Abstract
The Epstein-Barr Virus (EBV) productive cycle is initiated by the expression of the viral trans-activator EB1 (also called Zebra, Zta, or BZLF1), which belongs to the basic leucine zipper transcription factor family. We have previously identified the cellular NACos (nuclear and adherent junction complex components) protein ubinuclein (Ubn-1) as a partner for EB1, but the function of this complex has never been studied. Here, we have evaluated the consequences of this interaction on the EBV productive cycle and find that Ubn-1 overexpression represses the EBV productive cycle whereas Ubn-1 downregulation by short hairpin RNA (shRNA) increases virus production. By a chromatin immunoprecipitation (ChIP) assay, we show that Ubn-1 blocks EB1-DNA interaction. We also show that in epithelial cells, relocalization and sequestration of Ubn-1 to the tight junctions of nondividing cells allow increased activation of the productive cycle. We propose a model in which Ubn-1 is a modulator of the EBV productive cycle: in proliferating epithelial cells, Ubn-1 is nuclear and inhibits activation of the productive cycle, whereas in differentiated cells, Ubn-1 is sequestrated to tight junctions, thereby allowing EB1 to fully function in the nucleus.
Epstein-Barr virus (EBV) is a human gammaherpesvirus that infects more than 95% of the human adult population. It is the causative agent of infectious mononucleosis (30) and is associated with several human cancers, including Burkitt's lymphoma (BL), Hodgkin's disease (HD), nasopharyngeal carcinoma (NPC), gastric carcinoma, and immunoblastic lymphoma, in immunosuppressed individuals (58, 60, 61). After primary infection, EBV persists lifelong in a latent state in a subpopulation of resting memory B cells (4). A generally accepted model of viral persistence suggests that the virus uses its transforming abilities to activate newly infected resting B cells. Latency-associated viral proteins then provide surrogate signals to allow the newly activated cells to differentiate through the germinal center into the memory compartment (59). In memory B cells, expression of viral genes is shut down, which allows them to evade cytotoxic-T-cell immunosurveillance and achieve long-term persistence (32). Although the hallmark of latency is the absence of a complete viral productive cycle, EBV productively reactivates from latency in mucosal epithelia and infectious virions are shed continuously into the saliva of healthy carriers (28). This allows the efficient spread of the virus from host to host. Interestingly, there is strong evidence that both the latent and the productive forms of EBV infection are essential for the emergence of EBV-associated malignancies (33, 50).
In vivo, EBV reactivation has been linked to terminal differentiation of latently infected B cells into plasma cells (16, 39, 46, 47). In addition, in the case of oral hairy leukoplakia, a disease associated with the presence of EBV in the tongue's epithelial cells, the viral productive cycle takes place only in differentiated cells (8, 67). Up to now, the signals and molecular mechanisms that trigger the switch from latency to the productive cycle in vivo have remained unknown. In contrast, activation of the viral productive cycle in vitro has been well documented for cell lines established from EBV-positive BL biopsy specimens. The productive cycle can be induced by treating these infected cells with various agents, such as the tumor promoter 12-O-tetradecanoyl-phorbol-13-acetate (TPA), associated with sodium butyrate (BA) (70), anti-Ig (57), and tumor growth factor β (TGF-β) (20). In these models, it is now well established that these inducing agents activate the transcription of two immediate-early (IE) viral genes, the BZLF1 and BRLF1 genes, whose products are viral transcription factors called EB1 (or ZEBRA or Zta) and R (or Rta), respectively (13, 14, 15, 23, 29). EB1 is a bZip protein related to c-Jun and c-Fos that binds to the consensus AP1 motif as well as atypical AP1-like motifs known as Z-responsive elements (ZREs) (12, 21, 49, 62). Interestingly, EB1 is the only transcription factor known to preferentially activate the methylated form of its ZRE target (11, 18). R activates some early promoters through a direct binding mechanism but also activates some promoters indirectly, through effects on cellular transcription factors (1, 25, 27). EB1 and R activate the transcription of all the early genes, some of which are essential for viral DNA replication, a prerequisite for expression of most late genes, DNA encapsidation, and production of infectious virions (reviewed in reference 35).
We have previously characterized a ubiquitously expressed cellular protein, which we called ubinuclein (Ubn-1), through its interaction with EB1 (2). Ubn-1 can also interact with other transcription factors of the basic leucine zipper family, such as C/EBP and c-Jun. The N terminus of Ubn-1 is essential for nuclear localization of the protein, whereas its central domain is responsible for interaction with EB1's basic domain. In vitro, the Ubn-1-EB1 interaction inhibits EB1's binding to DNA (2). Ubn-1 transcripts are present in a wide variety of human adult, fetal, and tumor tissues (2, 7), and the protein is detected in the nuclei of cells throughout the human epidermis and also in cultured keratinocytes (2). More recently, we have demonstrated that Ubn-1 is a member of the NACos (nuclear adherent-junction complex components) protein family. This protein is localized mainly in the nuclei of isolated MDCK (Madin-Darby canine kidney) epithelial cells and undifferentiated keratinocytes but is relocalized to the tight junctions in confluent epithelial cells, polarized cells, or differentiated keratinocytes. In vivo, we observed that Ubn-1 is present in the tight junctions of both stratified and simple epithelia (3). As a NACos protein, Ubn-1 may participate in the regulation of gene expression (6), and interestingly, Ubn-1 has been found associated in a complex with HIRA (histone cell cycle regulation-defective homologue A) and H3.3 (histone H3 variant 3) (56). HIRA is required for the DNA replication-independent assembly of chromatin. More recently, Ubn-1 has also been identified as an ortholog of Saccharomyces cerevisiae Hpc2p and characterized as a novel regulator of fibroblast cell senescence (5, 7). Hence, Ubn-1 appears to act at the level of chromatin remodeling, and in this way, Ubn-1 may control gene expression.
Here, we have studied the consequences of the interaction between Ubn-1 and EB1 during the EBV productive cycle. We find that Ubn-1 can control the production of infectious virus. In HEK293 cells infected with EBV, Ubn-1 overexpression mediates a clear decrease in the level of virions produced, and inversely, repression of Ubn-1's expression ameliorates viral production. By using chromatin immunoprecipitation (ChIP) assays, we show that Ubn-1 blocks EB1-ZRE interaction in vivo. Ubn-1 overexpression also has a negative effect on AP1-induced promoters. In epithelial cells infected with EBV, the productive cycle appears to be more efficient when cells reach confluence, in accordance with relocalization of Ubn-1 to the tight junctions. We thus propose a model in which Ubn-1 contributes to the control of the EBV productive cycle in vivo depending on epithelial cell differentiation.
MATERIALS AND METHODS
Plasmids.
The EB1, Ubn-1, and R expression vectors, pRc/CMV-EB1, pCEP4t-Ubi, and pRc/CMV-R, respectively, have been described previously (3, 26, 27). Appropriate control vectors, pRc/CMV and pCEP4t (Invitrogen), were used in all experiments. The pRK5-BALF4 expression vector encodes the EBV gp110 protein and has previously been shown to enhance the infection efficiency of EBV virions derived from the B95-8 strain of EBV (a gift from W. Hammerschmidt) (45). The pIL6-Luc, pIL10-Luc, pLMP1-Luc, and pTK-Luc reporter plasmids have been described elsewhere (40, 63, 64). A TATA box-containing oligonucleotide was cloned into the pGL2-basic vector from Promega to create the pTATA-Luc reporter plasmid. In this vector, a concatemerized oligonucleotide bearing the TPA-responsive element TGAGTCA was cloned to generate the pTRE.TATA-Luc reporter plasmid (a gift from M. Castellazzi). In the pTP1.Gal4-Luc reporter plasmid, the luciferase gene is under the control of the LMP2 promoter, in which the EBNA2-responsive elements have been exchanged for 10 Gal4-binding sites. The Ubn-1 short hairpin RNA (shRNA) sequence (sh.Ubn) (CCGGGCCAGCTCAATCTCCAAACATCTCGAGATGTTTGGAGATTGAGCTGGCTTTTT) cloned in the pLKO1 plasmid was a gift from P. Adams (7). The shRNA-encoding lentiviral pseudoparticles were produced by the vectorology platform of IFR 128.
Cell lines.
HEK293 cells infected with the recombinant EBV virus (HEK293EBV), a gift from W. Hammerschmidt, have been described previously (17). The recombinant virus also encodes enhanced green fluorescent protein (eGFP) and the hygromycin B resistance gene. HEK293EBV cells were maintained in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS), penicillin-streptomycin, and hygromycin B (100 μg/ml; Invitrogen). HeLa cells were maintained in DMEM supplemented with 10% FBS and penicillin-streptomycin. AGSEBV cells (a gift from S. Kenney) (22) were maintained in F-12 medium containing 10% FBS, penicillin-streptomycin, and hygromycin B (100 μg/ml). Raji cells were maintained in RPMI 1640 medium supplemented with 10% FBS and penicillin-streptomycin. The MDCK cell line was purchased from Clontech (Palo Alto, CA) and grown according to the manufacturer's instructions in DMEM containing 10% FBS. These cells were infected with the recombinant EBVGFP virus and selected for hygromycin B resistance.
Viral titration.
Supernatants from HEK293EBV cells were harvested at 72 h posttransfection and filtered through a 0.45-μm-pore-size filter. Raji cells (2 × 105) were incubated in 0.5 ml of virus solution for 3 h at 37°C in a 24-well plate. Cells were then washed, resuspended in 1 ml of RPMI medium, and incubated for an additional 48 h at 37°C. GFP-expressing Raji cells were quantified by fluorescence-activated cell sorting (FACS) analysis.
Viral DNA analysis.
