Abstract
The oncogenic Merkel cell polyomavirus (MCPyV) infects humans worldwide, but little is known about the occurrence of viruses related to MCPyV in the closest phylogenetic relatives of humans, great apes. We analyzed samples from 30 wild chimpanzees and one captive gorilla and identified two new groups of polyomaviruses (PyVs). These new viruses are by far the closest relatives to MCPyV described to date, providing the first evidence of the natural occurrence of PyVs related to MCPyV in wild great apes. Similar to MCPyV, the prevalence of these viruses is relatively high (>30%). This, together with the fact that humans in West and Central Africa frequently hunt and butcher primates, may point toward further MCPyV-like strains spreading to, or already existing in, our species.
Polyomaviruses (PyVs) are known to infect a wide range of birds and mammals (31). This includes humans, from which eight PyVs have been identified to date, namely, BK virus (BKV) (17), JC virus (JCV) (41), KIPyV (2), WUPyV (19), Merkel cell polyomavirus (MCPyV) (15), human PyV 6 (HPyV6) and HPyV7 (47), and a Trichodysplasia spinulosa-associated PyV (TSV) (52). Primary PyV infection usually occurs in childhood and seems to result in lifelong persistence. In healthy humans, PyVs have not been associated with severe acute disease. However, PyV reactivation can cause severe diseases in the case of immunodeficiency (23).
MCPyV has been associated to (and named after) Merkel cell carcinoma (MCC), a rare but aggressive skin cancer (15). Since the first report of MCC in 1972 (under the former designation “trabecular carcinoma”) (51), its incidence has been increasing (46). MCC occurs worldwide, and its association with MCPyV was observed in studies from several continents (4, 18, 26, 56). Benign MCPyV infection seems to be both common (>40%) and geographically widespread (47).
MCPyV's closest relatives known to date have been identified from two nonhuman primate species: B-lymphotropic polyomavirus (LPV; from some African green monkeys (Chlorocebus aethiops) (7) and chimpanzee polyomavirus (ChPyV) from one chimpanzee (Pan troglodytes) (24). This raises the possibility that MCPyV is actually part of a primate-specific subgroup. However, little is known about the natural occurrence of viruses related to MCPyV in the closest relatives of humans, the African great apes.
MATERIALS AND METHODS
Sample collection and DNA isolation.
Necropsy samples (spleen, lymph nodes, bone marrow, thymus, lung, liver, intestine, muscle, heart, pancreas, blood, urine; n = 88) were collected from 29 chimpanzees (Pan troglodytes verus) from the Taï National Park in Côte d'Ivoire. The chimpanzees had died of anthrax (34), respiratory diseases (30), or other causes (35) between 2001 and 2009. Necropsy samples were also collected from one chimpanzee (Pan troglodytes schweinfurthii), which was found dead in the Budongo Forest area, Uganda, and one gorilla (Gorilla gorilla gorilla), which had been confiscated and died in the Projet Protection des Gorilles in the Republic of Congo. For all samples originating from Côte d'Ivoire, the sample collectors wore fully closed body protection suits and masks due to a history of ebola and anthrax in these populations and to avoid any contamination of samples with human pathogens. For the two great apes sampled in the Republic of Congo and Uganda, the sample collectors wore at a minimum single-use gloves and surgical masks (35). Permission for sample collection from wild primates was obtained from the authorities of national parks of each country, and tissue samples were exported with the appropriate CITES permissions from Côte d'Ivoire, Uganda, the Republic of Congo, and Germany. Importations took place according to German veterinary regulations for import of organic materials. All samples from Côte d'Ivoire were preserved in liquid nitrogen upon arrival at the research camps and were later transferred to −80°C at the Robert Koch Institute. Other samples were stored in RNAlater (Qiagen, Hilden, Germany). DNA was isolated using a DNeasy tissue kit (Qiagen).
PyV PCR and sequencing. (i) Generic PCR.
For identification of PyVs related to MCPyV, generic nested PCR targeting a short fragment of the VP1 gene (approximately 260 bp) was performed using two pairs of degenerate and deoxyinosine-substituted primers (see Table S1 in the supplemental material). All primers were designed to bind sites within the VP1 gene identified as being conserved among a wide range of PyVs. They were derived from the sequence of MCPyV (GenBank accession no. FJ173815) in order to bias amplification toward sequences from PyVs related to MCPyV. PCR was performed in a total volume of 25 μl with 0.4 μl AmpliTaq Gold (Applied Biosystems), 20 pmol of each primer, 200 μM deoxynucleoside triphosphates, 2 mM MgCl2, and 5% dimethyl sulfoxide. A T-Gradient thermocycler from Biometra was used with the following cycling conditions: 95°C for 12 min and 45 cycles of 95°C for 30 s, 46°C (1st round) or 50°C (2nd round) for 30 s, and 72°C for 2 min, followed by a 15-min final extension step at 72°C.
(ii) Long-distance PCR.
Nested specific primers were derived from the sequences amplified with the generic PCR (see Table S1 in the supplemental material). They were designed tail to tail for the amplification and sequencing of the remaining parts of the genomes. Nested long-distance PCR was performed with the TaKaRa-Ex PCR system, according to the instructions of the manufacturer (Takara Bio Inc., Japan).
(iii) Diagnostic PCR.
For the differential detection of chimpanzee PyVs, three primer sets were selected. The first set (VP1-ptv1) detects P. troglodytes verus PyV1a (PtvPyV1a) and PtvPyV1b, the second set (VP1-ptv2a/b) detects PtvPyV2a and -2b, and the third one (VP1-ptv2c) detects PtvPyV2c (see Table S1 in the supplemental material). They were used under the PCR conditions described above, except that AmpliTaq Gold was used at 0.2 μl/25-μl reaction volume. Cycling conditions were conducted as follows: 95°C for 12 min and 45 cycles of 95°C for 30 s; 55°C (VP1-ptv1), 58°C (VP1-ptv2a/b), or 60°C (VP1-ptv2c) for 30 s; and 72°C for 1 min, followed by a 10-min final extension step at 72°C.
(iv) Testing strategy.
