Abstract
The PB1-F2 protein of influenza A virus can contribute to viral pathogenesis of influenza virus strains. Of note, an N66S amino acid mutation in PB1-F2 has been shown to increase the pathogenesis associated with H5N1 Hong Kong/1997 and H1N1 Brevig Mission/1918 influenza viruses. To identify the mechanism of enhanced immunopathology, we evaluated the host response to two isogenic viruses that differ by a single amino acid at position 66 of the PB1-F2 protein. Various components of the adaptive immune response were ruled out as factors contributing to pathogenesis through knockout mouse studies. Transcriptional profiling of lungs from PB1-F2 66S-infected mice revealed an early delay in innate immune responses. In particular, enhanced activation of type I interferon (IFN) pathway genes, including IFN-β, RIG-I, and numerous interferon-inducible genes, was not observed until day 3 postinfection. The N66S mutant virus caused increased cellularity in the lungs, as a result of monocyte and neutrophil infiltration. Furthermore, numerous cytokines and chemokines related to monocyte and neutrophil migration and maturation were upregulated. The cellular infiltration and increased cytokine expression corresponded to increased PB1-F2 66S titer. These data suggest that PB1-F2 N66S may contribute to the delay of innate immune responses, allowing for unchecked viral growth and ultimately severe immunopathology observed in the lungs.
Highly pathogenic influenza A viruses have been widely studied due to their pandemic potential. Most pandemics have resulted in significant mortality and severe disease among the general population, the most memorable being the Spanish influenza pandemic of 1918 and 1919. To date, highly pathogenic avian influenza (HPAI) H5N1 virus poses a global health concern, with new cases continuing to emerge in the human population. The initial outbreak of avian influenza H5N1 virus in humans occurred in Hong Kong in 1997. H5N1 infection of aquatic birds and poultry has since become endemic in Southeast Asia, where the virus continues to evolve and potentially threaten a widespread pandemic (20). Isolates from the Hong Kong outbreak resulting in a fatal outcome, including A/Hong Kong/156/1997 H5N1, were shown to be lethal in mice (23). The mechanism of virulence for HPAI H5N1 viruses is based on their ability to cause immunopathogenesis. Host cytokine responses have been shown to exacerbate severe respiratory disease. During human infections with H5N1, patients are found to have increased cytokine levels and leukopenia (8). A study investigating the contribution of individual cytokines and chemokines during H5N1 infection demonstrated that interleukin-1 receptor (IL-1R) and tumor necrosis factor receptor (TNFR) can influence viral clearance and disease progression in a mouse model (30). A more recent study by Perrone et al. showed that mouse infection with H5N1 resulted in excessive macrophage and neutrophil infiltration in the lungs, as well as enhanced upregulation of cytokines, including interleukin-6 (IL-6) and gamma interferon (IFN-γ) (28).
Characterization of single-gene reassortants has led to a greater understanding of viral factors important for H5N1 virulence, particularly for the HA and PB2 proteins (3, 10, 15, 31). More recently, the PB1-F2 protein has been shown to increase pathogenesis of the 1918 pandemic influenza virus and a recombinant virus bearing the PB1 segment of the A/Hong Kong/156/1997 (H5N1) virus. Enhanced virulence resulted from a single polymorphism in the PB1-F2 gene, which is encoded by the +1 open reading frame in the PB1 gene. The presence of a serine at position 66 caused severe weight loss, increased viral titer, and elevated levels of IFN-γ and tumor necrosis factor alpha (TNF-α) in mouse lungs (7). The PB1-F2 protein may not be a critical virulence determinant for all pandemic viruses, as a recent study demonstrated that adding full-length PB1-F2 to the 2009 H1N1 pandemic virus did not significantly alter its pathogenicity (14).
Functional genomics has been used to evaluate host responses to 1918 pandemic influenza virus and H5N1 virus infection and to better elucidate the mechanisms of increased pathogenesis in mouse (9, 17) and macaque (1, 18) infection models. Recently, it was shown that early dysregulation of host innate immune pathways in 1918 pandemic influenza virus-infected macaques critically influenced later stages of immunopathology and contributed to the fatal outcome (6). Cilloniz and coworkers demonstrated that 1918 pandemic influenza and H5N1 A/Vietnam/1203/2004 (VN1203) viruses differentially regulated host pathways. In particular, 1918 pandemic influenza virus infection upregulated expression of inflammation- and cell death-related genes, including components of the inflammasome, while H5N1 VN1203 virus downregulated expression of these genes (6).
Induction of type I interferon is important for mounting antiviral defenses and modulating inflammatory responses. Alpha/beta interferon receptor (IFN-α/βR)-deficient mice had increased pathogenicity when infected with H5N1 A/Hong Kong/483/97 (HK/483) and A/Hong Kong/486/97 (HK/486) viruses (29). The interferon response was uncontrolled in IFN-αR1−/− mice infected with H5N1 VN1203 virus, which suggests that type I interferon receptor signaling was dispensable for H5N1 infection. This also suggests that the H5N1 virus induced expression of interferon genes through a distinct mechanism (5). In a cell culture model, IFN-α/βR−/− cells infected with 1918 pandemic influenza and H5N1 VN1203 viruses resulted in increased viral replication and decreased expression of antiviral response genes, including Stat1, PKR, and IFN-β. Yet, the absence of the type I receptor did not inhibit induction of inflammatory or apoptosis pathway-related gene expression (12). These studies support the notion that host inflammatory and interferon pathways play a key role in influenza virus pathogenesis and are differentially regulated by the 1918 pandemic influenza and H5N1 viruses.
Multiple functions have been proposed for the PB1-F2 protein. PB1-F2 has been shown to cause apoptosis through its interaction with the mitochondrial ANT3 and VDAC1 proteins (4, 11, 32, 34). Others have reported that PB1-F2 affects polymerase activity (24, 27). PB1-F2 is known to interact with PB1 within the nucleus (24, 26), and in the absence of PB1-F2, polymerase activity seems to be impaired (27). Studies by McAuley et al. further analyzed the effect of PB1-F2 on polymerase activity (27). This study demonstrated that abrogation of PB1-F2 expression caused a decrease in polymerase activity in a virus strain- and cell line-dependent manner (27). Of note, the authors observed no differences in viral load in mice infected with viruses defective in PB1-F2 expression (27). Another study by McAuley et al. demonstrated that PB1-F2 protein expression increases susceptibility to secondary Streptococcus pneumoniae infection by likely increasing the proinflammatory response in mice coinfected with mouse-adapted A/Puerto Rico/8/34 (H1N1) (PR8) virus (26). Further investigation is needed to elucidate the mechanism of PB1-F2 function, particularly in its importance to pathogenesis.
