Abstract
The rabies virus (RV) phosphoprotein (P) is a type I interferon (IFN) antagonist preventing both transcriptional induction of IFN and IFN-mediated JAK/STAT signaling. In addition, P is an essential cofactor of the viral polymerase and is required for encapsidation of viral RNA into nucleoprotein during replication. By site-directed mutagenesis, we have identified a domain of P required for efficient inhibition of IFN induction. Phosphoproteins lacking amino acids (aa) 176 to 181, 182 to 186, or 176 to 186 were severely compromised in counteracting phosphorylation of IRF3 and IRF7 by TBK1 or IKKi while retaining the full capacity of preventing nuclear import of activated STATs and of supporting virus transcription and replication. Recombinant RV carrying the mutated phosphoproteins (the SAD ΔInd1, SAD ΔInd2, and SAD ΔInd1/2 viruses) activated IRF3 and beta IFN (IFN-β) transcription in infected cells but still blocked STAT-mediated expression of IFN-stimulated genes. Due to a somewhat higher transcription rate, the SAD ΔInd1 virus activated IRF3 more efficiently than the SAD ΔInd2 virus. After intracerebral injection into mouse brains at high doses, the SAD ΔInd1 virus was completely apathogenic for wild-type (wt) mice, while the SAD ΔInd2 virus was partially attenuated and caused a slower progression of lethal rabies than wt RV. Neurovirulence of IFN-resistant RV thus correlates with the capacity of the virus to prevent activation of IRF3 and IRF7.
The type I interferon (IFN) system is an important host defense element against viruses with critical functions in both innate antiviral defense and in modulating adaptive immune responses. The expression of IFN is activated upon recognition of conserved pathogen-associated molecular patterns (PAMPs) by immune receptors, such as retinoic acid inducible gene I protein (RIG-I) and melanoma differentiation-associated gene product (MDA5) (for a review, see reference 42). Binding of viral triphosphate RNAs to these receptors activates downstream signaling, leading to activation of the nuclear factor-κB (NF-κB), interferon regulatory factor 3 (IRF3), and IRF7, if present, by the kinases TBK1 and IKKi (23). Apart from IFN genes, IRF3 controls transcription of several antiviral genes, leading to the establishment of an antiviral state in the infected cells (16, 35). This primary, cell-autonomous activation (40) goes along with secretion of IFN, which acts via the alpha/beta interferon (IFN-α/β) receptor (IFNAR), and JAK/STAT signaling in an autocrine and paracrine fashion to direct a huge secondary gene expression program, including antiviral, apoptotic survival, and immune genes (34, 44). By autocrine feedback to the infected cell, the pathogen-sensing and cell-autonomous activation is also further stimulated by IFN (35).
The ability of viruses to counteract IFN-mediated JAK/STAT signaling is known to be crucial for the establishment and maintenance of an infection. Viruses having nonfunctional JAK/STAT antagonists are attenuated in hosts with a functional IFN system, and hosts lacking a functional IFNAR are highly susceptible to infection even by severely attenuated viruses (for reviews, see references 17 and 35). More recently, it was found that inhibition of cell-autonomous activation is critical for viruses as well and that hosts with IFN induction pathways affected are more susceptible to viruses (42). Accordingly, even RNA viruses with a very limited coding capacity, such as the neurotropic rabies virus (RV) of the Rhabdoviridae family, have evolved functions to antagonize both IFN induction and JAK/STAT signaling pathways (3, 38).
The RV genome encodes five proteins, nucleoprotein (N), phosphoprotein (P), matrix protein (M), glycoprotein (G), and the polymerase (L), in the order 3′-N-P-M-G-L-5′. P is a multifunctional modular protein with diverse essential tasks during the viral life cycle and is therefore a prime target for the development of antivirals (6). Besides chaperoning N for specific encapsidation of viral RNA and acting as a cofactor of the RNA polymerase L (48), P is responsible for counteracting the host IFN system. On the one hand, P binds and retains phosphorylated STAT1 and STAT2 in the cytoplasm and thus prevents nuclear import of STATs and expression of IFN-stimulated genes (ISG) (5, 30, 45, 46). On the other hand, P prevents induction of type I IFN. In RV-infected cells, RIG-I is activated by viral triphosphate RNAs (11, 18); however, IRF3 phosphorylation, dimerization, and nuclear import are prevented in the presence of P (4). The importance of the IFN-antagonistic functions of P has been illustrated by recombinant RV expressing reduced amounts of P, either by shifting the P gene to a promoter-distal position from which it is transcribed poorly (4) or by using picornavirus internal ribosome entry site (IRES) elements to limit the translation rate of P (26). Apart from a modest defect in replication, such viruses have severe defects in their ability to counteract both IFN induction and IFN signaling and are therefore highly attenuated in vivo.
Preventing IFN induction and preventing STAT signaling are genetically independent functions of P. While the C terminus of P is required for STAT binding (5, 45), it is not required for interference with IRF3 activation. For this study, we have prepared diverse P mutants and have identified a small internal domain (amino acids [aa] 176 to 186) whose deletion abolishes the ability to efficiently prevent IRF activation while leaving the functions of P in replication and STAT binding unaffected. Recombinant RVs compromised exclusively in their capability to prevent IRF activation effectively stimulated antiviral defense and were attenuated after intracerebral injection in mice. This emphasizes the importance for neurotropic viruses to prevent cell-autonomous activation of innate immunity.
(This work was conducted by M.R. in partial fulfillment of the requirements for a Ph.D.)
MATERIALS AND METHODS
Cells and viruses.
HEp-2 cells (ATCC CCL-23) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% fetal calf serum (FCS) and antibiotics. HEK 293T cells were propagated in DMEM with 10% FCS, l-glutamine, and antibiotics, and BSR-T7/5 cells were propagated in MEM supplemented with 10% newborn calf serum. The recombinant RV SAD L16 virus comprising the consensus sequence of the attenuated SAD B19 vaccine strain was used as the wild-type (wt) RV (39). The SAD ΔPLP virus was described previously (4). Plasmids pSAD ΔInd1 and pSAD ΔInd2 were cloned by ligation of AvrII- and SnaBI-digested pSAD L16 and pCR3 PΔInd1 or PΔInd2, pSAD ΔInd1/2 was cloned by the exchange of the enhanced green fluorescent protein (eGFP) gene of pSAD ΔP-eGFP (A. Ghanem, unpublished data) with the P gene from pCR3 PΔInd1/2 by HpaI and NarI, and viruses were recovered from cDNA as described previously (4).
Serological reagents.
Commercial antibodies were obtained from the following manufacturers: anti-STAT1 p84/p91 (catalog no. sc-592), anti-STAT2 (sc-476), anti-ISGFγ p48 (IRF9) (sc-496), and anti-IRF3 (sc-9082) antibodies from Santa Cruz Biotechnology; anti-pY701-STAT1 from Cell Signaling; anti-pY689-STAT2 from Upstate Biotechnology; anti-pS386-IRF3 antibody from IBL; and anti-actin (19-32) and anti Flag M2 (F-3165) from Sigma. IFN-α A/D (catalog no. 11200-2) was purchased from PBL Biomedical Laboratories. For detection of RV nucleoproteins, the mouse monoclonal W239 was used. Polyclonal rabbit P antiserum P160-5 and the MxA antibody M134 were kindly provided by S. Finke (Riems, France) and O. Haller (Freiburg, Germany), respectively.
