Abstract
Elite controllers or suppressors (ES) are a group of HIV-1-infected individuals who maintain viral loads below the limit of detection of commercial assays for many years. The mechanisms responsible for this remarkable control are under intense study, with the hope of developing therapeutic vaccines effective against HIV-1. In this study, we addressed the question of the intrinsic susceptibility of ES CD4+ T cells to infection. While we and others have previously shown that CD4+ T cells from ES can be infected by HIV-1 isolates in vitro, these studies were confounded by exogenous activation and in vitro culture of CD4+ T cells prior to infection. In order to avoid the changes in chemokine receptor expression that have been associated with such exogenous activation, we infected purified CD4+ T cells directly after isolation from the peripheral blood of ES, viremic patients, and uninfected donors. We utilized a green fluorescent protein (GFP)-expressing proviral construct pseudotyped with CCR5-tropic or CXCR4-tropic envelope to compare viral entry using a fluorescence resonance energy transfer-based, single-round virus-cell fusion assay. The frequency of productive infection was also compared by assessing GFP expression. CD4+ T cells from ES were as susceptible as or more susceptible than cells from viremic patients and uninfected donors to HIV-1 entry and productive infection. The results of this physiological study strongly suggest that differences in HIV-1 entry and infection of CD4+ T cells alone cannot explain the elite control of viral replication.
Elite suppressors (ES) maintain control of HIV-1 infection without the use of antiretrovirals (5, 30, 36), despite the presence of ongoing viral replication and evolution (37). The mechanism by which they do this is unknown. Initial reports suggested that ES and long-term nonprogressors are able to control viremia due to defects in the infecting virus (2, 9, 13, 19, 22, 24, 27, 28, 42, 48). However, replication-competent (RC) virus has been isolated from ES (4, 7, 23, 26) and shown to be comparable in fitness to laboratory HIV-1 strains (7). Furthermore, full-genome sequence analysis of RC virus has not revealed large deletions or signature mutations in virus from ES (7). Thus, it appears that, in many cases, differences in the host account for the diverse pathogenesis observed in HIV-1-infected patients.
While significant attention has been paid to the role of cytotoxic T cells and immune activation in the control of HIV-1 replication, potential differences in the inherent susceptibility to infection of target cells have not been as carefully evaluated. The dramatic impact of the CCR5 Δ32 mutation on disease pathogenesis (18, 21, 29, 44) initially suggested that this mutation accounted for the ability of individuals to control infection, but this mutation is present in only a minor fraction of ES (11, 25, 35, 40).
We and others have previously shown, directly or indirectly, that CD4+ T cells from ES can be infected by both autologous (4, 7) and laboratory strain HIV-1 isolates (7, 14, 23, 26, 38, 47). However, previous infection assays have utilized in vitro activation of potential target cells to facilitate infection. Such activation can affect coreceptor expression (1, 8, 10) and cause cytokine and chemokine release, all of which may, in turn, impact the susceptibility to infection of the cells. For example, stimulation of CD4+ T cells with phytohemagglutinin and interleukin-2 results in upregulation of CXCR4 within 72 h (8). In contrast, stimulation with monoclonal antibodies to CD3 and CD28 results in downregulation of CCR5 (10). This downregulation correlates with a reduced susceptibility to infection of CD4+ T cells with CCR5-tropic (R5) virus and may contribute to the selection of CXCR4-tropic (X4) virus in vivo, as the frequency of CCR5-positive targets decreases (10).
In this study, we examined the susceptibility to infection of freshly isolated and purified CD4+ T cells without exogenous activation. We aimed to develop the most physiological system possible so as to more closely recapitulate HIV-1 infection in vivo. In addition to looking at infection with pseudotyped viruses as measured by green fluorescent protein (GFP) expression, we examined susceptibility to infection in terms of fusion of the virus to the target cell. We utilized both R5 and X4 pseudoviruses and characterized the expression of CCR5 and the baseline activation state of T cells from the different groups of patients.
MATERIALS AND METHODS
Subjects.
The clinical characteristics of the ES (n = 10) and viremic patients (n = 7) used in this study are shown in Table 1. Laboratory donors (n = 8) were used as controls.
TABLE 1.
Clinical characteristics of ES and viremic patients used in this study
| Subject | Yr of diagnosis | Last CD4+ T cell count/μl | No. of HIV-1 RNA copies/ml | CCR5 Δ32 genotyping |
|---|---|---|---|---|
| ES3* | 1991 | 728 | <50 | Wild type |
| ES4 | 1996 | 678 | <50 | Wild type |
| ES5 | 1990 | 839 | <50 | Wild type |
| ES6 | 1996 | 1,139 | <50 | Wild type |
| ES8 | 2003 | 602 | <50 | Wild type |
| ES9 | 1999 | 1,027 | <50 | Heterozygous Δ32 |
| ES18 | 1998 | 1,330 | <50 | Wild type |
| ES24 | 2009 | 2,033 | <50 | Wild type |
| ES25 | 2002 | 579 | <50 | Wild type |
| ES31 | 2006 | 758 | <50 | Wild type |
| Viremic 1 | 2000 | 383 | 55,987 | Wild type |
| Viremic 2 | 1990 | 348 | 12,481 | Wild type |
| Viremic 3 | 2006 | 475 | 8,190 | Wild type |
| Viremic 4 | 2010 | 490 | 155,000 | Wild type |
| Viremic 5 | 2003 | 213 | 105,662 | Wild type |
| Viremic 6 | 1997 | 413 | 19,681 | Wild type |
| Viremic 7 | 2009 | 970 | 22,220 | Wild type |
Infection assay.
