Abstract
Previous comparisons of different rabies virus (RV) strains suggested an inverse relationship between pathogenicity and the amount of glycoprotein produced in infected cells. In order to provide more insight into this relationship, we pursued an experimental approach that allowed us to alter the glycoprotein expression level without altering the glycoprotein sequence, thereby eliminating the contribution of amino acid changes to differences in viral virulence. To this end, we constructed an infectious clone of the highly pathogenic rabies virus strain CVS-N2c and replaced its cognate glycoprotein gene with synthetic versions in which silent mutations were introduced to replace wild-type codons with the most or least frequently used synonymous codons. A recombinant N2c variant containing the fully codon-optimized G gene and three variants carrying a partially codon-deoptimized G gene were recovered on mouse neuroblastoma cells and shown to express 2- to 3-fold more and less glycoprotein, respectively, than wild-type N2c. Pathogenicity studies in mice revealed the WT-N2c virus to be the most pathogenic strain. Variants containing partially codon-deoptimized glycoprotein genes or the codon-optimized gene were less pathogenic than WT-N2c but still caused significant mortality. We conclude that the expression level of the glycoprotein gene does have an impact on pathogenicity but is not a dominant factor that determines pathogenicity. Thus, strategies such as changes in codon usage that aim solely at altering the expression level of the glycoprotein gene do not suffice to render a pathogenic rabies virus apathogenic and are not a viable and safe approach for attenuation of a pathogenic strain.
According to estimates by the WHO, 55,000 human lives are lost per year as a result of bites inflicted by rabid animals (41). The vast majority of rabies cases occur in young children in Asia and Africa and can be attributed to infections with genotype 1 lyssaviruses, which are nonsegmented, negative-stranded RNA viruses that encode five structural proteins. While the correlates of immune protection are well defined and effective postexposure treatment has been developed, it is often not available, particularly in poor countries. If postexposure treatment is not administered and patients develop clinical signs of rabies, the outcome of the infection is almost always death (14, 25, 32).
While basically all mammals are susceptible to rabies, a comparatively small number of carnivores, mainly dogs, foxes, raccoons, raccoon dogs, skunks, and mongooses, serve as wildlife reservoirs. In addition, bats have been recognized as important vectors of rabies virus (RV) and of a large number of related lyssaviruses that occasionally cause rabies-like disease in humans. Bats also likely serve as a primary reservoir from which rabies spreads to terrestrial animals. Although eradication of rabies virus poses a formidable challenge (with respect to bats in particular), vaccination of the primary terrestrial vector species is the most effective method of controlling rabies (11, 29, 30). There is therefore a strong incentive for developing cheap and effective vaccines that are suitable for mass vaccination of wildlife carriers of rabies.
Since Louis Pasteur's first attempt to attenuate rabies virus, many approaches have been pursued to reduce the virulence of viruses, resulting in the development of live attenuated vaccines against a number of RNA viruses, including rabies virus itself. These traditional vaccines still play a prominent if not dominant role in the viral vaccine field, despite considerable and in some cases successful efforts to develop alternatives. However, replication-competent viral vectors carry the intrinsic risk of mutating or reverting to a more virulent phenotype. Considerable effort has therefore been expended on improving the safety of viral vectors. Among the most promising developments are single-cycle vectors, which are unable to produce infectious progeny virus in noncomplementing cells due to the lack of an essential gene and are therefore limited to one round of infection in the initial target cells of the vaccine recipient (7). Recently, a new method of attenuation based on large-scale changes in codon or codon-pair usage was successfully tested in poliovirus and has been proposed as a universal strategy for attenuation of many different RNA viruses (4, 26). This strategy, dubbed “attenuation by a thousand cuts,” promises safer vaccines that are less prone to revert to a pathogenic phenotype than traditional live vaccines.
In the case of rabies virus, safety is of prime concern because of the nearly 100% mortality resulting from infections with pathogenic virus strains. For this reason, only killed vaccines have been approved for use in humans. However, eradication of rabies from its wildlife reservoirs is currently feasible only through the use of live attenuated viruses (31). Thus, numerous approaches have been pursued to develop alternatives to traditional live rabies virus vaccines (33). Among these are recombinant viral vectors (such as canine adenovirus and poxviruses) that express the rabies glycoprotein (37, 40), which is the main protective antigen and the target of neutralizing antibodies (5, 42). Considerable effort has also been expended on improving the safety of live rabies virus vectors. These efforts include site-directed mutagenesis of the matrix and glycoprotein gene, insertions of proapoptotic and antiviral genes, gene deletions, and duplication of the glycoprotein gene (2, 8-10, 13, 17, 24, 28, 35, 43). The latter approach was formulated on the basis of the finding that there appears to be an inverse correlation between pathogenicity and glycoprotein expression level (23). This conclusion was largely derived from comparisons of different rabies virus strains. Later studies using recombinant rabies viruses that carry glycoprotein genes of other rabies virus strains were designed to verify this conclusion (21, 45). However, to date no study has addressed the relationship between glycoprotein expression level and pathogenicity using an approach that eliminates the effect on viral virulence of differences in glycoprotein sequences. Here we describe recombinant variants of the highly pathogenic rabies virus strain CVS-N2c that carry synthetic codon-optimized or -deoptimized versions of the N2c glycoprotein gene. Utilizing these variants, which have the same amino acid sequence but express different amounts of glycoprotein, we show that minor reductions in the glycoprotein level do not significantly change pathogenicity whereas larger changes do reduce pathogenicity to some extent but do not eliminate it.
MATERIALS AND METHODS
Virus growth and RNA purification.
