Abstract
Although human adenovirus type 5 (Ad5) has been widely studied, relatively little work has been done with other human adenovirus serotypes. The Ad5 E4orf6 and E1B55K proteins form Cul5-based E3 ubiquitin ligase complexes to degrade p53, Mre11, DNA ligase IV, integrin α3, and almost certainly other targets, presumably to optimize the cellular environment for viral replication and perhaps to facilitate persistence or latency. As this complex is essential for the efficient replication of Ad5, we undertook a systematic analysis of the structure and function of corresponding E4orf6/E1B55K complexes from other serotypes to determine the importance of this E3 ligase throughout adenovirus evolution. E4orf6 and E1B55K coding sequences from serotypes representing all subgroups were cloned, and each pair was expressed and analyzed for their capacity to assemble the Cullin-based ligase complex and to degrade substrates following plasmid DNA transfection. The results indicated that all formed Cullin-based E3 ligase complexes but that heterogeneity in both structure and function existed. Whereas Cul5 was present in the complexes of some serotypes, others recruited primarily Cul2, and the Ad16 complex clearly bound both Cul2 and Cul5. There was also heterogeneity in substrate specificity. Whereas all serotypes tested appeared to degrade DNA ligase IV, complexes from some serotypes failed to degrade Mre11, p53, or integrin α3. Thus, a major evolutionary pressure for formation of the adenovirus ligase complex may lie in the degradation of DNA ligase IV; however, it seems possible that the degradation of as-yet-unidentified critical targets or, perhaps even more likely, appropriate combinations of substrates plays a central role for these adenoviruses.
The human adenovirus type 5 (Ad5) early region 4 34-kDa product from open reading frame 6 (E4orf6) and the E1B55K protein have been known for some time to act in concert to carry out several important functions during the infectious cycle, including regulation of the activity and stability of p53 and, late in infection, the selective transport of viral mRNAs (2, 14, 18, 19, 37, 46, 51, 52, 56). Our group and others have shown that the cooperative functions of these Ad5 proteins, including the E4orf6-E1B55K interaction itself, appear to require formation of a Cul5-based E3 ubiquitin ligase complex (8). We showed that Ad5 E4orf6 recruits an E3 ubiquitin ligase complex containing the Cullin family member Cul5, Elongins B and C, and the RING protein Rbx1 (8, 20). E1B55K appears to associate with the E4orf6 protein only in the context of this complex, and it is believed to function as the substrate recruitment component, introducing specific proteins for ubiquitination and degradation by proteasomes (8, 11, 28). The formation and function of this complex are essential to permit efficient viral replication. At one time p53 was its only known substrate (10, 34, 36, 40, 47, 48); however, a growing list of additional targets is emerging, including the cellular proteins Mre11 (8, 49), DNA ligase IV (4), and integrin α3 (13), as well as two adeno-associated viral proteins (35). The basis for the role of the complex in late viral mRNA transport is still not known.
Many cellular and viral proteins are present in ligase complexes containing Elongins B and C and usually function to bind target proteins for ubiquitination and degradation (22-24, 29, 31). Such complexes are always associated with one of two members of the Cullin family, Cul2 or Cul5 (24, 29, 31). In most cases a single cellular or viral protein binds Elongins B and C and Cul 2 or Cul5 and recruits substrates for ubiquitination and degradation (22, 29); however, in the case of Ad5 E4orf6/E1B55K, E4orf6 protein forms the ligase complex whereas E1B55K seems to recruit substrates (8). Formation of Cul2/5-based complexes requires an interaction with Elongins B and C via a highly conserved “BC box” (23, 25). Curiously, E4orf6 from Ad5 (and, based on sequence homology, other Ad serotypes) contains three such BC boxes (BC1, BC2, and BC3), all of which are required for optimal degradation of p53 (8, 11, 28). Ad E4orf6 products are the only proteins known to contain multiple BC boxes, all of which conform to the generalized BC box consensus sequence, which is now considered to be [STP]LXXX[CSA]XXXΦ (31).
Cellular proteins selectively recruit either Cul2 or Cul5 via consensus sequences, termed Cul2 and Cul5 boxes (24, 31), as do certain viral products, including E7 from high-risk human papillomavirus (HPV) (Cul2) (21) and HIV Vif (Cul5) (55); however, neither of these motifs is present in the E4orf6 proteins from any Ad serotype (11, 28). Thus, it remains unclear how Ad E4orf6 proteins recruit Cullin family members.
The over 50 human Ad serotypes have been subdivided into six species (A to F) (15, 16) (also commonly termed subgroups [26]) with species B further subdivided into subspecies B1 and B2 based largely on antigenic and sequence similarity (3, 30). In the present report we have cloned appropriate cDNAs encoding E4orf6 and E1B55K from selective serotypes representing all six subgroups of human adenoviruses (including both B1 and B2) and analyzed them when expressed alone for their ability to form ligase complexes and to degrade known substrates of the Ad5 E4orf6/E1B55K complex. The results indicated significant heterogeneity both in selection of Cul5 or Cul2 and in substrate specificity.
MATERIALS AND METHODS
Cells and cell lines.
Human small cell carcinoma H1299 cells (ATCC CRL-5803), carrying a deletion of the p53 gene (33), were cultured in α minimal essential medium (Gibco) supplemented with 10% fetal calf serum (FCS; Gibco) containing 0.292 mg/ml l-glutamine (Gibco). The H1299 Cul5 knockdown cell line (H1299/Cul5KD) and H1299 control cells were described previously (8, 11) and maintained in the presence of 1 μg/ml puromycin. The H1299 Cul2 knockdown cell line (H1299/Cul2KD) was generated by cotransfection with plasmid DNAs expressing interfering RNA (RNAi) against Cul2, which was previously verified for Cul2 RNAi efficiency in transient-transfection studies, and with pcDNA3-puro (carrying the gene for puromycin resistance) at the ratio of 10:1, followed by selection in medium containing 2 μg/ml puromycin. Individual colonies were picked and expanded, and the level of expression of endogenous Cul2 was verified by Western blotting using antibody against Cul2. A clone that showed the greatest reduction in expression level of Cul2 was chosen for further experiments and maintained in the presence of 1 μg/ml puromycin.
Plasmids.
Hemagglutinin (HA)-Cul5, HA-Cul2 (38), and p53 (41) plasmid DNAs expressing human Cul5, Cul2, and p53, respectively, were described previously. cDNAs encoding E4orf6 and E1B55K proteins from each serotype studied were cloned by PCR using the respective viral genomic DNAs as templates with the primers indicated in Table 1. Sequences encoding the FLAG (E4orf6) and HA (E1B55K) tags were added to the primers. The PCR fragments were then cloned into the pcDNA3 vector using the restriction enzymes listed in Table 1. The Ad12 E1B55K plasmid was created by inserting an HA tag sequence into pcDNA3.1 Ad12 E1B54K plasmid (a gift from Roger Grand), and Ad5 pcDNA3 FLAG-E4orf6 was created by inserting a FLAG tag sequence into the Ad5 pcDNA3 E4orf6 plasmid DNA described previously (8), using PCR-based mutagenesis. As shown previously (39), an aberrantly spliced product is generated from the Ad5 E4orf6 sequence cloned into pcDNA3, as the 5′ donor site normally used to generate the E4orf6/7 product is utilized along with a 3′ receptor site in an alternative reading frame in the E4orf6 cDNA. As the same phenomenon was observed with the other serotypes, silent mutations were introduced by PCR mutagenesis in 5′ donor sites of each E4orf6 coding sequence to eliminate this aberrant spliced product. Primers utilized in all the PCRs described above are listed in Table 1.
