Abstract
High-titer autologous neutralizing antibody responses have been demonstrated during early subtype C human immunodeficiency virus type 1 (HIV-1) infection. However, characterization of this response against autologous virus at the monoclonal antibody (MAb) level has only recently begun to be elucidated. Here we describe five monoclonal antibodies derived from a subtype C-infected seroconverter and their neutralizing activities against pseudoviruses that carry envelope glycoproteins from 48 days (0 month), 2 months, and 8 months after the estimated time of infection. Sequence analysis indicated that the MAbs arose from three distinct B cell clones, and their pattern of neutralization compared to that in patient plasma suggested that they circulated between 2 and 8 months after infection. Neutralization by MAbs representative of each B cell clone was mapped to two residues: position 134 in V1 and position 189 in V2. Mutational analysis revealed cooperative effects between glycans and residues at these two positions, arguing that they contribute to a single epitope. Analysis of the cognate gp120 sequence through homology modeling places this potential epitope near the interface between the V1 and V2 loops. Additionally, the escape mutation R189S in V2, which conferred resistance against all three MAbs, had no detrimental effect on virus replication in vitro. Taken together, our data demonstrate that independent B cells repeatedly targeted a single structure in V1V2 during early infection. Despite this assault, a single amino acid change was sufficient to confer complete escape with minimal impact on replication fitness.
The UNAIDS organization estimates that in 2008 alone there were between 2.4 million and 3 million new infections of human immunodeficiency virus (HIV) worldwide, with over half of these occurring in sub-Saharan Africa (61). A vaccine to prevent these new infections is necessary; however, one major obstacle to this goal is the incredible genetic diversity of HIV type 1 (HIV-1) (15). This variation is categorized into viral subtypes and circulating recombinant forms, of which subtype C is essential to study because of its global preponderance and its dominance in sub-Saharan Africa (58). Within the viral genome, genetic variation is concentrated within the env gene, which encodes the surface unit gp120 and transmembrane unit gp41 (25, 28). These two glycoproteins are noncovalently linked and trimerize to form surface spikes on the virion. These trimers not only display the receptor (CD4) and coreceptor (CCR5 and/or CXCR4) binding sites for the virus but are also the main targets of neutralizing antibodies (NAbs) during an immune response (3, 4, 19, 57). HIV vaccine research has recently focused on defining epitopes in gp120 that are associated with neutralization breadth for use in an antibody-based vaccine. However, in early infection, NAb responses raised against the founder virus or a limited set of variants do not usually possess this desirable property and are readily escaped. Thus, a better understanding of the early NAb response during natural infection could lead to clues about how to improve Env immunogens and minimize the potential for escape.
It has been shown that early autologous antibody responses occur within the first few months in HIV-1 infection (1, 2, 6, 18, 31, 49, 65). In subtype C, this response has been shown to be of high potency but strain specific (7, 18, 31). Recent research has begun to illuminate how this NAb response develops. Moore et al. (41) demonstrated that the acute humoral response in four subtype C-infected individuals was quite narrowly targeted against the virus. The NAbs during the first year of infection in these South African subjects had only one or two different specificities, mainly targeting either the V1V2 region or the C3 region of gp120. Furthermore, our group reported that in two subtype C-infected individuals from Zambia, not only was the acute NAb response focused on one or a few regions of Env but the virus escaped by using multiple pathways. Rong et al. (53) demonstrated that in one subject, escape mainly occurred through mutations in the V3 to V5 region of gp120. The requirements for escape, however, changed in this subject over time, sometimes relying on cooperative effects between different regions, such as V1V2 and the gp41 ectodomain, confounding the identification of early NAb epitopes. In a second subject, escape was driven continuously over a 2-year period by changes in V1V2 involving sequence changes as well as potential glycan shifts. Two B cell hybridomas that produced neutralizing monoclonal antibodies (MAbs) were isolated from this individual, allowing a more detailed analysis of viral escape. A potential glycan addition in V2 was suggested to be the dominant escape pathway from these two MAbs. Thus, the potent NAb response in acute subtype C infection has been shown to involve only limited targets in gp120 (often V1V2) and to exert pressure on the virus that is easily escaped, sometimes requiring only a single amino acid change.
The nature of the antibodies that make up this polyclonal plasma response in early infection has not yet been elucidated. Here we expand on our knowledge of the B cell response and neutralization at the monoclonal antibody level during early subtype C infection. Using five MAbs isolated from peripheral memory B cells circulating in a subtype C-infected subject between 49 and 69 months postseroconversion, we show that the MAbs produced by these B cells reflect the plasma pool at 8 months postseroconversion. The MAbs represent antibodies produced from three individual B-cell clones that have undergone somatic hypermutation, and they rely on residues 134 and 189 in V1 and V2, respectively, to neutralize the virus. MAbs 13.6A, 6.4C, and 8.9D have similar but not identical requirements for neutralization; however, the virus appears to develop an efficient escape pathway, becoming resistant to all five MAbs with a single amino acid change in V2. Our present study demonstrates how these clonally distinct antibodies from early infection target a single epitope formed at the interface of V1 and V2 and how the virus escapes without an overt replication fitness cost.
MATERIALS AND METHODS
Env clones.
Details of the Zambia-Emory HIV Research Project (ZEHRP) cohort, sample collection, and processing have been described previously (11, 40, 53, 59). The Envs studied here were derived from a newly infected subject from this cohort (subject 205F) whose polyclonal antibody responses at the plasma level have been studied in detail (53). The Emory University Institutional Review Board and the University of Zambia School of Medicine Research Ethics Committee approved the informed consent and human subjects protocols. This subject did not receive antiretroviral therapy during the evaluation period. Single-genome PCR amplification and cloning of the Envs have been described previously (21, 53). The env genes were cloned into the cytomegalovirus-driven expression plasmids pcDNA 3.1/V5-His TOPO (Invitrogen), which were then used to generate viral pseudotypes. All Envs were derived from plasma or peripheral blood mononuclear cell (PBMC) DNA according to protocols previously described (31) and are subtype C. All amino acid sequences are based on subject 205F 0-month FPL Env 6.3 numbering.
PCR-based site mutagenesis and virus preparation.
To generate V1V2 mutations, site-directed mutagenesis was performed by PCR using two overlapping primers that contained the mutated sequence for each Env using a strategy described previously (51, 53). Briefly, the plasmid containing the 0-month FPL Env 6.3 env gene (plus the Rev- and partial Nef-coding sequences) was amplified using the following set of forward (F) and reverse (R) primer sequences (the mutated nucleotide is underlined, and the locations in HXB2 are provided): N134S F (positions 6613 to 6644; 5′-GCTGTAGCAATTATAGCAATTGTAATGATACC-3′) and N134S R (positions 6613 to 6644; 5′-GGTATCATTACAATTGCTATAATTGCTACAGC-3′), N134Q F (positions 6613 to 6644; 5′-GCTGTAGCAATTATCAGAATTGTAATGATACC-3′) and N134Q R (positions 6618 to 6659; 5′-GGCAGTACTATAGGTATCATTACAATTCTGATAATTGC-3′), N134T F (positions 6613 to 6644; 5′-GCTGTAGCAATTATACCAATTGTAATGATACC-3′) and N134T R (positions 6618 to 6659; 5′-GGCA>GTACTATAGGTATCATTACAATTGGTATAATTGC-3′), N132Q F (positions 6613 to 6644; 5′-GCTGTAGCCAGTATAACAATTGTAATGATACC-3′) and N132Q R (positions 6618 to 6659; 5′-GGCAGTACTATAGGTATCATTACAATTGTTATACTGGC-3′), N134T F (positions 6613 to 6644; 5′-GCTGTAGCAATTATACCAATTGTAATGATACC-3′) nd N134T R (positions 6618 to 6659; 5′-GGCAGTACTATAGGTATCATTACAATTGGTATAATTGC-3′), R189S F (positions 6779 to 6803; 5′-GCCTAATGATAGTAACTCTAGTGAGTATATATTA-3′) and R189S R (positions 6779 to 6803; 5′-TAATATATACTCACTAGAGTTACTATCATTAGGC-3′), and R189H F (positions 6779 to 6799; 5′-GCCTAATGATAGTAACTCTCACGAGTATAT-3′) and R189H R (positions 6787 to 6811; 5′-CAATTTATTAATATATACTCGTGAGAGTTAC-3′).
