Abstract
Spike (S) proteins, the defining projections of the enveloped coronaviruses (CoVs), mediate cell entry by connecting viruses to plasma membrane receptors and by catalyzing subsequent virus-cell membrane fusions. The latter membrane fusion requires an S protein conformational flexibility that is facilitated by proteolytic cleavages. We hypothesized that the most relevant cellular proteases in this process are those closely linked to host cell receptors. The primary receptor for the human severe acute respiratory syndrome CoV (SARS) CoV is angiotensin-converting enzyme 2 (ACE2). ACE2 immunoprecipitation captured transmembrane protease/serine subfamily member 2 (TMPRSS2), a known human airway and alveolar protease. ACE2 and TMPRSS2 colocalized on cell surfaces and enhanced the cell entry of both SARS S-pseudotyped HIV and authentic SARS-CoV. Enhanced entry correlated with TMPRSS2-mediated proteolysis of both S and ACE2. These findings indicate that a cell surface complex comprising a primary receptor and a separate endoprotease operates as a portal for activation of SARS-CoV cell entry.
Viruses exit from infected cells embedded with the energy required to enter new host cells. When viruses encounter new host cells, energy stored within metastable virus surface proteins is dissipated through protein refoldings and used to open the viruses and allow viral genomes to access the cell. This conversion from high-energy metastable to low-energy end stages is spatially and temporally regulated by a variety of triggers that are incorporated into the surface proteins. Depending on the virus, one or a combination of cell receptor bindings, protonations in the endosome, disulfide reductions, and proteolytic cleavages triggers viral protein refolding and opening. Insights into these activating conditions have advanced our understanding of virus-host interactions and have revealed new approaches for antiviral therapeutics.
These activating virus entry events can be further dissected through research with the human CoVs (HCoVs). The HCoVs are notable pathogens (27, 48), with one of them accounting for severe acute respiratory syndrome (SARS) (12, 24). Evolution of the CoVs in their protruding surface or spike (S) proteins can change virus-activating conditions and permit zoonoses (30, 40) and virulence changes. Unraveling S protein activations is therefore central to understanding HCoV tropism, ecology, and pathogenesis.
The S proteins include cell receptor-binding domains (RBDs) and virus-cell membrane fusion domains. Like other class I viral fusion proteins, the HCoV spikes require proteolytic priming to be activated (7). Notably, the majority of pathogenic HCoVs exit producer cells with unprimed S proteins (2, 34) and thus rely on target cell proteases for activation. Therefore, the HCoV cell entry factors on target cells include virus-binding agents (cell receptors) and also virus protein-cleaving agents (cell proteases).
SARS-CoV binds to its ectopeptidase receptor, angiotensin-converting enzyme 2 (ACE2), with very high affinity (44). ACE2 without ectopeptidase activity is also an efficient SARS-CoV receptor (30), and S proteins bind distant from the ACE2 enzyme pocket (28), making it clear that ACE2 is not a direct S-activating protease. There are, however, several proteases that can operate as SARS-CoV entry cofactors, including cathepsin L, elastase, trypsin, factor Xa, thermolysin, and plasmin (13, 31, 42). These are mostly soluble proteases, and it is not obvious how they might be retained in the vicinity of the ACE2 receptors. This question of protease subcellular locations and the timings of enzyme action is relevant because activating S protein endoproteolytic cleavages take place only after ACE2 engagement. Indeed, without prior ACE2 binding, these soluble proteases excessively cleave and inactivate virus spikes (31, 42). Given that the productive sequence is for S proteins to bind ACE2 and then undergo activating proteolysis, it is reasonable to suspect that the relevant proteases activating SARS-CoV entry might be anchored in the plasma membrane and juxtaposed near the ACE2 receptors.
Among the candidates for membrane-anchored virus-activating proteases are the type II transmembrane serine proteases (TTSPs), a family of serine proteases whose physiologic functions are just beginning to be discerned (9). TTSPs are expressed in the surface of airway epithelial cells (11, 21, 49) and thus can potentially be positioned appropriately at virus-cell junctions. Importantly, TTSPs are known to activate the entry of some respiratory viruses, including both seasonal and pathogenic human influenza viruses and human metapneumoviruses (8, 10, 41).
A recent report has implicated a particular TTSP, designated transmembrane protease/serine subfamily member 11a (TMPRSS11a), in the proteolysis of SARS-CoV S proteins (21). This valuable contribution stimulated our interest in cell entry cofactors and prompted questions concerning the transmembrane proteases, their substrate preferences, and their potential localization with the primary ACE2 receptors. In considering these questions, we made discoveries that relate to SARS-CoV and potentially other virus entry events.
MATERIALS AND METHODS
Cells.
293T and hACE2-293 cells, which were obtained from Shibo Jiang, New York Blood Center, were grown in Dulbecco's modified Eagle medium (DMEM) containing 10% heat-inactivated fetal bovine serum (FBS). All growth media were buffered with 0.01 M sodium HEPES (pH 7.4).
Plasmid DNAs.
TMPRSS2 cDNA containing a C-terminal FLAG epitope tag was PCR amplified using template pCMV-Sport6-TMPRSS2 (Open Biosystems) and the following primers: forward, 5′-GGAGAGCTCCACCATGGCTTTGAACTCAGGGTC-3′; reverse, 5′-CGACTCGAGCTACTTGTCATCGTCATCCTTGTAGTCTCCGCCGTCTGCCCTCAT-3′. Subsequently, TMPRSS2 cDNA was cloned into pCAGGS.MCS (35) between SacI and XhoI restriction sites. TMPRSS11a cDNA containing a C-terminal FLAG epitope tag was PCR amplified using template pCR-BluntII-TOPO-TMPRSS11a (Open Biosystems) and the following primers: forward, 5′-GGAGAATTCCACCATGATGTATCGGACAGTAGG-3′; reverse, 5′-CGACCCGGGCTACTTGTCATCGTCATCCTTGTAGTCTCCGATGCCTGTTTTTGAAG −3′. TMPRSS11d cDNA containing a C-terminal FLAG epitope tag was PCR amplified using template pCR-BluntII-TOPO-TMPRSS11d (Open Biosystems) and the following primers: forward, 5′-GGAGAATTCCACCATGTATAGGCCAGCACG-3′; reverse, 5′-CGACCCGGGCTACTTGTCATCGTCATCCTTGTAGTCTCCGATCCCAGTTTGT TGC-3′. Subsequently, TMPRSS11a and TMPRSS11d cDNAs were cloned into pCAGGS.MCS between EcoRI and XmaI restriction sites. The catalytically inactive pCAGGS-TMPRSS2(S441A)FLAG mutant was derived from pCAGGS-TMPRSS2FLAG using mutagenic primers and a site-directed mutagenesis protocol. (QuikChange XL, catalog no. 200519-5; Stratagene). Plasmids pcDNA3.1 SARS S and pcDNA3.1 ACEC9 were obtained from Michael Farzan, Harvard Medical School, Boston, MA. Plasmid pCDM8-NL63 S was obtained from Hyeryun Choe, Harvard Medical School, Boston, MA. pcDNA 3.1 Ebola Zaire virus glycoprotein (Ebo GP) and pHEF-VSV G (vesicular stomatitis virus glycoprotein [VSV G]) were obtained from Lijun Rong, University of Illinois, Chicago. pNL4.3-Luc R− E− was obtained from the NIH AIDS Research and Reference Program 3418. Plasmid pRL-TK, encoding Renilla luciferase, was purchased from Promega. Plasmids pCAGT7 and pT7EMC-Luc (36) were obtained from Richard Longnecker, Northwestern University Feinberg School of Medicine, Chicago, IL.
Cell-cell fusion assay.
