Abstract
Nonstructural protein 1 (NS1) is one of the major factors resulting in the efficient infection rate and high level of virulence of influenza A virus. Although consisting of only approximately 230 amino acids, NS1 has the ability to interfere with several systems of the host viral defense. In the present study, we demonstrate that NS1 of the highly pathogenic avian influenza A/Duck/Hubei/L-1/2004 (H5N1) virus interacts with human Ubc9, which is the E2 conjugating enzyme for sumoylation, and we show that SUMO1 is conjugated to H5N1 NS1 in both transfected and infected cells. Furthermore, two lysine residues in the C terminus of NS1 were identified as SUMO1 acceptor sites. When the SUMO1 acceptor sites were removed by mutation, NS1 underwent rapid degradation. Studies of different influenza A virus strains of human and avian origin showed that the majority of viruses possess an NS1 protein that is modified by SUMO1, except for the recently emerged swine-origin influenza A virus (S-OIV) (H1N1). Interestingly, growth of a sumoylation-deficient WSN virus mutant was retarded compared to that of wild-type virus. Together, these results indicate that sumoylation enhances NS1 stability and thus promotes rapid growth of influenza A virus.
A series of influenza A virus outbreaks in recent years has led to intensified interest in these zoonotic viruses. Influenza A viruses are negative-sense single-stranded RNA viruses of the family Orthomyxoviridae that are further subdivided according to their 16 hemagglutinin (HA) and 9 neuraminidase (NA) subtypes (13, 61). The natural reservoirs of influenza A viruses are aquatic birds, from which they are occasionally transmitted to other species, including humans, where they are responsible for annual epidemics and less frequent pandemics. The most recent pandemic, caused by the swine-origin influenza A virus (S-OIV) (H1N1), originated in the beginning of 2009 in Mexico and the United States and quickly spread around the world (7, 14, 62). Although not yet adapted to humans, highly pathogenic avian influenza (HPAI) viruses are also still of considerable concern (10, 11, 57). Understanding the pathogenesis of these newly emerged viruses is therefore mandatory.
The 10 or 11 viral proteins encoded by the influenza A virus genome include the multifunctional nonstructural protein NS1 (22). Past and recent studies have indicated that NS1 is involved not only in the regulation of transcription and translation of the viral genome but also in a variety of virus-host interactions (22). The most prominent among these functions is the role of NS1 as an interferon (IFN) antagonist. Virus lacking NS1 is unable to replicate efficiently in an IFN-competent system (3, 16, 32). NS1 antagonizes IFN by inhibiting IFN production and interfering with IFN-induced antiviral processes. The mechanisms involved in suppression of IFN production include, on one hand, inhibition of mRNA induction by interfering with the RIG-I/IRF signaling pathway (15, 38) and, on the other hand, inhibition of processing and translation of IFN mRNA. Thus, NS1 inhibits pre-mRNA splicing and polyadenylation by binding to U6 snRNA, CPSF30, and PABII (8, 43, 45, 48), and it inhibits nuclear export by targeting the mRNA export machinery and the nuclear pore complex (53). NS1 also mediates resistance to IFN-induced antiviral processes by targeting protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase (2′-5′-OAS), both of which are key regulators of transcription and translation of viral genes (35, 39, 40). It is reasonable to assume that modulation of NS1 by posttranslational modification is required to allow so many different functions for a protein that has an approximate molecular mass of <30 kDa. It has been demonstrated that NS1 proteins from certain strains are phosphorylated at their C termini (6, 21, 47). Although removal of a phosphorylation site by a threonine-alanine substitution at position 215 resulted in viral attenuation of A/Udorn/72 virus (21), the precise biological functions of NS1 phosphorylation remain unclear. Recently, NS1 was reported to be modified by ISG15, which appears to be an antiviral host response (58, 64). Only a little is known about other posttranslational modifications of NS1 and their functional implications.
The family of SUMO (small ubiquitin-like modifier) proteins was recently discovered as a group of reversible posttranslational protein modifiers. The four SUMO paralogs in mammals, SUMO1 to -4, share high structural homology with ubiquitin and are covalently attached to target proteins in a similar fashion (19). After proteolytic maturation, exposing a C-terminal Gly-Gly motif, SUMO proteins are attached via an isopeptide bond between the C terminus and a lysine residue of the target protein by a three-step enzymatic cascade. First, the SUMO-activating enzyme E1, which consists of a heterodimer of the Aos1/SAE1 and Uba2/SAE2 proteins and is equivalent to the ubiquitin-activating enzyme Uba1, activates and transfers SUMO to the conjugating enzyme E2. Second, Ubc9, the only known E2 conjugating enzyme for SUMO, interacts with and transfers SUMO to a lysine residue of the target protein. Third, the transfer of SUMO from Ubc9 to the target protein is further facilitated by several SUMO ligases (E3) (24). Sumoylation is a highly dynamic and reversible process which is accomplished by SUMO-specific proteases (SENPs) catalyzing the deconjugation of SUMO from its substrates (41). Ubiquitin and SUMO both regulate protein localization and activity by serving as adaptors for protein-protein interactions (19). Ubiquitin plays a central role in targeting proteins for proteolytic degradation by the proteasome. Moreover, ubiquitination is important for receptor endocytosis and for several processes in transcriptional control and DNA repair. Sumoylation not only has been implicated in a variety of mainly nuclear processes, such as regulation of nuclear transport and gene transcription, DNA repair, and chromatin remodeling, but also is involved in signal transduction events (17, 18, 20). More recently, it was reported that NS1 of an H1N1 influenza A virus may also be a target of sumoylation (46). However, an exact biochemical characterization including the modification site is still lacking. Moreover, it remains unclear whether sumoylation is a conserved feature of NS1 proteins among influenza A viruses and whether NS1 proteins of HPAI strains also carry this modification.
In the present study, we demonstrate that NS1 of an HPAI H5N1 virus is able to interact with human Ubc9 and, as a consequence, is posttranslationally modified by SUMO1. We further identify two C-terminal lysine residues, at positions 219 and 221, as those responsible for sumoylation of NS1. Mutation of these lysine residues reduces the stability of NS1 and its ability to suppress host protein expression and, furthermore, causes a retardation of virus growth. Our data also suggest that sumoylation of NS1 is a posttranslational modification common to most strains. Interestingly, however, NS1 from the newly emerging S-OIV (H1N1) is an exception, since the sumoylation motif is abrogated by a C-terminal deletion.
MATERIALS AND METHODS
Cell culture and transfection.
Human embryonic kidney 293T cells (HEK293T), HEK 293 cells, human cervical epithelial cancer cells (HeLa), Madin-Darby canine kidney cells (MDCK), and human lung adenocarcinoma epithelial A549 cells purchased from the ATCC were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (Gibco) at 37°C in a CO2 incubator.
Lipofectamine 2000 (Invitrogen) was applied for transient transfection following the manufacturer's procedure.
Plasmids and small interfering RNAs (siRNAs).
Viral cDNA of A/Duck/Hubei/L-1/2004 (H5N1) was kindly provided by Tianxian Li (Wuhan Institute of Virology, Chinese Academy of Science). Viral cDNAs of A/HK/213/03 (H5N1), A/HK/156/97 (H5N1), A/HK/486/97 (H5N1), A/Viet Nam/1194/2004 (H5N1), and A/HK/1073/99 (H9N2) were kindly provided by Bo-Jian Zheng (Hong Kong University). Viral cDNA of A/Chicken/Jiangsu/7/2002 (H9N2) was kindly provided by Ze Chen (Shanghai Institute of Biological Products). Viral cDNA from S-OIV A/Sichuan/1/2009 (H1N1) was kindly provided by Yuelong Shu (Chinese Center for Disease Control and Prevention), and the A/PR/8/34 (H1N1) NS gene in pHW2000 was kindly provided by Robert G. Webster (St. Jude Children's Research Hospital, University of Tennessee). A/WSN/33 (H1N1) viral genes in pHW2000 have been described previously (56).