The amplification conditions were 1 cycle of 95°C for 30 s and 18 cycles of 95°C for 30 s, 45°C for 1 min (the optimal annealing temperature was determined for each primer set), and 68°C for 9 min. The samples were then stored at 4°C. The 50-μl PCR mixtures contained 100 ng of each primer, 10 ng of the plasmid template, 0.2 mM deoxynucleoside triphosphate, and 1× reaction buffer. pfuUltra II Fusion Hotstart DNA polymerase (Stratagene) was used to generate the PCR amplicons, which were digested with DpnI to remove contaminating template DNA and then transformed into maximum-efficiency XL2-Blue ultracompetent cells (>5 × 109 CFU/μg DNA; Stratagene) so that the DNA volume did not exceed 5% of the cell volume. The entire transformation was plated onto LB-ampicillin agar plates, generally resulting in 10 to 50 colonies.
Colonies were inoculated into LB-ampicillin broth for overnight cultures, and the plasmid was prepared using a QIAprep spin miniprep kit. Each Env plasmid (1 μg) was cotransfected into 293T cells along with 2 μg of an Env-deficient subtype B proviral plasmid, pSG3ΔEnv, using the Fugene-6 transfection reagent, according to the manufacturer's instructions (Hoffmann-La Roche). Transfection supernatants were collected at 48 h posttransfection, clarified by low-speed centrifugation for 20 min, aliquoted into portions of 0.5 ml or less, and stored at −80°C. The titer of each pseudotyped virus stock was determined by infecting JC53-BL (Tzm-bl) cells with 5-fold serial dilutions of virus as described previously (11, 31). All env sequences were confirmed by nucleotide sequencing.
Generation of human monoclonal antibodies.
B cells from several viably frozen PBMC samples of subject 205F collected at 49 and 69 months after infection were inoculated with Epstein-Barr virus (EBV) and plated at low cell densities in multiple 96-well tissue culture plates containing irradiated, mature human macrophage feeder cells prepared from HIV-seronegative subjects as previously described (67). Two and 3 weeks later, culture fluids were screened for antibodies that were reactive by enzyme-linked immunosorbent assay (ELISA) and that could neutralize the founder autologous virus envelope 0-month FPB Env 1.1. B cells in antibody-positive wells were serially subcultured at increasing dilutions as described previously (50). Clones of antibody-producing B cell lines from subject 205F did not grow well enough to allow scaling up of MAb production; therefore, MAbs were produced by molecular cloning (20). RNA was isolated from antibody-positive cells and used in reverse transcription-PCRs to amplify VH and VL or VK genes for cloning into expression vectors (kindly provided by Yongjun Guan and George Lewis, University of Maryland). Subsequently, functional pairs of heavy- and light-chain vectors were cotransfected into 293T cells for MAb expression. MAbs were purified from culture supernatants by protein A affinity chromatography.
Screening ELISA for MAbs.
For antibody screening we used a reverse-capture immunoassay, as previously described (50). Briefly, B cell culture fluids were incubated in plates coated with goat anti-human IgG antibodies. Detergent (Triton X-100)-treated supernatant from cells transfected with plasmid expressing subject 205F 0-month FPB Env 1.1 gp160 was added to the wells. Bound HIV-1 Env was detected using a mixture of biotin-labeled human MAbs recognizing several nonoverlapping conserved sites in gp120 and gp41 (50). The wells were incubated with peroxidase-streptavidin, and the signal was developed with tetramethylbenzidine-H2O2 as the substrate. The color reaction was stopped with 1 M phosphoric acid, and the color was read as the optical density (OD)/absorbance at 450 nm. An excess concentration (>100 μg/ml) of human IgG was added to the dilution buffers to prevent the capture of the biotin-labeled human MAbs by the goat anti-human IgG, which would otherwise cause high background readings. OD readings greater than 1.500 were considered positive (they were usually >2.400), and the background OD was usually less than 0.200.
Neutralization screening assay for MAbs.
B cell culture fluids were screened for neutralizing activity in an adaptation of the single-cycle TZM-bl neutralization assay described previously (50). Briefly, supernatants from EBV-transformed B cells or transfected 293T cells were incubated with subject 205F 0-month FPB Env 1.1 pseudovirus in black 96-well culture plates. TZM-bl cells were added to a final concentration of 5 × 103 cells/well with 37.5 μg/ml DEAE-dextran, and the plates were incubated for 48 h. Neutralization was assessed by analyzing the amount of luciferase produced from the pseudovirus-infected cells. Luciferase was quantified using a commercially available kit (BriteGlo; Promega). Typically, neutralizing activity was evident when the reduction in the number of relative light units (RLU) was greater than 70% compared to the average number of RLU in the plate.
Pseudovirus inhibition assays.
Neutralization assays using a panel of five autologous monoclonal antibodies were performed using viral pseudotypes to infect JC53-BL (Tzm-bl) indicator cells using a luciferase readout as described previously (11, 31, 51-53). Briefly, 2,000 IU of pseudovirus was incubated for 1 h in Dulbecco minimal essential medium-10% fetal bovine serum (HyClone)-40 μg/ml DEAE-dextran with serial dilutions of monoclonal antibody, and subsequently, 100 μl was added to the indicator cells for a 48-h infection before being lysed and evaluated for luciferase activity.
Homology modeling of residues 134 and 189 in the V1V2 domain.
The subject 205F EnvPL6.3 gp120 sequence was modeled using the MODELLER program (14). The template for the homology model was a subtype C gp120 obtained by longtime all-atom molecular dynamics simulation using the CHARMM27 (37) potential in the NAMD program (45). This simulated gp120 was modeled using all known CD4-bound gp120 structures (Protein Data Bank [PDB] accession numbers 1G9M [29], 1RZK [24], 2B4C [23], 2NY7 [68], 3JWD [44], 3JWO [44], and 3LMJ [12]) as templates. In all of these structures, the core of gp120 was highly similar; however, it should be noted that 3LMJ was the only high-resolution structure of a subtype C gp120 sequence, while the rest were subtype B. Multiple templates were used because it has been shown that this creates high-quality homology models. In addition, each template has slightly different regions of gp120 resolved. Before modeling of these templates, they were arranged in the trimeric state, which has been resolved using cryoelectron microscopy (PDB accession number 3DNO [33]), in order to ensure that the hypervariable loops did not sterically clash with the neighboring monomers. During modeling, disulfide constraints were added for the conserved cysteines present in all gp120 sequences. All sequence alignments used for modeling templates were based on sequences in the HIV-1 database (www.hiv.lanl.gov). A 128-ns all-atom molecular dynamics simulation of the template described above was used to calculate the potential distance between N134 and R189.
Replication in CD4+ cells using an NL4.3 proviral cassette.
A panel of Envs was subcloned into a replication-competent NL4.3 backbone (34, 43) that we have used previously to evaluate replication of subtype C Envs (36). This system is amenable to accepting diverse env genes and facilitates substitution of virtually the entire coding region (only 36 and 6 amino acids at the N and C termini, respectively, are derived from NL4.3). PBMCs were isolated from the whole blood of a healthy, seronegative donor by Ficoll-Hypaque centrifugation. CD8-depleted PBMC cultures were prepared by negative selection using Dynabeads (Invitrogen). The CD4-enriched PBMCs were cultured in complete RPMI medium for 3 days in the presence of 3 μg/ml phytohemagglutinin (PHA) for activation prior to infection. Infected cultures were maintained in complete RPMI medium supplemented with 30 U/ml recombinant human interleukin-2 (IL-2; Roche) for up to 10 days. Every 2 days, 200 μl of supernatant was collected for p24 analysis (Perkin-Elmer), and this volume was replaced with fresh complete medium with IL-2. The subtype C Env MJ-4 was cloned into the NL4.3 backbone and used as a positive control for these experiments. The infectious subtype C proviral clone MJ-4 (contributed by Thumbi Ndung'u, Boris Renjifo, and Max Essex) (42) was obtained from the NIH AIDS Research and Reference and Reagent Program, Division of AIDS, NIAID, NIH.
