Abstract
The origin-binding domain (OBD) of simian virus 40 (SV40) large T-antigen (T-Ag) is essential for many of T-Ag's interactions with DNA. Nevertheless, many important issues related to DNA binding, for example, how single-stranded DNA (ssDNA) transits along the T-Ag OBD, have yet to be established. Therefore, X-ray crystallography was used to determine the costructure of the T-Ag OBD bound to DNA substrates such as the single-stranded region of a forked oligonucleotide. A second structure of the T-Ag OBD crystallized in the presence of poly(dT)12 is also reported. To test the conclusions derived from these structures, residues identified as being involved in binding to ssDNA by crystallography or by an earlier nuclear magnetic resonance study were mutated, and their binding to DNA was characterized via fluorescence anisotropy. In addition, these mutations were introduced into full-length T-Ag, and these mutants were tested for their ability to support replication. When considered in terms of additional homology-based sequence alignments, our studies refine our understanding of how the T-Ag OBDs encoded by the polyomavirus family interact with ssDNA, a critical step during the initiation of DNA replication.
The initiation of DNA replication in higher organisms depends upon numerous proteins and their associated protein-protein and protein-DNA interactions (reviewed in references 2, 23, and 31). DNA tumor viruses, in contrast, are able to initiate DNA replication with relatively few proteins. For instance, simian virus 40 (SV40) is able to catalyze the initiation of nascent-strand DNA synthesis from its origin with the virally encoded large T-antigen (T-Ag) and host-derived replication protein A (RPA), topoisomerase I, and polymerase α-primase complex (21, 47, 51). Indeed, it is anticipated that a molecular understanding of the initiation of DNA replication will result from further characterization of the structures of these proteins and their interactions with each other and with DNA (reviewed in references 5, 7, 14, and 42). Thus, SV40 and related viral model systems, such as the papillomaviruses (reviewed in reference 45), continue to serve as paradigms for the study of the initiation of DNA replication.
Because T-Ag initiates the replication process, there has been a considerable interest in T-Ag and its interactions with the SV40 origin of replication. Images from electron microscopy studies have provided many critical insights into the assembly of T-Ag into hexamers and double hexamers on the origin (10, 19, 32, 48, 49). A full-length T-Ag X-ray crystal structure is not yet available; however, structures are available for individual domains of T-Ag, including the N-terminal J domain (25), the origin-binding domain (OBD) (4, 30, 34, 35), and the C-terminal helicase domain (18, 29). Our focus has been on the structure of the T-Ag OBD and its interactions with DNA. In an initial study, we determined the structure of the T-Ag OBD by nuclear magnetic resonance (NMR) methods (30) and established that it is a monomer in solution. In subsequent crystallography studies, we determined the costructure of the T-Ag OBD bound in a site-specific manner to a GAGGC-containing duplex oligonucleotide (35). Furthermore, we solved a crystal structure of the T-Ag OBD in the absence of DNA, where it adopts an open ring structure containing six molecules per turn (34). This “lock-washer” or spiral hexamer structure contains a very basic channel that is thought to bind single-stranded DNA (ssDNA) during DNA unwinding (34) and a gap through which ssDNA may pass during the initiation process (10, 34). Assembly into the lock-washer structure positions the DNA binding A1 and B2 regions primarily on one face of the hexamer so that they can no longer engage the GAGGC sequences in the central region of the origin. This is one reason for the hypothesis that the lock-washer conformation forms after site-specific binding of T-Ag OBD monomers to the origin.
It has been reported that a significant portion of the ssDNA binding activity of T-Ag resides in the T-Ag OBD (36, 54). Indeed, the T-Ag OBD binds ssDNA with a greater affinity than non-sequence-specific duplex DNA (17, 41). Binding to ssDNA is mutually exclusive with site-specific binding to DNA containing GAGGC pentanucleotides (17), an indication that the same residues may be involved in both processes. Consistent with this hypothesis, we showed via NMR heteronuclear single-quantum coherence (HSQC) experiments that the binding site for ssDNA on the T-Ag OBD involves residues in the A1 and B2 loops which cluster on one surface of the spiral structure (40). However, the structure of the complex of ssDNA and the T-Ag OBD, either in its monomeric or hexameric form, remains unknown.
Hence, to extend the analyses of the interaction of ssDNA with the T-Ag OBD, we screened for crystal formation between the T-Ag OBD with a range of DNA templates containing regions of ssDNA. Herein, we report the costructure of the T-Ag OBD bound to the single-stranded region of an oligonucleotide serving as a mimic of a “forked DNA” structure. Of interest, this structure identified several T-Ag OBD residues not previously implicated in ssDNA binding. Therefore, we have mutated to alanines the T-Ag OBD residues that have been identified as being important for binding to ssDNA in either our crystallography or previous NMR studies. Upon purification, binding of the mutant T-Ag OBD proteins to ssDNA and forked DNA was analyzed by fluorescence anisotropy to determine the relative contributions of these residues to ssDNA binding. In addition, assays were conducted to determine the extent to which DNA replication was affected by these mutations. Finally, we also report the structure of the T-Ag OBD formed in the presence of poly(dT)12. In this structure, crystallographically related T-Ag OBDs arrange to form positively charged channels that could accommodate ssDNA; however, it is not clear if this crystallographic tetramer is biologically relevant.
MATERIALS AND METHODS
Molecular biology techniques. (i) Generation of mutants. (a) Generation of T-Ag OBD mutants.
Alanine substitutions were introduced at targeted positions using a QuikChange site-directed mutagenesis kit (Stratagene), plasmid pGEX TBD (24), and the oligonucleotides presented in Table S2 in the supplemental material and their complementary sequences. The thermocycling conditions consisted of an initial denaturation step at 95°C for 60s, followed by 12 cycles of denaturation at 95°C for 50s, annealing at 55°C for 60s, polymerase extension at 68°C for 11 min, and a final extension of 5 min. The composition of all mutant plasmids described in this report was confirmed by the dideoxy sequencing method at the Tufts Core Facility.
(b) Generation of mutations in pCMVneoT-Ag.
The ability of full-length T-Ag containing the desired mutations to catalyze DNA replication was analyzed as previously described (16; see also below). The mutations were introduced into the T-Ag-encoding plasmid pCMVneoT-Ag using a QuikChange kit, plasmid pCMVneoT-Ag (8), and the primers listed in Table S2 and their complementary sequences. The thermocycling conditions consisted of an initial denaturation step at 95°C for 60s, followed by 12 cycles of denaturation at 95°C for 50s, annealing at 55°C for 60s, polymerase extension at 68°C for 17 min, and a final extension at 68°C for 5 min.
(ii) Purification and storage of the T-Ag OBD mutants.