CD4+ T cells were isolated by negative selection from freshly isolated peripheral blood mononuclear cells (PBMCs) using the Miltenyi human CD4+ T cell isolation kit according to the manufacturer's instructions. To ensure removal of CD8+ T cells and NK cells, isolated CD4+ T cells were further purified using anti-CD8 and anti-CD16 antibodies and sheep anti-mouse secondary antibody conjugated to magnetic beads (Dynabeads; Invitrogen). Purity was confirmed using flow cytometry analysis with CD3-allophycocyanin (APC), CD4-APC-H7, CD8-fluorescein isothiocyanate (FITC), and CD16-phycoerythrin (PE; BD). Cells were then infected with X4 and R5 pseudotype NL43 virus in which GFP replaces most of the envelope gene, resulting in a virus capable of only a single round of infection (NL43-deltaEnvGFP) (49). Infection was via spinoculation (39) in a round-bottom 96-well plate for 1.5 h in RPMI 1640 medium supplemented with 10% fetal bovine serum. After infection, cells were placed in an incubator at 37°C for 72 h. After 72 h, cells were stained with CD3-PE, CD4-APC-H7, and HLA-DR-APC for 25 min on ice and fixed with 2% paraformaldehyde, and 50,000 events were analyzed on the fluorescence-activated cell sorter (FACS) Canto II. The percentage of lymphocytes was used as an indirect measure of viability to ensure that superinfection did not lead to cell death. To normalize the percentage of infection across patient samples, the percentage of CD3+ GFP+ cells was normalized to the percentage of CD4+ CD3+ cells after subtracting any background from negative controls.
Entry assay.
CD4+ T cells were isolated as described above for the infectivity assay. Viruses utilized were similar to what has been previously described (12): single-round infective X4 and R5 pseudotyped viruses were made which incorporate the enzyme β-lactamase fused to the viral accessory gene vpr. Equivalents of 177 ng and 227 ng of p24 were used to infect 100,000 CD4+ T cells with X4 and R5 pseudotyped viruses, respectively. To address whether a high multiplicity of infection artificially influenced the results, three lower concentrations of X4 virus (1.68, 0.96, and 0.48 ng p24/100,000 cells) and one lower concentration of R5 virus (2.7 ng p24/100,000 cells) were used to infect CD4+ T cells from a randomly selected subset of patients in this study. After spinoculation for 1.5 h as described for the infectivity assay, the viruses were allowed to fuse with the target cells for 2 h at 37°C. Target cells were washed once in RPMI 1640 medium supplemented with 10% fetal bovine serum and then incubated with a cell-permeating substrate, CCF2-AM (Invitrogen) for 1 h at room temperature. To stop any further fusion events, 1 μM T20 was added. After the cells were washed two times, they were incubated overnight in CO2-Independent Medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS). CCF2 is a fluorescence resonance energy transfer (FRET) substrate. The intact dye emits at 520 nm when excited by a 405-nm laser. Upon cleavage by the enzyme β-lactamase, the dye emits at 447 nm. This change in emission can be monitored by flow cytometric analysis. After approximately 12 h, the cells were stained with CD3-PE, CD4-APC-H7, and HLA-DR-APC for 25 min on ice, fixed with 2% paraformaldehyde, and analyzed on the FACS Canto II. The percentage of CD3+ cells that had undergone a fusion event was normalized to the percentage of CD3+ CD4+ cells as described above.
Three independent plasma and proviral envelope genes from two patients with progressive HIV-1 disease were cloned as described previously (3). Virus containing the Vpr-β-lactamase protein pseudotyped with these envelope genes was made by transfection as described above. Cell-free supernatant was obtained by spinning at 1,200 rpm for 10 min. Thirty-two nanograms of virus was used to infect 100,000 CD4+ T cells isolated from patients or healthy donors. The entry assay was performed as described above, and the results of three independent entry experiments were averaged.
Immunological analyses.
Whole PBMCs from each patient were frozen in 10% dimethyl sulfoxide (DMSO) and 90% FBS and then quick thawed at 37°C prior to CCR5 and activation marker analysis. Prior to staining with fluorescent conjugated antibodies, cells were treated with an Fc receptor blocking agent (Miltenyi) to minimize nonspecific binding. CCR5-APC (clone 3A9), CD3-PerCpCy5.5, CD4-APC-H7, and HLA-DR-FITC were used in combination for CCR5 expression analysis. CD3-PerCpCy5.5, CD4-APC-H7, HLA-DR-FITC, and CD38-PE were used in combination for CD38/HLA-DR expression analysis. All antibodies were from Becton Dickinson.
CCR5 genotyping.
DNA was isolated from PBMCs of each patient using the Gentra Puregene kit from Qiagen. CCR5 genotyping for the Δ32 mutation was determined by amplifying a portion of the gene as described previously (45), using primers CCR5-D32-F (5′CTTCATTACACCTGCAGCT3′) and CCR5-D32-R (5′TGAAGATAAGCCTCACAGCC3′).
Statistical analysis.
P values were derived from the Wilcoxon-Mann-Whitney rank-sum test unless otherwise stated. P values for correlations are calculated using the Analysis tool kit of Microsoft Excel 2007 assuming a linear regression model. A P value of <0.05 was chosen to represent statistical significance.
Investigators were blinded to the HIV-1 status of subjects when conducting all experiments, and the infectivity assay, the entry assay, and flow cytometry analysis were performed by different investigators.
RESULTS
Infection of unactivated, isolated CD4+ T cells.