Nucleotide sequence accession numbers.
The nucleotide sequences (either V1 to V4 or full length) from the present study have been deposited into GenBank under accession numbers GQ485415 to GQ485427.
RESULTS
Characterization of monoclonal antibodies isolated from a subtype C-infected patient and selected for neutralization activity against the founder Env.
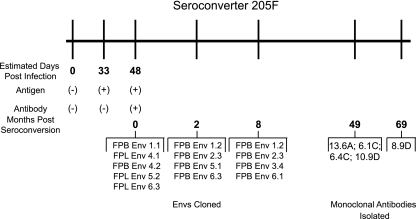
Here we investigated autologous neutralization at the single antibody level by generating monoclonal antibodies isolated from subject 205F, a subtype C-infected seroconverter in the ZEHRP cohort. This subject was identified at a time of antigen-positive, antibody-negative status, allowing a date of infection to be accurately estimated (Fig. 1). After seroconversion, Envs were cloned every 2 to 3 months from both the plasma and PBMCs of subject 205F and used to create a longitudinal panel of pseudoviruses against which autologous plasma was tested (53). Zero-month Envs refer to the first antibody-positive time point. At 49 and 69 months postseroconversion, viable PBMCs were collected from subject 205F and used for selection of memory B cells and production of monoclonal antibodies (Fig. 1). Specificity was tested by screening the antibodies for neutralization activity against a 0-month Env that was representative of the founder virus (53).
FIG. 1.
Timeline of seroconverter 205F from a Zambian cohort. A timeline was created to detail the stages of infection of a female seropositive subject from Zambia. Indicated are the number of days from estimated time of infection when 205F was antigen and/or antibody positive, the months when Envs were cloned out of plasma or PBMCs, and the times when the Ig genes were cloned out of memory B cells to produce monoclonal antibodies.
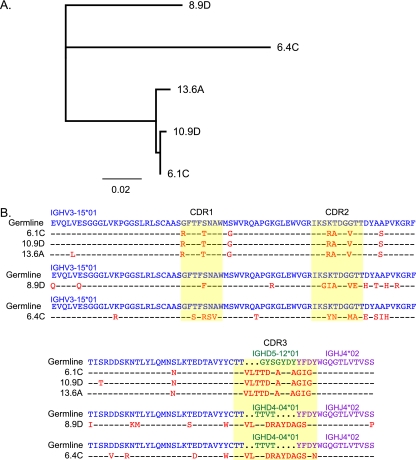
Five MAbs that neutralized the founder virus were recovered by cloning of Ig heavy and light chains. The sequences of the five MAbs revealed that all antibodies were IgG with a gamma heavy chain and a lambda light chain. Furthermore, analysis of the VH regions suggested that these MAbs arose from three distinct B cell clones and had undergone somatic hypermutation. A neighbor-joining tree of the nucleic acid sequences revealed three significant lineages, suggesting that MAbs 6.4C and 8.9D were clonally distinct from the three other MAbs (6.1C, 10.9D, and 13.6A) (Fig. 2A). All five MAbs were derived from heavy-chain alleles IGHV3-15*01 and IGHJ4*02 with MAbs 6.4C and 8.9D using IGHD4-04*01 and MAbs 6.1C, 10.9D, and 13.6A using IGHD5-12*01. An amino acid alignment highlighted the fact that although MAbs 6.1C, 10.9D, and 13.6A have nearly identical sequences, there were 1 to 2 amino acid differences between them, indicating somatic variation (Fig. 2B). Further analysis using the Joinsolver program confirmed that the latter three antibodies have the same DJ (D gene and J gene) joining sequence and the same number of n nucleotides (nontemplated junctional additions) at this junction, thus verifying that these are three somatic variants that arose from the same B cell clone (see Table S1 in the supplemental material). MAbs 6.4C and 8.9D were very similar in the CDR3 region; however, they did differ by 5 nucleotides (data not shown), and their DJ joining sequences and n nucleotides were different (see Table S1 in the supplemental material). Thus, we conclude that they arose from two distinct B cell clones, although we cannot distinguish between this inference and the possibility that they evolved over time after arising from the same B cell. All five antibodies had a heavy-chain CDR3 region composed of 16 amino acids, although the sequences differed according to the clone (Fig. 2B). The MAbs showed evidence of affinity maturation as well. The frequencies of mutations compared to the germ line sequence were 8.84% for MAb 8.9D and MAb 12.93% for 6.4C and ranged from 3.74 to 4.42% for the three MAb variants 6.1C, 10.9D, and 13.6A. These nonsilent mutations were preferentially found in the complementarity-determining regions (CDRs) (see Table S1 in the supplemental material). Thus, there appeared to be three genetically distinct B cell clones that were isolated from subject 205F.
FIG. 2.
Sequence analysis of MAbs isolated from seroconverter 205F. A neighbor-joining tree of the nucleotide sequences of the five MAbs was created using the Jukes-Cantor model in Geneious software (A). The scale is indicated. An amino acid alignment of the VH genes of these five MAbs plus the germ line sequences was generated using ClustalX in Geneious (B). Germ line V alleles were obtained from Joinsolver (http://joinsolver.niaid.nih.gov/) and are indicated in blue, germ line D alleles are indicated in green, and germ line J alleles are indicated in purple. CDRs are highlighted in yellow, while mutations from the germ line are in red.
The monoclonal antibodies likely arose before 8 months postseroconversion.
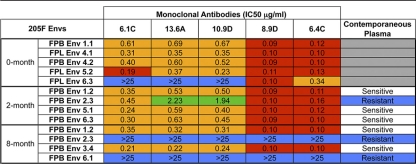
Previous studies suggested that two of these monoclonal antibodies arose during early infection of subject 205F (53). Here, all five monoclonal antibodies were tested for neutralization activity against a panel of Envs cloned from samples collected at between 0 and 8 months postseroconversion (Table 1). The MAbs neutralized four of the five 0-month Envs fairly potently, with 50% inhibitory concentrations (IC50s) ranging from 0.09 to 0.69 μg/ml. Surprisingly, the 0-month Env 6.3 was not neutralized by all of the MAbs. Three of the five antibodies could not reach 50% neutralization at the highest concentration tested (25 μg/ml); however, antibodies 6.4C and 8.9D did neutralize this Env, with IC50s being 0.34 and 0.1 μg/ml, respectively. At 2 months postseroconversion, all four Envs tested were sensitive to neutralization by the MAbs, although, again, MAbs 6.4C and 8.9D were the most potent antibodies, with all IC50s being under 0.2 μg/ml. By 8 months postseroconversion, two Envs were resistant to all five monoclonal antibodies, and this pattern of sensitivity and resistance to neutralization by the MAbs mirrored exactly the neutralization pattern of the contemporaneous 8-month plasma sample. The fact that 8-month Envs 2.3 and 6.1 were resistant to both plasma and MAbs while Envs 1.2 and 3.4 were sensitive to both suggests that these MAbs likely represent the antibodies circulating in the plasma near this time point after seroconversion.
TABLE 1.
Neutralization sensitivity of a longitudinal panel of subject 205F Env pseudoviruses to autologous monoclonal antibodies and contemporaneous plasmaa
Envs from time points relative to seroconversion are shown in the left-hand column. IC50s are indicated for each MAb-Env combination. The neutralization phenotype of each Env to contemporaneous subject 205F plasma is described in the right-hand column. Red indicates IC50 < 0.2 μg/ml; orange, 0.2 μg/ml < IC50 < 1.0 μg/ml; green, 1.0 μg/ml < IC50 < 25 μg/ml; and blue, IC50 > 25 μg/ml.
Glycosylation in V1 can confer sensitivity to monoclonal antibodies.
Previously it had been shown that V1V2 was the major determinant of resistance to neutralizing antibodies in subject 205F during the first 2 years of infection (53). Specifically, two residues, at position 134 in V1 and 189 in V2, that were each part of a potential N-linked glycosylation site were shown to influence neutralization sensitivity to MAbs 6.4C and 13.6A, but it was not clear whether the amino acid substitution or altered glycan motif at each site was responsible (position 189 was numbered position 197 elsewhere [53]). Here, the requirement for these specific residues and for N-linked glycosylation at both positions was more fully explored using a mutagenesis strategy to detect neutralization determinants of three MAbs representative of the three distinct B-cell lineages (MAbs 6.4C, 13.6A, and 8.9D). The 0-month Env 6.3 was used as the background Env for these experiments because it lacked potential glycosylation motifs at both of these positions.