First, the chimpanzee samples and the gorilla sample were tested with the generic PCR. Then, a core set of samples from which sufficient spleen and lymph node aliquots were available (21 samples from 15 P. t. verus chimpanzees) was tested with the generic PCR as well as all three diagnostic, PyV-specific PCRs (Table 1). A supplementary diagnostic PCR was also used for testing of nine additional P. t. verus chimpanzees and was conducted mainly with the VP1-ptv2c PCR only, since PtvPyV2c was the first virus discovered in the course of this study (43 samples from nine individuals) (data not shown). Additional testing of these samples with the other diagnostic PCRs was not possible because of sample limitations.
TABLE 1.
Generic and specific PCR detection of great ape PyVs
| Sample identifier | Organ | PCR result |
|||
|---|---|---|---|---|---|
| Generic PCR | Diagnostic PCR with primer set: |
||||
| VP1-ptv1 | VP1-ptv2a/b | VP1-ptv2c | |||
| P. t. verus 1 5740 | Spleen | —a | — | PtvPyV2a | — |
| P. t. verus 2 6446 | Lymph node | — | — | — | — |
| P. t. verus 3 2756 | Spleen | PtvPyV1b | PtvPyV1b | — | PtvPyV2c |
| P. t. verus 4 | |||||
| 2293 | Lymph node | — | — | — | — |
| 5744 | Lymph node | — | — | — | PtvPyV2c |
| 2221 | Spleen | — | — | — | — |
| 5743 | Spleen | — | — | — | PtvPyV2c |
| P. t. verus 5 5749 | Spleen | — | PtvPyV1a | — | — |
| P. t. verus 6 | |||||
| 3147 | Spleen | — | PtvPyV1a | — | PtvPyV2c |
| 6444 | Spleen | PtvPyV1a | PtvPyV1a | — | PtvPyV2c |
| P. t. verus 7 4699 | Spleen | — | — | — | — |
| P. t. verus 8 4579 | Spleen | PtvPyV2b | — | PtvPyV2b | — |
| P. t. verus 9 | |||||
| 6499 | Lymph node | PtvPyV2c | — | — | PtvPyV2c |
| 6498 | Spleen | — | — | — | — |
| P. t. verus 10 6501 | Spleen | PtvPyV1a | PtvPyV1a | PtvPyV2a | — |
| P. t. verus 11 6506 | Spleen | — | — | — | PtvPyV2c |
| P. t. verus 12 2757 | Spleen | — | — | — | — |
| P. t. verus 13 6433 | Lymph node | PtvPyV1b | — | — | — |
| P. t. verus 14 | |||||
| 6435 | Lymph node | — | PtvPyV1a | — | PtvPyV2c |
| 5779 | Spleen | — | — | — | — |
| P. t. verus 15 6452 | Spleen | — | — | — | — |
| P. t. schweinfurthii 5760 | Spleen | PtsPyV1 | NTb | NT | — |
| G. g. gorilla 5766 | Spleen | GggPyV1 | NT | NT | NT |
—, PCR negative.
NT, not tested.
Sequencing.
All PCR products were purified by using a PCR purification kit (Qiagen) and directly sequenced with a BigDye Terminator cycle sequencing kit (Applied Biosystems, Warrington, Great Britain) in a 377 DNA automated sequencer (Applied Biosystems).
Sequence analyses.
Phylogenetic analyses were performed so as to determine the positions of the newly identified strains in the Polyomaviridae family tree and decipher the relationships between the strains belonging to the clade of MCPyV-related PyVs. For that purpose, two alignments were generated and analyzed: (i) an amino acid alignment of the three main coding sequences (VP1, VP2, and large T), gathering most available polyomaviral genomes (i.e., a comprehensive sample of the overall genomic diversity of this viral family), and (ii) a nucleotide alignment of all distinct VP1 sequences from MCPyV-related PyVs (i.e., MCPyVs and the sequences depicted in this article) identified to date.
VP1, VP2, and large T amino acid sequence data set preparation and phylogenetic analysis.
VP1, VP2, and large T sequences were recovered from the complete genomes of polyomaviruses standing for the entire extent of the genetic diversity of the family (i.e., 4 avian polyomaviruses and 21 mammalian polyomaviruses, among which are the 8 known human polyomaviruses; see Table S2 in the supplemental material). The sequences from four MCPyVs were included in all data sets so as to account for the known diversity of the species (Asian and non-Asian MCPyV sequences were included [47]). The VP1 data set was completed with one sequence previously determined from a virus infecting a chimpanzee, but the complete genome of that virus had otherwise not been sequenced (24). Finally, all sequences obtained in the course of this study were added to the data sets, irrespective of their length.
Sequences were aligned in the SeaView program (version 4) (20) using Muscle software (13, 14) and on a web server using T-Coffee software (http://www.tcoffee.org/; 40). Both methods produced alignments of similar quality; however, the T-Coffee alignments were arbitrarily retained for further analyses. From each coding sequence alignment, a protein alignment of better quality was produced using the Gblocks server (http://molevol.cmima.csic.es/castresana/Gblocks_server.html) (8, 50). This resulted in removing blocks of the alignments where the hypothesis of homology was likely to be overoptimistic, a strategy that has been proved to lead to better estimation of phylogenetic trees (50). The automatically cured alignments were then manually edited in SeaView before being reduced to unique sequences using the FaBox program (55). The process resulted in three alignments: (i) VP1 with 38 unique sequences and 203 amino acids (aa); (ii) VP2 with 28 unique sequences and 63 aa; and (iii) large T with 32 unique sequences and 407 aa. From those data sets, a concatenation which ultimately was composed of an alignment of 673 positions, including 39 unique sequences, was assembled. Preliminary phylogenetic analyses of individual alignments supported similar topologies, in particular when it comes to our clade of interest (data not shown; individual alignments and trees are available upon request). Therefore, the concatenated alignment was used for all following analyses.