All NS1 genes were amplified with specific primers and were cloned into pcDNA-3′HA (Invitrogen). Subsequently, NS1 genes with the HA tag at the 3′ end were amplified by PCR and subcloned into pCAGGS vector, kindly provided by Jun-Ichi Miyazaki (Osaka University). The NS1 gene from S-OIV A/Sichuan/1/2009 (H1N1) was amplified with primers carrying a 5′ HA sequence and then cloned into pCAGGS vector so that the C-terminal deletion was kept intact. An NS1 mutant unable to bind to CPSF30 (NS1CPSFm) was obtained by using a QuikChange mutagenesis kit (Stratagene) on pCAGGS-NS1 with primers containing F103S and M106I mutations (31). Mutations of potential sumoylation sites on NS1 genes were introduced by performing one-step mutation PCR on pCAGGS-NS1, using primers containing the respective lysine (K)-to-arginine (R) mutation(s). All plasmids expressing NS1 were splice acceptor mutants in which the intron-exon junction of the CCAGGA sequence was replaced by CCCGGG to prevent production of spliced NS2 mRNA (2). The RNA-binding domain (RBD) (NS11-73) and the effector domain (NS174-225) of NS1 were cloned into pCAGGS vector with a 3′ HA tag. To restore the full-length C terminus of swine-origin NS1 without altering the NS2 sequence, a stop codon after K219 was mutated to arginine (23). The resulting gene was cloned into the pCAGGS vector together with an N-terminal HA tag (swNS1STOPmut). To generate recombinant virus, NS cDNA from A/Duck/Hubei/L-1/2004 (H5N1) was cloned into the pHW2000 vector. Sumoylation-deficient NS cDNA in the pHW2000 vector was obtained by one-step mutation PCR with primers containing K219E and K221E mutations, which kept the NS2 sequence intact.
The Saccharomyces cerevisiae expression plasmid pGAD-Ubc9 was obtained by amplifying Ubc9 from HeLa cell cDNA, which was then cloned into pGADT7 vector (Clontech), and the pUbc9-GST plasmid for prokaryotic expression was obtained by cloning the Ubc9 gene into pGEX4T-1 (GE). Ubc9 was also subcloned into pEGFP-C1 vector (Clontech) to obtain pEGFP-Ubc9. Human SUMO1, SUMO2, and SUMO3 were subcloned into pcDNA3 harboring cerulean (Cer; a variant of enhanced cyan fluorescent protein with a higher quantum yield and improved fluorescence lifetime [50]) to obtain Cer-SUMO1, Cer-SUMO2, and Cer-SUMO3 (30). Cer-SUMO1AA was obtained by one-step mutation PCR with primers encoding GG-to-AA mutations at the C terminus of SUMO1. SUMO1 was further subcloned into pCAGGS to obtain pCAGGS-SUMO1. Expression plasmids pcDNA3-SENP1 (SENP1) and pcDNA3-SENP1mut (SENP1mut) were described previously (9).
A luciferase reporter plasmid, pIFNβ-Luc, was constructed by amplifying the human IFN-β promoter (positions −320 to +21) from the HeLa cell genome and cloning it into the pGL3-Enhancer vector (Promega).
All plasmids were confirmed by sequence analysis.
Two 21-nucleotide human SENP1 siRNAs (si-1 [AACTACATCTTCGTGTACCTC] and si-2 [CTAAACCATCTGAATTGGCTC]) (9) and one 19-nucleotide siRNA against human Ubc9 (GGAGGAAAGACCACCCATT) were synthesized commercially (Ribobio Co., Ltd.).
Virus generation and infection.
Recombinant viruses were rescued by plasmid-based reverse genetics followed by two-round plaque purification and propagation on MDCK cells as previously described (26). Briefly, NS cDNA from A/Duck/Hubei/L-1/2004 (H5N1) or A/WSN/33 (H1N1) virus or NS cDNA expressing sumoylation-deficient NS1K219,221E was cotransfected with the remaining seven viral cDNAs from A/WSN/33 (H1N1) into HEK293T cells. The supernatant was then transferred onto MDCK cells at 72 h posttransfection. Virus multiplied from MDCK cells was further purified by a two-round plaque assay followed by multiplication. Viral RNA was extracted using a QIAamp viral RNA kit (Qiagen), followed by reverse transcription-PCR using Moloney murine leukemia virus reverse transcriptase (Invitrogen) and Uni12 primers (27). The viral cDNAs were then verified by commercial sequencing.
To examine the sumoylation of NS1 in infected cells, virus titers for infection were determined by plaque assay titration on A549 cells. Cells were transfected with siRNAs, followed by virus infection at a multiplicity of infection (MOI) of 5 for 1 h. Cell extracts were collected at 8 h postinfection and analyzed by Western blotting.
To stimulate IFN expression in the luciferase reporter assay, IFN-inducing Sendai virus (SeV) propagated in 10-day-old specific-pathogen-free (SPF) chicken embryos was used. The virus titer was measured by HA assay with 0.5% chicken red blood cells, and 10 HA units per well of SeV was used to infect HEK293T cells in 24-well plates.
Viral growth kinetics.
Confluent A549 cells were infected with wild-type A/WSN/33 (H1N1) virus (WSN-WT) or its sumoylation-deficient mutant (WSN-K219,221E) at an MOI of 0.1. Cell supernatants were harvested at the indicated times, and samples were titrated by plaque assay on MDCK cells.
Yeast two-hybrid assay.
The Matchmaker two-hybrid system 3 (Clontech) was used to identify the interaction(s) between NS1 and host proteins in vitro. The full-length A/Duck/Hubei/L-1/2004 (H5N1) NS1 gene was cloned in frame into pGBKT7 (Clontech) to generate the bait plasmid pGBK-NS1. However, because of strong self-activation of the pGBK-NS1 bait, we generated a series of C-terminal deletions of NS1 which were then tested for self-activation. pGBK-NS11-162, containing the N-terminal 162 amino acids (aa 1 to 162) of NS1 in pGBKT7, which did not show self-activation, was selected as the bait plasmid. The yeast strain AH109 transformed with the bait plasmid pGBK-NS11-162 was subsequently transformed with a human lymphocyte cDNA library in pACT2 (Clontech). SD/Trp-Leu yeast synthetic medium lacking tryptophan and leucine was used to select cotransformants. SD/Trp-Leu-His-Ade yeast synthetic medium lacking tryptophan, leucine, histidine, and adenine was used to select potential interactors. Colonies surviving on SD/Trp-Leu-His-Ade plates after 4 to 6 days at 30°C were further subjected to colony lift filter assay of β-galactosidase activity, and plasmid DNAs were prepared from positive colonies and sequenced for identification of cDNA clones.
The yeast strain SFY526, which contains a quantifiable lacZ reporter (kindly provided by George F. Gao, Institute of Microbiology, Chinese Academy of Sciences), was used for confirmation and calculation of the interaction strength between NS1 and human Ubc9. Yeast expression plasmids pGBK-NS11-162 and pGAD-Ubc9 were cotransformed into the yeast strain SFY526. A β-galactosidase colony lift filter assay using X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as the substrate and a liquid culture assay using ONPG (o-nitrophenyl-β-d-galactopyranoside) as the substrate were carried out according to the instructions in the Clontech Matchmaker manual and were repeated at least three times.
Protein expression, purification, and GST pull-down assay.
To purify glutathione S-transferase (GST) or GST-Ubc9 proteins, Escherichia coli strain BL21 was transformed with pGEX4T-1 or pUbc9-GST, and expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested after 12 h, resuspended in ice-cold phosphate-buffered saline (PBS), and homogenized by sonication. The lysates were cleared by centrifugation, and the supernatant was subjected to Sepharose 4B-glutathione resin (GE) for affinity purification. GST or GST-Ubc9 proteins were eluted from the column with 10 mM glutathione elution buffer.
For pull-down assays, equal amounts of purified GST or GST-Ubc9 were bound to Sepharose 4B-glutathione, and beads were mixed for 4 h at 4°C with 2 ml lysate from HEK293T cells expressing HA-tagged NS1 protein. The beads were washed three times with PBS. Bound proteins were eluted with SDS sample buffer and analyzed by SDS-PAGE and Western blotting.
Immunoprecipitation and Western blot analysis.
HEK293T cells were transfected with HA-tagged NS1 plasmids and collected at 36 h posttransfection. Cells were washed with cold PBS, lysed in RIPA lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA) supplemented with Complete protease inhibitor (Roche) and 10 mM N-ethylmaleimide (NEM; Sigma) at 4°C for 20 min, and then centrifuged at maximum speed in an Eppendorf microcentrifuge to remove cell debris. The supernatant was subjected to immunoprecipitation with anti-HA agarose (Sigma) at 4°C for 4 h. Subsequently, the beads were washed 5 times with RIPA buffer and eluted with SDS loading buffer, and eluates were separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes (Bio-Rad). NS1 proteins were detected with anti-HA polyclonal antibody (Sigma) and horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG secondary antibody (Southern Biotech). SUMO1 was detected with anti-SUMO1 monoclonal antibody (Zymed) and HRP-conjugated goat anti-mouse IgG secondary antibody (R&D). GST and Ubc9-GST were detected with anti-GST polyclonal antibody (Antibody Research Centre, Shanghai Institutes for Biological Sciences). Cer-SUMO was detected with anti-green fluorescent protein (anti-GFP) antibody (Antibody Research Centre, Shanghai Institute of Biological Science). Actin was detected with anti-actin polyclonal antibody (Sigma). Secondary antibodies were visualized with an enhanced chemiluminescence kit (RPN2132; GE).