HEK293EBV cells were transfected with an EB1 expression vector to activate the EBV productive cycle and with increasing amounts of the Ubn-1 expression vector. At 72 h posttransfection, DNA was prepared by the Hirt technique, digested with DpnI and BamHI, subjected to electrophoresis through a 0.7% agarose gel, transferred to a nylon N+ membrane (GE Healthcare), and probed with a randomly primed 32P-labeled EBV BRRF1 DNA probe. The replication efficiency of the EBV plasmid was quantified by scanning the Southern blot autoradiogram with a phosphorimager (Fuji FLA-5100).
DNA transfection.
HeLa or HEK293EBV cells were seeded at 1 × 106 cells per 100-mm-diameter petri dish 10 h prior to transfection. Transfections of HEK293EBV cells were performed by the calcium phosphate precipitation method as described previously (31). Transfections of HeLa cells for reporter assays were performed by use of PEI (Ozyme) according to the manufacturer's instructions. All transfections were performed in triplicate. Renilla luciferase assays were performed with a Renilla luciferase assay system (Promega).
Coimmunoprecipitation assays.
HEK293EBV cells transfected with expression vectors for EB1 and Ubn-1 were harvested from 100-mm dishes at 48 h posttransfection. Nuclear extracts were prepared from these cells by the method of Dignam et al. (19). Nuclear cell extracts were then incubated overnight with the Z125 anti-EB1 monoclonal antibody (MAb) (41) in Dignam buffer III supplemented with 140 mM NaCl and protease inhibitors (Roche Molecular Biochemicals) at 4°C, followed by a 15 min of incubation with 50 μl of PureProteome protein A magnetic beads (Millipore). Beads were then washed twice with Dignam buffer III supplemented with 140 mM NaCl and then 3 times with the same buffer supplemented with 1% Triton X-100. Immunopurified proteins were analyzed by Western blotting.
Immunoblotting.
Immunoblotting was performed as described previously (24). Briefly, cells were lysed in radioimmunoprecipitation assay (RIPA) buffer supplemented with protease inhibitors; equivalent amounts of proteins were then separated in sodium dodecyl sulfate-10% polyacrylamide electrophoresis gels. After being transferred and blocked, membranes were incubated 2 h at 20°C with the appropriate primary antibodies diluted in 1× Tris-buffered saline (TBS), 0.1% Tween 20 (TBS-T). The primary antibody dilutions were as follows: 1:250 anti-EB1 (Z125 monoclonal antibody) (41), 1:2,000 anti-Ubn-1 (ZAP1 monoclonal antibody) (3), 1:1,000 anti-gp350 (OT6) (69), and 1:250 anti-EB2 (31). The appropriate anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (GE Healthcare) were used at a dilution of 1:5,000 in 1× TBS-T for 1 h at room temperature. Bound antibodies were visualized by use of enhanced chemiluminescence (ECL) reagent (Pierce) according to the manufacturer's instructions.
Immunofluorescence studies.
For indirect immunofluorescence, cells were fixed with methanol for 10 min and further permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) for 5 min. Blocking was made with 0.2% gelatin in PBS for 1 h at room temperature, followed by incubation with the primary antibody at room temperature for 20 min in PBS-0.2% gelatin. Slides were washed three times with PBS, followed by incubation with the secondary antibody at room temperature for 20 min. Slides were washed three times with PBS, followed by DAPI (4′,6-diamidino-2-phenylindole) staining in PBS. After three washes with PBS, slides were mounted and studied using a fluorescence microscope (Axioplan 2; Zeiss). Images were processed with Photoshop 5.0 (Adobe Systems, San Jose, CA).
RT-PCR.
Total RNA was extracted using a Macherey-Nagel Nucleospin RNA/protein kit. Total RNA (2.5 μg) was reversed transcribed with oligo(dT)16 and 1 μl of Superscript II enzyme (Invitrogen) in a 20-μl reaction mixture at 42°C for 1 h. PCRs were performed using the GoTaq Hot Start polymerase (Promega) with a set of specific primer pairs. For quantification of PCR products, [α32P]dCTP (0.1μCi) was added to each PCR mixture, and the PCR products were fractionated on 6% polyacrylamide gels. Quantifications were made by means of a phosphorimager (Fuji FLA-5100). To monitor our reverse transcription-PCR (RT-PCR) experiments, we evaluated the endogenous expression of β-actin mRNA by RT-PCR. Amplification of a 690-bp DNA fragment corresponding to β-actin mRNA showed that no DNA contamination was present in our preparations. The primer sequences used are listed in Table 1 .
TABLE 1.
Primers used in this study
| Primer target (orientation) | Sequence |
|---|---|
| β-Actin (forward) | GCTGCGTGTGGCTCCCGAGGAG |
| β-Actin (reverse) | ATCTTCATTGTGCTGGGTGCCAG |
| Ubn-1 (forward) | CCTCTTTGATGGCTTCACCCTAC |
| Ubn-1 (reverse) | TGATTCCACCCAGACCCAAGTG |
| LMP1 (forward) | TGAACACCACCACGATGACT |
| LMP1 (reverse) | GTGCGCCTAGGTTTTGAGAG |
| BALF4 (forward) | GGAGTCGTAGGCAAATTGGA |
| BALF4 (reverse) | TCAAGAACCTGACGGAGCTT |
| BALF5 (forward) | AAGAGGCACTGGAGGATGTTGG |
| BALF5 (reverse) | AGGGCTACCCTGTGGGCTTTTTG |
| BMRF1 (forward) | CCAGACATACGGTCAGTCCA |
| BMRF1 (reverse) | TGCTTCACTTTCTTGGG |
| BDLF1 (forward) | CAGATTTGAAAGTGGTAG |
| BDLF1 (reverse) | TTATCTTAACCAGCAAGTG |
| BFRF3 (forward) | GGGAGGCTCAAAGAAGTTA |
| BFRF3 (reverse) | ATGAAGAAACAGAGGGGG |
| BdRF1 (forward) | CACTATCAGGTAACGCA |
| BdRF1 (reverse) | TCAAGCCAGCGTTTATTT |
| F19 promoter pZ (forward) | TGGGCTGTCTATTTTTGACACCAG |
| B20 promoter pZ (reverse) | CTTCAGCAAAGATAGCAAAGGTGG |
| F11 promoter pM (forward) | AAACTTAGTTCAGGTGTGCCATGC |
| B13 promoter pM (reverse) | GGGATGTAGTGCTGTCTTGACTGG |
ChIP.
The method described by Orlando et al. was used for chromatin immunoprecipitation (ChIP) (48). Briefly, 48 h after transfection, cells were fixed by the addition of formaldehyde to give a final concentration of 1% for 15 min. Cells were then washed twice with PBS, and crude nuclei were prepared and resuspended at a concentration of 1 × 107 nuclei/ml in lysis buffer (1% SDS, 10 mM EDTA, 50 mM Tris-Cl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride [PMSF]). The resulting solution was sonicated such that the average DNA fragment length was reduced to less than 1 kb. This solution was diluted 10-fold with immunoprecipitation buffer (1.2 mM EDTA, 0.01% SDS, 1.1% Triton, 16.7 mM Tris-Cl [pH 8.0], 167 mM NaCl, 1 mM PMSF) and precleared for 2 h with protein A-Sepharose beads preabsorbed with both bovine serum albumin (BSA) and sonicated single-stranded DNA. Immunoprecipitation was performed by incubation for 12 h at 4°C with monoclonal antibodies directed either to EB1 (MAb Z125 [44]), to Ubn-1 (MAb ZAP1 [3]), or to a control MAb. Immune complexes were collected by incubation with protein A-Sepharose beads, and washed extensively with low-salt buffer (0.1% SDS, 1% Triton, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 150 mM NaCl), high-salt buffer (0.1% SDS, 1% Triton, 2 mM EDTA, 20 mM Tris-HCl [pH 8.0], 500 mM NaCl), LiCl buffer (1% deoxycholate; 1% NP-40, 1 mM EDTA, 10 mM Tris-HCl [pH 8.0], 250 mM LiCl), and Tris-EDTA (TE) prior to elution from the beads with 300 μl of elution buffer (1% SDS, 0.1 M NaHCO3). Protein-DNA cross-links were reversed by heating to 65°C for 4 h. DNA fragments were purified by proteinase K digestion followed by a phenol-chloroform extraction and ethanol precipitation in the presence of a glycogen carrier. The immunoselected DNA fragments were dissolved in 50 μl of water. The presence of DNA was then evaluated using semiquantitative PCR with sets of primer pairs that hybridize along the different promoter regions analyzed (Table 1) in the presence of [α32P]dCTP (0.1 μCi). PCRs were performed using GoTaq Hot Start polymerase (Promega) with 1 μl of the solution of immunoselected DNA fragments, as well as with various dilutions of the DNA purified from the input chromatin, in order to generate a standard curve in a linear range. The PCR-amplified fragments (24 cycles) were then analyzed using 6% polyacrylamide gels and autoradiographed. Each PCR-amplified band was subsequently quantified with a phosphorimager (Fuji FLA-5100). The amount of DNA amplified from the control immunoprecipitated chromatin was systematically subtracted from the amount of DNA amplified from the specific immunoprecipitated chromatin and the results expressed as a percentage of immunoprecipitated DNA relative to the quantity of DNA present before immunoprecipitation (input).
RESULTS
Ubinuclein interferes with EB1-mediated induction of the EBV productive cycle in vivo.
