Abstract
Nef is an accessory protein critical for the ability of human and simian immunodeficiency viruses (HIV and SIV) to replicate efficiently in their respective hosts. Previous analyses of members of 15 different primate lentivirus lineages revealed a link between Nef function and the presence of a vpu gene. In particular, Nef proteins of all vpu-containing viruses had lost their ability to downmodulate the T cell (TCR-CD3) receptor. Here we examined Nef proteins from eight additional SIV lineages, including SIVgor, SIVwrc, SIVolc, SIVgri, SIVdrl, SIVlho, SIVden, and SIVasc, from western lowland gorillas, western red colobus monkeys, olive colobus monkeys, grivet monkeys, drills, L'Hoest's monkeys, Dent's mona monkeys, and red-tailed monkeys, respectively. We found that except for the nef gene of SIVdrl, all of them were efficiently expressed and modulated CD4, major histocompatibility complex class I (MHC-I), CD28, CXCR4, and Ii cell surface expression and/or enhanced viral infectivity and replication. Furthermore, the Nef proteins of SIVgri, SIVlho, SIVwrc, SIVolc, and SIVgor antagonized tetherin. As expected, the Nef protein of SIVgor, which carries vpu, failed to downmodulate CD3, whereas those of SIVwrc, SIVgri, SIVlho, and SIVasc, which lack vpu, were capable of performing this function. Surprisingly, however, the Nef protein of the vpu-containing SIVden strain retained the ability to downmodulate TCR-CD3, whereas that of SIVolc, which does not contain vpu, was unable to perform this function. Although the SIVden Vpu is about 20 amino acids shorter than other Vpu proteins, it degrades CD4 and antagonizes tetherin. Our data show that there are exceptions to the link between the presence of a vpu gene and nef alleles deficient in CD3 modulation, indicating that host properties also affect the selective pressure for Nef-mediated disruption of TCR-CD3 signaling. Our results are also further evidence that tetherin antagonism is a common function of primate lentivirus Nef proteins and that the resistance of human tetherin to Nef represents a relevant barrier to cross-species transmission of SIVs to humans.
One feature that distinguishes human and simian immunodeficiency viruses (HIV and SIV) from other retroviruses is that they encode several gene products that are not absolutely required for viral spread in cell culture but can dramatically alter the efficiency of viral replication and the course of disease progression in infected hosts in vivo (reviewed in references 2, 4, 29, 30, 35, and 40). Some of these “accessory” genes, i.e., vif, vpr, and nef, are present in the genomes of all primate lentiviruses. In contrast, a vpu gene is found only in HIV type 1 (HIV-1), in its chimpanzee and gorilla precursors, SIVcpz and SIVgor (18, 24, 44, 58), and in SIVgsn, SIVmus, SIVmon, and SIVden, infecting greater spot-nosed, mustached, mona, and Dent's mona monkeys (6, 11-14). Vpu was most likely acquired by a precursor of SIVs infecting Cercopithecus monkeys, with subsequent cross-species transmission and recombination events giving rise to other vpu-containing primate lentiviruses, such as SIVcpz, SIVgor, and HIV-1 (5, 51).
Vpu is expressed from a polycistronic message that also encodes Env during the late stages of the viral life cycle and has two major functions. First, it interferes with the transport of newly synthesized CD4 to the cell surface by targeting it for proteasomal degradation (8, 36, 62). Second, it promotes virion release by antagonizing an alpha interferon (IFN-α)-induced host restriction factor, named tetherin (also called BST2, CD317, or HM1.24), that tethers viral particles at the cell surface (41, 60). Notably, the multifunctional Nef protein also modulates CD4 cell surface expression and—in some lentiviruses—counteracts tetherin (26, 48, 63, 64). In contrast to Vpu, Nef is expressed at high levels early following cell infection and downmodulates CD4 by enhancing its internalization and lysosomal degradation (reviewed in references 2 and 31). Recently it has been shown that some SIVs that do not carry a vpu gene, but also SIVcpz and SIVgor (the vpu-containing precursors of HIV-1), use Nef to antagonize tetherin (26, 48, 63, 64). Nef also downmodulates major histocompatibility complex class I (MHC-I), CD28, and CXCR4, upregulates MHC-II-associated invariant chain (Ii), and enhances viral infectivity and replication (reviewed in references 2, 4, 30, and 35). The importance of the various Nef and Vpu activities in vivo is still largely unknown, although there is compelling evidence that a combination of functions allows HIV and SIV to replicate and spread efficiently in their respective hosts (reviewed in references 2, 4, and 30).
Vpu and Nef exhibit overlapping functions, and the presence of a vpu gene has been found to correlate with changes in Nef function. Specifically, data from a large number of primate lentiviruses revealed a striking concordance between the presence of a vpu gene and the inability of Nef to downmodulate CD3 (51). In fact, phylogenetic analyses strongly suggest that Nef-mediated downmodulation of T cell receptor-CD3 (TCR-CD3) was lost twice during primate lentivirus evolution: the first time after a vpu gene was acquired by an ancestor of SIVs now found in Cercopithecus monkeys and the second time after SIVrcm recombined with a vpu-containing precursor of SIVgsn/mus/mon/den in chimpanzees to become SIVcpz (5, 51).
The findings described above suggest that Vpu alleviates the need to maintain Nef-mediated TCR-CD3 downmodulation, possibly because an effective tetherin antagonist may allow efficient viral spread in the presence of higher levels of immune activation and thus reduce the selective pressure for suppression of T cell activation (28). However, this Nef function has been examined for only a fraction of the ∼40 African nonhuman primate species infected with primate lentiviruses (reviewed in references 20, 43, and 61). To obtain further insights into primate lentivirus accessory gene function, we examined nef alleles from eight additional SIV lineages, including two (SIVgor and SIVden) carrying a vpu gene. Surprisingly, one of these (SIVden) carried a vpu gene but retained Nef-mediated downregulation of TCR-CD3, while another strain (SIVolc), which lacked vpu, had lost this function. These findings suggest that other, yet-to-be-defined viral or host factors also affect the selective pressure on Nef-mediated modulation of T cell activation.
MATERIALS AND METHODS
nef and vpu alleles and proviral constructs.
SIV nef alleles from western red colobus (SIVwrc, Piliocolobus badius) and SIVolc from olive colobus (Procolobus verus) monkeys were PCR amplified from uncultured primate peripheral blood mononuclear cell (PBMC) DNA (Table 1). SIVwrc nef was amplified using the 5′ primer 5SIVwrcNIG (5′-GGATTTTGCTATAAGATGGGTGGAATCTTC-3′) and the 3′ primer 3SIVwrcNIG (5′-GCTGAAGCGGCACAAGTGACGCGTTGTTGC-3′). SIVolc nef was amplified in 2 rounds of PCR using the 5′ primer 5pSIVolcR1 (5′-GAGACTTACTATCACACCTGTGG-3′) and 3′ primer 3pSIVolcR1 (5′-GCTTTCGGTTTTGCCCTATAAAAG-3′) in the first round and the 5′ primer 5SIVolcNIG (5′-GGATTTTGCTATAAGATGGGATCAATTTGC-3′) and 3′ primer 3SIVolc-MluI (5′-CGCCTCTTAAGTGACGCGTATGTTCAG-3′) in the second round of PCR. SIVden, SIVgri, SIVdrl, SIVasc, and SIVlho sequences were derived from GenBank (Table 1) and chemically synthesized (GenScript, Piscataway, NJ). Molecular cloning of SIVgor has been described previously (58). Splice-overlap-extension PCR was used to replace the nef gene of HIV-1 (NL4-3-based) proviral constructs carrying functional nefs followed by an internal ribosome entry site (IRES) with the different primate lentivirus nef genes as described previously (51, 52). The integrity of all PCR-derived inserts was confirmed by sequence analysis. The control HIV-1 NL4-3-IRES-enhanced green fluorescent protein (eGFP) constructs expressing the NL4-3, NA7, and SIVmac239 Nefs or containing a disrupted nef gene (nef−) have been reported previously (50-52).
TABLE 1.