Cell-cell fusion was performed as described previously (32). Briefly, effector (293T) cells were transiently transfected with pCAG-T7 pol and pcDNA3.1-SARS S via calcium phosphate. Target cells were generated by cotransfection of 293T cells with pT7EMC-luc, which encodes firefly luciferase under T7 promoter control; pcDNA3.1-ACE2C9; and pCAGGS-TMPRSS2FLAG. At ∼24 h posttransfection (hptf), the target cells were quickly trypsinized and added to adherent effector cells in a 1:1 effector-to-target cell ratio. After an ∼3-h cocultivation period, luciferase activity was read as described above.
Pseudotyped virions and transductions.
To generate pseudotyped HIV particles, 293T cells were cotransfected via calcium phosphate with pNL4.3-Luc and the various envelope constructs. After 2 days, media were collected, clarified for 10 min at 2,000 × g, and then stored at −20°C. For transductions, HIV particles were concentrated into 293T cells by centrifugation at 1,600 × g for 2 h at 25°C, a process known as spinoculation. Subsequently, the inoculum was removed and replaced with DMEM-10% FBS. At 2 days posttransduction, the cells were rinsed with saline and dissolved in luciferase lysis buffer (Promega E397A). Luminescence was measured upon the addition of luciferase substrate (Promega E1501) using a Veritas microplate luminometer (Turner BioSystems).
Drug treatments during transductions.
Target 293T cells seeded into six-well plates were cotransfected with pcDNA3.1-ACE2C9 and pRL-TK along with pCAGGS-TMPRSS2FLAG or pCAGGS.MCS via calcium phosphate. At 2 days posttransfection, the cells were incubated with bafilomycin A1 (Sigma) at 300 nM or NH4Cl (Sigma) at 25 mM in complete DMEM for 1 h at 37°C. Parallel cultures were incubated in dimethyl sulfoxide (DMSO) and water, the vehicle controls for stock bafilomycin A1 and NH4Cl, respectively. After the 1-h incubation, HIV-SARS S particles in DMEM-10% FBS were spinoculated onto the target cells in the presence of endosomotropic agents. Following spinoculation, the cells were incubated at 37°C for another 27 h until lysis and evaluation of both Renilla and firefly luciferase accumulations (E1910; Promega).
Immunofluorescence microscopy.
293T cells seeded onto fibronectin-coated coverslips were cotransfected via polyethylenimine with a constant amount of pcDNA3.1-ACE2C9 and various doses of pCAGGS-TMPRSS2FLAG. At 24 hptf, the cells were fixed with 3.7% formaldehyde (Polysciences) in 0.1 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES, pH 6.8). Following blocking with 10% donkey serum, the coverslips were incubated with primary antibodies in phosphate-buffered saline (PBS) containing donkey serum. Cell surface ACE2 was detected using SARS-RBDFc, while TMPRSS2 was detected using rabbit anti-FLAG antibody (Sigma catalog no. F1804). The SARS-RBDFc protein contained a 15-amino-acid CD5 signal sequence and residues 318 to 510 of SARS spike in a modified pCEP4 vector. SARS-RBDFc protein was produced in 293-EBNA cells, harvested in serum-free medium, and purified using protein A-Sepharose beads (GE Healthcare). After thorough washing in PBS, the coverslips were incubated with goat anti-human secondary antibody conjugated to AlexaFluor488 (Molecular Probes) and goat anti-mouse secondary antibody conjugated to Cy5 (Jackson Immunoresearch). Nuclei were visualized with 4′,6-diamidino-2-phenylindole (DAPI) stain (Invitrogen). Coverslips were mounted on slides with Fluoro-Gel (EMS) and visualized using a Deltavision deconvolution microscope.
Immunoprecipitation (IP) and immunoblotting (IB).
293T cells were cotransfected via calcium phosphate with pcDNA3.1-ACE2, the pcDNA3.1 empty vector, the pCAGGS empty vector, pCAGGS-TMPRSS2FLAG, and pCAGGS-TMPRSS2 (S441A) FLAG in the indicated combinations. At 35 hptf, the cell monolayers were lysed in HNB buffer (25 mM Na HEPES, 150 mM NaCl, 0.01% bovine serum albumin) containing 0.5% NP-40, 0.5% sodium deoxycholate (DOC), and 0.1% protease inhibitor (Sigma P2714). Cell lysates were first clarified by centrifugation at 2,000 × g for 5 min, and then 150,000 cell equivalents were mixed with 0.9 μg anti-FLAG antibody (Sigma catalog no. F7425), 0.9 μg 1D4 antibody, or 0.9 μg mouse IgG and 0.06 ml protein G magnetic beads (NEB Corporation, Inc.) for 14 h at 4°C. Beads were rinsed three times with HNB buffer containing 0.5% NP-40 and 0.5% DOC. Proteins were eluted from beads by addition of electrophoresis sample buffer (0.125 M Tris [pH 6.8], 10% dithiothreitol, 2% sodium dodecyl sulfate [SDS], 10% sucrose, 0.004% bromophenol blue), heated at 95°C for 5 min, and subsequently subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE). SDS-gels were transferred to polyvinylidene difluoride membranes that were subsequently blocked for 1 h with 5% nonfat milk powder in TBS-T (25 mM Tris-HCl [pH 7.5], 140 mM NaCl, 2.7 mM KCl, 0.05% Tween 20). ACE2C9 proteins were detected with a 1:5,000 dilution of 1D4 antibody (obtained from Hyeryun Choe, Harvard Medical School, Boston, MA). TMPRSS2FLAG and TMPRSS2-S441AFLAG proteins were detected with rabbit anti-FLAG antibody (1:1,000 in TBS-T).
SARS-CoV infections.
For authentic SARS virus infections, 293T cells were transfected in duplicate via calcium phosphate with pcDNA3.1-ACE2C9 and each of the indicated pCAGGS-TTSPFLAG plasmids. At 20 hptf, cells were infected with HCoV-SARS (Urbani) at a multiplicity of infection (MOI) of 0.1. Subsequently, at 6 h postinfection (hpi), one set of infected samples was dissolved in Trizol (Invitrogen) and total cellular RNA was harvested. SARS CoV N and human glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene-specific RNAs were quantified by real-time PCR using an ABI Prism7700 thermocycler and software. The following primers were used: for SARS-CoV nucleocapsid (N) mRNA, forward primer 5′-ATATTAGGTTTTTACCCAGG-3′ and reverse primer 5′-CTTGCCCCATTGCGTCCTCC-3′; for the gene for human GAPDH, forward primer 5′-CCACTCCTCCACCTTTGAC-3′ and reverse primer 5′-ACCCTGTTGCTGTAGCCA-3′. The levels of SARS N gene amplicons were normalized to those of GAPDH amplicons.
The other set of infected cell cultures were harvested at 24 hpi, and protein lysates in electrophoresis sample buffer were evaluated by Western blotting for SARS N using rabbit anti-SARS N antibody (Imgenex IMG-549), for SARS S using anti-SARS S antibody (Imgenex IMG-541), and for β-actin using mouse anti-β-actin antibody (Sigma). All work with infectious SARS-CoV was performed in a biosafety level 3 laboratory at the University of Iowa.
RESULTS
Effect of TTSPs on pseudovirus transductions.