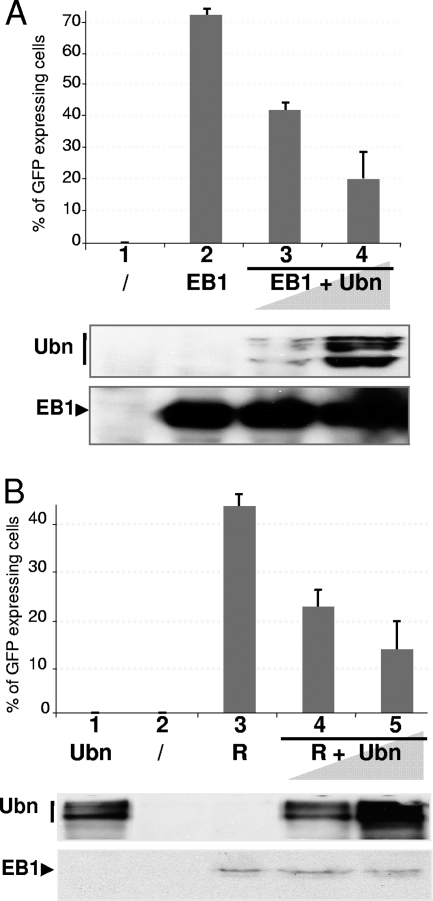
We have previously shown that Ubn-1 interacts with EB1's basic DNA-binding domain and abrogates EB1's binding to DNA in vitro (2). To determine whether Ubn-1 also impairs the function of EB1 in vivo, we analyzed the effect of Ubn-1 overexpression on the EBV productive cycle. For this, we used the HEK293EBV cell line (17). In this cell line, EBV is present in a latent state and ectopic expression of EB1 is sufficient to induce virus production. We evaluated the production of infectious EBV virions in this system, following transient expression of EB1, expressed either alone or together with increasing amounts of Ubn-1. The production of virus was evaluated by infection of Raji cells with the filtered HEK293EBV supernatant. Due to constitutive expression of GFP from the EBV recombinant genome, infected Raji cells appear bright green under UV light and can be quantified by FACS analysis. Expression of EB1 alone induced the production of a large number of EBV virions (Fig. 1A, lane 2), but when increasing amounts of Ubn-1 were coexpressed, the relative efficiency of virus production was reduced in a dose-dependent manner (Fig. 1A, lanes 3 and 4). This effect is not due to a decrease in EB1 levels, as can be seen by Western blot analysis (Fig. 1A, bottom panel). To confirm these results under conditions where EB1 would be present under more-physiological conditions, we took advantage of the fact that the other viral transcription factor, R, has been reported to induce expression of EB1 and consequently the EBV productive cycle in epithelial cells (68). We thus induced the EBV productive cycle in HEK293EBV cells by transfecting an R expression vector and evaluated the production of infectious EBV virions in the presence of increasing amounts of Ubn-1. As expected, untransfected HEK293EBV cells produced no virus (Fig. 1B, lane 2), nor did HEK293EBV cells transfected with an Ubn-1 expression plasmid (Fig. 1B, lane 1), demonstrating that Ubn-1 alone is not able to induce the EBV productive cycle. Expression of R alone induced the production of a large number of EBV virions (Fig. 1B, lane 3), but when increasing amounts of Ubn-1 were coexpressed, the efficiency of virus production was reduced in a dose-dependent manner (Fig. 1B, lanes 4 and 5). Again, Ubn-1 overexpression did not affect the level of EB1 expressed in HEK293EBV cells induced by R (Fig. 1B, bottom panel). These results strongly suggest that Ubn-1 can act as a negative regulator of the EBV productive cycle.
FIG. 1.
Ubinuclein overexpression inhibits virion production in HEK293EBV cells. (A) Raji cells (2.5 × 105) were incubated with 1 ml of filtered medium from HEK293EBV cells that had been either not transfected (lane 1), transfected with 0.5 μg of an EB1 expression plasmid (lane 2), or cotransfected with expression plasmids for both EB1 (0.5 μg) and increasing amounts of Ubn-1 (2 and 4 μg) (lanes 3 and 4). The amount of GFP-expressing Raji cells was determined by FACS analysis. The experiment was done 5 times, and the error bars represent standard deviations. (Lower panel) Immunoblot analysis for one representative experiment. Ubn-1 protein was detected using MAb ZAP1; EB1 protein was detected on the same membrane using MAb Z125. (B) Raji cells (2.5 × 105) were incubated with 1 ml of filtered medium from HEK293EBV cells that had been either not transfected (lane 2), transfected with 4 μg of expression plasmid for Ubn-1 (lane 1) or 1 μg of an R expression plasmid (lane 3), or cotransfected with expression plasmids for both R (1 μg) and increasing amounts of Ubn-1 (2 and 4 μg) (lanes 3 and 4). The number of GFP-expressing Raji cells was determined by FACS analysis. (Lower panel) Immunoblot analysis for one representative experiment.
Ubinuclein modulates expression of both early and late EBV genes.
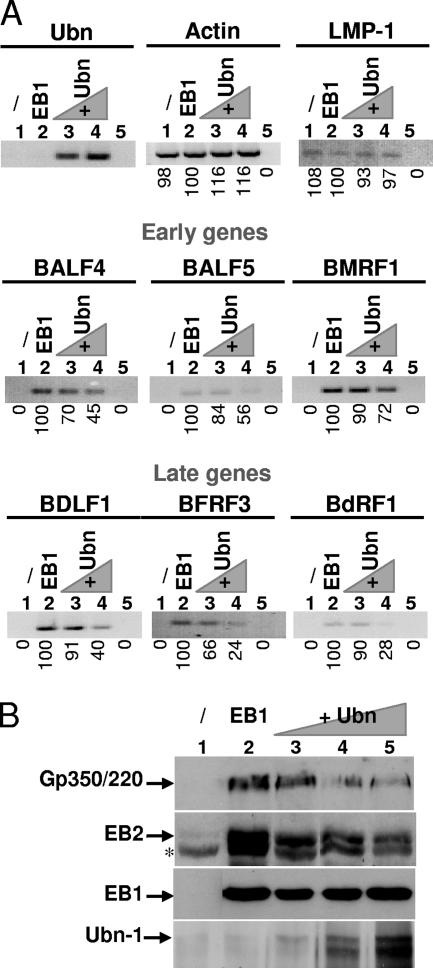
As a viral transcription factor, EB1 is required for activation of the EBV early genes (14), but EB1 is also directly implicated in the trans-activation of the lytic origin of replication (oriLyt) of the virus (52). In order to first investigate whether Ubn-1 directly affects viral gene expression, we analyzed the expression level of a selected batch of early and late EBV genes in the presence of overexpressed Ubn-1. After transfection of HEK293EBV cells with an EB1 expression vector and increasing amounts of an Ubn-1 expression vector, total cellular mRNA was purified and reverse transcribed, and the reverse transcribed products were amplified by semiquantitative PCR, using primer pairs specific for the different genes studied (Table 1). The products of these RT-PCRs were analyzed using agarose gels and quantified (Fig. 2A). β-Actin primers were used as a control to ensure that the same amount of RNA was used in each RT-PCR and that no DNA contamination was present in our mRNA preparations. The absence of DNA contamination was also confirmed by the absence of amplification when no reverse transcriptase was used (lane 5 of each panel). Overexpression of Ubn-1 mRNA was also monitored using RT-PCR (top left panel, lanes 3 and 4). The expression of the LMP1 gene, a gene normally expressed during latency, was not significantly altered when the productive cycle was induced by ectopic expression of EB1 (lane 2) or when Ubn-1 was overexpressed (lanes 3 and 4), demonstrating that Ubn-1 has no effect on the expression of this latent gene. As expected, expression of both the early genes (the BALF4, BALF5, and BMRF1 genes) and the late genes (the BDLF1, BFRF3, and BdRF1 genes) was efficiently induced when EB1 was expressed (compare lane 2 to lane 1 in each panel). Interestingly, Ubn-1 overexpression clearly downregulated the expression of the three early and three late EBV genes tested (lanes 3 and 4). At the higher levels of Ubn-1, expression of the early genes decreased between 28 and 55% and expression of the late genes between 60 and 76%. The decrease in early- and late-gene expression could also be visualized at the protein level, as can be seen by the immunoblot analysis for the early BMLF1 protein, EB2, and the late gp350/220 glycoprotein (Fig. 2B).
FIG. 2.
Ubinuclein modulates expression of both early and late EBV genes. (A) Total RNA from HEK293EBV cells that have been either not transfected (lane 1 of each panel), transfected with 0.5 μg of an EB1 expression plasmid (lane 2 of each panel), or cotransfected with expression plasmids for both EB1 (0.5 μg) and increasing amounts of Ubn-1 (2 and 4 μg) (lanes 3 and 4 of each panel) were purified, reverse transcribed, and analyzed by PCR using specific primers for actin (used to monitor the quality of mRNA purification and to normalize the amount of mRNA used under each condition), Ubn-1, the LMP-1 latent gene, the BALF4, BALF5, and BMRF1 early genes, and the BDLF1, BFRF3, and BdRF1 late genes. Lane 5 of each panel represents the results of a PCR amplification in the absence of reverse transcriptase. The results of PCR quantifications are indicated underneath each panel and are expressed relative to the amount of amplified DNA from the assay in which cells were transfected with an expression plasmid for EB1. (B) Immunoblot analysis of gp350/220, EB2, EB1, and Ubn-1 proteins expressed from HEK293EBV cells that have been either not transfected (lane 1), transfected with 0.5 μg of an EB1 expression plasmid (lane 2), or cotransfected with expression plasmids for both EB1 (0.5 μg) and increasing amounts of Ubn-1 (1, 2, and 4 μg) (lanes 3 and 4).