A protocol for the purification of amino acids (aa) 131 to 260 of the T-Ag OBD (OBD131-260) was previously reported (24, 35). The T-Ag OBD131-260 mutants were purified as described previously with the exception that the Sephacryl S-100 column was replaced with a Superdex 75 10/300 GL column. Upon purification, the T-Ag OBD131-260 mutants were stored in T-Ag storage buffer (20 mM Tris-HCl [pH 8.0], 50 mM NaCl, 1 mM EDTA, 0.1% β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, 0.2 μg of leupeptin per ml, 0.2 μg of antipain per ml, and 10% glycerol) at −80°C until ready for use.
(iii) Fluorescence anisotropy binding assays.
Fluorescence anisotropy-based binding assays employing the wild-type (wt) T-Ag OBD have been previously described (46). The reactions were conducted with the T-Ag OBD131-260 or mutant forms of the T-Ag OBD131-260 and with ssDNA or forked DNA substrates. The ssDNA probes were either 5′-fluorescein-TTTTTTTTTTTTTTTTTTTTTTTT (24 thymidine residues) or 5′-fluorescein-CCAGAATCATTCTTGCTGTAGAGC-3′. The forked DNA substrate was obtained by annealing the oligonucleotides 5′-fluorescein-CCAGAATCATTCTTGCTGTAGAGC-3′ and 3′-ATGACGCATAAGAACGACATCTCG-5′ as previously described (17). DNA binding assays were conducted using a binding buffer consisting of 20 mM Tris, pH 7.4, 50 mM NaCl, 1 mM dithiothreitol (DTT) and 0.01% NP-40.
(iv) Transient DNA replication assay.
A quantitative assay for studies of the replication of polyomavirus and papillomavirus was recently described (16). This assay was used to measure the replication ability of T-Ags containing point mutations at residues implicated in binding to ssDNA. Briefly, ∼5 × 104 C33A cells were transfected with three plasmids encoding, respectively, T-Ag, the minimal origin of DNA replication together with a firefly luciferase reporter gene, and Renilla luciferase. Replication of the origin-containing plasmid was quantified 72 h posttransfection by measuring the levels of firefly and Renilla luciferase activities using a Dual-Glo Luciferase assay system (Promega). Each T-Ag mutant was analyzed in duplicate in two separate experiments. Error bars represent the standard deviations.
(v) Western blotting assay.
Western blotting was performed via standard methods; extracts were prepared 24 h posttransfection. T-Ag proteins were detected using a mouse monoclonal anti-T-Ag antibody recognizing an epitope located within the N-terminal 82 amino acids of the protein (Sc-148; Santa Cruz). Tubulin was used as a loading control.
Biophysical techniques. (i) Crystallography. (a) DNA purification.
Synthetic oligonucleotides were made using the phosphoramidite method (Integrated DNA Technologies). The sequences used to form the 23-mer artificial fork were(5′ to 3′) ACTCCTCCGAAAAAACCTCCGGA and GAGGAGGCTTTTTTGGAGGCCTT. The sequence used for the single-strand crystallographic studies was poly(dT)12 (TTTTTTTTTTTT).
These oligonucleotides were purified via high-performance liquid chromatography (HPLC) by anion-exchange chromatography (DNAPak; Vydac, Inc.). The eluted DNA was lyophilized, desalted on a PD-10 column (GE, Inc.), and lyophilized again to dryness. The oligonucleotides were resuspended in annealing buffer (10 mM Tris, pH 7.5, 50 mM NaCl), and complementary strands were mixed in a 1:1 molar ratio and annealed by heating to 95°C for 3 min, followed by slow cooling to 4°C. The duplex DNA was stored at −20°C.
(b) Crystallization.
The T-Ag OBD131-260 and forked DNA complex was prepared in a 2:1.2 molar ratio. The T-Ag OBD131-260 and poly(dT)12 complex was prepared in a 1:2 molar ratio. The complexes were concentrated by ultrafiltration using a Vivaspin 10,000-molecular-weight-cutoff device to a protein concentration of ∼10 mg/ml, aliquoted in small volumes, quick-frozen in liquid nitrogen, and then stored at −80°C.
Crystals of the T-Ag OBD131-260-forked DNA complex were grown by vapor diffusion at 4°C by mixing 1 μl of complex with 1 μl of a reservoir solution (100 mM Tris, pH 8.5, 300 mM NH4Cl, 28% polyethylene glycol [PEG] 4000, 20 mM CaCl2).
Crystals of the T-Ag OBD131-260-poly(dT)12 complex were grown by vapor diffusion at 18°C by mixing 1 μl of complex with 1 μl of a reservoir solution (0.2 M thiocyanate [SCN], 17.5% PEG 3350, 5% glycerol).
(c) X-ray data collection and structure solution.
The crystals were transferred to a cryogenic solution using a microloop (Hampton Research Inc.) and then frozen in liquid nitrogen. The cryogenic solution used for the T-Ag OBD131-260-forked DNA complex consisted of 0.1 M Tris, pH 8.5, 300 mM NH4Cl, 30% PEG 4000, 20 mM CaCl2, and 15% ethylene glycol. The cryogenic solution used for the T-Ag OBD131-260-poly(dT)12 complex crystals was 0.2 M SCN, 25% PEG 3350, and 15% glycerol. X-ray data were collected at 100 K using an ADSC Quantum-315r, nine-quadrant, charge-coupled-device (CCD) detector at National Synchrotron Light Source (NSLS) beamline X-29 (Brookhaven National Laboratory, NY). The X-ray data were processed with the HKL2000 suite (38). Both structures, that of the T-Ag OBD131-260-forked DNA complex and that of T-Ag OBD131-260-poly(dT)12 complex, were solved by molecular replacement using apo-T-Ag OBD coordinates as a search model (RCSB Protein Data Bank [PDB] codes 2IF9 and 2FUF, respectively) and the program PHASER (33). The structures were refined using the program REFMAC (37) within the CCP4 suite of programs (9), and the program PHENIX (1). The resulting maps and models were visualized and improved with the molecular graphics program COOT (12) and PyMOL (11). All molecular graphics figures in this paper were made with PyMOL.
(ii) CD analyses of the purified T-Ag OBD131-260 and the T-Ag OBD131-260 mutants.
To confirm that the individual mutations do not change the overall fold of the proteins, wt and mutant T-Ag OBDs were analyzed by circular dichroism (CD). Spectra were recorded at 25°C, using a 1-mm-path-length cell in a Jasco J-810 spectropolarimeter. Each protein sample was at a concentration of 0.15 mg/ml in 5 mM sodium phosphate, 50 mM sodium fluoride, and 1 mM Tris (2-carboxyethyl)phosphine (TCEP), pH 7.0. CD data were saved in ASCII format and processed in Microsoft Excel. Ellipticity data were expressed as the mean residue molar ellipticity, or [θ]MRW, which was calculated as follows: [θ]MRW = (θobs × 100 × MRW)/c × l. In this formula MRW is the mean residue weight, calculated by dividing the molecular weight for T-Ag OBD131-260 (15,074 g/mole) by the number of amino acid residues (130 aa). θobs is the observed ellipticity expressed in degrees, c is the protein concentration in mg/ml (0.15 mg/ml), and l is the cell path length in cm (0.1 cm).