Model selection was performed on the concatenated data set using the ProtTest program (version 2.4) (1). Because we intended to use the software BEAST thereafter and this software currently implements only 6 substitution matrices (JTT, MtREV, Dayhoff, WAG, CpREV, and Blosum62), likelihoods were computed for models using these substitution matrixes. All model add-ons (+I, +G, or +I+G for rate heterogeneity modeling and +F for use of empirical equilibrium frequencies of amino acids) were activated. Base trees for calculations were maximum likelihood (ML) optimized (i.e., for each model, a tree was reconstructed using PhyML) (21). Likelihoods were compared according to the Akaike information criterion (AIC), resulting in selecting WAG+I+G as a model of amino acid evolution.
Phylogenetic analyses were performed in both ML and Bayesian frameworks under that model of amino acid evolution. ML analysis was performed on the PhyML web server (http://www.atgc-montpellier.fr/phyml/) (21, 22). Equilibrium frequencies, topology, and branch lengths were optimized; the starting tree was determined using the BioNJ program; and both nearest-neighbor interchange (NNI) and subtree pruning and regrafting (SPR) algorithms of tree search were used (keeping the best outcome). Branch robustness was assessed by performing nonparametric bootstrapping (500 replicates). Bayesian analyses were performed using BEAST (version 1.5.3) (12). Besides allowing modeling of the amino acid substitution process, BEAST also allows modeling of evolutionary rate variation and tree shape. Analyses were run under the assumption of a relaxed, uncorrelated lognormal clock and two speciation models (Yule process and birth-death model). Four independent runs totaling 8,000,000 generations were performed under each speciation model. Trees and numerical values taken by all parameters were sampled every 1,000 generations. The Tracer program (version 1.5) was used to check that individual runs had reached convergence, that independent runs converged on the same zones of parameter spaces, and that chain mixing was satisfactory (global effective sample size values, >100) (12). Tree samples were then gathered into a single file (after removal of a visually conservative 10% burn-in period) using the LogCombiner program (version 1.5.3; distributed with BEAST), and the information from approximately 7,200 trees was summarized onto the maximum clade credibility trees using the TreeAnnotator program (version 1.5.3; distributed with BEAST). Posterior probabilities were taken as a measure of branch robustness. Ninety-five percent highest posterior density (HPD) intervals for the maximum patristic distance observed for two clades of interest were also determined from the output of BEAST analyses.
VP1 nucleotide sequence data set preparation and phylogenetic analysis.
All available MCPyV VP1 sequences as well as all sequences generated for this study were aligned using the Muscle program, as implemented in SeaView. Given the good quality of the alignment, only minor manual editing of the nucleotide alignment was necessary. Reduction to unique sequences with Fabox resulted in an alignment of 1,182 positions comprising 25 sequences.
The nucleotide substitution model to which the data were a better fit was determined using the jModeltest program (version 0.1.1) (21, 44). Three substitution schemes (Jukes and Cantor [JC], Hasegawa, Kishino, and Yano [HKY], and global time reversible [GTR]) were examined along with rate variation (+I, +G, +I+G) and base frequency (+F) modeling. Base trees for calculations were maximum likelihood optimized (21). According to the AIC, comparisons of model likelihoods were more favorable to a GTR+G model.
Phylogenetic analyses were performed in both maximum likelihood and Bayesian frameworks under that model of nucleotide evolution. ML and Bayesian analyses were performed as described above, with the exceptions that in Bayesian analysis tree shape was modeled by a coalescent process (constant population size or exponential growth) and that two experiments were run according to each model for 4,000,000 generations total (accordingly, ca. 3,500 trees constituted the final samples).
For both data sets, ML and Bayesian methods globally supported congruent topologies with consistent branch supports (even though bootstrap [Bp] and posterior probability [pp] are not directly comparable [11]), as did different speciation and coalescent models. All .xml files (including sequence alignments) used for Bayesian analyses are available upon request. Figures summarizing phylogenetic analyses were drawn using the FigTree program (version 1.3.1; http://tree.bio.ed.ac.uk/software/figtree/).
Polyomaviral species delineation.
So as to put the extent of the genetic diversity of the newly identified ape strains in an appropriate context, we compared it to the known genetic diversity prevailing in four human polyomaviruses, all of which are currently considered constituting viral species (MCPyV, BKV, JCV, and WUPyV). For this, we took the VP1-coding sequence as a proxy of the global genomic divergence of polyomaviruses.
In addition to the VP1 nucleotide alignment, which already encompassed the known genetic diversity of MCPyV, three new data sets were assembled: (i) BKV sequences spanning the entire BKV genetic diversity were taken from a study of Krumbholz et al. (32). Representatives of the seven clades identified in this study were included, resulting in an alignment of 1,089 positions comprising 26 sequences. All sequences were unique. (ii) JCV sequences representing the overall genetic diversity of the species were taken from a study of Kitchen et al. (29). VP1 sequences from the 92 sequences chosen by these authors to compose four main regional groups (Asia, Europe, Japan, and the Americas) were gathered, resulting in an alignment of 1,065 positions. The latter was then reduced to the 64 unique sequences that it comprised. (iii) WUPyV sequences were drawn from a study of Bialasiewicz et al. (6). A total of 19 sequences representing the six clades described in this study were gathered, constituting an alignment of 1,110 positions. From those sequences, 10 could be identified to be unique.
For each data set, pairwise distances were first computed using SeaView. As pairwise distances are, in general, poor indicators of true evolutionary distances (36), we also computed patristic (tree) distances. For this, all data sets were analyzed along a common pipeline. The model of nucleotide substitution to which the data set was a best fit was first determined using jModeltest, as detailed above (for BKV, GTR+I; for JCV, GTR+I+G; for WUPyV, GTR+I). Then the maximum likelihood tree was determined using PhyML (as implemented in Seaview) under that model. Finally, patristic distances were computed from that tree using the software Patristic (16). From both pairwise and patristic distance matrices, maximum distances within known species were determined. Minimum patristic distances separating newly identified strains from their closest relatives were finally identified.
Provisional nomenclature and abbreviations of novel polyomaviruses.
Names and abbreviations for newly detected PyVs were formed from host genus and species names and numbered arbitrarily following the results of the species delineation, e.g., G. g. gorilla PyV1 (GggPyV1). They are listed in Table 2. Published PyVs that were used in the analyses are listed in Table S2 in the supplemental material.
TABLE 2.