TABLE 1.
Primers used in PCR-based mutagenesis for cloning cDNAs expressing E4orf6 and E1B55K of different serotypesa
| Subgroup (serotype) | Protein | Primer |
Restriction enzyme | |
|---|---|---|---|---|
| Direction | Sequence | |||
| A (Ad12) | E4orf6 | F | AAGGATCCATGGACTACAAGGACGACGATGACAAGACTACGCAGCGCGACAGACGGTATC | BamHI |
| R | AAGGATCCTCAGTGTCCATCAGCCGCC | |||
| E4orf6-fix | F | GTAATAGTCTGACTTTACATAATGTTTCCGAGGTAAGAGGCATTCCTTC | NA | |
| R | GAAGGAATGCCTCTTACCTCGGAAACATTATGTAAAGTCAGACTATTAC | |||
| E1B55K | F | GTCCAGTGTGGTGGAATTCATGTACCCATACGACGTCCCAGACTACGCTGAGCGAGAAATCCCACCTGAG | NA | |
| R | CTCAGGTGGGATTTCTCGCTCAGCGTAGTCTGGGACGTCGTATGGGTACATGAATTCCACCACACTGGAC | NA | ||
| B1 (Ad16) | E4orf6 | F | AAGAATTCATGGACTACAAGGACGACGATGACAAGACTACGTCAGGAAGCAACTCCATTATG | EcoRI |
| R | AAGAATTCTCATCTGGAAGAAGAACGATG | |||
| E4orf6-fix | F | GTGATACTATGACTATGCATAGTGTTTCCTGTGTTCGTGGTCTTCCTTGTTC | NA | |
| R | GAACAAGGAAGACCACGAACACAGGAAACACTATGCATAGTCATAGTATCAC | |||
| E1B55K | F | AAGAATTCATGTACCCATACGACGTCCCAGACTACGCTGATCCGCCAAACCCACTTCAG | EcoRI | |
| R | AATCTAGATTAGTCAGTTTCTTCACCACTAG | XbaI | ||
| B2 (Ad34) | E4orf6 | F | AAGGATCCATGGACTACAAGGACGACGATGACAAGACTACGTCAGGAAGCAACTCCATTATG | BamHI |
| R | AAGGATCCTCATCTGGAAGAAGAACGATG | |||
| E4orf6-fix | F | GTGATACTATGACTATGCATAGTGTTTCCTGTGTTCGTGGTCTTCCTTG | NA | |
| R | CAAGGAAGACCACGAACACAGGAAACACTATGCATAGTCATAGTATCAC | |||
| E1B55K | F | AAGAATTCATGTACCCATACGACGTCCCAGACTACGCTGATCCCGCAGACTCATTTCAG | EcoRI | |
| R | AATCTAGATTAGTCAGTTTCTTCTCCACTGG | XbaI | ||
| C (Ad5) | E4orf6 | F | GGTTTTGCTTCAGGAAATATGGACTACAAGGACGACGATGACAAGACTACGTCCGGCGTTCC | NA |
| R | GGAACGCCGGACGTAGTCTTGTCATCGTCGTCCTTGTAGTCCATATTTCCTGAAGCAAAACC | |||
| E4orf6-fix | F | GAATGTAACACTTTGACAATGCACAATGTTTCCTACGTGCGAGGTCTTCCCTGCAG | NA | |
| R | CTGCAGGGAAGACCTCGCACGTAGGAAACATTGTGCATTGTCAAAGTGTTACATTC | |||
| E1B55K | F | AAGAATTCATGTACCCATACGACGTCCCAGACTACGCTGAGCGAAGAAACCCATCTGAG | EcoRI | |
| R | AACTCGAGTCAATCTGTATCTTCATCGCTAGAG | XhoI | ||
| D (Ad9) | E4orf6 | F | AAGAATTCATGGACTACAAGGACGACGATGACAAGACTACGCAAACCGAGATTCAATCCAGC | EcoRI |
| R | AAGAATTCTCACAGTCTGGTGGAATGAG | |||
| E4orf6-fix | F | GTGATCATGCAGATTATCATAATGTTTCCTCTGTCCGTGGATTACCATGTG | NA | |
| R | CACATGGTAATCCACGGACAGAGGAAACATTATGATAATCTGCATGATCAC | |||
| E1B55K | F | AAGGATCCATGTACCCATACGACGTCCCAGACTACGCTGAGCCAGGACACCCAACTGAG | BamHI | |
| R | AATCTAGACTAATCTGTGTCCTCCCCAC | XbaI | ||
| E (Ad4) | E4orf6 | F | AAGGATCCATGGACTACAAGGACGACGATGACAAGACTACGTCCGGAAACAGCTCCATC | BamHI |
| R | AAGGATCCTTATCTGGAATAAGAGCGATG | |||
| E4orf6-fix | F | GATACCCTGTCCATGCACAATGTTTCCTCCGTGCGAGGATTGCCCTG | NA | |
| R | CAGGGCAATCCTCGCACGGAGGAAACATTGTGCATGGACAGGGTATC | |||
| E1B55K | F | AAGAATTCATGTACCCATACGACGTCCCAGACTACGCTGAGTCAAGAAACCCATTTCAGC | EcoRI | |
| R | AATCTAGACTAGTCAGATTCTTCCCCACTGG | XbaI | ||
| F (Ad40) | E4orf6 | F | AAGGATCCATGGACTACAAGGACGACGATGACAAGACTACGATGCAACGCGACAGATGG | BamHI |
| R | AAGGATCCTCAAGCTCTCCACGGGTTC | |||
| E4orf6-fix | F | GTGAACATTTGAATATGCATTATGTTTCCGAGGTGAGGTCTATTCCATC | NA | |
| R | GATGGAATAGACCTCACCTCGGAAACATAATGCATATTCAAATGTTCAC | |||
| E1B55K | F | AAGAATTCATGTACCCATACGACGTCCCAGACTACGCTGACCGCCCAAACTCATCTGTC | EcoRI | |
| R | AATCTAGATTAATCCTCATCGCTGGATTCG | XbaI | ||
Abbreviations: F, forward; R, reverse; NA, not applicable.
Antisera.
Anti-p53 pAb1801 hybridoma supernatants were prepared as described previously (41). HA epitopes were detected using anti-HA mouse monoclonal HA.11 (BabCO) or, for immunofluorescence (IF) with rat monoclonal 3F10 (Roche), FLAG epitopes with anti-FLAG M2 mouse monoclonal antibody peroxidase conjugate (Sigma), or, for IF with rabbit anti-FLAG (Sigma-Aldrich), Elongin C with mouse monoclonal antibody SIII p15 (Transduction Laboratories), Mre11 with rabbit polyclonal antiserum NB 100-142D3 (Novus Biologicals), DNA ligase IV with rabbit polyclonal antibody (AHP554) (Serotec), integrin α3A with mouse monoclonal 29A3 (Millipore), Cul5 with rabbit polyclonal anti-Cul5 antibody (H-300) (Santa Cruz Biotechnology), Cul2 with rabbit polyclonal anti-Cul2 antibody NBP1-02780 (Novus Biologicals), tubulin with rat monoclonal anti-α-tubulin antibody (YOL1/34) (Abcam), and actin with mouse monoclonal anti-actin C4 (Millipore).
