Abstract
Alpha interferon (IFN-α) is an approved medication for chronic hepatitis B. Gamma interferon (IFN-γ) is a key mediator of host innate and adaptive antiviral immunity against hepatitis B virus (HBV) infection in vivo. In an effort to elucidate the antiviral mechanism of these cytokines, 37 IFN-stimulated genes (ISGs), which are highly inducible in hepatocytes, were tested for their ability to inhibit HBV replication upon overexpression in human hepatoma cells. One ISG candidate, indoleamine 2,3-dioxygenase (IDO), an IFN-γ-induced enzyme catalyzing tryptophan degradation, efficiently reduced the level of intracellular HBV DNA without altering the steady-state level of viral RNA. Furthermore, expression of an enzymatically inactive IDO mutant did not inhibit HBV replication, and tryptophan supplementation in culture completely restored HBV replication in IDO-expressing cells, indicating that the antiviral effect elicited by IDO is mediated by tryptophan deprivation. Interestingly, IDO-mediated tryptophan deprivation preferentially inhibited viral protein translation and genome replication but did not significantly alter global cellular protein synthesis. Finally, tryptophan supplementation was able to completely restore HBV replication in IFN-γ- but not IFN-α-treated cells, which strongly argues that IDO is the primary mediator of IFN-γ-elicited antiviral response against HBV in human hepatocyte-derived cells.
Hepatitis B virus (HBV) is a hepatotropic virus in the family of Hepadnaviridae (18, 56) which has infected two billion people worldwide. Although most infected adults can resolve the infection, approximately 5 to 10% of adulthood infections and more than 90% of neonatal infections become lifelong persistent, which causes a significant public health burden currently affecting more than 350 million individuals worldwide (36, 41). Patients with chronic hepatitis B carry a high risk of cirrhosis and primary hepatocellular carcinoma (5, 36).
It is generally believed that the outcomes of HBV infection and disease pathogenesis are primarily determined by the host adaptive antiviral immune response (9). The resolution of HBV infection is associated with a robust, polyclonal viral antigen-specific cytotoxic T lymphocyte (CTL) response that kills and/or noncytopathically cures virally infected hepatocytes. The latter effect is primarily mediated by inflammatory cytokines, including gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α) (17, 18, 20, 21). Interestingly, although it has been shown that HBV can evade host innate immunity surveillance, as evidenced by its failure to induce the expression of interferon-stimulated genes (ISGs) in the liver of chimpanzees during the early phase of infection (66), the virus is sensitive to IFN-α/β-induced antiviral response in transgenic mice and in people (30, 49). For example, treatment of chronic hepatitis B with pegylated IFN-α for 48 weeks can achieve sustained virological response in 30 to 40% of the treated patients (29).
In efforts to elucidate the antiviral mechanism of IFNs, it has been demonstrated that IFN-α- and IFN-γ-induced antiviral responses either prevent the formation of pregenomic RNA (pgRNA)-containing capsids or promote the decay of replication-competent nucleocapsids in murine hepatocytes in culture and in transgenic mice in vivo (3, 67, 68). Recently, type III IFN (IFN-λ) has also been shown to inhibit HBV replication in cultured murine hepatocytes through a similar mechanism but with a weaker antiviral activity in transgenic mice (49, 54). Ironically, IFN treatment of human hepatoma-derived cells usually elicits no or a weaker antiviral response against HBV. Moreover, it remains unclear whether IFNs inhibit HBV replication in human hepatoma cells by targeting viral RNA transcription and/or disrupting viral DNA replication complexes (49, 53, 63).
To further elucidate the mechanism of IFNs against HBV in human hepatocyte-derived cells, we first demonstrated that IFN-α and IFN-γ did not alter the steady-state level of viral RNA but efficiently reduced the level of intracellular HBV DNA in HepG2-derived stable cell lines supporting HBV replication. To determine IFN-induced cellular proteins that mediate the antiviral response of the cytokines, we systematically examined 37 ISGs that are inducible in hepatocytes by IFN-α and/or IFN-γ for their ability to inhibit HBV replication upon overexpression in HepG2 cells. We identified two IFN-induced cellular enzymes, APOBEC3G and indoleamine 2,3-dioxygenase (IDO), which significantly reduced viral DNA but did not alter the level of viral RNA. Further mechanistic analyses provided strong evidence suggesting that the IDO-elicited antiviral response against HBV depends on induction of tryptophan deprivation, which preferentially inhibits viral protein translation but does not significantly alter global cellular protein synthesis in HepG2 cells. Intriguingly, we were able to show that tryptophan supplementation completely restored HBV replication in cells treated with IFN-γ but not with IFN-α. Our findings thus favor the hypothesis that IDO is the primary mediator of the IFN-γ-elicited antiviral response against HBV in human hepatocyte-derived cells. These results may shed light on the mechanism by which CTL and IFN-γ mediate immune responses to noncytolytically control HBV infection.
MATERIALS AND METHODS
Cell cultures and drugs.
HepG2 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM)/F-12 medium (Mediatech) supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and 100 μg/ml streptomycin. HepDE19 cells were maintained in the same way as HepG2 cells but with the addition of 1 μg/ml tetracycline and 400 μg/ml G418 (23). To initiate HBV replication in HepDE19 cells, tetracycline was withdrawn from the culture medium and the cells were cultured for the indicated time period. Recombinant human IFN-α2a and IFN-γ were purchased from PBL Biomedical Laboratories. l-Tryptophan (l-Trp) and 1-methyl-l-trp (1-MT) were purchased from Sigma-Aldrich.
Plasmid construction.
The construction strategy of plasmids expressing individual human ISGs, including IDO (listed in Table 1), was described in previous reports (33, 34), except for inducible nitric oxide synthase (iNOS). The iNOS gene was PCR amplified from hep-iNOS (15) (a gift from David Geller, University of Pittsburgh) and cloned into vector pcDNA5 (Invitrogen) restricted with AflII and NotI. All the ISGs contain a FLAG tag sequence fused to the N terminus, and their expression is under the control of a cytomegalovirus immediate-early (CMV-IE) promoter. The enzymatically inactive mutant of IDO, IDOH346A (37), was constructed by introducing a single amino acid mutation into the IDO expression plasmid with the QuikChange II site-directed mutagenesis kit (Stratagene). To construct an HBV core promoter (Cp) reporter plasmid, a fragment covering the HBV enhancer II (EnII) and Cp region (nucleotides 1400 to 1902; genotype ayw; GenBank accession number V01460) was PCR amplified from plasmid pHBV1.3 (25) and inserted into the SacI and HindIII restriction sites in pGL3-Basic vector (Promega). The generated plasmid was designated EnII/Cp-Luc, in which the expression of the firefly luciferase (FL) reporter gene is governed by the HBV core promoter. The primer sequences used in this study are available upon request. The sequences of all the DNA clones were confirmed by sequencing.
TABLE 1.