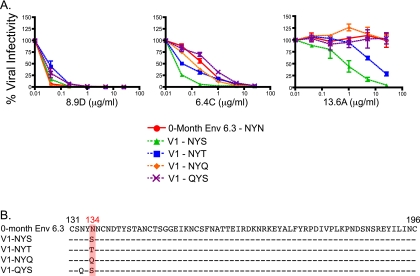
Env 6.3 contains the sequence NYN at positions 132 to 134 in V1 (Fig. 3B) and is highly sensitive to neutralization by MAb 8.9D, sensitive to 6.4C, and resistant to 13.6A (Fig. 3A). When the N134S mutation was introduced, thereby inserting the glycosylation motif NYS into V1, the Env remained sensitive to 8.9D and sensitivity to 6.4C increased, lowering the IC50 from 0.34 to 0.1 μg/ml. Most interestingly, Env 6.3 became fully sensitive to MAb 13.6A, indicating that either the serine or the glycan at this position might comprise a target for this antibody. The role of the potential glycan was further investigated by creating additional mutations that either did (NYT) or did not (NYQ, QYS) create glycosylation motifs in Env 6.3 (Fig. 3B). Sensitivity to MAb 8.9D did not appear to be affected by any of the mutations, and so all Envs remained potently neutralized. MAb 6.4C neutralized all Envs as well, regardless of glycan motif; however, the V1 NYS Env remained especially sensitive to neutralization compared to the other four Envs, despite the fact that the NYT variant also introduced a glycosylation site at this position. MAb 13.6A required a glycan motif (either NYS or NYT) for neutralization, while all other nonglycosylated mutations were resistant to this MAb. Once again, the V1-NYS Env was almost 1 log10 unit more sensitive than the V1-NYT Env, having IC50s of 0.77 and 7.19 μg/ml, respectively. The results for MAbs 6.4C and 13.6A suggest that although glycosylation status clearly plays a role in neutralization sensitivity, there might be differential efficiency of glycosylation at each sequon.
FIG. 3.
MAb neutralization of Env 6.3 and V1 mutants. Zero-month FPL Env 6.3 was used as a backbone for mutations at residues 134 and 132, and these Envs were assessed for sensitivity to MAbs 8.9D, 6.4C, and 13.6A using pseudoviruses (A). Each line on the graph represents an individual Env pseudovirus, listed in the key. Percent viral infectivity compared to that with no MAb is shown on the vertical axis and was calculated from luciferase units by dividing virus-infected wells in the presence of antibody by virus-infected wells in the absence of antibody. The concentration of each MAb (in μg/ml) is plotted on the horizontal axis on a log10 scale. Error bars represent the standard errors of the means of at least 2 independent experiments. An amino acid alignment of all Envs used is shown with a red box highlighting residue 134, and HXB2 standard numbering is indicated in black (B).
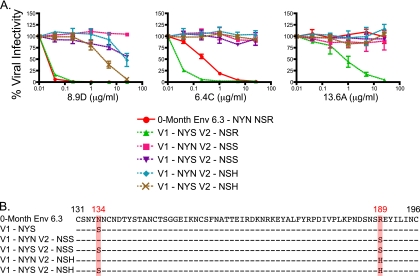
Amino acid sequence rather than glycosylation status in V2 defines neutralization sensitivity to MAbs.
As shown previously, MAb 8.9D potently neutralized 0-month Env 6.3 regardless of whether the N134S mutation (V1-NYS) was present. However, an R189S mutation, present in an 8-month escape variant, inserted the NSS glycan motif at the terminal end of V2 (Fig. 4B) and was sufficient to render the Env completely resistant to this MAb (Fig. 4A, Env V1-NYN V2-NSS). Surprisingly, glycosylation at this site was not necessary to induce resistance to MAb 8.9D. Substitution of a histidine for R189 resulted in a 200-fold increase in resistance to the MAb (Fig. 4A). Interestingly, the effect of mutating R189 was further modulated by the presence of the NYS glycan motif in V1. While the V1-NYN V2-NSS Env was resistant to 25 μg/ml of MAb 8.9D, it was neutralized 50% at the same concentration when the V1-NYS motif was present (Env V1-NYS V2-NSS). A similar result was seen when V1-NYN V2-NSH and V1-NYS V2-NSH were compared, with the V1 glycosylation motif increasing net sensitivity 10-fold. Collectively, these data suggest a cooperative effect between these two positions in neutralization by MAb 8.9D, with R189 acting as the dominant neutralization determinant. Thus, although R189 was necessary for potent neutralization by MAb 8.9D, changes at both residue 189 and residue 134 modulate sensitivity to this MAb.
FIG. 4.
MAb neutralization of Env 6.3 and V2 mutants. Zero-month FPL Env 6.3 was used as a backbone for mutations at residue 189 in conjunction with residue 134, and these Envs were assessed for sensitivity to MAbs 8.9D, 6.4C, and 13.6A using pseudoviruses (A). Each line on the graph represents an individual Env pseudovirus, listed in the key. Percent viral infectivity compared to that with no MAb is shown on the vertical axis and was calculated from luciferase units by dividing virus-infected wells in the presence of antibody by virus-infected wells in the absence of antibody. The concentration of each MAb (in μg/ml) is plotted on the horizontal axis on a log10 scale. Error bars represent the standard errors of the means of at least 2 independent experiments. An amino acid alignment of all Envs used is shown with a red box highlighting residues 134 and 189, and HXB2 standard numbering is indicated in black (B).
A somewhat different synergy between residues 134 and 189 was seen with MAb 6.4C. As shown above, the 0-month Env 6.3 was sensitive to neutralization by this MAb and changes at position 134 did increase neutralization sensitivity by 10-fold (Fig. 3A and 4A). Interestingly, substitutions at position 134 had no effect on neutralization in the context of the R189 mutations for MAb 6.4C. When either the V2-NSS mutation or the V2-NSH mutation was introduced, the Envs became completely resistant, irrespective of whether the V1 glycan motif was present or not. Thus, like MAb 8.9D, MAb 6.4C requires an R189 residue for neutralization, but in this case the requirement is absolute and loss of this residue resulted in complete resistance.
In contrast, MAb 13.6A exhibited an absolute requirement for the V1 glycosylation site and the R189 residue for neutralization. Thus, despite containing the V1 NYS glycan motif, Envs V1-NYS V2-NSS and V1-NYS-NSH were resistant to MAb 13.6A (Fig. 4A). Similarly, any Env that lacked the V1 glycosylation site was resistant as well (Fig. 3A and 4A). Therefore, the two residues at positions 134 and 189 play a key cooperative role in antibody neutralization by MAb 13.6A.
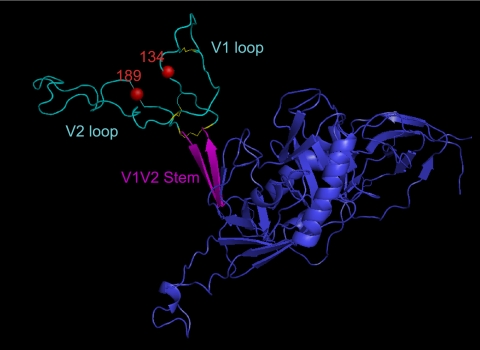
Residues 134 and 189 may contribute to a single epitope near the V1V2 stem.