Polyomaviruses detected in wild great apes
| Specimen no. | Virus name | Sequence present |
Host species | Origin of host | |
|---|---|---|---|---|---|
| 0.21-kb VP1 | 5.3-kb genome | ||||
| 6444 | PtvPyV1a | x | x | P. t. verus | Côte d'Ivoire |
| 6447 | PtvPyV1b | x | P. t. verus | Côte d'Ivoire | |
| 6520 | PtvPyV1b | x | x | P. t. verus | Côte d'Ivoire |
| 5760 | PtsPyV1 | x | P. t. schweinfurthii | Uganda | |
| 6512 | PtvPyV2a | x | x | P. t. verus | Côte d'Ivoire |
| 4579 | PtvPyV2b | x | P. t. verus | Côte d'Ivoire | |
| 5924 | PtvPyV2c | x | x | P. t. verus | Côte d'Ivoire |
| 5927 | PtvPyV2c | x | x | P. t. verus | Côte d'Ivoire |
| 6413 | PtvPyV2c | x | x | P. t. verus | Côte d'Ivoire |
| 5766 | GggPyV1 | x | x | G. g. gorilla | Republic of Congo |
Nucleotide sequence accession numbers.
PyV sequences determined in the course of this study have been deposited in GenBank under the following accession numbers: for PtvPyV1a, HQ385746; for PtvPyV1b, HQ326775 and HQ385747; for PtsPyV1, HQ326774; for PtvPyV2a, HQ385748; for PtvPyV2b, HQ326776; for PtvPyV2c, HQ385749 to HQ385751; and for GggPyV1, HQ385752.
RESULTS
A generic nested PCR strategy aimed at amplifying a fragment of the VP1 gene from MCPyV-related viral strains was employed to test necropsy samples of 25 wild chimpanzees and one gorilla. Fifteen chimpanzees (14/24 P. t. verus chimpanzees and 1/1 P. t. schweinfurthii chimpanzee) and the gorilla were PCR positive. In all cases, sequencing of PCR products confirmed their infection with PyVs. For 13 chimpanzees (12 P. t. verus chimpanzees and 1 P. t. schweinfurthii chimpanzee) and the gorilla, preliminary BLAST analyses gave MCPyV as a first hit (3). These MCPyV-like viruses are the focus of the present study (information on the other chimpanzee PyVs will be published elsewhere).
On the basis of the MCPyV-like VP1 sequences obtained using the first protocol, nested specific primers (see Table S1 in the supplemental material) were used for long-distance PCR amplification and sequencing of the remaining parts of the genomes. Seven complete genome sequences, six from chimpanzees (P. t. verus) and one from the gorilla (Table 2), could be determined. All genomes exhibited the typical set of PyV open reading frames (i.e., VP1, VP2, VP3, large T, and small T) and lacked any agnoprotein open reading frame.
Genome analysis of great ape PyVs.
A high overall degree of sequence similarity of the novel great ape PyVs to MCPyV, including in regions otherwise less conserved among PyVs, was observed (see Table S3 in the supplemental material). Analysis of the noncoding control region (NCCR), the most variable region of the PyV genome, revealed a high degree of similarity of the great ape PyVs to MCPyV. An important motif of this region is the DNA element GAGGC and its complement, GCCTC. Repeats of these motifs are considered large-T-antigen-binding sites (reviewed in reference 25). The highest number of these elements is found in the NCCRs of MCPyV isolates (n = 7 to 8), which are to be compared to those of simian virus 40 (SV40), BKV, JCV (n = 6), and LPV (n = 4). In MCPyV, these elements are also present in one or two overlapping, palindromic octamers which possibly affect binding of T-antigen hexamers and initiation of DNA replication (33). Interestingly, the novel African great ape PyVs also harbor eight or nine elements and three palindromic octamers (Fig. 1).
FIG. 1.
Noncoding control region of great ape PyVs. DNA sequences of the discovered great ape PyVs and a selection of published sequences from MCPyVs and other PyVs were compared using the ClustalW program (as implemented in the MacVector program, version 10.6) with additional manual adjustments. Base 1 corresponds to base 5316 of the MKL-1 isolate of MCPyV (GenBank accession no. FJ173815). Conserved regions are outlined with solid lines. The part of the alignment comprising the great ape PyVs and the MCPyVs is boxed with a dotted line. T-antigen-binding elements (GAGGC or the complement, GCCTC) are boxed with red (GAGGC) and blue (GCCTC) lines.
The large T proteins exhibit a high degree of conservation of functional domains (CR1, J, RB-binding, ori-binding, and Zn finger domains) (43). Additionally, a region of approximately 180 amino acids extending from the J domain to the ori-binding domain rich in serine, glutamine, and threonine and possibly affecting RB-binding function (25) is also highly conserved among the novel PyVs and MCPyVs but is absent from LPV, BKV, JCV, and SV40 (see Fig. S1 in the supplemental material).
In addition to its function as a virion structural protein, the major capsid protein VP1 plays an important role during the initial interaction of PyVs with host cells through its outfacing loops (BC, HI) (48, 49) and contains antigenic domains (39). Modifications in PyV VP1 loops have been shown to alter host specificity (38). Therefore, the striking similarity between the great ape PyVs and MCPyV in VP1, even in the highly variable loop regions, is particularly supportive of a close relationship between great ape PyVs and MCPyV (data not shown).
Phylogenetic analysis.