To examine the sumoylation of NS1 in infected cells, A549 cells were collected at 8 h postinfection. The cells were lysed directly in SDS loading buffer at 100°C for 10 min and immediately subjected to Western blot analysis using anti-NS1 polyclonal antibody (Antibody Research Centre, Shanghai Institute of Biological Science).
To block protein synthesis, HEK293 cells were treated at 36 h posttransfection with 100 μg/ml cycloheximide (CHX; Sigma). Cells were lysed at 0, 2, 4, 6, and 8 h post-CHX treatment and subjected to immunoprecipitation to pull down NS1 protein. NS1 amounts were analyzed by Western blotting with anti-HA antibody.
Luciferase reporter assay.
Firefly and Renilla luciferase activities were measured using a dual-luciferase kit (Promega) and a Veritas microplate luminometer (Turner Biosystems) with the Veritas 2.0.40 software package. HEK293T cells were seeded in 24-well plates 24 h before transfection. Fifty nanograms of the IFN-β promoter reporter plasmid pIFNβ-Luc expressing firefly luciferase, 1 ng of Renilla luciferase-expressing plasmid pSV40-Ren (pRL-SV40; Promega), and 1 μg of NS1-expressing plasmid were cotransfected into HEK293T cells by use of Lipofectamine 2000 (Invitrogen). Cells were infected with 10 HA units/well SeV to induce IFN expression at 16 h posttransfection. Cells were harvested an additional 16 h later and lysed by passive lysis buffer (Promega) for luciferase assay. The experiments were done at least twice, with at least duplicate samples in each study. Results are presented as means plus standard deviations.
Immunocytochemistry and confocal microscopy.
HeLa or A549 cells cultured on glass slides were fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100 in PBS. Cells were then immunolabeled with corresponding antibodies for 2 h. HA-tagged NS1 was detected with either anti-HA monoclonal antibody (Covance) or anti-HA polyclonal antibody (Sigma), and SUMO1 was detected with anti-SUMO1 monoclonal antibody (Zymed). Cells were washed 5 times with PBS and incubated with goat anti-mouse-Alexa 488 (Molecular Probes) together with goat anti-rabbit-Cy3 (Jackson) or goat anti-rabbit-Alexa 488 (Molecular Probes) together with goat anti-mouse-Cy3 (Jackson) for 1 h. DAPI (4′,6-diamidino-2-phenylindole; Sigma) was used for nuclear staining. All images were acquired on a Leica TCS SP2 confocal microscope (Leica Microsystems).
RESULTS
Identification of human Ubc9 as an H5N1 NS1-interacting protein in yeast two-hybrid assays.
To identify novel cellular interaction partners of the NS1 protein of an H5N1 HPAI virus, a yeast two-hybrid screening assay was applied by use of the NS1 gene of A/Duck/Hubei/L-1/2004 (H5N1) virus and a cDNA library from human lymphocytes. Initial experiments with full-length NS1 (225 aa) as a bait resulted in self-activation of NS1 cotransfected with empty vector pGADT7 (data not shown). Thus, a C-terminally truncated NS1 protein lacking the last 63 aa (NS11-162), which did not display self-activation, was used as the bait in the yeast two-hybrid assay. As shown in Fig. 1 A, an interaction between NS11-162 and the human SUMO E2 enzyme Ubc9 was observed in yeast strain AH109 growing on an SD/Trp-Leu-His-Ade plate. The interaction was further confirmed and quantified by β-galactosidase assay with yeast strain SFY526, which resulted in ∼20% of the signal for positive-control p53 and T antigen, indicating a medium interaction strength between NS11-162 and Ubc9 (Fig. 1B and C). These results indicate that human Ubc9 is able to interact with NS1.
FIG. 1.
A/Duck/Hubei/L-1/2004 (H5N1) NS1 is a new human Ubc9-interacting protein. (A) Yeast AH109 cells cotransformed with the indicated combinations of bait and prey plasmids were grown on SD/Trp-Leu medium to select for double-transformed cells (left) and then further grown on an SD/Trp-Leu-His-Ade plate for yeast two-hybrid interaction screening (right). 1, negative control (pGBK-Lam+pGAD-T); 2, positive control (pGBK-p53+pGAD-T); 3, pGBK+pGAD; 4, pGBK+pGAD-Ubc9; 5, pGBK-NS11-162+pGAD; 6, pGBK-NS11-162+ pGAD-Ubc9. (B) Colony lift filter assay. The indicated plasmid pairs were cotransfected into the yeast strain SFY526. Colonies grown on SD/Trp-Leu-His-Ade selection medium were transferred to a filter paper for β-galactosidase assay, using X-Gal as the substrate. (C) Liquid culture assay. Plasmid pairs were cotransfected into the yeast strain SFY526 as indicated, and β-galactosidase activity was quantified by measurements of the optical density at 600 nm, using ONPG as the substrate. Results shown are means for three independent experiments (arbitrary units). Error bars represent standard deviations.
NS1 and Ubc9 interact in vitro and colocalize in the nucleus.
To further verify the interaction between NS1 and human Ubc9, we performed GST pull-down experiments. GST-Ubc9 or GST was expressed in E. coli and incubated on glutathione-Sepharose columns with full-length NS1 from A/Duck/Hubei/L-1/2004 (H5N1) expressed in HEK293T cells. NS1 showed a strong interaction with GST-Ubc9, whereas it did not bind to GST alone (Fig. 2 A). NS1 consists of an RBD and an effector domain which are connected by a flexible linker. We thus asked whether either of the two domains would mediate the interaction with Ubc9. Again, GST pull-down experiments were performed, using GST-Ubc9 and both domains of NS1 expressed individually in 293T cells. Surprisingly, neither the RBD (aa 1 to 73) nor the effector domain (aa 74 to 225) of NS1 was able to bind to GST-Ubc9 (Fig. 2B). These results suggest that the interaction between NS1 and Ubc9 is dependent on an intact conformation of NS1 protein.
FIG. 2.
NS1 and Ubc9 interact in vitro and colocalize in the nucleus. (A) GST or Ubc9-GST was expressed in E. coli, bound to glutathione beads, and incubated with lysates from HEK293T cells expressing HA-tagged NS1 from A/Duck/Hubei/L-1/2004 (H5N1). Bound proteins were eluted and analyzed by SDS-PAGE and Western blotting with anti-HA or anti-GST antibody. Whole-cell lysates are shown as input. (B) GST pull-down experiments were done as described above, with lysates from 293T cells expressing either the HA-tagged RBD (NS11-73) or the HA-tagged effector domain (NS174-225) of NS1. Bound proteins were eluted and analyzed by SDS-PAGE and Western blotting with anti-HA antibody. (C) Colocalization of H5N1 NS1 and Ubc9. HeLa cells were cotransfected with pCAGGS-NS1 and pEGFP-Ubc9. Cells were fixed and permeabilized. NS1 was stained with mouse anti-HA monoclonal antibody followed by goat anti-mouse-Cy3 (red) as the secondary antibody. Nuclei were stained with DAPI (blue).
Having identified the interaction between NS1 and Ubc9, we wanted to know where the interaction occurs in mammalian cells. In infected cells, NS1 is located inside the nucleus throughout most of the infection cycle (36, 44). Similarly, the major fraction of Ubc9 is also found in the nucleus (52). Thus, we tested whether NS1 and Ubc9 proteins colocalize in the nucleus. HeLa cells were cotransfected with HA-tagged NS1 and Ubc9 fused to GFP. Immunocytochemistry revealed that Ubc9 was localized predominantly in the nucleus and that only a small proportion was localized in the cytoplasm, while NS1 was localized almost completely in the nucleus. An overlay of both channels indicated a strong colocalization between NS1 and Ubc9 (Fig. 2C). This suggests that the interaction between NS1 and Ubc9 proteins takes place mainly in the nucleus.
The H5N1 NS1 protein is sumoylated in vivo.