On the linear amino acid sequence, positions 134 and 189 occur in the N- and C-terminal regions, respectively, of the V1V2 domain. To gain insight into their position with respect to the three-dimensional gp120 and, in particular, the V1V2 loop, residues 134 and 189 were viewed on a composite, CD4-bound gp120 structure onto which the subject 205F Env 6.3 sequence was homology modeled. Figure 5 displays one possible conformation in which these residues coalesce at the interface of the V1 and V2 loops in a region that could be stabilized by multiple disulfide bonds, including an unusual pair of cysteines in the V1 loop that are found in most subject 205F Envs. This potential epitope is also located directly above the V1V2 stem, which is a highly conserved structure. The mean distance between the C-alpha atoms of N134 and R189 on the basis of the molecular dynamics simulation is 27 (±2) angstroms, which is consistent with the formation of an antibody epitope. For example, amino acids that comprise the epitope for MAb b12 can be as far apart as 37 Å (68). Thus, it is feasible that the subject 205F MAbs recognize slightly different versions of a novel conformational epitope at the interface of V1 and V2 comprised of these two residues and, in the case of MAb 13.6A, carbohydrate moieties. Furthermore, the variations in neutralization determinants of the three MAbs suggest that this region of V1V2 may exist in multiple conformations, or orientations, several of which elicited a B cell response.
FIG. 5.
Model of residues 134 and 189 in the V1V2 domain of 0-month FPL Env 6.3. Homology modeling was used (see Materials and Methods) to generate a three-dimensional structure of 0-month FPL Env 6.3 gp120 (in blue) and visualized in the MacPyMOL program. The V1V2 domain is shown in cyan, with disulfide bonds in this region highlighted in yellow. The V1V2 stem is labeled and shown in magenta. Residues 134 and 189 are represented by red spheres.
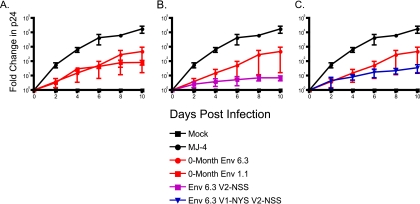
Amino acid changes at residues 134 and 189 do not overtly affect replication fitness in vitro.
To date, the question of whether specific autologous neutralization escape mutations exact a replication cost on the virus has not been explored. To investigate this possibility, the 0-month Env 1.1 and the 0-month Env 6.3 with the V1-NYS and V2-NSS motifs introduced singly and in combination were cloned into a replication-competent NL4.3 proviral backbone and used to infect PBMCs enriched for CD4 T cells by CD8 depletion. Supernatant was collected every 2 days, and viral p24 was quantified for a measure of viral replication for a 10-day period (Fig. 6). The subtype C provirus MJ-4 was used as a positive control for replication in CD4 T cells (42), and none of the Envs tested replicated as well as MJ-4. The 0-month Env 1.1, which contained the V1 to V4 region of the founder virus (53), replicated moderately well, while the 0-month Env 6.3, which resisted neutralization by MAb 13.6A due to its lack of a glycosylation motif in V1, replicated to similar if slightly better levels by day 10 (Fig. 6A). It is important to note, however, that small differences in replication are not apparent in this assay because of the large variation introduced by using ex vivo cells from three different donors. Thus, in this assay, the ability to escape MAb neutralization by losing a glycan sequon did not adversely affect replication. To completely escape neutralization by all three MAbs (MAbs 13.6A, 6.4C, and 8.9D), however, the 0-month Env 6.3 required a change at position R189, which in one 8-month escape virus was a serine (Fig. 4A). Interestingly, introducing this autologous escape mutation (NSS) into Env 6.3 had a detrimental effect on replication (Fig. 6B), a fact which may explain why the V1-NYN V2-NSS lineage is not seen circulating at later time points in seroconvertor 205F (53). Instead, the escape pathway utilized by later Envs was to create the R189S change while preserving the V1 glycosylation motif. Indeed, when the V2-NSS mutation was introduced into Env 6.3 in combination with V1-NYS, the virus replicated better than the V2-NSS Env, suggesting that the V1 NYS may in some way compensate for the V2-NSS replication cost (Fig. 6C). Thus, these results argue that the R189S mutation that confers resistance against autologous MAbs had only a minor effect on replication in this in vitro assay system in the context of the V1 glycosylation site, although it is important to note that this mutation was tested in the absence of other compensatory changes that may have been present in vivo.
FIG. 6.
Replication kinetics of parent and mutant Envs in PMBCs. Zero-month Envs 6.3 and 1.1 (A) as well as mutant Envs V2-NSS (B) and V1-NYS V2-NSS (C) were placed into a replication-competent NL4.3 backbone and used to infect CD8-depleted human PBMCs. Viral p24 antigen production (pg/ml) in the supernatant was measured by ELISA on day 10 and is plotted as the fold change over that on day 0 on the vertical axis on a log10 scale. The positive-control virus MJ-4 is shown in black circles, and the mock-infected negative control is shown by black squares. The error bars for each Env were calculated from three independent experiments using PBMCs from three different seronegative donors.
DISCUSSION
By 8 months postseroconversion, multiple B cell clones produced somatic variations of antibodies that neutralize the virus.
In HIV-1 infection, V1V2 has been shown to be a target of NAbs (26, 63), and multiple monoclonal antibodies isolated from seropositive subjects bind this region (16, 17, 22). This domain can confer escape from NAbs as well through either shielding effects or sequence change (47, 51, 54). Furthermore, V1V2 has been shown to confer strain specificity to the NAb response during simian-human immunodeficiency virus (SHIV) infection of rhesus macaques (30). Thus, it is not surprising that in addition to V5 and C3 in gp120, the V1V2 region is targeted by early antibodies during subtype C infection.
The current report is one of the first to reveal that in one subtype C-infected subject, the early NAb response is focused upon V1V2 but is actually comprised of multiple antibody specificities, all targeting the same structure within V1V2. Sequence analysis revealed that these five antibodies were mostly likely derived from three distinct B cell clones, although it is also possible that MAbs 6.4C and 8.9D evolved over time from a single B cell. All five showed variation from the germ line preferentially in the CDR, indicating somatic hypermutation. MAbs 6.4C and 8.9D were more highly mutated than the other three MAbs and also displayed greater breadth and potency against the autologous Envs. These observations suggest that MAbs 6.4C and 8.9D had undergone more extensive affinity maturation that increased their efficacy against autologous viral variants. Collectively, the five subject 205F MAbs neutralized all Envs from 0 and 2 months postseroconversion; however, by 8 months, the plasma escape variants were also refractory to neutralization by the MAbs. These data suggest that although these MAbs were isolated from B cells circulating 4 to 5 years after infection, they are representative of the major plasma pool at 8 months postseroconversion. Furthermore, what appeared to be a monospecific response against V1V2 in plasma is actually multiple B cell clones producing distinct neutralizing antibodies against this target.
Mutational analysis reveals that residues 134 and 189 contribute to a novel epitope near the V1V2 stem.
It has been well documented that V1V2 can contain neutralization and escape determinants from NAbs (41, 47, 51, 53, 54, 62), and in seroconvertor 205F, escape through sequence changes in V1V2 was demonstrated previously (53). Here we define a potential conformational epitope at the V1V2 interface in this subject that is glycan dependent in one case. The three MAbs from 205F evaluated in this study (MAbs 13.6A, 6.4C, and 8.9D) are dependent upon positions 134 and 189 in V1V2 for neutralization to various degrees, with the MAbs showing a decreasing dependence on glycosylation at residue N132 (13.6A > 6.4C > 8.9D). MAb 13.6A was the only one that absolutely required a glycan sequon (residues 132 to 134) in V1 for neutralization; however, the NYS and NYT glycosylation motifs were not equivalent in neutralization sensitivity, differing by 10-fold.
Overall, the results imply that the three MAbs recognize a common epitope formed by positions 134 and 189 at the interface of the V1V2 loop. It is possible that each MAb binds the epitope in a subtly different orientation and thus is influenced differentially by surrounding residues. This phenomenon would be similar to the variations seen in MAbs that recognize the CD4 binding site (9). A similar pattern was reported for a series of MAbs that were generated against SHIV162P in macaques (50). The region encompassing the V1V2 stem has been shown to affect both CD4 binding and formation of the bridging sheet, leading to the possibility that the subject 205F MAbs neutralize by a common mechanism in which they block gp120 interactions with CD4 or CCR5.
Glycosylation plays various roles in neutralization during early infection.