RESULTS
Structures of the T-Ag OBD with substrates containing ssDNA. (i) Costructure of the T-Ag OBD with a forked DNA substrate.
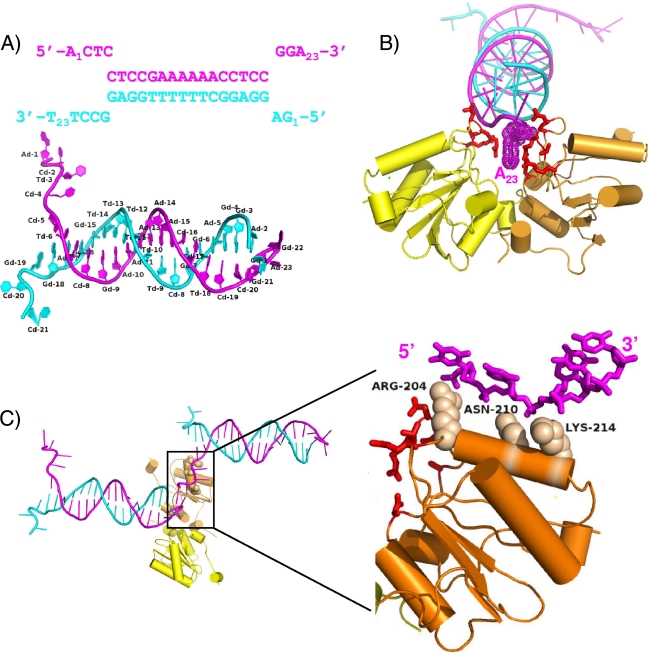
The 1.7-Å resolution crystal structure of T-Ag OBD131-260 in complex with a nonspecific forked DNA substrate is presented in Fig. 1. The X-ray data collection and refinement statistics are presented in Table S1 in the supplemental material. The refined structure has good geometry and an Rfactor and Rfree of 17.8% and 22.3%, respectively. The final model contains two molecules of T-Ag OBD131-260, each having a modified Cys-β-mercaptoethanol adduct, 1 acetate molecule, 3 calcium ions, 2 ethylene glycol molecules, 298 water molecules, and 43 out of 46 possible nucleic acid bases. As shown in Fig. 1A, the DNA is frayed at each end of the duplex. The frayed DNA ends are involved in stabilizing lattice contacts with neighboring DNA molecules. The DNA forms a continuous pseudo-double helix along one axis of the crystal, with ssDNA extending perpendicularly from this axis. Thus, the DNA provides a stable scaffold for the T-Ag OBDs.
FIG. 1.
The costructure of the SV40 T-Ag OBD bound to a forked DNA oligonucleotide. (A) The sequence of the forked DNA target used in crystallization indicating duplex DNA and ssDNA regions. The top strand is magenta, and the bottom strand is cyan. The first and last bases are numbered. Shown below, the labeled crystal structure of the DNA clearly depicts the frayed ends of the DNA. (B) Ribbon representation of T-Ag OBD interactions with the duplex portion of the DNA, including the 3′ terminal dA of the top strand. The T-Ag OBD molecules are shown in yellow and orange. The interacting protein residues are shown as red sticks. A Van der Waals dot surface is shown for the 3′ terminal adenine base to highlight the stacking interactions provided by the T-Ag OBDs. (C) Ribbon representation of T-Ag OBD interactions with single-stranded DNA. The T-Ag OBDs are colored as in panel B. A neighbor (symmetry-related) DNA molecule that interacts with the orange T-Ag OBD is shown at left. A close-up of the boxed region is shown (and rotated ∼90°) at right, where only the interacting ssDNA is shown for clarity. Van der Waals spheres are shown in tan for the side chains of the residues that interact with the ssDNA phosphate backbone.
The asymmetric unit contains two T-Ag OBD molecules bound to the forked DNA substrate (Fig. 1B). Superposition of the two crystallographically unique T-Ag OBDs in this structure shows that they have identical conformation (root mean square deviation [RMSD] of 0.22 Å over 133 Cαs). Interestingly, the overall conformation of each T-Ag OBD in this structure resembles that of the apo-OBD (DNA-free) conformer (34), not the GAGGC-bound T-Ag OBD conformer (35). Thus, this structure provides a view of how the previously defined “DNA-free” form of T-Ag OBD can interact with fork-containing DNA.
The two T-Ag OBDs are oriented with the A1 motifs (43) (residues 147 to 159) facing each other. The protein-protein interactions are depicted as a schematic in Fig. S1 in the supplemental material. The OBDs are positioned at one end of the duplex, near a junction formed with neighboring crystallographically related DNA molecules. Two different sets of protein-DNA interactions are observed involving the duplex portion of one forked DNA molecule and the ssDNA region of a neighboring DNA molecule, respectively. In the first set of interactions, the OBDs interact with the phosphate backbone at the junction of DNA strands that includes symmetry-related DNA molecules. The OBDs sandwich the terminal adenine base at the 3′ end of the top strand (adenine 23) through the A1 motif, primarily through a stacking interaction with R154 and, to a lesser extent, N227 (Fig. 1B). The residues involved in this first set of protein-DNA interactions include A1 motif residues A149, S152, N153, R154, and T155 and B2 motif residue R202 and N227 (shown in red in Fig. 1B). The second set of protein-DNA contacts shows how T-Ag OBD interacts with ssDNA (Fig. 1C). The T-Ag OBD interacts with the phosphate backbone of a neighboring (symmetry-related) ssDNA region (the 5′ end of the top strand having the sequence 5′ ACTC) (Fig. 1A) through the side chains of residues R204, N210, and K214. Specifically, the electron density shows that R204 is in an extended conformation and that it interacts with the phosphate oxygens through its guanidinium group. Next, the N210 makes a weak hydrogen bond with one phosphate oxygen and a water-mediated interaction with another. Lastly, the electron density for the side chain K214 is weak although it could clearly form a hydrogen bond with either of the oxygen atoms from adjacent phosphates.
(ii) Structure of the T-Ag OBD in the presence of poly(dT)12.
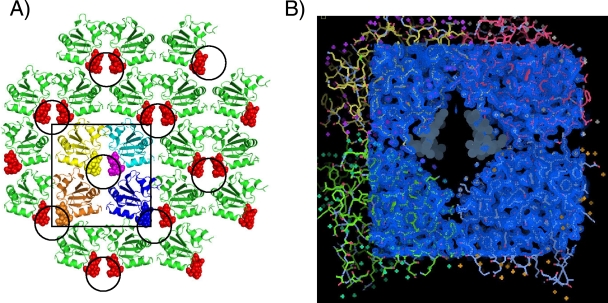
The 1.65-Å resolution crystal structure of T-Ag OBD cocrystallized in the presence of a 12-mer of poly(dT) is presented in Fig. 2. The X-ray data collection and refinement statistics are presented in Table S1 in the supplemental material.