List of 37 ISGs and their antiviral effects against HBV replication
| ISG | GenBank accession no. | Reduction of HBVa: |
|
|---|---|---|---|
| DNA | RNA | ||
| IFN-α inducible | |||
| APOBEC3G | NM_021822 | ++++ | − |
| PKR | AH008429 | ++++ | ++++ |
| ISG20 | NM_002201 | ++++ | ++++ |
| Viperin | AF442151 | ++ | ++ |
| Tetherin(Bst2) | NM_004335 | +++ | +++ |
| 1-8D (IFITM2) | NM_006435 | +++ | +++ |
| 1-8U (IFITM3) | NM_021034 | ++ | ++ |
| 9-27 (IFITM1) | NM_003641 | ++ | ++ |
| ISG12 | BN000227 | +++ | +++ |
| ISG15 | NM_005101 | + | + |
| USP18 | NM_017414 | + | + |
| UBE2L6 | NM_004223 | − | ND |
| MxA | NM_002462 | + | + |
| MxB | NM_002463 | − | ND |
| TRIM22 | X82200 | + | + |
| ADAR1 | NM_001111 | − | ND |
| ISG54 | NM_001547 | − | ND |
| ISG56 | NM_001548 | − | ND |
| GBP1 | NM_002053 | + | ND |
| IFI44 | NM_006417 | + | ND |
| IFI44L | NM_006820 | + | ND |
| PLSCR1 | NM_021105 | + | ND |
| PLSCR2 | NM_020359 | + | ND |
| GLYAT | BC009785 | − | ND |
| SAMHD1 | NM_015474 | + | ND |
| RIG-G | NM_001549 | + | ND |
| TRAFD1 | NM_006700 | + | ND |
| MAPK8 | NM_002750 | − | ND |
| FLJ20035 | AK000042 | + | ND |
| FLJ20637 | AK000644 | + | ND |
| FLJ38348 | AK095667 | + | ND |
| IFN-γ inducible | |||
| IDO | NM_002164 | ++++ | − |
| LAMP3 | NM_014398 | − | ND |
| iNOS | L09210 | − | ND |
| DCK | NM_000788 | + | ND |
| Nmi | U32849 | + | ND |
| NNMT | NM_006169 | + | ND |
Assays were done in at least triplicate. −, no reduction; +, <20% reduction; ++, 20 to 40% reduction; +++, 40 to 60% reduction; ++++, >60% reduction; ND, not determined.
Cell transfection and cytotoxicity assay.
Cells were seeded in a collagen-coated 6-well plate at a density of 1.2 × 106 cells per well in antibiotic-free complete DMEM/F-12 medium. After 6 h, each well was transfected with a total of 4 μg plasmid with Lipofectamine 2000 (Invitrogen) by following the manufacturer's directions. Transfected cells were harvested at the indicated time points. Cell viability was determined with the CellTiter 96 AQueous nonradioactive cell proliferation assay (Promega) by following the manufacturer's instructions.
Reporter assay.
HepG2 and HepDE19 cells were transfected using Lipofectamine 2000 (Invitrogen). For each transfection, 1 × 104 cells were cotransfected with 200 ng of plasmid EnII/Cp-Luc and 4 ng of plasmid pRL-CMV (Promega). Sixteen hours after transfection, cells were left untreated or treated with 1,000 IU/ml IFN-α2a or 100 ng/ml IFN-γ for 4 days, with treatment repeated every other day. Cells were then harvested and lysed. Luciferase activities in cell lysates were assayed using a dual-luciferase assay system (Promega) and measured by TopCount NXT.
Viral nucleic acid analysis.
Intracellular HBV core DNA and total cellular RNA were extracted as described previously (22, 23). For core DNA analysis, one-third of the DNA sample from each plate was resolved by electrophoresis into a 1.5% agarose gel and blotted onto a Hybond-XL membrane (GE Healthcare). For RNA analysis, 10 μg of total cellular RNA was resolved in a 1.5% agarose gel containing 2.2 M formaldehyde and transferred onto a Hybond-XL membrane in 20× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Membranes were probed with either an [α-32P]UTP (800 Ci/mmol; Perkin Elmer)-labeled minus- or plus-strand-specific full-length HBV riboprobe and exposed to a phosphorimager screen. Hybridization signals were quantified with QuantityOne software (Bio-Rad).
Reverse transcription (RT)-PCR analysis of ISG expression.
HepG2 cells were left untransfected or transfected with pHBV1.3. Sixteen hours later, cells were then left untreated or treated with 1,000 IU/ml of IFN-α or 100 ng/ml IFN-γ for 48 h and harvested. Total cellular RNA was extracted with TRIzol reagent by following the manufacturer's directions (Invitrogen). The first-strand cDNA was synthesized with oligo(dT)18 primer and Superscript III DNA polymerase (Invitrogen). To quantify mRNAs for ISGs and β-actin, 0.5 μl of each cDNA reaction mixture was amplified for 25 cycles with respective forward and reverse primers (sequence available upon request). PCR products were resolved by electrophoresis in a 1% agarose gel and stained with ethidium bromide.
Protein analysis.
For protein labeling, cells in one well of a 6-well plate were cotransfected with 2 μg of plasmid pHBV1.3 and 2 μg of control vector or plasmid expressing IDO. Cells were washed twice with warm phosphate-buffered saline (PBS) and labeled at 37°C for 1 h at various time points posttransfection with [35S]methonine (Perkin Elmer) to a final concentration of 100 μCi/ml. Cells were then lysed in 1 ml of lysis buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM EDTA, 1% NP-40, and 1× protease inhibitor cocktail (Qiagen). Thirty microliters of the clarified cell lysate was directly mixed with 10 μl of 4× Laemmli buffer and subjected to sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer onto a polyvinylidene difluoride (PVDF) membrane (Millipore). The remaining cell lysate was subjected to immunoprecipitation assay. Briefly, 500 μl of cell lysate was mixed with 20 μl of anti-FLAG M2 affinity gel (Sigma) or protein A/G Plus beads (Santa Cruz) that were preabsorbed with antibodies against HBcAg (Dako). The mixtures were incubated at room temperature for 2 h. Beads were washed four times with TBS buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.4) and boiled in 20 μl 1× Laemmli buffer. The clarified samples were subjected to a 12% SDS-PAGE and transferred onto a PVDF membrane (Millipore). The membranes were exposed to a phosphorimager screen, and radioactive signals were scanned and quantified with QuantityOne software (Bio-Rad).
For Western blot analysis, cells in one well of a 6-well plate were washed once with PBS buffer and lysed in 300 μl of 1× Laemmli buffer. Thirty microliters of the cell lysate was resolved in a 10% SDS-PAGE gel, and proteins were transferred onto an Immobilon PVDF-FL membrane (Millipore). The membranes were blocked with Western Breeze blocking buffer (Invitrogen) and probed with antibodies against FLAG tag (product no. F1804; Sigma), IDO (product no. SC-25808; Santa Cruz), TDO (a gift from Christine Miller, Johns Hopkins University) (44), HBcAg (24), HBsAg (product no. 20-HR20; Fitzgerald), and β-actin (product no. MAB1501R; Chemicon). Bound antibodies were revealed by IRDye secondary antibodies and visualized using the Li-COR Odyssey system.
RESULTS
IFN-α and IFN-γ inhibit HBV replication in human hepatocyte-derived cells.
There are several reports demonstrating that both IFN-α and IFN-γ are able to inhibit HBV replication in human hepatoma-derived cell lines, such as HepG2 and Huh7. However, it remains controversial whether the cytokine-induced antiviral response primarily targets viral RNA or DNA biosynthesis or degradation (28, 53). We reasoned that some of the discrepancy in the previous reports might be related to the different cell culture systems employed in their studies. Therefore, we set out to compare, side by side, the effects of IFN-α and IFN-γ on the steady-state levels of intracellular HBV RNA and DNA in HBV replicon plasmids, transiently transfected HepG2 cells, and a HepG2-derived HBV-replicating stable cell line, specifically HepDE19 cells (23).
Interestingly, as shown in Fig. 1, while both IFN-α and IFN-γ efficiently reduced HBV pgRNA and surface RNAs in HepG2 cells transiently transfected with plasmids transcribing the viral pgRNA under the direction of either the HBV authentic core promoter (pHBV1.3) or a CMV-IE promoter (pCMVHBV) (Fig. 1A and B, top), neither IFN-α nor IFN-γ affected the levels of HBV RNAs in HepDE19 cells (Fig. 1C, top). However, the cytokines efficiently reduced HBV DNA in both transiently and stably transfected HepG2 cells in a dose-dependent manner (Fig. 1A, B, and C, bottom). Such observation is consistent with previous reports showing that IFN reduced HBV promoter activity in the reporter gene transfection assay (63) but not in a stable cell line harboring the integrated tandem HBV dimer, in which the viral RNA expression is under the control of authentic viral promoters (28). These results thus imply that IFN-induced antiviral response selectively inhibits the transcription of HBV RNA from input plasmids in transiently transfected cells but not from the integrated HBV transgene in the stable cell line. To further confirm this notion and rule out the possibility that the observed difference on HBV RNA level is due to different cell lines (HepG2 versus HepDE19), we performed reporter assays by cotransfection of plasmids encoding HBV core promoter-directed firefly luciferase (EnII/Cp-Luc) and CMV-IE promoter-directed Renilla luciferase (pRL-CMV) into either HepG2 cells or HepDE19 cells and treated with IFN-α or IFN-γ, respectively. As shown in Fig. 1D, the results indicate that the IFNs significantly inhibited HBV core and CMV-IE promoter-driven transcription in both HepG2 and HepDE19 cells to a similar extent, suggesting that the effect of IFNs on transcription is a specific phenomenon for a transfected plasmid-based transcription template.