Studies of glycosylation patterns in gp120 are commonly performed on the sequence alone, with the addition of putative N-linked glycan motifs often being associated with neutralization resistance. Indeed, there is ample evidence for the role of the “glycan shield” as a robust mechanism of viral escape from NAb (8, 27, 32, 35, 39, 65, 66). Nevertheless, we provide here further data indicating that the reverse is also true. MAb 2G12 has long been identified as a broadly neutralizing antibody that is directed against glycans in gp120 (55, 56, 60), and other monoclonal antibodies that require a glycan for binding have been described (13, 63, 64). Our data offer another demonstration that antiglycan specificities occur naturally during infection and provide an example of how removing a glycosylation motif in V1 can result in autologous neutralization resistance. It is also important to note that the sequence changes associated with creating glycan motifs can themselves be important for neutralization (5, 50). Overall, changes in glycan motifs can affect neutralization in multiple ways, not all requiring glycosylation, making this an efficient scheme for the virus to utilize for NAb escape.
Replication kinetics of neutralization-resistant Envs are similar to those of neutralization-sensitive Envs.
In addition to reducing neutralization sensitivity, escape mutations in Env could potentially affect replication fitness and result in a lower viral load. One study found a negative correlation between autologous titer and viral load in chronic infection, but another demonstrated no correlation between these two parameters in long-term nonprogressors (10, 38). In subtype C infection, a slight dip in viral load was observed concurrently with the appearance of certain autologous NAb specificities (41). In vitro data have demonstrated no correlation between resistance to broadly neutralizing antibodies and replication (48) but has found that alanine mutations of targeted epitopes may result in loss of infectivity (46). To date, specific mutations that arise naturally during infection and confer resistance to autologous NAbs have not been examined for their effect on replication kinetics in vitro. Here we investigated the in vitro replication kinetics in CD4 T cells of Envs with and without the V2-NSS motif, which confers global autologous MAb escape. In the context of the 0-month Env 6.3, the V2-NSS motif had a detrimental effect on replication, suggesting that fitness cost may play a role in autologous escape pathways. However, HIV-1 uses multiple escape pathways and compensatory mutations to overcome this disadvantage. It is possible that further differences could have been detected in a more sensitive fitness competition assays; however, these data suggest that it is quite possible for Env to escape the humoral immune response without overt loss of replication capability.
Overall, this study describes a potential novel conformational epitope that is present in a subtype C-infected subject during early infection and that could be located at the interface of the V1 and V2 loops. This potential epitope was likely recognized by at least three different B cell receptors on the native Env trimer and elicited both glycan-dependent and -independent MAbs. Thus, there are complex and immunogenic structures formed by V1V2 on the native Env, and these may exist in slightly different conformations or be recognized in slightly different ways by distinct B cells. While we presently studied only one subject, these MAbs are likely to be representative of those that arise during the early neutralizing response in many HIV-1-infected subjects, as this response is often focused on only one or a few targets and is readily escaped. The findings provide new insight into why the early autologous NAb response is ineffective and fails to contain the virus. The results also illustrate the power of a single, strategically placed amino acid change in viral escape. The redundant and highly focused nature of the MAbs in this subject suggests that to elicit NAb breadth with an Env immunogen, it will be critical to present only the desired epitope, such as the CD4 binding site, and eliminate others that might divert the B cell response.
Supplementary Material
Acknowledgments
We thank the interns, staff, participants, and project management group at ZEHRP. We also gratefully acknowledge productive input from Joshy Jacob.
The work described in this paper was supported by NIH grant AI-58706. S.G. was supported by the collaboration for AIDS Vaccine Discovery (Bill and Melinda Gates Foundation) and LANL/DOE grant X9R8. A.S. was supported by CNLS.
Footnotes
Published ahead of print on 27 October 2010.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Aasa-Chapman, M. M., A. Hayman, P. Newton, D. Cornforth, I. Williams, P. Borrow, P. Balfe, and A. McKnight. 2004. Development of the antibody response in acute HIV-1 infection. AIDS 18:371-381. [DOI] [PubMed] [Google Scholar]
- 2.Arendrup, M., C. Nielsen, J. E. Hansen, C. Pedersen, L. Mathiesen, and J. O. Nielsen. 1992. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 5:303-307. [PubMed] [Google Scholar]
- 3.Barin, F., M. F. McLane, J. S. Allan, T. H. Lee, J. E. Groopman, and M. Essex. 1985. Virus envelope protein of HTLV-III represents major target antigen for antibodies in AIDS patients. Science 228:1094-1096. [DOI] [PubMed] [Google Scholar]
- 4.Binley, J. M., E. A. Lybarger, E. T. Crooks, M. S. Seaman, E. Gray, K. L. Davis, J. M. Decker, D. Wycuff, L. Harris, N. Hawkins, B. Wood, C. Nathe, D. Richman, G. D. Tomaras, F. Bibollet-Ruche, J. E. Robinson, L. Morris, G. M. Shaw, D. C. Montefiori, and J. R. Mascola. 2008. Profiling the specificity of neutralizing antibodies in a large panel of plasmas from patients chronically infected with human immunodeficiency virus type 1 subtypes B and C. J. Virol. 82:11651-11668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch, K. A., S. Rainwater, W. Jaoko, and J. Overbaugh. 2010. Temporal analysis of HIV envelope sequence evolution and antibody escape in a subtype A-infected individual with a broad neutralizing antibody response. Virology 398:115-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bunnik, E. M., L. Pisas, A. C. van Nuenen, and H. Schuitemaker. 2008. Autologous neutralizing humoral immunity and evolution of the viral envelope in the course of subtype B human immunodeficiency virus type 1 infection. J. Virol. 82:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bures, R., L. Morris, C. Williamson, G. Ramjee, M. Deers, S. A. Fiscus, S. Abdool-Karim, and D. C. Montefiori. 2002. Regional clustering of shared neutralization determinants on primary isolates of clade C human immunodeficiency virus type 1 from South Africa. J. Virol. 76:2233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chackerian, B., L. M. Rudensey, and J. Overbaugh. 1997. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J. Virol. 71:7719-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., Y. D. Kwon, T. Zhou, X. Wu, S. O'Dell, L. Cavacini, A. J. Hessell, M. Pancera, M. Tang, L. Xu, Z. Y. Yang, M. Y. Zhang, J. Arthos, D. R. Burton, D. S. Dimitrov, G. J. Nabel, M. R. Posner, J. Sodroski, R. Wyatt, J. R. Mascola, and P. D. Kwong. 2009. Structural basis of immune evasion at the site of CD4 attachment on HIV-1 gp120. Science 326:1123-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deeks, S. G., B. Schweighardt, T. Wrin, J. Galovich, R. Hoh, E. Sinclair, P. Hunt, J. M. McCune, J. N. Martin, C. J. Petropoulos, and F. M. Hecht. 2006. Neutralizing antibody responses against autologous and heterologous viruses in acute versus chronic human immunodeficiency virus (HIV) infection: evidence for a constraint on the ability of HIV to completely evade neutralizing antibody responses. J. Virol. 80:6155-6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Derdeyn, C. A., J. M. Decker, F. Bibollet-Ruche, J. L. Mokili, M. Muldoon, S. A. Denham, M. L. Heil, F. Kasolo, R. Musonda, B. H. Hahn, G. M. Shaw, B. T. Korber, S. Allen, and E. Hunter. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019-2022. [DOI] [PubMed] [Google Scholar]