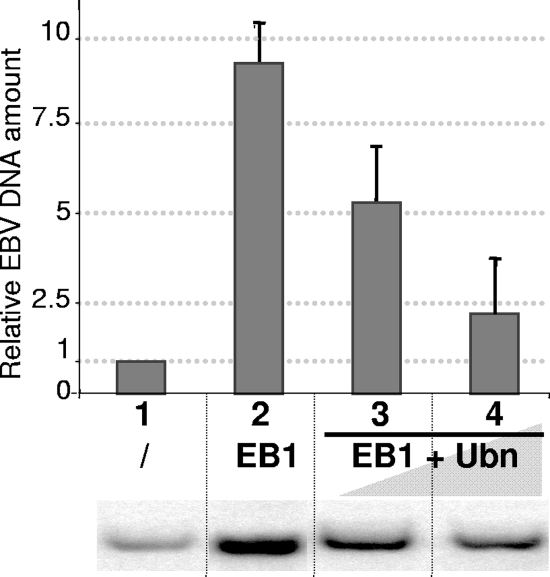
As shown in Fig. 3, in addition to its effect on expression of the EBV genes, Ubn-1 overexpression also had a strong effect on the efficiency of EBV's DNA replication during the productive cycle. In the absence of EB1, a small amount of EBV DNA was detected in the HEK293EBV cells (lane 1), whereas when EB1 was expressed, the amount of EBV DNA was increased 10 fold (lane 2), indicating that the viral genome was efficiently replicated. However, when Ubn-1 was coexpressed in these cells together with EB1, the amount of viral DNA replicated decreased in a dose-dependent manner (compare lanes 3 and 4 to lane 2). This decrease in the efficiency of EBV DNA replication is probably partly due to the weaker expression of the EBV early genes. However, since EB1 also acts as an origin-binding protein and directly transactivates oriLyt, the effect on DNA replication is likely to be also the result of a more direct effect on replication itself.
FIG. 3.
Ubinuclein overexpression inhibits viral DNA replication. Viral DNA extracted from HEK293EBV cells that had been either not transfected (lane 1), transfected with 0.5 μg of an EB1 expression plasmid (lane 2), or cotransfected with expression plasmids for both EB1 (0.5 μg) and increasing amounts of Ubn-1 (2 and 4 μg) (lanes 3 and 4) was quantified by Southern blotting using the BRRF1 probe. Fifty micrograms of total DNA obtained by Hirt extraction was cut by DpnI and BamHI and loaded in each lane. The results for one representative Southern blot are shown in the bottom panel. Quantification of the Southern blots is presented in the graph above as relative amounts of viral DNA observed after assignment of an arbitrary value of 1 to the quantified DNA present in the cells prior to reactivation of the productive cycle (lane 1). The experiment was done 3 times, and the error bars represent standard deviations.
Taken together, these results indicate that the downregulation of virion production following Ubn-1 overexpression is due to a combination of effects on early-gene expression, viral DNA replication, and late-gene expression, the last probably being only a consequence of the two precedent effects since EBV late-gene expression is known to be dependent on viral DNA replication.
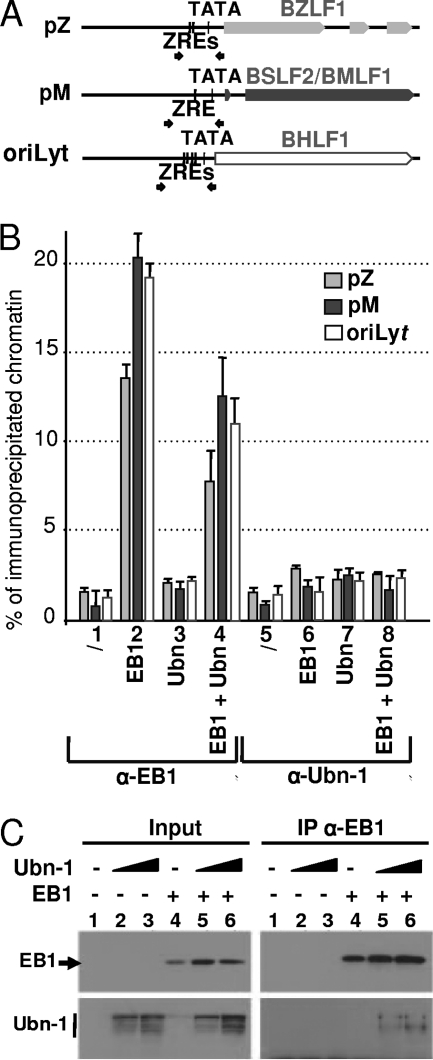
In vivo, ubinuclein overexpression reduces the interaction of EB1 with DNA. We have shown above that Ubn-1 interferes with the reactivation of the EBV productive cycle. We had previously shown that Ubn-1 inhibits the interaction between EB1 and its DNA-responsive elements (ZREs) in vitro (2). We thus speculated that in vivo, Ubn-1 could inhibit the production of infectious virus by interfering with EB1's binding to ZREs both on its viral target promoters and on the oriLyt region. In order to test this hypothesis, we assayed the EB1-DNA interaction in vivo by a chromatin immunoprecipitation (ChIP) assay, following cotransfection of latently infected HEK293EBV cells with a control vector or an EB1 expression plasmid together with (or without) an Ubn-1 expression plasmid. The chromatin was immunoprecipitated either with a monoclonal antibody directed against EB1 (MAb Z125) or with a monoclonal antibody directed against Ubn-1 (MAb ZAP1). We then measured the amount of specific immunoprecipitated DNA by semiquantitative PCR using primers encompassing ZRE sites present on the two viral promoters pM and pZ, which control expression of the BSLF2/BMLF1 and BZLF1 genes, respectively, and on the oriLyt region (Fig. 4A). Quantification of the amount of DNA amplified by PCR after immunoprecipitation of the chromatin was expressed as a percentage of the amount of input DNA used. The results presented in Fig. 4B show that when the cells were transfected with a control vector, immunoprecipitation of chromatin with MAb Z125 resulted only in a basic amplification of the pM, pZ, or oriLyt sequence (lane 1). In contrast, when EB1 was expressed, ChIP assays using MAb Z125 revealed striking increases in the amounts of pZ, pM, and oriLyt DNA present in the immunoprecipitated chromatin (lane 2). We found a consistent 14, 20, or 19% of the total pZ, pM, or oriLyt DNA, respectively, bound by EB1. However, when Ubn-1 was overexpressed, we observed a significative decrease in the amount of pZ, pM, or oriLyt DNA immunoprecipitated by MAb Z125 (down to 7.5, 12.5, or 11%, respectively) (lane 4). Interestingly, immunoprecipitation of chromatin with MAb ZAP1 did not allow any specific amplification of the pM, pZ, or oriLyt DNA under any conditions tested, even when Ubn-1 was overexpressed (lanes 5 to 8), indicating that Ubn-1 did not interact specifically on the two promoters tested or on the oriLyt region. These results indicate that in vivo, Ubn-1 reduces the amount of EB1 bound both to EB1-responsive promoters and to the EBV lytic origin of replication, oriLyt.
FIG. 4.
Ubinuclein overexpression inhibits EB1-DNA interaction. (A) Schematic representation of the BZLF1 and BSLF2/BMLF1 promoters and those for the oriLyt region. TATA, TATA box; ZRE, EB1-responsive element. Small arrows indicate the positions of the primers used for the PCR. (B) HEK293EBV cells that had been either not transfected (lanes 1 and 5), transfected with 0.5 μg of an EB1 expression plasmid (lanes 2 and 6) or 4 μg of a Ubn-1 expression plasmid (lanes 3 and 7), or cotransfected with expression plasmids for both EB1 (0.5 μg) and Ubn-1 (4 μg) (lanes 4 and 8) were fixed with paraformaldehyde. The chromatin was then immunoprecipitated using either Z125 MAb directed against EB1 (lanes 1 to 4) or Zap-1 MAb directed against Ubn-1 (lanes 5 to 8). Immunoprecipitated DNA was then quantified by PCR using specific primers located on the pM or pZ promoter or on the oriLyt region. The results are given as the percentage of coimmunoprecipitated DNA relative to the amount of input DNA. The results obtained for the pZ promoter are shown in light gray, those for the pM promoter in dark gray, and those for the oriLyt region in white. (C) Coimmunoprecipitation of Ubn-1 and EB1 in HEK293EBV cells. HEK293EBV cells were transfected with 0.5 μg of EB1 expression plasmids or 2 and 4 μg of Ubn-1 expression plasmid as indicated in the figure. EB1 was immunoprecipitated using MAb Z125 and protein A magnetic beads (Millipore), and the immunoprecipitated proteins were analyzed by immunoblotting using either MAb Z125 or MAb ZAP1 to reveal the presence of EB1 or Ubn1, respectively (right panels). The total amount of each protein present in the extract prior to immunoprecipitation was evaluated by analysis of aliquots (1/10 for EB1 and 1/50 for Ubn-1) of cell extracts by immunoblotting as shown in the left panels.
In order to confirm that this effect is due to a direct interaction between Ubn-1 and EB1, we performed a coimmunoprecipitation assay with HEK293EBV cells transfected with expression vectors for EB1 and Ubn-1. Immunoprecipitation from nuclear cell extracts was carried out using the anti-EB1 MAb Z125, and the amount of coimmunoprecipitated Ubn-1 was evaluated by immunoblotting using MAb ZAP1. As shown in Fig. 4C, Ubn-1 was indeed coimmunoprecipitated together with EB1 from cotransfected HEK293EBV cell extracts.
Taken together, these results indicate that Ubn-1 forms stable complexes with EB1 in cells and that when complexed with Ubn-1, EB1 is unable to interact with its target binding sites.