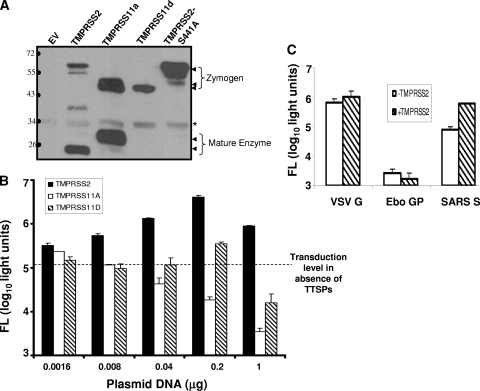
Three of the 17 known human TTSPs, TMPRSS2, TMPRSS11a, and TMPRSS11d (also known as human airway trypsin or HAT), were selected for study because they are expressed in human lungs and are known to activate cell entry of selected influenza virus strains (8, 10). On transfection of the three FLAG-tagged TTSP cDNAs into 293T cells, all synthesized FLAG-tagged proteins of the expected molecular weights (Fig. 1A). TTSPs are synthesized as inactive single-chain proenzymes (zymogens) and undergo self-cleavage into active forms during or after transport to cell surfaces (1, 33). Cell-associated C-terminal cleavage fragments were observed for TMPRSS11a and TMPRSS2 but not for TMPRSS11d, perhaps because the TMPRSS11d member sheds its peripheral enzymatic domain into the extracellular medium (49). To confirm that the cleaved forms of one TTSP, TMPRSS2, were indeed generated by autocatalytic activity, we generated an inactive mutant harboring a serine-to-alanine substitution at position 441 (S441A) (1). As expected, the TMPRSS2S441A mutant was only present as a full-length ∼70-kDa zymogen form (Fig. 1A).
FIG. 1.
TTSP expression and effect on HIV-SARS S entry. (A) 293T cells (106) were transfected with 1-μg samples of plasmids encoding the indicated FLAG-tagged TTSP constructs. Cells were lysed at 2 days posttransfection and proteins from 5 × 104−-cell aliquots were evaluated by IB with anti-FLAG antibodies. The asterisk denotes a nonspecific 34-kDa band. The values to the left are molecular sizes in kilodaltons. (B) 293T cells (106) were transfected with ACE2 (1 μg) along with the indicated amounts of TTSP plasmids. At 2 days posttransfection, cells were inoculated with HIV-SARS S and luciferase accumulations were evaluated 27 h later. The dotted line represents the transduction level in target cells transfected with ACE2 and the empty vector. (C) 293T cells cotransfected with equal amounts of either ACE2 plus the empty vector (white bars) or ACE2 plus TMPRSS2 (hatched bars) were inoculated with HIV-VSVG, -Ebo GP, or -SARS S pseudovirus at 2 days posttransfection. Luciferase accumulations were evaluated 27 h later. The error bars in panels B and C represent standard deviations (n = 3). The experiments were repeated three times with similar results. FL, firefly luciferase.
To reveal the effect of TTSPs on HCoV entry, we cotransfected 293T cells with constant amounts of ACE2C9 plasmid along with various doses of the different TTSPFLAG plasmid DNAs. After 2 days, these cells were transduced by HIV-SARS S pseudoviruses, as measured by the accumulation of a luciferase reporter gene product that is encoded in the HIV transducing vector. We selected 293T cells as targets because they support early HIV infection events and because they do not express endogenous TMPRSS2 or TMPRSS4 (1, 5, 10).
All three TTSPs enhanced SARS S-mediated pseudovirus entry. The augmenting effects varied as follows: TMPRSS2 > TMPRSS11d > TMPRSS11a (Fig. 1B). Peak effects of TMPRSS2 and TMPRSS11d were observed after transfection with 0.2 μg DNA and then declined at the higher 1-μg DNA dose; this was presumably due to toxicities generated by TTSP overexpression. HIV transductions mediated by VSV G and Ebo GP spikes were not affected by the presence of TMPRSS2 (Fig. 1C), indicating that this particular TTSP exhibits restricted, S-specific enhancing effects.
TMPRSS2 functions at the cell surface.
We focused on the most potent TTSP, TMPRSS2, and determined whether this protease activates SARS S for plasma membrane-localized fusion using a cell-cell fusion assay. Briefly, effector 293T cells were cotransfected with plasmids encoding SARS S and T7 RNA polymerase, while target 293T cells were cotransfected with plasmids encoding ACE2, TMPRSS2, and a luciferase reporter under the control of a T7 RNA polymerase promoter (see Fig. 2A). After a 3-h coculture of effector and target cells, cell-cell fusions, clearly discernible as microscopic ∼10- to 20-cell syncytia, were corroborated by luciferase assays, which indicated an ∼10-fold increase in membrane fusion (Fig. 2B). HCoV NL63 S protein-mediated membrane fusion was also enhanced by TMPRSS2, albeit less potently than SARS-CoV S (Fig. 2B). These data confirmed that the TMPRSS2 protease can exert its fusion-promoting effects on two somewhat distantly related HCoV S proteins and that these effects can take place at cell surfaces.
FIG. 2.
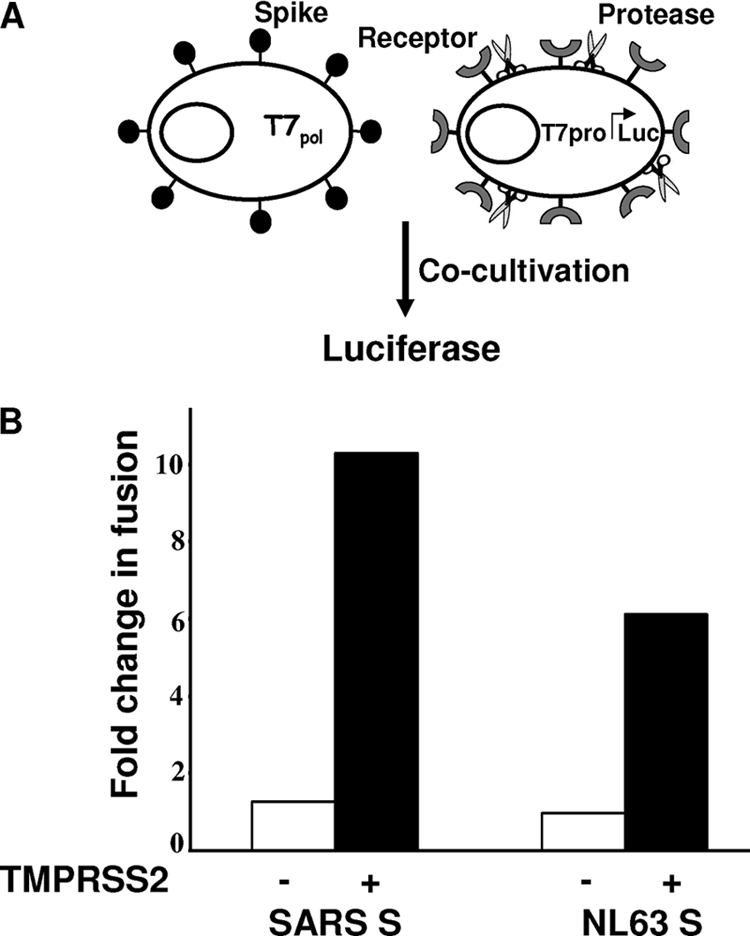
Effect of TMPRSS2 on SARS S-mediated membrane fusion. (A) Schematic diagram of cell-cell fusion. Effector 293T cells were generated by cotransfection of plasmids encoding the indicated spike constructs along pCAG-T7pol (1 μg DNA per 106 cells). Target 293T cells were generated by cotransfection of pcDNA3.1-ACE2 plus the pCAGGS empty vector (−) or pcDNA3.1-ACE2 plus pCAGGS-TMPRSS2 (+) together with pEMC-T7pro-luc. (B) Luciferase readings 3 h after cocultivation of effector and target cells (1:1 ratio) plotted as n-fold changes in fusion over negative controls without S proteins.
