Abstract
After recognizing and binding to its host cell, poliovirus (like other nonenveloped viruses) faces the challenge of translocating its genome across a cellular membrane and into the cytoplasm. To avoid entanglement with the capsid, the RNA must exit via a single site on the virion surface. However, the mechanism by which a single site is selected (from among 60 equivalents) is unknown; and until now, even its location on the virion surface has been controversial. To help to elucidate the mechanism of infection, we have used single-particle cryo-electron microscopy and tomography to reconstruct conformationally altered intermediates that are formed by the poliovirion at various stages of the poliovirus infection process. Recently, we reported icosahedrally symmetric structures for two forms of the end-state 80S empty capsid particle. Surprisingly, RNA was frequently visible near the capsid; and in a subset of the virions, RNA was seen on both the inside and outside of the capsid, caught in the act of exiting. To visualize RNA exiting, we have now determined asymmetric reconstructions from that subset, using both single-particle cryo-electron microscopy and cryo-electron tomographic methods, producing independent reconstructions at ∼50-Å resolution. Contrary to predictions in the literature, the footprint of RNA on the capsid surface is located close to a viral 2-fold axis, covering a slot-shaped area of reduced density that is present in both of the symmetrized 80S reconstructions and which extends by about 20 Å away from the 2-fold axis toward each neighboring 5-fold axis.
In its role as the intermediate that links one round of infection with the next, a virus particle protects the viral genome during passage from cell to cell and from host to host, it specifically recognizes and binds to target cells, and it delivers the viral genome into the appropriate compartment in the target cell. For enveloped viruses, which have their own external membranes, fusion of the viral membrane with a host membrane presents a conceptually simple mechanism for delivery of the genome or nucleoprotein into the cytoplasm. For nonenveloped viruses, the viral particle must provide the machinery necessary for either the entire virion, a nucleoprotein complex, or the viral genome to cross a membrane. This process remains poorly understood. Poliovirus provides an excellent model system for probing the mechanisms used for genome translocation. As the type member of the Picornavirus family and the etiological agent of poliomyelitis, poliovirus has been well characterized biochemically and genetically (42), its cell entry pathways have been well characterized (5, 15, 30, 52), and a number of cell entry intermediates have been identified and are accessible for structural studies (2-4, 7, 8, 18, 34, 38, 42, 55, 56).
The capsid of the mature poliovirion (160S particle) consists of 60 copies of each of the four coat proteins VP1, VP2, VP3, and VP4 (which is myristolated at its amino-terminal glycine [13]) and encloses a 7.5-kbp positive-sense RNA genome. The outer surface of the capsid has a number of major features, including star-shaped mesas at its 5-fold axes, 3-fold propeller-like protrusions, canyon-like depressions surrounding each of the 5-fold mesas, and depressions at the 2-fold axes (30, 31).
Poliovirus infection is initiated when the virus binds to the host-cell-surface poliovirus receptor (called Pvr or CD155) (41), triggering a conformational change of the native capsid into an altered particle called the A particle or 135S particle (18, 19). The 135S particle has been shown to be expanded by about 4% (2, 7), is infectious (16, 33), and is believed to be a productive intermediate in viral entry (30, 33). This conformational change results in the externalization of the small myristoylated capsid protein, VP4 (18), and of the amino-terminal extension of VP1 (which includes a conserved amphipathic helix) (23). Both of these externalized polypeptides then associate with membranes (17, 23). In subsequent steps, the viral genome is released from the capsid and translocated across a membrane (probably an endosomal membrane [5]) to gain access to the cytoplasm, leaving behind an end-state empty capsid shell (called the 80S particle). The trigger for RNA release and the mechanism of genome translocation are both poorly understood (30, 52).