CVS-N2c was grown on NA cells at 34°C, and virus particles were recovered from culture supernatants by first pelleting cell debris at 1,500 × g for 10 min. The virus was then concentrated and purified by centrifugation at 120,000 × g for 60 min twice through 20% sucrose in TEN buffer (100 mM Tris-HCl at pH 7.6, 100 mM NaCl, 10 mM EDTA). RNA was isolated from purified virions by the use of an RNeasy kit (Qiagen) according to the instructions of the manufacturer and used for reverse transcription-PCR (RT-PCR) amplification and sequence determination of the viral genome.
Sequence determination of the N2C strain.
A nearly full-length sequence (GenBank accession number DQ875050) with close to 100% homology to the N2c glycoprotein sequence (GenBank accession number AF042823) (22) was identified by database searches and utilized to design primers for RT-PCR amplification of viral cDNA by the use of a One-Step RT-PCR kit (Invitrogen) and for use with a commercial rapid amplification of cDNA ends (RACE) kit (Invitrogen) to determine the terminal sequences of the N2c strain. A full-length sequence was compiled by direct sequencing of PCR fragments and deposited in the GenBank database under accession number 362716.
Plasmid constructs.
A full-length infectious clone was assembled by stepwise cloning of five cDNA fragments into a Bluescript SK(−) vector (Stratagene) containing a cytomegalovirus (CMV) promoter, T7 promoter hammerhead ribozyme, and hepatitis delta virus ribozyme (19) following published strategies (15, 18). PCR mutagenesis was used to flank the glycoprotein gene with XmaI and NheI sites. The wild-type (WT) glycoprotein gene was then replaced with synthetic variants that were codon-deoptimized or codon-optimized over the entire length of the gene, apart from a few nucleotide positions that were left unchanged to preserve restriction sites that are present in the wild-type sequence. The D40N chimera was constructed by replacing the PvuII-SphI fragment (codons 43 to 240) with the corresponding region of the codon-deoptimized gene. The D40C chimera was constructed by replacing the fragment extending from the single BsmBI site to the 3′ end of the wild-type gene (codons 322 to 524) with the corresponding region of the codon-deoptimized gene, and the D80 chimera was constructed by replacing both fragments with the codon-deoptimized regions. For transient expression in mammalian cells, the full-length glycoprotein genes were inserted into the pCAGGS vector (27). Codon optimization and codon deoptimization were performed using the mouse codon usage table available at www.kazusa.or.jp and Upgene (12) or GeneDesigner (DNA2.0, Menlo Park, CA) program packages.
Recovery of infectious viruses and amplification of virus.
Infectious virus was recovered as described in reference 43 except that 0.5 μg of a plasmid expressing T7 RNA polymerase was included in the transfection mixture and NA mouse neuroblastoma cells were used instead of BSR cells to recover and amplify recombinant virus. NA cells were grown in RPMI medium containing 5% fetal calf serum (FCS) or in Cellgro serum-free medium for preparation of viral stocks. Titration of viral stocks was performed using 96-well plates by infecting 3 to 4 × 104 NA cells with 10-fold serial dilutions of virus stock for 48 to 60 h at 34°C. Cells were fixed with 80% acetone, stained with fluorescein isothiocyanate (FITC)-labeled monoclonal antibody (MAb) against the nucleoprotein for several hours at 37°C, and examined under a fluorescence microscope.
One-step and multistep growth analysis of viruses.
NA cell monolayers were infected at a multiplicity of infection (MOI) of 0.05 for multistep growth curves and at an MOI of 5 for single-step growth curves. After 1 h of incubation at 37°C, the inoculum was removed, cells were washed three times with phosphate-buffered saline (PBS), and 3 ml of fresh serum-free medium was added. Supernatants were harvested at the time points indicated in the figures, and virus titers were determined in triplicate on NA monolayers as described previously.
Mouse pathogenicity studies.
Groups of 5 to 10 male C57/B6 mice 6 to 10 weeks of age were inoculated intramuscularly with 5 to 10 × 105 or 5 × 106 focus-forming units (FFU) of recombinant virus diluted in PBS. The mice were monitored over 4 weeks for clinical signs of rabies and weight loss. Any mouse that lost more than 20% of its preinfection body weight was euthanized. Survival curve comparisons were performed using Prism software (GraphPad Software, San Diego, CA), and statistical significance was determined using the log-rank (Mantel-Haenszel) test.
Western blotting.
Mouse neuroblastoma cells were infected at an MOI of 2 to 5 at 34°C. At 24 to 48 h postinfection, the cells were washed once in PBS and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate [SDS], 1× protease inhibitor cocktail [Sigma]) on ice for 30 min. The suspension was transferred to a microcentrifuge tube and spun for 10 min at 16,000 × g to remove cell debris. Proteins were separated by SDS-10% polyacrylamide gel electrophoresis (SDS-10% PAGE) and transferred to a nitrocellulose membrane (Whatman, Minnetonka, MN). Blots were blocked for 1 h in 5% dry milk powder in Tris-buffered saline (TBS) (pH 7.4). After being blocked, blots were washed twice using a 0.05% TBS-Tween 20 solution and incubated with polyclonal rabbit anti-RV G antibody overnight at 4°C. Blots were then washed four times with 0.1% TBS-Tween. Secondary goat anti-rabbit or goat anti-mouse horseradish peroxidase-conjugated antibodies (Jackson ImmunoResearch, West Grove, PA) (diluted 1:50,000) were added, and blots were incubated for 1 h at room temperature. Blots were washed four times with 0.1% TBS-Tween and washed once with PBS (pH 7.4). Chemiluminescence analysis using WestPico substrate (Thermo Fisher Scientific, Rockford, IL) was performed as instructed by the vendor.