DNA transfections, cell lysis, and preparation of samples for degradation assays and immunoprecipitations.
Cells growing in 6-well plates for degradation assays or 100-mm-diameter dishes for binding assays were transfected with plasmid DNAs using Lipofectamine 2000 (Invitrogen), as described by the manufacturer. The amount of Lipofectamine 2000 used was calculated based on a ratio of 3 μl of transfection reagent per 1 μg of DNA. For the p53 degradation assays, 0.25 μg of pcDNA3-p53, 2 μg of the appropriate pcDNA3-FLAG E4orf6, and 1.75 μg of the appropriate pcDNA3-HA E1B55K were transfected for 24 h. When required, the total amount of DNA was made up by addition of pcDNA3 empty vector DNA. For the Mre11 and DNA ligase IV degradation, cells were transfected as previously except without the p53 plasmid. For integrin α3 degradation, cells were first either mock infected or infected with Ad5LacZ (5) diluted in infection medium (0.2 mM CaCl2, 0.2 mM MgCl2, and 2% serum in phosphate-buffered saline [PBS]) at a multiplicity of infection (MOI) of 10 PFU/cell for 90 min, followed by DNA transfection as described for Mre11 and DNA ligase IV, except at 48 h postinfection (p.i.). Cells were lysed for 15 min on ice with NP-40 lysis buffer (50 mM Tris-HCl, pH 8, containing 150 mM NaCl, 5 mM EDTA, 0.15% NP-40, 2 mM dithiothreitol [DTT], 4 mM NaF, 2 mM NaPP, 500 μM Na3VO4, 200 μg/ml phenylmethylsulfonyl fluoride [PMSF], 2 μg/ml aprotinin, 5 μg/ml leupeptin) followed by three cycles of freezing-thawing and then two rounds of sonication at 100% power output for 30 s with a VCX130 sonicator (Sonics & Materials). Aliquots of cell extracts containing 15 to 50 μg of protein were examined by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
For binding assays with E4orf6 or E1B55K, cells in 100-mm dishes were transfected with different quantities of plasmid DNAs for each of the seven serotypes in order to attempt to obtain comparable levels of protein expression. Nevertheless, there remains some unavoidable variability in expression with individual cDNAs. To ensure that our results were reliable and that the conclusions were unequivocally correct, every experiment was repeated several times (4 to 6 and even more in some cases). For these reasons, it was not possible to make specific comparisons on the relative level of binding between all the serotypes. Rather it was more appropriate to make semiquantitative conclusions, which should be sufficient for the purposes of this study. pcDNA3 empty vector DNA was added to each preparation to yield a total of 9 μg of DNA transfected for E4orf6 or 8 μg for E1B55K. In addition, 2 μg of plasmid DNAs expressing HA-Cul5 or HA-Cul2 was also cotransfected in respective binding assays. Cells were then lysed for 20 min on ice with the immunoprecipitation lysis buffer (20 mM Tris-HCl, pH 7.5, containing 150 mM NaCl, 2 mM EDTA, 1% Triton X-100, 5% glycerol, 2 mM DTT, 4 mM NaF, 2 mM NaPP, 500 μM Na3VO4, 200 μg/ml PMSF, 2 μg/ml aprotinin, 5 μg/ml leupeptin). Aliquots of cell extracts containing 400 to 1,000 μg of protein (constant within each experiment) were used for immunoprecipitation using 1 μl of mouse monoclonal anti-HA antibody (HA.11) (BabCO), 1 μl of rabbit polyclonal antibody anti-HA (HA.11) (BabCO), or 1.5 μl of rabbit polyclonal anti-FLAG antibody (Sigma) followed by incubation with 50% protein G and 50% protein A agarose (Upstate). The beads were extensively washed in the lysis buffer and eluted, and bound proteins were examined by SDS-PAGE.
Western blotting.
Proteins were examined by Western blotting, essentially as described previously (8, 11). Briefly, proteins separated by SDS-PAGE were transferred to polyvinylidene difluoride (PVDF) membranes and blocked using 5% nonfat dry milk in Tris-buffered saline-Tween (TBST). Membranes were incubated with the indicated primary antibodies in TBS-1% Tween with 1% nonfat dry milk, followed by appropriate peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories). Polypeptides on membranes were then visualized by enhanced chemiluminescence with ECL Plus Western blotting reagents (Amersham-Pharmacia Biotech).
Immunofluorescence.
Cells grown on coverslips in 6-well dishes were transfected as described with 0.5 μg of HA-Cul2 or 0.75 μg HA-Cul5 with up to 3 μg of FLAG-E4orf6 for 24 h. Cells were then fixed for 15 min with 4% paraformaldehyde and permeabilized for 15 min with 0.5% Triton X-100 in PBS. Following washes in PBS, immunofluorescence was performed with rabbit FLAG antibodies and rat HA antibodies for 2 h at room temperature in a humidity chamber. The secondary antibodies used were anti-rat antibody conjugated to Alexa 594 dye and anti-rabbit antibody conjugated to Alexa 488 dye (Molecular Probes). Images were taken on an LSM3 confocal microscope with a 63× objective with the LSM 4.2 Image browser software.
RESULTS
Cloning of E4orf6 and E1B55K cDNAs of representative serotypes.
Ad5 is by far the most highly studied human adenovirus and has served as a model for the more than 50 identified serotypes. A high degree of homology exists in the sequences encoding the corresponding E4orf6 and E1B55K products of these various serotypes (data not shown). (Note that there are some small variations in the molecular masses of the E1B55K protein among serotypes; however, for simplicity we will refer to all as E1B55K.) Thus, it seemed reasonable to believe that E4orf6/E1B55K pairs from all subgroups form E3 ubiquitin ligase complexes similar to that in Ad5 (8, 11, 38). To examine this possibility, we studied E4orf6 and E1B55K products from serotypes representative of each of the six human adenovirus subgroups, including one from both subgroups B1 and B2 (Table 2 ). In all cases it was necessary to clone the appropriate coding sequences from full-length viral DNAs using a PCR-based approach, as most had not been studied previously (see Materials and Methods and Table 1 for details). In the case of E4orf6 cDNAs, modified forms were generated to eliminate a splice variant found previously to be produced with Ad5 (39) and predicted for the other serotypes. All were introduced into pcDNA3 plasmid DNA and shown to produce stable products upon transfection into H1299 cells (see, for example, Fig. 1A and B).
TABLE 2.
Human adenovirus serotypes
| Subgroup | Serotype(s) | Model serotype |
|---|---|---|
| A | 12, 18, 31 | 12a |
| B1 | 3, 7, 16, 21, 50 | 16b |
| B2 | 11, 14, 34, 35 | 34c |
| C | 1, 2, 5, 6 | 5d |
| D | 8, 9, 10, 13, 15, 17, 19, 20, 22-30, 32, 33, 36-39, 42-49, 51 | 9e |
| E | 4 | 4f |
| F | 40, 41 | 40g |
Ad12, NCBI accession no. X73487; E4orf6, positions 31436 to 32311; E1B55K, positions 1847 to 3295.
Ad16, NCBI accession no. AY601636; E4orf6, positions 32800 to 33696; E1B55K, positions 1903 to 3381.
Ad34, NCBI accession no. AY737797; E4orf6, positions 32060 to 32959; E1B55K, positions 1916 to 3400.
Ad5, NCBI accession no. AC_000008; E4orf6, positions 33193 to 34077; E1B55K, positions 2019 to 3509.