Plasmids and transfection.
The described pCR3-RV P plasmid (4) was used for PCR-based in vitro mutagenesis to generate short deletions within the P-coding region, yielding the PΔ176-181aa and PΔ182-186aa mutants. For the mutant lacking both motifs (PΔ176-186aa mutant), a two-insert ligation into pCR3 was carried out. The primer sequences and detailed cloning steps are available from the authors upon request. The ΔRIG-I plasmid was kindly provided by T. Fujita (50). The Flag-tagged MDA5 1-350 (ΔMDA5) construct was generated by PCR amplification from pcDNA3.1-Flag-MDA5. pCR3 Fl-TBK1 was generated by PCR amplification from pCR3-Ig-TBK1.
For dual-luciferase reporter gene assays, HEK 293T cells were seeded in 24-well plates and transfected the next day at 60 to 70% confluence with 100 ng per well of p125-luc (Stratagene) by using Lipofectamine 2000. In all experiments, 1 ng of pCMV-RL encoding Renilla luciferase was cotransfected as an internal control. For reporter gene assays using P cDNA transfection, HEK 293T cells were cotransfected with 0.2 μg empty vector or RV P expression constructs and 0.2 μg ΔRIG-I, ΔMDA5, or TBK1 expression vector by using Lipofectamine 2000. Twenty-four hours after transfection, cell lysates were prepared and subjected to reporter gene assays and Western blot analysis.
For quantitative reverse transcription-PCR (qRT-PCR) experiments, HEK 293T cells were seeded in 12-well plates and transfected the following day at 60 to 70% confluence with 0.4 μg empty vector or RV P expression constructs and 0.4 μg empty vector or ΔRIG-I per well using Lipofectamine 2000. Total RNA was extracted by using the RNeasy minikit (Qiagen). IFN-β cDNA levels were measured with the LightCycler 2.0 (Roche).
For Western blot analysis of IRF3 dimerization and phosphorylation, HEK 293T cells were transfected in 6-well plates at 60 to 70% confluence with 5 μg of Fl-TBK1 or Fl-IKKi and 10 μg of P-expressing plasmid or empty vector DNA as indicated. After 24 h, cell extracts were prepared and were analyzed by nondenaturing or SDS-PAGE and Western blotting.
The minigenome transfections were carried out on BSR-T7/5 cells, seeded in 6-well plates, and transfected the next day at 90% confluence with 4 μg pSDI-luc, 5 μg pTIT-N, 2.5 μg pTIT-L, 10 ng pRL, and 2.5 μg of the P expression plasmids per well by using CaPO4 (Stratagene). Forty-eight hours after transfection, cell lysates were prepared and subjected to luciferase reporter gene assays and SDS-PAGE and Western blotting for expression control.
Cell culture infection.
For reporter gene assays in virus-infected cells, HEK 293T or BSR cells were infected at a multiplicity of infection (MOI) of 3 at 6 h posttransfection with p125-luc or pISREluc (Stratagene) and pCMV-RL. After an additional 24 h, cell extracts were harvested and analyzed using the Dual-Luciferase reporter system (Promega) in a luminometer (Centro 960; Berthold) according to the supplier's instructions. For quantitative RT-PCR experiments, HEK 293T cells were seeded in 12-well plates and infected at an MOI of 3 the following day at 60 to 70% confluence. RNA was extracted by using the RNeasy minikit (Qiagen) and subjected to RT-PCR. IFN-β cDNA levels were measured with the LightCycler 2.0 (Roche). For growth curve analysis of recombinant RVs, an MOI of 0.05 was applied with HEp-2 and BSR-T7/5 cells. For Western blot analysis of protein levels and immunofluorescence microscopy in HEp-2 cells, an MOI of 1 was used.
Western blots.
Cell extracts were prepared by treatment with cell lysis buffer (62.5 mM Tris, 2% SDS, 10% glycerol, 6 M urea, 5% β-mercaptoethanol, 0.01% bromophenol blue, 0.01% phenol red). Proteins were resolved by 10% SDS-PAGE. For native PAGE, cell extracts from infected or transfected cell cultures were prepared as described previously (4). The cells from a confluent well of a 6-well plate were lysed in 100 μl lysis buffer. After centrifugation (14,000 rpm, 4°C, 10 min), 10 μl of the lysate was loaded on the 6% native gel. After gel separation by SDS-PAGE or native PAGE, the probes were transferred to either a nitrocellulose or polyvinylidene difluoride (PVDF) membrane (Schleicher & Schuell) by using a semidry blotter (Peq-Lab). Membranes were incubated overnight at 4°C with primary antibodies. Protein signals were visualized with horseradish peroxidase-conjugated secondary antibodies and the ECL system (Perkin Elmer) by exposure to X-ray films or by digital detection in the Fusin FX device (Vilber Lourmat, Germany).
Northern blotting.
Total RNA from infected cells was isolated with the RNeasy minikit (Qiagen) according to the manufacturer's instructions. Northern blotting and hybridizations with [α-32P]dCTP-labeled cDNAs were performed as described previously (9). Hybridization signals were quantified using a phosphorimaging device (Molecular Dynamics Storm) and analyzed with Image Quant 5.0 software.
Immunofluorescence microscopy.
HEp-2 cells were seeded on glass coverslips and were infected with RV at an MOI of 1 for 24 h. Cells were fixed using 3% paraformaldehyde for 20 min at room temperature and were permeabilized in 0.5% Triton X-100 in phosphate-buffered saline (PBS). After incubation with primary anti-IRF3 antibodies (1:100 in PBS, 45 min, 37°C), the specimens were incubated with fluorescence-labeled secondary antibodies at a dilution of 1:200 in PBS for 1 h at 37°C (goat anti-rabbit Alexa Flour 488 and anti-mouse tetramethylrhodamine, both from Molecular Probes). Nuclear chromatin was stained by adding a final concentration of 0.5 μM TO-PRO-3-iodide (Molecular Probes) to the secondary antibodies.
Confocal laser-scanning microscopy was performed with a Zeiss LSM510 Meta laser system using a Zeiss Axiovert200 microscope. Excitation of Alexa Fluor 488, tetramethylrhodamine, and TO-PRO-3-iodide occurred at wavelengths of 488 nm, 543 nm, and 633 nm, respectively. To avoid cross talk, the individual channels were scanned sequentially.
Mouse infection experiments.
Wild-type 129 mice and transgenic mice on the 129 background were originally obtained from M. Aguet (31) and were bred at the Institute of Immunology, Friedrich Loeffler Institute, Tübingen, Germany. Transgenic knockout mice were kept in the specific-pathogen-free (SPF) facility of the institute. Sex-matched, 5- to 8-week-old wt 129 mice and IFNAR−/− or IFNGR−/− mice (31) were infected intracerebrally into the left hemisphere with 105 focus-forming units (FFU) in 20 μl, and newborn mice received 10 μl. The animals were observed daily three times and scored for the appearance of neurological signs on an arbitrary scale of 1 to 3 (level 1 for slight neurological signs, such as beginning ataxia and slightly reduced motility; level 2 for increased neurological signs, such as trembling and/or disorientation after tail spinning; level 3 for severe signs of disease, such as ruffled fur, hunched position, and inability to move). Animals scored twice at level 3 or at level 2 at noon and level 3 in the afternoon were immediately sacrificed according to the German Animal Protection law, and serum and organs were preserved. The number of mice and experimental design were approved by local authorities (Regional Board; permission no. FLI 223/05; 35/9185.81-6).