It is believed that the primary function of Ubc9 is to mediate the transfer of SUMO to target proteins (24). We therefore examined whether NS1 protein from the H5N1 subtype is a target of SUMO modification. To address this question, we first assessed the subcellular localization of NS1 and SUMO1 by immunocytochemistry with cells cotransfected with HA-tagged NS1 and SUMO1. As we expected, a strong colocalization was observed between NS1 and SUMO1 in nuclei (Fig. 3 A). Second, we examined sumoylation of NS1 in transfected cells. HEK293T cells were cotransfected with HA-tagged NS1 and Cer-SUMO1, and the expression of Cer-SUMO1 was monitored by fluorescence microscopy prior to sumoylation analysis. A strong inhibition of Cer-SUMO1 expression was observed in NS1-cotransfected cells, which was most likely caused by the mRNA-interfering properties of NS1 protein (see Fig. S1 in the supplemental material). To overcome this, an NS1 mutant (NS1CPSFm) which contained two point mutations within the CPSF30-binding sites and thereby lost the ability to inhibit cellular pre-mRNA processing (31) was used and showed no significant reduction of SUMO expression (see Fig. S1). The G-protein-interacting protein phosducin (Phd), recently identified to be sumoylated, was used as a positive control for sumoylation (30). HA-tagged NS1, NS1CPSFm, or Phd was enriched by immunoprecipitation with anti-HA antibody after quick lysis of the cells in the presence of NEM to prevent desumoylation. Proteins were analyzed by SDS-PAGE and subsequent Western blot detection with anti-HA antibody. For cells not expressing Cer-SUMO1, NS1 and Phd were detected as bands of ∼30 kDa and ∼33 kDa, respectively. However, with cells coexpressing Cer-SUMO1, a second band was detected for NS1- and Phd-transfected cells, at ∼70 kDa and ∼73 kDa, respectively (Fig. 3B, left panel). The shifts in molecular mass of ∼40 kDa can be explained by modification of NS1 and Phd with Cer-SUMO1. Bands of ∼70 kDa and ∼73 kDa were also observed when the same samples were probed again with anti-SUMO1 antibody, confirming that they were sumoylated forms of NS1 and Phd (Fig. 3B, right panel). Remarkably, the ∼70-kDa band of NS1 was more pronounced in NS1CPSFm-expressing cells than in NS1-expressing cells, reflecting inhibition of Cer-SUMO1 expression by NS1. To ensure maximal sumoylation, NS1CPSFm was therefore used in the following experiments.
FIG. 3.
Sumoylation of A/Duck/Hubei/L-1/2004 (H5N1) NS1 protein in vivo. (A) Colocalization of NS1 and SUMO1. HeLa cells were cotransfected with pCAGGS-NS1 and pCAGGS-SUMO1. Cells were fixed and permeabilized. NS1 protein was stained with anti-HA-tag rabbit polyclonal antibody, and SUMO1 was stained with mouse anti-SUMO1 monoclonal antibody. Goat anti-rabbit-Alexa 488 (green) and goat anti-mouse-Cy3 (red) were used as secondary antibodies. Nuclei were stained with DAPI (blue). (B) Sumoylation of H5N1 NS1. HEK293T cells were transfected with expression plasmids for HA-tagged NS1, NS1CPSFm, Phd, and Cer-SUMO1, as indicated. (Left) NS1 and Phd were immunoprecipitated with anti-HA agarose (IP), and precipitated proteins were further analyzed by SDS-PAGE and Western blotting (WB) with anti-HA polyclonal antibody. (Right) The samples were probed again with anti-SUMO1 monoclonal antibody. Western blots with anti-actin or anti-HA antibody from whole-cell lysates are shown as loading controls (Input). (C) HEK293T cells were transfected with H5N1 NS1CPSFm together with Cer-SUMO1 or Cer-SUMO1AA. Cells were lysed, and NS1 was immunoprecipitated with anti-HA agarose. NS1 was detected as described above. Western blots with anti-GFP (WB: Cerulean) or anti-actin antibody for whole-cell lysates are shown as Cer-SUMO1 expression and loading controls (Input). The positions of molecular mass standards are marked on the left.
A SUMO1 mutant which cannot be attached to target proteins (Cer-SUMO1AA) further verified the sumoylation of NS1. The ∼70-kDa sumoylated form of NS1 was detectable only when NS1CPSFm and Cer-SUMO1, not NS1CPSFm and Cer-SUMO1AA, were coexpressed (Fig. 3C). Taken together, the data confirm that NS1 of the H5N1 subtype is a target for sumoylation with SUMO1.
Deconjugation of SUMO1 from NS1 is regulated by SENP1.
Sumoylation is a highly dynamic process which is mediated by several proteases (SENPs) specifically cleaving SUMO from substrate proteins. Among these, SENP1 is the predominant protease for the cleavage of SUMO1 (49). We thus tested whether SENP1 is involved in SUMO1 modification of NS1. As shown in Fig. 4 A, when SENP1 was coexpressed with Cer-SUMO1 and NS1CPSFm, the sumoylated band of NS1 was strongly reduced, while in cells coexpressing a functionally deficient SENP1 protein (SENP1mut), the ∼70-kDa band representing sumoylated NS1 was affected little. This result indicates that SENP1 is involved in the SUMO1 modification of NS1CPSFm and cleaves SUMO1 from NS1CPSFm.
FIG. 4.
Deconjugation of SUMO1 from A/Duck/Hubei/L-1/2004 (H5N1) NS1 is regulated by SENP1. (A) HEK293T cells were transfected with expression plasmids for NS1CPSFm, Cer-SUMO1, SENP1, or SENP1mut, as indicated, and total DNA levels were adjusted by adding vector plasmid. NS1 was immunoprecipitated (IP) with anti-HA agarose, and precipitated proteins were further analyzed by SDS-PAGE and Western blotting (WB) with anti-HA polyclonal antibody. (B) Modification of H5N1 NS1 protein with endogenous SUMO1. HEK293T cells were cotransfected with NS1CPSFm, SENP1, or SENP1mut, as indicated. Immunoprecipitation and detection of NS1 were performed as described above. Western blots with anti-SUMO1 or anti-actin antibody for whole-cell lysates are shown as input. The positions of molecular mass standards are marked on the left.
Detection of sumoylated proteins is often difficult owing to the rapid cycle of sumoylation and desumoylation, which ultimately results in low steady-state levels of modified proteins. Nevertheless, it was of interest to find out whether NS1 is also modified by endogenous SUMO1. To accomplish this, we took advantage of the above-mentioned desumoylation of NS1 by SENP1. NS1CPSFm protein was immunoprecipitated from cells expressing HA-tagged NS1CPSFm and analyzed by Western blotting. In addition to the 30-kDa band of NS1, a band of approximately 43 kDa was detected, in accordance with an 11- to 15-kDa mass shift caused by endogenous SUMO1. Coexpression of SENP1 strongly reduced the appearance of this additional band, whereas SENP1mut did not (Fig. 4B). These data show that NS1 is indeed modified with endogenous SUMO1 in mammalian cells and that this reaction is reversed by SENP1.
NS1 is sumoylated in infected cells.
In order to address whether the sumoylation of H5N1 NS1 protein occurs naturally in infected cells, A/Duck/Hubei/L-1/2004 (H5N1) NS cDNA was introduced into an A/WSN/33 (H1N1) background to generate a recombinant virus expressing H5N1 NS1 protein (r-H5N1-NS virus) that can be operated under biosafety level 2 conditions. siRNAs specific for human Ubc9 and SENP1 were used to detect endogenous sumoylation of NS1 protein. A549 cells were transfected with siRNAs as indicated, followed by infection with r-H5N1-NS virus at 24 h posttransfection. Extracts from cells collected at 8 h postinfection were analyzed by immunoblotting with anti-NS1 antibody. As shown in Fig. 5, a band of ∼28 kDa representing untagged NS1 protein and an additional, ∼43-kDa band corresponding to NS1-SUMO were detected in cells transfected with control siRNA (NC). The ∼43-kDa sumoylated NS1 protein was clearly increased in cells transfected with the two SENP1-specific siRNAs. In contrast, for mock-infected cells and cells transfected with Ubc9-specific siRNA, the ∼43-kDa band was not detectable. These data further show that NS1 is also sumoylated in infected cells by endogenous SUMO, which is regulated by Ubc9 and SENP1.
FIG. 5.
Sumoylation of A/Duck/Hubei/L-1/2004 (H5N1) NS1 in infected cells. A549 cells were transfected with the indicated siRNA for 24 h, followed by infection with r-H5N1-NS virus (MOI = 5) or mock infection. At 8 h postinfection, cells were lysed in SDS loading buffer containing NEM, followed by SDS-PAGE and Western blotting with antibodies against NS1 or actin. The positions of molecular mass standards are marked on the left. NC, control siRNA.
The SUMO1 acceptor sites are located at the C terminus of H5N1 NS1.