Ad9, NCBI accession no. AJ854486; E4orf6, positions 32279 to 33157; E1B55K, positions 1876 to 3363.
Ad4, NCBI accession no. NC_003266; E4orf6, positions 33270 to 34169; E1B55K, positions 1905 to 3356.
Ad40, NCBI accession no. NC_001454; E4orf6, positions 31876 to 32745; E1B55K, positions 1719 to 3149.
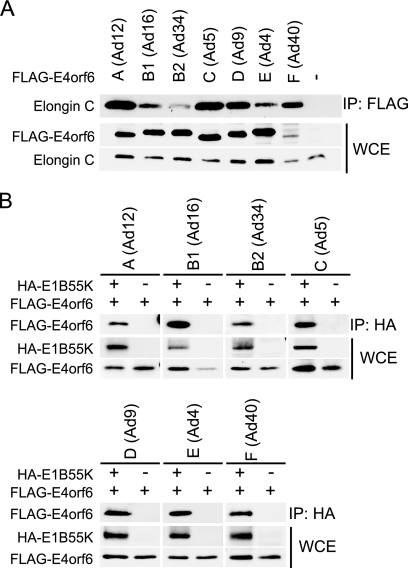
FIG. 1.
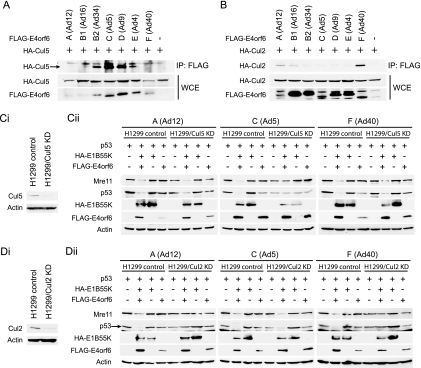
Formation of the E4orf6/E1B55K E3 ligase complex in different serotypes. (A) Association of E4orf6 with Elongin C. H1299 cells were transfected with plasmid DNAs expressing FLAG-tagged E4orf6 of different serotypes. Immunoprecipitates (IP) were prepared using anti-FLAG antibodies and analyzed by Western blotting using anti-Elongin C antibodies. Whole-cell extracts (WCE) were also immunoblotted using anti-FLAG (E4orf6) or anti-Elongin C antibodies. (B) Association of E1B55K with E4orf6. H1299 cells were transfected with plasmid DNAs expressing FLAG-E4orf6 and HA-E1B55K of different serotypes as indicated. Immunoprecipitates obtained using anti-HA (E1B55K) antibodies were immunoblotted using anti-FLAG antibodies, and whole-cell extracts were immunoblotted with anti-HA (E1B55K) and anti-FLAG (E4orf6) antibodies. In both panels A and B the serotype (with subgroup) has been indicated at the top.
E4orf6 products from all serotypes generate complexes with Elongin C and E1B55K.
To determine if, as with Ad5, the E4orf6 proteins from all selected serotypes form complexes with Elongin C and E1B55K, extracts from H1299 cells that had been transfected with plasmid DNAs expressing the various FLAG-E4orf6 products were immunoprecipitated using anti-FLAG antibody and then analyzed by Western blotting using anti-Elongin C antibodies. The levels of E4orf6 and Elongin C in whole-cell extracts were determined by Western blotting using anti-FLAG or anti-Elongin C antibodies. Due to the often variable expression levels of E4orf6 (and E1B55K), it should be noted that for this type of study, several repetitions (a minimum of 4 to 6) were routinely carried out and the results presented reflect the clear consensus outcome for each serotype. Figure 1 A shows that in all cases, reasonable levels of E4orf6 expression were observed, although in this particular study somewhat less was seen with Ad40. In the many studies that we have done, the E4orf6 products of Ad12 and Ad40 were present at consistently lower levels than were those with other serotypes, even using large amounts of plasmid DNA. This effect may occur because these proteins are intrinsically more unstable. Importantly, Fig. 1A also shows that significant levels of Elongin C coimmunoprecipitated with E4orf6 from all serotypes. It should be noted that in control studies no interaction was observed between Elongin C and unrelated FLAG-tagged proteins (data not shown). Similar studies conducted using anti-Elongin B antibodies indicated that this species was also present in complex with E4orf6 (data not shown). These results indicated that E4orf6 proteins from all serotypes form complexes with Elongins B and C. To determine if E1B55K products, like that of Ad5, bind to such complexes, a similar study was conducted except that cells were cotransfected with plasmid DNAs expressing both FLAG-E4orf6 and HA-E1B55K. Figure 1B shows that with all serotypes E1B55K coimmunoprecipitated with E4orf6. With the Ad5 serotype, it is known that E1B55K does not bind directly to E4orf6 but rather to the assembled ligase complex (8). Thus, unless complex assembly is drastically different with the other serotypes, these data suggest that with all of the serotypes studied, the E4orf6 products formed complexes containing Elongins B and C and E1B55K as was the case with Ad5.
Not all Ad E4orf6/E1B55K E3 ubiquitin ligases degrade p53.
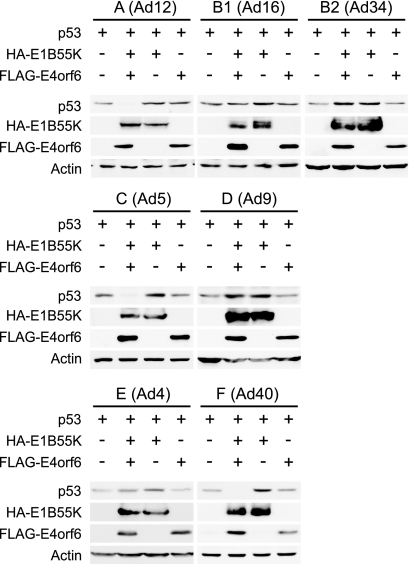
The first known substrate of the E4orf6/E1B55K complex of Ad5 was p53 (8, 10, 34, 36, 38, 40, 47-49). To determine if the E4orf6/E1B55K complexes from other serotypes are functional as E3 ubiquitin ligases and if they also degrade p53, studies were carried out in a fashion similar to those of Fig. 1B using p53-null H1299 cells, except that cells were also cotransfected with plasmid DNA expressing human p53. The levels of exogenous p53 present in whole-cell extracts harvested at 24 h posttransfection, as well as those of E4orf6 and E1B55K, were determined by Western blotting using appropriate antibodies. Figure 2 shows that in the case of Ad5 and as described previously (8, 38), degradation of p53 was evident from the virtual absence of this protein in extracts from cells expressing both E4orf6 and E1B55K. Highly comparable results were also evident with E4orf6 and E1B55K products of Ad12 (subgroup A) and Ad40 (subgroup F), suggesting that p53 was also a substrate with these serotypes. Such was not the case with other serotypes. Figure 2 shows that with Ad16 (subgroup B1), Ad34 (subgroup B2), Ad9 (subgroup D), and Ad4 (subgroup E), little p53 degradation was evident, indicating that the E4orf6/E1B55K ligases from some serotypes did not degrade p53 efficiently.
FIG. 2.
Degradation of p53 by E4orf6/E1B55K ligase complexes from different serotypes. H1299 cells were transfected with plasmid DNAs encoding human p53, FLAG-E4orf6, and HA-E1B55K as indicated, and at 24 h posttransfection whole-cell extracts were immunoblotted for p53, FLAG (E4orf6), and HA (E1B55K) using appropriate antibodies. Actin levels were assessed by Western blotting as loading controls, and the serotype (with subgroup) has been indicated at the top of each set. Degradation of p53 was determined by the decrease of p53 in the presence of both E4orf6 and E1B55K.