FACS analysis.
Monolayers of NA cells grown in six-well plates were infected at an MOI of 5 and incubated for 24 to 72 h at 34°C. Cells were resuspended and washed once in fluorescence-activated cell sorter (FACS) buffer (PBS, 2% bovine serum albumin [BSA], 0.02% sodium azide). After centrifugation, cells were fixed with 2% paraformaldehyde for 20 min at 4°C and washed three times with FACS buffer. To stain for surface glycoprotein, fixed cells were incubated with human anti-RV G CR57 MAb (6) diluted 1:400 in FACS buffer followed by Dylight 649-conjugated goat anti-human antibody (Jackson Immunoresearch, West Grove, PA) diluted 1:300. For staining of intracellular protein, cells were incubated with FITC-conjugated monoclonal antibody against RV strain N (Fujirebio, Malvern, PA) diluted 1:200 in Cytoperm buffer (BD Biosciences) for 30 min at 4°C and washed twice with Cytoperm and once with FACS buffer. Flow cytometry was performed on a FACSCalibur system (Becton Dickinson). Results were analyzed using FlowJo software (TreeStar, Ashland, OR).
RNA isolation and Northern blotting.
Monolayers of NA cells grown in six-well plates were infected at an MOI of 5 and incubated for 48 h at 37°C. Cells were washed once in PBS, and RNA was isolated using an RNeasy Mini kit following the instructions of the manufacturer (Qiagen, Valencia, CA). For Northern blotting, 4 μg of total RNA was denatured with glyoxal for 30 min at 50°C, separated on a 0.9% agarose gel in MOPS (morpholinepropanesulfonic acid) buffer, and transferred to a positively charged nylon membrane by the use of reagents provided in a NorthernMax-Gly kit (Ambion, Austin, TX). After UV cross-linking was performed, the membrane was hybridized at 48°C with heat-denatured DNA probes at a concentration of 5 pM. The Pseudogene and M probes were chemically labeled using a Psoralen-Biotin kit from Ambion. Posthybridization treatment, incubation with streptavidin-alkaline phosphatase (streptavidin-AP) conjugate, and incubation with CDP-Star substrate were carried out following the instructions provided with a Brightstar BioDetect kit from Ambion.
Nucleotide sequence accession number.
A full-length sequence of strain N2C was compiled by direct sequencing of PCR fragments and deposited in the GenBank database under accession number 362716.
RESULTS
To address the relationship between glycoprotein level and pathogenesis, we decided to utilize an infectious clone of the CVS-N2c strain. The rationale for doing so was threefold. (i) The N2c glycoprotein has been extensively used to demonstrate the crucial role of the glycoprotein for pathogenicity, neurotropism, and neuroinvasiveness (21, 23). (ii) The conclusion that pathogenicity inversely correlates with glycoprotein expression level was originally derived by comparing N2c and the closely related B2c variant of the CVS-24 RV strain. (iii) N2c is a highly pathogenic and neurotropic virus and is therefore better suited to analysis of attenuation of pathogenic RV than the already highly attenuated SAD and NC-HL strains, which served as a genetic background in earlier studies of the RV glycoprotein and its role in pathogenicity (21, 34, 36).
Construction of the infectious clone was aided by the availability of a nearly full-length sequence of the CVS-24 strain in the PubMed database and was accomplished by stepwise cloning of cDNA fragments into an existing vector that carries promoters and ribozyme sequences for generation of an exact copy of the antigenomic RNA. To be able to distinguish recombinant N2c from CVS-N2c, an AflII site upstream of the glycoprotein gene in CVS-N2c was replaced with an XmaI site (Fig. 1A). Infectious virus (rN2c) was recovered in mouse neuroblastoma cells and confirmed to be as pathogenic in mice as CVS-N2c after peripheral inoculation (Fig. 1B and C).
FIG. 1.
Genomic map and pathogenicity of wild-type CVS-N2c virus and the recombinant rN2c virus recovered on mouse neuroblastoma cells. (A) A full-length infectious clone of CVS-N2c virus was constructed by stepwise assembly of cDNA fragments, utilizing the five singular restriction sites shown in the genomic map of rN2C. An AflII site upstream of the glycoprotein gene in CVS-N2c was replaced with an XmaI site for cloning purposes and as a marker for identification of the recombinant virus. (B and C) Groups of 10 Swiss Webster mice 6 to 8 weeks of age were inoculated intramuscularly with 1 × 106 infectious particles of CVS-N2c or the recombinant rN2C virus and monitored daily for weight loss (B) and mortality (C). Mice that lost more than 20% of their initial weight or showed hind leg paralysis were euthanized. Mean weight curves are shown for each day postinfection until day 10, when all of the CVS-N2c-infected mice (n = 10) and 9 of 10 mice infected with rN2c succumbed to disease. One mouse in the rN2c group survived the infection without showing signs of disease by day 14.
In order to alter the amount of glycoprotein produced in cells infected with rN2c, we designed a codon-optimized and a codon-deoptimized synthetic version of the N2c glycoprotein gene. To verify that the synthetic genes were functional and that the changes in codon usage would have the desired effect, we first carried out transient transfections in NA cells. As expected, transfection of the codon-optimized construct resulted in increased expression compared to wild-type gene results, whereas the codon-deoptimized gene showed decreased expression compared to the WT gene (Fig. 2). In fact, the deoptimized gene showed such low expression that we decided to construct two chimeras that were only partially codon-deoptimized in the 5′ and 3′ half of the gene, respectively, and a third variant that combined the two codon-deoptimized stretches to generate a chimera (rN2c-D80) that was deoptimized over approximately 80% of the length of the glycoprotein gene. As expected, all three chimeric genes showed higher expression than the fully deoptimized gene but lower expression than the wild-type gene.