- 12.Diskin, R., P. M. Marcovecchio, and P. J. Bjorkman. 2010. Structure of a clade C HIV-1 gp120 bound to CD4 and CD4-induced antibody reveals anti-CD4 polyreactivity. Nat. Struct. Mol. Biol. 17:608-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doores, K. J., and D. R. Burton. 2010. Variable loop glycan dependency of the broad and potent HIV-1 neutralizing antibodies PG9 and PG16. J. Virol. 84:10510-10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eswar, N., B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian, M. Y. Shen, U. Pieper, and A. Sali. 2006. Comparative protein structure modeling using Modeller, unit 5-6. In Current protocols in bioinformatics. John Wiley & Sons, Inc., New York, NY. [DOI] [PMC free article] [PubMed]
- 15.Gaschen, B., J. Taylor, K. Yusim, B. Foley, F. Gao, D. Lang, V. Novitsky, B. Haynes, B. H. Hahn, T. Bhattacharya, and B. Korber. 2002. Diversity considerations in HIV-1 vaccine selection. Science 296:2354-2360. [DOI] [PubMed] [Google Scholar]
- 16.Gorny, M. K., J. P. Moore, A. J. Conley, S. Karwowska, J. Sodroski, C. Williams, S. Burda, L. J. Boots, and S. Zolla-Pazner. 1994. Human anti-V2 monoclonal antibody that neutralizes primary but not laboratory isolates of human immunodeficiency virus type 1. J. Virol. 68:8312-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorny, M. K., L. Stamatatos, B. Volsky, K. Revesz, C. Williams, X. H. Wang, S. Cohen, R. Staudinger, and S. Zolla-Pazner. 2005. Identification of a new quaternary neutralizing epitope on human immunodeficiency virus type 1 virus particles. J. Virol. 79:5232-5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gray, E. S., P. L. Moore, I. A. Choge, J. M. Decker, F. Bibollet-Ruche, H. Li, N. Leseka, F. Treurnicht, K. Mlisana, G. M. Shaw, S. S. Karim, C. Williamson, and L. Morris. 2007. Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J. Virol. 81:6187-6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gray, E. S., N. Taylor, D. Wycuff, P. L. Moore, G. D. Tomaras, C. K. Wibmer, A. Puren, A. DeCamp, P. B. Gilbert, B. Wood, D. C. Montefiori, J. M. Binley, G. M. Shaw, B. F. Haynes, J. R. Mascola, and L. Morris. 2009. Antibody specificities associated with neutralization breadth in plasma from human immunodeficiency virus type 1 subtype C-infected blood donors. J. Virol. 83:8925-8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guan, Y., M. M. Sajadi, R. Kamin-Lewis, T. R. Fouts, A. Dimitrov, Z. Zhang, R. R. Redfield, A. L. DeVico, R. C. Gallo, and G. K. Lewis. 2009. Discordant memory B cell and circulating anti-Env antibody responses in HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:3952-3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haaland, R. E., P. A. Hawkins, J. Salazar-Gonzalez, A. Johnson, A. Tichacek, E. Karita, O. Manigart, J. Mulenga, B. F. Keele, G. M. Shaw, B. H. Hahn, S. A. Allen, C. A. Derdeyn, and E. Hunter. 2009. Inflammatory genital infections mitigate a severe genetic bottleneck in heterosexual transmission of subtype A and C HIV-1. PLoS Pathog. 5:e1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Honnen, W. J., C. Krachmarov, S. C. Kayman, M. K. Gorny, S. Zolla-Pazner, and A. Pinter. 2007. Type-specific epitopes targeted by monoclonal antibodies with exceptionally potent neutralizing activities for selected strains of human immunodeficiency virus type 1 map to a common region of the V2 domain of gp120 and differ only at single positions from the clade B consensus sequence. J. Virol. 81:1424-1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, C. C., M. Tang, M. Y. Zhang, S. Majeed, E. Montabana, R. L. Stanfield, D. S. Dimitrov, B. Korber, J. Sodroski, I. A. Wilson, R. Wyatt, and P. D. Kwong. 2005. Structure of a V3-containing HIV-1 gp120 core. Science 310:1025-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, C. C., M. Venturi, S. Majeed, M. J. Moore, S. Phogat, M. Y. Zhang, D. S. Dimitrov, W. A. Hendrickson, J. Robinson, J. Sodroski, R. Wyatt, H. Choe, M. Farzan, and P. D. Kwong. 2004. Structural basis of tyrosine sulfation and VH-gene usage in antibodies that recognize the HIV type 1 coreceptor-binding site on gp120. Proc. Natl. Acad. Sci. U. S. A. 101:2706-2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunter, E. 1997. Viral entry and receptors, p. 71-121. In J. M. Coffin and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory, Plainview, NY. [PubMed]
- 26.Kayman, S. C., Z. Wu, K. Revesz, H. Chen, R. Kopelman, and A. Pinter. 1994. Presentation of native epitopes in the V1/V2 and V3 regions of human immunodeficiency virus type 1 gp120 by fusion glycoproteins containing isolated gp120 domains. J. Virol. 68:400-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koch, M., M. Pancera, P. D. Kwong, P. Kolchinsky, C. Grundner, L. Wang, W. A. Hendrickson, J. Sodroski, and R. Wyatt. 2003. Structure-based, targeted deglycosylation of HIV-1 gp120 and effects on neutralization sensitivity and antibody recognition. Virology 313:387-400. [DOI] [PubMed] [Google Scholar]
- 28.Korber, B., B. Gaschen, K. Yusim, R. Thakallapally, C. Kesmir, and V. Detours. 2001. Evolutionary and immunological implications of contemporary HIV-1 variation. Br. Med. Bull. 58:19-42. [DOI] [PubMed] [Google Scholar]
- 29.Kwong, P. D., R. Wyatt, S. Majeed, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Structure 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 30.Laird, M. E., T. Igarashi, M. A. Martin, and R. C. Desrosiers. 2008. Importance of the V1/V2 loop region of simian-human immunodeficiency virus envelope glycoprotein gp120 in determining the strain specificity of the neutralizing antibody response. J. Virol. 82:11054-11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li, B., J. M. Decker, R. W. Johnson, F. Bibollet-Ruche, X. Wei, J. Mulenga, S. Allen, E. Hunter, B. H. Hahn, G. M. Shaw, J. L. Blackwell, and C. A. Derdeyn. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J. Virol. 80:5211-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li, Y., B. Cleveland, I. Klots, B. Travis, B. A. Richardson, D. Anderson, D. Montefiori, P. Polacino, and S. L. Hu. 2008. Removal of a single N-linked glycan in human immunodeficiency virus type 1 gp120 results in an enhanced ability to induce neutralizing antibody responses. J. Virol. 82:638-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, J., A. Bartesaghi, M. J. Borgnia, G. Sapiro, and S. Subramaniam. 2008. Molecular architecture of native HIV-1 gp120 trimers. Nature 455:109-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohrengel, S., F. Hermann, I. Hagmann, H. Oberwinkler, L. Scrivano, C. Hoffmann, D. von Laer, and M. T. Dittmar. 2005. Determinants of human immunodeficiency virus type 1 resistance to membrane-anchored gp41-derived peptides. J. Virol. 79:10237-10246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lynch, R. M., R. Rong, B. Li, T. Shen, W. Honnen, J. Mulenga, S. Allen, A. Pinter, S. Gnanakaran, and C. A. Derdeyn. 2010. Subtype-specific conservation of isoleucine 309 in the envelope V3 domain is linked to immune evasion in subtype C HIV-1 infection. Virology 404:59-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.MacKerell, J. A. 1998. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102:3586-3616. [DOI] [PubMed] [Google Scholar]
- 38.Mahalanabis, M., P. Jayaraman, T. Miura, F. Pereyra, E. M. Chester, B. Richardson, B. Walker, and N. L. Haigwood. 2009. Continuous viral escape and selection by autologous neutralizing antibodies in drug-naive human immunodeficiency virus controllers. J. Virol. 83:662-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCaffrey, R. A., C. Saunders, M. Hensel, and L. Stamatatos. 2004. N-linked glycosylation of the V3 loop and the immunologically silent face of gp120 protects human immunodeficiency virus type 1 SF162 from neutralization by anti-gp120 and anti-gp41 antibodies. J. Virol. 78:3279-3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna, S. L., G. K. Muyinda, D. Roth, M. Mwali, N. Ng'andu, A. Myrick, C. Luo, F. H. Priddy, V. M. Hall, A. A. von Lieven, J. R. Sabatino, K. Mark, and S. A. Allen. 1997. Rapid HIV testing and counseling for voluntary testing centers in Africa. AIDS 11(Suppl. 1):S103-S110. [PubMed] [Google Scholar]