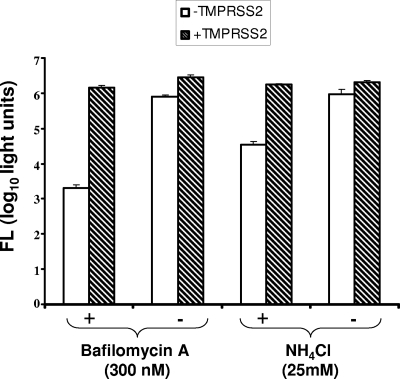
Endosomal cathepsin L is a known S-activating protease (6, 42). To determine whether TMPRSS2 at the target cell surface might nullify any requirements for this and related acid pH-dependent proteases during SARS S-mediated virus entry, the endosomal H+/ATPase inhibitor bafilomycin A1 or the endosomotropic weak base NH4Cl was applied to target cells immediately before and during HIV-SARS S inoculation. These inhibitors of endosomal acidification potently suppressed S-mediated transductions (Fig. 3), with 2- and 3-log10 reductions by NH4Cl and bafilomycin A1, respectively. Suppressions were eliminated by TMPRSS2 (Fig. 3), indicating that TMPRSS2 fully activates SARS S-mediated entry (∼1,000-fold) when acid pH-dependent protease(s) is absent. The findings in Fig. 3 thus reveal the indiscriminate nature of SARS S entry functions, with either a cell surface neutral-pH TMPR serine protease or an endosomal acid-pH-requiring (likely cathepsin) cysteine protease operating as an entry catalyst.
FIG. 3.
Effect of TMPRSS2 on HIV-SARS S entry into drug-treated cells. 293T cells were cotransfected (1 μg per 106 cells) with ACE2 plus the empty vector (white bars) or with ACE2 plus TMPRSS2 (hatched bars). One day later, and 1 h prior to transduction with HIV-SARS S pseudoviruses, cells were incubated with bafilomycin A1 (300 nM) or NH4Cl (25 mM). Vehicle controls were DMSO and water for bafilomycin A1 and NH4Cl, respectively. HIV-SARS S particles were then concentrated onto cells by a 2-h spinoculation. Bafilomycin and NH4Cl remained on cells during and after spinoculation, until 6 h posttransduction, at which time cells were rinsed and replenished with fresh medium. Luciferase accumulations were determined at 28 h posttransduction. Error bars represent standard deviations (n = 3). The experiment was repeated three times with similar results. FL, firefly luciferase.
SARS S protein cleavage by TMPRSS2.
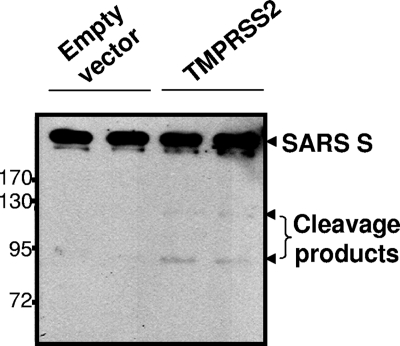
We assumed that the S proteins would be the relevant TTSP substrates for enhancing virus entry. Proteolyses at two positions, designated S1-S2 and S2′, are necessary to enhance S-mediated membrane fusions (4), and there are arginines at both of these cleavage sites that could constitute TTSP substrates (21). To determine whether incoming S proteins on pseudoviruses might be cleaved specifically by TMPRSS2, we generated target hACE2-293 cells with or without TMPRSS2FLAG and adsorbed HIV-SARS S particles by spinoculation. After rinsing steps, cells and bound viruses were incubated for 1 h at 37°C and lysed and S proteins were visualized by IB. There was slight but convincing evidence of TMPRSS2-specific SARS S cleavage, as indicated by the presence of C-terminal ∼120- and ∼85-kDa S fragments (Fig. 4). The extent of proteolysis was admittedly low; however, it is certainly conceivable that this very small proportion of S proteins undergoing cleavage represents the activated S proteins operating in pseudovirus entry.
FIG. 4.
Cleavage of SARS S proteins during virus entry. HIV-SARS S pseudoviruses were spinoculated onto target hACE2-293 cells transfected 2 days earlier with the pCAGGS empty vector or pCAGGS-TMPRSS2 (1 μg per 106 cells, in duplicate). After spinoculation, cells were incubated at 37°C for 1 h to allow the cleavage of S proteins by cell surface TMPRSS2. After removal of unbound pseudovirions and washing, cells were lysed and evaluated by IB for S proteins using anti-C9 tag antibody. The values to the left are molecular sizes in kilodaltons.
ACE2 protein cleavage by TMPRSS2.
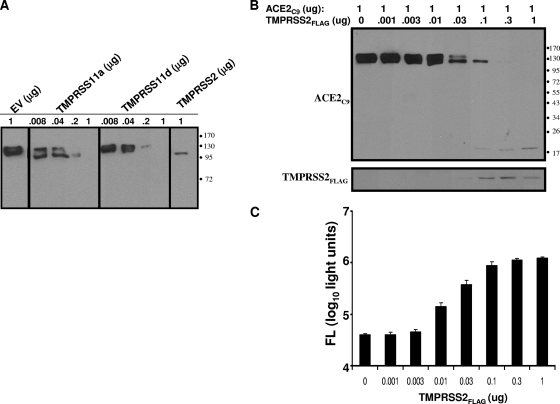
The discovery that S proteins were targeted by TMPRSS2 led us to consider whether the integral membrane ACE2 proteins might also be substrates. Thus, we introduced graded doses of the TTSP, plasmids along with constant amounts of pcDNA3.1-ACE2C9, into 293T cells and evaluated the expressed proteins 2 days later. A striking finding was that all three TTSPs targeted ACE2 for distinctive proteolysis (Fig. 5A). In a separate experiment, 293T cultures with gradually increasing levels of TMPRSS2 revealed doses ultimately eliminating full-length ACE2, leaving a slightly smaller ∼115-kDa (perhaps underglycosylated) form and an ∼20-kDa C-terminal ACE2 fragment (Fig. 5B). Remarkably, these cultures with undetectable complete ACE2 were ∼30-fold more susceptible to HIV-S transduction than controls (Fig. 5C), suggesting that few receptors can mediate efficient entry, provided that the relevant protease is nearby to activate incoming virions. These findings raised additional questions about the levels and distributions of ACE2 and TMPRSS2 on susceptible cell surfaces.
FIG. 5.
Effect of TTSP expression on ACE2 and HIV-SARS S transductions. (A) Cells (106) transfected with a constant 1 μg of ACE2 and increasing doses of the indicated TTSPsor pCAGGS empty vector (EV) were analyzed at 2 days posttransfection by IB for the C9 epitope appended to the ACE2 C terminus. (B) Cells transfected with 1 μg of ACE2C9 and the indicated amounts of TMPRSS2 were analyzed at 2 days posttransfection with anti-C9 and anti-FLAG tag antibodies, respectively. (C) Parallel unlysed cell cultures were transduced with HIV-SARS S pseudoviruses, and luciferase accumulations were measured at 40 h posttransduction. FL, firefly luciferase. The values to the right of panels A and B are molecular sizes in kilodaltons.
ACE2 associations with TMPRSS2.