RESULTS
An internal domain of RV P is important for efficient inhibition of IRF3 and IRF7 phosphorylation by the kinases TBK1 and IKKi.
RV P is known to interfere with transcriptional activation of IFN by inhibiting phosphorylation of IRF3 and IRF7 by TBK1 and IKKi (4) but is unable to prevent IKKα-mediated activation of IRF7 through TLR7 signaling in pDC (33). In addition, it prevents IFN-α/β- and IFN-γ-stimulated JAK/STAT signaling by binding to activated STAT1 and STAT2. Deletion of the 10 C-terminal residues of P (aa 288 to 297) was previously found to abolish binding of P to STATs, while inhibition of IRF3 phosphorylation was not affected, indicating two independent genetic functions (5, 45).
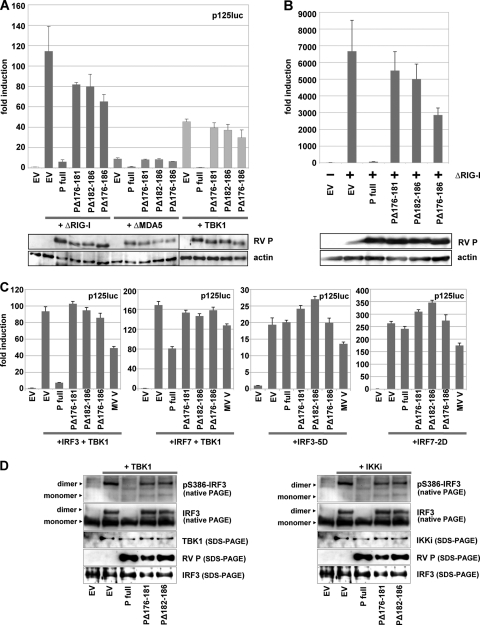
To identify the RV P domains required for inhibition of IRF3 activation, P mutants with short internal deletions were screened for their ability to interfere with the expression of IFN-β promoter-controlled firefly luciferase (FL) from p125-luc in transfected HEK 293T cells. While wt P almost completely prevented FL activity stimulated by constitutively active N-terminal domains of RIG-I and MDA5 (the ΔRIG-I and ΔMDA5 mutants, respectively) and by TBK1 (19-, 9-, and 75-fold inhibition, respectively), two mutants, the PΔ176-181 (PΔInd1) and PΔ182-186 (PΔInd2) mutants, were identified which allowed higher levels of FL activity, although some residual inhibitory effect was apparent in comparison with cells transfected with empty vector plasmids (Fig. 1A). Notably, even a construct combining the two deletions, the PΔ176-186 (PΔInd1/2) mutant, did not lose the residual inhibitory activity; rather, a somewhat improved interference was suggested. Quantitative RT-PCR experiments from RIG-I-stimulated cells revealed effective transcription of IFN-β mRNA in the presence of the PΔInd1 and PΔInd2 mutants (Fig. 1B). Activation of cells by transfection of a ΔRIG-I-coding plasmid resulted in a 760-fold accumulation of IFN-β mRNA (100%). While wt P almost completely abolished transcription to less than 1%, the PΔ176-181 and PΔ182-186 mutants were inefficient in inhibition, resulting in the accumulation of IFN-β mRNA to 83% and 75% of the control, respectively. Again, an improved inhibitory activity of the PΔ176-186 mutant (43% of control) was suggested.
FIG. 1.
RV P mutants with decreased ability to inhibit IFN-β transcription. HEK 293T cells were transfected with full-length RV P coding plasmids or the indicated P mutants using Lipofectamine 2000. (A) Dual-luciferase reporter gene assays were performed 24 h p.i. with p125-luc and pCMV-Renilla, and plasmids encoding ΔRIG-I, ΔMDA5, or TBK1. (B) ΔRIG-I and P constructs were expressed from transfected plasmids, and IFN-β mRNA levels were analyzed by qRT-PCR 24 h posttranscription. Averages of results from two experiments are depicted, with error bars representing standard deviations. Expression levels were controlled by SDS-PAGE by using antibodies against RV P and actin. (C) Dual-luciferase assay from cells transfected with IRF3 plus TBK1 (150 ng each), IRF7 plus TBK1 (150 ng each), IRF3-5D (300 ng), or IRF7-2D (300 ng) and expressing the indicated P constructs. (D) Phosphorylation and dimerization of IRF3 were analyzed for HEK 293T cells cotransfected with TBK1 (left) or IKKi (right) and with RV P expression vectors as indicated. Cell lysates were prepared 24 h posttransfection and submitted to native PAGE analysis. Only wt RV P was able to efficiently prevent IRF3 phosphorylation and dimerization.
As indicated by further reporter gene experiments in which the kinase TBK1 was coexpressed with IRF3 or IRF7 from transfected plasmids, RV P is able to interfere with TBK1-mediated activation not only of IRF3 (4) but also of IRF7, although its inhibitory activity was markedly less pronounced in comparison to IRF3 (Fig. 1C). Notably, the PΔInd mutants have lost the ability (Fig. 1C) to interfere with both IRF3 and IRF7 activation by TBK1. In addition, the activity of phosphomimetic forms of the transcription factors, IRF3-5D and IRF7-2D, was not affected by any of the RV P constructs, while the measles virus V protein showed a partial inhibition, as was observed previously (33).
In order to validate the results of the reporter gene assays and to demonstrate that the mutant phosphoproteins have lost the ability to interfere with the activation of the transcription factor IRF3, dimerization and phosphorylation of IRF3 was examined by native gel electrophoresis and Western blot experiments. Expression of both TBK1 and IKKi from transfected plasmids resulted in efficient phosphorylation of S386 of IRF3 and in IRF3 dimerization (Fig. 1D), which was almost completely prevented by coexpression of wt P. In contrast, in the presence of the PΔInd1 and PΔInd2 mutants, IRF3 phosphorylation and dimerization was readily detected, although a reduced accumulation of activated IRF3 compared to that of the control (EV) was indicated in the case of TBK1 stimulation (Fig. 1D). Taken together, the PΔ176-181 and PΔ182-186 mutants have lost most of the inhibitory activity of wt P in preventing activation of IRF3 by TBK1 and IKKi and induction of IFN-β mRNA transcription.
PΔInd proteins support viral growth.