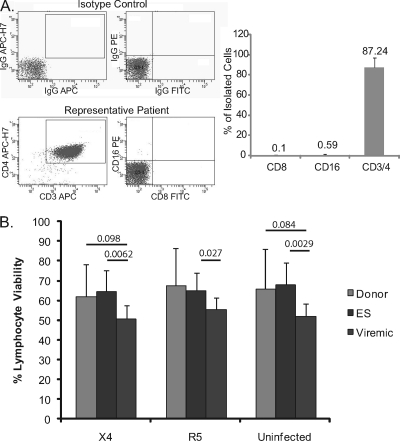
To compare susceptibilities to infection of cells from ES, viremic progressors, and uninfected donors in the most physiological manner possible, we infected freshly isolated CD4+ T cells without exogenous activation. CD4+ T cells underwent double purification to eliminate the possibility of confounding effects due to contaminating CD8+ T cells or NK cells (Fig. 1 A). We also compared the percentage of viable lymphocytes in uninfected samples and samples infected with X4 or R5 virus to ensure that superinfection was not decreasing viability (Fig. 1B). Infection of cells had no effect on viability in any patient or donor subset, although viremic patients had lower viability than ES and uninfected donors overall (P = 0.002 and 0.084, respectively, using Student's t test).
FIG. 1.
(A) Purity of CD4+ T cells was determined by flow cytometric analysis to ensure depletion of CD8+ T cells and NK cells. Data shown are averages of patient cohorts with standard deviations. Less than 1% CD8 or CD16 contamination was usually observed after purification. (B) Viability as determined by the percentage of cells in the lymphocyte forward and side scatter gates after infection with X4 and R5 virus and for uninfected controls. Data shown are averages of patient cohorts, i.e., uninfected donors, ES, and viremic patients.
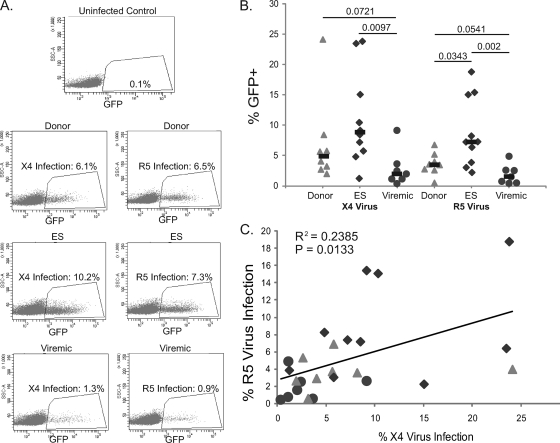
We utilized an X4 envelope (NL4-3) and an R5 envelope (SF162) to package the same viral vector. In this vector, env was partially replaced by the GFP coding sequence, making the virus capable of only a single round of replication and permitting enumeration of infected cells by flow cytometric analysis, as described in Materials and Methods. Figure 2 A shows representative infection data for patients from each subset. As summarized in Fig. 2B, we observed significant infection of unstimulated CD4+ T cells from uninfected donors, ES, and viremic patients with both X4 and R5 viruses. The percent infection was actually higher for CD4+ T cells from ES and uninfected donors than for CD4+ T cells from viremic patients, and this difference was still significant after normalization for the difference in baseline lymphocyte viability (data not shown). Interestingly, the percent infection with R5 virus also correlated with the percent infection with X4 virus (Fig. 2C).
FIG. 2.
Infection of freshly isolated CD4+ T cells with X4 and R5 pseudotyped viruses. (A) Representative infection data for uninfected donors, ES, and viremic patients. (B) Percent infection of CD3+ CD4+ T cells as measured by GFP expression. The percentage of GFP+ CD3+ cells was normalized to the percentage of CD4+ CD3+ cells in the uninfected controls for each patient. P values were derived from the Wilcoxon-Mann-Whitney rank-sum test. The median is indicated by a black bar. (C) Correlation between the percentage of cells infected with R5 virus and the percentage of cells infected with X4 virus.
Viral entry.
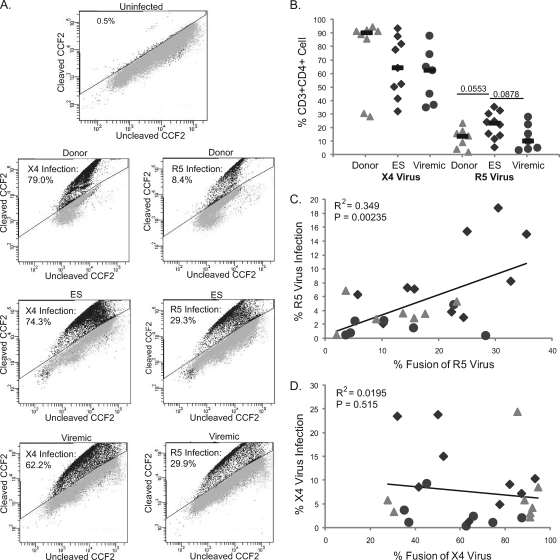
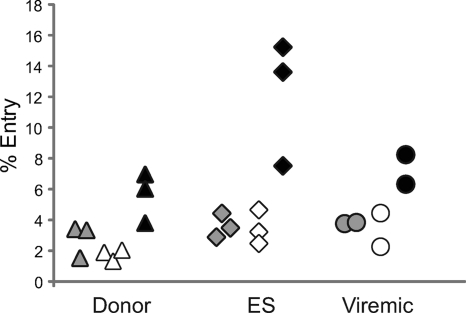
Having evaluated infection in terms of viral protein expression, we examined viral entry into CD4+ T cells from the same cohort of uninfected donors, ES, and viremic patients by using a previously described viral fusion assay (12). Data for representative patients from each subgroup are shown in Fig. 3 A. As summarized in Fig. 3B, there was a trend toward a higher level of fusion of R5 virus with CD4+ T cells from ES compared to CD4+ T cells from viremic patients and uninfected donors. Interestingly, there was a trend toward a higher rate of X4 virus entry in cells from uninfected donors compared to cells from ES and viremic patients (Fig. 3B). The trend may be due to the fact that uninfected donors have generally lower levels of cellular activation and higher levels of naïve CD4+ T cells than ES or viremics (unpublished observations), and CXCR4 is expressed on naïve T cells whereas CCR5 generally is not (8, 33). Thus, this may result in fusion of virus to naïve cells that did not then result in productive infection, as measured by GFP expression.