In the current study, we used two isogenic viruses, differing only at amino acid position 66 of the PB1-F2 protein, to evaluate the increased immunopathogenesis resulting from the N66S mutation (7). Through the use of recombinant A/WSN/33 viruses containing PB1 of Hong Kong/156/1997 (H5N1) virus, termed WH and WH N66S as previously reported (7), we show that mice infected with the WH N66S virus exhibit delayed activation of interferon-stimulated gene (ISG) expression early in infection. Together with increased infiltration of monocytes and neutrophils and elevated cytokine and chemokine levels in the lungs, we speculate that PB1-F2 N66S association with postponed innate immune responses dictates later stages of immunopathology. Our work suggests a new function for PB1-F2 where the N66S mutation allows PB1-F2 to modulate type I interferon responses, leading to increased disease severity.
MATERIALS AND METHODS
Viruses.
The viruses used in all experiments described were generated for work reported in a previous publication (7). Briefly, the viruses were rescued using a 12-plasmid system; 7 of the viral RNA segments were derived from the A/WSN/33 (H1N1) virus. The PB1 segment is from the A/Hong Kong/156/1997 (H5N1) virus. The WH N66S virus contains a single nucleotide mutation that results in an amino acid change from asparagine to serine at position 66 in the PB1-F2 coding region. The amino acid sequence of the PB1 protein is unchanged by this mutation.
Wild-type and knockout mouse infections.
Six-to 8-week-old female mice were obtained from the Jackson Laboratory and kept in a barrier facility. C57BL/6J mice, strain 000664, were used as wild-type (wt) mice. Knockout mice used are as follows: CD4−/− mice, strain 002663, B6.129S2-Cd4tm1Mak/J; CD8−/− mice, strain 002665, B6.129S2-Cd8atm1Mak/J; IFN-γ−/− mice, strain 002287, B6.129S7-Ifngtm1Ts/J; and Rag1−/− mice, strain 002096, B6.129S7-Rag1tm1Mom/J. Mice were inoculated with 30 μl of virus in phosphate-buffered saline (PBS) containing penicillin-streptomycin and bovine serum albumin (PBS-BA-PS). A total of 104 PFU of virus was given in all inoculations. Control mice were given PBS-BA-PS. Ten mice per group were used for monitoring weight loss. Mice were weighed every day, and percentage of initial weight was calculated. Any mouse that lost 25% of initial body weight was euthanized using CO2. To determine viral replication in the lungs, lungs were collected from 3 mice per group on days 1, 2, 3, 4, and 5 after infection. The lungs were homogenized in PBS using a Dounce homogenizer and processed for determination of virus titer. Virus titers in the supernatant of lung homogenates were determined by plaque assay with MDCK cells.
Lung pathology sections.
Whole lungs were harvested from mice on day 5 postinoculation and washed in PBS. The whole lung was then fixed in formaldehyde. The lungs were then embedded in paraffin, cut into 6-μm sections, and stained with hematoxylin and eosin using standard protocols (21). Pictures were taken on a Zeiss Axioplan 2 microscope using the 10× objective.
Flow cytometry.
Cells were collected from infected lungs by using a collagenase and DNase digestion protocol. Briefly, the lung was removed and washed in PBS for 30 s. The lungs were then placed in Miltenyi C tubes containing 5 ml of RPMI medium with 0.7 Wünsch units/ml of Liberase Blendzyme 3 (Roche 11814184001) and 10 U/ml DNase (Roche 04716728001). The lungs were homogenized using protocol E on the Miltenyi gentleMACS dissociator. The samples were incubated for 30 min at 37°C and homogenized again after incubation. The homogenized lung was passed through a 70-μm cell strainer, and red blood cells were removed using a red blood cell lysis buffer. Cells were then prepared for staining by being washed in PBS containing 0.2% fetal bovine serum and blocked using mouse Fc Block (BD 553141). Cell surface staining was then done using the following antibodies from eBioscience: CD11c-phycoerythrin (PE) (1:200), CD11b-fluorescein isothiocyanate (FITC) (1:100), CD11b-allophycocyanin (APC) (1:100), Ly6G-FITC (1:50), and major histocompatibility complex class II (MHC-II)-APC (1:100). Appropriate isotype controls were also performed.
Cytokine and chemokine ELISAs.
To measure cytokine and chemokine levels in the lungs, mouse lungs were harvested, placed in 1 ml of PBS, and homogenized using a Potter-Elvehjem homogenizer. Lung homogenates were then spun for 15 min at 13,000 rpm in microcentrifuge tubes to pellet debris. Supernatants were removed and filtered through a 0.45-μm filter plate. Then, 50 μl of the filtered homogenates was used in duplicate in a Millipore Milliplex map kit, following the kit protocol provided, to determine protein levels of macrophage inflammatory protein 1β (MIP-1β), RANTES, keratinocyte-derived chemokine (KC), monocyte chemotactic protein 1 (MCP-1), IL-6, IL-1β, TNF-α, IFN-γ, macrophage colony-stimulating factor (M-CSF), and granulocyte colony-stimulating factor (G-CSF). A traditional enzyme-linked immunosorbent assay (ELISA) was performed (using triplicates for each sample) using a Thermo Scientific mouse IFN-β colorimetric ELISA kit (424001).
Microarray analysis and bioinformatics.
Gene expression changes in mock- and virus-infected mouse lungs were evaluated by microarray from two independent experiments. Total RNA was isolated from three individual animals at 12 h and 1, 3, and 5 days postinfection (dpi) per virus group. As a mock control, animals were inoculated with PBS, and total RNA was isolated from mouse lungs (n = 2 mice) per time point. Probe labeling and microarray slide hybridization for each biological replicate were performed using a whole mouse genome microarray kit (G4122A; Agilent Technologies). Lung gene expression array data are represented as the in silico pool for time-matched mock-infected animals (n = 2) compared to the in silico pool for each infection condition (n = 3) at each time point. All data were entered into a custom-designed relational Oracle 9i backed relational database, Expression Array Manager, and were then uploaded into Rosetta Resolver system 7.2 (Rosetta Biosoftware) and Spotfire Decision site 8.1. Primary expression microarray data are available at http://viromics.washington.edu, in accordance with the proposed minimum information about a microarray experiment (MIAME) standards.