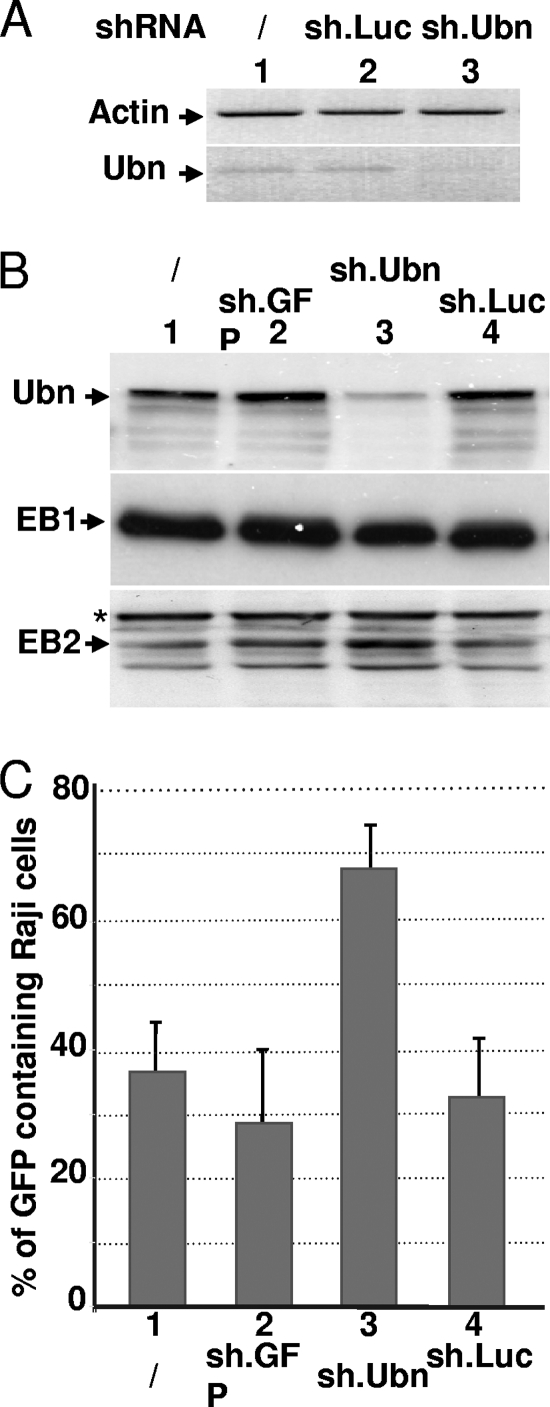
Ubinuclein knockdown boosts the production of viral particles.
Overexpression of ubinuclein leads to a decrease in the number of viral particles produced by HEK293EBV cells. We then tested the consequence of Ubn-1 knockdown on the EBV productive cycle. For this, HEK293EBV cells were infected with lentiviral particles allowing expression of Ubn-1-specific shRNA (sh.Ubn) or control shRNAs (sh.GFP and sh.Luc). As shown in Fig. 5A, at 8 days postinfection, the shRNA directed against Ubn-1 largely decreased Ubn-1 mRNA expression (Fig. 5A, lane 3) whereas the sh.Luc shRNA had no effect (Fig. 5A, lane 2). The level of Ubn-1 protein itself was also substantially decreased (Fig. 5B, lane 3). We then quantified the number of viral particles produced after induction of the EBV productive cycle by ectopic expression of EB1 in HEK293EBV cells treated or not treated with the anti-Ubn-1 shRNA. Interestingly, the number of virions produced by the HEK293EBV cells doubled when Ubn-1 expression was downregulated (Fig. 5C). We checked that this increase was not due to an unspecific increase in the amount of ectopically EB1 protein expressed (Fig. 5B, middle panel). However, we observed an increase in the amount of the early BMLF1 protein, EB2, whose gene expression is known to be directly upregulated by EB1 (Fig. 5B, bottom panel), which correlates with the increase in viral production. From these results, we can conclude that the presence of endogenous Ubn-1 in the cells limits the efficiency of viral production.
FIG. 5.
Ubinuclein knockdown allows an increased production of virions. HEK293EBV cells were infected with lentiviruses coding for an Ubn-1-specific shRNA (sh.Ubn) or an unspecific shRNA, either sh.GFP or sh.Luc, and then transfected with an expression plasmid for EB1. The relative levels of both Ubn-1 mRNA and the actin control mRNA were measured by RT-PCR (A), and the relative amounts of Ubn-1, EB1, and BMLF1 proteins were evaluated by immunoblotting using specific antibodies (B). Supernatants from HEK293EBV cells infected with the shRNA-carrying lentiviruses and transfected by an EB1 expression vector were collected and used to infect Raji cells. The number of GFP-expressing Raji cells was determined by FACS analysis (C). The experiment was done 3 times, and the error bars represent standard deviations.
Ubinuclein also interferes with AP1 trans-activation of various promoters.
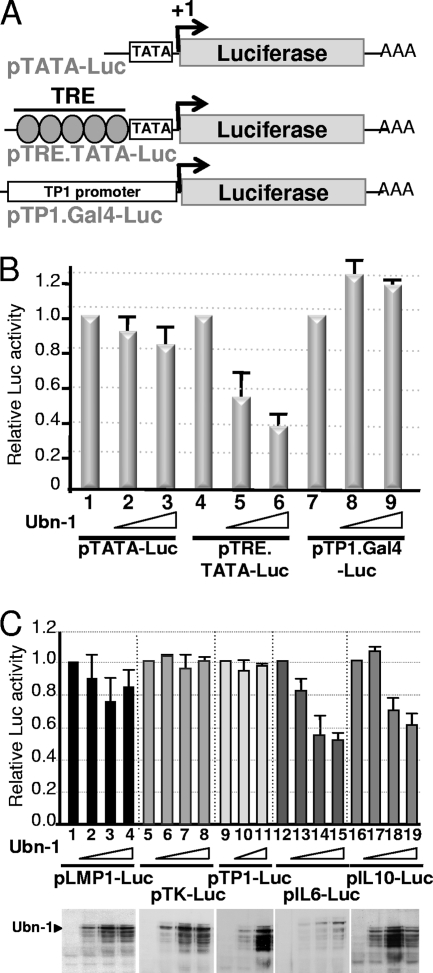
We had previously shown that Ubn-1 interacts with the basic domain of another b-ZIP protein, c-Jun, known to participate in AP1 complexes (2). Since Ubn-1 is a negative regulator of EB1-induced genes, we asked whether it could also interfere with c-Jun in the activation of reporter genes dependent upon AP1. In order to test this hypothesis, we transfected HeLa cells with luciferase reporter plasmids in which the luciferase gene was placed under the control of a minimal promoter (a TATA box), with or without the presence of upstream AP1-binding sites (Fig. 6A). The results presented in Fig. 6B show that Ubn-1 overexpression has only a weak effect on the minimal promoter construct (pTATA-Luc), which can be explained by the presence of an AP1-binding site further upstream in the plasmid sequence. However, when five AP1-binding sites were added immediately upstream of the minimal promoter (pTRE.TATA-Luc), the luciferase activity decreased substantially in the presence of overexpressed Ubn-1. In contrast, pTP1.Gal4, a construct containing the luciferase reporter gene under the control of an artificial promoter, which does not contain any AP1-binding sites, was unaffected by Ubn-1 overexpression. We then tested the effect of Ubn-1 overexpression on a selection of viral and cellular promoters, among which pIL6 and pIL10 are known to bear AP1-binding sites. HeLa cells were cotransfected with the different luciferase reporter constructs together with increasing amounts of Ubn-1 expression vector. Forty-eight hours after transfection, the luciferase activity was measured and the amount of Ubn-1 expressed in the cells was monitored by Western blotting. The results presented in Fig. 6C show that the level of luciferase expressed from promoters not regulated by AP1, like the EBV LMP1 or TP1 latent promoter or the herpes simplex virus type 1 (HSV-1) TK promoter, was not significantly affected by Ubn-1 overexpression (lanes 1 to 11). In contrast, transcription induced from two cellular promoters (interleukin 6 [IL-6] and IL-10), known to be activated by AP1, was decreased in a dose-dependent manner by overexpression of Ubn-1 (lanes 12 to 19). These results clearly show that the cellular protein c-Jun, like EB1, can be a target for Ubn-1.
FIG. 6.
Ubinuclein interferes with AP1 trans-activation. (A) Schematic representation of the luciferase reporter constructs. TATA, TATA box; AAA, poly(A) site; TRE, AP-1-responsive element; +1, transcription initiation site. (B) HeLa cells were transfected with the various reporter constructs represented in panel A, together with no or increasing amounts of Ubn-1 expression plasmid. The luciferase activities presented in the histogram are given as relative values, using the results for the assay performed in the absence of Ubn-1 as a reference. (C) HeLa cells were transfected with reporter constructs carrying the luciferase-coding sequence under the control of various viral and cellular promoters as indicated in the figure, together with no or increasing amounts of Ubn-1 expression plasmid. The luciferase activities presented in the histogram are given as relative values, using the results for the assay performed in the absence of Ubn-1 as a reference. (Lower panel) Representative immunoblots from transfected HeLa cell extracts incubated with MAb ZAP1. The experiments were done 4 times, and the error bars represent standard deviations.
Ubinuclein inhibits spontaneous reactivation of the EBV productive cycle in latently infected epithelial cells.