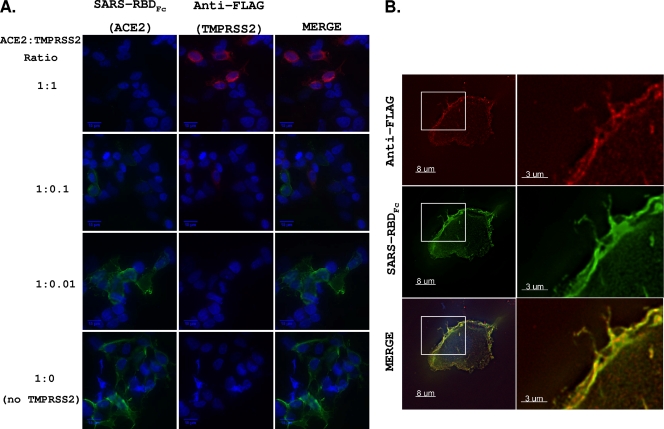
We hypothesized that ACE2 and TMPRSS2 interact in cellular exocytic pathways and at cell surfaces and that cleavage of ACE2 is a result of these interactions. This was first evaluated using immunofluorescence assays. Here we cosynthesized ACE2C9 and TMPRSS2FLAG in 293T cells, fixed cells without permeabilization, and then used a human IgG1 Fc-tagged form of the SARS S RBD to detect ACE2 and a mouse anti-FLAG antibody to detect TMPRSS2. The confocal images (Fig. 6A) indicated that cells expressing surface TMPRSS2 typically did not have ACE2. Thus, it was likely that TMPRSS2 cleaved ACE2 before it reached the cell surface or shortly afterwards. On slides of cells transfected with small amounts of TMPRSS2 (1:0.1 and 1:0.01 ACE2-to-TMPRSS2 plasmid ratios), rare cells were identified with both ACE2 and TMPRSS2 (Fig. 6B). In these cells, the receptor and protease colocalized on the plasma membrane, although the levels of each were decreased compared to those in cells expressing either protein alone. Therefore, the majority of cells expressing TMPRSS2 had undetectable ACE2, but when TMPRSS2 was scarce, ACE2 localized to TMPRSS2-containing regions.
FIG. 6.
Cellular localization of TMPRSS2 and ACE2. (A) Cells were cotransfected with ACE2 and TMPRSS2 at various ACE2-TMPRSS2 ratios. At 24 hptf, cells were fixed without permeabilization and incubated with anti-FLAG and SARS-RBDFc. Anti-human IgG (green) and anti-mouse IgG (red) secondary antibodies were used to detect bound SARS-RBDFc and anti-FLAG antibodies, respectively. All images were obtained using identical acquisition times and display levels. (B) A representative ACE2+ TMPRSS2+ cell from the 1:0.1 ACE2-TMPRSS2 DNA ratio.
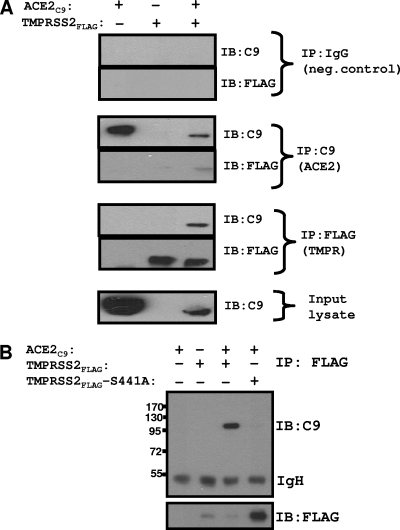
To further evaluate ACE2-TMPRSS2 interactions, we used an IP-IB assay. Transfected 293T cells were dissolved in a buffer containing Nonidet P-40 and DOC; this detergent formulation is known to completely lyse cells while preserving selected membrane protein interactions (37). From these lysates, ACE2C9 and TMPRSS2FLAG were captured on magnetic protein G beads using mouse anti-C9 and rabbit anti-FLAG antibodies, respectively, and then proteins were detected by IB using the same epitope-specific antibodies. The results (Fig. 7A) revealed specific co-IP of both TMPRSS2 and ACE2 by either the C9 or FLAG antibodies. As there was digestion of the ∼130-kDa ACE2 form in these cultures (Fig. 5), the predominant immunoprecipitated form of ACE2 was ∼115 kDa. We attempted to determine whether the complete ∼130-kDa ACE2 might be coimmunoprecipitated with the catalytically inactive TMPRSS2-S441A. While the anti-FLAG IPs efficiently adhered both the wild type and the inactive mutant TMPRSS2(S441A)FLAG, co-IP of ACE2 was only observed with the wild-type form (Fig. 7B). These data indicated that enzymatic activity was required for TMPRSS2 association with ACE2.
FIG. 7.
Interaction between TMPRSS2 and ACE2. (A) 293T cells transfected with pCAGGS-TMPRSS2FLAG (0.5 μg per 106 cells) and pcDNA3.1-ACE2C9 (1 μg per 106 cells), individually or in combination, were lysed and subsequently incubated with rabbit anti-FLAG, mouse anti-C9, or mouse IgG antibody on protein G magnetic beads. Eluted proteins were analyzed by IB using the indicated antibodies. (B) Lysates from 293T cells transfected with pCAGGS-TMPRSS2FLAG (0.5 μg per 106 cells) or pCAGGS-TMPRSS2(S441A)FLAG (0.5 μg per 106 cells), individually or in combination with pcDNA3.1-ACE2C9 (1 μg per 106 cells), were subjected to IPs using rabbit anti-FLAG antibody. Eluted proteins were analyzed by IB using mouse anti-C9 antibody. The values to the left are molecular sizes in kilodaltons.
ACE2-TMPRSS2 associations in cis and in trans.
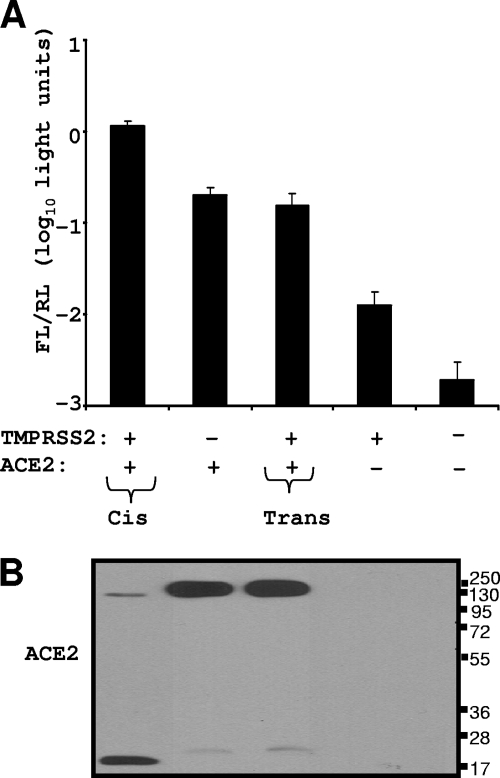
Some TTSPs, notably HAT, will cleave cellular substrates both in cis and in trans, i.e., when presented from neighboring cells (3). This is because some TTSPs shed their catalytic C-terminal domains into medium (45, 49), thereby creating paracrine proteolytic activities. We sought to determine whether the TMPRSS2 that fosters SARS S entry and cleaves ACE2 in cis would do the same in trans. To this end, a mixed population of target cells was generated, with half being ACE2+ and the other half TMPRSS2+. These cells were both challenged with SARS S pseudotype viruses and evaluated for ACE2 cleavage. If TMPRSS2 extended its effect broadly, then the mixed populations would be highly susceptible to S-mediated entry and would also have cleaved ACE2. Unlike the condition in which ACE2 and TMPRSS2 were expressed in the same cells, the expression of these two proteins on separate cells did not foster virus entry at a level above that observed when only ACE2 was expressed (Fig. 8A). In addition, the ACE2 proteins were not proteolyzed when TMPRSS2 was on separate cells, even though the TMPRSS2+ and ACE2+ cells were contacting each other in the monolayers (Fig. 8B). This was in striking contrast to the ACE2 digestion during cis presentation of TMPRSS2. These data suggest that the relevant in vivo targets are those in which both the ACE2 and TMPRSS2 entry factors are simultaneously present in the same cells and that extracellularly shed or adjacent TMPRSS2 has no effect on infection.