Statistically significant differences in gene expression between infection groups at each time point postinfection were determined by one-way analysis of variance (ANOVA) (P < 0.05). Differentially expressed genes were filtered to include only genes whose expression had changed 2-fold relative to the level in the mock-infected group (P < 0.01) in at least one of the strains being compared at a given time point postinfection. Transcriptional data were analyzed using Ingenuity Pathways Analysis (IPA; Ingenuity Systems). This software examines RNA expression data in the context of known biological functions and pathways, mapping each gene identifier in a data set to its corresponding gene object in the Ingenuity Pathways Knowledge Base. For all analyses, IPA-generated P values were calculated using Fisher's exact test to identify biological functions and pathways that were most significant. The assigned P values are a measure of the likelihood that the association between a set of focus molecules and a given function or pathway is not due to random chance. The focus molecules within IPA-generated networks represent nodes, and the edges that define the biological relationship between two nodes are represented as a line. All edges are supported by at least one published reference or from canonical information stored in the Ingenuity Pathways Knowledge Base. Arrows pointing between nodes signify different actions (e.g., binding). Arrows pointing back to the same node indicate self-regulation.
Quantitative reverse transcription-PCR (RT-PCR).
Quantitative real-time PCR was performed on RNA analyzed by microarray to validate gene expression changes. Total RNA samples were treated with DNA-free DNase and removal reagents (Ambion, Inc., Austin, TX). Reverse transcription was performed using a QuantiTect reverse transcription kit (Qiagen Inc., Valencia, CA). Primer and probe sets for each of the target sequences were chosen from the Applied Biosystems assays-on-demand product list. Each target was run in triplicate on an ABI 7500 real-time PCR system. Stromal cell-derived factor 2 (Sdf2) was used to normalize each target within Applied Biosystems Sequence Detections software version 1.3. Gene expression of Sdf2 was unchanged in all samples, treatment conditions, and time points as determined by microarray and hence was used for normalization. Quantification of normalized targets was calculated using the threshold cycle (2−ΔΔCT) method (22).
Ethics statement.
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee at the Mount Sinai School of Medicine (permit number 03-0058). All inoculation was performed under ketamine-xylazine anesthesia, and all efforts were made to minimize suffering.
RESULTS
The adaptive immune response is not responsible for pathogenesis.
We previously showed that amino acid position 66 in the PB1-F2 protein of influenza A virus is important for pathogenesis (7). Wild-type C57BL/6 mice were inoculated individually with WH or WH N66S reassortant virus containing the PB1 gene from A/Hong Kong/156/97 (H5N1) virus in the genetic background of A/WSN/33 virus, and pathogenesis was measured by weight loss, viral titer, and cytokine levels in the lungs. The WH virus has been shown to be a suitable model virus to study the effect of PB1-F2 on influenza virus pathogenesis (35). The WH and WH N66S viruses differ by a single amino acid at position 66 in PB1-F2, which does not change the primary sequence of the PB1 protein. Mice infected with the N66S mutant virus exhibit weight loss and increased IFN-γ levels at around day 5 postinfection (7). We thus sought to elucidate the mechanism for this finding in the present work.
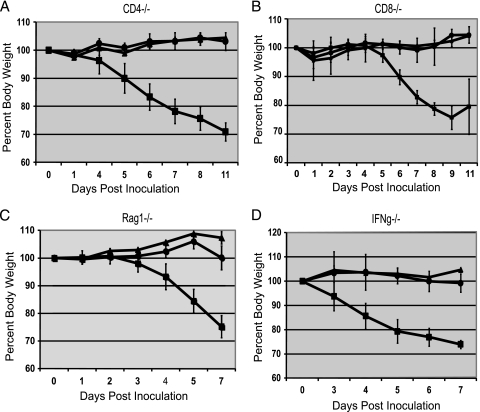
To investigate whether the enhanced virulence attributed to the PB1-F2 N66S mutation involved the adaptive immune response, knockout mice deficient in T-cell (CD4−/− or CD8−/− mice), B- and T-cell (Rag1−/− mice), or IFN-γ (IFN-γ−/−) production were infected with 104 PFU of WH or WH N66S virus, and weight loss was measured. The Rag1−/− mice were used to evaluate the entire adaptive immune response, as these mice have no functional B cells or T cells. As shown in Fig. 1, the infection of mice with WH N66S resulted in significant morbidity, whereas WH-infected mice showed minimal weight changes. The presence of a weight loss trend for both WH and WH N66S viruses in these knockout mice that was similar to that seen in wild-type mice (7) indicated that none of these cell types or molecules were essential to the pathogenesis mechanism. Were CD4 or CD8 T cells or IFN-γ important to pathogenesis, we would have expected the WH N66S-infected mice to recover from infection and the knockout mice to experience decreased morbidity. Pathogenicity in the knockout mice pointed to a mechanism that occurs earlier during infection, when the innate immune response is involved.
FIG. 1.
Morbidity in immune cell knockout mice. Mice were inoculated with 104 PFU of either WH (circles) or WH N66S (squares) virus or PBS-BA-PS (triangles) as a control. Weight was monitored and plotted as percent weight upon inoculation. Error bars indicate standard deviations.
Differential regulation of interferon response genes by WH and WH N66S viruses.
To further investigate the mechanism by which PB1-F2 N66S enhances pathogenesis, we evaluated the host response in mice individually infected with WH or WH N66S virus. Mouse lungs were collected at 1, 3, and 5 dpi and transcriptional profiles assessed by microarray. Statistically significant differences in gene expression between the two virus groups were determined by one-way ANOVA (P < 0.05) at each time point (Table 1) and then further filtered to include genes whose expression changed at least 2-fold (P < 0.01) relative to that for time-matched mock-infected animals. Gene expression data were analyzed using Ingenuity Pathways Analysis to identify cellular pathways impacted by PB1-F2 from H5N1. At 1 dpi, there was enhanced expression of interferon-stimulated antiviral response genes, including IFIT-1, IFN-β, Stat1, and Mx1, and genes associated with interferon regulatory factor (IRF) activation, such as RIG-I, LGP2, ZBP1, and Irf7, in WH-infected mouse lungs compared to levels in WH N66S-infected lungs (Table 2). By 3 dpi, WH N66S strongly activated a greater number of interferon-regulated genes than did WH. The dysregulation of host responses increased through the infection course, with the magnitude of WH N66S-induced transcriptional changes surpassing that observed for WH on day 5 postinfection. Based on these findings, we speculate that key changes in the host response begin around day 1 postinfection, which implicates an important role for innate immune signaling during infection. Further, the delay in activation of type I interferon signaling genes during WH N66S infection at day 1 postinfection suggests that the N66S mutation may modulate PB1-F2 function to either directly or indirectly perturb the interferon response. This early event could be detrimental to the host, ultimately leading to a fatal outcome in mice. We speculate that the differential gene expression distinguishing WH and WH N66S viruses early in infection is likely to contribute to the immunopathological outcome at later stages.