- 41.Moore, P. L., N. Ranchobe, B. E. Lambson, E. S. Gray, E. Cave, M. R. Abrahams, G. Bandawe, K. Mlisana, S. S. Abdool Karim, C. Williamson, and L. Morris. 2009. Limited neutralizing antibody specificities drive neutralization escape in early HIV-1 subtype C infection. PLoS Pathog. 5:e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ndung'u, T., B. Renjifo, and M. Essex. 2001. Construction and analysis of an infectious human immunodeficiency virus type 1 subtype C molecular clone. J. Virol. 75:4964-4972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neumann, T., I. Hagmann, S. Lohrengel, M. L. Heil, C. A. Derdeyn, H. G. Krausslich, and M. T. Dittmar. 2005. T20-insensitive HIV-1 from naive patients exhibits high viral fitness in a novel dual-color competition assay on primary cells. Virology 333:251-262. [DOI] [PubMed] [Google Scholar]
- 44.Pancera, M., S. Majeed, Y. E. Ban, L. Chen, C. C. Huang, L. Kong, Y. D. Kwon, J. Stuckey, T. Zhou, J. E. Robinson, W. R. Schief, J. Sodroski, R. Wyatt, and P. D. Kwong. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc. Natl. Acad. Sci. U. S. A. 107:1166-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phillips, J. C., R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kale, and K. Schulten. 2005. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26:1781-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pietzsch, J., J. F. Scheid, H. Mouquet, F. Klein, M. S. Seaman, M. Jankovic, D. Corti, A. Lanzavecchia, and M. C. Nussenzweig. 2010. Human anti-HIV-neutralizing antibodies frequently target a conserved epitope essential for viral fitness. J. Exp. Med. 207:1995-2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinter, A., W. J. Honnen, Y. He, M. K. Gorny, S. Zolla-Pazner, and S. C. Kayman. 2004. The V1/V2 domain of gp120 is a global regulator of the sensitivity of primary human immunodeficiency virus type 1 isolates to neutralization by antibodies commonly induced upon infection. J. Virol. 78:5205-5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quakkelaar, E. D., E. M. Bunnik, F. P. van Alphen, B. D. Boeser-Nunnink, A. C. van Nuenen, and H. Schuitemaker. 2007. Escape of human immunodeficiency virus type 1 from broadly neutralizing antibodies is not associated with a reduction of viral replicative capacity in vitro. Virology 363:447-453. [DOI] [PubMed] [Google Scholar]
- 49.Richman, D. D., T. Wrin, S. J. Little, and C. J. Petropoulos. 2003. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc. Natl. Acad. Sci. U. S. A. 100:4144-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robinson, J. E., K. Franco, D. H. Elliott, M. J. Maher, A. Reyna, D. C. Montefiori, S. Zolla-Pazner, M. K. Gorny, Z. Kraft, and L. Stamatatos. 2010. Quaternary epitope specificities of anti-HIV-1 neutralizing antibodies generated in rhesus macaques infected by the simian/human immunodeficiency virus SHIVSF162P4. J. Virol. 84:3443-3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rong, R., F. Bibollet-Ruche, J. Mulenga, S. Allen, J. L. Blackwell, and C. A. Derdeyn. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J. Virol. 81:1350-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rong, R., S. Gnanakaran, J. M. Decker, F. Bibollet-Ruche, J. Taylor, J. N. Sfakianos, J. L. Mokili, M. Muldoon, J. Mulenga, S. Allen, B. H. Hahn, G. M. Shaw, J. L. Blackwell, B. T. Korber, E. Hunter, and C. A. Derdeyn. 2007. Unique mutational patterns in the envelope alpha 2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J. Virol. 81:5658-5668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rong, R., B. Li, R. M. Lynch, R. E. Haaland, M. K. Murphy, J. Mulenga, S. A. Allen, A. Pinter, G. M. Shaw, E. Hunter, J. E. Robinson, S. Gnanakaran, and C. A. Derdeyn. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog. 5:e1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sagar, M., X. Wu, S. Lee, and J. Overbaugh. 2006. Human immunodeficiency virus type 1 V1-V2 envelope loop sequences expand and add glycosylation sites over the course of infection, and these modifications affect antibody neutralization sensitivity. J. Virol. 80:9586-9598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders, R. W., M. Venturi, L. Schiffner, R. Kalyanaraman, H. Katinger, K. O. Lloyd, P. D. Kwong, and J. P. Moore. 2002. The mannose-dependent epitope for neutralizing antibody 2G12 on human immunodeficiency virus type 1 glycoprotein gp120. J. Virol. 76:7293-7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Scanlan, C. N., R. Pantophlet, M. R. Wormald, E. Ollmann Saphire, R. Stanfield, I. A. Wilson, H. Katinger, R. A. Dwek, P. M. Rudd, and D. R. Burton. 2002. The broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2G12 recognizes a cluster of alpha1→2 mannose residues on the outer face of gp120. J. Virol. 76:7306-7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scheid, J. F., H. Mouquet, N. Feldhahn, M. S. Seaman, K. Velinzon, J. Pietzsch, R. G. Ott, R. M. Anthony, H. Zebroski, A. Hurley, A. Phogat, B. Chakrabarti, Y. Li, M. Connors, F. Pereyra, B. D. Walker, H. Wardemann, D. Ho, R. T. Wyatt, J. R. Mascola, J. V. Ravetch, and M. C. Nussenzweig. 2009. Broad diversity of neutralizing antibodies isolated from memory B cells in HIV-infected individuals. Nature 458:636-640. [DOI] [PubMed] [Google Scholar]
- 58.Taylor, B. S., M. E. Sobieszczyk, F. E. McCutchan, and S. M. Hammer. 2008. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 358:1590-1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Trask, S. A., C. A. Derdeyn, U. Fideli, Y. Chen, S. Meleth, F. Kasolo, R. Musonda, E. Hunter, F. Gao, S. Allen, and B. H. Hahn. 2002. Molecular epidemiology of human immunodeficiency virus type 1 transmission in a heterosexual cohort of discordant couples in Zambia. J. Virol. 76:397-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trkola, A., M. Purtscher, T. Muster, C. Ballaun, A. Buchacher, N. Sullivan, K. Srinivasan, J. Sodroski, J. P. Moore, and H. Katinger. 1996. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 70:1100-1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.UNAIDS. 2009. AIDS epidemic update. World Health Organization, Geneva, Switzerland.
- 62.Walker, L. M., S. K. Phogat, P. Y. Chan-Hui, D. Wagner, P. Phung, J. L. Goss, T. Wrin, M. D. Simek, S. Fling, J. L. Mitcham, J. K. Lehrman, F. H. Priddy, O. A. Olsen, S. M. Frey, P. W. Hammond, S. Kaminsky, T. Zamb, M. Moyle, W. C. Koff, P. Poignard, and D. R. Burton. 2009. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science 326:285-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walker, L. M., M. D. Simek, F. Priddy, J. S. Gach, D. Wagner, M. B. Zwick, S. K. Phogat, P. Poignard, and D. R. Burton. 2010. A limited number of antibody specificities mediate broad and potent serum neutralization in selected HIV-1 infected individuals. PLoS Pathog. 6:e1001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Warrier, S. V., A. Pinter, W. J. Honnen, M. Girard, E. Muchmore, and S. A. Tilley. 1994. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68:4636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 66.Wolk, T., and M. Schreiber. 2006. N-glycans in the gp120 V1/V2 domain of the HIV-1 strain NL4-3 are indispensable for viral infectivity and resistance against antibody neutralization. Med. Microbiol. Immunol. 195:165-172. [DOI] [PubMed] [Google Scholar]
- 67.Xiang, S. H., N. Doka, R. K. Choudhary, J. Sodroski, and J. E. Robinson. 2002. Characterization of CD4-induced epitopes on the HIV type 1 gp120 envelope glycoprotein recognized by neutralizing human monoclonal antibodies. AIDS Res. Hum. Retrovir. 18:1207-1217. [DOI] [PubMed] [Google Scholar]
- 68.Zhou, T., L. Xu, B. Dey, A. J. Hessell, D. Van Ryk, S. H. Xiang, X. Yang, M. Y. Zhang, M. B. Zwick, J. Arthos, D. R. Burton, D. S. Dimitrov, J. Sodroski, R. Wyatt, G. J. Nabel, and P. D. Kwong. 2007. Structural definition of a conserved neutralization epitope on HIV-1 gp120. Nature 445:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.