FIG. 2.
The crystal structure of the SV40 T-Ag OBD grown in the presence of poly(dT)12 substrate. (A) View of the crystallographic lattice. The arrangement of T-Ag OBD molecules in the lattice is shown as ribbon diagrams, and the poorly modeled A1 motif residues are shown as spheres. The disordered A1 residues occur within a solvent channel that is made up of four T-Ag OBD molecules, highlighted by the square, and the four T-Ag OBDs are shown in yellow, cyan, brown, and blue. The circles indicate the location of the solvent channels in the crystal structure. This arrangement positions two A1 motifs inside the channel; the other two participate in neighboring channels. (B) Electron density map of the T-Ag OBD crystallographic tetramer. This view shows a 2Fo-Fc (observed and calculated structure factor amplitudes, respectively) electron density map (blue) covering four T-Ag OBD molecules, shown in cyan, green, yellow, and red. The position of the disordered A1 residues are shown as gray dot surfaces and occur within a positively charged solvent channel.
This crystal form contains one molecule of T-Ag OBD per asymmetric unit. The structure has been refined to good geometry, with an Rfactor and Rfree of 16.6% and 19.5%, respectively. Residues 148 to 154 within the DNA binding A1 motif are disordered (hence, not well modeled), as seen in Fig. 2B. This disorder is unusual as this region has been ordered in all previous crystal structures (4, 34, 35), both in the presence and absence of DNA. Unfortunately, we were also unable to model any ssDNA in this structure. However, we infer that DNA is present in this structure as this new crystal form occurs only in the presence of ssDNA. Furthermore, these crystals fluoresce when soaked with a DNA-sensitive fluorescent dye (SYBR Gold; Invitrogen) (data not shown), suggesting the presence of ssDNA. Inspection of the crystal lattice reveals that the disordered A1 loop lies in a positively charged channel formed by four symmetry related T-Ag OBDs (Fig. 2A).
This arrangement positions two A1 motifs in the central portion of the channel (Fig. 2A). In addition, other residues (165, 166, 169, 170, 172 to 174, 176, 186, 188, 191, 193, 201, 202, 204, 227, 228, 243, and 245) are solvent accessible and also line this channel. Some of these residues have been implicated in DNA binding, for example, those in or near the B2 motif (H201, R202, and R204). The channel is oblong, and the diameter of the channel is consistent with the size of ssDNA (∼12 Å, in the shortest dimension). Finally, the T-Ag OBD131-260 residues at the interfaces of the crystallographically related molecules are presented in Fig. S2 in the supplemental material. It is noted that these protein-protein interfaces are not symmetric.
As mentioned earlier, the previous crystal structures of the T-Ag OBD clustered into two conformations: an apo (DNA-free) form (34) and a DNA-bound form (35). In these previous structures, the largest conformational change upon binding DNA occurs in the A1 motif, which acts as a toggle switch having an “on” (DNA bound) and “off” (DNA free) conformation (35). F151 is at the apex of this loop, having the greatest conformational change upon binding DNA. In addition, in all previous crystal structures of the T-Ag OBD, the A1 motif has always been well ordered, whether in the free or bound form. Thus, in this structure with ssDNA, we infer that the A1 motif is changing conformation between these states and interacting with the ssDNA although we do not have structural details of this interaction.
Determining the relative ability of the T-Ag-OBD mutants to bind ssDNA. (i) Purification and characterization of the T-Ag OBD mutants.
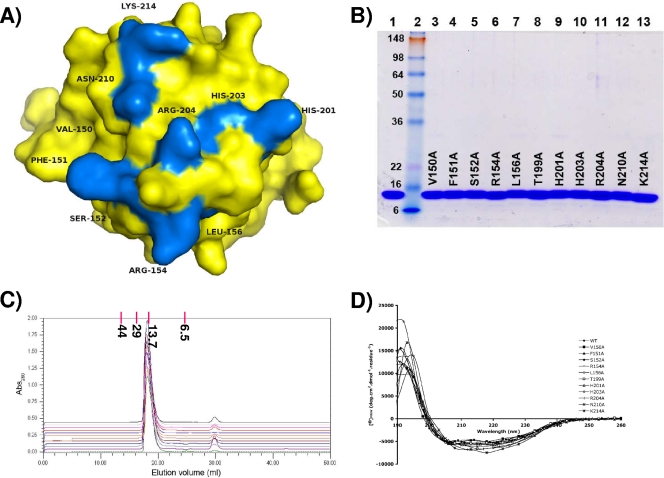
The results from our current crystallography experiments and from our previous NMR studies (40) implicated a number of residues in the T-Ag OBD as potential binders to ssDNA (i.e., residues 150 to 152, 154, 156, 199, 201, 203, 204, 210, and 214). The relative positions of these residues on a surface representation of the T-Ag OBD is presented in Fig. 3A; it is apparent that these residues cluster on one surface of the molecule in a region that contains the DNA binding A1 (aa 147 to 159) and B2 motifs (aa 203 to 207) (43, 55). However, it is not clear if particular residues are necessary for binding to ssDNA. Therefore, the codons encoding the residues potentially engaged in binding to ssDNA were mutated to alanines, and the individual T-Ag OBDs were purified using previously described protocols (Materials and Methods).
FIG. 3.
Residues implicated in binding to ssDNA by either X-ray crystallography or NMR methods and therefore mutated to alanine. (A) T-Ag OBD is shown as a surface representation in yellow. The residues mutated to alanine are shown in blue and labeled. These include A1 residues 150 to 152, 154, and 156, B2 residues 203 and 204, and additional residues 199, 201, 210, and 214. (B) SDS-PAGE analysis of purified T-Ag OBD samples containing the indicated point mutations. As a control, an aliquot of wt T-Ag OBD131-260 (5.0 μg) was loaded in lane 1. Lane 2 contains See Blue Plus 2 prestained protein size markers (10 μl; Invitrogen); the molecular masses of the individual markers are indicated in kDa. Aliquots (5.0 μg) of the individual T-Ag OBD131-260 mutants are displayed in lanes 3 to 13; the particular mutation present in a given T-Ag OBD molecule is indicated. The gel is stained with Coomassie blue. (C) Evidence that, relative to wt T-Ag OBD131-260, the individual T-Ag OBD131-260 mutants are not structurally altered. Chromatograms are shown of the wt T-Ag OBD131-260 and the T-Ag OBD131-260 mutants eluted from a Superdex 75 10/300 GL column in T-Ag storage buffer (see Materials and Methods). The baselines of the chromatograms are offset on the y axis (absorbance at 280 nm [Abs280]) in order to separate them. The wt T-Ag OBD131-260 chromatogram (green) is at the bottom of the Abs280 axis, followed by (in ascending order, to the top of the series) the mutants V150A, F151A, S152A, R154A, L156A, T199A, H201A, H203A, R204A, N210A, and K214A. Elution position and molecular mass (in kDa) of gel filtration standards are indicated at the top of the graph. (D) CD analyses of wt T-Ag OBD131-260 and the T-Ag OBD131-260 mutants. The spectra associated with the individual mutants are identified on the figure.