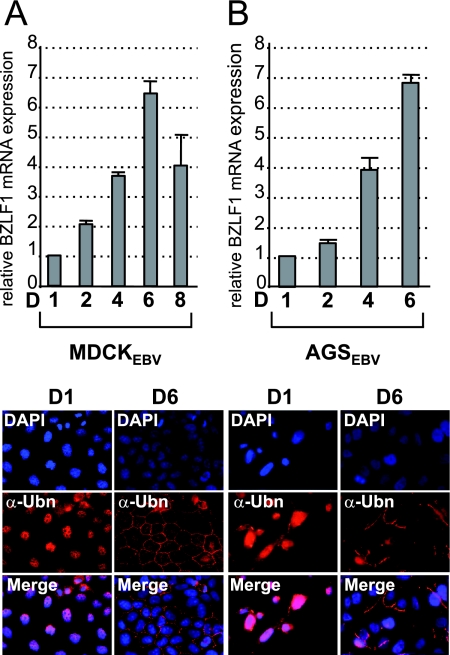
We have shown above that the ubiquitously expressed cellular protein Ubn-1 is a potent inhibitor of the EBV productive cycle. Interestingly, we had previously demonstrated (3) that Ubn-1 has a different localization depending on the cell's proliferation or differentiation state (nuclear in proliferating or undifferentiated cells and localized in the tight junctions in confluent polarized or differentiated cells). In addition, it has been reported that in vivo, the EBV productive cycle takes place in differentiated epithelial cells (8, 39, 67). In such cells, Ubn-1 is localized exclusively in the tight junctions (2). We thus hypothesized that if Ubn-1 plays a role in the regulation of the EBV productive cycle, EBV-infected epithelial cells should enter more efficiently into the productive cycle when they grow to confluence, because at this time, Ubn-1 will be sequestered outside the cell nucleus. To test this hypothesis, we used MDCK cells, a cell model which we have used previously to demonstrate the delocalization of Ubn-1 from the nucleus of the cells in sparse culture, to the tight junctions when cells become confluent (3). We thus infected MDCK cells with the EBVGFP recombinant virus, and after selection of the infected cells, we analyzed by RT-PCR the expression of the BZLF1 immediate-early gene (encoding EB1) in cells collected at different stages of confluence. The same assay was also done with another EBV-positive epithelial cell line derived from a gastric carcinoma, the AGSEBV cell line (22). It should be noted that MDCKEBV cells are not able to support a complete EBV productive cycle, because the BALF5 gene, encoding the EBV DNA polymerase, is not expressed in these cells after reactivation of the viral productive cycle (data not shown). We then purified the total mRNA and analyzed the expression level of the BZLF1 mRNA by semiquantitative PCR at different time points after plating the cells. The results are presented in Fig. 7A. When MDCKEBV cells were plated at low density, a low but constitutive level of expression of the BZLF1 mRNA was clearly detected. This basic level of BZLF1 mRNA was given an arbitrary value of 1. Interestingly, when the cells grew to confluence, expression of the BZLF1 mRNA increased significantly until day 6 (Fig. 7A, lanes 2 to 6). This increase correlated with a relocalization of Ubn-1 to the tight junctions (Fig. 7A, bottom panels, compare lanes D6 to lanes D1). At day 8, however, the expression level of BZLF1 mRNA decreased, probably because of the drastic reduction of cell proliferation and cell death at high density. In contrast to the MDCKEBV cells, the AGSEBV cells stably infected with the EBV B95-8 strain support lytic infection (37). In these cells, as in the MDCKEBV cells, BZLF1 mRNA expression is clearly detected when cells are plated at low density (Fig. 7B, lane 1), and again, when the cell culture grew to reach confluence, the expression level of BZLF1 mRNA increased dramatically (Fig. 7B, lanes 2 to 6). We have also looked by RT-PCR at the expression of another lytic gene, the BMRF1 gene, and found that its expression level also increased along with that of the BZLF1 mRNA (data not shown). At day 6, when the amount of BZLF1 mRNA was largest, Ubn-1 was found to be totally relocalized to the tight junctions (Fig. 7, bottom panels). After day 6, likely because the AGSEBV cells are actively producing virions, most of the cells lysed and the culture died.
FIG. 7.
Correlation between ubinuclein localization and activation of the EBV productive cycle. Nonconfluent MDCKEBV cells (A) or AGSEBV cells (B) were plated in petri dishes. Cells were collected at different time points (D, day), and expression of the BZLF1 gene was monitored by semiquantitative RT-PCR. The results are presented as the amount of [32P]dCTP incorporation, relative to that obtained at day 1, which has been given an arbitrary value of 1. The experiments were done 4 times, and the error bars represent standard deviations. (Bottom panels) Localization of Ubn-1 was monitored by immunofluorescence staining at day 1 (nonconfluent cells) and day 6 (confluent cells). Cells were labeled using the anti-Ubn (α-Ubn) MAb ZAP1 (red); nuclei were visualized using DAPI (blue).
Taken together, these results demonstrate the existence of a tight correlation between cell density, Ubn-1 localization, and BZLF1 expression in EBV-infected epithelial cells.
DISCUSSION
Ubn-1, first described as a nuclear protein, is now classified in the NACos (nuclear adhesion complexes) protein family (3) since Ubn-1 is localized in the nuclei of proliferating cells but sequestered to the tight junctions of nonproliferating cells. Characterized for its capacity to interact in vitro with the basic domains of both cellular (c-Jun and C/EBP) and viral (EB1 from EBV) members of the basic leucine zipper family of proteins, Ubn-1 has been shown to interfere in vitro with their DNA-binding function. In the present study, we demonstrate that Ubn-1 is able to modulate the EBV productive cycle in vivo. When overexpressed in HEK293EBV cells, Ubn-1 inhibits binding of EB1 to its target promoters, thereby reducing both expression of the EBV early genes and EBV DNA replication during the viral productive cycle. Consequently, Ubn-1's overexpression lowers the production of viral particles. In contrast, when Ubn-1 expression is downregulated by shRNA, viral particle production increases. Interestingly, in epithelial cells latently infected by EBV, the amount of EBV immediate-early-gene expression increases with the level of confluence of the cell culture, suggesting that in subconfluent cells, EB1 produced at a low level is inactive because of the presence of Ubn-1 in the cell nuclei. However, when cells become confluent and Ubn-1 is relocalized from the nucleus to the tight junctions, EB1 is free to activate its target promoters as well as its own promoter.
Epithelial cell proliferation and differentiation are regulated by cell-to-cell interaction. Apical cell-cell junctions participate in these processes by using different types of shuttling proteins, such as the NACos, which exhibit both nuclear and junctional localization. Tight junctions are an example of such cell-cell junctions, and several signaling complexes have been identified to associate with them (43). In general, expression of NACos in tight junctions suppresses proliferation and allows differentiation in a coordinated manner with adherent junctions and extracellular matrix adhesion. When localized in the nucleus, some of these tight junction components have been shown to affect different signaling and transcriptional pathways (6, 42). Ubn-1 function is not as yet well characterized. However, three recent reports show that Ubn-1 forms a complex with the histone chaperone protein HIRA (5, 7, 56). In addition, Banumathy et al. have shown that Ubn-1 is a regulator of cell senescence in fibroblasts (7). Taken together, these observations argue in favor of a role for Ubn-1 in the regulation of gene expression linked to cell proliferation such as that described for ZONA/B (38). In this work, we have shown that Ubn-1 is indeed able to control expression of various viral and cellular genes. However, as shown by chromatin immunoprecipitation, Ubn-1 does not appear to directly interact with the promoters of early EBV genes. Rather, Ubn-1 interacts directly and specifically with the DNA-binding domain of transcription factors such as c-Jun or EB1. This interaction inhibits the binding of these factors to their target recognition sequences in vitro (2) and in vivo (this report). Hence, Ubn-1 could be a negative regulator of gene expression via its interaction with DNA-binding factors, but it could also be implicated in chromatin remodeling via its interaction with the histone chaperone HIRA.
We have shown that Ubn-1 not only alters gene expression induced by the viral transcription factor EB1 but also controls AP1-induced gene expression. Interestingly, many genes under the control of the AP1 complex have been implicated in cell differentiation and proliferation (51). This is reminiscent of the observation that another NACos protein, ZO2 (zonula occludens 2), is able to relocalize c-Jun to the tight junctions and in this way controls expression of some of c-Jun's target genes (9, 36).
Ubn-1 is a potent inhibitor of EB1's and AP1's function. In vitro experiments using electrophoretic mobility shift assays and in vivo observation by chromatin immunoprecipitation suggest that this inhibition is due to an interaction between Ubn-1 and EB1 that results in the loss of EB1 DNA-binding activity, without the need for Ubn-1 to bind DNA. Hence, in the nucleus, Ubn-1, similarly to the retinoic acid receptor (RAR) (53, 54, 66), could modulate reactivation of latent EBV infection by a direct protein-protein interaction not only with EB1 but also with c-Jun or other cellular proteins of the b-ZIP family. Several reports have shown that AP1 complexes or C/EBP is implicated in the regulation of BZLF1 expression (34, 55, 65). Since some of these proteins can interact with Ubn-1, which is able to repress their DNA-binding activity (2), we speculate that in EBV-infected cells, endogenous Ubn-1 might also regulate induction of BZLF1 expression. In this way, Ubn-1 could be a negative regulator of the EBV productive cycle both by inhibiting BZLF1 expression induced by AP1 complexes and by interfering with transcriptional activation of early-gene expression by EB1. However, it is important to note that activation of the EBV productive cycle is not dependent just on absence of Ubn-1 from the cell nucleus. In effect, it has been shown that the switch between latent and productive EBV infection in B cells is mediated by the cellular transcription factor XBP-1, which plays an essential role in cell differentiation (10). Ubn-1 may participate in the maintenance of the EBV latent cycle, but removing Ubn-1 is certainly not sufficient for reactivation. EBV reactivation requires cell differentiation and transcriptional activation of the BZLF1 and BRLF1 promoters by XBP-1 in B cells or a yet unknown factor in epithelial cells.