FIG. 2.
N2C glycoprotein genes with altered codon usage (A) and their expression levels in NA cells (B). A fully codon-optimized version (Opt) or deoptimized version (D100) of the N2C glycoprotein gene was generated by introduction of 384 or 440 silent mutations over the entire length of the gene to replace wild-type codons with the most or least frequently used synonymous codon where necessary. Additional partially codon-deoptimized genes were generated by replacing codons 43 to 240 and codons 322 to 524 of the wild-type gene with the corresponding fragments of the fully deoptimized version. The synthetic genes were placed under the control of a chicken actin promoter and their expression levels monitored by Western blotting of NA cell lysates obtained at 48 h posttransfection. The lysates were probed with antibodies against actin and rabies virus glycoprotein (G). Two different exposure times are shown for the G probe. Control lysate from NA cells transfected with a vector expressing red fluorescent protein (RFP) was analyzed in parallel. Numbers in panel A indicate codon positions in the glycoprotein gene.
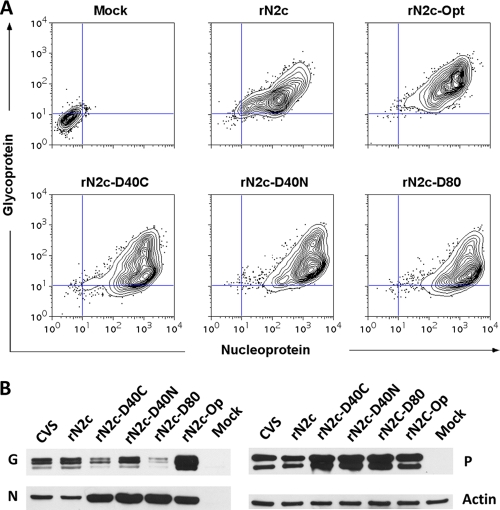
To analyze the effect of the changes in codon usage on protein expression in infected cells, we transferred the synthetic glycoprotein genes into a full-length infectious clone of the N2c strain in which the glycoprotein gene was flanked by XmaI and NheI sites to allow for easy exchange of the wild-type gene. The engineered rN2c virus was confirmed to be phenotypically indistinguishable from CVS-N2c in terms of protein expression and growth kinetics (Fig. 3A and B; see also Fig. 5B). Infectious virus was recovered on mouse neuroblastoma cells from all of the synthetic recombinants except for that which carried the fully deoptimized gene. Growth curves demonstrated that the codon-deoptimized recombinants grew more slowly than wild-type virus, whereas the codon-optimized variant grew with kinetics similar to those of wild-type N2c (Fig. 3C and D). The deoptimized variants also spread more slowly, as revealed by staining of infected NA monolayers (Fig. 4). Interestingly, the staining performed with monoclonal antibody against the nucleoprotein showed a stronger signal for the deoptimized viruses compared to wild-type virus results.
FIG. 3.
Growth kinetics of N2c strains on mouse neuroblastoma cells. Multistep (A, C, and D) or one-step (B) growth curves of the CVS-N2c and recombinant N2c strains are shown. NA cells were infected at an MOI of 5 (one-step) or 0.01 (multistep) and incubated at 34°C. Aliquots of tissue culture supernatant were collected and subjected to titration in triplicate on NA cells. The recombinant N2c strain encoding WT-G grew with kinetics similar to those of CVS-N2c (A and B), whereas the N2c strains containing codon-deoptimized glycoprotein genes grew more slowly (D). The N2c strain carrying the codon-optimized gene grew with kinetics similar to those of CVS-N2C but reached slightly higher titers at later times postinfection (C). Growth curves are representative of the results of two independent experiments.
FIG. 4.
Viral spread in mouse neuroblastoma cells. NA cells were infected with N2c virus strains at an MOI of 0.005 for 60 h at 34°C, stained with FITC-conjugated monoclonal antibody against RV nucleoprotein, and examined under a fluorescence microscope. Wild-type N2c and the recombinant virus carrying the codon-optimized glycoprotein gene formed significantly larger foci than the recombinants containing codon-deoptimized glycoprotein genes. NA cells infected with wild-type virus also showed dimmer green fluorescence than cells infected with the codon-deoptimized variants, indicating lower intracellular accumulation of nucleoprotein. For each virus, three representative images taken from different areas of the same slide are shown.
To evaluate the expression level of the glycoprotein, we performed flow cytometric analysis of infected NA cells after subjecting them to staining with monoclonal antibodies against the glycoprotein G (Fig. 5A). As expected, we observed an increase in the glycoprotein level in cells infected with the codon-optimized N2c strain compared to wild-type N2c levels, whereas glycoprotein expression was lower in cells infected with codon-deoptimized variants of N2c. Simultaneous staining with antibody against N confirmed that all cells were infected. To verify the difference in glycoprotein expression and to assess expression of other viral proteins, we also performed Western blotting using lysates from infected NA cells (Fig. 5B). As with the result obtained by immunofluorescence staining of infected cells, we observed a significant increase in the expression level of the nucleoprotein in the deoptimized variants. Notably, the expression level of P was also increased in the deoptimized variants, while the glycoprotein expression was lower. An increase in intracellular protein accumulation was also observed in cells infected with the codon-optimized variant. However, the increase was generally lower than the increase seen in cells infected with the deoptimized viruses.