Electrophysiology and mutational experiments have shown that the externalization of VP4 and of the amino terminus of VP1 is associated with the formation of channels in membranes (17, 49, 50) and, furthermore, that point mutations in threonine 28 of VP4 can either eliminate (T28G) or alter (T28V, T28S) the ability to form channels and either eliminate (T28G) or slow (T28V, T28S) the kinetics of productive RNA release (17). These observations have led to the hypothesis that the viral polypeptides insert into host cell membranes during infection and rearrange to form channels that permit the viral genome to pass through the membrane, thereby gaining access to the cytoplasm (7, 17, 49, 50).
Speculation about the sites of externalization of the viral peptides and of the viral genome began soon after the structures of mature rhinovirus and poliovirus were determined crystallographically 25 years ago (31, 44). In both structures there is a solvent-filled channel running along each 5-fold axis. This channel is closed off at the outer surface of the capsid by polypeptide loops and on the inner surface by a plug that is formed by five intertwined copies of the amino terminus of VP3, forming a parallel beta tube (31, 44). In poliovirus this tube is flanked on its inner surface by five copies of a three-stranded beta sheet in which the outermost two strands come from a beta hairpin at the amino terminus of VP4 and the innermost strand comes from residues at the extreme amino terminus of VP1 (20). The presence of this channel, together with its proximity to peptide segments that were known to be externalized upon receptor attachment, and analogies with other viruses led to a model in which both the peptides and the viral RNA are externalized via the channel at the 5-fold axis (25, 45). At that time, an alternative model for the egress of polypeptides was proposed, based on an analogy with the externalization of the amino-terminal extensions of capsid proteins in expanded states of the topologically similar T=3 plant viruses (26, 32, 43, 47) and on genetic and biochemical studies of mutations that affect cell entry and capsid stability in poliovirus (14, 39, 54). In the latter model, the peptides were proposed to exit from the base of the canyon and then proceed along the outer surface toward the 5-fold peak (43, 47). Both models suggested that five copies of each of the externalized peptides would interact in some way to form a pore in the membrane that was contiguous with one of the 5-fold channels, thus providing a way for RNA to be released from the virion at a 5-fold axis of symmetry. No data yet exist to specify what specific structural roles VP4 and the amino terminus of VP1 might play in forming pores and serving as membrane anchors. However, both the electrophysiology data (cited above) and the greater sequence conservation of VP4 suggest that its role in pore formation may be the more central (17, 49, 50).
To further elucidate various steps along the infection pathway, cryo-electron microscopy (cryo-EM) reconstructions have been determined for a number of cell entry intermediates of poliovirus and rhinoviruses, and their resolutions have been improved over time (2, 3, 7, 28, 38). Structures of the complexes of polioviruses and major-group rhinoviruses with the ectodomains of their respective receptors have confirmed earlier models that suggested that the canyon is the receptor-binding site and have begun to suggest how receptor binding might lead to receptor-induced conformational rearrangements (3, 56). Cryo-EM and cryo-electron tomography structures (cryo-ET) of a poliovirus-receptor-membrane complex (using a novel receptor-decorated liposome model [51]) confirmed that initial receptor binding brings the surface of the 5-fold mesa into close proximity with the membrane and appears to produce an outward distortion of the outer leaflet of the membrane in its area of closest approach to the virus particle (4, 8).
Structures have also been determined for the soluble 135S and 80S particles of poliovirus, formed by heating the virus at 50°C (135S) or 56°C (80S) in hypotonic buffers, and for the 80S particles of rhinovirus 14 and 16, formed by exposing virus to acidic pH. All of the biological and immunological evidence that is currently available indicates that the particles prepared in vitro and used for structural studies are indistinguishable from the particles that are released from the cell surface during infection (6, 53). These structures have allowed the models for peptide release and genome release to be extended and refined (7, 38) and indeed have confirmed that VP1 exits from the particle surface at the base of the canyon and climbs up the side of the 5-fold mesa. However, contrary to the assumptions of the earlier models, the 10-Å structures of the poliovirus 135S and 80S particles show that the amino end of the amino-terminal extension of VP1 does not remain associated with the mesa. Instead, it forms an alpha-helical bridge that stretches across the canyon and binds to the large EF loop of VP2, a surface projection that appears as a 3-fold propeller blade (7, 38).