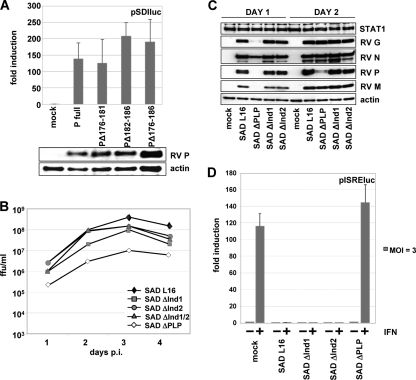
To see whether the deletions in the PΔInd mutants would affect the functions of P during viral RNA synthesis, their ability to rescue a cDNA-directed RV minigenome into functional RNPs and to support its replication and gene expression was tested. Five plasmids encoding RV N, L, P or P deletion mutants, RL, and the minigenome plasmid pSDI-luc (10) were cotransfected into BSR-T7/5 cells expressing T7 RNA polymerase to yield minigenome-like T7 polymerase transcripts. As suggested by dual-luciferase assays, the PΔInd1, -2, or -1/2 mutants did support encapsidation of T7 transcripts by N-P complexes, replication of RNPs by N-L-P complexes, and transcription of FL mRNAs by L-P complexes (Fig. 2A). This strongly suggested that the functions of P during viral RNA synthesis were not substantially impeded by the deletion of residues 176 to 186.
FIG. 2.
(A) RV P mutants support gene expression from the RV pSDI-luc minigenome. BSR-T7/5 cells were transfected with empty vector, RV P, or the indicated P mutants along with plasmids encoding RV L, RV N, and Renilla luciferase. Luciferase activities and protein expression were determined at 48 h posttransfection. (B) Infectious titers of SAD PΔInd mutant viruses from supernatants of BSR-T7/5 cells infected at an MOI of 0.1. (C) Production of virus proteins in BSR-T7/5 cells infected at an MOI of 1. Cell extracts were harvested at days 1 and 2 after infection and submitted to Western blot analysis. Accumulation of viral proteins is comparable in cells infected with the SAD PΔInd and wt SAD L16 viruses, while protein accumulation is delayed in the SAD ΔPLP mutant. (D) Ability of SAD PΔInd viruses to inhibit IFN signaling. BSR-T7/5 cells were transfected with an ISRE luciferase reporter plasmid, infected at an MOI of 3, and stimulated at 24 h p.i. with 1,000 IU/ml of IFN-α A/D. Luciferase induction by IFN is observed only in mock- and SAD ΔPLP mutant-infected cells.
Indeed, viable recombinant RV carrying PΔInd1, PΔInd2, and PΔInd1/2 genes (SAD ΔInd1, SAD ΔInd2, and SAD ΔInd1/2 viruses, respectively) could be rescued from cDNA. Moreover, growth kinetics of the mutant viruses in BSR-T7/5 cells, which do not express IFN in response to RIG-I, were identical to those of the parental SAD L16 virus. After infection at an MOI of 0.1, infectious supernatant virus production reached approximately 108 FFU/ml at day 3 postinfection (p.i.) (Fig. 2B). In contrast, and as observed before, the SAD ΔPLP mutant, which expresses little full-length P from a 3′-terminal position (4), showed a marked delay in propagation and yielded 10- to 15-fold lower infectious titers. Similar growth of the SAD ΔInd1, SAD ΔInd2, and SAD L16 viruses was also indicated by Western blot experiments, revealing comparable high-level accumulation of viral N, P, M, and G proteins. In contrast, SAD ΔPLP protein accumulation was delayed (see Fig. 2C, day 1), probably by limiting concentrations of P.
SAD ΔInd viruses inhibit IFN-mediated JAK/STAT signaling.
To corroborate that the internal deletions in the P genes of the SAD ΔInd1 and SAD ΔInd2 viruses do not affect the ability of the viruses to prevent IFN-mediated induction of ISGs, dual-luciferase reporter assays were performed with BSR-T7/5 cells. Cells infected at an MOI of 3 were treated 24 h p.i. with 1,000 IU/ml IFN-α A/D, and ISRE-controlled FL activities were determined 24 h later. In mock-infected cells and in cells infected with the SAD ΔPLP mutant, IFN-α A/D treatment resulted in an approximately 120-fold induction of FL from the ISRE plasmid. In cells infected with the SAD L16, SAD ΔInd1, and SAD ΔInd2 viruses, however, IFN-mediated activation was almost completely abolished (Fig. 2D), revealing the full capacity of the ΔInd mutants in counteracting JAK/STAT signaling.
SAD ΔInd viruses grow in IFN-competent cells in spite of IRF3 activation and induction of IFN.
The recombinant viruses were then tested for their growth phenotype in IFN-competent HEp-2 cells, in which the SAD ΔPLP virus is known to be completely attenuated and not able to grow. In contrast to the SAD ΔPLP virus, however, both the SAD ΔInd1 and SAD ΔInd2 viruses amplified and produced infectious titers only 10-fold below those of wt SAD L16 after infection at an MOI of 0.1 (Fig. 3A).
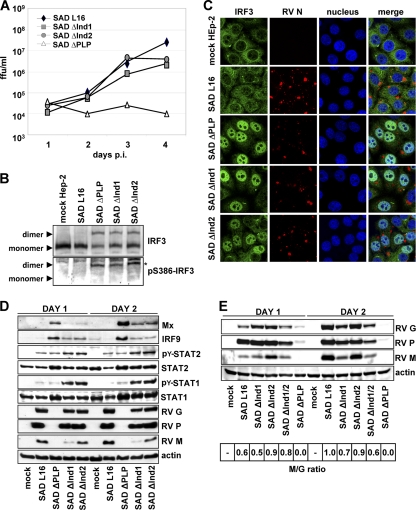
FIG. 3.
(A) Growth of SAD ΔInd mutant viruses on IFN-competent HEp-2 cells infected at an MOI of 0.05. While SAD ΔPLP virus is completely attenuated, the SAD ΔInd1 and SAD ΔInd2 mutants were able to amplify similar to SAD L16. (B) Activation of IRF3 in HEp-2 cells infected with the indicated viruses at an MOI of 1 for 24 h. Native PAGE and Western blot analysis revealed phosphorylated and dimerized IRF3 in the mutant viruses. (C) Nuclear localization of IRF3 in HEp-2 cells infected with SAD ΔInd viruses. Twenty-four hours after infection at an MOI of 1, HEp-2 cells were stained with antibodies specific for IRF3 and RV and with To-PRO-3 dye to delineate the nucleus. (D) Western blot analysis of HEp-2 cells infected with the indicated viruses at an MOI of 1 for 24 and 48 h. The indicated viral proteins, interferon-stimulated genes (MxA, IRF9), and the phosphorylation status of STATs were monitored using respective antibodies. For details, see Results. (E) Western blot analysis of HEp-2 cells infected with SAD PΔInd mutant viruses at an MOI of 1. The lysates were prepared 1 and 2 days after infection. Viral proteins were analyzed with respect to their expression levels using respective antibodies. The ratio of RV M relative to RV G (bottom; wt RV set to 1.0) was calculated using the Bio 1D software of the Fusion Molecular Imaging device.
To address the activation status of IRF3 in virus-infected cells, extracts from HEp-2 cells infected at an MOI of 1 were subjected to native polyacrylamide gel electrophoresis. While in wt SAD L16-infected cells only nonphosphorylated IRF3 monomers were detected, infection with the SAD ΔPLP, SAD ΔInd1, and SAD ΔInd2 viruses led to dimerization and S386 phosphorylation of IRF3 to a similar extent (Fig. 3B). This correlated with nuclear translocation of IRF3, as illustrated by fluorescence microscopy (Fig. 3C). While in mock- and wt SAD L16-infected cells IRF3 was predominantly located in the cytoplasm, the bulk of IRF3 showed a nuclear localization in cells infected with the SAD ΔPLP mutant and the two SAD ΔInd mutants.