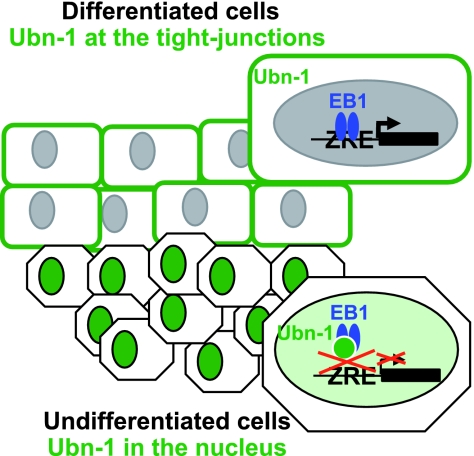
In addition, it is now well established that the EBV productive cycle takes place in differentiated cells (39), and it has been suggested from studies made with biopsy specimens from oral hairy leukoplakia, an AIDS-associated lesion characterized by high-level EBV replication in the tongue, that BZLF1 expression could be regulated by the degree of epithelial cell differentiation (8, 67). Interestingly, we show here that spontaneous expression of the EBV BZLF1 gene is also regulated by cell culture confluence conditions, in accordance with Ubn-1 localization. Together with our previous demonstration that Ubn-1 is sequestered to the tight junctions in differentiated cells (3), these observations led us to propose a model in which Ubn-1 controls the EBV productive cycle according to the differentiation status of epithelial cells (Fig. 8). In undifferentiated cells from the basal epithelium, Ubn-1 is nuclear and is free to interact with c-Jun and with the small amount of EB1 that is expressed. By interacting with the DNA-binding domains of both c-Jun and EB1, Ubn-1 can inhibit the association of these proteins with DNA and repress activation of the productive cycle. In differentiated cells of the squamus epithelium, however, Ubn-1 is sequestered to the tight junctions and BZLF1 expression is activated by a yet-unknown mechanism probably linked to the differentiation of the cells. In the absence of Ubn-1, EB1 would then be free to activate its target genes, allowing completion of a full productive cycle. Ubn-1 should thus be considered an important modulator of the EBV productive cycle in epithelial cells.
FIG. 8.
Model of control of the EBV productive cycle in epithelial cells. In undifferentiated cells, ubinuclein localized in the cell nucleus controls reactivation of the EBV productive cycle by inhibiting both AP1- and EB1-mediated gene expression, whereas in differentiated cells, ubinuclein is sequestered to the tight junctions so that EB1 is free to activate the productive cycle. ZRE, EB1-responsive elements.
Acknowledgments
We thank W. Hammerschmidt for the gift of the HEK293EBV cells, S. Kenney for the AGSEBV cells, and M. Castellazzi for the pTATA-Luc and pTRE-TATA-Luc constructs. PLATIM, FACS, and the lentivirus vector production facilities of IFR 128 are also acknowledged. We thank R. Buckland for editing the manuscript prior to submission.
This work was supported by the Institut National de la Santé et de la Recherche Médicale (INSERM), the Association pour la Recherche contre le Cancer (grant numbers 4914 and 3420), the Ligue Régionale Contre le Cancer (Comité du Rhône) (grant number 05.10.23), the Agence Nationale pour la Recherche (grant numbers RPV06120CSA and RPV09056CSA), the Pôle de Competitivité Lyon-Biopôle, and the Réseau INSERM Herpesvirus et Cancer. E.M. is a CNRS scientist.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Adamson, A. L., D. Darr, E. Holley-Guthrie, R. A. Johnson, A. Mauser, J. Swenson, and S. Kenney. 2000. Epstein-Barr virus immediate-early proteins BZLF1 and BRLF1 activate the ATF2 transcription factor by increasing the levels of phosphorylated p38 and c-Jun N-terminal kinases. J. Virol. 74:1224-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aho, S., M. Buisson, T. Pajunen, Y. W. Ryoo, J. F. Giot, H. Gruffat, A. Sergeant, and J. Uitto. 2000. Ubinuclein, a novel nuclear protein interacting with cellular and viral transcription factors. J. Cell Biol. 148:1165-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aho, S., J. Lupo, P. A. Coly, A. Sabine, M. Castellazzi, P. Morand, A. Sergeant, E. Manet, V. Boyer, and H. Gruffat. 2009. Characterization of the ubinuclein protein as a new member of the nuclear and adhesion complex components (NACos). Biol. Cell 101:319-334. [DOI] [PubMed] [Google Scholar]
- 4.Babcock, G. J., L. L. Decker, M. Volk, and D. A. Thorley-Lawson. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395-404. [DOI] [PubMed] [Google Scholar]
- 5.Balaji, S., L. M. Iyer, and L. Aravind. 2009. HPC2 and ubinuclein define a novel family of histone chaperones conserved throughout eukaryotes. Mol. Biosyst. 5:269-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balda, M. S., and K. Matter. 2009. Tight junctions and the regulation of gene expression. Biochim. Biophys. Acta 1788:761-767. [DOI] [PubMed] [Google Scholar]
- 7.Banumathy, G., N. Somaiah, R. Zhang, Y. Tang, J. Hoffmann, M. Andrake, H. Ceulemans, D. Schultz, R. Marmorstein, and P. D. Adams. 2009. Human UBN1 is an ortholog of yeast Hpc2p and has an essential role in the HIRA/ASF1a chromatin-remodeling pathway in senescent cells. Mol. Cell. Biol. 29:758-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Becker, J., U. Leser, M. Marschall, A. Langford, W. Jilg, H. Gelderblom, P. Reichart, and H. Wolf. 1991. Expression of proteins encoded by Epstein-Barr virus trans-activator genes depends on the differentiation of epithelial cells in oral hairy leukoplakia. Proc. Natl. Acad. Sci. U. S. A. 88:8332-8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Betanzos, A., M. Huerta, E. Lopez-Bayghen, E. Azuara, J. Amerena, and L. Gonzalez-Mariscal. 2004. The tight junction protein ZO-2 associates with Jun, Fos and C/EBP transcription factors in epithelial cells. Exp. Cell Res. 292:51-66. [DOI] [PubMed] [Google Scholar]
- 10.Bhende, P. M., S. J. Dickerson, X. Sun, W. H. Feng, and S. C. Kenney. 2007. X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 81:7363-7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bhende, P. M., W. T. Seaman, H. J. Delecluse, and S. C. Kenney. 2004. The EBV lytic switch protein, Z, preferentially binds to and activates the methylated viral genome. Nat. Genet. 36:1099-1104. [DOI] [PubMed] [Google Scholar]
- 12.Chang, Y.-N., D. L.-Y. Dong, G. S. Hayward, and D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chevallier-Greco, A., H. Gruffat, E. Manet, A. Calender, and A. Sergeant. 1989. The Epstein-Barr virus (EBV) DR enhancer contains two functionally different domains: domain A is constitutive and cell specific, domain B is transactivated by the EBV early protein R. J. Virol. 63:615-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV) encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. U. S. A. 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford, D. H., and I. Ando. 1986. EB virus induction is associated with B-cell maturation. Immunology 59:405-409. [PMC free article] [PubMed] [Google Scholar]
- 17.Delecluse, H. J., T. Hilsendegen, D. Pich, R. Zeidler, and W. Hammerschmidt. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dickerson, S. J., Y. Xing, A. R. Robinson, W. T. Seaman, H. Gruffat, and S. C. Kenney. 2009. Methylation-dependent binding of the Epstein-Barr virus BZLF1 protein to viral promoters. PLoS Pathog. 5:e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fahmi, H., C. Cochet, Z. Hmama, P. Opolon, and I. Joab. 2000. Transforming growth factor beta 1 stimulates expression of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA by an indirect mechanism which requires the MAPK kinase pathway. J. Virol. 74:5810-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng, W. H., R. J. Kraus, S. J. Dickerson, H. J. Lim, R. J. Jones, X. Yu, J. E. Mertz, and S. C. Kenney. 2007. ZEB1 and c-Jun levels contribute to the establishment of highly lytic Epstein-Barr virus infection in gastric AGS cells. J. Virol. 81:10113-10122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grogan, E., H. Jenson, J. Countryman, L. Heston, L. Gradoville, and G. Miller. 1987. Transfection of a rearranged viral DNA fragment, WZhet, stably converts latent Epstein-Barr viral infection to productive infection in lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 84:1332-1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gruffat, H., J. Batisse, D. Pich, B. Neuhierl, E. Manet, W. Hammerschmidt, and A. Sergeant. 2002. Epstein-Barr virus mRNA export factor EB2 is essential for production of infectious virus. J. Virol. 76:9635-9644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gruffat, H., E. Manet, A. Rigolet, and A. Sergeant. 1990. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 18:6835-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruffat, H., E. Manet, and A. Sergeant. 2002. MEF2-mediated recruitment of class II HDAC at the EBV immediate early gene BZLF1 links latency and chromatin remodeling. EMBO Rep. 3:141-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gruffat, H., and A. Sergeant. 1994. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 22:1172-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hadinoto, V., M. Shapiro, C. C. Sun, and D. A. Thorley-Lawson. 2009. The dynamics of EBV shedding implicate a central role for epithelial cells in amplifying viral output. PLoS Pathog. 5:e1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hardwick, J. M., P. M. Lieberman, and S. D. Hayward. 1988. A new Epstein-Barr virus transactivator, R, induces expression of a cytoplasmic early antigen. J. Virol. 62:2274-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henle, W. 1972. Role of Epstein-Barr virus in infectious mononucleosis and malignant lymphomas in man. Fed. Proc. 31:1674. [PubMed] [Google Scholar]
- 31.Hiriart, E., G. Farjot, H. Gruffat, M. V. Nguyen, A. Sergeant, and E. Manet. 2003. A novel nuclear export signal and a REF interaction domain both promote mRNA export by the Epstein-Barr virus EB2 protein. J. Biol. Chem. 278:335-342. [DOI] [PubMed] [Google Scholar]
- 32.Hochberg, D., J. M. Middeldorp, M. Catalina, J. L. Sullivan, K. Luzuriaga, and D. A. Thorley-Lawson. 2004. Demonstration of the Burkitt's lymphoma Epstein-Barr virus phenotype in dividing latently infected memory cells in vivo. Proc. Natl. Acad. Sci. U. S. A. 101:239-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong, G. K., M. L. Gulley, W. H. Feng, H. J. Delecluse, E. Holley-Guthrie, and S. C. Kenney. 2005. Epstein-Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J. Virol. 79:13993-14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang, J., G. Liao, H. Chen, F. Y. Wu, L. Hutt-Fletcher, G. S. Hayward, and S. D. Hayward. 2006. Contribution of C/EBP proteins to Epstein-Barr virus lytic gene expression and replication in epithelial cells. J. Virol. 80:1098-1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Israel, B. F., and S. C. Kenney. 2005. EBV lytic infection. Robertson ES, Philadelphia, PA.