Overview of newly analyzed SIV nef allelesa
| Clone | Group/subtype | Species/subspecies/origin | Size of nef (bp) | Source | GenBank accession no. |
|---|---|---|---|---|---|
| CP2139 | SIVgor | Western lowland gorilla (G. g. gorilla) | 633 | Fecal viral RNA | FJ424866 |
| 98CI04 | SIVwrc | Western red colobus (Piliocolobus badius) | 702 | PBMC proviral DNA | AM713177 |
| 97CI12 | SIVolc | Olive colobus (Procolobus verus) | 558 | PBMC proviral DNA | FM165201 |
| gri-1 | SIVgri | Grivet monkey (Chlorocebus aethiops) | 672 | Proviral DNA, CEMss cells | M58410 |
| 1FAO | SIVdrl | Drill (Mandrillus leucophaeus) | 711 | Proviral DNA, Molt4 cl. 8 cells | AY159321 |
| SIVlho | SIVlho | L'Hoest monkeys (C. lhoesti) | 624 | Proviral DNA, CEMss cells | AF075269 |
| NA | SIVden | Dent's mona monkey (C. mona denti) | 747 | Plasma viral RNA | AJ580407 |
| NA | SIVasc | Red-tailed monkey (C. ascanius schmidti) | 681 | PBMC proviral DNA | FN994812 |
Abbreviations: NA, not available; G., Gorilla; C., Cercopithecus.
Cell culture and virus stocks.
Jurkat and 293T cells were cultured as described previously (51, 52). Briefly, 293T cells were maintained in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal bovine serum. PBMCs from healthy human donors were isolated using lymphocyte separation medium (Biocoll separating solution; Biochrom), stimulated for 3 days with phytohemagglutinin (PHA) (1 μg/ml), and cultured in RPMI-1640 medium with 10% fetal calf serum (FCS) and 10 ng/ml interleukin 2 (IL-2) prior to infection. To generate viral stocks, 293T cells were either transfected with the proviral HIV-1 constructs alone (to measure viral infectivity or replication) or cotransfected with a plasmid (pHIT-G) expressing the vesicular stomatitis virus G protein (VSV-G) for flow cytometric analyses (51, 52). The medium was changed after overnight incubation, and virus was harvested 24 h later. Residual cells in the supernatants were pelleted, and the supernatants were stored at −80°C. Virus stocks were quantified using a p24 antigen capture assay provided by the NIH AIDS Research and Reference Reagent Program.
Transduction and flow cytometry.
Jurkat T cells, THP1 cells, or PBMCs were transduced with HIV-1 (NL4-3) constructs coexpressing eGFP and various nef alleles, and CD4, TCR-CD3, MHC-I, CD28, CXCR4, CD25, CD69, Ii, and eGFP expression was measured as described previously (51, 52). For quantification of Nef-mediated modulation of specific surface molecules, the levels of receptor expression (red fluorescence) were determined for cells expressing a specific range of eGFP. The extent of downmodulation (n-fold) was calculated by dividing the mean fluorescence intensity (MFI) obtained for cells infected with the nef− NL4-3 control viruses by the corresponding values obtained for cells infected with viruses coexpressing Nef and eGFP.
Western blot.
To monitor Vpu expression, 293T cells were transfected with 5 μg of vector DNA coexpressing eGFP and AU-1-tagged Vpus or proviral constructs. Two days posttransfection, cells were harvested and lysed in RIPA (1% NP-40, 0.5% Na-DOC, 0.1% SDS, 0.15 M NaCl, 50 mM Tris-HCl [pH 7.4], and 5 mM EDTA), and cell lysates were separated in 16% SDS-polyacrylamide (PAA) gels in a Tris-Tricine buffer system. To reduce the oligomerization of Vpu, the SDS concentration in the sample buffer was increased to 1%. After gel electrophoresis, proteins were transferred onto polyvinylidene difluoride (PVDF) membranes and probed with AU-1 antibody (MMS-130P), followed by enhanced chemiluminescence detection.
CD8 fusions containing the cytoplasmic domain of the CD3-ζ chain.
The generation of a vector expressing CD8 fused to the cytoplasmic domain of the human CD3-ζ chain has been described previously (57). To replace the human CD3-ζ gene with the simian version, the rhesus macaque CD3-ζ gene (GenBank accession number DQ437670) was chemically synthesized (GenScript). Subsequently, splice-overlap-extension PCR using the primers p5-huCD8 (5′-CCTTCTAGACTAAAGATGGCCTTACCAGTG-3′) and p3-huCD8 (5′-GCTGAACTTCGCTCTGTGGTTGCAG-3′) and primers p5-rhζ (5′-CTGCAACCACAGAGCGAAGTTCAGC-3′) and p3-rhζ (5′-GATCCGACGCGTTTAGCGAGGGGGCAG-3′) was performed to fuse the gene encoding the extracellular part of human CD8 to the rhesus-derived sequence encoding the CD3-ζ cytoplasmic domain. The PCR-derived fragment was verified by sequence analysis. To assess species-specific effects of Nef on CD3, 293T cells were cotransfected with NL4-3-based proviral constructs coexpressing Nef and eGFP together with pCGCG vectors expressing CD8 fusion containing the human or macaque CD3-ζ chains. For quantification of CD8 surface expression, cells were stained with phycoerythrin (PE)-conjugated monoclonal antibody RPA-T8 (BD Biosciences). Fluorescence intensity on eGFP-positive cells was measured by flow cytometry and compared to that on cells transfected with a nef-defective HIV-1 control construct (100%).
NFAT assay.
Jurkat cells stably transfected with an NFAT-dependent reporter gene vector (17) were either left uninfected or transduced with HIV-1 Nef-eGFP constructs expressing various nef alleles. Except for those cells used as controls, cultures were treated with PHA (1 μg/ml; Murex). Luciferase activity was measured and n-fold induction determined by calculating the ratio of measured relative light units (RLU) of treated samples to that of untreated samples, as described previously (50).
Viral infectivity.
Virus infectivity was determined using P4-CCR5 and TZM-bl cells as described previously (39). Briefly, the cells were sown out in 96-well dishes in a volume of 100 μ1 and infected after overnight incubation with virus stocks, containing 1 ng of p24 antigen, produced by transfected 293T cells. Two days postinfection, viral infectivity was detected using the Gal screen kit from Tropix as recommended by the manufacturer. β-Galactosidase activities were quantified as relative light units per second using the Orion microplate luminometer.
Virus spread in PBMCs.
To assess the ability of Nef to promote viral spread, 2 × 105 prestimulated PBMCs per well were sown in 48-well dishes and infected with 293T cell-derived virus stocks containing 1 ng of p24 antigen. Aliquots of the cells were obtained at 3, 5, and 7 days postinfection, and the number of virally infected eGFP+ cells was determined by flow cytometric analysis.
CD4 degradation.
To determine the effect of Vpu on CD4 cell surface expression, 293T cells were transfected by the calcium phosphate method in triplicate with 1 μg of a CD4 expression vector and 5 μg of pCGCG eGFP/Vpu constructs expressing eGFP alone or together with Vpu. Two days posttransfection, CD4 expression was examined by fluorescence-activated cell sorting (FACS) analysis, as described previously (48).
Tetherin antagonism.
To determine the capability of Nef and Vpu to antagonize tetherin, 293T cells were seeded in six-well plates and transfected with 2 μg of NL4-3 ΔVpu/ΔNef/eGFP or 500 ng Vpu or Nef expression plasmid and different dilutions of tetherin expression vectors (100, 50, and 25 ng). A pCGCG vector expressing eGFP only was used to equalize the DNA concentrations. At 2 days posttransfection, supernatants were harvested and analyzed for the release of p24 antigen and infectious virus as described previously (48).
Phylogenetic analysis.
Phylogenetic trees of Nef amino acid sequences were inferred by the Bayesian method (23) implemented in the MrBayes software program, v3.2.1, using a mixed model (45) and gamma distributed rates at sites with 1 million generations. The average standard deviation of split frequencies was 0.007. Virus strains used for analysis were HIV-1 NL4-3, HIV-1 NA7, SIVcpz GAB2, SIVcpz TAN1, SIVrcm15, SIVrcm GB1, HIV-2 BEN, HIV-2 60415k2, SIVsmm FFm1, SIVagm TAN1, SIVagm TAN18, SIVagm SAB1, SIVmon NG1, SIVmon CML1, SIVmus CMS1085, SIVgsn CM166, SIVdeb CM40, SIVdeb CM5, SIVblu 3-1, SIVsyk 44, SIVsyk 51, SIVtal CM266, SIVtal CM8023, and SIVsun Mbolok1.
Statistical methods.
The activities of nef alleles were compared using a two-tailed Student t test. The PRISM software package, version 4.0 (Abacus Concepts, Berkeley, CA), was used for all calculations.