FIG. 3.
Fusion of X4 and R5 pseudotyped viruses to freshly isolated CD4+ T cells. (A) Representative entry data for uninfected donors, ES, and viremic patients. (B) Percentage of cells undergoing fusion as measured by flow cytometry, normalized to the number of CD3+ CD4+ cells in the uninfected controls for each patient as described for infection. P values were derived from the Wilcoxon-Mann-Whitney rank-sum test. The median is indicated by a black bar. (C) Correlation between productive infection (percent GFP+) and fusion of R5 pseudotyped virus and (D) X4 pseudotyped virus for all patients.
There was a significant correlation between viral entry and productive infection for R5 virus (Fig. 3C) in spite of the fact that different viruses (with the same env gene) were used for the assays. No such correlation between X4 virus entry and infection was seen (Fig. 3D), possibly due to infection of naïve T cells expressing CXCR4, as described above.
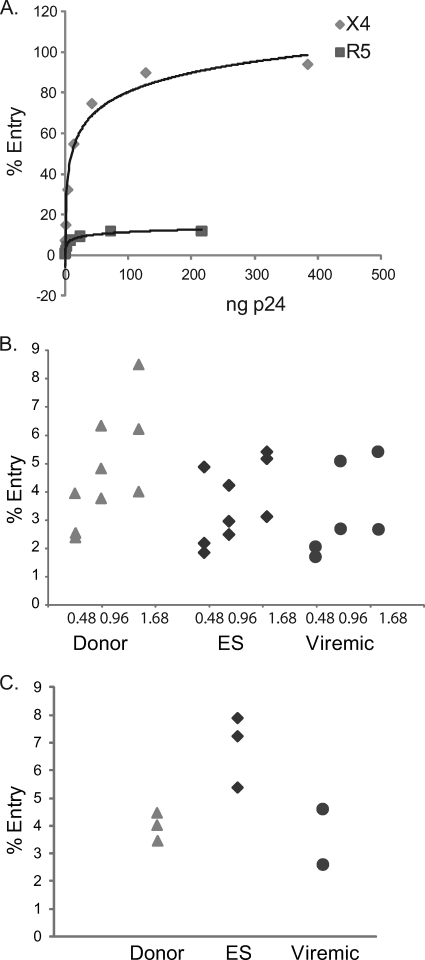
The high level of entry we observed raised the question of whether we were missing subtle differences in viral fusion events by using too large a viral inoculum. To address this question, we titrated the amount of virus in order to achieve a level of viral entry that was comparable to the levels of productive infection we observed in Fig. 2. As shown in Fig. 4 A, there was a correlation between the concentration of virus used and the amount of viral entry seen for both the R5 and X4 pseudotyped viruses. Using cells from a subset of patients randomly selected from each group, we found that using lower concentrations of virus resulted in patterns of infection with the X4 (Fig. 4B) and R5 (Fig. 4C) viruses that were very similar to the patterns seen when larger viral inoculums were used.
FIG. 4.
Titration of X4 and R5 pseudotyped viruses. (A) Serial dilutions of X4 and R5 pseudotyped viruses were used to infect CD4+ T cells isolated from a healthy donor. CD4+ T cells (n = 100,000) from a randomly selected subset of uninfected donors (triangles), ES (diamonds), and viremic patients (circles) were infected with 1.68, 0.96, or 0.48 ng p24 equivalents of X4 virus (B) or 2.7 ng p24 equivalents of R5 virus (C). Each symbol represents cells from an individual patient that were infected with different concentrations of virus.
We also tested the hypothesis that the use of env genes from laboratory strains resulted in very efficient viral entry, thereby masking subtle differences between patient groups. A prior study has shown that env can significantly affect simian immunodeficiency virus replication in a postentry step (34). env genes cloned from patients with progressive HIV disease were thus used to generate pseudotyped viruses as previously described (3). Cells from a subset of patients from each group were infected with these pseudotyped viruses. As shown in Fig. 5, the pattern of viral entry by viruses pseudotyped with env from three different primary HIV-1 isolates was similar to the pattern seen with infection by laboratory strains. Taken together, these data indicate that both viral fusion to target cells and productive infection are functional in ES.
FIG. 5.
Infection with pseudotyped viruses utilizing patient-derived env. CD4+ T cells (n = 100,000) isolated from a randomly selected subset of HIV-1-negative donors (triangles), ES (diamonds), and viremic patients (circles) were infected with 32 ng p24 equivalents of virus pseudotyped with three independent plasma and proviral envelope genes from two patients with progressive HIV-1 disease. Each symbol represents cells from an individual patient which were infected with one of three different viruses pseudotyped with env from three different primary HIV-1 isolates (black, white, or gray).
CCR5 cell surface expression and cellular activation.
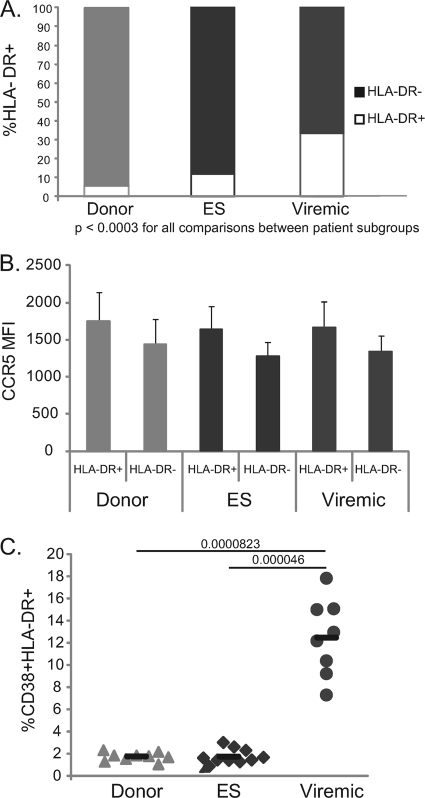
Trends seen in viral fusion with R5 virus were similar to those seen upon measurement of the expression of GFP. We therefore sought to determine whether differences in coreceptor expression might account for differences between patient groups. None of the patients or donors were homozygous for the delta 32-bp deletion (Table 1). In addition, there were no significant differences in the percentage of CD4+ T cells that were CCR5+ in the three patient groups (data not shown). We thus examined CCR5 expression on HLA-DR+ (activated) and HLA-DR− (unactivated) CD3+ CD4+ PBMCs in order to determine whether differences in the density of this coreceptor could explain the differences in susceptibility to infection.