TABLE 1.
Numbers of differentially expressed genes in mouse lungs that distinguish WH and WH N66Sa
| Characteristic | No. of genes |
|---|---|
| Unique at day 1 postinfection | 1,776 |
| Unique at day 3 postinfection | 3,085 |
| Unique at day 5 postinfection | 2,299 |
| Different between days 1 and 3 | 927 |
| Different between days 3 and 5 | 767 |
| Different between days 1 and 5 | 434 |
| Different on all 3 days | 65 |
Differential expression for WH- and WH N66S-infected lungs compared to time-matched mock-infected lungs on each day postinfection was determined by one-way ANOVA (P < 0.05). Samples from animal 3 were omitted from the statistical analysis for WH on days 1 and 5 postinfection, due to poor quality control measures.
TABLE 2.
Canonical pathways distinguishing WH and WH N66S at days 1, 3, and 5 postinfectiona
| Day | Ingenuity canonical pathway | P value |
|---|---|---|
| 1 | Interferon signaling | 9.60E−17 |
| Activation of IRF by cytosolic pattern recognition receptors | 4.78E−15 | |
| Role of pattern recognition receptors in recognition of bacteria and viruses | 3.55E−09 | |
| Role of RIG-I-like receptors in antiviral innate immunity | 4.96E−07 | |
| Retinoic acid-mediated apoptosis signaling | 3.96E−06 | |
| 3 | Role of pattern recognition receptors in recognition of bacteria and viruses | 7.80E−19 |
| Activation of IRF by cytosolic pattern recognition receptors | 1.97E−15 | |
| Communication between innate and adaptive immune cells | 6.41E−15 | |
| Dendritic cell maturation | 1.17E−14 | |
| Cross talk between dendritic cells and natural killer cells | 1.40E−14 | |
| 5 | Role of pattern recognition receptors in recognition of bacteria and viruses | 5.17E−05 |
| Acute-phase response signaling | 1.53E−04 | |
| Atherosclerosis signaling | 2.70E−04 | |
| Interferon signaling | 6.70E−04 | |
| Dendritic cell maturation | 7.74E−04 |
Top canonical pathways were determined using Ingenuity Pathways Analysis of genes that were differentially expressed, as determined by ANOVA (P < 0.05), at each day postinfection. Fisher's exact text was used to calculate P values associated with each pathway.
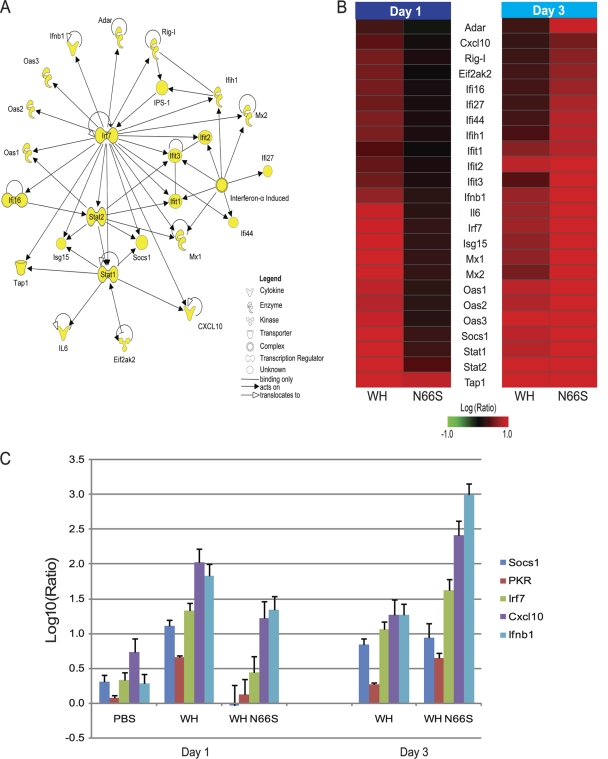
Further investigation of top functional categories using IPA resulted in a top scoring network containing genes related to innate immune responses. The molecular network diagram shown in Fig. 2 A depicts genes as the nodes, and their direct relationships are symbolized by the connections. With the exception of beta interferon promoter stimulator 1 (IPS-1), the essential mitochondrial adaptor protein for type I IFN signaling, all nodes represent genes differentially expressed between infection groups at both day 1 and day 3 postinfection. There is a high degree of connectivity among interferon response genes, with transcription factor Irf7 serving as a central hub in the network. Interferon-stimulated genes, including Mx1 and Mx2, ISG15 ubiquitin-like modifier, OAS, PKR, and RIG-I, strongly emphasize differential regulation of interferon signaling during infection. Gene expression changes of the molecules represented in the network diagram are shown in Fig. 2B. At day 1 postinfection, the interferon-regulated gene panel shows enhanced gene expression in WH-infected lungs compared to that in WH N66S-infected lungs. By day 3 postinfection, transcriptional expression is upregulated for the interferon-stimulated genes (ISGs) to a greater extent in WH N66S-infected lungs than in WH-infected lungs.
FIG. 2.
Interferon-stimulated response genes distinguish WH and WH N66S infection groups at days 1 and 3 postinfection. (A) Functional analysis of differentially expressed genes distinguishing WH and WH N66S infection groups on days 1 and 3 postinfection. The IPA network diagram shows direct functional relationships between genes involved in antimicrobial response, inflammatory response, and infection mechanism. Nodes depict genes in the network, and solid lines represent direct relationships between nodes. For signaling pathways, an arrow pointing from one node to the next signifies an activation event, for example, binding or phosphorylation. Nodes with arrows or connections pointing to themselves indicate self-binding or autoregulation events. (B) Transcription of genes related to antimicrobial response, inflammatory response, and infection mechanism from WH- and WH N66S-infected mice at 1 and 3 dpi. Two-dimensional clustering of differentially expressed genes within the categories of antimicrobial response, inflammatory response, and infection mechanism is represented. The average expression level for each group is shown as the log10 (ratio) expression relative to that for a pool of RNA extracted from lungs of time-matched mock-infected mice (n = 2). Red and green denote increases and decreases, respectively, in expression level relative to that for the uninfected reference. Clustering was performed with the hierarchical unweighted-pair group method using average linkages (UPGMA), with Euclidean distance similarity measure using an average-value ordering function. (C) Transcript levels for interferon response-related genes. Cellular transcript levels at days 1 and 3 postinfection in mouse lungs infected with WH or WH N66S virus, as measured by quantitative real-time PCR. Average log10 (ratio) values were calculated by averaging CT values from three technical replicates in each case. Data represent averages of three biological replicates from two independent experiments. Sdf2 was used as an internal control for each assay. PBS-inoculated lungs at day 3 postinfection were set as the baseline. Error bars indicate standard deviations.