An SDS-PAGE (12% acrylamide) gel of the 11 mutant proteins analyzed in this study is presented in Fig. 3B. Given that all of the substitutions were introduced into surface residues, it is highly unlikely that significant conformational changes resulted from any of these mutations. This conclusion is supported by the gel filtration results (Fig. 3C) and CD analyses (Fig. 3D) of the individual proteins.
(ii) Establishing the relative binding affinity of the mutant T-Ag OBDs for substrates containing ssDNA.
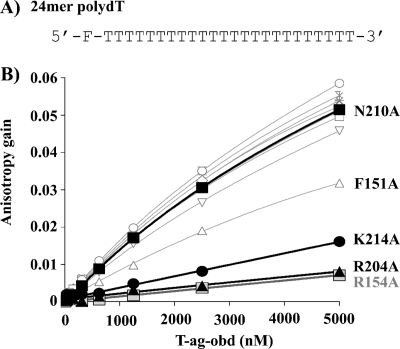
The relative affinity of the T-Ag OBD mutants for ssDNA was measured by fluorescence anisotropy as previously described (reference 17 and references therein). The results of experiments conducted with poly(dT)24 are presented in Fig. 4. Inspection of Fig. 4B establishes that two of the three residues identified by the X-ray costructure as contacting the sugar-phosphate backbone of the ssDNA portion of the forked DNA substrate (i.e., B2 residue R204 and K214) are critical for binding to poly(dT)24. In addition, A1 residue R154 is also critical for binding to this single-stranded substrate. A1 residue F151 also appears to play a significant role in binding to single-stranded DNA. In contrast, the other residues (i.e., V150, S152, L156, T199, H201, H203, and the crystallographically identified residue N210) are not important for binding to poly(dT)24. It should be noted that in contrast to our results with the H201A mutant, it was recently reported that T-Ags containing the H201F and H201N mutants were slightly defective in terms of their ability to bind a 55-mer nonspecific ssDNA (15). Finally, virtually identical results were obtained when these experiments were repeated with a 24-mer having the sequence 5′-fluorescein-CCAGAATCATTCTTGCTGTAGAGC-3′ (data not shown).
FIG. 4.
Binding of T-Ag OBD proteins to poly(dT) DNA measured by fluorescence anisotropy. (A) Sequence of the 24-nucleotide poly(dT) probe used in this study. (B) Measurement of the T-Ag OBD-poly(dT) interaction via fluorescent anisotropy. Binding isotherms were measured in triplicate with 15 nM single-stranded DNA probe and increasing concentrations of the wild-type T-Ag-OBD (○) or of the V150A (□), F151A (Δ), S152A (▿), R154A ( ), L156A (+), T199A (⋄), H201A (*), H203A (×), R204A (▴), N210A (▪), or K214A (•) mutant protein. Curves were obtained by a nonlinear regression and fitting to a one-binding-site equilibrium. Error bars are not visible on the graph as they are smaller than the size of the symbols. Kds (dissociation constants) were not calculated for these binding curves because they are not saturated although the relative affinities may be compared.
), L156A (+), T199A (⋄), H201A (*), H203A (×), R204A (▴), N210A (▪), or K214A (•) mutant protein. Curves were obtained by a nonlinear regression and fitting to a one-binding-site equilibrium. Error bars are not visible on the graph as they are smaller than the size of the symbols. Kds (dissociation constants) were not calculated for these binding curves because they are not saturated although the relative affinities may be compared.
(iii) Establishing the relative binding affinity of the mutant T-Ag-OBDs for a forked DNA substrate.
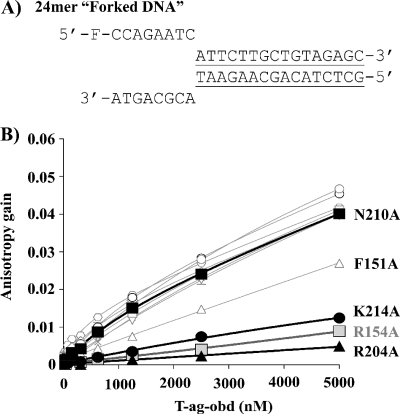
A forked DNA substrate (that does not contain a GAGGC duplex region) was used in the crystallography studies that identified residues involved in ssDNA binding. Therefore, the binding studies were repeated with a similar model of a forked DNA substrate (Fig. 5A). Of interest, very similar results were obtained with this forked substrate as with the poly(dT)24; i.e., residues R154 (A1), R204 (B2), and K214 are critical for binding, F151 has an intermediate phenotype, but the rest of the residues do not appear to play important roles in this process (Fig. 5B). These results may simply reflect that the T-Ag OBD has a ∼10-fold higher affinity for ssDNA than for nonspecific duplex DNA (17, 40); thus, only the interactions with the single-stranded region of the molecule are detected. In summary, these studies confirm the important role that T-Ag OBD residues F151, R154, R204, and K214 play in ssDNA binding.
FIG. 5.
Binding of T-Ag OBD proteins to forked DNA measured by fluorescence anisotropy. (A) Nucleotide sequence of the forked DNA probe used in this study. The double-stranded region is underlined. (B) Measurement of the T-Ag OBD-forked DNA interaction via fluorescent anisotropy. Binding isotherms were measured in triplicate with 15 nM probe and increasing concentrations of the wild-type T-Ag OBD (○) or of the V150A (□), F151A (▵), S152A (▿), R154A (gray square), L156A (+), T199A (⋄), H201A (*), H203A (×), R204A (▴), N210A (▪), or K214A (•) mutant protein. Curves were obtained by a nonlinear regression and fitting to a one-binding-site equilibrium. Error bars are not visible on the graph as they are smaller than the size of the symbols.
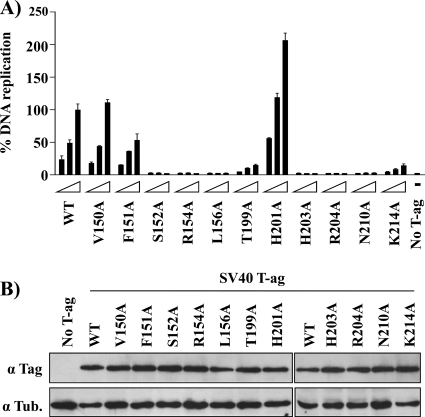
Testing the ability of T-Ag molecules harboring the mutations implicated in binding to ssDNA to catalyze DNA replication.