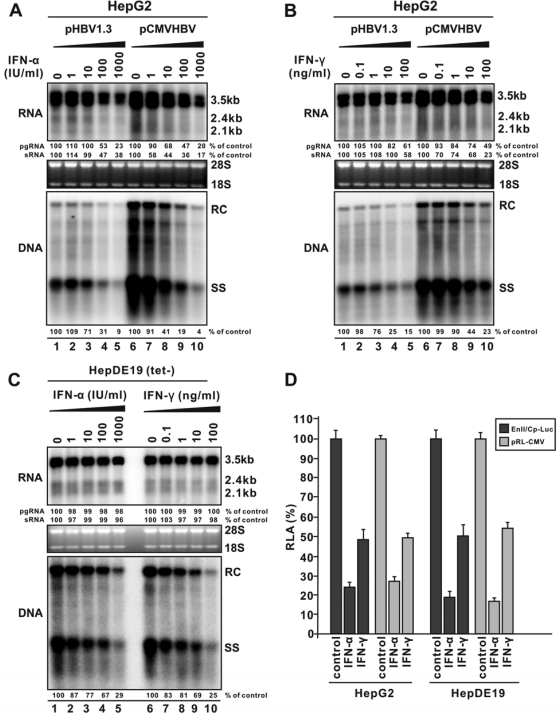
FIG. 1.
Inhibitory effects of IFNs against HBV replication in human hepatocyte-derived cells. (A) IFN-α inhibits HBV replication in transfected cells. HepG2 cells were transfected with 4 μg of pHBV1.3 containing 1.3-mer-genome-length HBV sequences or 4 μg of pCMVHBV, in which viral pgRNA transcription is under the control of the CMV-IE promoter. Sixteen hours posttransfection, cells were left untreated or treated with serial dilutions of IFN-α, as indicated. Cells were harvested at day 5 posttransfection, and the levels of viral RNA and core-associated DNA were determined by Northern (top) and Southern (bottom) blot hybridization, respectively. For RNA analysis, each lane was loaded with 10 μg of total RNA and probed with a genome-length, plus-strand-specific HBV riboprobe. Ribosomal RNAs (28S and 18S) were presented as loading controls. The positions of HBV pgRNA (3.5 kb) and surface RNAs (sRNA; 2.4 kb and 2.1 kb) are indicated. For DNA analysis, HBV core DNA was probed with a genome-length, minus-strand-specific HBV riboprobe. The positions of relaxed circular (RC) and single-stranded (SS) DNAs are indicated. The relative viral pgRNA, sRNA, or total DNA replicative intermediate level in each sample is expressed as the percentage of RNA or DNA in untreated cells (lanes 1 and 6) and is indicated underneath each of the blots. (B) IFN-γ inhibits HBV replication in transfected cells. HepG2 cells were transfected in the same way as described above, followed by the treatment with IFN-γ at the indicated concentrations. Five days later, viral nucleic acids were quantified as described above. (C) IFNs inhibit HBV replication in a stable cell line. HepDE19 cells were cultured in the presence of 1 μg/ml tetracycline (tet) until confluent, followed by the withdrawal of tet. Cells were then left untreated or treated with serial doses of IFN-α or IFN-γ. Five days later, cells were harvested, and viral DNA and RNA were analyzed as described above. (D) IFNs inhibit viral promoter activity in transfected cells. HepG2 cells and HepDE19 cells (tet−) were seeded in a 96-well plate and cotransfected with 200 ng of EnII/Cp-Luc and 4 ng of pRL-CMV. Sixteen hours posttransfection, cells were left untreated or treated with IFN-α (1,000 IU/ml) or IFN-γ (100 ng/ml). Four days after transfection, cells were harvested, and luciferase activities were determined as described in Materials and Methods. The plotted relative luciferase activity (RLA) represents the means ± standard deviations (SD; n = 4 experiments) of the ratios of absorbance obtained from wells treated with IFNs to that obtained from wells that were untreated.
Thus, these results allowed us to conclude that the specific antiviral activity of both IFN-α and IFN-γ is to inhibit HBV replication in human hepatoma cells by posttranscriptionally reducing viral DNA replication. Such an effect is observed only in HBV stable cell lines but could not be clearly revealed in the transiently transfected cells due to a primary strong inhibition of IFNs on HBV RNA transcription from the input plasmids.
Identification of IFN-induced cellular proteins that inhibit HBV DNA replication.
In our previous efforts to identify ISGs that mediate the antiviral effects of IFNs against hepatitis C virus and dengue virus, we established a library of ISG expression plasmids, including more than 30 ISGs that are highly induced by IFN-α in Huh7 cells and 6 ISGs that are commonly induced by IFN-γ in several cell types, including macrophages and hepatocytes (27, 33, 34) (Table 1). By taking advantage of this resource, with the addition of two IFN-γ-inducible enzymes, specifically IDO and iNOS (15, 31), we set out to identify ISGs that mediate the effects of IFNs on HBV DNA replication in HepG2 cells.
HepG2 cells were cotransfected with pHBV1.3 and a control plasmid expressing chloramphenicol acetyltransferase (pcDNA5CAT; Invitrogen) or plasmids expressing individual ISGs at a 1:1 ratio. Five days later, cells were harvested and the expression of each individual ISG was confirmed by Western blot analysis with an antibody against a FLAG epitope that is engineered at the N terminus of the ISG polypeptides (data not shown). The culture media were collected for cytotoxicity assay with the Promega cell proliferation assay kit and revealed no toxicity of any ISG upon expression (data not shown). Intracellular HBV DNA was analyzed by Southern blot hybridization. As shown in Table 1, among the 37 ISGs tested, overexpression of 7 ISGs reduced HBV DNA by more than 40% compared with that of cells cotransfected with a control plasmid expressing chloramphenicol acetyltransferase (CAT). To examine if the ISGs affect HBV RNA transcription from the input pHBV1.3 plasmid DNA, the levels of HBV RNA in the above 7 ISG-expressing plasmids cotransfected cells were assayed by Northern blot hybridization. Interestingly, except for two IFN-induced cellular enzymes, APOBEC3G and IDO, the other 5 ISGs potently reduced the level of HBV RNA (Table 1).