As anticipated, viremic patients had a higher percentage of CD3+ CD4+ HLA-DR+ lymphocytes than did those in the ES and uninfected patient groups (Fig. 6 A). We then examined the mean fluorescence intensity (MFI) of CCR5 staining on CCR5+ lymphocytes from CD3+ CD4+ HLA-DR+ and CD3+ CD4+ HLA-DR− cellular subsets from each patient in our cohort (Fig. 4B). We found no significant difference in the MFI of CCR5 on CD4+ T cells from ES versus CD4+ T cells from viremic patients or uninfected donors (Fig. 6B), and there was no correlation between CCR5 expression and susceptibility to infection or viral fusion (data not shown). HLA-DR+ cells expressed higher surface levels of CCR5 than HLA-DR− cells (Fig. 6B). These differences were not statistically significant, however.
FIG. 6.
CCR5 expression and HLA-DR expression on PBMCs from patient subgroups. (A) Average frequencies of HLA-DR-positive and HLA-DR-negative CD3+ CD4+ cells from each patient subgroup. (B) Average MFI of CCR5 expression on CCR5+ cells. Gating is on CD3+ CD4+ lymphocytes and then on HLA-DR+ and HLA-DR− cells for each patient subgroup. (C) Percentage of CD3+ CD4+ T cells that express both CD38 and HLA-DR. The median percentage is indicated by a black bar. P values were derived from the Wilcoxon-Mann-Whitney rank-sum test.
Considering the apparently higher susceptibility to infection of CD4+ T cells from ES than those from viremic patients, we also examined the level of coexpression of CD38 and HLA-DR on CD3+ CD4+ PBMCs in our cohort to determine whether higher activation levels of T cells in our ES might account for the disparity in infection. Our data, however, showed that viremic patients had significantly higher CD4+ CD38+ HLA-DR+ T cells than did ES or uninfected donors (Fig. 6C), in agreement with previous studies (20).
DISCUSSION
More than a decade of rigorous research has led to the conclusion that in many cases, ES control HIV-1 infection through unique host mechanisms and not by virtue of infection by deficient virus (36). Identification of effective cytotoxic T lymphocyte responses in ES has suggested that the adaptive immune response is responsible for the control of viremia (6, 17, 31, 32, 41). However, the relatively low viral loads seen in acute infection (16) have suggested that the earliest possible stages of HIV-1 infection may play a determining role in disease pathogenesis. This observation suggests that innate immunity, or intrinsic resistance of CD4+ T cells to infection, may play a key role in the early control of HIV-1 replication in ES.
In this study, we compared the susceptibilities to infection of CD4+ T cells from ES, viremic progressors, and uninfected donors in the most physiological way possible. Because exogenous activation affects coreceptor expression levels, we utilized CD4+ T cells that were isolated and infected within hours of phlebotomy. We vigorously ensured that no contaminating CD8+ T cells or NK cells would inhibit infection. Our data show that CD4+ T cells from ES are at least as susceptible to infection as those isolated from viremic patients and uninfected donors. This is true for both R5 and X4 pseudotyped viruses. We examined susceptibility to infection in terms of GFP expression (Fig. 2) and in terms of viral fusion to the patients' cells (Fig. 3). Despite the fact that these experiments used viral vectors which were identical only in their envelope, there was a strong correlation between productive infection as determined by GFP expression and fusion with R5 virus.
One limitation of our study is that we used pseudotyped viruses that are only capable of a single cycle of infection. While this leads to a very quantitative measurement of viral infection, it does not measure the events in the viral life cycle that occur after protein (GFP) expression. It is thus possible that there are differences in HIV-1 budding from CD4+ T cells from ES versus those from viremic patients and uninfected donors.
Without the ability to compare samples from ES and viremic patients before and after infection, it is difficult to separate the causes of viremic control from the consequences of differing levels of viremia. While our data suggest that the CD4+ T cells of ES are, in fact, more susceptible to infection than those of viremic patients, there are potentially confounding factors that must be considered. Cells from viremic progressors may have been altered by the presence of high levels of viremia in their blood, and it is inevitable that some small portion of the cells we utilized in our study were already infected by autologous HIV-1 isolates. It is also possible that the cells that were most susceptible to infection in viremic patients have already been selectively infected and depleted in vivo, and thus the remaining CD4+ T cells in these patients are less susceptible to infection. Alternatively, infected cells from ES may have slower turnover than those from viremics. A recent study found that memory cells from ES persist longer than those from HIV-1-infected individuals, due, at least in part, to differences in the FOX03a pathway (46). It is possible that this phenomenon also aids in the survival of HIV-1-infected cells in ES in vitro, resulting in an apparently higher percentage of infected cells in ES than in viremic subjects.
This is the first study to compare HIV-1 fusion and productive infection of unstimulated CD4+ T cells from ES and viremic patients. The finding that CD4+ T cells from ES are no less susceptible to infection than those from viremic patients is a significant step toward understanding the processes by which ES control viremia. The finding implies that inherent differences in the infected cell cannot explain elite control of HIV-1 infection and that it may be possible to develop effective therapeutic vaccines or therapies that derive from host immune responses to HIV-1.