Having determined that influenza virus NS1 protein is posttranslationally modified by SUMO1, we tried to identify the modification sites in NS1. Sequence analysis revealed that A/Duck/Hubei/L-1/2004 (H5N1) NS1 contains 13 lysine residues, 2 of which are located in sumoylation consensus motifs (ψKXD/E; ψ is a hydrophobic residue and X is any amino acid), at positions 77 to 80 (LKMP) and 117 to 120 (IKMD) (sequence numbering is according to the PR8 NS1 sequence) (17). However, since more and more proteins have been identified to be sumoylated at nonconsensus sites (29), systematic one-by-one mutations of all 13 individual lysine residues were introduced into NS1CPSFm to include all possible modification sites, and the ability of these mutants to be sumoylated was investigated. Since neighboring lysine residues may substitute for each other as SUMO acceptors, two double mutants (NS1CPSFmK108,110R and NS1CPSFmK219,221R) were also established. Indeed, mutation of the lysine residues in the two sumoylation consensus sequences (NS1CPSFmK78R and NS1CPSFmK115R) did not result in reduced sumoylation compared to that with NS1CPSFm (Fig. 6 A). Of all single mutants, only NS1CPSFmK221R displayed a significant decrease of sumoylation (Fig. 6A). The mutation of K221 together with the nearby residue K219 completely abolished sumoylation of NS1CPSFm, while the K219R mutation itself was not able to affect sumoylation levels (Fig. 6A). The sumoylation sites at positions 219 and 221 were further examined in wild-type NS1 to exclude any possible influence of the CPSF30-binding-site mutations in NS1CPSFm. NS1 or NS1K219,221R was cotransfected with either Cer-SUMO1 or Cer-SUMO1AA. Again, only NS1 was modified by Cer-SUMO1, while NS1K219,221R lost this modification (Fig. 6B). These data indicate that K221 most likely is the major SUMO1 acceptor site of NS1, whereas K219 may substitute for it.
FIG. 6.
A/Duck/Hubei/L-1/2004 (H5N1) NS1 is sumoylated at lysine 219 and lysine 221. (A) Mapping of lysine residues responsible for sumoylation of H5N1 NS1. HEK293T cells were transfected with expression plasmids encoding H5N1 NS1CPSFm or mutants containing lysine (K)-to-arginine (R) substitutions at the indicated positions. Cer-SUMO1 or Cer-SUMO1AA was cotransfected as shown. Cells were lysed, and NS1 was immunoprecipitated with anti-HA agarose. NS1 was detected in Western blots with anti-HA polyclonal antibody. (B) Wild-type H5N1 NS1 or NS1K219,221R was coexpressed with Cer-SUMO1 or Cer-SUMO1AA, as indicated, and NS1 was visualized as described above. The positions of molecular mass standards are marked on the left.
In addition to SUMO1, two other paralogs, SUMO2 and SUMO3, exist that are ubiquitously expressed and are conjugated to proteins in a similar fashion. We thus tested whether the above-identified sumoylation sites could also serve as acceptor sites for SUMO2 or SUMO3. Indeed, upon cotransfection of SUMO2 or SUMO3 together with NS1, a high-molecular-weight form was detected that was compatible with sumoylated NS1. However, SUMO2- and SUMO3-modified NS1 levels were much lower than SUMO1-modified NS1 levels. NS1K219,221R was not sumoylated when coexpressed with SUMO2 or SUMO3, indicating that SUMO2 and SUMO3 are also conjugated to K219 and K221 (see Fig. S2 in the supplemental material).
Sumoylation enhances NS1 protein stability.
NS1 interferes with processing, nuclear export, and translation of host mRNA, resulting in decreased production of antiviral cytokines, including IFN (8, 43, 45, 48). To test whether sumoylation of NS1 affects this important function, we measured gene expression levels in the absence or presence of different NS1 mutants of A/Duck/Hubei/L-1/2004 (H5N1) virus. A reporter plasmid expressing firefly luciferase under the control of the human IFN-β promoter (pIFNβ-Luc) was used to measure infection-induced IFN-β production, while a second reporter plasmid expressing Renilla luciferase under the control of the constitutive simian virus 40 (SV40) promoter (pSV40-Ren) was applied to measure host expression in general. Empty vector or plasmids expressing NS1, NS1CPSFm, or their sumoylation-deficient mutants were cotransfected with pIFNβ-Luc and pSV40-Ren. SeV, which causes strong IFN induction without interfering with the host expression system, was applied to stimulate IFN-β production. As shown in Fig. 7 A, IFN and general gene expression was strongly inhibited by wild-type NS1, while sumoylation-deficient NS1K219,221R lost part of the blocking ability. Moreover, NS1CPSFm, which was unable to inhibit the pre-mRNA cleavage and polyadenylation factor CPSF30, slightly suppressed host gene expression, while expression of its sumoylation-deficient mutant, NS1CPSFmK219,221R, had no effect on general gene expression. Since sumoylation often impairs protein degradation, we further examined the expression levels of NS1 proteins from cell extracts of the above samples. Amounts of the sumoylation-deficient NS1K219,221R and NS1CPSFmK219,221R mutants were reduced compared to those of the sumoylation-competent NS1 and NS1CPSFm proteins (Fig. 7B). These observations suggest that sumoylation-deficient NS1 has reduced stability, resulting in an impaired ability to inhibit host gene expression.
FIG. 7.
Increased stability of wild-type NS1 (from A/Duck/Hubei/L-1/2004 [H5N1]) compared to the sumoylation-deficient NS1K219,221R mutant. (A) SUMO1 conjugation of A/Duck/Hubei/L-1/2004 (H5N1) NS1 protein inhibits reporter gene activity. A firefly luciferase reporter plasmid under the control of the IFN-β promoter (pIFNβ-Luc) and a Renilla luciferase plasmid (PRL-SV40) under the control of a constitutive SV40 promoter (pSV40-Ren) were cotransfected with the indicated NS1-expressing plasmids into HEK293T cells. After 16 h, cells were infected with 10 HA units/well SeV or mock infected. Cell extracts were collected at 16 h postinfection, and the luciferase activities were measured. Results are given as percentages of the luciferase activity in SeV-infected vector-expressing cells (considered 100%) and represent the averages for three independent experiments, with standard deviations. *, statistically significant (P < 0.05) by Student's t test. (B) Cell extracts from panel A were diluted 5 times and further analyzed by Western blotting (WB) with anti-HA or anti-actin antibody. (C) HEK293T cells were cotransfected with pCAGGS-SUMO1 and NS1 or NS1K219,221R, as indicated. Thirty-six hours after transfection, cells were treated with 100 μg/ml CHX for the indicated periods. NS1 proteins were immunoprecipitated with anti-HA agarose (IP: HA), and precipitated proteins were further analyzed by SDS-PAGE and Western blotting with anti-HA polyclonal antibody to detect NS1 proteins (WB: HA). Western blots of whole-cell lysates with anti-actin antibody are shown as a loading control (Input). The positions of molecular mass standards are marked on the left. (D) Western blots as described for panel C were analyzed by densitometry. Means for three independent experiments (+ standard deviations) are shown.
To further demonstrate that the reduced levels of sumoylation-deficient NS1 were due to lower stability, protein synthesis was blocked with CHX and the steady-state levels of NS1 protein expressed before CHX treatment were monitored (30). As expected, a significant amount of wild-type NS1 was sumoylated (Fig. 7C, left panel), whereas in cells expressing NS1K219,221R, only the unmodified form of NS1 was detectable (Fig. 7C, right panel). Quantification by densitometry revealed that the levels of NS1 did not change significantly after 8 h with intact sumoylation. In contrast, the amounts of NS1K219,221R were decreased after 8 h, by nearly 75% (Fig. 7D). Thus, the CHX experiment supports the concept that sumoylation stabilizes NS1.
SUMO1 modification of NS1 protein is observed with different influenza virus subtypes and strains.
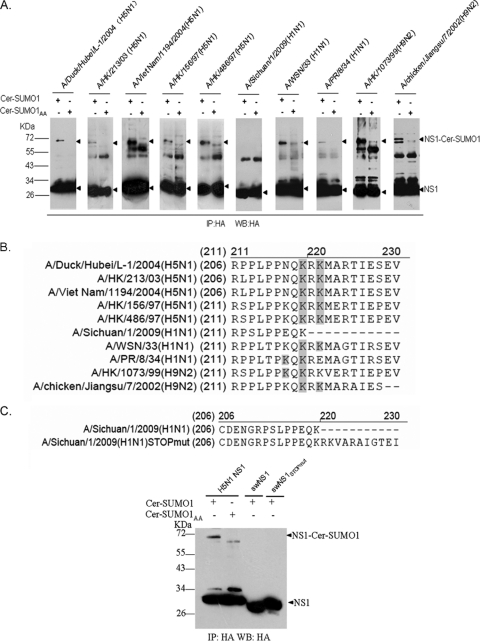
We tested the sumoylation of NS1 proteins from several avian and human isolates which have the potential to cause cross-species infection and exhibit high pathogenicity. HA-tagged NS1 proteins from several H5N1, H1N1, and H9N2 strains were coexpressed with Cer-SUMO1 or Cer-SUMO1AA in HEK293T cells. Subsequently, NS1 was immunoprecipitated and detected in Western blots with HA antibodies. With the exception of A/Sichuan/1/2009 (H1N1) NS1, all NS1 proteins showed a strong ∼30-kDa band and an ∼70-kDa band when coexpressed with Cer-SUMO1 but not with Cer-SUMO1AA (Fig. 8 A). Thus, all tested NS1 proteins were modified by SUMO1, except for NS1 from A/Sichuan/1/2009 (H1N1). We further examined all of the SUMO1 acceptor sites on these individual NS1 proteins (Fig. 8B; see Fig. S3 in the supplemental material) and found that K219 and K221 or K219 and K217 are responsible for SUMO1 modification of these NS1 proteins.