RESULTS
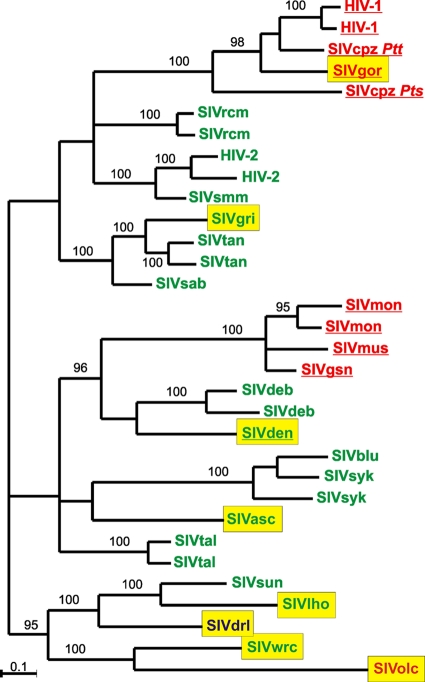
In this work, we studied nef alleles from eight SIV lineages that have not been previously examined: SIVgor from the western lowland gorillas (Gorilla gorilla gorilla), SIVwrc from western red colobus monkeys (Piliocolobus badius), SIVolc from olive colobus monkeys (Procolobus verus), SIVgri from grivet monkeys (Chlorocebus aethiops), SIVdrl from drills (Mandrillus leucophaeus), SIVlho from L'Hoest's monkeys (Cercopithecus lhoesti lhoesti), SIVden from a Dent's mona monkey (Cercopithecus denti), and SIVasc from a red-tailed monkey (Cercopithecus ascanius schmidti) (summarized in Table 1). Phylogenetic examination confirmed their authenticity (Fig. 1). Together with 30 SIV and HIV nef genes from 15 different primate species investigated in previous studies (39, 51), they represent the majority of primate lentivirus lineages currently characterized (summarized in Table S1 in the supplemental material). As expected from published data (12-14, 22, 32, 58), SIVgor Nef clustered with those of HIV-1 and SIVcpz, SIVgri Nef with SIVtan and SIVsab, infecting other African green monkey (AGM) species, and SIVdrl with SIVlho and SIVsun (Fig. 1). The Nef amino acid sequences of SIVwrc and SIVolc found in western red and olive colobus monkeys also clustered together but were only distantly related to one another and to those of other SIVs. The SIVasc Nef was most closely related to those of SIVsyk. Interestingly, Nef of the vpu-containing SIVden strain was more closely related to Nefs of SIVdeb strains (which do not have vpu) than to Nefs derived from SIVgsn, SIVmus, and SIVmon, which also encode Vpu (14) (Fig. 1). Altogether, the Nef amino acid sequences were highly variable, and the lengths of the open reading frames ranged from 558 (SIVolc) to 747 (SIVden) bp (summarized in Table 1).
FIG. 1.
Evolutionary relationships among primate lentivirus Nef sequences. Nef alleles newly analyzed in the present study are highlighted with yellow boxes, and those of vpu-containing viruses are underlined. Nef alleles that downmodulate CD3 are shown in green, those that are selectively defective in CD3 downmodulation in red, and those that are generally poorly functional in blue. The tree was midpoint routed and inferred by the Bayesian method (23) implemented in MrBayes v3.2.1 as described in Methods. Numbers on branches are percent estimated posterior probabilities. Only those 95% and above are shown.
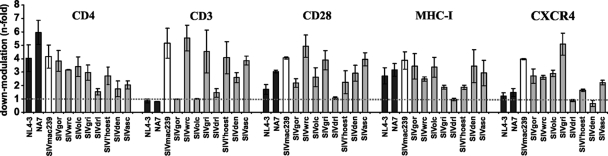
To assess the potencies of SIVgor, SIVwrc, SIVolc, SIVgri, SIVdrl, SIVlho, SIVden, and SIVasc Nefs in downmodulating CD3, CD4, CD28, CXCR4, and MHC-I, we transduced human PBMCs with proviral HIV-1 NL4-3 constructs coexpressing the various Nef proteins and eGFP and analyzed them by flow cytometry. Most Nef proteins downmodulated CD4, CD28, CXCR4, and MHC-I. The exception was the SIVdrl Nef protein, which did not downmodulate CXCR4 and MHC-I and had only very modest effects on CD4 and CD3 (Fig. 2). In agreement with this poor activity, the SIVdrl Nef was not expressed at detectable levels (data not shown). Like the HIV-1 NL4-3 and NA7 nef alleles, that of SIVgor failed to downmodulate TCR-CD3, although it was capable of modulating CD4, CD28, CXCR4, and MHC-I (Fig. 2). The SIVmac239, SIVwrc, SIVgri, SIVlho, SIVden, and SIVasc Nefs downmodulated TCR-CD3 as well as other receptors investigated (Fig. 2). Surprisingly, the nef allele of the Vpu-encoding SIVden was capable of downmodulating CD3 (2.59 ± 0.38-fold), albeit less efficiently than those of SIVmac, SIVwrc, SIVgri, SIVlho, and SIVasc (3.86-fold to 5.54-fold) (Fig. 2). It was also unexpected that the SIVolc Nef protein did not downmodulate TCR-CD3 (Fig. 2), since it is derived from a virus that does not encode Vpu (28). The SIVolc nef open reading frame is substantially shorter (558 bp) than those of other primate lentiviruses (between 621 and 792 bp), including that of its closest relative, SIVwrc (702 bp) (Table 1). To exclude the possibility that the small size and the lack of CD3 modulation were allele specific, we analyzed a total of eight different SIVolc nef genes derived from two independent PCRs. All had the same size, and none of them modulated TCR-CD3 cell surface expression (data not shown).
FIG. 2.
Modulation of cellular receptors by primate lentivirus nef alleles. PBMCs were transduced with HIV-1 NL4-3 Nef-eGFP constructs coexpressing the indicated nef alleles and eGFP and assayed for surface expression of CD4, CD3, CD28, MHC-I, and CXCR4. The HIV-1 NL4-3, NA7, and SIVmac239 nef alleles have been functionally characterized in previous studies (3, 50) and are shown for comparison. Nef-mediated modulation of the indicated cellular receptors was calculated by dividing the mean fluorescence intensity obtained on cells coexpressing eGFP and the different nef alleles by the mean fluorescence obtained for cells transduced with a control HIV-1 construct containing a disrupted nef gene. The same ranges of eGFP expression were used in all calculations. Values give averages ± SD derived from three independent experiments.
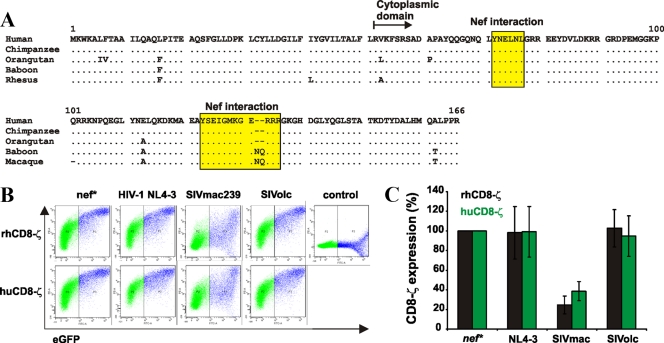
Previous studies showed that the capability of different primate lentivirus Nefs to downmodulate CD4, CD3, CXCR4, and MHC-I from the cell surface is usually species independent (21, 50, 51). However, we considered the possibility that the SIVolc Nef may downmodulate TCR-CD3 in simian but not human cells, since the cytoplasmatic domain of the CD3-ζ chain that interacts with Nef differs in four to six amino acid positions in this region (Fig. 3 A). In particular, we wondered whether a two-amino-acid (NQ) insertion may influence Nef binding since it is located in a region shown to interact with the SIVmac239 Nef protein (49). We thus analyzed CD8-CD3-ζ fusion proteins containing the cytoplasmatic domains of the human and rhesus TCR-ζ chains. We selected this approach since SIVmac and HIV-2 Nefs have been shown to interact specifically with the cytoplasmatic domain of CD3-ζ and this interaction is sufficient to cause downmodulation of TCR-CD3 and other receptors fused to this domain (49, 57). The rhesus CD3-ζ sequence was selected because rhesus macaques and olive colobus monkeys belong to the same family (Cercopithecidae) and because the CD3-ζ sequence of the olive colobus is not available. 293T cells were cotransfected with constructs expressing Nef and the CD8-CD3-ζ fusions, and the cell surface expression of CD8 was measured by flow cytometric analysis 2 days later. We found that SIVmac239 Nef reduced the surface expression of CD8 fused to both human- and rhesus-derived CD3-ζ cytoplasmic regions whereas SIVolc and HIV-1 Nefs were generally inactive (Fig. 3B and C). Thus, the inability of SIVolc Nefs to downmodulate TCR-CD3 is not limited to the human ortholog.