Our data also show that there is no significant difference in the levels of CCR5 expression on the surface of CD4+ T cells from ES and those from uninfected donors or viremic patients and that, in fact, the expression level of this coreceptor varies only within a very narrow range across all subjects. Additionally, we show that while CD38 and/or HLA-DR expression on CD4+ T cells may be an excellent surrogate for predicting disease progression (15, 43), it does not play a direct role in determining the susceptibility of CD4+ T cells to infection in vitro.
Acknowledgments
We thank Ferdynand Kos of the Johns Hopkins Human Immunology core for FACS analysis.
This work was supported by NIH grant R01 AI080328 (J.N.B.) and the Howard Hughes Medical Institute (R.F.S.).
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Agrawal, L., X. Lu, J. Qingwen, Z. VanHorn-Ali, I. V. Nicolescu, D. H. McDermott, P. M. Murphy, and G. Alkhatib. 2004. Role for CCR5Delta32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4+ cells. J. Virol. 78:2277-2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander, L., E. Weiskopf, T. C. Greenough, N. C. Gaddis, M. R. Auerbach, M. H. Malim, S. J. O'Brien, B. D. Walker, J. L. Sullivan, and R. C. Desrosiers. 2000. Unusual polymorphisms in human immunodeficiency virus type 1 associated with nonprogressive infection. J. Virol. 74:4361-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey, J. R., K. G. Lassen, H. C. Yang, T. C. Quinn, S. C. Ray, J. N. Blankson, and R. F. Siliciano. 2006. Neutralizing antibodies do not mediate suppression of human immunodeficiency virus type 1 in elite suppressors or selection of plasma virus variants in patients on highly active antiretroviral therapy. J. Virol. 80:4758-4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bailey, J. R., K. O'Connell, H. C. Yang, Y. Han, J. Xu, B. Jilek, T. M. Williams, S. C. Ray, R. F. Siliciano, and J. N. Blankson. 2008. Transmission of HIV-1 from a patient who developed AIDS to an elite suppressor. J. Virol. 82:7395-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, B. M., B. L. Block, A. C. Rothchild, and B. D. Walker. 2009. Elite control of HIV infection: implications for vaccine design. Expert Opin. Biol. Ther. 9:55-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Betts, M. R., M. C. Nason, S. M. West, S. C. De Rosa, S. A. Migueles, J. Abraham, M. M. Lederman, J. M. Benito, P. A. Goepfert, M. Connors, M. Roederer, and R. A. Koup. 2006. HIV nonprogressors preferentially maintain highly functional HIV-specific CD8+ T cells. Blood 107:4781-4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blankson, J. N., J. R. Bailey, S. Thayil, H. C. Yang, K. Lassen, J. Lai, S. K. Gandhi, J. D. Siliciano, T. M. Williams, and R. F. Siliciano. 2007. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J. Virol. 81:2508-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bleul, C. C., L. Wu, J. A. Hoxie, T. A. Springer, and C. R. Mackay. 1997. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc. Natl. Acad. Sci. U. S. A. 94:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calugi, G., F. Montella, C. Favalli, and A. Benedetto. 2006. Entire genome of a strain of human immunodeficiency virus type 1 with a deletion of nef that was recovered 20 years after primary infection: large pool of proviruses with deletions of env. J. Virol. 80:11892-11896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carroll, R. G., J. L. Riley, B. L. Levine, Y. Feng, S. Kaushal, D. W. Ritchey, W. Bernstein, O. S. Weislow, C. R. Brown, E. A. Berger, C. H. June, and D. C. St Louis. 1997. Differential regulation of HIV-1 fusion cofactor expression by CD28 costimulation of CD4+ T cells. Science 276:273-276. [DOI] [PubMed] [Google Scholar]
- 11.Casado, C., S. Colombo, A. Rauch, R. Martinez, H. F. Gunthard, S. Garcia, C. Rodriguez, J. Del Romero, A. Telenti, and C. Lopez-Galindez. 2010. Host and viral genetic correlates of clinical definitions of HIV-1 disease progression. PLoS One 5:e11079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cavrois, M., C. De Noronha, and W. C. Greene. 2002. A sensitive and specific enzyme-based assay detecting HIV-1 virion fusion in primary T lymphocytes. Nat. Biotechnol. 20:1151-1154. [DOI] [PubMed] [Google Scholar]
- 13.Deacon, N. J., A. Tsykin, A. Solomon, K. Smith, M. Ludford-Menting, D. J. Hooker, D. A. McPhee, A. L. Greenway, A. Ellett, C. Chatfield, V. A. Lawson, S. Crowe, A. Maerz, S. Sonza, J. Learmont, J. S. Sullivan, A. Cunningham, D. Dwyer, D. Dowton, and J. Mills. 1995. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 270:988-991. [DOI] [PubMed] [Google Scholar]
- 14.Gandhi, S. K., J. D. Siliciano, J. R. Bailey, R. F. Siliciano, and J. N. Blankson. 2008. Role of APOBEC3G/F-mediated hypermutation in the control of human immunodeficiency virus type 1 in elite suppressors. J. Virol. 82:3125-3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giorgi, J. V., L. E. Hultin, J. A. McKeating, T. D. Johnson, B. Owens, L. P. Jacobson, R. Shih, J. Lewis, D. J. Wiley, J. P. Phair, S. M. Wolinsky, and R. Detels. 1999. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J. Infect. Dis. 179:859-870. [DOI] [PubMed] [Google Scholar]