FIG. 8.
Sumoylation of NS1 proteins from different subtypes and strains. (A) HEK293T cells were cotransfected with plasmids encoding HA-tagged NS1 proteins from different influenza A virus subtypes and strains together with Cer-SUMO1 or Cer-SUMO1AA. Cells were lysed, and NS1 was immunoprecipitated (IP) with anti-HA agarose. Precipitated proteins were further analyzed by SDS-PAGE and Western blotting (WB) with anti-HA polyclonal antibody. The positions of molecular mass standards are marked on the left. (B) Schematic representation of the C-terminal amino acid sequences of NS1 proteins from different influenza A virus strains. Lysine (K) residues (shaded) indicate SUMO1 modification sites for each NS1 protein. (C) (Top) Schematic representation of the C-terminal amino acid sequences of swNS1 and swNS1STOPmut. (Bottom) Swine-origin NS1 (swNS1) and its extended form (swNS1STOPmut) are not sumoylated. HEK293T cells were transfected with H5N1 NS1 (from A/Duck/Hubei/L-1/2004) or with swNS1 (from A/Sichuan/1/2009 [H1N1]) or its C-terminally extended form, swNS1STOPmut, together with Cer-SUMO1 or Cer-SUMO1AA, as indicated. Cells were lysed, and NS1 was immunoprecipitated with anti-HA agarose. NS1 was detected with anti-HA antibody. The positions of molecular mass standards are marked on the left.
Interestingly, in comparing the amino acid sequences of the different strains, a C-terminal deletion of 11 aa was found in NS1 from A/Sichuan/1/2009 (H1N1), which may contribute to the lack of sumoylation for this protein (Fig. 8B). To test this hypothesis, the C terminus of A/Sichuan/1/2009 (H1N1) NS1 was restored by mutating a stop codon after K219 (swNS1STOPmut) (Fig. 8C, top panel). Nonetheless, no sumoylation of swNS1STOPmut could be observed when it was coexpressed with Cer-SUMO1 (Fig. 8C, bottom panel). Thus, it appears that properties other than the truncated C terminus alone prevent sumoylation of A/Sichuan/1/2009 (H1N1) NS1. Notably, for swine-origin NS1, an N-terminal HA tag was used to exclude any interference of the tag with possible sumoylation sites at the truncated C terminus. N-terminally tagged H5N1 NS1 was still sumoylated, excluding the possibility that the position of the tag in general may alter sumoylation of NS1 (data not shown). Taken together, these data suggest that sumoylation is a posttranslational modification occurring with many strains, with the exception of the newly emerged S-OIV A/Sichuan/1/2009 (H1N1) virus.
Sumoylation of NS1 protein accelerates virus replication.
To analyze the effects of NS1 sumoylation on virus replication, attempts have been made to generate recombinant viruses containing NS1 proteins with mutated sumoylation sites. When K219R and K221R mutations were introduced into NS1, we were unable to rescue WSN and H5N1 viruses. The failure to rescue virus with the K219R mutation was also observed in a previous study and may have been due to a missense mutation introduced into NS2 (64). However, when we replaced the lysine residues with glutamic acid (E), which leaves the NS2 protein intact, we obtained a WSN virus containing NS1 with the K219E and K221E mutations. In A549 cells infected with this virus (WSN-K219,221E), NS1 was not sumoylated, while after infection with wild-type virus, SENP1-dependent sumoylation of NS1 protein was clearly detected (Fig. 9 A). We further compared growth curves for mutant and wild-type viruses. Figure 9B shows that the WSN-K219,221E virus grew to lower titers than the WSN-WT virus. The differences in growth rates were small but reproducible and were most pronounced early in replication. Thus, it appears that NS1 sumoylation accelerates replication of WSN virus.
FIG. 9.
Sumoylation of K219 and K221 of NS1 accelerates virus replication. (A) A549 cells were transfected with the indicated siRNA for 24 h, followed by infection with wild-type WSN-WT (A/WSN/33) or a recombinant WSN virus expressing the NS1K219,221E mutant protein (WSN-K219,221E) at an MOI of 5. At 8 h postinfection, cells were lysed in SDS loading buffer containing NEM, followed by SDS-PAGE and Western blotting with anti-NS1 or anti-actin antibody. The positions of molecular mass standards are marked on the left. (B) A549 cells were infected with either WSN-WT or WSN-K219,221E virus at an MOI of 0.1, and virus production at the indicated times postinfection was determined by plaque assay on MDCK cells. Means for three independent experiments (+ standard deviations) are shown. **, P < 0.01.
DISCUSSION
As cell invaders and usurpers of the cellular machinery, many viruses exploit posttranslational modification systems of their hosts for their own purposes. Besides phosphorylation, glycosylation, and ubiquitination, an increasing number of viral proteins have been reported to interact with the host sumoylation system (4, 63). Some viral proteins inhibit sumoylation of host proteins. For instance, the human cytomegalovirus (HCMV) IE1 protein was reported to prevent sumoylation of the cellular PML protein (34), the adenovirus Gam1 protein inhibits the sumoylation cascade by binding to the E1 heterodimer (5), and the herpes simplex virus (HSV) ICP0 protein reduces sumoylation of PML and other substrates, probably by recruiting the SUMO protease SENP1 (1). On the other hand, multiple viral proteins exploit the sumoylation system of the host to modulate their own functions. Viral proteins from several important pathogens, including the severe acute respiratory syndrome (SARS) coronavirus and HIV, are found to be sumoylated (4). More recently, two genome-wide screening studies revealed that components of the host sumoylation machinery are involved in the replication of influenza A viruses (33, 55). Furthermore, NS1 protein from influenza A/PR/8/34 virus was recently reported to be sumoylated in a SUMO1 overexpression system (46). However, it was not known if NS1 sumoylation is a general phenomenon with all influenza A viruses, and identification of the modification site and elucidation of the function of sumoylation were also lacking.
Using a yeast two-hybrid system, we identified human Ubc9, the human E2 enzyme involved in SUMO modification, as a host protein interacting with the NS1 protein of an H5N1 virus. Further analysis revealed that NS1 from the H5N1 virus was modified by SUMO1, not only in transfected but also in infected cells, indicating that the NS1 protein is a bona fide target of sumoylation. As is the case with other sumoylated proteins, sumoylation of NS1 appears to be highly reversible, since the SUMO protease SENP1 effectively removed SUMO1 from NS1. To identify the sumoylation sites, we exchanged all lysine residues of NS1 for alanine. Interestingly, mutations of lysines at positions 78 and 115, which matched with sumoylation consensus motifs, did not abolish sumoylation of NS1. Instead, we were able to demonstrate that two lysine residues at the C terminus serve as SUMO1 acceptor sites. The K221R mutation strongly impaired sumoylation, whereas only the double mutation K219R/K221R completely abolished sumoylation of NS1. Since both residues are in close proximity to each other, it appears that K221 is the preferential sumoylation site and that K219 may compensate when K221 is mutated. Similar observations have been made in other systems (29, 59). Since the C terminus of NS1 is an evolutionarily highly variable region, a comparison among different human and avian influenza A virus strains was performed. We showed that sumoylation is conserved among most influenza A virus NS1 proteins with acceptor sites at positions K219 and K221, while K217 and K219 serve as acceptor sites in only some strains. The PR8 NS1 protein has a K221E mutation which cannot be sumoylated. The A/HK/1073/99 virus NS1 protein has an M222V mutation which may restrict sumoylation at K221. K217 might therefore replace K221 as the acceptor site in these viruses. Interestingly, NS1 from the newly emerged S-OIV H1N1 strain, lacking 11 C-terminal amino acids, was not sumoylated. A likely explanation would be that the truncation disrupts C-terminal sumoylation sites within the protein. However, restoration of the C terminus did not restore sumoylation. Thus, most likely, other features of A/Sichuan/1/2009 (H1N1) NS1 may contribute to its lack of sumoylation. It also has to be pointed out that the NS1 truncation observed with S-OIV is not unique. A comparative analysis has shown that about 20% of 7,141 NS1 proteins from many different subtypes display C-terminal deletions of the same or even a larger size (12). Whether all of these NS1 proteins lack sumoylation or whether they are sumoylated at another site remains to be seen in future studies.