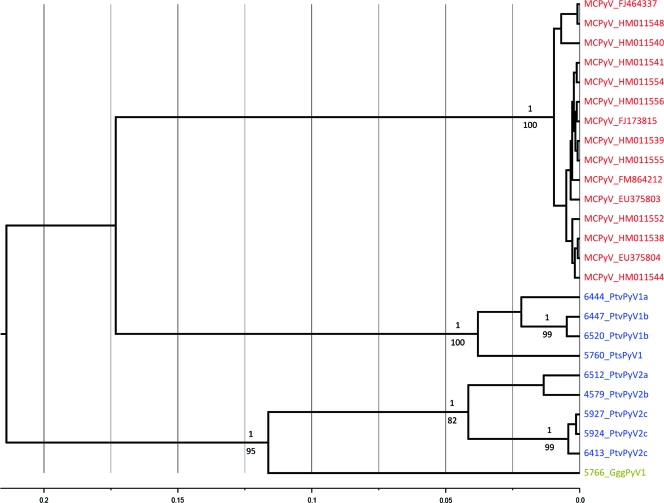
Phylogenetic trees inferred from an alignment of concatenated VP1, VP2, and large T sequences (673 aa) comprising most known PyVs evidenced the close relationship of wild great ape PyVs to MCPyV, which altogether formed a highly supported monophyletic group (Bp and pp values, 100 and 1, respectively) (Fig. 2). Within this group, the maximum patristic distance was estimated to stand between 0.119 and 0.270 aa substitution per site (95% HPD interval), while the maximum patristic distance to members of the sister clade comprising primate polyomavirus sequences was assessed as lying at between 0.539 and 0.910 aa substitution per site (95% HPD interval) (Fig. 2). Of note, MCPyV was firmly established as monophyletic (Bp, 91; pp, 1; Fig. 2).
FIG. 2.
Bayesian chronogram deduced from the analysis of a 673-amino-acid concatenation of VP1, VP2, and large T sequences. PyVs identified from human hosts are in red, PyVs from chimpanzees in blue, and PyV from a gorilla in green. Ninety-five percent HPD intervals of maximum patristic distances are indicated in parentheses for two clades (lines are drawn to the corresponding nodes). The clade formed by MCPyVs and the newly described ape PyVs is highlighted. Statistical support for branches is given where Bp values are ≥70 and pp values are ≥0.95. Bp values are shown below the branches, and pp values are shown above the branches (pp values are those obtained from analyses performed under the Yule model of speciation). The scale axis is in number of amino acid substitution per site. This chronogram was rooted using a relaxed clock. A maximum likelihood analysis of the same data set concluded a similar topology, and thus, the results are not shown here. It is noted that when only VP1 sequences were considered, ChPyV was found to be the sister taxon of the MCPyV-related PyV clade.
Phylogenetic trees were also inferred from a nucleotide alignment of 1,182 bp so as to refine the picture of MCPyV-like virus relationships (all known unique MCPyV sequences were included). This confirmed the MCPyV monophyly (Bp, 100; pp, 1) and found support for the existence of at least two clades of wild great ape PyVs (Fig. 3). The most basal one comprised sequences from the gorilla and chimpanzees of the subspecies P. t. verus only (GggPyV1 and PtvPyV2 clade). GggPyV1 appeared to have diverged first from all other sequences of this clade (Bp, 82; pp, 1; Fig. 3). All other sequences (PtvPyV1 and PtsPyV1 clade) identified from chimpanzees (P. t. verus and P. t. schweinfurthii) belonged to another clade which was found to be in sistership with the MCPyV clade (Fig. 3). Within this clade, the sequence determined from a P. t. schweinfurthii individual was always found to be the first to diverge, though with moderate statistical support.
FIG. 3.
Bayesian chronogram deduced from the analysis of a 1,182-nucleotide VP1 alignment. PyVs identified from human hosts are in red, PyVs from chimpanzees in blue, and PyV from a gorilla in green. Statistical support for branches is given where Bp values are ≥70 and pp values are ≥0.95. Bp values are shown below the branches, and pp values are shown above the branches (pp values are those obtained from analyses performed under the constant-population-size model of coalescence). The scale axis is in number of nucleotide substitution per site. This chronogram was rooted using a relaxed clock (midpoint rooting of the maximum likelihood tree identified the same root). A maximum likelihood analysis of the same data set concluded a similar topology, and thus, the results are not shown here.
Maximum VP1 patristic distances within recognized human polyomavirus species varied from 0.016 (MCPyV) to 0.216 (BKV) nucleotide substitution per site (Table 3). All strains belonging to the clades PtvPyV1/PtsPyV1, PtvPyV2, and GggPyV1 exhibited minimum patristic distances from the other clades greater than 0.216 (respective minima, 0.354, 0.272, and 0.272 nucleotide substitution per site, respectively; Table 4). Within those species, subclades were defined that gather sequences whose patristic distances do not outscore the maximum patristic distance observed between MCPyVs (PtvPyV1a and -b, PtsPyV1, PtvPyV2a to -c).
TABLE 3.
Maximum pair-wise and patristic distances observed in four human polyomaviral species
| Virus | Maximum pairwise distance | Maximum patristic distance | Most distantly related sequences (patristic distances) |
|---|---|---|---|
| MCPyV | 0.016 | 0.016 | MCC350 and 10b (Asian clade), GenBank accession nos. NC_010277 and HM011540 |
| WUV | 0.023 | 0.054 | O342 (IIIb) and FZ18 (Ia), GenBank accession nos. GU296362 and FJ890981 |
| JCV | 0.036 | 0.141 | Italy03 and Peru01, GenBank accession nos. AB074582 and AB081023 |
| BKV | 0.075 | 0.216 | KOM7 (18) and CAP-m13 (28), GenBank accession nos. AB211388 and AY682230 |
TABLE 4.
Minimum patristic distance involving the new great ape PyVs
| PyV | Minimum patristic distance | Closest relative(s) (patristic distance) | Minimum patristic distance to other groups of sequences |
|---|---|---|---|
| 6444_PtvPyV1a | 0.043a | 6520_PtvPyV1b | 0.354b |
| 6447_PtvPyV1b | 0.010 | 6520_PtvPyV1b | |
| 6520_PtvPyV1b | 0.010 | 6447_PtvPyV1b | |
| 5760_PtsPyV1c | 0.078 | 6520_PtvPyV1b | |
| 6512_PtvPyV2a | 0.028 | 4579_PtvPyV2b | 0.272 |
| 4579_PtvPyV2b | 0.028 | 6512_PtvPyV2a | |
| 5924_PtvPyV2c | 0.002 | 5927_PtvPyV2c | |
| 5927_PtvPyV2c | 0.002 | 5924_PtvPyV2c | |
| 6413_PtvPyV2c | 0.006 | 5927_PtvPyV2c | |
| 5766_GggPyV1 | 0.272 | 6512_PtvPyV2a | 0.272 |
Values outscoring MCPyV maximum patristic distance are italicized.