FIG. 5.
Viral protein expression in mouse neuroblastoma cells. (A) NA cells were infected at an MOI of 5 for 40 h at 34°C, fixed, and stained with monoclonal antibody against RV-G followed by staining with fluorescently labeled secondary antibody and FITC-labeled monoclonal antibody against the RV. (B) NA cells were infected at a MOI of 5 for 36 h at 34°C and analyzed by Western blotting with polyclonal antiserum against RV G, N, and P and a monoclonal antibody against actin.
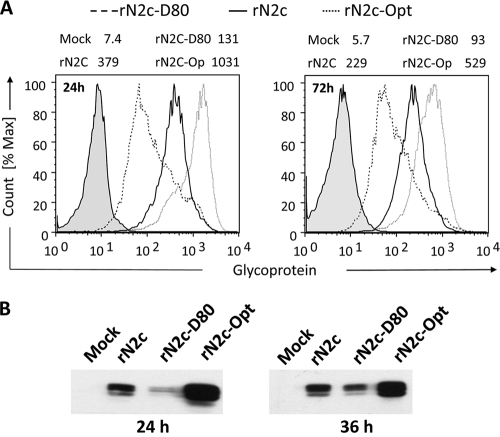
To quantify the amounts of glycoprotein produced by the different viruses, we performed flow cytometric analysis on cells that were infected for 24 or 72 h. Surface staining with anti-G MAb revealed a 2- to 3-fold reduction of G glycoprotein levels in cells infected with deoptimized viruses compared to WT results and a 2-fold increase in glycoprotein in cells infected with the codon-optimized virus (Fig. 6A). The difference in the amount of glycoprotein expression compared to wild-type virus levels was highest at early times after infection. At later times, the expression level gradually approached that of the wild-type virus. This was also confirmed by Western blotting (Fig. 6B).
FIG. 6.
Viral protein expression in mouse neuroblastoma cells. (A) NA cells were infected at an MOI of 5 for 24 and 72 h at 34°C, fixed, stained with monoclonal antibody against the RV glycoprotein, and analyzed by flow cytometry. Numbers at the top of the panels indicate mean geometric fluorescence results. The analysis shows a 3-fold difference in glycoprotein expression between N2c (containing the wild-type G gene) and the rN2C-D80 and rN2C-Opt variants at 24 h postinfection. At 72 h postinfection, the difference was approximately 2-fold. (B) Western blot analysis of glycoprotein expression. NA cells were infected with N2c virus strains at an MOI of 5 at 34°C. Cell lysate was prepared 24 and 36 h postinfection and analyzed by Western blotting with polyclonal antiserum against RV-G.
The observed changes in protein expression levels could potentially have been caused by changes in translation efficiency or by changes in transcript levels. To distinguish between these two possibilities, we performed Northern blot analysis on RNA isolated from NA cells at 52 h postinfection (Fig. 7). Genomic RNA and glycoprotein mRNA were detected using a probe for the pseudogene that is located downstream of the glycoprotein gene on the same mRNA. The pseudogene sequence was left unaltered in the codon-modified viruses, allowing detection of the codon-modifed G mRNAs by the use of a wild-type pseudogene probe. Our results show that replication was not negatively affected, as the amount of genomic RNA produced by the codon-altered viruses was very similar to the amount produced by wild-type virus. Likewise, transcription was not negatively affected in either of the codon-altered variants. In contrast, we did observe a slight increase in G mRNA in the 80%-deoptimized virus. This increase was most likely due to increased stability of the mRNA, as transcription levels of the N and M genes were unaltered (data not shown for N mRNA). These results indicate that the changes in protein expression level observed in the codon-altered viruses were largely due to altered translation efficiencies.
FIG. 7.
Northern blot analysis of viral RNA production in mouse neuroblastoma cells. (A) Total RNA was isolated at 48 h postinfection and probed with a 450-nucleotide probe specific for the pseudogene region that is located between G and L and transcribed as part of the G messenger RNA. (B) Northern analysis of total RNA with a probe specific for the pseudogene and a second probe specific for the matrix protein mRNA.
To evaluate the pathogenicity of the recombinant viruses, C57BL/6 mice were infected intramuscularly with 5 to 10 × 105 infectious particles and monitored daily for signs of rabies and weight loss. Mice infected with wild-type virus started losing weight on day 5 postinfection, after which their health deteriorated rapidly (Fig. 8). By day 7 to 8, most animals showed severe signs of disease and hind leg paralysis. By day 10, a total of 85% of the mice had to be euthanized, as their body weight had dropped below 80% of the initial weight and the animals had reached the terminal stage of disease. Very similar mortality rates were observed in mice that were infected with the D40N deoptimized variant. However, disease progression was slightly delayed compared to that seen with wild-type virus-infected mice, and the maximum mortality rate of 80% was reached only on day 16. A similar clinical picture (but a somewhat lower mortality level of 70%) was observed in mice infected with the D40C variant. Mice that were infected with the D80 variant displayed the most pronounced delay in disease progression and the lowest mortality rate, with 68% of animals succumbing to the infection. Of note, 1 out of 60 mice infected with the D80 virus lost weight transiently, starting at day 10 with signs of ataxia, but started to recover on day 19. No transient weight loss or recovery was observed in any of the other groups of infected mice.
FIG. 8.