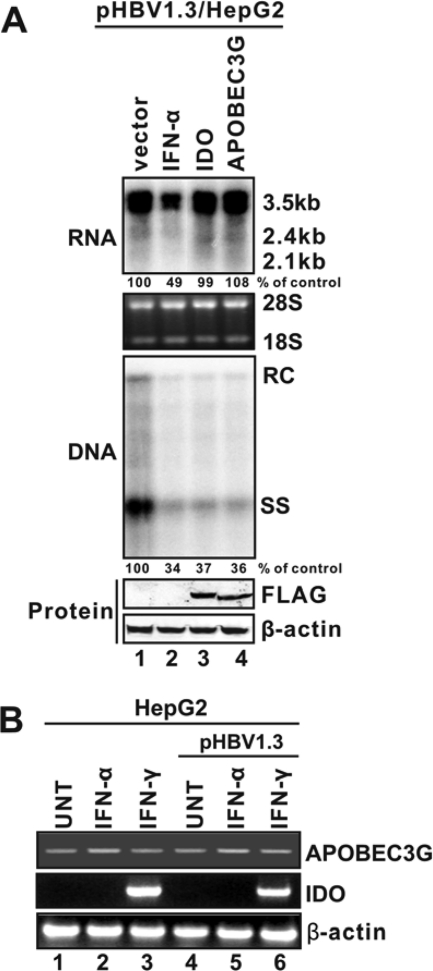
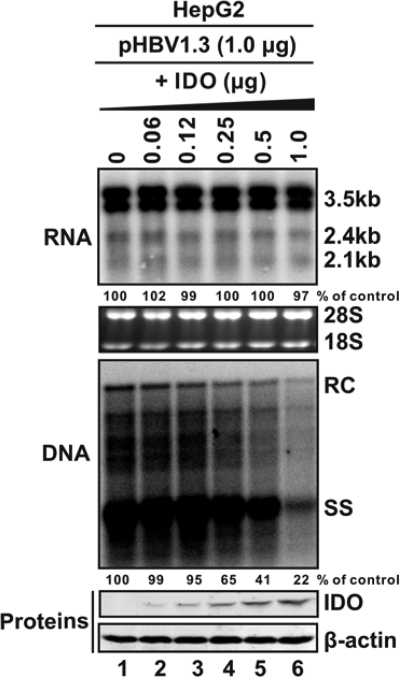
As demonstrated in Fig. 2, while IFN-α reduced both HBV RNA and DNA in pHBV1.3-transfected HepG2 cells, cotransfection of either APOBEC3G or IDO did not apparently affect HBV RNA but dramatically reduced HBV DNA (Fig. 2A, top and middle). Moreover, we demonstrated that IDO can be efficiently induced by IFN-γ but not IFN-α in HepG2 cells under the condition without or with HBV replication (Fig. 2B). Furthermore, similar to observations in HepDE19 cells treated with IFN-γ, expression of IDO dose-dependently inhibited HBV DNA replication in HepG2 cells, without altering the level of HBV RNA at any concentration tested (Fig. 3).
FIG. 2.
Inhibition of HBV replication by APOBEC3G and IDO. (A) APOBEC3G and IDO predominantly inhibit HBV DNA replication without reducing viral RNA levels. HepG2 cells were cotransfected with 2 μg of pHBV1.3 and 2 μg of control vector (lanes 1 and 2), 2 μg of plasmids that express IDO (lane 3), or APOBEC3G (lane 4). Sixteen hours later, one set of cells transfected with pHBV1.3 plus the control vector was treated with IFN-α (1,000 IU/ml). Cells were harvested at day 5 posttransfection, and viral RNA and core DNA were analyzed by Northern (top) and Southern (middle) blot assays, respectively. The relative viral pgRNA or total DNA level in each sample is expressed as the percentage of pgRNA or total DNA level in the control sample (lane 1) and is presented underneath each of the blots. Expression of APOBEC3G and IDO was revealed by Western blot analysis with FLAG antibodies. The levels of β-actin served as a loading control (bottom). (B) Induction profiles of APOBEC3G and IDO under IFN treatment. HepG2 cells were left untransfected or transfected with 2 μg of pHBV1.3. Sixteen hours later, cells were left untreated or treated with IFN-α (1,000 IU/ml) or IFN-γ (100 ng/ml) for 48 h. Total cellular RNA was extracted by TRIzol and followed by cDNA synthesis, and the levels of APOBEC3G and IDO mRNA were determined by semiquantitative RT-PCR assay as described in Materials and Methods. RT-PCR products of β-actin mRNA served as loading controls.
FIG. 3.
IDO inhibits HBV DNA replication in a dose-dependent manner. HepG2 cells were cotransfected with 1 μg of pHBV1.3 and 1 μg of control vector (lane 1), 1 μg of IDO plasmid (lane 6), or serially decreasing amounts of IDO plasmid plus the control vector (1 μg in total) (lanes 2 to 5). Cells were harvested at day 5 posttransfection, and viral RNA and core DNA were analyzed by Northern (top) and Southern (middle) blot assays, respectively. The relative viral pgRNA or total DNA level in each sample is expressed as the percentage of pgRNA or total DNA level in the control sample (lane 1) and is presented underneath each of the blots. Dose-dependent expression of IDO was revealed by Western blot analysis with FLAG antibodies, with the levels of β-actin serving as a loading control (bottom).
While the five ISGs that reduce HBV RNA could be the cellular proteins that mediate the observed decline of viral RNA in the transfected cells under IFN treatment, we are more interested in the ISGs that target HBV DNA replication, which represent a more authentic antiviral effect of the cytokines, as suggested by our studies described in the previous section. Since the antiviral mechanism of APOBEC3G against HBV has been well studied (45, 46), we set out to further elucidate the antiviral mechanism of IDO and determine its role in IFN-induced antiviral response against HBV.
IDO inhibited HBV DNA replication via tryptophan deprivation.
IDO is a heme-containing enzyme that catalyzes the conversion of l-tryptophan into NAD through the kynurenine pathway, which is the major metabolic route of tryptophan degradation in humans (52, 61). Therefore, we first investigated whether the antiviral function of IDO depends upon its enzymatic activity and is mediated by tryptophan metabolism. To this end, HepG2 cells were transfected with pHBV1.3 in combination with a control vector expressing CAT or a plasmid expressing either wild-type or enzymatically inactive mutant IDO that contains an amino acid substitution in heme binding sites, specifically IDOH346A (37). When indicated, wild-type IDO-cotransfected cells were cultured with an excess amount of l-tryptophan or an IDO inhibitor, 1-MT. As shown in Fig. 4 and consistent with the results presented above, expression of wild-type IDO efficiently reduced HBV DNA without reducing the steady-state level of HBV RNA (Fig. 4, top and middle, lane 3). Interestingly, IDO expression dramatically reduced the levels of HBV precore, core, and envelope (L/M/S) proteins (Fig. 4, bottom, lane 3). These results suggest that the reduction of HBV DNA in IDO-transfected cells is secondary to the reduced viral protein synthesis, especially the core protein, which serves as an essential component for the formation of HBV nucleocapsid (56).
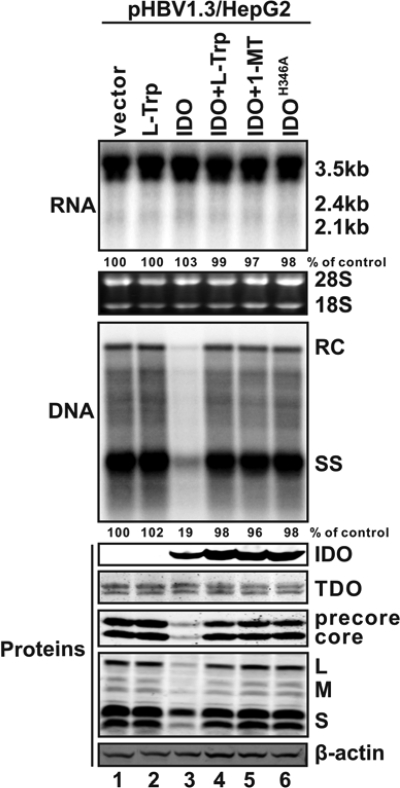
FIG. 4.
IDO inhibits HBV replication via tryptophan deprivation. HepG2 cells were cotransfected with 2 μg of pHBV1.3 and 2 μg of control plasmid (lanes 1 and 2), 2 μg of IDO plasmid (lanes 3 to 5), or 2 μg of an enzymatically inactive IDO mutant (IDOH346A; lane 6). Sixteen hours after transfection, cells were left untreated (lanes 1, 3, and 6) or treated with 100 μg/ml l-tryptophan (l-Trp; lanes 2 and 4) or 100 μg/ml 1-methyl-l-trp (1-MT; lane 5). Cells were harvested at day 5 posttransfection, and viral RNAs and core DNA were analyzed by Northern (top) and Southern (middle) blot assays, respectively. The relative viral pgRNA or total DNA levels in each sample are expressed as the percentage of pgRNA or total DNA levels in the control sample (lane 1) and are presented underneath each of the blots. Expression of the indicated viral and host proteins was analyzed by Western blot analysis with antibodies described in Materials and Methods, and the levels of β-actin served as loading controls (bottom).