Values outscoring the maximum patristic distance observed in a human PyV (i.e., 0.216 substitution per site for BKV) are in boldface.
Prevalence of great ape PyVs.
Since diverse groups of PyVs were identified in chimpanzees, three specific PCR tests (directed against PtvPyV1a/b, PtvPyV2a/b, or PtvPyV2c; see Table S1 in the supplemental material) were used to get insight into their respective prevalence and to determine the extent of coinfection. All PCR products were sequenced to check for proper clade and subclade identification. In the core sample set (21 samples from 15 individuals), PtvPyV1 was detected in 6/15 individuals, PtvPyV2a/b was detected in 3/15 individuals, and PtvPyV2c was detected in 6/15 individuals, corresponding to prevalence rates of approximately 40%, 20%, and 40%, respectively. Four of 15 individuals were coinfected (2 with PtvPyV1a/2c, 1 with PtvPyV1b/2c, and 1 with PtvPyV1a/2a; Table 1). Furthermore, PtvPyV2c-specific PCR was performed on nine additional individuals, among which three were found to be positive (not listed). Together with the data from the core set (Table 1), PtvPyV2c was thus detected in 9/24 individuals (38%).
DISCUSSION
Here PyVs infecting wild African great apes were discovered and shown to be closely related to human MCPyV using genome analysis (Fig. 1; see Fig. S1 and Table S3 in the supplemental material) and phylogenetic analysis (Fig. 2 and 3). These findings are particularly interesting since they convincingly demonstrate that MCPyV stems from a (so far) primate-specific and even ape-specific group of PyVs.
Though this might at first be seen as an argument in favor of the cospeciation hypothesis (42), two diverging interpretations of our phylogenetic trees can be made that end up with contrasting conclusions about the processes at play along MCPyV-related PyV evolution. If three distinct MCPyV-related PyVs whose present descendants would be MCPyV, PtvPyV2/GggPyV1, and Ptv/PtsPyV1, respectively, are assumed to have been infecting the last common ancestor of African great apes, then the fact that along two of the corresponding evolutionary lineages (those leading to PtvPyV2/GggPyV1 and Ptv/PtsPyV1) species-specific patterns are not contradicted would indeed be consistent with the codivergence hypothesis. On the contrary, if the last common ancestor of all African great apes is assumed to have been infected with only one MCPyV-related PyV (the last common ancestor of all MCPyV-related PyVs), then strict codivergence is an unsatisfactory explanation for the observed phylogenetic pattern (i.e., a bad overlap of viral and host phylogenies) and some degree of host switching has to be assumed as well. Our data do not allow favoring one hypothesis over the other. Therefore, the question of the modalities of evolution of PyVs related to MCPyV remains largely open.
The finding of new viral strains that cannot be identified directly as belonging to a recognized species raises the question of their taxonomic status (i.e., are they likely to represent new viral species?). Viral species delineation is often controversial (54), including among the Polyomaviridae (31). In particular, when only sequences are available, it is impossible to define any objective threshold beyond which genetic divergence undoubtedly reflects the existence of separate viral species (54). Here we find that three entities—two clades (Ptv/PtsPyV1 and PtvPyV2) and a single sequence (GggPyV1)—present minimum patristic distances to their closest outgroup relatives greater than the maximum patristic distance observed within BKV, the human polyomavirus with the highest degree of genetic diversity known thus far. On the basis of this observation, we propose to provisionally consider those three entities as separate viral species (or at least as separate taxa of the same rank as BKV). This should obviously not be taken as implying that they have different biological properties, whose characterization will be required to truly install those groups of viruses as valid viral species (45, 54).
This being said, it is striking that PyVs related to MCPyV circulating in chimpanzees are, from a genetic standpoint, much more diverse than the human MCPyVs described so far (and are actually even more diverse than any human PyV), all the more so since nearly the entire extent of this diversity could be found in a single P. t. verus chimpanzee community of the Taï National Park, Côte d'Ivoire. This could be interpreted as indicating that West Africa (and possibly Central Africa) is a hot spot of MCPyV-related PyV diversity. These data may point toward a higher degree of genetic diversity of MCPyV in humans worldwide and specifically in West and Central Africa, where conclusive studies with human populations are missing (47).
Though our analyses do not provide direct evidence that MCPyV was transmitted from apes to humans, the presence of PyVs related to MCPyV in wild great apes is also significant with respect to human health. SV40 and LPyV are much more distantly related to MCPyV than PtvPyV1, PtvPyV2, or GggPyV1. However, they seem to infect humans, as suggested by their identification in human samples (7, 10, 27, 37). If the assumption is made that the closer that two species are the higher that the likelihood of successful transmission of their respective pathogens is (the preferential host switching hypothesis) (9), then transmission of great ape PyVs to sympatric humans must be considered. Besides, the practical determinants of a successful transmission to human populations are already in place. First, according to our data set, PtvPyV is infecting chimpanzees (including coinfections with various strains) at a high prevalence rate, at least in the Taï Forest, Côte d'Ivoire. Second, nonhuman primates (including chimpanzees and gorillas) still represent an important proportion of the bush meat consumed in West and Central Africa (ca. 12%) (5). Hunting and butchering of bush meat have been shown to provide the major routes of pathogen transmission to humans (e.g., human immunodeficiency viruses) (28, 53). This could well apply to PyVs related to MCPyV.
The wide variety of MCPyV-related viruses in African great apes presented here calls for larger studies to unravel the diversity of PyVs related to MCPyV currently circulating in nonhuman primates of West and Central Africa (and more particularly in apes) as well as in local human populations. It is reasonable to assume that such studies will reveal unprecedented levels of PyV diversity, which will provide a sound basis for a better assessment of both PyV natural history and the risk that nonhuman primate PyVs really pose.