Until recently, the mechanism of RNA release (during the 135S-to-80S transition) has been largely a matter of conjecture. We can infer that the RNA must exit via a single site on the virion surface, to avoid entanglement with the capsid (particularly as entanglement has never been observed in electron micrographs), though the mechanism by which a single site is selected (from among 60 equivalents) is unknown. All models presented to date have assumed that the RNA is released from the channel at the 5-fold axes (2, 3, 7, 8, 25, 27, 28, 30, 42, 45). However, in the icosahedrally constrained 10-Å structures of both the poliovirus 135S and 80S particles (7, 38), the apparent intactness and stability of the 5-fold mesa argues against the 5-fold axis being the site of RNA egress, given that the diameter of the opening, as seen in those structures, would be insufficient to accommodate RNA, even if the “plug” formed by the intertwined amino termini of VP3 was displaced. Moreover, both structures revealed significant thinning between 2-fold-related pentamers in the vicinity of the 2-fold axes. Most convincingly, large holes (easily sufficient to accommodate RNA) were seen at and near the 2-fold axes in the atomic model of the late-80S structure. This coincided with an open hole in the reconstruction, when viewed at a contour level that left most of the remainder of the capsid intact. This evidence was suggestive, but not definitive, as a number of other openings were present, particularly in the interfaces between protomers. Furthermore, the behavior of the capsid structure in the immediate vicinity of the unique site of RNA exit is likely to be different from what we see in the icosahedral average, which is dominated by the remainder of the capsid.
In the course of solving icosahedrally symmetric cryo-EM structures for the poliovirus end-state 80S empty capsid particle (7, 38), we were surprised to find that RNA was frequently visible near the capsid and that in a subset of about 5% of the sampled virions, RNA was seen on both the inside and outside of the capsid, apparently caught in the act of exiting. This was an exciting development, as images of viral RNA release had never previously been reported. We were able to improve the resolution to ∼10 Å by classifying the projected images into two groups: an early 80Se particle that was more prevalent in the population after a shorter heating time and a late 80Sl particle that was seen more often when the heating time was increased. The amount of RNA density remaining in the interior appears to be continuously variable in both classes, suggesting that release is gradual. Of the 5% subset of particles clearly caught in the act, almost all belonged to the 80Se class. Our interpretation was that the 80Se class may represent particles in which exiting RNA is still engaged with the capsid machinery and traversing the capsid, while the 80Sl class (in which much of the capsid resembles the 135S form more closely in structure) represents particles with the RNA disengaged, possibly after nuclease cleavage. More than two structural classes may be present, but at the current resolution, we could not distinguish them.
The present report addresses the question of what we can learn about the details of RNA release from an asymmetric cryo-EM reconstruction, based on the 540-particle caught-in-the-act subset, and independently from cryo-electron tomographic reconstructions of a similarly prepared sample. In each projected particle image or subtomogram, preliminary orientation parameters are first determined from an icosahedrally symmetric calculation, and in a second stage, the symmetry is broken by choosing 1 of the 60 symmetry-equivalent orientations. Both methods have yielded similar information, at about 50-Å resolution, concerning the footprint of the RNA on the virion surface, which demonstrates that RNA is released from an asymmetric site at the base of the canyon near a particle 2-fold axis and not at the channel at the 5-fold axes, as suggested by previous models. Additionally, the demonstrated success of the methodology provides us with a blueprint for resolving the molecular details of the RNA-capsid interaction in future experiments.
MATERIALS AND METHODS
Sample preparation.