- 36.Jaramillo, B. E., A. Ponce, J. Moreno, A. Betanzos, M. Huerta, E. Lopez-Bayghen, and L. Gonzalez-Mariscal. 2004. Characterization of the tight junction protein ZO-2 localized at the nucleus of epithelial cells. Exp. Cell Res. 297:247-258. [DOI] [PubMed] [Google Scholar]
- 37.Jones, R. J., S. Dickerson, P. M. Bhende, H. J. Delecluse, and S. C. Kenney. 2007. Epstein-Barr virus lytic infection induces retinoic acid-responsive genes through induction of a retinol-metabolizing enzyme, DHRS9. J. Biol. Chem. 282:8317-8324. [DOI] [PubMed] [Google Scholar]
- 38.Kavanagh, E., M. Buchert, A. Tsapara, A. Choquet, M. S. Balda, F. Hollande, and K. Matter. 2006. Functional interaction between the ZO-1-interacting transcription factor ZONAB/DbpA and the RNA processing factor symplekin. J. Cell Sci. 119:5098-5105. [DOI] [PubMed] [Google Scholar]
- 39.Laichalk, L. L., and D. A. Thorley-Lawson. 2005. Terminal differentiation into plasma cells initiates the replicative cycle of Epstein-Barr virus in vivo. J. Virol. 79:1296-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahot, S., A. Sergeant, E. Drouet, and H. Gruffat. 2003. A novel function for the Epstein-Barr virus transcription factor EB1/Zta: induction of transcription of the hIL-10 gene. J. Gen. Virol. 84:965-974. [DOI] [PubMed] [Google Scholar]
- 41.Manet, E., C. Allera, H. Gruffat, I. Mikaelian, A. Rigolet, and A. Sergeant. 1993. The acidic activation domain of the Epstein-Barr virus transcription factor R interacts in vitro with both TBP and TFIIB and is cell-specifically potentiated by a proline-rich region. Gene Expr. 3:49-59. [PMC free article] [PubMed] [Google Scholar]
- 42.Matter, K., and M. S. Balda. 2007. Epithelial tight junctions, gene expression and nucleo-junctional interplay. J. Cell Sci. 120:1505-1511. [DOI] [PubMed] [Google Scholar]
- 43.Matter, K., and M. S. Balda. 2003. Signalling to and from tight junctions. Nat. Rev. Mol. Cell Biol. 4:225-236. [DOI] [PubMed] [Google Scholar]
- 44.Mikaelian, I., E. Drouet, V. Marechal, G. Denoyel, J. C. Nicolas, and A. Sergeant. 1993. The DNA-binding domain of two bZIP transcription factors, the Epstein-Barr virus switch gene product EB1 and Jun, is a bipartite nuclear targeting sequence. J. Virol. 67:734-742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Neuhierl, B., R. Feederle, W. Hammerschmidt, and H. J. Delecluse. 2002. Glycoprotein gp110 of Epstein-Barr virus determines viral tropism and efficiency of infection. Proc. Natl. Acad. Sci. U. S. A. 99:15036-15041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niedobitek, G., A. Agathanggelou, H. Herbst, L. Whitehead, D. H. Wright, and L. S. Young. 1997. Epstein-Barr virus (EBV) infection in infectious mononucleosis: virus latency, replication and phenotype of EBV-infected cells. J. Pathol. 182:151-159. [DOI] [PubMed] [Google Scholar]
- 47.Niedobitek, G., A. Agathanggelou, N. Steven, and L. S. Young. 2000. Epstein-Barr virus (EBV) in infectious mononucleosis: detection of the virus in tonsillar B lymphocytes but not in desquamated oropharyngeal epithelial cells. Mol. Pathol. 53:37-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Orlando, V., H. Strutt, and R. Paro. 1997. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods 11:205-214. [DOI] [PubMed] [Google Scholar]
- 49.Petosa, C., P. Morand, F. Baudin, M. Moulin, J. B. Artero, and C. W. Muller. 2006. Structural basis of lytic cycle activation by the Epstein-Barr virus ZEBRA protein. Mol. Cell 21:565-572. [DOI] [PubMed] [Google Scholar]
- 50.Rickinson, A. B., J. E. Jarvis, D. H. Crawford, and M. A. Epstein. 1974. Observations on the type of infection by Epstein-Barr virus in peripheral lymphoid cells of patients with infectious mononucleosis. Int. J. Cancer. 14:704-715. [DOI] [PubMed] [Google Scholar]
- 51.Santaguida, M., K. Schepers, B. King, A. J. Sabnis, E. C. Forsberg, J. L. Attema, B. S. Braun, and E. Passegue. 2009. JunB protects against myeloid malignancies by limiting hematopoietic stem cell proliferation and differentiation without affecting self-renewal. Cancer Cell 15:341-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schepers, A., D. Pich, J. Mankertz, and W. Hammerschmidt. 1993. cis-acting elements in the lytic origin of DNA replication of Epstein-Barr virus. J. Virol. 67:4237-4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sista, N. D., C. Barry, K. Sampson, and J. Pagano. 1995. Physical and functional interaction of the Epstein-Barr virus BZLF1 transactivator with the retinoic acid receptors RAR alpha and RXR alpha. Nucleic Acids Res. 23:1729-1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sista, N. D., J. S. Pagano, W. Liao, and S. Kenney. 1993. Retinoic acid is a negative regulator of the Epstein-Barr virus protein (BZLF1) that mediates disruption of latent infection. Proc. Natl. Acad. Sci. U. S. A. 90:3894-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speck, S. H., T. Chatila, and E. Flemington. 1997. Reactivation of the Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 5:399-405. [DOI] [PubMed] [Google Scholar]
- 56.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51-61. [DOI] [PubMed] [Google Scholar]
- 57.Takada, K., and Y. Ono. 1989. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 63:445-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thompson, M. P., and R. Kurzrock. 2004. Epstein-Barr virus and cancer. Clin. Cancer Res. 10:803-821. [DOI] [PubMed] [Google Scholar]
- 59.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 60.Thorley-Lawson, D. A., and M. J. Allday. 2008. The curious case of the tumour virus: 50 years of Burkitt's lymphoma. Nat. Rev. Microbiol. 6:913-924. [DOI] [PubMed] [Google Scholar]
- 61.Thorley-Lawson, D. A., and A. Gross. 2004. Persistence of the Epstein-Barr virus and the origins of associated lymphomas. N. Engl. J. Med. 350:1328-1337. [DOI] [PubMed] [Google Scholar]
- 62.Urier, G., M. Buisson, P. Chambard, and A. Sergeant. 1989. The Epstein-Barr virus early protein EB1 activates transcription from different responsive elements including AP-1 binding sites. EMBO J. 8:1447-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Waltzer, L., P. Y. Bourillot, A. Sergeant, and E. Manet. 1995. RBP-J.κ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 23:4939-4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waltzer, L., M. Perricaudet, A. Sergeant, and E. Manet. 1996. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-J.κ-EBNA2-activated transcription by inhibiting the binding of RBP-J.κ to DNA. J. Virol. 70:5909-5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu, F. Y., H. Chen, S. E. Wang, C. M. ApRhys, G. Liao, M. Fujimuro, C. J. Farrell, J. Huang, S. D. Hayward, and G. S. Hayward. 2003. CCAAT/enhancer binding protein alpha interacts with ZTA and mediates ZTA-induced p21(CIP-1) accumulation and G(1) cell cycle arrest during the Epstein-Barr virus lytic cycle. J. Virol. 77:1481-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang-Yen, H. F., X. K. Zhang, G. Graupner, M. Tzukerman, B. Sakamoto, M. Karin, and M. Pfahl. 1991. Antagonism between retinoic acid receptors and AP-1: implications for tumor promotion and inflammation. New Biol. 3:1206-1219. [PubMed] [Google Scholar]
- 67.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, A. B. Rickinson, et al. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zalani, S., E. Holley-Guthrie, and S. Kenney. 1996. Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U. S. A. 93:9194-9199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang, J. X., H. L. Chen, Y. S. Zong, K. H. Chan, J. Nicholls, J. M. Middeldorp, J. S. Sham, B. E. Griffin, and M. H. Ng. 1998. Epstein-Barr virus expression within keratinizing nasopharyngeal carcinoma. J. Med. Virol. 55:227-233. [DOI] [PubMed] [Google Scholar]
- 70.Zur Hausen, H., F. J. O'Neil, U. K. Freese, and E. Hecker. 1978. Persisting oncogenic herpesvirus induced by the tumor promoter TPA. Nature 27:373-375. [DOI] [PubMed] [Google Scholar]