, and S. Dales. 1984. Investigation of vaccinia virus DNA replication employing a conditional lethal mutant defective in DNA. Virology 132:436-444. [DOI] [PubMed] [Google Scholar]
- 26.Pogo, B. G., M. O'Shea, and P. Freimuth. 1981. Initiation and termination of vaccinia virus DNA replication. Virology 108:241-248. [DOI] [PubMed] [Google Scholar]
- 27.Prangishvili, D., H. P. Arnold, D. Gotz, U. Ziese, I. Holz, J. K. Kristjansson, and W. Zillig. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387-1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prangishvili, D., R. A. Garrett, and E. V. Koonin. 2006. Evolutionary genomics of archaeal viruses: unique viral genomes in the third domain of life. Virus Res. 117:52-67. [DOI] [PubMed] [Google Scholar]
- 29.Schramm, B., and J. K. Locker. 2005. Cytoplasmic organization of POXvirus DNA replication. Traffic 6:839-846. [DOI] [PubMed] [Google Scholar]
- 30.Steinfeldt, T., T. Finsterbusch, and A. Mankertz. 2006. Demonstration of nicking/joining activity at the origin of DNA replication associated with the rep and rep' proteins of porcine circovirus type 1. J. Virol. 80:6225-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Timchenko, T., F. de Kouchkovsky, L. Katul, C. David, H. J. Vetten, and B. Gronenborn. 1999. A single rep protein initiates replication of multiple genome components of faba bean necrotic yellows virus, a single-stranded DNA virus of plants. J. Virol. 73:10173-10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vadivukarasi, T., K. R. Girish, and R. Usha. 2007. Sequence and recombination analyses of the geminivirus replication initiator protein. J. Biosci. 32:17-29. [DOI] [PubMed] [Google Scholar]
- 33.van Mansfeld, A. D., H. A. van Teeffelen, P. D. Baas, and H. S. Jansz. 1986. Two juxtaposed tyrosyl-OH groups participate in phi X174 gene A protein catalysed cleavage and ligation of DNA. Nucleic Acids Res. 14:4229-4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vega-Rocha, S., I. J. Byeon, B. Gronenborn, A. M. Gronenborn, and R. Campos-Olivas. 2007. Solution structure, divalent metal and DNA binding of the endonuclease domain from the replication initiation protein from porcine circovirus 2. J. Mol. Biol. 367:473-487. [DOI] [PubMed] [Google Scholar]
- 35.Vega-Rocha, S., B. Gronenborn, A. M. Gronenborn, and R. Campos-Olivas. 2007. Solution structure of the endonuclease domain from the master replication initiator protein of the nanovirus faba bean necrotic yellows virus and comparison with the corresponding geminivirus and circovirus structures. Biochemistry 46:6201-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vestergaard, G., M. Haring, X. Peng, R. Rachel, R. A. Garrett, and D. Prangishvili. 2005. A novel rudivirus, ARV1, of the hyperthermophilic archaeal genus Acidianus. Virology 336:83-92. [DOI] [PubMed] [Google Scholar]
- 37.Vestergaard, G., S. A. Shah, A. Bize, W. Reitberger, M. Reuter, H. Phan, A. Briegel, R. Rachel, R. A. Garrett, and D. Prangishvili. 2008. Stygiolobus rod-shaped virus and the interplay of crenarchaeal rudiviruses with the CRISPR antiviral system. J. Bacteriol. 190:6837-6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.