Based on IDO's ability to metabolize the essential amino acid tryptophan, it is very likely that the observed inhibition of HBV protein synthesis and thus DNA replication is due to the IDO-catalyzed tryptophan depletion. In support of this hypothesis, an IDO enzymatic inhibitor, 1-MT, completely abolished IDO's antiviral activity (Fig. 4, lane 5); expression of an enzymatically inactive IDOH346A failed to inhibit HBV protein synthesis and DNA replication (Fig. 4, lane 6), thus suggesting that IDO's antiviral activity relies on its catalytic function. Furthermore, supplementation of tryptophan into the culture medium completely restored viral protein expression and subsequently rescued HBV DNA replication in IDO-expressing cells (Fig. 4, lane 4). Collectively, the above data clearly favor the notion that IDO inhibits HBV DNA replication through heme-catalyzed tryptophan deprivation and not via tryptophan metabolites, such as kynurenine and NAD.
Interestingly, a noninducible tryptophan metabolic enzyme, tryptophan 2,3-dioxygenase (TDO), is constitutively expressed in hepatocytes as well as in skin cells (44, 52, 61). Therefore, it was of interest to ask whether the endogenous TDO is involved in the tryptophan depletion-mediated antiviral response. While Western blot analysis indeed revealed the expression of two TDO isoforms in HepG2 cells (Fig. 4, middle), tryptophan supplementation in the absence of IDO expression did not further enhance the viral protein expression and DNA replication (Fig. 4, lane 2). This result indicates that endogenous TDO is not a limiting factor for HBV replication in cultured cells.
IDO preferentially inhibited HBV but not cellular protein synthesis.
It has been shown that IDO-induced local tryptophan deprivation in vivo usually results in T lymphocyte growth inhibition or apoptosis (12, 43). However, we did not observe any cytotoxic effect in HepG2 cultures transfected with a plasmid expressing IDO (data not shown). In addition, the fact that overexpression of IDO does not affect the steady-state level of HBV pgRNA, for which the half-life is less than 12 h in cell cultures (11, 68, 69), plus the lack of effect on the steady-state content of cellular proteins, such as TDO and β-actin (Fig. 4, bottom, lane 3), strongly suggested that expression of IDO in HepG2 cells did not significantly affect the host cellular transcription and translation machinery but preferentially inhibited HBV protein and DNA biosynthesis. One possible explanation for such a selective antiviral effect is that confluent HepG2 cells are less sensitive to amino acid starvation than HBV, possibly due to the viral replication putting a greater demand on the amino acid pool and tryptophan in particular, since it is the rarest essential amino acid in humans.
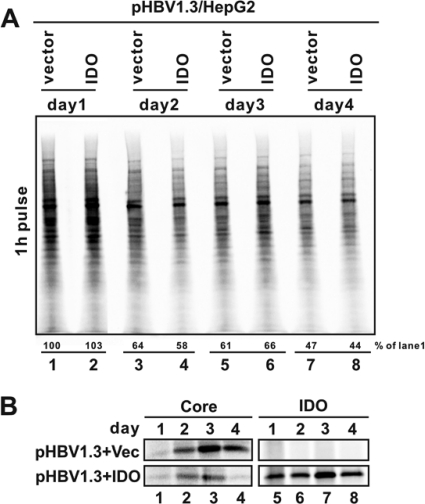
To investigate this possibility, we performed a pulse-labeling experiment to determine the impact of IDO expression on viral and host cellular protein translation activity. As shown in Fig. 5A, compared with control vector-transfected cells, IDO expression did not significantly reduce the global cellular protein synthesis at any time point examined. The gradual reduction of global protein translation over time is most likely due to the cell density-dependent growth arrest (4, 58). To determine the effect of IDO on HBV protein translation, HBV core protein and IDO synthesis were determined by immunoprecipitation of the indicated proteins with specific antibodies followed by electrophoresis and phosphorimaging analysis. As shown in Fig. 5B, the translation activity of HBV core protein increased gradually and reached the peak at day 3 after transfection in control plasmid-transfected cells (top left), which was similar to the kinetics of IDO expression (bottom right). However, the rate of core protein synthesis was dramatically reduced in the presence of IDO at days 2 to 4 (bottom left). These results thus imply that IDO expression preferentially inhibits the de novo synthesis of viral but not global host cellular proteins under the selected experimental condition.
FIG. 5.
IDO expression does not alter global protein synthesis but preferentially inhibits viral protein translation. (A) Cellular protein synthesis is not blocked by IDO-mediated tryptophan starvation. HepG2 cells were cotransfected with 2 μg of pHBV1.3 and 2 μg of control vector (lanes 1, 3, 5, and 7) or IDO plasmid (lanes 2, 4, 6, and 8). Sixteen hours later, one pair of transfected cells (lanes 1 and 2, minus and plus IDO, respectively) were pulse labeled for 1 h with 35S-Met and harvested as samples of day 1. Afterward, the other three pairs of cells were pulse labeled for 1 h and harvested at days 2, 3, and 4 posttransfection. To determine total cellular protein translation activity, cell lysate samples were prepared as described in Materials and Methods and resolved in SDS-PAGE, followed by transfer onto a PVDF membrane and exposure to a phosphorimager screen (Bio-Rad). The relative level of labeled proteins in each sample is expressed as the percentage of radioactivity in the control sample (lane 1) and is presented underneath. (B) IDO expression blocked de novo synthesis of viral core protein. Cell lysates obtained from the above experiment were subjected to immunoprecipitation with bead-bounded antibodies against HBcAg and FLAG-tagged IDO, respectively. Precipitated proteins were resolved in SDS-PAGE and transferred onto a PVDF membrane, and 35S-labeled HBV core protein (left) and IDO (right) were revealed by autoradiography.
IDO is a primary mediator of the IFN-γ-induced antiviral response against HBV.
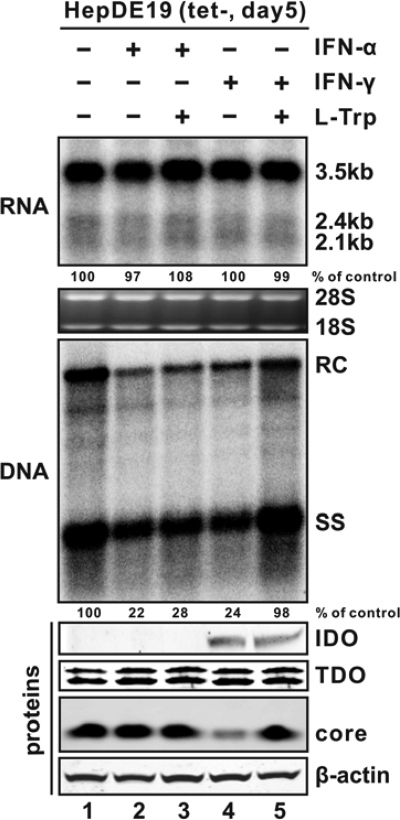
As described above, IFN-γ treatment reduces HBV DNA without altering the steady-state level of viral RNA in HepDE19 cells (Fig. 1C). Interestingly, expression of IDO, an IFN-γ-inducible protein in HepG2 cells (Fig. 2B), elicits a similar antiviral effect. These observations prompted us to investigate the physiological role of IDO in IFN-γ-induced antiviral response in HepDE19 cells. For this purpose, we attempted to examine whether tryptophan supplementation could abrogate the antiviral function of IFNs in HepDE19 cells. As shown in Fig. 6, while neither IFN-α nor IFN-γ further induced TDO expression, IDO expression was profoundly upregulated under IFN-γ treatment but not by IFN-α stimulation. Upon the IDO induction by IFN-γ, HBV core protein expression was significantly inhibited but remained unchanged in IFN-α-treated cells (Fig. 6, bottom). The absence of precore protein in HepDE19 cells was due to the start codon of the precore open reading frame (ORF) having been artificially mutated in the integrated HBV genome during cell line establishment (23). Although both cytokines dramatically inhibited HBV DNA replication without reducing the steady-state content of viral RNA (Fig. 6, top), tryptophan supplementation did not abolish the antiviral effect of IFN-α; remarkably, IFN-γ-elicited antiviral response, including the reduction of viral core protein expression and subsequent DNA replication, was completely attenuated by tryptophan supplementation (Fig. 6, middle). Hence, our results strongly support the notion that IDO is a primary, if not the only, ISG that mediates the antiviral effect of IFN-γ against HBV in human hepatocyte-derived cells.