As predicted, infection of cells with the SAD ΔPLP virus, which is deficient in interfering with both IFN induction and IFN signaling, led to IFN production as indicated by strong induction of the ISGs MxA and IRF9 and of detectable induction of STAT1 and STAT2 (Fig. 3D). ISG induction was also observed for SAD ΔInd1 and SAD ΔInd2 mutant-infected cells, although at substantially lower levels. Most importantly, in SAD ΔInd1 and ΔInd2 mutant-infected cells, a prominent accumulation of tyrosine phosphorylated STAT1 (pY-STAT1) and a less pronounced accumulation of pY-STAT2 (Fig. 3B) were observed. Such accumulation is typical of SAD L16-infected cells treated with exogenous IFN and is caused by reduced recycling of activated STATs owing to P-mediated retention in the cytoplasm (5). Accumulation of pY-STAT in SAD ΔInd mutant-infected cells therefore indicates not only expression of endogenous IFN but also the capability of the PΔInd1 and PΔInd2 mutants to interfere with STAT nuclear import. In SAD L16-infected cells, the low levels of pY-STATs indicate the lack of substantial IFN induction.
Western blot analysis of the infected HEp-2 cell lysates further confirmed a minor attenuation of viral protein accumulation in the SAD ΔInd1 and SAD ΔInd2 viruses compared to that of SAD L16 and a lack of SAD ΔPLP protein accumulation, as was expected from the growth curves. Notably, while the ratios of G and P appeared similar, a reduced level of the matrix protein (M) was suggested in SAD ΔInd1 virus-infected cells (Fig. 3D). To follow this up, Western blot analysis was performed using a digital readout for quantitative comparison of chemiluminescence signals (Fig. 3E). Additionally, we included the double-deletion SAD ΔInd1/2 mutant, which grew like the single-deletion mutants in HEp-2 cells and which activated IRF3 similarly (not shown). Indeed, after 2 days, comparable ratios of M and G of 1.0 and 0.9 were determined for wt RV and the SAD ΔInd2 mutant, respectively (Fig. 3E). Both the SAD ΔInd1 and SAD ΔInd1/2 mutants showed reduced levels of M (ratios of 0.7 and 0.6, respectively). This indicated a correlation of the deletion of aa 176 to 181 and the levels of M.
Taken together, the data indicate that in spite of unhindered IRF3 activation and IFN induction, SAD ΔInd mutants are able to grow in HEp-2 cells because of their ability to keep expression of antiviral ISGs below critical levels.
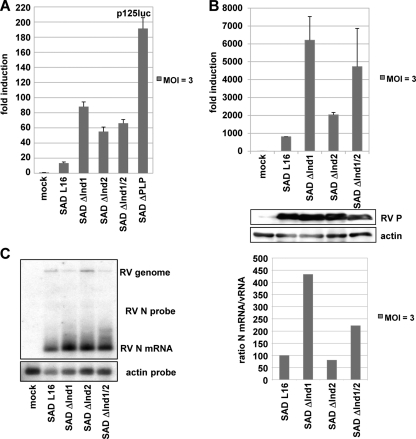
To quantify induction of IFN-β in cells infected with the recombinant viruses, HEK 293T cells transfected with p125-luc and the control plasmid pCMV-RL were infected 6 h later at an MOI of 3. Luciferase activities as determined at 24 h and 48 h p.i. indicated modest activation of the IFN-β promoter at 1 day p.i. and only a minor increase over time in wt SAD L16-infected cells (Fig. 4A). In contrast, infection with the mutant viruses resulted in a stronger activation at day 1 p.i. (4- to 7-fold of wt virus levels for the SAD ΔInd1 viruses and 15-fold for the SAD ΔPLP mutant). While increased IFN-β promoter activation by the SAD ΔInd1 and ΔInd2 mutants was predicted, the observation of differential induction, in spite of comparable antagonistic activity of the phosphoproteins and similar growth rates, was appealing.
FIG. 4.
(A and B) IFN induction in 293T cells after infection with mutant RV (MOI of 3). (A) Dual-luciferase assay from cells infected for 24 h. (B) qRT-PCR for IFN-β mRNA at 24 h p.i. Infection and P expression were controlled by Western blot analysis. (C) Transcription analysis of recombinant viruses. BSR-T7/5 cells were infected with the indicated viruses at an MOI of 3. RNA was harvested 24 h postinfection and analyzed by Northern blot hybridization with an N probe. The ratio between N gene mRNA and the genome RNA was calculated using ImageQuant and presented as the percentage of the wt SAD L16 ratio.
To confirm differential activation of IFN-β mRNA transcription, quantitative RT-PCR experiments were performed. RNA extracted from HEK 293T cells infected for 24 h with the SAD L16, SAD ΔInd1, SAD ΔInd2, and SAD ΔInd1/2 viruses was evaluated. Low levels of IFN-β mRNA were detectable in cells infected with wt SAD L16, and a moderate 2-fold increase was observed with the SAD ΔInd2 mutant (2- to 3-fold compared to that of the wt). However, infection with the SAD ΔInd1 mutant stimulated transcription almost 8-fold better than SAD L16 and 3-fold better than the SAD ΔInd2 mutant, although comparable levels of P were present, indicating similar infection levels (Fig. 4B). Infection with the SAD PΔInd1/2 double mutant resulted in an intermediate, approximately 4-fold induction over that of the wt virus.
As the phosphoproteins of the SAD ΔInd1 and -2 mutants showed comparable activities of counteracting IRF3 activation (see Fig. 1), it was of interest to reveal the basis for higher IFN induction by the SAD ΔInd1 mutant. Northern blot analysis of total RNA from virus-infected cells confirmed transcription of correctly sized N mRNA and viral RNA (vRNA) in cells infected with all viruses and indicated the absence of defective interfering particles, which are known to contribute to RIG-I activation. However, particularly the SAD ΔInd1 mutant appeared to produce more N mRNA than the SAD L16 virus in the presence of less vRNA (Fig. 4C). Measurement of the ratio between N mRNA and vRNA by ImageQuant indicated an approximately 4-fold-higher transcription rate (N/vRNA) of the SAD ΔInd1 mutant than that of SAD L16, while the SAD ΔInd2 mutant was intermediate and more similar to SAD L16, as was observed for IFN-β mRNA induction (Fig. 4C). A higher transcription rate for the SAD ΔInd1 mutant was also suggested by Northern blotting with RNA from infected 293T cells (not shown).
Attenuation of IFN-inducing viruses in vivo.
The above in vitro experiments demonstrated a specific loss of function of the SAD ΔInd1 and SAD ΔInd2 viruses in preventing IRF3 phosphorylation and activation in virus-infected cells, while the functions in preventing JAK/STAT signaling and in supporting virus replication were not affected. To specifically assess the contribution of IRF3 activation to host anti-RV defense, mouse experiments were performed. Five- to 8-week-old wt 129 mice and knockout mice on the 129 background lacking a functional IFN-α receptor (IFNAR−/−) or IFN-γ receptor (IFNGR−/−) (31) were intracerebrally injected with 105 FFU of the SAD ΔPLP, SAD ΔInd1, and SAD ΔInd2 mutants and the wt SAD L16 virus.