Degradation of Mre11 and DNA ligase IV by various Ad serotypes.
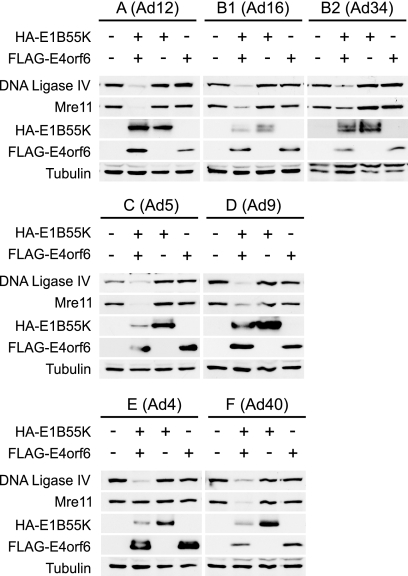
As both Mre11 and DNA ligase IV have been identified as substrates for the Ad5 E4orf6/E1B55K ligase, studies were conducted to determine if ligases from other Ad serotypes also degrade them. An experiment similar to that described in Fig. 2 was conducted at 24 h posttransfection in the absence of p53, and the disappearance of endogenous Mre11 and DNA ligase IV was determined by Western blotting of whole-cell extracts using appropriate antibodies. Figure 3 shows that the E4orf6/E1B55K ligases from all serotypes induced significant or extensive loss of DNA ligase IV, indicating that all appeared to degrade this protein fairly efficiently. Such was not the case with Mre11. Figure 3 shows that whereas the ligases from Ad5, Ad12, and Ad40 induced an almost total loss of Mre11, those from Ad9, Ad16, and Ad34 were considerably less efficient and that of Ad4 had no detectable effect on Mre11 levels. As the analyses of DNA ligase IV and Mre11 levels were conducted in the same cell extracts, these results clearly indicated that the ligases from some serotypes were much more efficient in degrading DNA ligase IV, and at least in one case, Ad4, Mre11 did not appear to be a substrate.
FIG. 3.
Degradation of Mre11 and DNA ligase IV by E4orf6/E1B55K ligase complexes from different serotypes. A study similar to that shown in Fig. 2 was conducted, except that no p53 was present and levels of endogenous Mre11 and DNA ligase IV were determined by Western blotting using appropriate antibodies. Tubulin levels were assessed by Western blotting as loading controls, and the serotype (with subgroup) has been indicated at the top. Degradation of DNA ligase IV and Mre11 was determined by the decrease of these species in the presence of both E4orf6 and E1B55K.
Degradation of integrin α3 by various Ad serotypes.
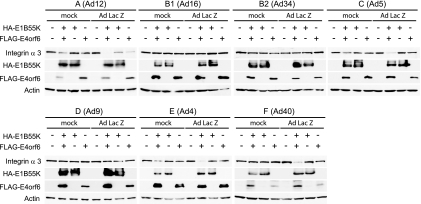
As we have recently found that integrin α3 is also degraded in response to the expression of Ad5 E4orf6/E1B55K, this substrate was also tested with the ligase complexes of the other serotypes. For reasons that still remain unclear, degradation of integrin α3 with Ad5 is much enhanced after infection by wild-type virus, or in the case of expression of E4orf6 and E1B55K alone from plasmid DNAs, by coinfection with AdLacZ, a control Ad5 viral vector that expresses almost exclusively LacZ under the control of the cytomegalovirus (CMV) promoter (13). Indeed we have found that expression of E4 genes by AdLacZ is limited to less than 5% of levels seen with wild-type virus (E. Querido and P. E. Branton, unpublished data). Thus, H1299 cells were transfected with plasmid DNAs expressing E4orf6 and E1B55K from the range of serotypes directly following infection with AdLacZ at an MOI of 10. Cells were harvested at 48 h posttransfection and analyzed for expression levels of integrin α3. As shown in Fig. 4 and as published previously (13), degradation of integrin α3 by Ad5 was more efficient following infection with control viral vector (AdLacZ lanes). Similar results were seen with the serotypes from group A (Ad12), E (Ad4), and F (Ad40). In contrast, little or no degradation was observed with serotypes from group B1 (Ad16), B2 (Ad34), and D (Ad9). Thus, as was the case with p53 and Mre11, integrin α3 is not a conserved substrate for all serotypes.
FIG. 4.
Degradation of integrin α3 by E4orf6/E1B55K ligase complexes from different serotypes. A study similar to that shown in Fig. 3 was performed, except that in some cases cells were infected with AdLacZ viral vector prior to transfection with plasmid DNAs encoding FLAG-E4orf6 and HA-E1B55K as indicated. The levels of endogenous integrin α3 were determined by Western blotting using appropriate antibodies. Actin levels were assessed by Western blotting as loading controls, and the serotype (with subgroup) has been indicated at the top. Degradation of integrin α3 was determined by the decrease of these species in the presence of both E4orf6 and E1B55K.
Cullin specificity of E4orf6/E1B55K ligase complexes from various serotypes.
Previous studies have indicated that Ad5 E4orf6 forms ligase complexes predominantly with Cul5, as only extremely low levels of Cul2 were evident (38) (C. Y. Cheng and P. Blanchette, unpublished data). To determine if E4orf6 proteins from other serotypes maintain this Cul5 specificity, coimmunoprecipitation studies similar to those described above were conducted using cells expressing FLAG-E4orf6 from the various serotypes and HA-Cul5 or HA-Cul2. The presence of Cullins was detected by Western blotting using anti-HA antibodies of immunoprecipitates prepared using anti-FLAG antibodies. Figure 5 A shows that whereas E4orf6 from Ad34, Ad5, and Ad9 interacted efficiently with Cul5, somewhat lower levels were observed with Ad16 and Ad4, and little or none was evident with E4orf6 from Ad12 and Ad40. Figure 5B shows that Cul2 was present with E4orf6 from Ad12, Ad16, and Ad40, but little was evident in the cases of Ad34, Ad5, Ad9, and Ad4. These results suggested that E4orf6 proteins from Ad34, Ad5, Ad9, and Ad4 form ligase complexes that contain primarily Cul5 and that those from Ad12 and Ad40 form ligase complexes that contain primarily Cul2, whereas Ad16 E4orf6 appears to interact with both at significant levels. If indeed the complexes from Ad12 and Ad40 rely on Cul2, one would predict that degradation of Mre11 and p53 with these serotypes, which both degrade these substrates, should not be inhibited in H1299/Cul5KD cells in which Cul5 has been knocked down by constitutive expression of Cul5-specific RNAi (Fig. 5Ci). To test this possibility, studies similar to those shown in Fig. 2 and 3 were conducted in both H1299 control and H1299/Cul5KD cells. Figure 5Cii shows that with Ad5, a significant decrease in Mre11 and p53 degradation was evident in H1299/Cul5KD cells, as shown previously (11), whereas no reduction occurred with E4orf6/E1B55K from Ad12 and Ad40. These data were consistent with the use of Cul2 by these two serotypes.
FIG. 5.