FIG. 6.
IDO mediates IFN-γ-elicited inhibition of HBV replication. HepDE19 cells were cultured in medium containing 1 μg/ml tet until confluent, after which the cells were cultured in tet-free medium without treatment (lane 1), treated with 1,000 IU/ml IFN-α alone (lane 2) or in combination with 100 μg/ml l-Trp (lane 3), or treated with 100 ng/ml IFN-γ alone (lane 4) or with supplementation of 100 μg/ml l-Trp (lane 5). Cells were harvested at day 5 after tet withdrawal, and viral RNA and core DNA levels were analyzed by Northern (top) and Southern (middle) blot assays, respectively. The relative viral pgRNA or total DNA level in each sample is expressed as the percentage of pgRNA or total DNA level in the control sample (lane 1) and is presented underneath each of the blots. Induction of endogenous IDO by IFN-γ was confirmed by Western blot analysis with IDO-specific antibodies (Santa Cruz). Expression of TDO and HBV core protein was analyzed by Western blot analysis with the antibodies described in Materials and Methods. The levels of β-actin served as a loading control (bottom).
DISCUSSION
Type I IFNs, represented by IFN-α/β, are the principal mediators of host innate antiviral defense in vertebrates (13, 19). IFN-α is an FDA-approved therapeutic agent for chronic hepatitis B (32, 50). IFN-γ, produced by activated T lymphocytes and natural killer cells, is a mediator of the adaptive antiviral response and plays a key role in noncytolytic inhibition of HBV replication during the clearance phase of acute HBV infection by viral antigen-specific CTL (9, 19). Although the antiviral activity and mechanism of IFN-α and IFN-γ against HBV have been well documented in murine hepatocyte cultures and a transgenic mouse model in vivo, the antiviral mechanism of the cytokines against HBV in human hepatocytes is not well understood. To resolve this issue, we first clarified a long-standing controversy on whether IFNs target HBV RNA or DNA biosynthesis and/or degradation in human hepatoma cells. Our results clearly demonstrated that both IFN-α and IFN-γ inhibited HBV replication in human hepatoma cells by posttranscriptionally reducing viral DNA replication. However, due to the fact that IFNs potently inhibit HBV RNA transcription from input plasmids, the effects of the cytokines on HBV DNA replication could not be directly revealed in transiently transfected hepatoma cells. Although it is possible that the observed inhibition of IFNs on plasmid-based transcription may mimic the cytokines' effects on HBV covalently closed circular DNA (cccDNA)-dependent transcription, the fact that IFNs inhibit both HBV core promoter- and CMV-IE promoter-driven transcription to a similar extent strongly suggests that the observed transcriptional inhibition is a nonspecific, even artificial effect in transiently transfected cells. Hence, we consider that inhibition of HBV DNA replication, rather than RNA transcription, is the primary target of IFNs in human hepatocyte-derived cells. Accordingly, we took a forward genetic approach to identify IFN-induced cellular proteins that may mediate the inhibitory effect of IFN-α and IFN-γ on HBV DNA replication.
Taking advantage of our ISG expression library, which also includes the three ISGs, namely APOBEC3G (45, 46), myxovirus resistance A (MxA) (16), and tripartite motif-containing 22 (TRIM22) (14), that are reportedly able to inhibit HBV replication in human hepatocyte-derived cells, we first performed a cotransfection assay in HepG2 cells to identify individual ISGs that are able to reduce the intracellular level of HBV DNA. After ruling out the ISGs that reduce HBV RNA, we discovered two ISGs, APOBEC3G and IDO, that significantly reduced HBV DNA but did not affect the steady-state level of HBV RNA and thus most likely inhibited HBV DNA replication. While we were able to confirm the anti-HBV effect of APOBEC3G, the antiviral effects of MxA and TRIM22 could not be conclusively demonstrated in our assay.
APOBEC3G is an IFN-α-inducible cytidine deaminase that inhibits the replication of human immunodeficiency virus (HIV) and other retroviruses by inducing deoxycytidine to deoxyuridine hypermutations in viral cDNA and thus impairing the coding and replicative capacity of the viruses (26). The editing activity of APOBEC3G on HBV DNA has also been observed in previous studies, albeit at a lower efficiency (7, 47, 60, 62). Moreover, APOBEC3G can be packaged into HBV nucleocapsids and inhibit HBV DNA reverse transcription in a deaminase-independent manner (45, 46). However, because APOBEC3G is not efficiently induced in HepG2 cells by either IFN-α or IFN-γ (Fig. 2B), in conjunction with the previous reports showing that APOBEC3G is not an essential effector in the IFN-elicited anti-HBV response (35, 51), its role in mediating the antiviral effects of IFNs against HBV remains unclear.
On the contrary, IDO can be efficiently induced in a variety of cell types, including hepatocytes, by IFN-γ in vitro and in vivo (31, 42). As mentioned above, IDO is a cytosolic heme-containing enzyme catalyzing the oxidation of l-tryptophan into N-formylkynurenine, the first and rate-limiting step in the kynurenine pathway. Therefore, IDO plays a central role in the regulation of tryptophan metabolism upon induction by IFN-γ (61). In fact, tryptophan is the least abundant essential amino acid in mammals, and deprivation of tryptophan can lead to numerous pathophysiological and immunological consequences (61). Most importantly, IDO is considered a key immunoregulatory enzyme to induce and maintain peripheral immune tolerance by induction of tryptophan deprivation, which results in T cell growth inhibition and apoptosis (12, 43). Such a strategy is also utilized by many tumors to develop immune subversion and appears to be the mechanism of maternal acceptance of the semiallogeneic fetus (38, 39).
Not surprisingly, IDO, upon induction by IFN-γ, has potent antimicrobial and antiviral activities against a variety of intracellular pathogens, including toxoplasma (10), chlamydia (8), streptococci (40), human cytomegalovirus (6), herpes simplex virus (1, 2), measles virus (48), and HIV (59). In all these cases, the antimicrobial function of IDO is mediated by tryptophan deprivation. In this study, we demonstrated for the first time that IDO noncytolytically inhibited HBV DNA replication, via preferential inhibition of HBV, but not host cellular, protein biosynthesis. Most strikingly, the antiviral state of IFN-γ in HepDE19 cells is single-handedly built up by the induction of IDO. Given the fact that tryptophan is the rarest essential amino acid in human cells, it is conceivable that a slight depletion of intracellular tryptophan reservoir could result in a drastic decrease of viral protein production, especially in confluent cells at the stationary growth phase, where the viral biosynthesis operates optimally to support virus replication (57). In addition, accumulation of uncharged tryptophanal tRNA may serve as a stress signal to activate GCN2 kinase, which in turn phosphorylates eukaryotic translation initiation factor 2 alpha (eIF2α) to further suppress protein translation (65).
Although TDO is a constitutively expressed hepatic tryptophan degradation enzyme, we have shown that it is not a cellular factor that limits HBV replication. What is more, while IDO is highly induced by IFN-γ in hepatocyte-derived cells, TDO is not further induced by IFNs. This is consistent with previous reports that the endogenous TDO removes only the excess amount of tryptophan to maintain the tryptophan level in the blood instead of causing tryptophan starvation (55). Such different roles of TDO and IDO in tryptophan homeostasis are due mainly to IDO having a much higher substrate affinity than TDO, with a Km value of 10 μM for IDO versus 222 μM for TDO (52). In agreement with this, we have also observed that even overexpression of TDO had a marginal antiviral effect against HBV replication in transfected HepG2 cells (data not shown).