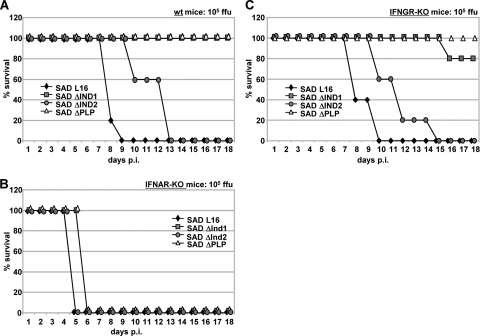
Notably, the two SAD ΔInd viruses differed dramatically in their pathogenicities. All IFNAR−/− mice succumbed to infection by any of the viruses, although a 1-day delay was already observed for the SAD ΔInd1 and SAD ΔPLP mutants (Fig. 5B). In wt mice, SAD L16 caused death within 9 days, while infection with the SAD ΔInd2 mutant required 13 days for killing of all mice (Fig. 5A). In contrast, the SAD ΔInd1 mutant was completely apathogenic such that all mice survived without developing symptoms. Similarly, in IFNGR−/− mice, the SAD ΔInd1 mutant was severely attenuated and killed one out of five injected mice, while all survived injection with the SAD ΔPLP mutant (Fig. 5C). In terms of survival rate, the degree of attenuation of the SAD ΔInd1 mutant is therefore similar to that of the SAD ΔPLP mutant, in spite of the marked growth defects in vitro of the latter and in spite of a growth rate similar to that of the SAD ΔInd2 mutant, which appears to induce less IFN than the SAD ΔInd1 mutant. This indicates that the level of IRF activation and IFN induction is the major attenuating factor for these viruses and that the SAD ΔInd1 and SAD ΔInd2 mutants are above and below a critical threshold, respectively.
FIG. 5.
Attenuation in vivo of SAD ΔInd viruses. wt mice (A), IFNAR−/− mice (B), and IFNGR−/− mice (C) were intracranially infected with 105 FFU of the indicated viruses, and survival was monitored. Whereas IFNAR−/− mice rapidly succumbed to the infection with either RV, the SAD PΔInd2 mutant caused a delayed disease course, and the SAD ΔInd1 mutant was completely apathogenic in wt mice.
DISCUSSION
Successful restraining of the host IFN system by a virus appears to require multiple means, including the spoiling of both IFN induction and IFN effector mechanisms (for a comprehensive review, see reference 35). As we have shown previously, the multifunctional RV P combines both arms. It counteracts IRF3 and IRF7 activation by TBK-1 in virus-infected cells and prevents import of activated STAT1 and STAT2 into the nucleus (4, 5). In addition, P is an essential virus protein critically required during numerous steps throughout the viral life cycle, including viral RNA synthesis. To dissect individual functions of P, in order to create proteins and recombinant RV with single, defined defects in IFN escape and to assess the contribution of either of these IFN antagonistic functions to the course of RV infection in vivo, in vitro mutagenesis was performed. We here have identified residues whose deletion impaired the ability of P to interfere with IRF3 phosphorylation, while leaving other functions of P intact.
RV P is a modular protein which interacts with itself to form oligomers, with soluble nucleoprotein (N0) during N-RNA encapsidation, and with both N-RNA and L during RNA synthesis. The respective binding sites have been mapped to the 19 N-terminal aa (L), an N-terminal stretch (N0), a central region (oligomerization), and an extended C-terminal moiety of P (N-RNA) (7, 8, 14, 15, 21, 28, 29, 32, 49). The C-terminal domain of P was also reported to bind the promyelocytic leukemia protein (2), and the 10 C-terminal amino acids are required for STAT binding (5, 45). A short conserved motif in the N-terminal part of P (aa 139 to 151) mediates strong binding to dynein light chain 1 (DLC1/LC8) (20, 36), which might be involved in modulating viral primary transcription in neuronal cells (43) but not in axonal transport of RV (24). Here, mutagenesis of RV P identified the residues 176 to 186 as important for interference with IRF3 activation in RV-infected cells (see Fig. 1). This domain is located in a part of the protein predicted to be structurally disordered (15) and directly adjacent to a region (aa 61 to 175) previously reported to be nonessential for basic transcription of RV minigenomes (21). As observed here, this nonessential region can be extended to aa 186.
It must be noted that while the deletion of aa 176 to 186 did strongly affect the interference of P with IRF activation, it is uncertain whether these residues are directly involved in binding targets of the signaling complex. Expression of P fragments containing this domain did not detectably interfere with IRF3 (not shown). Moreover, the deletion of either aa 176 to 181 (ΔInd1 mutant) or aa 181 to 186 (ΔInd2 mutant) resulted in similar strong defects with respect to IRF3 inhibition, while the combined deletion of the entire domain in the PΔInd1/2 mutant appeared to have a rather less pronounced defect. It can be speculated that the deletions rather affect specific protein folding required for interference with IRF3 activation.
Notably, the deletions did not seriously affect the functions of P in transcription and replication, as indicated by minigenome experiments and the recovery of viable RV. In BSR-T7/5 cells, which have an endogenous defect in IRF3 activation such that not only induction of IFN but also possible effects of activated IRF3 on other target genes are circumvented (1, 16), the SAD ΔInd1, SAD ΔInd2, and SAD ΔInd1/2 mutants were almost indistinguishable in terms of growth kinetics and virus production and were highly similar to the parental SAD L16. The viruses were equally capable of interfering with IFN-mediated ISG induction, as indicated in reporter gene assays involving treatment of infected cells with IFN (Fig. 2). Robust and similar inhibition of the JAK/STAT pathway was also illustrated in virus-infected HEp-2 cells where infection with the SAD ΔInd1 and SAD ΔInd2 mutants led to accumulation of P-Y-STAT as a result of STAT retention in the cytoplasm and impaired STAT recycling (5). Accordingly, the SAD ΔInd1 and ΔInd2 mutants are viruses defective only with respect to inhibition of IRF3 activation and IFN induction but otherwise equipped with the full capacity of IFN resistance and virus propagation.
Infection experiments with the IFN-competent HEp-2 cell cultures revealed a clear growth advantage of the SAD ΔInd1 and SAD ΔInd2 mutants over the SAD ΔPLP mutant, which is not able to counteract IFN signaling and which has severe growth defects because of limiting P amounts. While the latter was not able to amplify in HEp-2 cells, the former reached final titers only 10-fold below those of SAD L16. This must be attributed at least partially to the capacity to prevent IFN signaling, although the SAD ΔPLP mutant may in addition suffer from induction of higher levels of IFN in the initially infected cells and a more rapid establishment of an antiviral state in noninfected neighboring cells. Most intriguing, however, was the observation of similar growth of the ΔInd mutants and SAD L16, indicating that cell-autonomous activation of HEp-2 cells does not contribute markedly to the establishment of an antiviral state.