Cullin specificity of the various E3 ligase complexes. (A and B) H1299 cells were cotransfected with plasmid DNAs encoding FLAG-E4orf6 of different serotypes and either HA-Cul5 (A) or HA-Cul2 (B). Immunoprecipitations were carried out using anti-FLAG (E4orf6) antibody, and immunoprecipitates were immunoblotted using anti-HA (Cul5 or Cul2) antibodies. Whole-cell extracts were also immunoblotted using anti-HA or anti-FLAG antibodies. (Ci) Cul5 levels in H1299 control and H1299/Cul5KD cells. The levels of endogenous Cul5 present in H1299 control cells and H1299/Cul5KD cells were analyzed by Western blotting using anti-Cul5 antibodies, as described in Materials and Methods. (Cii) Dependence of p53 and Mre11 degradation by various E3 ligase complexes on Cul5. Mre11 and p53 degradation assays were conducted as described for Fig. 2, except that levels of both exogenous p53 and endogenous Mre11 were determined by Western blotting using appropriate antibodies, and studies were conducted in both H1299 control and H1299/Cul5KD cells, as indicated. Actin levels were assessed by Western blotting as loading controls, and the serotype (with subgroup) has been indicated at the top. (Di) Cul2 levels in H1299 control and H1299/Cul2KD cells. The levels of endogenous Cul2 present in H1299 control cells and H1299/Cul2KD cells were analyzed by Western blotting using anti-Cul2 antibodies. (Dii) Dependence of p53 and Mre11 degradation by various E3 ligase complexes on Cul2. A study identical to that shown in panel Ci was conducted using H1299 control cells and H1299/Cul2KD cells.
To confirm that Cul2 functioned in the E4orf6/E1B55K ligases of Ad12 and Ad40, we generated a new H1299/Cul2KD cell line in which Cul2 levels were reduced by constitutive expression of Cul2-specific RNAi (Fig. 5Di) and studied again the degradation of p53 and Mre11 by Ad5, Ad12, and Ad40 ligase complexes. Figure 5Dii shows that indeed the degradation of p53 and Mre11 was significantly reduced in H1299/Cul2KD cells, whereas with Ad5 degradation remained efficient.
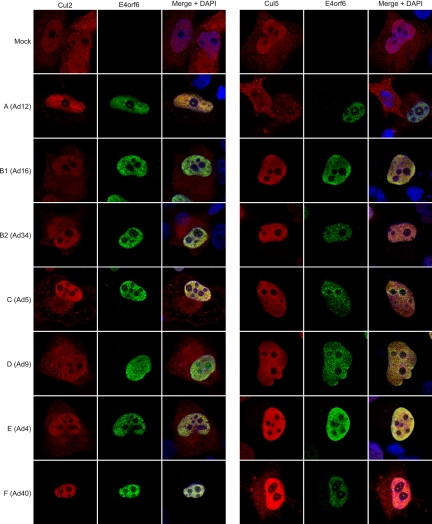
Additional evidence for recruitment of Cul2 or Cul5 was obtained by comparing the abilities of E4orf6 proteins of different serotypes to relocalize either Cul2 or Cul5. H1299 cells grown on coverslips were transfected with FLAG-tagged E4orf6 of different serotypes in combination with either HA-Cul2 or HA-Cul5 for 24 h. An immunofluorescence assay was then performed on the fixed and permeabilized cells using antibodies to the FLAG and HA tag. As shown in Fig. 6, the localization of Cul2 and Cul5 in the absence of E4orf6 includes both the nucleus and the cytoplasm (Mock row). Thus, a relocalization of either Cul2 or Cul5 is characterized by the transition of the cytoplasmic form of the Cullin to the nucleus. Complementing the results obtained in Fig. 5, a better relocalization of Cul2 was observed for Ad12 and Ad40, whereas relocalization of Cul5 was observed for the other serotypes. It should be noted that relocalization of Cul2 by Ad12 and Ad40 did not appear to be as efficient as that of Cul5 by most of the other serotypes, most likely because E4orf6 is present at somewhat lower levels with these serotypes. A less efficient relocalization of Cul5 by Ad16 was also observed even though high levels of E4orf6 were present, a finding that was perhaps consistent with the earlier observation that Ad16 appears to interact with both Cul5 and Cul2. Together, results from Fig. 5 and 6 confirmed that with Ad12 and Ad40 ligase complexes containing Cul2 and not Cul5 were utilized.
FIG. 6.
Cullin specificity of the various E3 ligase complexes as seen by Cullin relocalization. H1299 cells were cotransfected with plasmid DNAs encoding FLAG-E4orf6 of different serotypes and either HA-Cul2 (left panels) or HA-Cul5 (right panels). The immunofluorescence assay was carried out using rabbit anti-FLAG (E4orf6) antibody and rat anti-HA (Cul2 or Cul5) antibody. DAPI, 4′,6-diamidino-2-phenylindole.
DISCUSSION
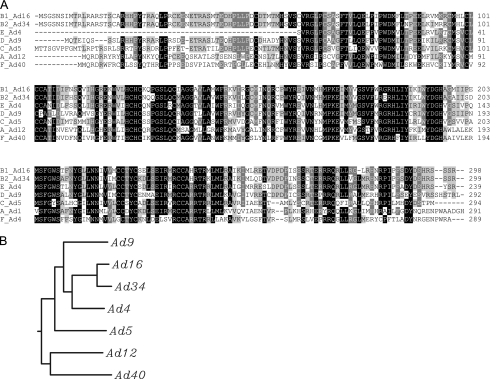
The present studies have indicated that the E4orf6 proteins of representative serotypes from all subgroups of human adenoviruses form Cullin- and Elongin B/C-based E3 ubiquitin ligase complexes that also contain E1B55K. In our earlier work we found that the ligase complex formed by Ad5 appeared to contain predominantly or perhaps exclusively Cul5 (38); however, our continuing studies have led us to believe that a spectrum of Cullin specificity probably exists with E4orf6 proteins from various serotypes. Several form primarily Cul5-based ligases (Ad5/subgroup C, Ad34/B2, Ad9/D, and Ad4/E); others form primarily Cul2-based ligases (Ad12/A and Ad40/F). Another group has also found a similar difference in Cullin specificity between Ad5 and Ad12 (7) (A. S. Turnell and R. J Grand, personal communication). Nevertheless, it is uncertain whether or not these interactions are entirely Cullin specific, and thus, some level of association with both Cul5 and Cul2 may occur with all species. Studies employing cell lines with levels of either Cul5 or Cul2 reduced through constitutive expression of appropriate RNAi species clearly indicated dependence on either Cul5 or Cul2 with E4orf6 from these serotypes for the degradation of p53 and Mre11. With E4orf6 from Ad16/B1, the case was found to be somewhat different. This protein was seen to bind fairly efficiently with both Cul5 and Cul2; however, it was seen to only efficiently relocalize Cul5. Thus, we have still not clearly resolved whether Ad16 forms a truly functional complex with Cul2. An alignment of the predicted protein sequences of the E4orf6 proteins from all serotypes used in this study is shown in Fig. 7 A, with the most highly conserved amino acids highlighted. Figure 7B shows the evolutionary tree of E4orf6 sequences from the serotypes that we have examined. Ad12 and Ad40, which both predominantly bind Cul2, are highly related and the most divergent from those of other serotypes, suggesting that the ability to bind Cul2 preferentially may have evolved separately. Based on this evolutionary homology, E4orf6 from Ad16, which seems to bind both Cul2 and Cul5 fairly efficiently, is most closely related to products of predominantly Cul5-binding species, suggesting that its dual specificity (and that of possibly other serotypes that we have not examined) may be the result of as-yet-unidentified modifications in sequence. At present it is also unclear if the properties of these serotypes are typical of all members of the various subgroups or if heterogeneity also exists among individual members of subgroups as well. Although the finding of heterogeneity in Cullin selection was perhaps unexpected, it is not yet understood how Ad5 E4orf6 preferentially selects Cul5, as it lacks a typical Cul5 box. In fact, none of the adenovirus E4orf6 sequences contain either Cul5 or Cul2 boxes, suggesting that Cullin selection occurs via a different process or binding sequence. E4orf6 coding sequences from all serotypes are highly similar, and thus, it may be possible to identify the Cul5/Cul2 binding sites within the nonhomologous regions, and such studies are now under way in our group.