To confirm this finding, we analyzed mRNA levels of IFN-β and selected ISGs, such as Irf7, PKR, Cxcl10, and Socs1, represented in the IPA network diagram by quantitative real-time PCR (Fig. 2C). The cellular transcript levels mirror the genomics findings (Fig. 2C). There is enhanced expression of these cellular transcripts in WH-infected mouse lungs compared to that in WH N66S-infected lungs at day 1 postinfection, and by day 3 postinfection, WH N66S induced greater mRNA levels than did WH, with the exception of Socs1. Based on these results, it seems that the early interferon response is suppressed during infection with the WH N66S virus. The data furthermore suggest that the PB1-F2 protein with the N66S mutation suppresses the antiviral response early during infection (1 dpi), allowing for uncontrolled replication of the virus in the lungs at later time points, which then results in the cytokine and chemokine upregulation and cell infiltration.
Cytokine dysregulation in WH N66S-infected mouse lungs.
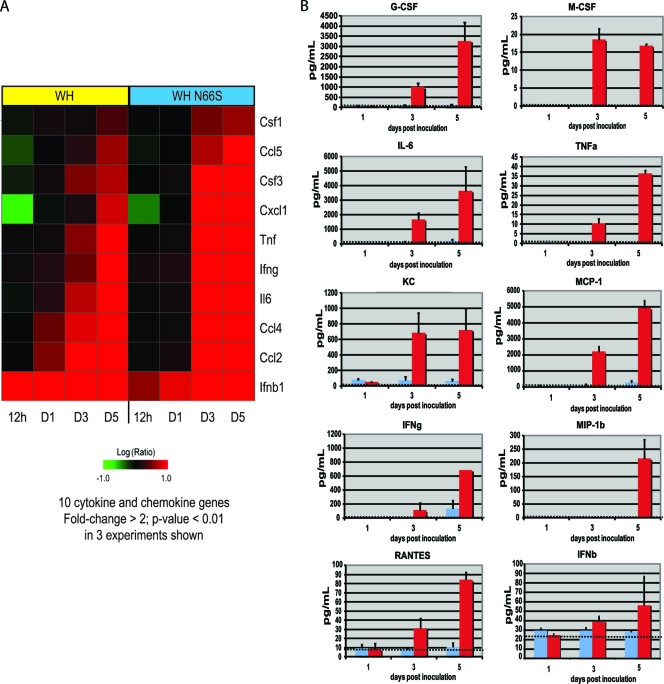
Gene expression profiles for the panel of 10 chemokine and cytokine molecules tested were assessed to confirm that transcriptional changes correlated with protein expression. Gene expression differences of at least 2-fold (P < 0.01) at each time point are shown for each infection group (Fig. 3A). At 12 hpi, IFN-β was the sole cytokine from the analyzed panel that showed upregulated gene expression in both WH- and WH N66S-infected mouse lungs, although WH virus induced IFN-β gene expression to a greater extent. Additionally, gene expression for MCP-1 and MIP-1β, corresponding to Ccsl2 and Ccl4, respectively, was upregulated in WH-infected mouse lungs at day 1 postinfection, whereas levels of these genes were relatively unchanged in WH N66S-infected lungs. By day 3 postinfection, WH N66S induced a stronger activation of the cytokine and chemokine genes examined, which were upregulated to a greater extent than after WH infection. The upregulated gene expression was sustained through day 5 postinfection for both WH and WH N66S infection groups (Fig. 3A). Several cytokines, such as MCP-1 (Ccl2), show upregulation at the transcriptional level on day 1 postinfection (Fig. 3A) that was not detected in the multiplex ELISA, which measures protein levels (Fig. 3B). This could be due to the higher sensitivity of the transcriptional profiling method than of the ELISA technique. Alternatively, it is possible that the mRNA of the cytokine can be detected earlier than the protein product. This is supported by the detectable protein levels of MCP-1 at later time points postinfection (Fig. 3B).
FIG. 3.
Cytokine and chemokine levels in infected mice. (A) Gene expression profiles for the examined cytokine and chemokine panel are represented. The average expression level for each group is shown as the log10 (ratio) expression relative to that for a pool of RNA extracted from lungs of time-matched mock-infected mice (n = 2). Red and green denote increases and decreases, respectively, in expression level relative to that for the mock-infected reference. (B) Mice were inoculated with PBS-BA-PS (dotted line as baseline) as a control or 104 PFU of either WH (blue) or WH N66S (red) virus. Mouse lungs were collected on days 1, 3, and 5 postinoculation and homogenized. MIP-1β, RANTES, TNF-α, M-CSF, KC, IFN-γ, G-CSF, MCP-1, and IL-6 levels in lung homogenate were measured using a Millipore Milliplex ELISA kit. IFN-β was measured using a standard ELISA. Each harvested lung homogenate was 1 ml. If the baseline is not visible, the cytokine level for the PBS-BA-PS group was 0. Error bars indicate standard deviations.
Severe respiratory disease resulting from highly pathogenic influenza virus infections is typically associated with elevated levels of proinflammatory cytokines and chemokines (8, 28, 33). To assess whether WH N66S infection causes enhanced cytokine and chemokine levels in the lungs, we examined a panel of 10 chemokines and cytokines using a multiplex ELISA and a traditional ELISA for IFN-β. Infection with WH N66S caused an increase in all of the cytokines and chemokines tested (Fig. 3B). At day 3 postinfection, there was an upregulation of KC, IL-6, and M-CSF. KC is important for monocyte trafficking into the lung, and M-CSF is necessary for macrophage maturation and activation. IL-6 is known to be secreted early during the acute response to infection. All three of these molecules seem to be first responders to the infection, creating the environment to allow increased cellularity in the lungs. Also on day 3 postinfection, TNF-α, G-CSF, and MCP-1 appear in the lung milieu. These molecules reinforce the trafficking of cells into the lung and aid monocyte maturation by upregulating other antiviral cytokines. On day 5, there was full upregulation of multiple cytokines and chemokines, such as MIP-1β, RANTES, IFN-β, and IFN-γ.