Pathogenicity of N2c strains. Mice were infected intramuscularly with 5 to 10 × 105 infectious particles and their mortality rates recorded daily. Statistical analysis using the Mantel-Haenszel log-rank test showed a statistically significant difference from wild-type rN2C results (n = 80) for the rN2C-D80 virus (P = 0.0087; n = 60), the rN2C-D40C strain (P = 0.0096; n = 60), and the codon-optimized variant (P = 0.0388; n = 64) but no significant difference for the rN2C-D40N virus (P = 0.3024; n = 40).
Based on earlier studies, a significant reduction in pathogenicity was expected in mice that were infected with the codon-optimized variant. However, the rN2c-Op virus still caused a high (70%) mortality rate that was only slightly reduced compared to rN2c results and similar to the mortality observed for the D40C and D80 codon-deoptimized viruses. However, unlike the latter, no significant delay in disease progression was observed, which was a result very similar to that seen with mice infected with wild-type virus.
Statistical analysis of the survival data revealed significantly lower pathogenicity compared to wild-type N2c for the D80, D40C, and codon-optimized virus, whereas no significant difference in pathogenicity compared to wild-type virus results was found for the D40N variant. At higher doses, the differences between the viruses were not significant and no reduction in pathogenicity was observed for either the codon-optimized or the codon-deoptimized rN2c-D80 virus compared to wild-type virus (Fig. 9).
FIG. 9.
Pathogenicity of N2c strains in C57/B6 mice. Mice were infected intramuscularly with 5 × 106 infectious particles and monitored daily for weight loss and clinical signs of rabies. Mice that lost more than 20% of their initial body weight or displayed paralysis were euthanized. No statistically significant difference in mortality from the wild-type virus (n = 10) was observed for either the codon-deoptimized rN2C-D80 strain (P = 0.3511; n = 10) or the codon-optimized rN2C-Op virus (P = 0.432; n = 10).
DISCUSSION
The present study was undertaken to address two questions. First, we wanted to revisit the hypothesis that pathogenicity of RV inversely correlates with glycoprotein expression level by the use of an experimental approach in which only the amount of G is varied. Even though RV G expression levels and pathogenicity were analyzed in previous studies, interpretation of these earlier results is complicated by simultaneous changes in other parameters such as the G amino acid sequence and gene number (21, 23), which contribute significantly to the pathogenicity of RV. Second, we wanted to examine whether large-scale changes in codon usage that are aimed at reducing protein expression are able to attenuate RV. This strategy, which was first employed to attenuate poliovirus, holds considerable promise, as it greatly reduces or eliminates the risk that an attenuated virus would revert to a pathogenic phenotype (4, 26). However, the general applicability of this strategy for RNA viruses is still questionable.
Our data show that neither an increase nor a decrease of the amount of G renders a pathogenic rabies virus apathogenic, although it does reduce pathogenicity to some extent. Several conclusions can be drawn from our results. First, it appears that the amount of G produced by the N2c strain is at a level that confers maximum pathogenicity. This is an interesting and important observation. Although we have not extended our studies to other viral isolates, we speculate that the same likely holds true for many field strains. Second, our data show that reducing the amount of glycoprotein by large-scale codon deoptimization does not result in a safe vaccine candidate. This strategy is also unlikely to further attenuate existing vaccine strains. Rather, it will most likely decrease their attenuation, since a hallmark of these strains is increased viral glycoprotein synthesis. Third, our data show that, in addition, increases in glycoprotein level alone do not suffice to render a pathogenic virus apathogenic.
Our results are in clear contrast to results obtained with poliovirus, which is attenuated almost 100-fold by codon deoptimization of the capsid genes (26). There are several factors that likely contribute to this difference. In the case of poliovirus, attenuation of capsid protein synthesis simultaneously reduces the amount of the viral nonstructural proteins, thereby resulting in a general reduction of replication and viral protein synthesis (26). This is not the case for RV. Reducing the amount of RV-G does not reduce the amount of other viral proteins; rather, it leads to an increase in the amount of N, P, and M. Second, poliovirus is a fast-replicating virus that overwhelms host defenses with speed. In stark contrast to poliovirus, rabies is a slowly replicating virus that aims to preserve host cell function and relies on mechanisms other than speed to overcome host defenses (1, 3, 16, 38, 39). It remains to be seen to what extent codon deoptimization of viral genes other than G attenuates rabies virus. Since attenuation of RV is generally correlated with increased replication and transcription, faster growth, and higher titers, we would expect attenuation to be less pronounced than is the case with poliovirus.
Increased viral protein expression in cells infected with codon-deoptimized variants clearly points to regulatory mechanisms that impact protein synthesis and intracellular accumulation. The existence of such mechanisms is also suggested by the fact that, in the context of an infectious virus, the reduction in the amount of G that is caused by codon deoptimization of the glycoprotein gene is much less than the reduction observed after transient transfection of the same genes into NA cells. A direct role of G in the regulation of transcription and replication has not yet been reported. However, given the important role of G in virus release, a smaller amount of G would reduce the amount of viral protein that is released from infected cells in the form of viral particles (20). This would result in increased accumulation of intracellular viral proteins and could explain the changes observed in cells infected with codon-deoptimized viruses.
Significant effort has been expended on further increasing the attenuation and safety of live rabies virus vectors by introduction of additional copies of the glycoprotein gene. In light of the data presented here, it would appear that the additional attenuation observed in these studies could be due in part to the increased length of the viral genome and a reduction in the amount of L due to the insertion of additional transcription start-stop signals. It remains to be clarified how much of the attenuation is really due to an increase in glycoprotein expression.
In summary, our data provide strong evidence that the glycoprotein expression level does play a role in pathogenicity but is not a dominant determining factor. Construction of a live attenuated rabies virus is therefore not achieved by solely targeting the expression level of the glycoprotein gene but necessitates the use of multiple attenuation markers and a multigenic approach (44).