- 16.Goujard, C., M. L. Chaix, O. Lambotte, C. Deveau, M. Sinet, J. Guergnon, V. Courgnaud, C. Rouzioux, J. F. Delfraissy, A. Venet, L. Meyer, and the Agence Nationale de Recherche sur le Sida PRIMO Study Group. 2009. Spontaneous control of viral replication during primary HIV infection: when is “HIV controller” status established? Clin. Infect. Dis. 49:982-986. [DOI] [PubMed] [Google Scholar]
- 17.Hersperger, A. R., F. Pereyra, M. Nason, K. Demers, P. Sheth, L. Y. Shin, C. M. Kovacs, B. Rodriguez, S. F. Sieg, L. Teixeira-Johnson, D. Gudonis, P. A. Goepfert, M. M. Lederman, I. Frank, G. Makedonas, R. Kaul, B. D. Walker, and M. R. Betts. 2010. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 6:e1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 19.Huang, Y., L. Zhang, and D. D. Ho. 1998. Characterization of gag and pol sequences from long-term survivors of human immunodeficiency virus type 1 infection. Virology 240:36-49. [DOI] [PubMed] [Google Scholar]
- 20.Hunt, P. W., J. Brenchley, E. Sinclair, J. M. McCune, M. Roland, K. Page-Shafer, P. Hsue, B. Emu, M. Krone, H. Lampiris, D. Douek, J. N. Martin, and S. G. Deeks. 2008. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J. Infect. Dis. 197:126-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ioannidis, J. P., P. S. Rosenberg, J. J. Goedert, L. J. Ashton, T. L. Benfield, S. P. Buchbinder, R. A. Coutinho, J. Eugen-Olsen, T. Gallart, T. L. Katzenstein, L. G. Kostrikis, H. Kuipers, L. G. Louie, S. A. Mallal, J. B. Margolick, O. P. Martinez, L. Meyer, N. L. Michael, E. Operskalski, G. Pantaleo, G. P. Rizzardi, H. Schuitemaker, H. W. Sheppard, G. J. Stewart, I. D. Theodorou, H. Ullum, E. Vicenzi, D. Vlahov, D. Wilkinson, C. Workman, J. F. Zagury, T. R. O'Brien, and International Meta-Analysis of HIV Host Genetics. 2001. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann. Intern. Med. 135:782-795. [DOI] [PubMed] [Google Scholar]
- 22.Iversen, A. K., E. G. Shpaer, A. G. Rodrigo, M. S. Hirsch, B. D. Walker, H. W. Sheppard, T. C. Merigan, and J. I. Mullins. 1995. Persistence of attenuated rev genes in a human immunodeficiency virus type 1-infected asymptomatic individual. J. Virol. 69:5743-5753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Julg, B., F. Pereyra, M. J. Buzon, A. Piechocka-Trocha, M. J. Clark, B. M. Baker, J. Lian, T. Miura, J. Martinez-Picado, M. M. Addo, and B. D. Walker. 2010. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin. Infect. Dis. 51:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirchhoff, F., T. C. Greenough, D. B. Brettler, J. L. Sullivan, and R. C. Desrosiers. 1995. Brief report: absence of intact nef sequences in a long-term survivor with nonprogressive HIV-1 infection. N. Engl. J. Med. 332:228-232. [DOI] [PubMed] [Google Scholar]
- 25.Lambotte, O., F. Boufassa, Y. Madec, A. Nguyen, C. Goujard, L. Meyer, C. Rouzioux, A. Venet, J. F. Delfraissy, and the SEROCO-HEMOCO Study Group. 2005. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin. Infect. Dis. 41:1053-1056. [DOI] [PubMed] [Google Scholar]
- 26.Lamine, A., A. Caumont-Sarcos, M. L. Chaix, A. Sáez-Cirión, C. Rouzioux, J. F. Delfraissy, G. Pancino, and O. Lambotte. 2007. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study). AIDS 21:1043-1045. [DOI] [PubMed] [Google Scholar]
- 27.Mariani, R., F. Kirchhoff, T. C. Greenough, J. L. Sullivan, R. C. Desrosiers, and J. Skowronski. 1996. High frequency of defective nef alleles in a long-term survivor with nonprogressive human immunodeficiency virus type 1 infection. J. Virol. 70:7752-7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michael, N. L., G. Chang, L. A. d'Arcy, P. K. Ehrenberg, R. Mariani, M. P. Busch, D. L. Birx, and D. H. Schwartz. 1995. Defective accessory genes in a human immunodeficiency virus type 1-infected long-term survivor lacking recoverable virus. J. Virol. 69:4228-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michael, N. L., L. G. Louie, A. L. Rohrbaugh, K. A. Schultz, D. E. Dayhoff, C. E. Wang, and H. W. Sheppard. 1997. The role of CCR5 and CCR2 polymorphisms in HIV-1 transmission and disease progression. Nat. Med. 3:1160-1162. [DOI] [PubMed] [Google Scholar]
- 30.Migueles, S. A., and M. Connors. 2010. Long-term nonprogressive disease among untreated HIV-infected individuals: clinical implications of understanding immune control of HIV. JAMA 304:194-201. [DOI] [PubMed] [Google Scholar]
- 31.Migueles, S. A., A. C. Laborico, W. L. Shupert, M. S. Sabbaghian, R. Rabin, C. W. Hallahan, D. Van Baarle, S. Kostense, F. Miedema, M. McLaughlin, L. Ehler, J. Metcalf, S. Liu, and M. Connors. 2002. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 3:1061-1068. [DOI] [PubMed] [Google Scholar]
- 32.Migueles, S. A., C. M. Osborne, C. Royce, A. A. Compton, R. P. Joshi, K. A. Weeks, J. E. Rood, A. M. Berkley, J. B. Sacha, N. A. Cogliano-Shutta, M. Lloyd, G. Roby, R. Kwan, M. McLaughlin, S. Stallings, C. Rehm, M. A. O'Shea, J. Mican, B. Z. Packard, A. Komoriya, S. Palmer, A. P. Wiegand, F. Maldarelli, J. M. Coffin, J. W. Mellors, C. W. Hallahan, D. A. Follman, and M. Connors. 2008. Lytic granule loading of CD8+ T cells is required for HIV-infected cell elimination associated with immune control. Immunity 29:1009-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo, H., S. Monard, H. Pollack, J. Ip, G. Rochford, L. Wu, J. Hoxie, W. Borkowsky, D. D. Ho, and J. P. Moore. 1998. Expression patterns of the HIV type 1 coreceptors CCR5 and CXCR4 on CD4+ T cells and monocytes from cord and adult blood. AIDS Res. Hum. Retroviruses 14:607-617. [DOI] [PubMed] [Google Scholar]