Although PΔInd1 and PΔInd2 proteins were equal in their capacity to support viral growth and inhibition of JAK/STAT signaling, the recombinant SAD ΔInd1 and SAD Ind1/2 viruses stimulated higher IFN production than the SAD ΔInd2 mutant. This suggested stronger stimulation of the RIG-I pathway, which is activated by RV triphosphate RNAs (18). As indicated by Northern blot experiments, the SAD ΔInd1 and SAD ΔInd1/2 mutants produced more mRNAs per genome. The reason for this is unclear, but an apparent correlation of the lack of aa 176 to 181 shared by these two viruses and reduced levels of the viral matrix protein (Fig. 3E), which has previously been shown to regulate the balance of transcription and replication (13), may provide the first hints for explanation of the phenotype. Whether mutations in the RV P might directly affect RV M levels or whether this is an effect caused by alerted cells remains to be elucidated. RV mRNAs represent the overwhelming amount of total viral RNA in infected cells, and the further increase of mRNAs in SAD ΔInd1 mutant-infected cells is obviously correlated with stronger IFN induction. Although a recent publication has indicated a major role of the genome RNA of Sendai virus in activation of RIG-I (37), the present findings point toward a major role of subgenomic RNAs.
The ability of P to sufficiently thwart the host IFN system is critical for survival of RV in the brain of infected animals. This was first demonstrated by recent experiments involving recombinant RV expressing limited amounts of P, owing to picornavirus IRES-dependent translation of P (26). Like the SAD ΔPLP mutant, these viruses are defective in counteracting both IFN induction and IFN signaling. As illustrated, for example, by the lack of protection even against the SAD ΔPLP mutant in IFNAR−/− mice (Fig. 5B), an essential determinant of successful virus control in vivo is the IFN produced, which provides a paracrine alarm signal to uninfected neighboring cells.
The importance of functional STAT inhibition for RV virulence was recently supported by the finding that attenuation of an RV vaccine strain, NI-CE, is due partially to a failure of its phosphoprotein to retain STAT in the cytoplasm (19). There is evidence, however, that IFN-independent activation mechanisms are in addition needed for full antiviral activity against rhabdoviruses. IPS-1-KO mice, in which IRF3 cannot be activated by infection, were highly susceptible to vesicular stomatitis virus (VSV) (25, 41) despite normal systemic levels of type I IFNs, probably induced by pDCs through TLR7/9-dependent mechanisms. Moreover, virulence of recombinant RV expressing IFN-β was only modestly attenuated (12), and attenuated RV strains activate innate immunity in the brain more strongly than virulent viruses (22, 47). Thus, an important contribution of cell-autonomous activation of IRF3 to the control of virus infection, probably by activation of antiviral genes and/or by labeling infected cells for killing by NK cells or by apoptosis, is suggested (1, 16, 40).
To investigate the importance of blocking IRF3 activation and IFN induction on the outcome of RV infection, we performed mouse experiments with the SAD ΔInd1 and SAD ΔInd2 mutants, whose P had an equal loss of activity in blocking IRF3 activation and the same competence in blocking the JAK/STAT pathway. As predicted from the complete attenuation of the IFN-inducing and IFN-sensitive SAD ΔPLP mutant in IFN-competent cell cultures, injection of 105 FFU of the SAD ΔPLP mutant into the brain of wt mice did not cause symptoms, disease, or death, while the same dose of wt SAD L16 led to death of the mice within a few days. The SAD ΔInd2 mutant caused the typical rabies symptoms, both in wt and IFNGR−/− mice, but in both cases the course of disease was delayed by several days compared to that of wt RV, illustrating a protective role of IRF3 activation and IFN induction. Moreover, the SAD ΔInd1 mutant was even completely attenuated and did not cause symptoms in wt mice and was severely attenuated in IFNGR−/− mice.
Accordingly, in the case of injection of the SAD ΔInd1 mutant into the brain of mice, activation of IRF3 and induction of IFN is sufficient to make the virus visible to the host defense system and, in spite of fully competent JAK/STAT signal antagonists, to trigger a fully protective response. Activation of host cells by the SAD ΔInd2 mutant seems to be below a critical threshold required for protective innate immunity under these stringent conditions, although an improved host response, as illustrated by prolonged survival, is already obvious. In the case of the SAD ΔInd1 mutant, the critical threshold is exceeded through increased production of RNA transcripts activating RIG-I, such as 5′ pppRNA leader RNA. This may indicate that in addition to conserving the full capacity of P in counteracting the IFN system, a limited production of immune-stimulating RNAs may be critical for RV in order not to exceed the antagonistic capacity of P to prevent JAK/STAT signaling. Indeed, increased IFN induction was observed for RV with mutations not only in the P gene, as shown here, but also in the N gene, probably by affecting proper N-RNA packaging (27), and in the M gene (our unpublished data), which is involved in adjusting transcription rates of RV (13). IRF-activating and IFN-inducing RVs which are largely resistant to the antiviral effects of IFN, like the SAD ΔInd1 and SAD ΔInd2 mutants described here, may be interesting for the development of self-adjuvanted live vaccines.
Acknowledgments
We thank Nadin Hagendorf and Uli Wulle for perfect technical assistance and A. Ghanem for help with the minigenome experiments. MxA antibody was kindly provided by O. Haller and G. Kochs, P160-5 by S. Finke, and cDNAs by S. Akira, T. Fujita, and J. Hiscott.
This work was supported by the Deutsche Forschungsgemeinschaft through GraKo 1202, “Oligonucleotides in cell biology and therapy,” and SFB 455, “Viral functions and immune modulation.”
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Andersen, J., S. VanScoy, T. F. Cheng, D. Gomez, and N. C. Reich. 2008. IRF-3-dependent and augmented target genes during viral infection. Genes Immun. 9:168-175. [DOI] [PubMed] [Google Scholar]
- 2.Blondel, D., et al. 2002. Rabies virus P and small P products interact directly with PML and reorganize PML nuclear bodies. Oncogene 21:7957-7970. [DOI] [PubMed] [Google Scholar]
- 3.Brzózka, K., and K. K. Conzelmann. 2009. IFN escape of rhabdoviruses, p. 211-227. In A. Brasier and A. García-Sastre (ed.), Cellular signaling and innate immune responses to RNA virus infections. ASM Press, Washington, DC.