FIG. 3.
Interaction of Nef with the human and rhesus macaque TCR-CD3-ζ chains. (A) Alignment of the human CD3 sequence with those of other primate species. Dots represent amino acid identity; dashes represent gaps. The two domains previously proposed to interact with Nef (49) are indicated. (B) Expression levels of CD8 fusions containing CD3-ζ cytoplasmic domains of human (hu) or rhesus macaque (rh) origin. 293T cells were transiently cotransfected with pCGCG vectors expressing eGFP alone (control) or together with CD8-CD3-ζ fusion proteins and proviral HIV-1 constructs containing the nef alleles or a disrupted nef gene (nef*). The cells were stained for CD8 at 2 days posttransfection. (C) Levels of CD8 cell surface expression in the presence of the indicated nef alleles compared to those measured in the absence of Nef (100%). Values give averages ± SD derived from two independent experiments. Similar results were obtained with 5 different SIVolc nef alleles.
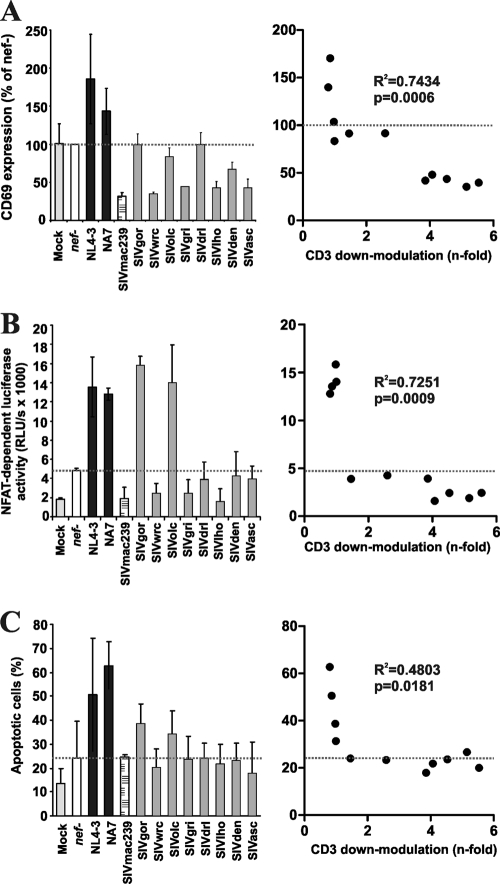
It has been shown that Nef proteins which downmodulate TCR-CD3 suppress T cell activation whereas those that do not perform this function have little inhibitory effect or may even render the infected cells hyperresponsive to stimulation (3, 17, 51). To assess whether this was also true for the newly analyzed SIV Nefs, we measured the cell surface expression levels of the early activation marker CD69 on virally infected PBMCs upon PHA stimulation. As shown in Fig. 4 A, all Nef proteins that downmodulated TCR-CD3 suppressed the induction of CD69 expression. Consistent with its lower activity in CD3 modulation, the SIVden Nef protein was less effective in inhibiting CD69 induction than all remaining Nefs that downmodulate CD3 (Fig. 4A). We also transduced Jurkat T cells stably transfected with the luciferase reporter gene under the control of an NFAT-dependent promoter (16) with the proviral HIV-1 eGFP-Nef constructs and examined their responsiveness to activation. T cells infected with a nef-defective HIV-1 construct showed about 3-fold enhanced levels of NFAT activity upon PHA stimulation compared to mock-infected cells (Fig. 4B). This increase was further enhanced by expression of NL4-3, NA7, SIVgor, and SIVolc Nefs that lack the CD3 downmodulation function (Fig. 4B). In contrast, SIV Nefs that remove TCR-CD3 from the cell surface usually suppressed the induction of NFAT-dependent luciferase activity (Fig. 4B). Nef-mediated suppression of T cell activation by downmodulation of TCR-CD3 was also associated with reduced levels of apoptotic virally infected T cells (Fig. 4C). Notably, relatively modest efficiencies of CD3 downmodulation were sufficient to suppress NFAT activation and apoptosis (Fig. 4B and C, right panels).
FIG. 4.
Modulation of T cell activation and apoptosis by primate lentiviral nef alleles. (A) CD69 expression levels on PBMCs infected with HIV-1 constructs coexpressing eGFP and the indicated nef alleles are shown relative to those measured on T cells transduced with the nef defective (nef−) control virus (100%). (B) Analysis of Jurkat cells stably transfected with an NFAT-dependent reporter gene following transduction with the indicated HIV-1 Nef-eGFP constructs and subsequent stimulation with PHA. “Mock” specifies uninfected control cells. (C) Percentages of Annexin V+ apoptotic cells in uninfected cultures or in HIV-1-infected cells expressing eGFP alone (nef−) or the indicated nef alleles are indicated. Values shown in the left in panels A to C give averages ± SD derived from three independent experiments. The right panels indicate the correlation between the levels of CD69 expression (A), PHA-induced levels of NFAT-dependent luciferase activities (B), or the percentages of apoptotic eGFP+ HIV-1-infected PBMCs and the efficiency of Nef-mediated CD3 downmodulation (C) (n = 11).
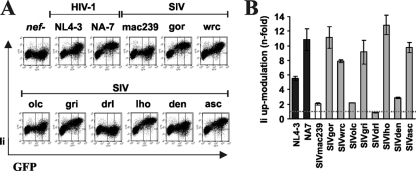
Previous data have shown that Nef proteins from HIV-1 and HIV-2 upregulate the expression of the invariant chain (Ii) at the cell surface, most likely to interfere with MHC-II antigen presentation (52, 55). To determine whether other primate lentiviral Nefs also modulate Ii, we used the human monocytic leukemia THP-1 cell line, which shares many properties with monocyte-derived macrophages and expresses high levels of MHC-II (59). THP-1 cells infected with proviral constructs expressing the HIV-1 NL4-3 and NA7, as well as the SIVgor, SIVwrc, SIVgri, SIVlho, and SIVasc Nefs, showed about 8- to 12-fold-enhanced levels of Ii surface expression (Fig. 5 A and B). In comparison, the SIVmac239, SIVolc, and SIVden nef alleles had only modest effects on Ii, and the SIVdrl Nef was inactive. Our finding that several highly divergent SIV Nefs were as active as those of HIV-1 in modulating Ii surface expression supports that the capability to modulate MHC-II-restricted antigen presentation is a conserved property of primate lentiviruses.
FIG. 5.
Modulation of the MHC-II-associated invariant chain (Ii) by SIV nef alleles. (A) Surface expression of Ii on THP-1 cells infected with HIV-1 Nef-eGFP constructs containing the indicated intact nef gene or a disrupted one. Ranges of eGFP expression levels used to calculate receptor modulation are indicated by black bars. (B) Quantitative assessment of Nef-mediated upmodulation of Ii on THP-1 cells. Values give averages ± SD derived from three independent experiments.
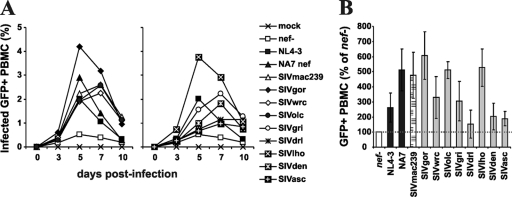
In addition to manipulating the functions of T cells and antigen-presenting cells (APCs), Nef also directly enhances the infectivity of viral particles (1, 10, 38, 39). To assess the abilities of the various SIV Nef proteins to enhance virion infectivity, we infected P4-CCR5 indicator cells (16) with virus stocks derived from 293T cells transiently transfected with the different proviral constructs. The results showed that the NL4-3, NA7, SIVmac, SIVgor, SIVwrc, SIVolc, and SIVden Nefs enhanced virion infectivity between 9- and 15-fold (Fig. 6). In comparison, the SIVasc and SIVdrl Nefs were inactive, and the SIVgri and SIV lho Nefs showed an intermediate phenotype. It is known that the abilities of Nef to enhance virion infectivity and to stimulate viral replication in primary T cells do not correlate with one another but represent independent Nef functions (34). Thus, we next investigated whether the different SIV nef alleles promote viral spread in human PBMCs. We found that all Nef proteins enhanced viral spread, albeit with different efficiencies (Fig. 7 A). The HIV-1 NA7, SIVmac239, SIVgor, SIVolc, and SIVlho Nefs were highly effective (Fig. 7B). In comparison, the SIVdrl, SIVden, and SIVasc Nefs were poorly active in the PBMC assay, and the NL4-3, SIVwrc, and SIVgri nef alleles showed an intermediate phenotype.