It is apparent from Fig. 4 and 5 that several of the T-Ag OBD mutants analyzed in this study are defective for binding to ssDNA, a property that suggests that they would be defective for DNA replication. In the context of full-length T-Ag, several of these residues were previously mutated (reviewed in reference 40) but not to alanines, while others (e.g., N210) were never tested for their ability to support DNA replication. Therefore, the ssDNA binding mutations were introduced into T-Ag (Materials and Methods), and their ability to support DNA replication was analyzed (Fig. 6). As expected from previous studies using mutant T-Ags with different substitutions at these positions (reviewed in reference 40), alanine mutations at basic residues implicated in binding to ssDNA (i.e., R154A, R204A, and K214A) are also defective in DNA replication. Regarding aromatic residue F151, its mutation to alanine generated a molecule having approximately half the replication activity of wild-type T-Ag. However, the F151Y mutation was previously demonstrated to be replication defective in vivo (44, 55), thus confirming that it plays a significant role in viral replication. In addition, a number of residues not implicated in binding to ssDNA (i.e., S152, L156, T199, H203, and N210) are nevertheless important for DNA replication, which is additional evidence that they play multiple roles in DNA replication that are distinct from binding to ssDNA (reviewed in reference 40). For example, previous mutational studies of full-length T-Ag in the A1 and B2 elements (i.e., mutations A149G, F159Y, and H203N) revealed that these residues were necessary for binding to ssDNA containing helicase substrates (44). Finally, it is of interest that, relative to the wild type, the H201A mutant had an increased level of DNA replication. Consistent with this observation, it was recently reported that full-length T-Ags containing the H201F and H201N mutations have higher levels of replication than the wild type (15). The physical basis for the enhancement of replication observed upon mutation of residue H201 is not currently understood.
FIG. 6.
DNA replication activity of large T-Ag mutant proteins. (A) Transient DNA replication activities of the indicated wild-type or mutant T-Ag proteins in C33A cells. DNA replication activities were measured by determining the ratio of firefly (Fluc-ori plasmid) to Renilla (Rluc control plasmid) luciferase activity as described in Materials and Methods. Replication activities are reported as a percentage of the Fluc/Rluc ratio obtained with the largest amount of wild-type T-Ag expression vector. Cells transfected with vector only (No T-Ag) were used as a negative control. (B) Expression of T-Ag proteins. Western blot analysis of total protein extracts prepared from transfected C33A cells expressing the wild-type or the indicated T-Ag mutant proteins. α, anti.
DISCUSSION
The T-Ag origin-binding domain plays multiple roles in DNA replication. Reflecting this functional diversity, the T-Ag OBD is highly dynamic in terms of the different structures and oligomeric forms that it can adopt. Indeed, the two crystal structures presented here further demonstrate how this small domain can reorganize to interact with different DNA substrates and generate alternative complexes with new protein-protein surfaces. The costructure with the forked DNA substrate revealed novel protein-DNA interactions through the A1 motifs as well as revealing how residues R204, N210, and K214 can interact with the backbone of ssDNA. The structure of T-Ag OBD grown in the presence of ssDNA showed another possible mode of assembly of the T-Ag OBDs that could accommodate ssDNA within positively charged solvent channels. In this structure, residues in the DNA binding A1 motif were disordered (in particular, F151), implying that multiple conformations of this loop exist in the crystal.
Our previous NMR study of the purified T-Ag OBD with poly(dT)25 suggested that residues involved in binding to ssDNA were primarily from the A1 and B2 regions (40). The greatest chemical shift changes occurred at R204 within the B2 motif. The structure of the T-Ag OBD bound to a single-stranded region of a forked DNA oligonucleotide (Fig. 1C) confirmed the importance of B2 residue R204 and provided further evidence that residues beyond the B2 region (i.e., N210A and K214A) are important in binding to ssDNA. Extending these structure-based experiments, the fluorescence anisotropy studies confirmed that A1 residues F151 and R154 and B2 residue R204 and residue K214 play very important roles in ssDNA binding. Finally, differences between the residues needed for binding to ssDNA (Fig. 4) and forked DNA substrates (Fig. 5) were not detected in this study.
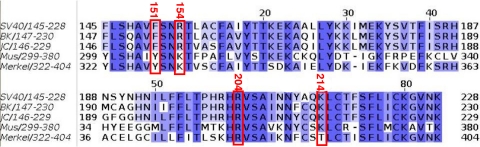
The question of what amino acid residues in the single-stranded DNA binding proteins from eukaryotic (3) and prokaryotic organisms (6, 39) are optimal for interactions with ssDNA has been previously addressed. The research has established that there are two primary types of interactions with DNA: aromatic residues stack with individual bases while electrostatic interactions occur between side chains and both the phosphate backbone and particular bases. This theme of protein-ssDNA interactions being mediated by aromatic and electrostatic interactions has been noted in related studies (e.g., PriB-ssDNA [20]). Thus, our conclusions that binding of the T-Ag OBD to ssDNA is dependent upon basic (i.e., R154, R204, and K214) and aromatic (i.e., F151) residues are consistent with these previous studies. Also of interest is the evidence presented in Fig. 7 that these residues are highly conserved among the polyomaviruses. Thus, the routing of ssDNA through the T-Ag OBDs encoded by these viruses may be highly conserved.
FIG. 7.
Sequence alignment of several T-Ag OBDs. The programs Clustal W (27) and Jalview (50) were used to calculate and display the sequence alignment of several representative T-Ag OBDs. The amino acids are colored by sequence identity (a color gradient from dark blue, indicating 100% identity, to white, indicating 0% identity). The red boxes indicate the location of the four residues identified as being important in ssDNA binding in this study; the SV40 T-Ag amino acid numbers are also presented. The sequences used in this alignment are from the polyomavirus members SV40, BK, JC, mouse (Mus), and Merkel cell carcinoma.