Acknowledgments
This work was supported by a Center for Neuroanatomy and Neurotropic Viruses grant to Peter Strick (Center for Neuroscience, University of Pittsburgh) (USPHS-NCRR P40RR018604, subcontract to M.J.S.) and the NIAID NIH AI082325 to M.J.S.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Brzózka, K., S. Finke, and K. K. Conzelmann. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cenna, J., M. Hunter, G. S. Tan, A. B. Papaneri, E. P. Ribka, M. J. Schnell, P. A. Marx, and J. P. McGettigan. 2009. Replication-deficient rabies virus-based vaccines are safe and immunogenic in mice and nonhuman primates. J. Infect. Dis. 200:1251-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chelbi-Alix, M. K., A. Vidy, J. El Bougrini, and D. Blondel. 2006. Rabies viral mechanisms to escape the IFN system: the viral protein P interferes with IRF-3, Stat1, and PML nuclear bodies. J. Interferon Cytokine Res. 26:271-280. [DOI] [PubMed] [Google Scholar]
- 4.Coleman, J. R., D. Papamichail, S. Skiena, B. Futcher, E. Wimmer, and S. Mueller. 2008. Virus attenuation by genome-scale changes in codon pair bias. Science 320:1784-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cox, J. H., B. Dietzschold, and L. G. Schneider. 1977. Rabies virus glycoprotein. II. Biological and serological characterization. Infect. Immun. 16:754-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dietzschold, B., M. Gore, P. Casali, Y. Ueki, C. E. Rupprecht, A. L. Notkins, and H. Koprowski. 1990. Biological characterization of human monoclonal antibodies to rabies virus. J. Virol. 64:3087-3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dudek, T., and D. M. Knipe. 2006. Replication-defective viruses as vaccines and vaccine vectors. Virology 344:230-239. [DOI] [PubMed] [Google Scholar]
- 8.Etessami, R., K. K. Conzelmann, B. Fadai-Ghotbi, B. Natelson, H. Tsiang, and P. E. Ceccaldi. 2000. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: an in vitro and in vivo study. J. Gen. Virol. 81:2147-2153. [DOI] [PubMed] [Google Scholar]
- 9.Faber, M., M. L. Faber, A. Papaneri, M. Bette, E. Weihe, B. Dietzschold, and M. J. Schnell. 2005. A single amino acid change in rabies virus glycoprotein increases virus spread and enhances virus pathogenicity. J. Virol. 79:14141-14148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Faber, M., R. Pulmanausahakul, S. S. Hodawadekar, S. Spitsin, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2002. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 76:3374-3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freuling, C., A. Vos, N. Johnson, A. R. Fooks, and T. Muller. 2009. Bat rabies—a Gordian knot? Berl. Munch. Tierarztl. Wochenschr. 122:425-433. [PubMed] [Google Scholar]
- 12.Gao, W., A. Rzewski, H. Sun, P. D. Robbins, and A. Gambotto. 2004. UpGene: application of a web-based DNA codon optimization algorithm. Biotechnol. Prog. 20:443-448. [DOI] [PubMed] [Google Scholar]
- 13.Gomme, E. A., E. J. Faul, P. Flomenberg, J. P. McGettigan, and M. J. Schnell. 2010. Characterization of a single-cycle rabies virus-based vaccine vector. J. Virol. 84:2820-2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hemachudha, T. 1994. Human rabies: clinical aspects, pathogenesis, and potential therapy. Curr. Top. Microbiol. Immunol. 187:121-143. [DOI] [PubMed] [Google Scholar]
- 15.Inoue, K., Y. Shoji, I. Kurane, T. Iijima, T. Sakai, and K. Morimoto. 2003. An improved method for recovering rabies virus from cloned cDNA. J. Virol. Methods 107:229-236. [DOI] [PubMed] [Google Scholar]
- 16.Ito, N., G. W. Moseley, D. Blondel, K. Shimizu, C. L. Rowe, Y. Ito, T. Masatani, K. Nakagawa, D. A. Jans, and M. Sugiyama. 2010. The role of interferon-antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84:6699-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito, N., M. Sugiyama, K. Yamada, K. Shimizu, M. Takayama-Ito, J. Hosokawa, and N. Minamoto. 2005. Characterization of M gene-deficient rabies virus with advantages of effective immunization and safety as a vaccine strain. Microbiol. Immunol. 49:971-979. [DOI] [PubMed] [Google Scholar]
- 18.Le Mercier, P., Y. Jacob, K. Tanner, and N. Tordo. 2002. A novel expression cassette of lyssavirus shows that the distantly related Mokola virus can rescue a defective rabies virus genome. J. Virol. 76:2024-2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McGettigan, J. P., K. Naper, J. Orenstein, M. Koser, P. M. McKenna, and M. J. Schnell. 2003. Functional human immunodeficiency virus type 1 (HIV-1) Gag-Pol or HIV-1 Gag-Pol and env expressed from a single rhabdovirus-based vaccine vector genome. J. Virol. 77:10889-10899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mebatsion, T., M. Konig, and K. K. Conzelmann. 1996. Budding of rabies virus particles in the absence of the spike glycoprotein. Cell 84:941-951. [DOI] [PubMed] [Google Scholar]