FIG. 6.
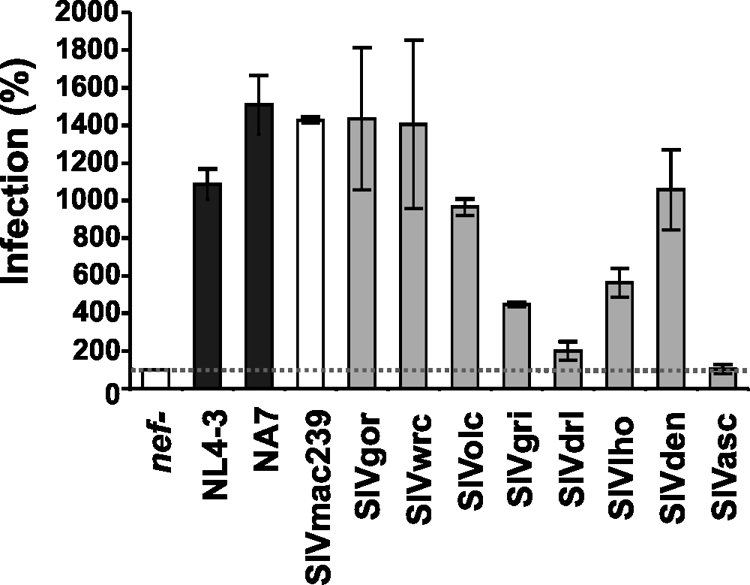
Enhancement of viral infectivity by primate lentivirus nef alleles. Infectivity of HIV-1 NL4-3 variants containing the indicated nef alleles. P4-CCR5 reporter cells were infected with HIV-1 NL4-3 IRES-eGFP constructs containing the indicated nef genes or a disrupted nef allele. Infections were carried out in triplicate with two different virus stocks containing 1 ng p24 antigen. Infectivity is shown relative to that of the recombinant virus containing the NL4-3 nef allele. Similar results were obtained in two independent experiments.
FIG. 7.
Enhancement of viral replication by primate lentivirus nef alleles. (A) Replication kinetics of recombinant NL4-3 variants containing the indicated nef alleles in PBMCs. Infections were carried out using virus stocks containing 1 ng p24 antigen. The numbers of virally infected GFP+ cells were measured by flow cytometric analysis. Similar results were obtained in two independent experiments. (B) Cumulative numbers of HIV-1-infected GFP+ cells detected in PBMC cultures at days 3, 5, 7, and 10 postinfection. Shown are averages ± SD derived from three independent infections.
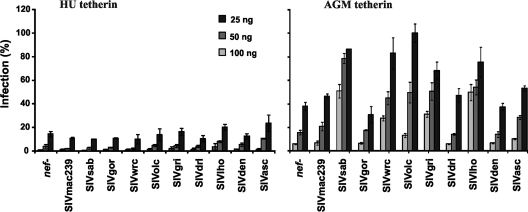
Recently it has been shown that some SIVs that do not possess a vpu gene, but also SIVcpz and SIVgor, which carry vpu, use their Nef proteins to antagonize the host restriction factor tetherin (BST-2) (26, 48, 64). Since it is not known to what extent primate lentiviruses use Nef to counteract tetherin, we cotransfected 293T cells with HIV-1 NL4-3 constructs containing defective vpu and nef genes as well as pCG vectors expressing AU-1-tagged versions of different Nef proteins at different doses of human or African green monkey tetherin. We selected these two tetherin variants because those of other primate hosts, including those of the SIVs analyzed in the present study, are not available for analysis. None of the Nef proteins analyzed was capable of antagonizing human tetherin (Fig. 8, left). This was expected, because the human protein contains a deletion in its cytoplasmic domain that renders it resistant to Nef (26, 48, 64). In contrast, SIVsab Nef (26, 27, 64), as well as the Nef proteins of SIVgri, SIVlho, SIVolc, and SIVwrc, increased the release of infectious virions in the presence of AGM tetherin (Fig. 8, right). Consistent with previous results, the SIVmac239 and SIVgor Nefs were only poor antagonists of AGM tetherin, although they counteract the tetherin variants found in their own macaque and gorilla hosts (48, 64). Such species specificity may also explain why the SIVasc Nef did not antagonize AGM tetherin. Altogether, these data show that tetherin antagonism by Nef is a common feature of primate lentiviruses.
FIG. 8.
Tetherin antagonism by primate lentivirus Nef proteins. Infectious virus yield from 293T cells cotransfected with the proviral HIV-1 NL4-3 ΔVpu ΔNef construct (48) containing disrupted vpu and nef genes (2 μg) and pCGCG vectors expressing the indicated nef alleles (500 ng), in combination with the indicated quantities of plasmids expressing the HU or AGM tetherins. Shown are average values derived from triplicate infections of TZM-bl indicator cells. All values are shown relative to those obtained in the absence of the tetherin expression vector (100%).
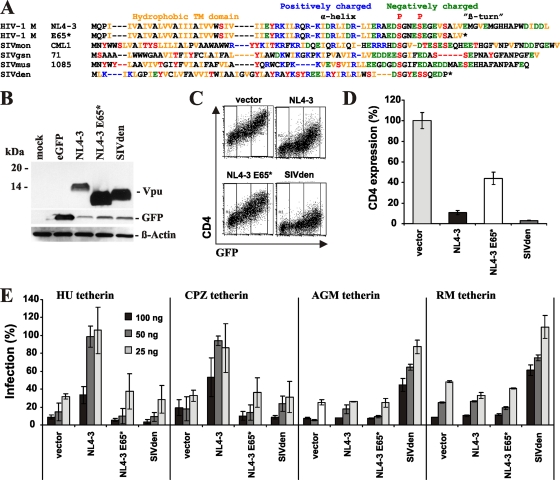
One possible explanation for why the SIVden Nef protein has not lost its TCR-CD3 downmodulation function is that its Vpu protein is prematurely truncated (Fig. 9 A) and may thus be unstable and/or nonfunctional. To assess this, we first examined the expression of an AU1-tagged version of the SIVden Vpu protein. For comparison, we also generated a C-terminally truncated mutant NL4-3 Vpu (E65*) protein. Western blot analysis showed that the SIVden and NL4-3 E65* Vpus were efficiently expressed (Fig. 9B). To determine whether these Vpus prevented cell surface expression of CD4, we cotransfected 293T cells with vectors coexpressing Vpu and eGFP (or eGFP alone for the control) together with a human CD4 expression construct. In the absence of Vpu, cells coexpressed CD4 and eGFP at high and correlating levels (Fig. 9C). However, upon coexpression of the wild-type NL4-3 and SIVden Vpus, CD4 surface expression was strongly decreased. In contrast, the NL4-3 E65* Vpu protein had only a modest effect (Fig. 9C). Quantitative analyses showed that the SIVden Vpu protein blocked CD4 cell surface expression almost entirely (Fig. 9D). Thus, the C-terminal truncation does not impair the capability of the SIVden Vpu to degrade the CD4 receptor.
FIG. 9.
Functional activity of the SIVden vpu allele. (A) Alignment of HIV-1 and SIV Vpu sequences. The hydrophobic transmembrane (TM) domain, the central charged region, the positions of two serine phosphorylation sites, and a β-turn motif in the NL4-3 Vpu protein are indicated. Dashes indicate gaps introduced to optimize the alignment. (B) Expression of the HIV-1 and SIVden Vpu proteins. 293T cells were transfected with expression plasmids encoding the indicated AU1-tagged Vpu proteins and eGFP. Mock-transfected cells were used as negative controls; β-actin and eGFP expression levels were analyzed to control for loading and transfection efficiency (lower panels). (C) Suppression of CD4 surface expression by the SIVden Vpu. FACS analysis of 293T cells cotransfected with a CD4 expression vector and pCGCG plasmids expressing eGFP alone (vector) or together with the indicated vpu alleles is shown. (D) Levels of CD4 cell surface expression relative to those measured in cells transfected with the eGFP-only control vector (100%). Shown are average values ± SD derived from three experiments. (E) Effects of the NL4-3, NL4-3 E65*, and SIVden Vpu on infectious virus release in the presence of human (HU), chimpanzee (CPZ), AGM, or rhesus macaque (RM) tetherin. 293T cells were cotransfected with HIV-1 ΔVpu NL4-3 (2 μg) and pCGCG vectors expressing eGFP alone or together with Vpu (500 ng) and the indicated amounts of tetherin expression construct (25, 50, or 100 ng). Viral supernatants were obtained 2 days later and used to measure the quantity of infectious HIV-1 in the culture supernatants by infecting TZM-bl indicator cells. Shown are average values ± SD (n = 3) of infectious virion yield relative to those obtained in the absence of the tetherin expression vector (100%). The results were confirmed in two independent experiments.