There is a growing body of evidence suggesting the physiologic antiviral function of IDO under the condition of HBV natural infection. It has been shown that the recovery from acute infection of woodchuck hepatitis virus correlated with the elevated level of IFN-γ and increased intrahepatic expression of IDO (64). What's more, administration of HBV-specific CTLs into an acute hepatitis B transgenic mouse model resulted in enhanced IDO expression in hepatocytes and upregulation of serum kynurenine levels (31). These evidences, together with IDO's immunoregulatory and antiviral functions, thus favor a notion that IDO plays a dual role in virus-host interaction. On one hand, intrahepatic IDO induction by IFN-γ inhibits HBV replication; on the other hand, IDO-induced tryptophan deprivation dampens CTL killing of virally infected hepatocytes and thus limits the cytopathic effects of adaptive antiviral immune response. Hence, IDO may play a critical role in balancing the cytopathic and noncytopathic antiviral mechanisms of host immune response and thus determining the pathogenesis and outcomes of HBV infection (21).
Taken together, our work presented herein has identified two ISGs that potently inhibit HBV DNA replication in human hepatocyte-derived cells. More importantly, we provide compelling evidence suggesting that IDO is a primary, if not the only, mediator of IFN-γ-induced antiviral response against HBV in HepG2 cells. Our work thus provides valuable insight for better understanding of host antiviral immunity and HBV pathogenesis.
Acknowledgments
We thank William Mason (Fox Chase Cancer Center) and Pamela Norton for critical reading of the manuscript.
This work was supported by NIH grant AI088424 (to Haitao Guo) and the Hepatitis B Foundation through an appropriation of the Commonwealth of Pennsylvania. Work in Jiming Zhang's laboratory was supported by grants from the National Science and Technology Major Project of China (2008ZX10002-006, 2009ZX10603). Richeng Mao received scholarships from the Hepatitis B Foundation and Chinese Scholarship Council. Haitao Guo and Ju-Tao Guo are a Bruce Witte fellow and a scholar of the Hepatitis B Foundation, respectively.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Adams, O., et al. 2004. Inhibition of human herpes simplex virus type 2 by interferon gamma and tumor necrosis factor alpha is mediated by indoleamine 2,3-dioxygenase. Microbes Infect. 6:806-812. [DOI] [PubMed] [Google Scholar]
- 2.Adams, O., et al. 2004. Role of indoleamine-2,3-dioxygenase in alpha/beta and gamma interferon-mediated antiviral effects against herpes simplex virus infections. J. Virol. 78:2632-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, A. L., K. E. Banks, M. Pontoglio, M. Yaniv, and A. McLachlan. 2005. Alpha/beta interferon differentially modulates the clearance of cytoplasmic encapsidated replication intermediates and nuclear covalently closed circular hepatitis B virus (HBV) DNA from the livers of hepatocyte nuclear factor 1α-null HBV transgenic mice. J. Virol. 79:11045-11052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baenziger, N. L., C. H. Jacobi, and R. E. Thach. 1974. Regulation of protein synthesis during density-dependent growth inhibition of BHK21-13 cells. J. Biol. Chem. 249:3483-3488. [PubMed] [Google Scholar]
- 5.Block, T. M., H. Guo, and J. T. Guo. 2007. Molecular virology of hepatitis B virus for clinicians. Clin. Liver Dis. 11:685-706, vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bodaghi, B., et al. 1999. Role of IFN-gamma-induced indoleamine 2,3 dioxygenase and inducible nitric oxide synthase in the replication of human cytomegalovirus in retinal pigment epithelial cells. J. Immunol. 162:957-964. [PubMed] [Google Scholar]
- 7.Bonvin, M., et al. 2006. Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43:1364-1374. [DOI] [PubMed] [Google Scholar]
- 8.Carlin, J. M., E. C. Borden, and G. I. Byrne. 1989. Interferon-induced indoleamine 2,3-dioxygenase activity inhibits Chlamydia psittaci replication in human macrophages. J. Interferon Res. 9:329-337. [DOI] [PubMed] [Google Scholar]
- 9.Chisari, F. V., M. Isogawa, and S. F. Wieland. 2010. Pathogenesis of hepatitis B virus infection. Pathol. Biol. (Paris) 58:258-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daubener, W., et al. 2001. Restriction of Toxoplasma gondii growth in human brain microvascular endothelial cells by activation of indoleamine 2,3-dioxygenase. Infect. Immun. 69:6527-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehlers, I., et al. 2004. Functional characterization of the interaction between human La and hepatitis B virus RNA. J. Biol. Chem. 279:43437-43447. [DOI] [PubMed] [Google Scholar]
- 12.Fallarino, F., et al. 2002. T cell apoptosis by tryptophan catabolism. Cell Death Differ. 9:1069-1077. [DOI] [PubMed] [Google Scholar]
- 13.Fensterl, V., and G. C. Sen. 2009. Interferons and viral infections. Biofactors 35:14-20. [DOI] [PubMed] [Google Scholar]
- 14.Gao, B., Z. Duan, W. Xu, and S. Xiong. 2009. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50:424-433. [DOI] [PubMed] [Google Scholar]
- 15.Geller, D. A., et al. 1993. Molecular cloning and expression of inducible nitric oxide synthase from human hepatocytes. Proc. Natl. Acad. Sci. U. S. A. 90:3491-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordien, E., et al. 2001. Inhibition of hepatitis B virus replication by the interferon-inducible MxA protein. J. Virol. 75:2684-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guidotti, L. G., et al. 1994. Cytotoxic T lymphocytes inhibit hepatitis B virus gene expression by a noncytolytic mechanism in transgenic mice. Proc. Natl. Acad. Sci. U. S. A. 91:3764-3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guidotti, L. G., and F. V. Chisari. 2006. Immunobiology and pathogenesis of viral hepatitis. Annu. Rev. Pathol. 1:23-61. [DOI] [PubMed] [Google Scholar]
- 19.Guidotti, L. G., and F. V. Chisari. 2001. Noncytolytic control of viral infections by the innate and adaptive immune response. Annu. Rev. Immunol. 19:65-91. [DOI] [PubMed] [Google Scholar]
- 20.Guidotti, L. G., et al. 1996. Intracellular inactivation of the hepatitis B virus by cytotoxic T lymphocytes. Immunity 4:25-36. [DOI] [PubMed] [Google Scholar]
- 21.Guidotti, L. G., et al. 1999. Viral clearance without destruction of infected cells during acute HBV infection. Science 284:825-829. [DOI] [PubMed] [Google Scholar]
- 22.Guo, H., et al. 2009. Activation of pattern recognition receptor-mediated innate immunity inhibits the replication of hepatitis B virus in human hepatocyte-derived cells. J. Virol. 83:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo, H., et al. 2007. Characterization of the intracellular deproteinized relaxed circular DNA of hepatitis B virus: an intermediate of covalently closed circular DNA formation. J. Virol. 81:12472-12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo, H., R. Mao, T. M. Block, and J. T. Guo. 2010. Production and function of the cytoplasmic deproteinized relaxed circular DNA of hepadnaviruses. J. Virol. 84:387-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo, H., et al. 2007. Regulation of hepatitis B virus replication by the phosphatidylinositol 3-kinase-akt signal transduction pathway. J. Virol. 81:10072-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. S., and M. T. Liddament. 2004. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 4:868-877. [DOI] [PubMed] [Google Scholar]
- 27.Hayashi, J., R. Stoyanova, and C. Seeger. 2005. The transcriptome of HCV replicon expressing cell lines in the presence of alpha interferon. Virology 335:264-275. [DOI] [PubMed] [Google Scholar]
- 28.Hayashi, Y., and K. Koike. 1989. Interferon inhibits hepatitis B virus replication in a stable expression system of transfected viral DNA. J. Virol. 63:2936-2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoofnagle, J. H., E. Doo, T. J. Liang, R. Fleischer, and A. S. Lok. 2007. Management of hepatitis B: summary of a clinical research workshop. Hepatology 45:1056-1075. [DOI] [PubMed] [Google Scholar]