- 21.Morimoto, K., H. D. Foley, J. P. McGettigan, M. J. Schnell, and B. Dietzschold. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6:373-381. [DOI] [PubMed] [Google Scholar]
- 22.Morimoto, K., D. C. Hooper, H. Carbaugh, Z. F. Fu, H. Koprowski, and B. Dietzschold. 1998. Rabies virus quasispecies: implications for pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 95:3152-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morimoto, K., D. C. Hooper, S. Spitsin, H. Koprowski, and B. Dietzschold. 1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73:510-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morimoto, K., Y. Shoji, and S. Inoue. 2005. Characterization of P gene-deficient rabies virus: propagation, pathogenicity and antigenicity. Virus Res. 111:61-67. [DOI] [PubMed] [Google Scholar]
- 25.Mrak, R. E., and L. Young. 1994. Rabies encephalitis in humans: pathology, pathogenesis and pathophysiology. J. Neuropathol. Exp. Neurol. 53:1-10. [DOI] [PubMed] [Google Scholar]
- 26.Mueller, S., D. Papamichail, J. R. Coleman, S. Skiena, and E. Wimmer. 2006. Reduction of the rate of poliovirus protein synthesis through large-scale codon deoptimization causes attenuation of viral virulence by lowering specific infectivity. J. Virol. 80:9687-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 28.Pulmanausahakul, R., M. Faber, K. Morimoto, S. Spitsin, E. Weihe, D. C. Hooper, M. J. Schnell, and B. Dietzschold. 2001. Overexpression of cytochrome C by a recombinant rabies virus attenuates pathogenicity and enhances antiviral immunity. J. Virol. 75:10800-10807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupprecht, C. E., J. Barrett, D. Briggs, F. Cliquet, A. R. Fooks, B. Lumlertdacha, F. X. Meslin, T. Muler, L. H. Nel, C. Schneider, N. Tordo, and A. I. Wandeler. 2008. Can rabies be eradicated? Dev. Biol. (Basel) 131:95-121. [PubMed] [Google Scholar]
- 30.Rupprecht, C. E., C. A. Hanlon, and D. Slate. 2006. Control and prevention of rabies in animals: paradigm shifts. Dev. Biol. (Basel) 125:103-111. [PubMed] [Google Scholar]
- 31.Rupprecht, C. E., C. A. Hanlon, and D. Slate. 2004. Oral vaccination of wildlife against rabies: opportunities and challenges in prevention and control. Dev. Biol. (Basel) 119:173-184. [PubMed] [Google Scholar]
- 32.Schnell, M. J., J. P. McGettigan, C. Wirblich, and A. Papaneri. 2010. The cell biology of rabies virus: using stealth to reach the brain. Nat. Rev. Microbiol. 8:51-61. [DOI] [PubMed] [Google Scholar]
- 33.Schnell, M. J., G. S. Tan, and B. Dietzschold. 2005. The application of reverse genetics technology in the study of rabies virus (RV) pathogenesis and for the development of novel RV vaccines. J. Neurovirol. 11:76-81. [DOI] [PubMed] [Google Scholar]
- 34.Shimizu, K., N. Ito, T. Mita, K. Yamada, J. Hosokawa-Muto, M. Sugiyama, and N. Minamoto. 2007. Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res. 123:154-160. [DOI] [PubMed] [Google Scholar]
- 35.Shoji, Y., S. Inoue, K. Nakamichi, I. Kurane, T. Sakai, and K. Morimoto. 2004. Generation and characterization of P gene-deficient rabies virus. Virology 318:295-305. [DOI] [PubMed] [Google Scholar]
- 36.Takayama-Ito, M., N. Ito, K. Yamada, M. Sugiyama, and N. Minamoto. 2006. Multiple amino acids in the glycoprotein of rabies virus are responsible for pathogenicity in adult mice. Virus Res. 115:169-175. [DOI] [PubMed] [Google Scholar]
- 37.Tordo, N., A. Foumier, C. Jallet, M. Szelechowski, B. Klonjkowski, and M. Eloit. 2008. Canine adenovirus based rabies vaccines. Dev. Biol. (Basel) 131:467-476. [PubMed] [Google Scholar]
- 38.Vidy, A., M. Chelbi-Alix, and D. Blondel. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79:14411-14420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vidy, A., J. El Bougrini, M. K. Chelbi-Alix, and D. Blondel. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weyer, J., C. E. Rupprecht, and L. H. Nel. 2009. Poxvirus-vectored vaccines for rabies—a review. Vaccine 27:7198-7201. [DOI] [PubMed] [Google Scholar]
- 41.WHO. 2005. WHO expert consultation on rabies, p. 1-88 and back cover. WHO technical report series, no. 931. WHO Press, Geneva, Switzerland. [PubMed]
- 42.Wiktor, T. J., E. Gyorgy, D. Schlumberger, F. Sokol, and H. Koprowski. 1973. Antigenic properties of rabies virus components. J. Immunol. 110:269-276. [PubMed] [Google Scholar]
- 43.Wirblich, C., G. S. Tan, A. Papaneri, P. J. Godlewski, J. M. Orenstein, R. N. Harty, and M. J. Schnell. 2008. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J. Virol. 82:9730-9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, K., N. Ito, M. Takayama-Ito, M. Sugiyama, and N. Minamoto. 2006. Multigenic relation to the attenuation of rabies virus. Microbiol. Immunol. 50:25-32. [DOI] [PubMed] [Google Scholar]
- 45.Yan, X., M. Prosniak, M. T. Curtis, M. L. Weiss, M. Faber, B. Dietzschold, and Z. F. Fu. 2001. Silver-haired bat rabies virus variant does not induce apoptosis in the brain of experimentally infected mice. J. Neurovirol. 7:518-527. [DOI] [PubMed] [Google Scholar]