Next, we examined whether the highly divergent SIVden Vpu protein antagonizes tetherin. Since the tetherin variant found in Dent's mona monkeys, the natural host of SIVden, is not available, we used tetherins derived from humans, chimpanzees (CPZ), AGMs, and rhesus macaques (RMs). To test whether the SIVden and control Vpus were capable of counteracting tetherins from these species, we measured infectious virus yields from 293T cells following cotransfection of a vpu-deleted (ΔVpu) HIV-1 proviral construct with Vpu and tetherin expression plasmids. In agreement with the results of a previous study (63), our data showed that the SIVden Vpu protein efficiently counteracted monkey tetherins but was inactive against the human- and CPZ-derived restriction factors (Fig. 9E). In contrast, the NL4-3 control Vpu protein had the opposite phenotype, and the truncated NL4-3 E65* Vpu mutant was largely inactive. These results are in agreement with published data showing that Vpu antagonizes tetherin in a species-specific manner (19, 37, 48, 63). Moreover, SIVden Vpu, despite its C-terminal truncation, is highly active in CD4 degradation and tetherin antagonism.
DISCUSSION
In the present study, we expanded previous functional analyses of nef and vpu alleles from diverse primate lentiviruses (21, 26, 39, 48, 51, 52, 63, 64). We showed that the Nef proteins of SIVgor, SIVwrc, SIVolc, SIVgri, SIVlho, SIVden, and SIVasc modulate the surface expression of human CD4, CD28, MHC-I, and Ii molecules and enhance viral spread and infectivity. Consistent with previous data (39, 51), these results reinforce the view that nef alleles from highly divergent primate lentiviruses do not require adaptive changes to perform most of their activities in human cells. The exception is Nef-mediated antagonism of tetherin. It has been shown previously that the Nef proteins of SIVagm, SIVsmm, SIVmac, SIVcpz, and SIVgor antagonize the tetherins of their particular hosts but are all inactive against human tetherin because of a unique five-amino-acid deletion in the cytoplasmic domain (26, 48, 64). Here we have shown that the SIVwrc, SIVolc, SIVgri, and SIVlho Nefs also counteract simian tetherins but have no activity against the human restriction factor (Fig. 9). It is thus clear that Nef-mediated tetherin antagonism is a fundamental property of primate lentiviruses (see Table S1 in the supplemental material), and the resistance of human tetherin to primate lentivirus Nefs seems to represent a significant barrier to zoonotic transmissions. Unfortunately, this hurdle is not insurmountable, since HIV-1 and HIV-2 managed to switch from Nef to Vpu or Env, respectively, to regain anti-tetherin activity in the new human host (reviewed in references 15, 29, and 47).
It has been reported that some primate lentiviruses, such as SIVagm, did not evolve tetherin antagonists because they do not induce chronic immune activation, and thus expression of type I interferons and tetherin, in their natural simian hosts (33). In contrast, we and others have shown that the SIVagm and SIVsmm Nef proteins are tetherin antagonists (26, 48, 64), although these viruses cause neither chronic inflammation nor disease in their natural African green monkey and sooty mangabey hosts (reviewed in references 42 and 54). Moreover, as summarized in Table S1 in the supplemental material, it is now clear that most primate lentiviruses have evolved tetherin antagonists, suggesting that they encounter this restriction factor during natural infection. One possible explanation for this seeming controversy is that primate lentiviruses that induce high levels of inflammation may need particularly effective tetherin antagonists. In accordance with this, the Vpu proteins of pathogenic HIV-1 strains seem to be more effective in counteracting tetherin than the Nef proteins of the nonpathogenic SIVagm and SIVsmm strains (26, 33, 48, 63, 64). However, it should be noted that the efficiency of tetherin antagonism of primate lentiviruses in primary cells and its relevance in vivo (46) remain largely unknown, since most data thus far are limited to transient transfection assays.
To date, nef alleles from all groups of HIV-1 (M, N, O, and P), HIV-2 groups A and B, and about 20 different SIVs have been functionally analyzed. The results from these studies have shown that some Nef activities, such as downmodulation of CD4 and MHC-I, as well as enhancement of virion infectivity and stimulation of virus replication, are conserved among most or all primate lentiviruses (summarized in Table S1 in the supplemental material). These Nef functions facilitate viral immune evasion and promote viral spread directly. Thus, it seems plausible that they are always advantageous for the virus and thus are generally preserved. In contrast, the HIV and SIV Nef proteins show fundamental differences in activities affecting the responsiveness of virally infected T cells to stimulation via the TCR-CD3 complex (see Table S1). Most primate lentivirus Nefs efficiently downmodulate both TCR-CD3 and the CD28 costimulatory factor from the surfaces of virally infected T cells to prevent their interaction with antigen-presenting cells and responsiveness to stimulation (Fig. 4) (3, 51). The advantage for the virus is most likely a longer time period of virus production due to the suppression of apoptosis and thus an increased life span of the infected cells, as well as a suppression of overall immune activation (28). In contrast, the Nef proteins of HIV-1 and its direct SIV progenitors have no effect on CD3 and are poorly active in CD28 downmodulation (3, 51). As a consequence, these Nef proteins do not suppress the stimulation of virally infected CD4+ T cells via the TCR-CD3 complex (3, 17, 51). The acquisition of a vpu gene seems to reduce the selective pressure for suppression of T cell activation, because its presence was associated with the inability of Nef to downmodulate TCR-CD3 in all primate lentiviruses previously analyzed (51). Furthermore, phylogenetic analyses indicate that the SIV precursors of HIV-1 lost this Nef function twice when they acquired a vpu gene (51). Vpu, as an effective virion release factor and tetherin antagonist, may thus have favored the emergence of viruses that enhance rather than block T cell activation and thus proviral transcription because it allows them to cope with the higher activation of the host immune system (reviewed in reference 28). Differences in Nef-mediated downmodulation of TCR-CD3 and CD28 may help primate lentiviruses to compensate for species-specific differences in the responsiveness of T cells to activation and thus in viral gene expression. Since different primate species may respond to SIV infection differently (7, 25, 53) and the host environment may also affect Nef function (9), it is perhaps not surprising that the presence of vpu and lack of Nef-mediated TCR-CD3 downmodulation are not generally linked. A strong inflammatory response of Dent's mona monkeys to SIV infection may explain why this virus maintained the capability to downmodulate TCR-CD3 from the cell surface (albeit with relatively low efficiency), although it expresses a functional Vpu protein. On the other hand, SIVolc may have lost its capability to remove CD3 from the surface to achieve levels of T cell activation sufficient for viral replication. Studies of these viruses in their natural primate hosts will be required to address these possibilities.
Early data suggested that the vpu gene originated from a common ancestor of SIVgsn, SIVmus, and SIVmon after the divergence of SIVsyk and SIVdeb and was subsequently transferred to chimpanzees and humans by zoonotic viral transmissions and a recombination event (5, 51). However, the finding that the vpu-containing SIVden strain is more closely related to SIVdeb (which does not carry a vpu gene) than to SIVgsn, SIVmus, and SIVmon (Fig. 1) (14) is at odds with this hypothesis. It remains elusive whether SIVs found in some Cercopithecus species lost their vpu gene during evolution (14) or whether SIVden acquired its vpu gene more recently by recombination with an as yet unknown SIV strain, and both possibilities seem plausible.
The current data add to a comprehensive set of Nef proteins that differ in their capability to downmodulate TCR-CD3 (see Table S1 in the supplemental material). Unfortunately, inspection of their alignment does not allow pinpointing of the sequence variations responsible for the differential capability of primate lentiviral Nef proteins to modulate TCR-CD3. As shown in Fig. S1, the Nef amino acid sequences are highly divergent, and the two groups that differ in their CD3 downmodulation capabilities do not exhibit any striking differences. Previous analyses of the SIVmac239 Nef protein have shown that the CD3 downmodulation activity maps to the central core region of Nef and involves cooperative binding of Nef and CD3-zeta to bind AP-2 (49, 57). In agreement with these data, we identified amino acid substitutions in the core region of SIVsmm Nef that selectively affect the CD3 downmodulation function (J. Schmökel and F. Kirchhoff, unpublished data). However, the critical residues are not conserved in other SIV Nefs that downmodulate TCR-CD3. Furthermore, changes in the flexible N-proximal region of the SIVdeb Nef protein also selectively disrupt CD3 downmodulation (data not shown). Thus, similarly to downmodulation of MHC-I (56), the effect on CD3 may involve different domains in various primate lentivirus Nef proteins.