- 4.Brzózka, K., S. Finke, and K. K. Conzelmann. 2005. Identification of the rabies virus alpha/beta interferon antagonist: phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 79:7673-7681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzózka, K., S. Finke, and K. K. Conzelmann. 2006. Inhibition of interferon signaling by rabies virus phosphoprotein P: activation-dependent binding of STAT1 and STAT2. J. Virol. 80:2675-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Castel, G., et al. 2009. Peptides that mimic the amino-terminal end of the rabies virus phosphoprotein have antiviral activity. J. Virol. 83:10808-10820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chenik, M., K. Chebli, Y. Gaudin, and D. Blondel. 1994. In vivo interaction of rabies virus phosphoprotein (P) and nucleoprotein (N): existence of two N-binding sites on P protein. J. Gen. Virol. 75:2889-2896. [DOI] [PubMed] [Google Scholar]
- 8.Chenik, M., M. Schnell, K. K. Conzelmann, and D. Blondel. 1998. Mapping the interacting domains between the rabies virus polymerase and phosphoprotein. J. Virol. 72:1925-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Conzelmann, K. K., J. H. Cox, and H. J. Thiel. 1991. An L (polymerase)-deficient rabies virus defective interfering particle RNA is replicated and transcribed by heterologous helper virus L proteins. Virology 184:655-663. [DOI] [PubMed] [Google Scholar]
- 10.Conzelmann, K. K., and M. Schnell. 1994. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J. Virol. 68:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, S., et al. 2008. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol. Cell 29:169-179. [DOI] [PubMed] [Google Scholar]
- 12.Faul, E. J., C. N. Wanjalla, J. P. McGettigan, and M. J. Schnell. 2008. Interferon-beta expressed by a rabies virus-based HIV-1 vaccine vector serves as a molecular adjuvant and decreases pathogenicity. Virology 382:226-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finke, S., R. Mueller-Waldeck, and K. K. Conzelmann. 2003. Rabies virus matrix protein regulates the balance of virus transcription and replication. J. Gen. Virol. 84:1613-1621. [DOI] [PubMed] [Google Scholar]
- 14.Fu, Z. F., Y. Zheng, W. H. Wunner, H. Koprowski, and B. Dietzschold. 1994. Both the N- and the C-terminal domains of the nominal phosphoprotein of rabies virus are involved in binding to the nucleoprotein. Virology 200:590-597. [DOI] [PubMed] [Google Scholar]
- 15.Gerard, F. C., et al. 2009. Modular organization of rabies virus phosphoprotein. J. Mol. Biol. 388:978-996. [DOI] [PubMed] [Google Scholar]
- 16.Grandvaux, N., et al. 2002. Transcriptional profiling of interferon regulatory factor 3 target genes: direct involvement in the regulation of interferon-stimulated genes. J. Virol. 76:5532-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller, O., G. Kochs, and F. Weber. 2007. Interferon, Mx, and viral countermeasures. Cytokine Growth Factor Rev. 18:425-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hornung, V., et al. 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science 314:994-997. [DOI] [PubMed] [Google Scholar]
- 19.Ito, N., et al. 2010. The role of interferon-antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 84:6699-6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob, Y., H. Badrane, P. E. Ceccaldi, and N. Tordo. 2000. Cytoplasmic dynein LC8 interacts with lyssavirus phosphoprotein. J. Virol. 74:10217-10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacob, Y., E. Real, and N. Tordo. 2001. Functional interaction map of lyssavirus phosphoprotein: identification of the minimal transcription domains. J. Virol. 75:9613-9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, N., et al. 2006. Lyssavirus infection activates interferon gene expression in the brain. J. Gen. Virol. 87:2663-2667. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., and S. Akira. 2008. Toll-like receptor and RIG-I-like receptor signaling. Ann. N. Y. Acad. Sci. 1143:1-20. [DOI] [PubMed] [Google Scholar]
- 24.Klingen, Y., K. K. Conzelmann, and S. Finke. 2008. Double-labeled rabies virus: live tracking of enveloped virus transport. J. Virol. 82:237-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar, H., et al. 2006. Essential role of IPS-1 in innate immune responses against RNA viruses. J. Exp. Med. 203:1795-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marschalek, A., et al. 2009. Attenuation of rabies virus replication and virulence by picornavirus internal ribosome entry site elements. J. Virol. 83:1911-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Masatani, T., et al. 2010. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J. Virol. 84:4002-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mavrakis, M., A. A. McCarthy, S. Roche, D. Blondel, and R. W. Ruigrok. 2004. Structure and function of the C-terminal domain of the polymerase cofactor of rabies virus. J. Mol. Biol. 343:819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mavrakis, M., et al. 2006. Rabies virus chaperone: identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 349:422-429. [DOI] [PubMed] [Google Scholar]
- 30.Moseley, G. W., et al. 2009. Dual modes of rabies P-protein association with microtubules: a novel strategy to suppress the antiviral response. J. Cell Sci. 122:3652-3662. [DOI] [PubMed] [Google Scholar]
- 31.Muller, U., et al. 1994. Functional role of type I and type II interferons in antiviral defense. Science 264:1918-1921. [DOI] [PubMed] [Google Scholar]
- 32.Olivier, D., R. Assenberg, J. M. Grimes, and H. Bourhy. 2010. The structure of the nucleoprotein binding domain of lyssavirus phosphoprotein reveals a structural relationship between the N-RNA binding domains of Rhabdoviridae and Paramyxoviridae. RNA Biol. 7:322-327. [DOI] [PubMed] [Google Scholar]
- 33.Pfaller, C. K., and K. K. Conzelmann. 2008. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365-12373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Platanias, L. C. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat. Rev. Immunol. 5:375-386. [DOI] [PubMed] [Google Scholar]
- 35.Randall, R. E., and S. Goodbourn. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1-47. [DOI] [PubMed] [Google Scholar]
- 36.Raux, H., A. Flamand, and D. Blondel. 2000. Interaction of the rabies virus P protein with the LC8 dynein light chain. J. Virol. 74:10212-10216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rehwinkel, J., et al. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell 140:397-408. [DOI] [PubMed] [Google Scholar]
- 38.Rieder, M., and K. K. Conzelmann. 2009. Rhabdovirus evasion of the interferon system. J. Interferon Cytokine Res. 29:499-510. [DOI] [PubMed] [Google Scholar]
- 39.Schnell, M. J., T. Mebatsion, and K. K. Conzelmann. 1994. Infectious rabies viruses from cloned cDNA. EMBO J. 13:4195-4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stetson, D. B., and R. Medzhitov. 2006. Antiviral defense: interferons and beyond. J. Exp. Med. 203:1837-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun, Q., et al. 2006. The specific and essential role of MAVS in antiviral innate immune responses. Immunity 24:633-642. [DOI] [PubMed] [Google Scholar]
- 42.Takeuchi, O., and S. Akira. 2008. MDA5/RIG-I and virus recognition. Curr. Opin. Immunol. 20:17-22. [DOI] [PubMed] [Google Scholar]
- 43.Tan, G. S., M. A. Preuss, J. C. Williams, and M. J. Schnell. 2007. The dynein light chain 8 binding motif of rabies virus phosphoprotein promotes efficient viral transcription. Proc. Natl. Acad. Sci. U. S. A. 104:7229-7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Boxel-Dezaire, A. H., M. R. Rani, and G. R. Stark. 2006. Complex modulation of cell type-specific signaling in response to type I interferons. Immunity 25:361-372. [DOI] [PubMed] [Google Scholar]
- 45.Vidy, A., M. Chelbi-Alix, and D. Blondel. 2005. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 79:14411-14420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vidy, A., J. El Bougrini, M. K. Chelbi-Alix, and D. Blondel. 2007. The nucleocytoplasmic rabies virus P protein counteracts interferon signaling by inhibiting both nuclear accumulation and DNA binding of STAT1. J. Virol. 81:4255-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang, Z. W., et al. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J. Virol. 79:12554-12565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whelan, S. P., J. N. Barr, and G. W. Wertz. 2004. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:61-119. [DOI] [PubMed] [Google Scholar]
- 49.Yang, J., et al. 1998. The specificity of rabies virus RNA encapsidation by nucleoprotein. Virology 242:107-117. [DOI] [PubMed] [Google Scholar]
- 50.Yoneyama, M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730-737. [DOI] [PubMed] [Google Scholar]