FIG. 8.
ACE2-TMPRSS2 associations in cis and in trans. Four 293T cell populations were transfected with a Renilla luciferase (RL) control plasmid along with the empty vector (EV), TMPRSS2, and ACE2 plasmids, either alone or together, as indicated. One day later, cell populations were cocultivated as indicated by plus signs. FL, firefly luciferase. (A) After a 20-h cocultivation period, cells were transduced with HIV-SARS S. FL/RL was measured at 27 h posttransduction. Error bars represent standard deviations (n = 3). The experiment was repeated three times with similar results. (B) After a 20-h cocultivation period, cells were lysed and evaluated for ACE2 cleavage by IB using anti-C9 tag antibody. The values to the right are molecular sizes in kilodaltons.
Effect of TTSPs on SARS-CoV infections.
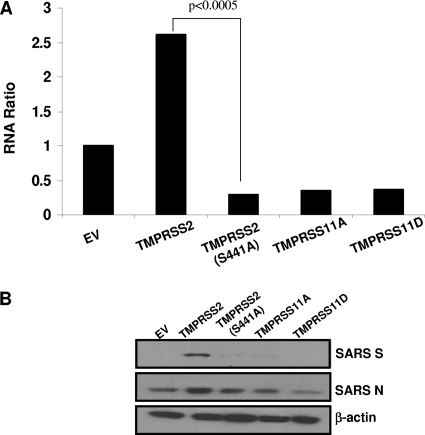
To determine whether TMPRSS2 also increased cell susceptibility to authentic SARS-CoV infections, we challenged transfected 293T cells with authentic SARS-CoV (Urbani strain). We evaluated SARS N RNA levels at 6 hpi by quantitative reverse transcription-PCR (Fig. 9A), as well as SARS S and N protein levels at 24 hpi by IB (Fig. 9B). There was 9-fold more SARS N RNA in cells expressing TMPRSS2 than in cells expressing enzymatically inactive TMPRSS2 (S441A) [P < 0.0005]. These viral RNAs were translated to generate significantly more N proteins in TMPRSS2+ cells (Fig. 9B). Additionally, there was significantly more S protein in TMPRSS2+ cells. These findings, notably the comparisons of proteolytically active and inactive TMPRSS2s, allowed us to assign natural infection-enhancing activity to proteolysis.
FIG. 9.
Effect of TTSPs on SARS-CoV infections. Cells transfected with ACE2 and each TTSP were challenged with SARS-CoV at an MOI of 0.1. (A) SARS-CoV N- and human GAPDH gene-specific RNAs were quantified by real-time PCR, and levels of SARS N gene amplicons were normalized to those of GAPDH amplicons. Data are plotted as the ratio of each RNA to the empty vector (EV) RNA. Statistical analysis was performed using Student's t test for unpaired samples on threshold cycle values (n = 3). The experiment was performed twice. (B) SARS S and N and β-actin protein levels were evaluated at 24 hpi by IB using anti-SARS S, anti-SARS N, and anti-β-actin antibodies.
DISCUSSION
Viral proteins are proteolytically processed to provide structural flexibility to nonenveloped and enveloped viruses, notably in the surface glycoproteins extending from virion membranes. Surface glycoproteins facilitating virus-cell membrane fusions are synthesized and maintained in precursor intermediate folding states, and proteolysis permits the refolding and energy release required to create stable virus-cell linkages and membrane coalescence. Here we evaluated the proteolytic priming of the SARS-CoV S proteins that function in virus-cell binding and in virus-cell membrane fusion. As this priming takes place after S proteins bind their receptors (31, 42) and involves multiple stepwise endoproteolytic cleavages (4), we asked whether relevant proteases might be complexed with the SARS receptor, making for rapid and efficient activation of virus entry.
The TTSPs are displayed on cell surfaces and could thus potentially link to ACE2, the SARS-CoV receptor. We began by evaluating several TTSPs for activation of S protein entry into 293T cells and found that TMPRSS2, a member of the Hepsin/TMPRSS subfamily (9), was potent in enhancing S-mediated entry, more so than TMPR11a or TMPR11d (HAT). The TMPRSS2 levels providing ∼10-fold augmentation of SARS S-mediated entry were similar to those found endogenously in Calu3 lung-like cells (T.H.-S. and T. G., unpublished data). Notably, TMPRSS2 is expressed in epithelial cells lining the nose, trachea, and distal airways, including alveoli and type II pneumocytes, as determined by in situ hybridization studies (11). As such, TMPRSS2 may be a relevant protease for lower-airway SARS-CoV infections.
The TMPRSS2-mediated enhancement of SARS virus entry was accompanied by remarkable changes in ACE2. At low doses, TMPRSS2 converted ACE2 from an ∼130-kDa to an ∼115-kDa form, a change we believe comes either from ACE2 deglycosylation or from a failure to initially glycosylate the six predicted N-glycan sites in the ACE2 ectodomain (46). TMPRSS2 and the related hepsin are known to interfere with protein N glycosylation by an unknown mechanism (5), and so it is possible that under- or nonglycosylated membrane proteins are common to cells expressing various TTSPs. Increasing amounts of TMPRSS2 converted the detectable portion of ACE2 to an ∼20-kDa C-terminal fragment. We did not identify the sister ectodomain fragment in culture medium but did readily detect soluble ACE2 liberated from cells by tumor necrosis factor alpha-converting enzyme (TACE), an unrelated protease known to cleave ACE2 (20, 26). It is possible that TTSPs cleave at multiple arginines and lysines throughout ACE2, leaving only short peptides (17). At any rate, this ACE2 diminution amounts to “shedding,” a well-known phenomenon thus far attributed only to TACE. On the basis of the ∼20-kDa fragment mobility in Fig. 4B, the TMPRSS2 cleaves more N terminally than TACE (19, 20), and indeed, we have found that TACE-resistant ACE2 (20) is subject to TMPRSS2 shedding (data not shown), indicating distinct protease targets. That ACE2 shedding is achieved by at least two divergent proteases suggests multiple posttranslational regulations of this ectoenzyme, testifying to the importance of ACE2 regulation in SARS pathogenesis (25) and lung homeostasis in general (18). ACE2 diminution in response to TMPRSS2 was readily apparent by immunofluorescence microscopy as well. Indeed, the inverse relationships between TMPRSS2 and ACE2 levels on cell surfaces (Fig. 6A) again suggested that TMPRSS2 associates with ACE2 during or after exocytic transport and that TMPRSS2 occludes, degrades, and/or sheds the vast majority of the ACE2 that is capable of binding to viral S proteins. The findings also highlight the miniscule, virtually undetectable ACE2 levels that will support SARS virus entry as long as the TMPRSS2 protease is available. In concert with this view, 293T cells, which are thought to have little, if any, ACE2 (34), were made susceptible to SARS S entry by TMPRSS2 alone (Fig. 8B). The readily detectable host susceptibility determinant here is the protease, not the primary virus receptor.
The fact that TMPRSS2 degraded ACE2 made it challenging to microscopically identify cells in which both of these proteins were present. However, when rare ACE2+ TMPRSS2+ cells were found, clear colocalizations were immediately evident. We expected these colocalizations, first because our data argue that infecting viruses depend simultaneously on these two proteins and second because both proteins may partition into lipid microdomains. ACE2 is a known lipid raft-embedded protein (14), and TMPRSS2 is set apart from other TTSPs by its longer, potentially palmitoylated 84-residue cytoplasmic tail (9), which may confer positioning into lipid rafts. The SARS-CoV-susceptible cell may be defined by tendencies of ACE2 and TMPRSS2 to congregate together.