Substantial progress has been made in understanding the functions of the primate lentivirus Vpu and Nef proteins, and the results suggest that differences in the functions of these two accessory proteins may play relevant roles in the pathogenesis and spread of HIV-1 (reviewed in references 2, 4, 28-30, and 35). Nonetheless, our knowledge is still far from complete. For some SIV strains, only single nef or vpu alleles are available for analysis. Another caveat is that most studies were performed in human-derived cells because primary cells of the natural hosts of most SIVs are not available for experimentation. These analyses have shown that most Nef functions and Vpu-mediated degradation of CD4 are largely species independent (39, 51). However, the capability of the viral accessory proteins to antagonize host restriction factors is usually highly specific, and some functions may be missed in human-derived cells (19, 26, 37, 41, 48, 63, 64). Thus, the analysis of primary SIV strains in relevant target cells from their respective host species, as well as studies in primate models of pathogenic and nonpathogenic SIV infection, is required to better understand the relevance of specific Vpu and Nef functions for viral pathogenesis and transmission in vivo.
Supplementary Material
Acknowledgments
We thank Jan Muench for support, Martha Mayer and Veronika Bach for expert technical assistance, Michel J. Tremblay for Jurkat cells stably transfected with an NFAT-dependent reporter gene vector, and Dré van der Merwe and Kristina Wohllaib for critical reading of the manuscript.
S.S. was supported by a grant from the Agence National de Recherche sur le SIDA (ANRS). This work was supported by the Deutsche Forschungsgemeinschaft and grants from the NIH (R01 AI50529 and R01 AI58715).
Footnotes
Published ahead of print on 10 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aiken, C., and D. Trono. 1995. Nef stimulates human immunodeficiency virus type 1 proviral DNA synthesis. J. Virol. 69:5048-5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arhel, N. J., and F. Kirchhoff. 2009. Implications of Nef: host cell interactions in viral persistence and progression to AIDS. Curr. Top. Microbiol. Immunol. 339:147-175. [DOI] [PubMed] [Google Scholar]
- 3.Arhel, N., M. Lehmann, K. Clauss, G. U. Nienhaus, V. Piguet, and F. Kirchhoff. 2009. The inability to disrupt the immunological synapse between infected human T cells and APCs distinguishes HIV-1 from most other primate lentiviruses. J. Clin. Invest. 119:2965-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariën, K. K., and B. Verhasselt. 2008. HIV Nef: role in pathogenesis and viral fitness. Curr. HIV Res. 6:200-208. [DOI] [PubMed] [Google Scholar]
- 5.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 6.Barlow, K. L., A. O. Ajao, and J. P. Clewley. 2003. Characterization of a novel simian immunodeficiency virus (SIVmonNG1) genome sequence from a mona monkey (Cercopithecus mona). J. Virol. 77:6879-6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosinger, S. E., Q. Li, S. N. Gordon, N. R. Klatt, L. Duan, L. Xu, N. Francella, A. Sidahmed, A. J. Smith, E. M. Cramer, M. Zeng, D. Masopust, J. V. Carlis, L. Ran, T. H. Vanderford, M. Paiardini, R. B. Isett, D. A. Baldwin, J. G. Else, S. I. Staprans, G. Silvestri, A. T. Haase, and D. J. Kelvin. 2009. Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. J. Clin. Invest. 119:3556-3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bour, S., U. Schubert, and K. Strebel. 1995. The human immunodeficiency virus type 1 Vpu protein specifically binds to the cytoplasmic domain of CD4: implications for the mechanism of degradation. J. Virol. 69:1510-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carl, S., T. C. Greenough, M. Krumbiegel, M. Greenberg, J. Skowronski, J. L. Sullivan, and F. Kirchhoff. 2001. Modulation of different human immunodeficiency virus type 1 Nef functions during progression to AIDS. J. Virol. 75:3657-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowers, M. Y., C. A. Spina, T. J. Kwoh, N. J. Fitch, D. D. Richman, and J. C. Guatelli. 1994. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 68:2906-2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, E. A., E. F. Terwilliger, J. G. Sodroski, and W. A. Haseltine. 1988. Identification of a protein encoded by the vpu gene of HIV-1. Nature 334:532-534. [DOI] [PubMed] [Google Scholar]
- 12.Courgnaud, V., B. Abela, X. Pourrut, E. Mpoudi-Ngole, S. Loul, E. Delaporte, and M. Peeters. 2003. Identification of a new simian immunodeficiency virus lineage with a vpu gene present among different cercopithecus monkeys (C. mona, C. cephus, and C. nictitans) from Cameroon. J. Virol. 77:12523-12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. H. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dazza, M. C., M. Ekwalanga, M. Nende, K. B. Shamamba, P. Bitshi, D. Paraskevis, and S. Saragosti. 2005. Characterization of a novel vpu-harboring simian immunodeficiency virus from a Dent's mona monkey (Cercopithecus mona denti). J. Virol. 79:8560-8571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. T., R. Serra-Moreno, R. K. Singh, and J. C. Guatelli. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fenard, D., G. Lambeau, E. Valentin, J. C. Lefebvre, M. Lazdunski, and A. Doglio. 1999. Secreted phospholipases A(2), a new class of HIV inhibitors that block virus entry into host cells. J. Clin. Invest. 104:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fortin, J. F., C. Barat, Y. Beausejour, B. Barbeau, and M. J. Tremblay. 2004. Hyper-responsiveness to stimulation of human immunodeficiency virus-infected CD4+ T cells requires Nef and Tat virus gene products and results from higher NFAT, NF-kappaB, and AP-1 induction. J. Biol. Chem. 279:39520-39531. [DOI] [PubMed] [Google Scholar]
- 18.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 19.Gupta, R. K., S. Hue, T. Schaller, E. Verschoor, D. Pillay, and G. J. Towers. 2009. Mutation of a single residue renders human tetherin resistant to HIV-1 Vpu-mediated depletion. PLoS Pathog. 5:e1000443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 21.Hrecka, K., T. Swigut, M. Schindler, F. Kirchhoff, and J. Skowronski. 2005. Nef proteins from diverse groups of primate lentiviruses downmodulate CXCR4 to inhibit migration to the chemokine stromal derived factor 1. J. Virol. 79:10650-10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu, J., W. M. Switzer, B. T. Foley, D. L. Robertson, R. M. Goeken, B. T. Korber, V. M. Hirsch, and B. E. Beer. 2003. Characterization and comparison of recombinant simian immunodeficiency virus from drill (Mandrillus leucophaeus) and mandrill (Mandrillus sphinx) isolates. J. Virol. 77:4867-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huelsenbeck, J. P., and F. Ronquist. 2001. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 24.Huet, T., R. Cheynier, A. Meyerhans, G. Roelants, and S. Wain-Hobson. 1990. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature 345:356-359. [DOI] [PubMed] [Google Scholar]
- 25.Jacquelin, B., V. Mayau, B. Targat, A. S. Liovat, D. Kunkel, G. Petitjean, M. A. Dillies, P. Roques, C. Butor, G. Silvestri, L. D. Giavedoni, P. Lebon, F. Barré-Sinoussi, A. Benecke, and M. C. Müller-Trutwin. 2009. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J. Clin. Invest. 119:3544-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jia, B., R. Serra-Moreno, W. Neidermyer, A. Rahmberg, J. Mackey, I. B. Fofana, W. E. Johnson, S. Westmoreland, and D. T. Evans. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from West African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirchhoff, F. 2009. Is the high virulence of HIV-1 an unfortunate coincidence of primate lentiviral evolution? Nat. Rev. Microbiol. 7:467-476. [DOI] [PubMed] [Google Scholar]
- 29.Kirchhoff, F. 2010. Immune evasion and counteraction of restriction factors by HIV-1 and other primate lentiviruses. Cell Host Microbe 8:55-67. [DOI] [PubMed] [Google Scholar]
- 30.Kirchhoff, F., M. Schindler, A. Specht, N. Arhel, and J. Munch. 2008. Role of Nef in primate lentiviral immunopathogenesis. Cell Mol. Life Sci. 65:2621-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lama, J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr. HIV Res. 1:167-184. [DOI] [PubMed] [Google Scholar]