Cell infiltration characterization.
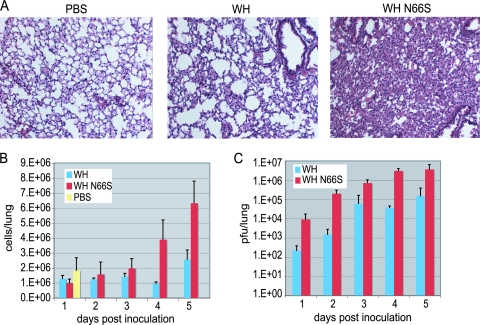
The upregulation of cytokine and chemokine genes and protein expression indicates that there was a significant innate immune response occurring in the lungs that could cause monocyte migration into the lung. To investigate this, lung histology and lung cell flow cytometry were conducted on infected mice. Figure 4A shows hematoxylin and eosin staining of lung sections taken from mice on day 5 postinoculation. Mice inoculated with WH N66S showed the greatest infiltration of cells in their lungs. The WH-infected mice also showed a marginal level of cell infiltration compared to that for PBS-inoculated mice but not to the same extent as that seen in WH N66S-infected mice. Next, we focused on characterizing the infiltration on days 1 to 5 by using flow cytometry to count the total number of cells in the lung. Figure 4B shows the total number of cells in the lung over the course of infection, and the increase in cell number corresponds with the histopathology observed at day 5 postinfection. The infiltration of cells also seems to occur as the viral titer increases in the lungs of mice (Fig. 4C).
FIG. 4.
Lung titer and histology in WH- and WH N66S-infected mice. Mice were inoculated with PBS-BA-PS as a control or 104 PFU of either WH or WH N66S virus. Mouse lungs were collected on days 1 to 5 postinoculation. (A) Lung sections were taken on day 5 postinfection and stained with hematoxylin and eosin. Cell infiltration was observed using bright-field microscopy at ×10 magnification of 6-μm sections. (B) Cell counts were taken using cell counting beads and flow cytometry. (C) Lung titers were analyzed and graphed as PFU/lung. Error bars indicate standard deviations.
Monocytes and neutrophils are two immune cell types that initially respond to infection, migrating to the site of infection. These cells are marked by the presence of CD11b/CD11c or Ly6G, respectively, on the cell surface. In the lungs of infected mice, there was an increase in monocytes starting at day 3 postinfection and remaining until day 5 (Fig. 5A and B). Starting on day 4, there was also an increase in the number of neutrophils (Fig. 5C and D). The number of dendritic cells (DCs) increased over days 1 to 5, as expected during infection, although the percentage of DCs decreased (Fig. 5E and F). This was due to the increase in number of monocytes and neutrophils in the lung (Fig. 5A and C), which is concomitant with an increase in the total number of cells in the lung (Fig. 4B). The increase in monocytes, neutrophils, and DCs corresponds to an increased response to the infection, but the observed increase in monocytes and neutrophils was higher in WH N66S-infected mice than in WH-infected mice. The cellular response and increase in cytokines and chemokines that occur in the WH N66S-infected mice appear to be a result of the high viral titer in these mice. It is also possible that the differences in cytokine and chemokine levels in WH N66S-infected mice cause increased cellularity in the lungs. The increased viral titers at later points during infection could occur because of a suppressed interferon response or a decreased stimulation of the interferon pathway, which is present early during infection.
FIG. 5.
Lung cell populations in infected mice. Mice were inoculated with PBS-BA-PS as a control or 104 PFU of either WH or WH N66S virus. Mouse lungs were collected on days 1 to 5 postinoculation. (A and B) Monocytes in the lung cell population were stained using fluorescent antibodies. CD11c+ CD11b+ cells were considered monocytes. (C and D) Neutrophils in the lung cell population were stained using fluorescent antibodies. Ly6C+ cells were considered neutrophils. (E and F) Dendritic cells in the lung cell population were stained using fluorescent antibodies. CD11chigh MHC-II+ CD11b− cells were considered dendritic cells. Percentages of total cells and absolute cell numbers are shown. For panels A through E, PBS samples were averaged from all days and plotted on the graphs on day 1. Error bars indicate standard deviations.
DISCUSSION
It has become increasingly clear that influenza A viral proteins have multiple functions during infection. Our group and others have demonstrated that PB1-F2 protein from influenza A virus has multiple activities both in vitro and in vivo, including apoptosis induction, enhancement of secondary bacterial pneumonia, and a capacity to increase pathogenicity and induce a proinflammatory response (7, 25, 26, 34, 35). However, the mechanism of PB1-F2's role in pathogenesis has not been fully described. There has been speculation that the apoptotic function of PB1-F2 is responsible for increased pathogenesis (34, 35). This possibility was investigated in the present work by performing a terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay for apoptosis on whole lung homogenates, but we observed no difference in apoptosis induction caused by the WH and WH N66S viruses (data not shown). However, these results do not eliminate the possibility that the N66S mutation may have an effect on apoptosis in a specific cell type. Increased apoptosis in the presence of PB1-F2 was shown to be specific to immune cells in a previous publication (4). PB1-F2 N66S may contribute to this effect by increasing apoptosis even more than wt PB1-F2. However, it is likely that the significant infiltration of cells into the lungs and reduction in air space are what causes the mice to succumb to viral infection.
To determine the mechanism for the increased immunopathogenesis caused by the PB1-F2 N66S-expressing virus, the innate and adaptive immune responses were studied in detail. We have shown that elimination of key components of the adaptive immune response did not change the level of pathogenesis caused by the WH N66S virus. We expect the virulence of the WH N66S virus to decrease when an important part of the pathogenesis mechanism is eliminated. Instead, we found that regardless of what cell type (CD4−/−, CD8−/−, Rag1−/−) or cytokine (IFN-γ−/−) was knocked out, the WH N66S virus maintained its pathogenesis. While we cannot completely rule out a role for adaptive immune responses in mediating WH N66S-induced pathogenesis, our work suggests that the increase in pathogenesis results from events occurring during the acute-phase response to influenza virus infection. Inflammation and activation of proinflammatory pathways during influenza virus infection can either contribute to a protective phenotype or have a detrimental consequence to the host. Here, we report that mice infected with WH N66S virus show enhanced immunopathology, driving significant cellular accumulation in the lungs that is not observed for WH-infected mice. Infiltration of multiple leukocyte populations, including monocytes and neutrophils, is seen beginning at day 3 postinfection for WH N66S-infected mice. This increased lung cellularity is concomitant with enhanced levels of cytokines and chemokines, specifically chemoattractant molecules responsible for homing of leukocytes into lungs during infection. These observations suggest that the N66S mutation in PB1-F2 likely alters the innate immune response that may be directly or indirectly mediated by PB1-F2.