Of the four SUMO paralogs, SUMO1 has been studied best and has been implicated in several processes of protein localization, interaction, stability, and activity, resulting in the regulation of mitosis, gene transcription, DNA repair, and subnuclear targeting (42, 54, 60). The almost identical paralogs SUMO2 and SUMO3 have been associated functionally with cellular stress responses, yet the exact physiological differences among SUMO1 and SUMO2/3 are unknown (51). Whether the recently identified SUMO4 protein shares similar properties with the other SUMO members is largely unclear. Despite its presence on the transcriptional level in a small number of tissues, endogenous SUMO4 protein has not been detected yet, and its conjugation to target proteins in vivo is questionable (37). Many viral proteins seem to be modified preferentially by SUMO1 (4). Consistent with this observation, we provide evidence that NS1 is preferentially sumoylated with SUMO1. Although upon overexpression SUMO2 and SUMO3 were also attached to NS1 at the same lysine residues as those used by SUMO1, the extent of SUMO2/3 modification was much weaker than that of SUMO1 modification. In transfected and, more importantly, in influenza A virus-infected cells, knockdown of SENP1 strongly increased the sumoylated form of NS1. Among the six desumoylating enzymes, SENP1 preferentially cleaves SUMO1 from target proteins. Together, these data suggest that NS1 is modified predominantly with SUMO1 in vivo.
Sumoylation of NS1 has functional consequences. Our data indicate that it enhances the stability of the NS1 protein. It is reasonable to assume that the increased NS1 stability is responsible for an enhanced impediment of host gene expression which is also dependent on NS1 sumoylation, as shown here. So far, we have not been able to link these effects to any of the functional motifs at the C terminus of NS1, including the C-terminal nuclear localization signal of NS1 (NLS2), composed of amino acids K219, R220, R224, K229, R231, and R232 (36); the nucleolar localization signal (NoLS), found in some strains, that overlaps with NLS2 (36); the SH3-interacting motif formed by P212, P215, and K217, which appears to be involved in the activation of host cell signaling (25); and the PDZ-binding motif at positions 227 to 230, which was found to be a determinant of pathogenicity (28). Furthermore, mutation of the sumoylation sites affects neither nuclear and nucleolar localization, the SH3 interaction of NS1, nor the interaction with CPSF30 (see Fig. S4A to C in the supplemental material). Whether the PDZ domain is affected by sumoylation is unknown.
Finally, we show here that sumoylation of NS1 accelerates the replication rate and slightly increases virus titers in WSN-infected cells. Although these effects are not dramatic, they are particularly interesting in the light of recent observations indicating that NS1 is also modified by ISG15 conjugation after IFN is induced (58, 64). Remarkably, one of these studies showed that ISG15 conjugation occurs at C-terminal lysines, which are also the sumoylation sites, and that ISG15 conjugation of NS1 results in virus attenuation (58). Thus, it appears that virus growth is up- and downregulated by sumoylation and ISG15 conjugation of NS1, respectively, and that the balance between both modifications may play an important role in the outcome of infection.
Supplementary Material
Acknowledgments
We thank Otto Haller (Freiburg University, Germany) for reviewing the manuscript and for constructive suggestions.
This work was supported by grants from the European Union (FLUINNATE SP5B-CT-2006-044161), the Deutsche Forschungsgemeinschaft (SFB 593 TP B1), CAS (KSCX2-YW-R-161), the National Ministry of Science and Technology (20072714) and the National Natural Science Foundation of China (30950002, 30623003, 30721065, 30801011, 30870126, and 90713044), the Science and Technology Commission of Shanghai Municipality (08DZ2291703, 088014199, and 08431903004), the National Science and Technology Major Project (2008ZX10002-01, 2008ZX10004-002, and 2009ZX10004-105), the National 973 Key Project (2007CB512404), SPHRF (SPHRF2008001 and SPHRF2009001), the National 863 Project (2006AA02A247), and the Li Kha Shing Foundation.
Footnotes
Published ahead of print on 3 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Bailey, D., and P. O'Hare. 2002. Herpes simplex virus 1 ICP0 co-localizes with a SUMO-specific protease. J. Gen. Virol. 83:2951-2964. [DOI] [PubMed] [Google Scholar]
- 2.Basler, C. F., A. H. Reid, J. K. Dybing, T. A. Janczewski, T. G. Fanning, H. Zheng, M. Salvatore, M. L. Perdue, D. E. Swayne, A. Garcia-Sastre, P. Palese, and J. K. Taubenberger. 2001. Sequence of the 1918 pandemic influenza virus nonstructural gene (NS) segment and characterization of recombinant viruses bearing the 1918 NS genes. Proc. Natl. Acad. Sci. U. S. A. 98:2746-2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, M., A. Garcia-Sastre, E. Carnero, H. Pehamberger, K. Wolff, P. Palese, and T. Muster. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boggio, R., and S. Chiocca. 2006. Viruses and sumoylation: recent highlights. Curr. Opin. Microbiol. 9:430-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boggio, R., R. Colombo, R. T. Hay, G. F. Draetta, and S. Chiocca. 2004. A mechanism for inhibiting the SUMO pathway. Mol. Cell 16:549-561. [DOI] [PubMed] [Google Scholar]
- 6.Bornholdt, Z. A., and B. V. Prasad. 2006. X-ray structure of influenza virus NS1 effector domain. Nat. Struct. Mol. Biol. 13:559-560. [DOI] [PubMed] [Google Scholar]
- 7.CDC. 2009. Swine influenza (H1N1) infection in two children. MMWR Morb. Mortal. Wkly. Rep. 58:400-402. [PubMed] [Google Scholar]
- 8.Chen, Z., Y. Li, and R. M. Krug. 1999. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 18:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng, J., X. Kang, S. Zhang, and E. T. Yeh. 2007. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell 131:584-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claas, E. C. J. 1998. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 351:472-477. [DOI] [PubMed] [Google Scholar]
- 11.De Jong, J. C., E. C. Claas, A. D. Osterhaus, R. G. Webster, and W. L. Lim. 1997. A pandemic warning? Nature 389:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundon, W. G., and I. Capua. 2009. A closer look at the NS1 of influenza virus. Viruses 1:1057-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier, R. A., V. Munster, A. Wallensten, T. M. Bestebroer, S. Herfst, D. Smith, G. F. Rimmelzwaan, B. Olsen, and A. D. Osterhaus. 2005. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J. Virol. 79:2814-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraser, C., C. A. Donnelly, S. Cauchemez, W. P. Hanage, M. D. Van Kerkhove, T. D. Hollingsworth, J. Griffin, R. F. Baggaley, H. E. Jenkins, E. J. Lyons, T. Jombart, W. R. Hinsley, N. C. Grassly, F. Balloux, A. C. Ghani, N. M. Ferguson, A. Rambaut, O. G. Pybus, H. Lopez-Gatell, C. M. Alpuche-Aranda, I. B. Chapela, E. P. Zavala, D. M. Guevara, F. Checchi, E. Garcia, S. Hugonnet, and C. Roth. 2009. Pandemic potential of a strain of influenza A (H1N1): early findings. Science 324:1557-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 17.Geiss-Friedlander, R., and F. Melchior. 2007. Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell. Biol. 8:947-956. [DOI] [PubMed] [Google Scholar]
- 18.Gill, G. 2005. Something about SUMO inhibits transcription. Curr. Opin. Genet. Dev. 15:536-541. [DOI] [PubMed] [Google Scholar]
- 19.Gill, G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046-2059. [DOI] [PubMed] [Google Scholar]
- 20.Girdwood, D. W., M. H. Tatham, and R. T. Hay. 2004. SUMO and transcriptional regulation. Semin. Cell Dev. Biol. 15:201-210. [DOI] [PubMed] [Google Scholar]
- 21.Hale, B. G., A. Knebel, C. H. Botting, C. S. Galloway, B. L. Precious, D. Jackson, R. M. Elliott, and R. E. Randall. 2009. CDK/ERK-mediated phosphorylation of the human influenza A virus NS1 protein at threonine-215. Virology 383:6-11. [DOI] [PubMed] [Google Scholar]
- 22.Hale, B. G., R. E. Randall, J. Ortin, and D. Jackson. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359-2376. [DOI] [PubMed] [Google Scholar]