- 32.Liegeois, F., B. Lafay, P. Formenty, S. Locatelli, V. Courgnaud, E. Delaporte, and M. Peeters. 2009. Full-length genome characterization of a novel simian immunodeficiency virus lineage (SIVolc) from olive Colobus (Procolobus verus) and new SIVwrcPbb strains from Western Red Colobus (Piliocolobus badius badius) from the Tai Forest in Ivory Coast. J. Virol. 83:428-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim, E. S., and M. Emerman. 2009. Simian immunodeficiency virus SIVagm from African green monkeys does not antagonize endogenous levels of African green monkey tetherin/BST-2. J. Virol. 83:11673-11681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lundquist, C. A., M. Tobiume, J. Zhou, D. Unutmaz, and C. Aiken. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J. Virol. 76:4625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malim, M. H., and M. Emerman. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388-398. [DOI] [PubMed] [Google Scholar]
- 36.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCp, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565-574. [DOI] [PubMed] [Google Scholar]
- 37.McNatt, M. W., T. Zang, T. Hatziioannou, M. Bartlett, I. B. Fofana, W. E. Johnson, S. J. Neil, and P. D. Bieniasz. 2009. Species-specific activity of HIV-1 Vpu and positive selection of tetherin transmembrane domain variants. PLoS. Pathog. 5:e1000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miller, M. D., M. T. Warmerdam, I. Gaston, W. C. Greene, and M. B. Feinberg. 1994. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J. Exp. Med. 179:101-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munch, J., D. Rajan, M. Schindler, A. Specht, E. Rucker, F. J. Novembre, E. Nerrienet, M. C. Muller-Trutwin, M. Peeters, B. H. Hahn, and F. Kirchhoff. 2007. Nef-mediated enhancement of virion infectivity and stimulation of viral replication are fundamental properties of primate lentiviruses. J. Virol. 81:13852-13864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neil, S., and P. Bieniasz. 2009. Human immunodeficiency virus, restriction factors, and interferon. J. Interferon Cytokine Res. 29:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425-430. [DOI] [PubMed] [Google Scholar]
- 42.Paiardini, M., I. Pandrea, C. Apetrei, and G. Silvestri. 2009. Lessons learned from the natural hosts of HIV-related viruses. Annu. Rev. Med. 60:485-495. [DOI] [PubMed] [Google Scholar]
- 43.Pandrea, I., D. L. Sodora, G. Silvestri, and C. Apetrei. 2008. Into the wild: simian immunodeficiency virus (SIV) infection in natural hosts. Trends Immunol. 29:419-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plantier, J. C., M. Leoz, J. E. Dickerson, F. De Oliveira, F. Cordonnier, V. Lemée, F. Damond, D. L. Robertson, and F. Simon. 2009. A new human immunodeficiency virus derived from gorillas. Nat. Med. 15:871-872. [DOI] [PubMed] [Google Scholar]
- 45.Ronquist, F., and J. P. Huelsenbeck. 2003. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 10:1572-1574. [DOI] [PubMed] [Google Scholar]
- 46.Rotger, M., K. K. Dang, J. Fellay, E. L. Heinzen, S. Feng, P. Descombes, K. V. Shianna, D. Ge, H. F. Günthard, D. B. Goldstein, and A. Telenti, Swiss HIV Cohort Study, and Center for HIV/AIDS Vaccine Immunology. 2010. Genome-wide mRNA expression correlates of viral control in CD4+ T-cells from HIV-1-infected individuals. PLoS Pathog. 6:e1000781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sauter, D., A. Specht, and F. Kirchhoff. 2010. Tetherin: holding on and letting go. Cell 141:392-398. [DOI] [PubMed] [Google Scholar]
- 48.Sauter, D., M. Schindler, A. Specht, W. N. Landford, J. Munch, K. A. Kim, J. Votteler, U. Schubert, F. Bibollet-Ruche, B. F. Keele, J. Takehisa, Y. Ogando, C. Ochsenbauer, J. C. Kappes, A. Ayouba, M. Peeters, G. H. Learn, G. Shaw, P. M. Sharp, P. Bieniasz, B. H. Hahn, T. Hatziioannou, and F. Kirchhoff. 2009. Tetherin-driven adaptation of Vpu and Nef function and the evolution of pandemic and nonpandemic HIV-1 strains. Cell Host Microbe 6:409-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schaefer, T. M., I. Bell, M. E. Pfeifer, M. Ghosh, R. P. Trible, C. L. Fuller, C. Ashman, and T. A. Reinhart. 2002. The conserved process of TCR/CD3 complex down-modulation by SIV Nef is mediated by the central core, not endocytic motifs. Virology 302:106-122. [DOI] [PubMed] [Google Scholar]
- 50.Schindler, M., J. Schmokel, A. Specht, H. Li, J. Munch, M. Khalid, D. L. Sodora, B. H. Hahn, G. Silvestri, and F. Kirchhoff. 2008. Inefficient Nef-mediated downmodulation of CD3 and MHC-I correlates with loss of CD4+ T cells in natural SIV infection. PLoS. Pathog. 4:e1000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schindler, M., J. Munch, O. Kutsch, H. Li, M. L. Santiago, F. Bibollet-Ruche, M. C. Muller-Trutwin, F. J. Novembre, M. Peeters, V. Courgnaud, E. Bailes, P. Roques, D. L. Sodora, G. Silvestri, P. M. Sharp, B. H. Hahn, and F. Kirchhoff. 2006. Nef-mediated suppression of T cell activation was lost in a lentiviral lineage that gave rise to HIV-1. Cell 125:1055-1067. [DOI] [PubMed] [Google Scholar]
- 52.Schindler, M., S. Wurfl, P. Benaroch, T. C. Greenough, R. Daniels, P. Easterbrook, M. Brenner, J. Munch, and F. Kirchhoff. 2003. Down-modulation of mature major histocompatibility complex class II and up-regulation of invariant chain cell surface expression are well-conserved functions of human and simian immunodeficiency virus nef alleles. J. Virol. 77:10548-10556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silvestri, G., A. Fedanov, S. Germon, N. Kozyr, W. J. Kaiser, D. A. Garber, H. McClure, M. B. Feinberg, and S. I. Staprans. 2005. Divergent host responses during primary simian immunodeficiency virus SIVsm infection of natural sooty mangabey and nonnatural rhesus macaque hosts. J. Virol. 79:4043-4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sodora, D. L., J. S. Allan, C. Apetrei, J. M. Brenchley, D. C. Douek, J. G. Else, J. D. Estes, B. H. Hahn, V. M. Hirsch, A. Kaur, F. Kirchhoff, M. Muller-Trutwin, I. Pandrea, J. E. Schmitz, and G. Silvestri. 2009. Toward an AIDS vaccine: lessons from natural simian immunodeficiency virus infections of African nonhuman primate hosts. Nat. Med. 15:861-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stumptner-Cuvelette, P., S. Morchoisne, M. Dugast, G. S. Le, G. Raposo, O. Schwartz, and P. Benaroch. 2001. HIV-1 Nef impairs MHC class II antigen presentation and surface expression. Proc. Natl. Acad. Sci. U. S. A. 98:12144-12149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swigut, T., A. J. Iafrate, J. Münch, F. Kirchhoff, and J. Skowronski. 2000. Simian and human immunodeficiency virus Nef proteins use different surfaces to down-regulate class I major histocompatibility antigen expression. J. Virol. 74:5691-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swigut, T., M. Greenberg, and J. Skowronski. 2003. Cooperative interactions of simian immunodeficiency virus Nef, AP-2, and CD3-zeta mediate the selective induction of T-cell receptor-CD3 endocytosis. J. Virol. 77:8116-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takehisa, J., M. H. Kraus, A. Ayouba, E. Bailes, H. F. Van, J. M. Decker, Y. Li, R. S. Rudicell, G. H. Learn, C. Neel, E. M. Ngole, G. M. Shaw, M. Peeters, P. M. Sharp, and B. H. Hahn. 2009. Origin and biology of simian immunodeficiency virus in wild-living western gorillas. J. Virol. 83:1635-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 60.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Van Heuverswyn, F., and M. Peeters. 2007. The origins of HIV and implications for the global epidemic. Curr. Infect. Dis. Rep. 9:338-346. [DOI] [PubMed] [Google Scholar]
- 62.Willey, R. L., F. Maldarelli, M. A. Martin, and K. Strebel. 1992. Human immunodeficiency virus type 1 Vpu protein induces rapid degradation of CD4. J. Virol. 66:7193-7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang, S. J., L. A. Lopez, H. Hauser, C. M. Exline, K. G. Haworth, and P. M. Cannon. 2010. Anti-tetherin activities in Vpu-expressing primate lentiviruses. Retrovirology 7:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, F., S. J. Wilson, W. C. Landford, B. Virgen, D. Gregory, M. C. Johnson, J. Munch, F. Kirchhoff, P. D. Bieniasz, and T. Hatziioannou. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.