- 34.Mori, K., D. J. Ringler, and R. C. Desrosiers. 1993. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J. Virol. 67:2807-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navis, M., I. Schellens, D. van Baarle, J. Borghans, P. van Swieten, F. Miedema, N. Kootstra, and H. Schuitemaker. 2007. Viral replication capacity as a correlate of HLA B57/B5801-associated nonprogressive HIV-1 infection. J. Immunol. 179:3133-3143. [DOI] [PubMed] [Google Scholar]
- 36.O'Connell, K. A., J. R. Bailey, and J. N. Blankson. 2009. Elucidating the elite: mechanisms of control in HIV-1 infection. Trends Pharmacol. Sci. 30:631-637. [DOI] [PubMed] [Google Scholar]
- 37.O'Connell, K. A., T. P. Brennan, J. R. Bailey, S. C. Ray, R. F. Siliciano, and J. N. Blankson. 2010. Control of HIV-1 in elite suppressors despite ongoing replication and evolution in plasma virus. J. Virol. 84:7018-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Connell, K. A., Y. Han, T. M. Williams, R. F. Siliciano, and J. N. Blankson. 2009. Role of natural killer cells in a cohort of elite suppressors: low frequency of the protective KIR3DS1 allele and limited inhibition of human immunodeficiency virus type 1 replication in vitro. J. Virol. 83:5028-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O'Doherty, U., W. J. Swiggard, and M. H. Malim. 2000. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74:10074-10080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pereyra, F., M. M. Addo, D. E. Kaufmann, Y. Liu, T. Miura, A. Rathod, B. Baker, A. Trocha, R. Rosenberg, E. Mackey, P. Ueda, Z. Lu, D. Cohen, T. Wrin, C. J. Petropoulos, E. S. Rosenberg, and B. D. Walker. 2008. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 197:563-571. [DOI] [PubMed] [Google Scholar]
- 41.Sáez-Cirión, A., C. Lacabaratz, O. Lambotte, P. Versmisse, A. Urrutia, F. Boufassa, F. Barre-Sinoussi, J. F. Delfraissy, M. Sinet, G. Pancino, A. Venet, and the Agence Nationale de Recherches sur le Sida EP36 HIV Controllers Study Group. 2007. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc. Natl. Acad. Sci. U. S. A. 104:6776-6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salvi, R., A. R. Garbuglia, A. Di Caro, S. Pulciani, F. Montella, and A. Benedetto. 1998. Grossly defective nef gene sequences in a human immunodeficiency virus type 1-seropositive long-term nonprogressor. J. Virol. 72:3646-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa, A. E., J. Carneiro, M. Meier-Schellersheim, Z. Grossman, and R. M. Victorino. 2002. CD4 T cell depletion is linked directly to immune activation in the pathogenesis of HIV-1 and HIV-2 but only indirectly to the viral load. J. Immunol. 169:3400-3406. [DOI] [PubMed] [Google Scholar]
- 44.Stewart, G. J., L. J. Ashton, R. A. Biti, R. A. Ffrench, B. H. Bennetts, N. R. Newcombe, E. M. Benson, A. Carr, D. A. Cooper, and J. M. Kaldor. 1997. Increased frequency of CCR-5 delta 32 heterozygotes among long-term non-progressors with HIV-1 infection. The Australian Long-Term Non-Progressor Study Group. AIDS 11:1833-1838. [DOI] [PubMed] [Google Scholar]
- 45.Trecarichi, E. M., M. Tumbarello, K. de Gaetano Donati, E. Tamburrini, R. Cauda, C. Brahe, and F. D. Tiziano. 2006. Partial protective effect of CCR5-Delta 32 heterozygosity in a cohort of heterosexual Italian HIV-1 exposed uninfected individuals. AIDS Res. Ther. 3:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Grevenynghe, J., F. A. Procopio, Z. He, N. Chomont, C. Riou, Y. Zhang, S. Gimmig, G. Boucher, P. Wilkinson, Y. Shi, B. Yassine-Diab, E. A. Said, L. Trautmann, M. El Far, R. S. Balderas, M. R. Boulassel, J. P. Routy, E. K. Haddad, and R. P. Sekaly. 2008. Transcription factor FOXO3a controls the persistence of memory CD4(+) T cells during HIV infection. Nat. Med. 14:266-274. [DOI] [PubMed] [Google Scholar]
- 47.Wang, B., W. B. Dyer, J. J. Zaunders, M. Mikhail, J. S. Sullivan, L. Williams, D. N. Haddad, G. Harris, J. A. Holt, D. A. Cooper, M. Miranda-Saksena, R. Boadle, A. D. Kelleher, and N. K. Saksena. 2002. Comprehensive analyses of a unique HIV-1-infected nonprogressor reveal a complex association of immunobiological mechanisms in the context of replication-incompetent infection. Virology 304:246-264. [DOI] [PubMed] [Google Scholar]
- 48.Yamada, T., and A. Iwamoto. 2000. Comparison of proviral accessory genes between long-term nonprogressors and progressors of human immunodeficiency virus type 1 infection. Arch. Virol. 145:1021-1027. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, H., Y. Zhou, C. Alcock, T. Kiefer, D. Monie, J. Siliciano, Q. Li, P. Pham, J. Cofrancesco, D. Persaud, and R. F. Siliciano. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J. Virol. 78:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]