Supplementary Material
Acknowledgments
We thank the Ivorian authorities for long-term support, especially the Ministry of the Environment and Forests, as well as the Ministry of Research, the directorship of the Taï National Park, the Swiss Research Center in Abidjan, the Uganda Wildlife Authority, and the Uganda National Council for Science and Technology for granting us permission to conduct this research. We also thank the CITES authorities of Côte d'Ivoire, Uganda, Republic of Congo, and Germany for permission. We are grateful to S. Schenk, S. Metzger, and the field assistants of the Taï chimpanzee project and T. King and C. Chamberlain of the Aspinall Foundation for sample collection; F. Babweteera and R. Wittig for their support; and D. Wevers, S. Liebmann, C. Walter, N. Yasmum, S. Köndgen, A. Blasse, and C. Hedemann for technical support in the laboratory.
The Budongo Conservation Field Station receives core funding from the Royal Zoological Society of Scotland. This work was supported by the Robert Koch Institute and the Max Planck Society.
Footnotes
Published ahead of print on 3 November 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Abascal, F., R. Zardoya, and D. Posada. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104-2105. [DOI] [PubMed] [Google Scholar]
- 2.Allander, T., K. Andreasson, S. Gupta, A. Bjerkner, G. Bogdanovic, M. A. Persson, T. Dalianis, T. Ramqvist, and B. Andersson. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Becker, J. C., R. Houben, S. Ugurel, U. Trefzer, C. Pfohler, and D. Schrama. 2009. MC polyomavirus is frequently present in Merkel cell carcinoma of European patients. J. Investig. Dermatol. 129:248-250. [DOI] [PubMed] [Google Scholar]
- 5.Bennett, E. L., E. Blencowe, K. Brandon, D. Brown, R. W. Burn, G. Cowlishaw, G. Davies, H. Dublin, J. E. Fa, E. J. Milner-Gulland, J. G. Robinson, J. M. Rowcliffe, F. M. Underwood, and D. S. Wilkie. 2007. Hunting for consensus: reconciling bushmeat harvest, conservation, and development policy in West and Central Africa. Conserv. Biol. 21:884-887. [DOI] [PubMed] [Google Scholar]
- 6.Bialasiewicz, S., R. Rockett, D. W. Whiley, Y. Abed, T. Allander, M. Binks, G. Boivin, A. C. Cheng, J. Y. Chung, P. E. Ferguson, N. M. Gilroy, A. J. Leach, C. Lindau, J. W. Rossen, T. C. Sorrell, M. D. Nissen, and T. P. Sloots. 2010. Whole-genome characterization and genotyping of global WU polyomavirus strains. J. Virol. 84:6229-6234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brade, L., N. Muller-Lantzsch, and H. zur Hausen. 1981. B-lymphotropic papovavirus and possibility of infections in humans. J. Med. Virol. 6:301-308. [DOI] [PubMed] [Google Scholar]
- 8.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 9.Charleston, M. A., and D. L. Robertson. 2002. Preferential host switching by primate lentiviruses can account for phylogenetic similarity with the primate phylogeny. Syst. Biol. 51:528-535. [DOI] [PubMed] [Google Scholar]
- 10.Delbue, S., S. Tremolada, E. Branchetti, F. Elia, E. Gualco, E. Marchioni, R. Maserati, and P. Ferrante. 2008. First identification and molecular characterization of lymphotropic polyomavirus in peripheral blood from patients with leukoencephalopathies. J. Clin. Microbiol. 46:2461-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Douady, C. J., F. Delsuc, Y. Boucher, W. F. Doolittle, and E. J. Douzery. 2003. Comparison of Bayesian and maximum likelihood bootstrap measures of phylogenetic reliability. Mol. Biol. Evol. 20:248-254. [DOI] [PubMed] [Google Scholar]
- 12.Drummond, A. J., and A. Rambaut. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 7:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar, R. C. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng, H., M. Shuda, Y. Chang, and P. S. Moore. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fourment, M., and M. J. Gibbs. 2006. PATRISTIC: a program for calculating patristic distances and graphically comparing the components of genetic change. BMC Evol. Biol. 6:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner, S. D., A. M. Field, D. V. Coleman, and B. Hulme. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253-1257. [DOI] [PubMed] [Google Scholar]
- 18.Garneski, K. M., A. H. Warcola, Q. Feng, N. B. Kiviat, J. H. Leonard, and P. Nghiem. 2009. Merkel cell polyomavirus is more frequently present in North American than Australian Merkel cell carcinoma tumors. J. Investig. Dermatol. 129:246-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaynor, A. M., M. D. Nissen, D. M. Whiley, I. M. Mackay, S. B. Lambert, G. Wu, D. C. Brennan, G. A. Storch, T. P. Sloots, and D. Wang. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gouy, M., S. Guindon, and O. Gascuel. 2010. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 27:221-224. [DOI] [PubMed] [Google Scholar]
- 21.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 22.Guindon, S., F. Lethiec, P. Duroux, and O. Gascuel. 2005. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 33:W557-W559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, M., J. R. Abend, S. F. Johnson, and M. J. Imperiale. 2009. The role of polyomaviruses in human disease. Virology 384:266-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johne, R., D. Enderlein, H. Nieper, and H. Muller. 2005. Novel polyomavirus detected in the feces of a chimpanzee by nested broad-spectrum PCR. J. Virol. 79:3883-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson, E. M. 2010. Structural evaluation of new human polyomaviruses provides clues to pathobiology. Trends Microbiol. 18:215-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katano, H., H. Ito, Y. Suzuki, T. Nakamura, Y. Sato, T. Tsuji, K. Matsuo, H. Nakagawa, and T. Sata. 2009. Detection of Merkel cell polyomavirus in Merkel cell carcinoma and Kaposi's sarcoma. J. Med. Virol. 81:1951-1958. [DOI] [PubMed] [Google Scholar]