FIG. 7.
(A) E4orf6 protein sequences. An alignment of the E4orf6 protein sequences was made using the Clustal W program. The shading is based on the percent identity as analyzed by Jalview (alignment editor program). (B) Phylogenetic relationship of E4orf6 sequences from various human adenovirus serotypes. The protein sequences of E4orf6 proteins of the seven serotypes were analyzed using the multiple alignment program CLUSTAL W. The figure illustrates the relationship between these E4orf6 proteins using the dendrogram with branch length option provided by this program.
Another perhaps surprising result was the failure of ligase complexes from some serotypes to degrade p53, Mre11, or integrin α3 (summarized in Table 3 ). Recent studies by other groups have suggested that in addition to Ad5, Ad12 infection also targets p53 (6, 53), Mre11 (6), and DNA ligase IV (7) for degradation. Degradation of p53 was believed to be vital to prevent early apoptotic death of infected cells and thus poor progeny yields due to the activation and stabilization of p53 by the adenovirus E1A product (12, 42, 45). In addition, Mre11 and DNA ligase IV were believed to be of major importance in preventing formation of viral genome concatemers, thus enhancing packaging of progeny DNA (32, 49). It should be noted, however, that Ad5 (and thus perhaps all adenoviruses) has multiple ways to block p53 activity (43), and Mre11 is also functionally inactivated by relocalization by E4orf3 (1, 27). In fact, the effectiveness of the ligase may lie in its ability to degrade a sufficient collection of substrates to establish conditions suitable enough for efficient progeny production or to enhance viral persistence or latency. For example, DNA ligase IV was found to be the only substrate that was significantly degraded by all serotypes tested, and its degradation, even in the absence of a significant reduction in Mre11 levels, may be sufficient to prevent formation of genomic DNA concatemers, as is suggested by results from Stracker et al. (49). We observed that Mre11 did not appear to be degraded by Ad4, unlike what was suggested by previous studies by another group (50); however, the degradation of Mre11 observed by these researchers was in the context of a full Ad4 infection whereas our studies employed Ad4 E4orf6 and E1B55K expressed alone. Thus, it is possible that other viral gene products may also regulate Mre11 levels.
TABLE 3.
Summary of results
| Subgroup (serotype) | Degradation ofa: |
Cul2/5 selectivity | |||
|---|---|---|---|---|---|
| p53 | Mre11 | DNA ligase IV | Integrin α3 | ||
| A (Ad12) | +++ | +++ | ++ | +++ | Predominantly Cul2 |
| B1 (Ad16) | − | + | ++ | − | Both Cul2 and Cul5 |
| B2 (Ad34) | − | + | ++ | − | Predominantly Cul5 |
| C (Ad5) | +++ | +++ | ++ | +++ | Predominantly Cul5 |
| D (Ad9) | − | + | ++ | − | Predominantly Cul5 |
| E (Ad4) | − | − | ++ | +++ | Predominantly Cul5 |
| F (Ad40) | +++ | +++ | ++ | +++ | Predominantly Cul2 |
+++ to −, most to least degradation, respectively.
The ligase complexes from serotype B1, B2, and D subgroups were unable to degrade integrin α3 even following infection by the AdLacZ viral vector. This finding may not be totally surprising, as the AdLacZ vector has an Ad5 backbone. Since at least the B subgroup viruses use a different cellular receptor (17) for viral entry than that used by Ad5, it is possible that these viruses might require infection with a viral vector from the same subgroup. Integrin α3 functions as a coreceptor for Ad5 entry (44), and we are currently investigating the relationship between the degradation of integrin α3 and cell surface receptors utilized by various adenovirus serotypes.
Our present results suggest that the E4orf6/E1B55K ligases of the different subgroups did not appear to evolve to target p53, Mre11, or integrin α3. Of more importance may be the degradation of as-yet-unidentified targets. The Ad5 E4orf6/E1B55K ligase complex is required for the regulation of late viral mRNA transport to optimize formation of high levels of progeny virions (9, 54). It is not known if this function of the ligase has been conserved across all adenovirus serotypes; however, the search for potential substrates or actions of the ligase complex with respect to the control of late viral mRNA remains a major priority of our group.
Acknowledgments
We thank Roger Grand and Andrew Turnell (University of Birmingham, Birmingham, United Kingdom) for supplying vector pcDNA3.1 Ad12E1B54K and for generously discussing some of their results on Cullin specificity prior to publication.
This work was supported by grants from the Canadian Institutes of Health Research and Fonds de la recherche en santé du Québec (P.E.B./P.B.). P.B. had an FRSQ postdoctoral fellowship during part of this work.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Araujo, F. D., T. H. Stracker, C. T. Carson, D. V. Lee, and M. D. Weitzman. 2005. Adenovirus type 5 E4orf3 protein targets the Mre11 complex to cytoplasmic aggresomes. J. Virol. 79:11382-11391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, A., and V. Mautner. 1994. Phylogenetic relationships among adenovirus serotypes. Virology 205:438-452. [DOI] [PubMed] [Google Scholar]
- 4.Baker, A., K. J. Rohleder, L. A. Hanakahi, and G. Ketner. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034-7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bett, A. J., L. Prevec, and F. L. Graham. 1993. Packaging capacity and stability of human adenovirus type 5 vectors. J. Virol. 67:5911-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blackford, A. N., R. K. Bruton, O. Dirlik, G. S. Stewart, A. M. Taylor, T. Dobner, R. J. Grand, and A. S. Turnell. 2008. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J. Virol. 82:7640-7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackford, A. N., R. N. Patel, N. A. Forrester, K. Theil, P. Groitl, G. S. Stewart, A. M. Taylor, I. M. Morgan, T. Dobner, R. J. Grand, and A. S. Turnell. 2010. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. U. S. A. 107:12251-12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blanchette, P., C. Y. Cheng, Q. Yan, G. Ketner, D. A. Ornelles, T. Dobner, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619-9629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blanchette, P., K. Kindsmuller, P. Groitl, F. Dallaire, T. Speiseder, P. E. Branton, and T. Dobner. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cathomen, T., and M. D. Weitzman. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407-11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, C. Y., P. Blanchette, and P. E. Branton. 2007. The adenovirus E4orf6 E3 ubiquitin ligase complex assembles in a novel fashion. Virology 364:36-44. [DOI] [PubMed] [Google Scholar]
- 12.Chiou, S. K., and E. White. 1997. p300 binding by E1A cosegregates with p53 induction but is dispensable for apoptosis. J. Virol. 71:3515-3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dallaire, F., P. Blanchette, P. Groitl, T. Dobner, and P. E. Branton. 2009. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dobner, T., N. Horikoshi, S. Rubenwolf, and T. Shenk. 1996. Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272:1470-1473. [DOI] [PubMed] [Google Scholar]