- 23.Hale, B. G., J. Steel, B. Manicassamy, R. A. Medina, J. Ye, D. Hickman, A. C. Lowen, D. R. Perez, and A. Garcia-Sastre. 2010. Mutations in the NS1 C-terminal tail do not enhance replication or virulence of the 2009 pandemic H1N1 influenza A virus. J. Gen. Virol. 91:1737-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hay, R. T. 2005. SUMO: a history of modification. Mol. Cell 18:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Heikkinen, L. S., A. Kazlauskas, K. Melen, R. Wagner, T. Ziegler, I. Julkunen, and K. Saksela. 2008. Avian and 1918 Spanish influenza A virus NS1 proteins bind to Crk/CrkL Src homology 3 domains to activate host cell signaling. J. Biol. Chem. 283:5719-5727. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann, E., J. Stech, Y. Guan, R. G. Webster, and D. R. Perez. 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146:2275-2289. [DOI] [PubMed] [Google Scholar]
- 28.Jackson, D., M. J. Hossain, D. Hickman, D. R. Perez, and R. A. Lamb. 2008. A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc. Natl. Acad. Sci. U. S. A. 105:4381-4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, M. J., I. V. Chia, and F. Costantini. 2008. SUMOylation target sites at the C terminus protect Axin from ubiquitination and confer protein stability. FASEB J. 22:3785-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klenk, C., J. Humrich, U. Quitterer, and M. J. Lohse. 2006. SUMO-1 controls the protein stability and the biological function of phosducin. J. Biol. Chem. 281:8357-8364. [DOI] [PubMed] [Google Scholar]
- 31.Kochs, G., A. Garcia-Sastre, and L. Martinez-Sobrido. 2007. Multiple anti-interferon actions of the influenza A virus NS1 protein. J. Virol. 81:7011-7021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochs, G., I. Koerner, L. Thiel, S. Kothlow, B. Kaspers, N. Ruggli, A. Summerfield, J. Pavlovic, J. Stech, and P. Staeheli. 2007. Properties of H7N7 influenza A virus strain SC35M lacking interferon antagonist NS1 in mice and chickens. J. Gen. Virol. 88:1403-1409. [DOI] [PubMed] [Google Scholar]
- 33.Konig, R., S. Stertz, Y. Zhou, A. Inoue, H. H. Hoffmann, S. Bhattacharyya, J. G. Alamares, D. M. Tscherne, M. B. Ortigoza, Y. Liang, Q. Gao, S. E. Andrews, S. Bandyopadhyay, P. De Jesus, B. P. Tu, L. Pache, C. Shih, A. Orth, G. Bonamy, L. Miraglia, T. Ideker, A. Garcia-Sastre, J. A. Young, P. Palese, M. L. Shaw, and S. K. Chanda. 2010. Human host factors required for influenza virus replication. Nature 463:813-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee, H. R., D. J. Kim, J. M. Lee, C. Y. Choi, B. Y. Ahn, G. S. Hayward, and J. H. Ahn. 2004. Ability of the human cytomegalovirus IE1 protein to modulate sumoylation of PML correlates with its functional activities in transcriptional regulation and infectivity in cultured fibroblast cells. J. Virol. 78:6527-6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li, S., J. Y. Min, R. M. Krug, and G. C. Sen. 2006. Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349:13-21. [DOI] [PubMed] [Google Scholar]
- 36.Melen, K., L. Kinnunen, R. Fagerlund, N. Ikonen, K. Y. Twu, R. M. Krug, and I. Julkunen. 2007. Nuclear and nucleolar targeting of influenza A virus NS1 protein: striking differences between different virus subtypes. J. Virol. 81:5995-6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meulmeester, E., and F. Melchior. 2008. Cell biology: SUMO. Nature 452:709-711. [DOI] [PubMed] [Google Scholar]
- 38.Mibayashi, M., L. Martinez-Sobrido, Y. M. Loo, W. B. Cardenas, M. Gale, Jr., and A. Garcia-Sastre. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514-524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo(A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. U. S. A. 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Min, J. Y., S. Li, G. C. Sen, and R. M. Krug. 2007. A site on the influenza A virus NS1 protein mediates both inhibition of PKR activation and temporal regulation of viral RNA synthesis. Virology 363:236-243. [DOI] [PubMed] [Google Scholar]
- 41.Mukhopadhyay, D., and M. Dasso. 2007. Modification in reverse: the SUMO proteases. Trends Biochem. Sci. 32:286-295. [DOI] [PubMed] [Google Scholar]
- 42.Muller, S., A. Ledl, and D. Schmidt. 2004. SUMO: a regulator of gene expression and genome integrity. Oncogene 23:1998-2008. [DOI] [PubMed] [Google Scholar]
- 43.Nemeroff, M. E., S. M. Barabino, Y. Li, W. Keller, and R. M. Krug. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol. Cell 1:991-1000. [DOI] [PubMed] [Google Scholar]
- 44.Newby, C. M., L. Sabin, and A. Pekosz. 2007. The RNA binding domain of influenza A virus NS1 protein affects secretion of tumor necrosis factor alpha, interleukin-6, and interferon in primary murine tracheal epithelial cells. J. Virol. 81:9469-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noah, D. L., K. Y. Twu, and R. M. Krug. 2003. Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307:386-395. [DOI] [PubMed] [Google Scholar]
- 46.Pal, S., J. M. Rosas, and G. Rosas-Acosta. 2010. Identification of the non-structural influenza A viral protein NS1A as a bona fide target of the Small Ubiquitin-like MOdifier by the use of dicistronic expression constructs. J. Virol. Methods 163:498-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Privalsky, M. L., and E. E. Penhoet. 1981. The structure and synthesis of influenza virus phosphoproteins. J. Biol. Chem. 256:5368-5376. [PubMed] [Google Scholar]
- 48.Qiu, Y., M. Nemeroff, and R. M. Krug. 1995. The influenza virus NS1 protein binds to a specific region in human U6 snRNA and inhibits U6-U2 and U6-U4 snRNA interactions during splicing. RNA 1:304-316. [PMC free article] [PubMed] [Google Scholar]
- 49.Reverter, D., and C. D. Lima. 2004. A basis for SUMO protease specificity provided by analysis of human Senp2 and a Senp2-SUMO complex. Structure 12:1519-1531. [DOI] [PubMed] [Google Scholar]
- 50.Rizzo, M. A., G. H. Springer, B. Granada, and D. W. Piston. 2004. An improved cyan fluorescent protein variant useful for FRET. Nat. Biotechnol. 22:445-449. [DOI] [PubMed] [Google Scholar]
- 51.Saitoh, H., and J. Hinchey. 2000. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 275:6252-6258. [DOI] [PubMed] [Google Scholar]
- 52.Saitoh, H., M. D. Pizzi, and J. Wang. 2002. Perturbation of SUMOlation enzyme Ubc9 by distinct domain within nucleoporin RanBP2/Nup358. J. Biol. Chem. 277:4755-4763. [DOI] [PubMed] [Google Scholar]
- 53.Satterly, N., P. L. Tsai, J. van Deursen, D. R. Nussenzveig, Y. Wang, P. A. Faria, A. Levay, D. E. Levy, and B. M. Fontoura. 2007. Influenza virus targets the mRNA export machinery and the nuclear pore complex. Proc. Natl. Acad. Sci. U. S. A. 104:1853-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seeler, J. S., and A. Dejean. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell. Biol. 4:690-699. [DOI] [PubMed] [Google Scholar]
- 55.Shapira, S. D., I. Gat-Viks, B. O. Shum, A. Dricot, M. M. de Grace, L. Wu, P. B. Gupta, T. Hao, S. J. Silver, D. E. Root, D. E. Hill, A. Regev, and N. Hacohen. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stech, J., H. Garn, M. Wegmann, R. Wagner, and H. D. Klenk. 2005. A new approach to an influenza live vaccine: modification of the cleavage site of hemagglutinin. Nat. Med. 11:683-689. [DOI] [PubMed] [Google Scholar]
- 57.Subbarao, K. 1998. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279:393-396. [DOI] [PubMed] [Google Scholar]
- 58.Tang, Y., G. Zhong, L. Zhu, X. Liu, Y. Shan, H. Feng, Z. Bu, H. Chen, and C. Wang. 2010. Herc5 attenuates influenza A virus by catalyzing ISGylation of viral NS1 protein. J. Immunol. 184:5777-5790. [DOI] [PubMed] [Google Scholar]
- 59.Tseng, C. H., T. S. Cheng, C. Y. Shu, K. S. Jeng, and M. M. Lai. 2010. Modification of small hepatitis delta virus antigen by SUMO protein. J. Virol. 84:918-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verger, A., J. Perdomo, and M. Crossley. 2003. Modification with SUMO. A role in transcriptional regulation. EMBO Rep. 4:137-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Webster, R. G., W. J. Bean, O. T. Gorman, T. M. Chambers, and Y. Kawaoka. 1992. Evolution and ecology of influenza A viruses. Microbiol. Rev. 56:152-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.WHO. 2009. New influenza (H1N1) virus infections: global surveillance summary, May 2009. Wkly. Epidemiol. Rec. 84:173-179. [PubMed] [Google Scholar]
- 63.Wilson, V. G., and D. Rangasamy. 2001. Viral interaction with the host cell sumoylation system. Virus Res. 81:17-27. [DOI] [PubMed] [Google Scholar]
- 64.Zhao, C., T. Y. Hsiang, R. L. Kuo, and R. M. Krug. 2010. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 107:2253-2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.