- 27.Kean, J. M., S. Rao, M. Wang, and R. L. Garcea. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog. 5:e1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keele, B. F., F. Van Heuverswyn, Y. Li, E. Bailes, J. Takehisa, M. L. Santiago, F. Bibollet-Ruche, Y. Chen, L. V. Wain, F. Liegeois, S. Loul, E. M. Ngole, Y. Bienvenue, E. Delaporte, J. F. Brookfield, P. M. Sharp, G. M. Shaw, M. Peeters, and B. H. Hahn. 2006. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science 313:523-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kitchen, A., M. M. Miyamoto, and C. J. Mulligan. 2008. Utility of DNA viruses for studying human host history: case study of JC virus. Mol. Phylogenet. Evol. 46:673-682. [DOI] [PubMed] [Google Scholar]
- 30.Kondgen, S., H. Kuhl, P. K. N′Goran, P. D. Walsh, S. Schenk, N. Ernst, R. Biek, P. Formenty, K. Matz-Rensing, B. Schweiger, S. Junglen, H. Ellerbrok, A. Nitsche, T. Briese, W. I. Lipkin, G. Pauli, C. Boesch, and F. H. Leendertz. 2008. Pandemic human viruses cause decline of endangered great apes. Curr. Biol. 18:260-264. [DOI] [PubMed] [Google Scholar]
- 31.Krumbholz, A., O. R. Bininda-Emonds, P. Wutzler, and R. Zell. 2009. Phylogenetics, evolution, and medical importance of polyomaviruses. Infect. Genet. Evol. 9:784-799. [DOI] [PubMed] [Google Scholar]
- 32.Krumbholz, A., P. Wutzler, and R. Zell. 2008. The non-coding region of BK subtype II viruses. Virus Genes 36:27-29. [DOI] [PubMed] [Google Scholar]
- 33.Kwun, H. J., A. Guastafierro, M. Shuda, G. Meinke, A. Bohm, P. S. Moore, and Y. Chang. 2009. The minimum replication origin of Merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J. Virol. 83:12118-12128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leendertz, F. H., H. Ellerbrok, C. Boesch, E. Couacy-Hymann, K. Matz-Rensing, R. Hakenbeck, C. Bergmann, P. Abaza, S. Junglen, Y. Moebius, L. Vigilant, P. Formenty, and G. Pauli. 2004. Anthrax kills wild chimpanzees in a tropical rainforest. Nature 430:451-452. [DOI] [PubMed] [Google Scholar]
- 35.Leendertz, F. H., G. Pauli, K. Maetz-Rensing, W. Boardmann, C. Nunn, H. Ellerbrok, S. A. Jensen, S. Junglen, and C. Boesch. 2006. Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biol. Conserv. 131:325-337. [Google Scholar]
- 36.Lefebure, T., C. J. Douady, M. Gouy, and J. Gibert. 2006. Relationship between morphological taxonomy and molecular divergence within Crustacea: proposal of a molecular threshold to help species delimitation. Mol. Phylogenet. Evol. 40:435-447. [DOI] [PubMed] [Google Scholar]
- 37.Martini, F., A. Corallini, V. Balatti, S. Sabbioni, C. Pancaldi, and M. Tognon. 2007. Simian virus 40 in humans. Infect. Agents Cancer 2:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mezes, B., and P. Amati. 1994. Mutations of polyomavirus VP1 allow in vitro growth in undifferentiated cells and modify in vivo tissue replication specificity. J. Virol. 68:1196-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murata, H., B. Teferedegne, L. Sheng, A. M. Lewis, Jr., and K. Peden. 2008. Identification of a neutralization epitope in the VP1 capsid protein of SV40. Virology 381:116-122. [DOI] [PubMed] [Google Scholar]
- 40.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 41.Padgett, B. L., D. L. Walker, G. M. ZuRhein, R. J. Eckroade, and B. H. Dessel. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257-1260. [DOI] [PubMed] [Google Scholar]
- 42.Perez-Losada, M., R. G. Christensen, D. A. McClellan, B. J. Adams, R. P. Viscidi, J. C. Demma, and K. A. Crandall. 2006. Comparing phylogenetic codivergence between polyomaviruses and their hosts. J. Virol. 80:5663-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pipas, J. M. 1992. Common and unique features of T antigens encoded by the polyomavirus group. J. Virol. 66:3979-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Posada, D. 2008. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25:1253-1256. [DOI] [PubMed] [Google Scholar]
- 45.Pringle, C. R. 1991. The 20th Meeting of the Executive Committee of the ICTV. Virus species, higher taxa, a universal virus database and other matters. Arch. Virol. 119:303-304. [Google Scholar]
- 46.Pulitzer, M. P., B. D. Amin, and K. J. Busam. 2009. Merkel cell carcinoma: review. Adv. Anat. Pathol. 16:135-144. [DOI] [PubMed] [Google Scholar]
- 47.Schowalter, R. M., D. V. Pastrana, K. A. Pumphrey, A. L. Moyer, and C. B. Buck. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stehle, T., and S. C. Harrison. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stehle, T., Y. Yan, T. L. Benjamin, and S. C. Harrison. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160-163. [DOI] [PubMed] [Google Scholar]
- 50.Talavera, G., and J. Castresana. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56:564-577. [DOI] [PubMed] [Google Scholar]
- 51.Toker, C. 1972. Trabecular carcinoma of the skin. Arch. Dermatol. 105:107-110. [PubMed] [Google Scholar]
- 52.van der Meijden, E., R. W. Janssens, C. Lauber, J. N. Bouwes Bavinck, A. E. Gorbalenya, and M. C. Feltkamp. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromised patient. PLoS Pathog. 6:e1001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Heuverswyn, F., Y. Li, C. Neel, E. Bailes, B. F. Keele, W. Liu, S. Loul, C. Butel, F. Liegeois, Y. Bienvenue, E. M. Ngolle, P. M. Sharp, G. M. Shaw, E. Delaporte, B. H. Hahn, and M. Peeters. 2006. Human immunodeficiency viruses: SIV infection in wild gorillas. Nature 444:164. [DOI] [PubMed] [Google Scholar]
- 54.Van Regenmortel, M. H. 2010. Logical puzzles and scientific controversies: the nature of species, viruses and living organisms. Syst. Appl. Microbiol. 33:1-6. [DOI] [PubMed] [Google Scholar]
- 55.Villesen, P. 2007. FaBox: an oline tool for FASTA sequences. Mol. Ecol. Notes 7:965-968. [Google Scholar]
- 56.Wieland, U., C. Mauch, A. Kreuter, T. Krieg, and H. Pfister. 2009. Merkel cell polyomavirus DNA in persons without Merkel cell carcinoma. Emerg. Infect. Dis. 15:1496-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.