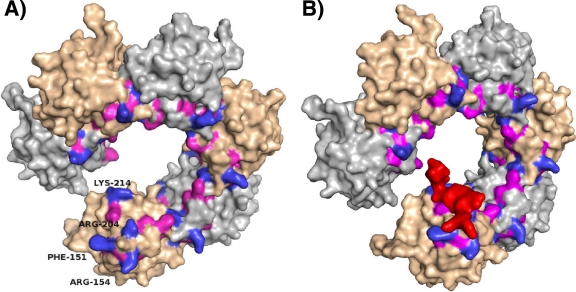
Currently, the structural details of assembly of T-Ag onto origin DNA and subsequent unwinding are not clear. T-Ag double-hexamer formation at the origin of replication is a complicated process whereby T-Ag generates ssDNA via its β-hairpin motif located in the helicase domain and assembles into opposing hexamers around a single strand of DNA (26, 29, 41). To date, no high-resolution structure exists of this process. Our studies focus on the role of the T-Ag OBD in these processes, and our structures are a snapshot of the initiation process. In particular, the question arises as to the “path” taken by the DNA during replication. We previously proposed that in the context of T-Ag, the T-Ag OBD is in a dynamic equilibrium between monomer and hexamer forms (26, 35). The T-Ag OBD forms a hexameric open-ring structure in the crystal, having a positively charged channel that could accommodate two opposing strands of ssDNA as well as providing a gap through which ssDNA could exit (34). Recent electron microscopy studies of full-length T-Ag have provided additional evidence for an open-ring structure of the T-Ag OBDs (10). Figure 8 A depicts the relative positions of the critical ssDNA binding residues identified in this study mapped onto the lock-washer hexameric structure. It is apparent that on the gap-proximal T-Ag OBD monomer, residues F151, R154, R204, and K214 (shown in blue) are completely accessible while they are less accessible (indeed, F151 is completely buried) in the other subunits. The T-Ag OBD is connected to the helicase domain by a flexible linker, and hence much of the surface of the T-Ag OBD hexamer remains solvent accessible even in the context of full-length T-Ag. In addition, the T-Ag OBD is likely to be highly dynamic, forming and unforming, so even which T-Ag OBD acts as the gap-proximal subunit (where all ssDNA critical residues are exposed) may change. The accessible basic residues R154, R204, and K214 would presumably be involved in electrostatic interactions with the sugar-phosphate backbone of DNA while the aromatic group of F151 would stack with the bases, and the A1 motif can toggle between the two known conformations. Thus, these residues are arranged in a manner that suggests a path for one single strand of DNA over the T-Ag OBD surface. A model depicting the transit of ssDNA over the surface of the lock-washer is shown in Fig. 8B. The second strand of DNA could also pass through the inner surface of the T-Ag OBD hexamer making less specific electrostatic interactions (34). Complementary to our results, a recent study (15) conducted with full-length T-Ag investigated the role of particular residues lining the inner surface of the lock-washer conformation of the T-Ag OBD on binding to ssDNA (i.e., E177, K178, Y179, H201, Y211, and K214). Binding to ssDNA was modestly affected by conservative mutations and to a greater extent by nonconservative mutations (particularly K178E and H201N).
FIG. 8.
ssDNA binding mutations mapped onto the surface of the T-Ag OBD spiral hexamer. (A) The spiral hexamer of T-Ag OBD generated from the crystal structure (RCSB PDB code 2FUF) is shown as a solvent-accessible surface representation. The individual subunits are shown in alternating tan and gray. Residues that were mutated in this study are shown as blue or magenta. The blue residues are critical for binding ssDNA and are labeled in the terminal subunit. The magenta residues are not critical for ssDNA binding and are not labeled. (B) Depiction of ssDNA transiting over the lock-washer structure of T-Ag OBD via the path described herein. This figure was made by superposition of coordinates of the T-Ag OBD of the forked DNA complex onto the T-Ag OBD of the lock-washer structure. The resulting position of the short sequence of ssDNA (5 bases) is shown in red and displayed as a surface representation. This figure illustrates the relative size of ssDNA to the channel.
In addition to binding to ssDNA, the T-Ag OBD is involved in other interactions critical for initiation of DNA replication. For instance, the interaction between T-Ag and the single-stranded DNA binding protein RPA is mediated via the T-Ag OBD and domains within the RPA 70-kDa subunit (52; reviewed in references 13 and 53). Therefore, it is of interest that a positively charged patch in the T-Ag OBD containing the residues R154, R202, and R204 is important for both the interaction with the RPA 70-kDa subunit (22) and with ssDNA. How the same T-Ag OBD residues can bind both the 70-kDa subunit of RPA and ssDNA is not currently clear. One possibility is that as RPA expands from its 8- to 10-nt binding mode to its 30-nt mode, it no longer binds to the T-Ag OBD, a process that would promote the next cycle of T-Ag OBD interactions with ssDNA (22). However, additional studies are needed to address this issue and to extend our structural understanding of the interactions that occur between the T-Ag OBD and its cellular binding partners during initiation of DNA replication.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health to P.A.B. (9R01GM55397) and from the Canadian Institutes for Health Research to J.A. A.F.-T. holds a studentship from the Fonds de la Recherche en Santé du Québec (FRSQ).
We thank Howard Robinson at beamline X29 at the National Synchrotron Light Source, Brookhaven National Laboratory, for collection of the X-ray data.
Footnotes
Published ahead of print on 27 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Adams, P. D., P. V. Afonine, G. Bunkoczi, V. B. Chen, I. W. Davis, N. Echols, J. J. Headd, L. W. Hung, G. J. Kapral, R. W. Grosse-Kunstleve, A. J. McCoy, N. W. Moriarty, R. Oeffner, R. J. Read, D. C. Richardson, J. S. Richardson, T. C. Terwilliger, and P. H. Zwart. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bell, S. P., and A. Dutta. 2002. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 71:333-374. [DOI] [PubMed] [Google Scholar]
- 3.Bochkarev, A., R. A. Pfuetzner, A. M. Edwards, and L. Frappier. 1997. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature 385:176-181. [DOI] [PubMed] [Google Scholar]
- 4.Bochkareva, E., D. Martynowski, A. Seitova, and A. Bochkarev. 2006. Structure of the origin-binding domain of simian virus 40 large T antigen bound to DNA. EMBO J. 25:5961-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 6.Bujalowski, W., and T. M. Lohman. 1989. Negative co-operativity in Escherichia coli single strand binding protein-oligonucleotide interactions. J. Mol. Biol. 207:269-288. [DOI] [PubMed] [Google Scholar]
- 7.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 8.Campbell, K. S., K. P. Mullane, I. A. Aksoy, H. Stubdal, J. M. Pipas, P. A. Silver, T. M. Roberts, B. S. Schaffhausen, and J. A. DeCaprio. 1997. DnaJ/hsp40 chaperone domain of SV40 large T antigen promotes efficient viral DNA replication. Genes Dev. 11:1098-1110. [DOI] [PubMed] [Google Scholar]
- 9.Collaborative Computational Project, Number 4.1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr 50:760-763. [DOI] [PubMed] [Google Scholar]
- 10.Cuesta, I., R. Nunez-Ramirez, S. H. W. Scheres, D. Gai, X. S. Chen, E. Fanning, and J. M. Carazo. 2010. Conformational rearrangements of SV40 large T antigen during early replication events. J. Mol. Biol. 397:1276-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLano, W. L. 2002. The PyMOL molecular graphics system. Delano Scientific, Palo Alto, CA.