We investigated host responses to WH and WH N66S infection by using transcriptional profiling of infected mouse lungs at 12 h and 1, 3, and 5 days postinfection. This revealed differential regulation of innate immune responses, supporting a suppression of interferon-stimulated genes in WH N66S-infected mice compared to the level in WH-infected mice (Fig. 2). Hence, a virus containing a single amino acid mutation in PB1-F2 found in highly pathogenic viruses allows for increased immunopathology by downregulating the early interferon response. Further studies are needed to determine whether the N66S mutation in the PB1-F2 protein of other influenza viruses also leads to a decrease in the interferon response. Based on the finding that PB1-F2 66S increases virulence in both the 1918 pandemic influenza virus and an H5N1 virus, as shown in our previous work (7), we feel that the suppression of early innate responses may be a general feature of PB1-F2 66S proteins derived from different strains. Overall, this work suggests a novel function of PB1-F2 derived from a highly pathogenic influenza virus strain in contributing to enhanced pathogenesis through a mechanism that targets the host innate immune pathways early in infection.
To broaden our view of infection and determine how early differential regulation of interferon responses by WH and WH N66S viruses can be detected, we conducted a separate experiment that included a 12-h time point. The transcriptional profiles of the two viruses were similar at 12 h postinfection, which indicates that differences seen on day 1 postinfection are probably not due to different titers of the virus inocula. Based on the observation that the downregulation of innate immune genes by the WH N66S virus occurs at this early time point, we speculate that critical events begin by day 1 postinfection.
We have shown that the WH N66S virus, possessing only a single amino acid substitution in the PB1-F2 protein compared to its isogenic counterpart, causes a delayed expression of innate immune pathway genes. This suggests that there may be a blockade in the sensing capabilities for membrane-bound or cytosolic pathogen recognition receptors responsible for detecting viral RNA and transmitting downstream signals to initiate induction of interferon and proinflammatory responses. The downregulation of interferon-related genes by the PB1-F2 N66S protein may occur via two mechanisms. PB1-F2 N66S could be a traditional interferon antagonist, binding to a molecule in the type I interferon pathway and reducing the production of interferon. With the impairment of interferon expression, there would then be a reduction of interferon-stimulated genes compared to that found with wt infection. Alternatively, PB1-F2 N66S could inhibit a transcription factor that is common among interferon-stimulated genes. This inhibition by PB1-F2 N66S would either prevent the transcription factor from binding the promoter complex or block the action of the promoter complex.
A recent study by Le Goffic et al. reports that PB1-F2 proteins of certain influenza virus strains increase the expression of IFN during viral infection (19). The authors suggest that the PB1-F2-mediated IFN induction is dependent on the mitochondrial antiviral signaling protein (MAVS, also known as IPS-1, Cardif, or VISA) and the NF-κB transcription factor. This interferon-inducing effect of PB1-F2 was found in epithelial cells only. Future studies are necessary to sort out whether this pro/anti-interferon effect is tissue specific or whether these effects are strain or backbone specific.
We believe that PB1-F2 proteins of highly pathogenic influenza viruses, such as the WH N66S virus, may have evolved to reduce the property of IFN induction and thus contribute to increased virulence. The N66S mutation in the PB1-F2 protein may specifically mediate inhibition of innate immune responses through a newly gained direct physical interaction with key modulators of IFN signaling responses. The interaction with IFN signaling mediators may not occur with PB1-F2 proteins derived from low-pathogenic strains or may have lower affinity. Alternatively, the N66S mutation may result in PB1-F2 adopting an entirely different function. It is probable that the N66S mutation could alter the function of PB1-F2, because the N66S mutation is located in an exposed portion of the α-helix in the C-terminal region of PB1-F2. The C-terminal region is important for other functions of PB1-F2, such as apoptosis, and the α-helix is known to be the only secondary structure of the molecule (2, 11, 34). However, a molecular mechanism for the decreased IFN induction by PB1-F2 N66S has yet to be uncovered.
The transcriptional profiles observed for WH N66S at days 3 and 5 postinfection are representative of an infection involving significant cell infiltration and extreme upregulation of cytokines and chemokines. The cytokines and chemokines with increased protein levels at day 3 were also shown to have increased gene expression on the same day. This not only validates the microarray data but also indicates that the upregulation is due to increased gene expression and not just increased translation or stored protein release.
Previous publications indicate that the PB1-F2 proteins vary in ability to contribute to pathogenesis. Here, we suggest that this may be due to point mutations that can enhance or decrease the function of PB1-F2. The microarray data compiled show an important suppression of interferon-stimulated genes early during infection, allowing for higher viral titers and increased pathogenesis later during infection. What this demonstrates is that initially the PB1-F2 N66S protein is able to suppress the early antiviral response, which allows for unchecked viral growth. This increased viral growth then triggers an overwhelming inflammatory response that overloads the lung environment with cells. Our data support this mechanism through the timeline of events occurring; on day 1, there was a suppression of interferon signaling that would allow for increased viral replication, and on day 3, there were increased levels of proinflammatory genes and high expression of proinflammatory molecules. The excessive infiltration of cells then makes it difficult for the mice to maintain lung function, and they eventually succumb to infection. Alternatively, not only the increase in viral titer but also the increase in cytokine and chemokine levels may be responsible for WH N66S-mediated virulence.
Recent studies have identified additional viral proteins besides the well-characterized IFN antagonist NS1 that limit the host IFN response (13, 16). Graef et al. show that the polymerase protein PB2 interacts with the MAVS protein at the mitochondria and thus inhibits the induction of IFN (13). These studies highlight that the IFN antagonism strategy of influenza viruses is more complex than previously thought. Our study supports the hypothesis that the PB1-F2 protein has anti-interferon activity in addition to previously published functions, and future studies should unravel the molecular mechanism of PB1-F2 N66S-mediated interferon suppression. The importance of studying this protein from influenza A viruses is essential to understanding virulence in different influenza A virus strains.