The results of co-IP analyses supported the view of convening ACE2 and TMPRSS2 ligands. While catalytically inactive TMPRSS2 (S441A) did not coprecipitate ACE2, wild-type TMPRSS2 did, and this ACE2-TMPRSS2 connection was striking given that the overall ACE2 levels were low in the presence of the active TMPRSS2 protease. These findings indicated an unusually stable tethering of the active enzyme, but not the inactive zymogen, with one of its substrates. It will be interesting and valuable to identify the spectrum of TTSP substrates in the cell.
TMPRSS2 and related cell surface proteases are known to shed their enzyme-active domains into extracellular environments (49), and exogenous serine proteases such as trypsin and elastase are known to activate SARS S proteins (31). Thus, we asked whether TMPRSS2 might operate in “trans” to activate SARS S-mediated entry into adjacent ACE2+ cells. We found no evidence for this—adjacent TMPRSS2 did not influence SARS S-mediated entry. Corroborating this finding, we further demonstrated that TMPRSS2 did not digest ACE2 when the two membrane proteins were expressed in adjacent cells. Therefore, we suggest that the cells most susceptible to SARS-CoV infection are those in which ACE2 and TTSPs are simultaneously present and that at least the TMPRSS2 on ACE2-negative cells has limited paracrine activities on ACE2 or SARS-CoV infections.
The ACE2-TMPRSS2 complexes operating in cis to strengthen virus entry might remain throughout the infection cycle, thus raising questions about their effects on the S proteins synthesized and incorporated into infective progeny viruses. For example, S proteins might be cleavage activated by TMPRSS2 during their transport in exocytic pathways, analogous to influenza virus HA0 activation (8) and to numerous furin-cleaved viral glycoproteins (23). To date, we have no evidence for TTSP-mediated S protein cleavage-activation within authentic SARS-CoV-infected cells. However, this issue deserves further consideration, particularly in certain in vivo environments with high localized TTSP densities. It is conceivable that SARS-CoV spikes are prematurely proteolyzed and inactivated in these contexts. Interestingly, late-stage SARS disease, which is known to correlate with low virus infectivities (39), may nonetheless include virus particle productions, which could elicit proinflammatory responses characteristic of late-stage SARS immunopoathologies (47).
SARS-CoV appears to have remarkable resourcefulness, as either endosomal acidophilic proteases or cell surface TMPRSS2 could provide equivalent ∼1,000-fold-enhanced entry. Are these two divergent protease activities completely redundant, or are there contexts in which TTSPs are specifically required? Our hypothesis is that certain in vivo infections require TTSPs; this can be addressed pharmacologically by infecting human airway epithelial cultures (43) in the presence of type-specific protease inhibitors (38) and genetically by infecting mice lacking selected transmembrane proteases (22). We suggest that TTSPs are crucial in vivo entry cofactors because they activate S at cell surfaces and thus support rapid virus dissemination via syncytia (Fig. 2). Furthermore, TTSPs may provide a certain “immediacy” of S activation that can quickly tether incoming viruses to cells via their S protein membrane fusion domains. In contrast, endosome-localized activations are delayed until after subcellular virus transport (31) and demand that viruses remain attached to ACE2 receptors for much longer times. Therefore, TTSPs may especially support CoVs having low affinities for their receptors, for example, zoonotic CoVs entering new host organisms bearing orthologous receptors (30). Viruses entering these new hosts bind loosely to receptors (30), and the benefits of rapidly triggered S protein activation might be in creating additional S-based connections before viruses are eluted away. Zoonotic CoVs may thus evolve affinities for those receptors that are associated with S-activating TTSPs, as might be inferred by the existence of the SARS-CoV and HCoV NL63, whose divergent S proteins both bind to ACE2 receptors (15, 16, 29). Virus evolution and adaptation for particular receptors may have much to do with associated activating proteolytic enzymes.
Acknowledgments
We are especially indebted to Edward Campbell (Loyola University) for expert advice on microscopy. We thank Chandra Tangudu and Anna Mielech for technical assistance and helpful discussions.
This work was supported by NIH grant AI060030.
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Afar, D. E., I. Vivanco, R. S. Hubert, J. Kuo, E. Chen, D. C. Saffran, A. B. Raitano, and A. Jakobovits. 2001. Catalytic cleavage of the androgen-regulated TMPRSS2 protease results in its secretion by prostate and prostate cancer epithelia. Cancer Res. 61:1686-1692. [PubMed] [Google Scholar]
- 2.Arpin, N., and P. J. Talbot. 1990. Molecular characterization of the 229E strain of human coronavirus. Adv. Exp. Med. Biol. 276:73-80. [DOI] [PubMed] [Google Scholar]
- 3.Beaufort, N., D. Leduc, H. Eguchi, K. Mengele, D. Hellmann, T. Masegi, T. Kamimura, S. Yasuoka, F. Fend, M. Chignard, and D. Pidard. 2007. The human airway trypsin-like protease modulates the urokinase receptor (uPAR, CD87) structure and functions. Am. J. Physiol. Lung Cell. Mol. Physiol. 292:L1263-L1272. [DOI] [PubMed] [Google Scholar]
- 4.Belouzard, S., V. C. Chu, and G. R. Whittaker. 2009. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 106:5871-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertram, S., I. Glowacka, P. Blazejewska, E. Soilleux, P. Allen, S. Danisch, I. Steffen, S. Y. Choi, Y. Park, H. Schneider, K. Schughart, and S. Pohlmann. 2010. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 84:10016-10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch, B. J., W. Bartelink, and P. J. Rottier. 2008. Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J. Virol. 82:8887-8890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch, B. J., R. van der Zee, C. A. de Haan, and P. J. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Böttcher, E., T. Matrosovich, M. Beyerle, H. D. Klenk, W. Garten, and M. Matrosovich. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J. Virol. 80:9896-9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bugge, T. H., T. M. Antalis, and Q. Wu. 2009. Type II transmembrane serine proteases. J. Biol. Chem. 284:23177-23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaipan, C., D. Kobasa, S. Bertram, I. Glowacka, I. Steffen, T. S. Tsegaye, M. Takeda, T. H. Bugge, S. Kim, Y. Park, A. Marzi, and S. Pohlmann. 2009. Proteolytic activation of the 1918 influenza virus hemagglutinin. J. Virol. 83:3200-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donaldson, S. H., A. Hirsh, D. C. Li, G. Holloway, J. Chao, R. C. Boucher, and S. E. Gabriel. 2002. Regulation of the epithelial sodium channel by serine proteases in human airways. J. Biol. Chem. 277:8338-8345. [DOI] [PubMed] [Google Scholar]