- 12.Emsley, P., B. Lohkamp, W. G. Scott, and K. Cowtan. 2010. Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66:486-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fanning, E., V. Klimovich, and A. R. Nager. 2006. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34:4126-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 15.Foster, E. C., and D. T. Simmons. 2010. The SV40 large T-antigen origin binding domain directly participates in DNA unwinding. Biochemistry 49:2087-2096. [DOI] [PubMed] [Google Scholar]
- 16.Fradet-Turcotte, A., G. Morin, M. Lehoux, P. A. Bullock, and J. Archambault. 2010. Development of quantitative and high-throughput assays of polyomavirus and papillomavirus DNA replication. Virology 399:65-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fradet-Turcotte, A., C. Vincent, S. Joubert, P. A. Bullock, and J. Archambault. 2007. Quantitative analysis of the binding of simian virus 40 large T antigen to DNA. J. Virol. 81:9162-9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gai, D., D. Li, C. V. Finkielstein, R. D. Ott, P. Taneja, E. Fanning, and X. S. Chen. 2004. Insights into the oligomeric states, conformational changes and helicase activities of SV40 large tumor antigen. J. Biol. Chem. 279:38952-38959. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Lorenzo, M. G., M. Valle, L. E. Donate, C. Gruss, C. O. S. Sorzano, M. Radermacher, J. Frank, and J. M. Carazo. 2003. Large T antigen on the SV40 origin of replication: a 3D snapshot prior to DNA replication. EMBO J. 22:6205-6213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, C.-Y., C.-H. Hsu, Y.-J. Sun, H.-N. Wu, and C.-D. Hsiao. 2006. Complexed crystal structure of replication restart primosome protein PriB reveals a novel single-stranded DNA-binding mode. Nucleic Acids Res. 34:3878-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ishimi, Y., A. Claude, P. Bullock, and J. Hurwitz. 1988. Complete enzymatic synthesis of DNA containing the SV40 origin of replication. J. Biol. Chem. 263:19723-19733. [PubMed] [Google Scholar]
- 22.Jiang, X., V. Klimovich, A. I. Arunkumar, E. B. Hysinger, Y. Wang, R. D. Ott, G. D. Guler, B. Weiner, W. J. Chazin, and E. Fanning. 2006. Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 25:5516-5526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, A., and M. O'Donnell. 2005. Cellular DNA replicases: components and dynamics at the replication fork. Annu. Rev. Biochem. 74:283-315. [DOI] [PubMed] [Google Scholar]
- 24.Joo, W. S., X. Luo, D. Denis, H. Y. Kim, G. J. Rainey, C. Jones, K. R. Sreekumar, and P. A. Bullock. 1997. Purification of the SV40 T-antigen DNA binding domain and characterization of its interactions with the SV40 origin. J. Virol. 71:3972-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, H.-Y., B. Y. Ahn, and Y. Cho. 2001. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. EMBO J. 20:295-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, A., G. Meinke, D. K. Reese, S. Moine, P. J. Phelan, A. Fradet-Turcotte, J. Archambault, A. Bohm, and P. A. Bullock. 2007. Model for T-antigen-dependent melting of the simian virus 40 core origin based on studies of the interaction of the beta-hairpin with DNA. J. Virol. 81:4808-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 28.Reference deleted.
- 29.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 30.Luo, X., D. G. Sanford, P. A. Bullock, and W. W. Bachovchin. 1996. Structure of the origin specific DNA binding domain from simian virus 40 T-antigen. Nat. Struct. Biol. 3:1034-1039. [DOI] [PubMed] [Google Scholar]
- 31.Masai, H., S. Matsumoto, Z. You, N. Yoshizawa-Sugata, and M. Oda. 2010. Eukaryotic chromosome DNA replication: where, when, and how? Annu. Rev. Biochem. 79:89-130. [DOI] [PubMed] [Google Scholar]
- 32.Mastrangelo, I. A., P. V. C. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 33.McCoy, A. J., R. W. Grosse-Kunstleve, L. C. Storoni, and R. J. Read. 2005. Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61:458-464. [DOI] [PubMed] [Google Scholar]
- 34.Meinke, G., P. A. Bullock, and A. Bohm. 2006. The crystal structure of the T-antigen origin binding domain. J. Virol. 80:4304-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meinke, G., P. J. Phelan, S. Moine, E. Bochkareva, A. Bochkarev, P. A. Bullock, and A. Bohm. 2007. The crystal structure of the SV40 T-antigen origin binding domain in complex with DNA. PloS Biol. 5:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr, I. J., M. P. Fairman, B. Stillman, and Y. Gluzman. 1989. Large T-antigen mutants define multiple steps in the initiation of simian virus 40 DNA replication. J. Virol. 63:4181-4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 38.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307-326. [DOI] [PubMed] [Google Scholar]
- 39.Raghunathan, S., A. G. Kozlov, T. M. Lohman, and G. Waksman. 2000. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature 7:648-652. [DOI] [PubMed] [Google Scholar]
- 40.Reese, D. K., G. Meinke, A. Kumar, S. Moine, K. Chen, J. L. Sudmeier, W. Bachovchin, A. Bohm, and P. A. Bullock. 2006. Analyses of the interaction between the origin binding domain from simian virus 40 T-antigen and single stranded DNA provides insights into DNA unwinding and initiation of DNA replication. J. Virol. 80:12248-12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reese, D. K., K. R. Sreekumar, and P. A. Bullock. 2004. Interactions required for binding of simian virus 40 T antigen to the viral origin and molecular modeling of initial assembly events. J. Virol. 78:2921-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons, D. T. 2000. SV40 large T antigen functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 43.Simmons, D. T., G. Loeber, and P. Tegtmeyer. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simmons, D. T., K. Wun-Kim, and W. Young. 1990. Identification of simian virus 40 T-antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stenlund, A. 2003. Initiation of DNA Replication: Lessons from viral initiator proteins. Nat. Rev. 4:777-785. [DOI] [PubMed] [Google Scholar]
- 46.Titolo, S., E. Welchner, P. W. White, and J. Archambault. 2003. Characterization of the DNA-binding properties of the origin-binding domain of SV40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsurimoto, T., M. P. Fairman, and B. Stillman. 1989. Simian virus 40 DNA replication in vitro: identification of multiple stages of initiation. Mol. Cell. Biol. 9:3839-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valle, M., X. S. Chen, L. E. Donate, E. Fanning, and J. M. Carazo. 2006. Structural basis for the cooperative assembly of large T antigen on the origin of replication. J. Mol. Biol. 357:1295-1305. [DOI] [PubMed] [Google Scholar]
- 49.Valle, M., C. Gruss, L. Halmer, J. M. Carazo, and L. E. Donate. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waterhouse, A. M., J. B. Procter, D. M. Martin, M. Clamp, and G. J. Barton. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189-1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg, D. H., and T. J. Kelly. 1989. Requirement for two DNA polymerases in the replication of simian virus 40 DNA in vitro. Proc. Natl. Acad. Sci. U. S. A. 86:9742-9746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weisshart, K., P. Taneja, and E. Fanning. 1998. The replication protein A binding site in simian virus 40 (SV40) T antigen and its role in the initial steps of SV40 DNA replication. J. Virol. 72:9771-9781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wold, M. S. 1997. Replication protein A: a Heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66:61-92. [DOI] [PubMed] [Google Scholar]
- 54.Wu, C., R. Roy, and D. T. Simmons. 2001. Role of single-stranded DNA binding activity of T antigen in simian virus 40 DNA replication. J. Virol. 75:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wun-Kim, K., R. Upson, W. Young, T. Melendy, B. Stillman, and D. T. Simmons. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67:7608-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.