Acknowledgments
We thank Jean Chang for technical assistance, Sean Proll for microarray analysis support, and Marcus Korth for review and helpful discussions of the manuscript.
This research was supported by Public Health Service grant P01AI058113 from the National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Baskin, C. R., et al. 2009. Early and sustained innate immune response defines pathology and death in nonhuman primates infected by highly pathogenic influenza virus. Proc. Natl. Acad. Sci. U. S. A. 106:3455-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruns, K., et al. 2007. Structural characterization and oligomerization of PB1-F2, a proapoptotic influenza A virus protein. J. Biol. Chem. 282:353-363. [DOI] [PubMed] [Google Scholar]
- 3.Chen, L. M., C. T. Davis, H. Zhou, N. J. Cox, and R. O. Donis. 2008. Genetic compatibility and virulence of reassortants derived from contemporary avian H5N1 and human H3N2 influenza A viruses. PLoS Pathog. 4:e1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen, W., et al. 2001. A novel influenza A virus mitochondrial protein that induces cell death. Nat. Med. 7:1306-1312. [DOI] [PubMed] [Google Scholar]
- 5.Cilloniz, C., et al. 2010. Lethal dissemination of H5N1 influenza virus is associated with dysregulation of inflammation and lipoxin signaling in a mouse model of infection. J. Virol. 84:7613-7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cilloniz, C., et al. 2009. Lethal influenza virus infection in macaques is associated with early dysregulation of inflammatory related genes. PLoS Pathog. 5:e1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conenello, G. M., D. Zamarin, L. A. Perrone, T. Tumpey, and P. Palese. 2007. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 3:1414-1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong, M. D., et al. 2005. Fatal avian influenza A (H5N1) in a child presenting with diarrhea followed by coma. N. Engl. J. Med. 352:686-691. [DOI] [PubMed] [Google Scholar]
- 9.Fornek, J. L., et al. 2009. A single-amino-acid substitution in a polymerase protein of an H5N1 influenza virus is associated with systemic infection and impaired T-cell activation in mice. J. Virol. 83:11102-11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, Y., et al. 2009. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 5:e1000709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibbs, J. S., D. Malide, F. Hornung, J. R. Bennink, and J. W. Yewdell. 2003. The influenza A virus PB1-F2 protein targets the inner mitochondrial membrane via a predicted basic amphipathic helix that disrupts mitochondrial function. J. Virol. 77:7214-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodman, A. G., et al. 2010. The alpha/beta interferon receptor provides protection against influenza virus replication but is dispensable for inflammatory response signaling. J. Virol. 84:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graef, K. M., et al. 2010. The PB2 subunit of the influenza virus RNA polymerase affects virulence by interacting with the mitochondrial antiviral signaling protein and inhibiting expression of beta interferon. J. Virol. 84:8433-8445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hai, R., et al. 2010. PB1-F2 expression by the 2009 pandemic H1N1 influenza virus has minimal impact on virulence in animal models. J. Virol. 84:4442-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatta, M., P. Gao, P. Halfmann, and Y. Kawaoka. 2001. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 293:1840-1842. [DOI] [PubMed] [Google Scholar]
- 16.Iwai, A., et al. 2010. Influenza A virus polymerase inhibits type I interferon induction by binding to interferon β promoter stimulator 1. J. Biol. Chem. 285:32064-32074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kash, J. C., et al. 2006. Genomic analysis of increased host immune and cell death responses induced by 1918 influenza virus. Nature 443:578-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobasa, D., et al. 2007. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature 445:319-323. [DOI] [PubMed] [Google Scholar]
- 19.Le Goffic, R., et al. 2010. Influenza A virus protein PB1-F2 exacerbates IFN-beta expression of human respiratory epithelial cells. J. Immunol. 185:4812-4823. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., et al. 2010. Continued evolution of H5N1 influenza viruses in wild birds, domestic poultry, and humans in China from 2004 to 2009. J. Virol. 84:8389-8397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lillie, R. D. 1965. Histopathologic technic and practical histochemistry, 3rd ed., p. 715. McGraw-Hill Book Co., New York, NY.
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 23.Lu, X., et al. 1999. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J. Virol. 73:5903-5911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mazur, I., et al. 2008. The proapoptotic influenza A virus protein PB1-F2 regulates viral polymerase activity by interaction with the PB1 protein. Cell. Microbiol. 10:1140-1152. [DOI] [PubMed] [Google Scholar]
- 25.McAuley, J. L., et al. 2010. PB1-F2 proteins from H5N1 and 20 century pandemic influenza viruses cause immunopathology. PLoS Pathog. 6:e1001014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McAuley, J. L., et al. 2007. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe 2:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAuley, J. L., K. Zhang, and J. A. McCullers. 2010. The effects of influenza A virus PB1-F2 protein on polymerase activity are strain specific and do not impact pathogenesis. J. Virol. 84:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Perrone, L. A., J. K. Plowden, A. Garcia-Sastre, J. M. Katz, and T. M. Tumpey. 2008. H5N1 and 1918 pandemic influenza virus infection results in early and excessive infiltration of macrophages and neutrophils in the lungs of mice. PLoS Pathog. 4:e1000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szretter, K. J., et al. 2009. Early control of H5N1 influenza virus replication by the type I interferon response in mice. J. Virol. 83:5825-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Szretter, K. J., et al. 2007. Role of host cytokine responses in the pathogenesis of avian H5N1 influenza viruses in mice. J. Virol. 81:2736-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wasilenko, J. L., et al. 2008. NP, PB1, and PB2 viral genes contribute to altered replication of H5N1 avian influenza viruses in chickens. J. Virol. 82:4544-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamada, H., R. Chounan, Y. Higashi, N. Kurihara, and H. Kido. 2004. Mitochondrial targeting sequence of the influenza A virus PB1-F2 protein and its function in mitochondria. FEBS Lett. 578:331-336. [DOI] [PubMed] [Google Scholar]
- 33.Yuen, K. Y., et al. 1998. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet 351:467-471. [DOI] [PubMed] [Google Scholar]
- 34.Zamarin, D., A. Garcia-Sastre, X. Xiao, R. Wang, and P. Palese. 2005. Influenza virus PB1-F2 protein induces cell death through mitochondrial ANT3 and VDAC1. PLoS Pathog. 1:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zamarin, D., M. B. Ortigoza, and P. Palese. 2006. Influenza A virus PB1-F2 protein contributes to viral pathogenesis in mice. J. Virol. 80:7976-7983. [DOI] [PMC free article] [PubMed] [Google Scholar]