- 30.Isogawa, M., M. D. Robek, Y. Furuichi, and F. V. Chisari. 2005. Toll-like receptor signaling inhibits hepatitis B virus replication in vivo. J. Virol. 79:7269-7272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwamoto, N., et al. 2009. Upregulation of indoleamine 2,3-dioxygenase in hepatocyte during acute hepatitis caused by hepatitis B virus-specific cytotoxic T lymphocytes in vivo. Liver Int. 29:277-283. [DOI] [PubMed] [Google Scholar]
- 32.Jafri, S. M., and A. S. Lok. 2010. Antiviral therapy for chronic hepatitis B. Clin. Liver Dis. 14:425-438. [DOI] [PubMed] [Google Scholar]
- 33.Jiang, D., et al. 2008. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J. Virol. 82:1665-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang, D., et al. 2010. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84:8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jost, S., P. Turelli, B. Mangeat, U. Protzer, and D. Trono. 2007. Induction of antiviral cytidine deaminases does not explain the inhibition of hepatitis B virus replication by interferons. J. Virol. 81:10588-10596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang, T. J. 2009. Hepatitis B: the virus and disease. Hepatology 49:S13-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Littlejohn, T. K., O. Takikawa, R. J. Truscott, and M. J. Walker. 2003. Asp274 and his346 are essential for heme binding and catalytic function of human indoleamine 2,3-dioxygenase. J. Biol. Chem. 278:29525-29531. [DOI] [PubMed] [Google Scholar]
- 38.Liu, X., R. C. Newton, S. M. Friedman, and P. A. Scherle. 2009. Indoleamine 2,3-dioxygenase, an emerging target for anti-cancer therapy. Curr. Cancer Drug Targets 9:938-952. [DOI] [PubMed] [Google Scholar]
- 39.Lob, S., and A. Konigsrainer. 2009. Role of IDO in organ transplantation: promises and difficulties. Int. Rev. Immunol. 28:185-206. [DOI] [PubMed] [Google Scholar]
- 40.MacKenzie, C. R., U. Hadding, and W. Daubener. 1998. Interferon-gamma-induced activation of indoleamine 2,3-dioxygenase in cord blood monocyte-derived macrophages inhibits the growth of group B streptococci. J. Infect. Dis. 178:875-878. [DOI] [PubMed] [Google Scholar]
- 41.McMahon, B. J. 2005. Epidemiology and natural history of hepatitis B. Semin. Liver Dis. 25(Suppl. 1):3-8. [DOI] [PubMed] [Google Scholar]
- 42.Mellor, A. L., and D. H. Munn. 2004. IDO expression by dendritic cells: tolerance and tryptophan catabolism. Nat. Rev. Immunol. 4:762-774. [DOI] [PubMed] [Google Scholar]
- 43.Mellor, A. L., and D. H. Munn. 1999. Tryptophan catabolism and T-cell tolerance: immunosuppression by starvation? Immunol. Today 20:469-473. [DOI] [PubMed] [Google Scholar]
- 44.Miller, C. L., et al. 2004. Expression of the kynurenine pathway enzyme tryptophan 2,3-dioxygenase is increased in the frontal cortex of individuals with schizophrenia. Neurobiol. Dis. 15:618-629. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen, D. H., S. Gummuluru, and J. Hu. 2007. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 81:4465-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nguyen, D. H., and J. Hu. 2008. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J. Virol. 82:6852-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noguchi, C., et al. 2005. G to A hypermutation of hepatitis B virus. Hepatology 41:626-633. [DOI] [PubMed] [Google Scholar]
- 48.Obojes, K., O. Andres, K. S. Kim, W. Daubener, and J. Schneider-Schaulies. 2005. Indoleamine 2,3-dioxygenase mediates cell type-specific anti-measles virus activity of gamma interferon. J. Virol. 79:7768-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pagliaccetti, N. E., E. N. Chu, C. R. Bolen, S. H. Kleinstein, and M. D. Robek. 2010. Lambda and alpha interferons inhibit hepatitis B virus replication through a common molecular mechanism but with different in vivo activities. Virology 401:197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perrillo, R. 2009. Benefits and risks of interferon therapy for hepatitis B. Hepatology 49:S103-S111. [DOI] [PubMed] [Google Scholar]
- 51.Proto, S., J. A. Taylor, S. Chokshi, N. Navaratnam, and N. V. Naoumov. 2008. APOBEC and iNOS are not the main intracellular effectors of IFN-gamma-mediated inactivation of hepatitis B virus replication. Antiviral Res. 78:260-267. [DOI] [PubMed] [Google Scholar]
- 52.Rafice, S. A., N. Chauhan, I. Efimov, J. Basran, and E. L. Raven. 2009. Oxidation of l-tryptophan in biology: a comparison between tryptophan 2,3-dioxygenase and indoleamine 2,3-dioxygenase. Biochem. Soc. Trans. 37:408-412. [DOI] [PubMed] [Google Scholar]
- 53.Rang, A., S. Gunther, and H. Will. 1999. Effect of interferon alpha on hepatitis B virus replication and gene expression in transiently transfected human hepatoma cells. J. Hepatol. 31:791-799. [DOI] [PubMed] [Google Scholar]
- 54.Robek, M. D., B. S. Boyd, and F. V. Chisari. 2005. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79:3851-3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schmidt, S. K., et al. 2009. Antimicrobial and immunoregulatory properties of human tryptophan 2,3-dioxygenase. Eur. J. Immunol. 39:2755-2764. [DOI] [PubMed] [Google Scholar]
- 56.Seeger, C., and W. S. Mason. 2000. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 64:51-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sells, M. A., A. Z. Zelent, M. Shvartsman, and G. Acs. 1988. Replicative intermediates of hepatitis B virus in HepG2 cells that produce infectious virions. J. Virol. 62:2836-2844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stoker, M. G., and H. Rubin. 1967. Density dependent inhibition of cell growth in culture. Nature 215:171-172. [DOI] [PubMed] [Google Scholar]
- 59.Suh, H. S., et al. 2007. Astrocyte indoleamine 2,3-dioxygenase is induced by the TLR3 ligand poly(I:C): mechanism of induction and role in antiviral response. J. Virol. 81:9838-9850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Suspene, R., et al. 2005. Extensive editing of both hepatitis B virus DNA strands by APOBEC3 cytidine deaminases in vitro and in vivo. Proc. Natl. Acad. Sci. U. S. A. 102:8321-8326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thackray, S. J., C. G. Mowat, and S. K. Chapman. 2008. Exploring the mechanism of tryptophan 2,3-dioxygenase. Biochem. Soc. Trans. 36:1120-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turelli, P., B. Mangeat, S. Jost, S. Vianin, and D. Trono. 2004. Inhibition of hepatitis B virus replication by APOBEC3G. Science 303:1829. [DOI] [PubMed] [Google Scholar]
- 63.Tur-Kaspa, R., et al. 1990. Alpha interferon suppresses hepatitis B virus enhancer activity and reduces viral gene transcription. J. Virol. 64:1821-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y., et al. 2004. Interferon-gamma-associated responses to woodchuck hepatitis virus infection in neonatal woodchucks and virus-infected hepatocytes. J. Viral Hepat. 11:404-417. [DOI] [PubMed] [Google Scholar]
- 65.Wek, R. C., H. Y. Jiang, and T. G. Anthony. 2006. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 34:7-11. [DOI] [PubMed] [Google Scholar]
- 66.Wieland, S. F., and F. V. Chisari. 2005. Stealth and cunning: hepatitis B and hepatitis C viruses. J. Virol. 79:9369-9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wieland, S. F., A. Eustaquio, C. Whitten-Bauer, B. Boyd, and F. V. Chisari. 2005. Interferon prevents formation of replication-competent hepatitis B virus RNA-containing nucleocapsids. Proc. Natl. Acad. Sci. U. S. A. 102:9913-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu, C., et al. 2010. Interferons accelerate decay of replication-competent nucleocapsids of hepatitis B virus. J. Virol. 84:9332-9340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhou, T., et al. 2006. Hepatitis B virus e antigen production is dependent upon covalently closed circular (ccc) DNA in HepAD38 cell cultures and may serve as a cccDNA surrogate in antiviral screening assays. Antiviral Res. 72:116-124. [DOI] [PubMed] [Google Scholar]