- 15.Echavarria, M. 2008. Adenoviruses in immunocompromised hosts. Clin. Microbiol. Rev. 21:704-715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fauquet, C. M., M. A. Mayo, J. Maniloff, U. Desselberger, and L. A. Ball (ed.). 2005. Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 17.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez, R. A., and S. J. Flint. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507-4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harada, J. N., A. Shevchenko, A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huh, K., X. Zhou, H. Hayakawa, J. Y. Cho, T. A. Libermann, J. Jin, J. W. Harper, and K. Munger. 2007. Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 81:9737-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwai, K., K. Yamanaka, T. Kamura, N. Minato, R. C. Conaway, J. W. Conaway, R. D. Klausner, and A. Pause. 1999. Identification of the von Hippel-Lindau tumor-suppressor protein as part of an active E3 ubiquitin ligase complex. Proc. Natl. Acad. Sci. U. S. A. 96:12436-12441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamura, T., D. Burian, Q. Yan, S. L. Schmidt, W. S. Lane, E. Querido, P. E. Branton, A. Shilatifard, R. C. Conaway, and J. W. Conaway. 2001. Muf1, a novel Elongin BC-interacting leucine-rich repeat protein that can assemble with Cul5 and Rbx1 to reconstitute a ubiquitin ligase. J. Biol. Chem. 276:29748-29753. [DOI] [PubMed] [Google Scholar]
- 24.Kamura, T., K. Maenaka, S. Kotoshiba, M. Matsumoto, D. Kohda, R. C. Conaway, J. W. Conaway, and K. I. Nakayama. 2004. VHL-box and SOCS-box domains determine binding specificity for Cul2-Rbx1 and Cul5-Rbx2 modules of ubiquitin ligases. Genes Dev. 18:3055-3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kamura, T., S. Sato, D. Haque, L. Liu, W. G. Kaelin, Jr., R. C. Conaway, and J. W. Conaway. 1998. The Elongin BC complex interacts with the conserved SOCS-box motif present in members of the SOCS, ras, WD-40 repeat, and ankyrin repeat families. Genes Dev. 12:3872-3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knipe, D. M., P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.). 2007. Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 27.Liu, Y., A. Shevchenko, A. Shevchenko, and A. J. Berk. 2005. Adenovirus exploits the cellular aggresome response to accelerate inactivation of the MRN complex. J. Virol. 79:14004-14016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo, K., E. Ehrlich, Z. Xiao, W. Zhang, G. Ketner, and X. F. Yu. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J. 21:1742-1750. [DOI] [PubMed] [Google Scholar]
- 29.Luo, K., Z. Xiao, E. Ehrlich, Y. Yu, B. Liu, S. Zheng, and X. F. Yu. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc. Natl. Acad. Sci. U. S. A. 102:11444-11449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madisch, I., G. Harste, H. Pommer, and A. Heim. 2005. Phylogenetic analysis of the main neutralization and hemagglutination determinants of all human adenovirus prototypes as a basis for molecular classification and taxonomy. J. Virol. 79:15265-15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahrour, N., W. B. Redwine, L. Florens, S. K. Swanson, S. Martin-Brown, W. D. Bradford, K. Staehling-Hampton, M. P. Washburn, R. C. Conaway, and J. W. Conaway. 2008. Characterization of Cullin-box sequences that direct recruitment of Cul2-Rbx1 and Cul5-Rbx2 modules to Elongin BC-based ubiquitin ligases. J. Biol. Chem. 283:8005-8013. [DOI] [PubMed] [Google Scholar]
- 32.Mathew, S. S., and E. Bridge. 2007. The cellular Mre11 protein interferes with adenovirus E4 mutant DNA replication. Virology 365:346-355. [DOI] [PubMed] [Google Scholar]
- 33.Mitsudomi, T., S. M. Steinberg, M. M. Nau, D. Carbone, D. D'Amico, S. Bodner, H. K. Oie, R. I. Linnoila, J. L. Mulshine, J. D. Minna, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171-180. [PubMed] [Google Scholar]
- 34.Moore, M., N. Horikoshi, and T. Shenk. 1996. Oncogenic potential of the adenovirus E4orf6 protein. Proc. Natl. Acad. Sci. U. S. A. 93:11295-11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nayak, R., K. D. Farris, and D. J. Pintel. 2008. E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J. Virol. 82:3803-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nevels, M., S. Rubenwolf, T. Spruss, H. Wolf, and T. Dobner. 2000. Two distinct activities contribute to the oncogenic potential of the adenovirus type 5 E4orf6 protein. J. Virol. 74:5168-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Querido, E., H. Chu-Pham-Dang, and P. E. Branton. 2000. Identification and elimination of an aberrant splice product from cDNAs encoding the human adenovirus type 5 E4orf6 protein. Virology 275:263-266. [DOI] [PubMed] [Google Scholar]
- 40.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Querido, E., M. R. Morrison, H. Chu-Pham-Dang, S. W. Thirlwell, D. Boivin, and P. E. Branton. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Querido, E., J. G. Teodoro, and P. E. Branton. 1997. Accumulation of p53 induced by the adenovirus E1A protein requires regions involved in the stimulation of DNA synthesis. J. Virol. 71:3526-3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roulston, A., R. C. Marcellus, and P. E. Branton. 1999. Viruses and apoptosis. Annu. Rev. Microbiol. 53:577-628. [DOI] [PubMed] [Google Scholar]
- 44.Salone, B., Y. Martina, S. Piersanti, E. Cundari, G. Cherubini, L. Franqueville, C. M. Failla, P. Boulanger, and I. Saggio. 2003. Integrin alpha3beta1 is an alternative cellular receptor for adenovirus serotype 5. J. Virol. 77:13448-13454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuelson, A. V., and S. W. Lowe. 1997. Selective induction of p53 and chemosensitivity in RB-deficient cells by E1A mutants unable to bind the RB-related proteins. Proc. Natl. Acad. Sci. U. S. A. 94:12094-12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarnow, P., Y. S. Ho, J. Williams, and A. J. Levine. 1982. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell 28:387-394. [DOI] [PubMed] [Google Scholar]
- 47.Shen, Y., G. Kitzes, J. A. Nye, A. Fattaey, and T. Hermiston. 2001. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 75:4297-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16:349-357. [DOI] [PubMed] [Google Scholar]
- 49.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348-352. [DOI] [PubMed] [Google Scholar]
- 50.Stracker, T. H., D. V. Lee, C. T. Carson, F. D. Araujo, D. A. Ornelles, and M. D. Weitzman. 2005. Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J. Virol. 79:6664-6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teodoro, J. G., and P. E. Branton. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weinberg, D. H., and G. Ketner. 1986. Adenoviral early region 4 is required for efficient viral DNA replication and for late gene expression. J. Virol. 57:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wienzek, S., J. Roth, and M. Dobbelstein. 2000. E1B 55-kilodalton oncoproteins of adenovirus types 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J. Virol. 74:193-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Woo, J. L., and A. J. Berk. 2007. Adenovirus ubiquitin-protein ligase stimulates viral late mRNA nuclear export. J. Virol. 81:575-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao, Z., Y. Xiong, W. Zhang, L. Tan, E. Ehrlich, D. Guo, and X. F. Yu. 2007. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J. Mol. Biol. 373:541-550. [DOI] [PubMed] [Google Scholar]
- 56.Yew, P. R., and A. J. Berk. 1992. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 357:82-85. [DOI] [PubMed] [Google Scholar]