- 12.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 13.Du, L., R. Y. Kao, Y. Zhou, Y. He, G. Zhao, C. Wong, S. Jiang, K. Y. Yuen, D. Y. Jin, and B. J. Zheng. 2007. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem. Biophys. Res. Commun. 359:174-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glende, J., C. Schwegmann-Wessels, M. Al-Falah, S. Pfefferle, X. Qu, H. Deng, C. Drosten, H. Y. Naim, and G. Herrler. 2008. Importance of cholesterol-rich membrane microdomains in the interaction of the S protein of SARS-coronavirus with the cellular receptor angiotensin-converting enzyme 2. Virology 381:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofmann, H., K. Pyrc, L. van der Hoek, M. Geier, B. Berkhout, and S. Pohlmann. 2005. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. U. S. A. 102:7988-7993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hofmann, H., G. Simmons, A. J. Rennekamp, C. Chaipan, T. Gramberg, E. Heck, M. Geier, A. Wegele, A. Marzi, P. Bates, and S. Pohlmann. 2006. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J. Virol. 80:8639-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hooper, J. D., J. A. Clements, J. P. Quigley, and T. M. Antalis. 2001. Type II transmembrane serine proteases. Insights into an emerging class of cell surface proteolytic enzymes. J. Biol. Chem. 276:857-860. [DOI] [PubMed] [Google Scholar]
- 18.Imai, Y., K. Kuba, S. Rao, Y. Huan, F. Guo, B. Guan, P. Yang, R. Sarao, T. Wada, H. Leong-Poi, M. A. Crackower, A. Fukamizu, C. C. Hui, L. Hein, S. Uhlig, A. S. Slutsky, C. Jiang, and J. M. Penninger. 2005. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature 436:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwata, M., J. E. Silva Enciso, and B. H. Greenberg. 2009. Selective and specific regulation of ectodomain shedding of angiotensin-converting enzyme 2 by tumor necrosis factor alpha-converting enzyme. Am. J. Physiol. Cell Physiol. 297:C1318-C1329. [DOI] [PubMed] [Google Scholar]
- 20.Jia, H. P., D. C. Look, P. Tan, L. Shi, M. Hickey, L. Gakhar, M. C. Chappell, C. Wohlford-Lenane, and P. B. McCray, Jr. 2009. Ectodomain shedding of angiotensin converting enzyme 2 in human airway epithelia. Am. J. Physiol. Lung Cell. Mol. Physiol. 297:L84-L96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kam, Y. W., Y. Okumura, H. Kido, L. F. Ng, R. Bruzzone, and R. Altmeyer. 2009. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One 4:e7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim, T. S., C. Heinlein, R. C. Hackman, and P. S. Nelson. 2006. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 26:965-975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 24.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 25.Kuba, K., Y. Imai, S. Rao, H. Gao, F. Guo, B. Guan, Y. Huan, P. Yang, Y. Zhang, W. Deng, L. Bao, B. Zhang, G. Liu, Z. Wang, M. Chappell, Y. Liu, D. Zheng, A. Leibbrandt, T. Wada, A. S. Slutsky, D. Liu, C. Qin, C. Jiang, and J. M. Penninger. 2005. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 11:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lambert, D. W., M. Yarski, F. J. Warner, P. Thornhill, E. T. Parkin, A. I. Smith, N. M. Hooper, and A. J. Turner. 2005. Tumor necrosis factor-alpha convertase (ADAM17) mediates regulated ectodomain shedding of the severe-acute respiratory syndrome-coronavirus (SARS-CoV) receptor, angiotensin-converting enzyme-2 (ACE2). J. Biol. Chem. 280:30113-30119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee, N., D. Hui, A. Wu, P. Chan, P. Cameron, G. M. Joynt, A. Ahuja, M. Y. Yung, C. B. Leung, K. F. To, S. F. Lui, C. C. Szeto, S. Chung, and J. J. Sung. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1986-1994. [DOI] [PubMed] [Google Scholar]
- 28.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 29.Li, W., M. J. Moore, N. Vasilieva, J. Sui, S. K. Wong, M. A. Berne, M. Somasundaran, J. L. Sullivan, K. Luzuriaga, T. C. Greenough, H. Choe, and M. Farzan. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, W., C. Zhang, J. Sui, J. H. Kuhn, M. J. Moore, S. Luo, S. K. Wong, I. C. Huang, K. Xu, N. Vasilieva, A. Murakami, Y. He, W. A. Marasco, Y. Guan, H. Choe, and M. Farzan. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 24:1634-1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuyama, S., M. Ujike, S. Morikawa, M. Tashiro, and F. Taguchi. 2005. Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc. Natl. Acad. Sci. U. S. A. 102:12543-12547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McShane, M. P., and R. Longnecker. 2005. Analysis of fusion using a virus-free cell fusion assay. Methods Mol. Biol. 292:187-196. [DOI] [PubMed] [Google Scholar]
- 33.Miyake, Y., M. Yasumoto, S. Tsuzuki, T. Fushiki, and K. Inouye. 2009. Activation of a membrane-bound serine protease matriptase on the cell surface. J. Biochem. 146:273-282. [DOI] [PubMed] [Google Scholar]
- 34.Moore, M. J., T. Dorfman, W. Li, S. K. Wong, Y. Li, J. H. Kuhn, J. Coderre, N. Vasilieva, Z. Han, T. C. Greenough, M. Farzan, and H. Choe. 2004. Retroviruses pseudotyped with the severe acute respiratory syndrome coronavirus spike protein efficiently infect cells expressing angiotensin-converting enzyme 2. J. Virol. 78:10628-10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 36.Okuma, K., M. Nakamura, S. Nakano, Y. Niho, and Y. Matsuura. 1999. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology 254:235-244. [DOI] [PubMed] [Google Scholar]
- 37.Opstelten, D. J., M. J. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Otlewski, J., F. Jelen, M. Zakrzewska, and A. Oleksy. 2005. The many faces of protease-protein inhibitor interaction. EMBO J. 24:1303-1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sheahan, T., B. Rockx, E. Donaldson, A. Sims, R. Pickles, D. Corti, and R. Baric. 2008. Mechanisms of zoonotic severe acute respiratory syndrome coronavirus host range expansion in human airway epithelium. J. Virol. 82:2274-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shirogane, Y., M. Takeda, M. Iwasaki, N. Ishiguro, H. Takeuchi, Y. Nakatsu, M. Tahara, H. Kikuta, and Y. Yanagi. 2008. Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J. Virol. 82:8942-8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons, G., D. N. Gosalia, A. J. Rennekamp, J. D. Reeves, S. L. Diamond, and P. Bates. 2005. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. U. S. A. 102:11876-11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sims, A. C., R. S. Baric, B. Yount, S. E. Burkett, P. L. Collins, and R. J. Pickles. 2005. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J. Virol. 79:15511-15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sui, J., W. Li, A. Murakami, A. Tamin, L. J. Matthews, S. K. Wong, M. J. Moore, A. S. Tallarico, M. Olurinde, H. Choe, L. J. Anderson, W. J. Bellini, M. Farzan, and W. A. Marasco. 2004. Potent neutralization of severe acute respiratory syndrome (SARS) coronavirus by a human mAb to S1 protein that blocks receptor association. Proc. Natl. Acad. Sci. U. S. A. 101:2536-2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Szabo, R., and T. H. Bugge. 2008. Type II transmembrane serine proteases in development and disease. Int. J. Biochem. Cell Biol. 40:1297-1316. [DOI] [PubMed] [Google Scholar]
- 46.Tipnis, S. R., N. M. Hooper, R. Hyde, E. Karran, G. Christie, and A. J. Turner. 2000. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J. Biol. Chem. 275:33238-33243. [DOI] [PubMed] [Google Scholar]
- 47.Tseng, C. T., L. A. Perrone, H. Zhu, S. Makino, and C. J. Peters. 2005. Severe acute respiratory syndrome and the innate immune responses: modulation of effector cell function without productive infection. J. Immunol. 174:7977-7985. [DOI] [PubMed] [Google Scholar]
- 48.van der Hoek, L. 2007. Human coronaviruses: what do they cause? Antivir. Ther. 12:651-658. [PubMed] [Google Scholar]
- 49.Yasuoka, S., T. Ohnishi, S. Kawano, S. Tsuchihashi, M. Ogawara, K. Masuda, K. Yamaoka, M. Takahashi, and T. Sano. 1997. Purification, characterization, and localization of a novel trypsin-like protease found in the human airway. Am. J. Respir. Cell Mol. Biol. 16:300-308. [DOI] [PubMed] [Google Scholar]