The Mahoney strain of type 1 poliovirus was grown in HeLa cell suspension, harvested by centrifugation, and released from the cell pellets by freeze-thaw lysis. The virus was then purified from the cell lysate by CsCl density gradient fractionation, as described previously (16, 38, 46). To obtain 80S end-state empty capsids, purified mature virions were heated for 4 to 60 min at 56°C in 20 mM Tris buffer (pH 7.4)-50 mM NaCl-2 mM CaCl2. Heat treatment incubation times varied for different experiments, with shorter heating times producing more particles in the act of RNA release and with longer incubation times producing emptier particles. For the cryo-ET studies, samples heat treated for 4 to 10 min were diluted to 0.2 mg/ml and deposited on Quantifoil holey carbon grids and mixed with a solution of colloidal gold. The grid was blotted and then plunge-frozen in liquid ethane using an FEI Vitrobot apparatus.
Cryo-electron tomography.
The samples were transferred under liquid nitrogen to the cryo-transfer system of a Polara G2 field emission gun microscope (FEI) equipped with a Gatan imaging filter (GIF) 2000 energy filter and operated at an acceleration voltage of 300 kV under low-dose conditions. The specimen was tilted in increments of 1° over a total angular range of −70 to +70 degrees. The total dose accumulated was 140 e−/Å2 per tilt series. A total of 10 tilt series were recorded on a Gatan 2K × 2K charge-coupled device camera at a magnification of 34,000 and a defocus of 6 μm.
Image processing.
Images in each tilt series were aligned using the colloidal gold particles, and tomographic three-dimensional reconstructions were calculated by weighted back-projection, using the IMOD program package (36). Individual tomograms measured 2,048 by 2,048 by 256 pixels, with a 4.2-Å pixel size. Virus particles were selected and windowed in 128-pixel cubes. A strongly low-pass-filtered map of the 160S poliovirus atomic model (31) was used initially to align the subtomograms. Subsequently, the alignment parameters were refined iteratively. After each step, the 80% of the particles that had the highest cross-correlation coefficients were icosahedrally averaged; their sum was used as a reference for the next iteration. During the alignment, artifacts due to the missing wedge were avoided by comparing Fourier coefficients only in those regions of reciprocal space where measurable Fourier coefficients were present (4). (Simple visual comparisons would not have been reliable.) All image processing was carried out using scripts written in SPIDER (22).
The resolution of the final tomographic map was estimated by calculating the Fourier shell correlation (FSC) between two maps, each based on a randomly selected half of the complete set of subtomograms. At a correlation level of 0.5, the resolution was estimated to be 40 Å. We then filtered the map to 45 Å, roughly corresponding to the first zero in the contrast transfer function (at 6.0-μm defocus).
Cryo-electron microscopy.
For single-particle reconstructions, images were recorded using a Gatan cryo-holder on a Tecnai (FEI) F30 microscope at 200 kV, with a magnification of ×59,000 and a dose of 15 to 20 e−/Å2. Images were recorded on Kodak SO-163 films, with an underfocus range of 0.9 to 3.0 μm. Micrographs were scanned using a Zeiss SCAI microdensitometer with a raster size of 7 μm and were binned twice to a final pixel size of 2.32 Å/pixel.
To calculate an asymmetric reconstruction, we chose a subset of 30 particles and computed individual icosahedrally symmetrically averaged maps. We estimated in each case the resolution by FSC (compared with the 45-Å-resolution reconstruction obtained using the entire data set) in order to verify the quality of each icosahedral alignment. Six particles showing a resolution worse than 50 Å were excluded from further analysis. To align the remaining capsids with one another, we chose the single-orientation alternative, out of the 60 symmetry equivalents, that best superimposed the putative RNA densities and then averaged the resulting oriented maps in an asymmetric fashion. In every case, the orientation (selected visually) had yielded 1 of the 3 highest correlation coefficients out of 60.
Single-particle analysis.
Individual virus particles for which RNA was visibly in contact with the capsid were selected using the program Signature (12). In the cryo-EM analysis, images were corrected for the contrast transfer function (CTF) using the program Bshow (29). Orientation search was done by projection matching, using the program PFT2 (1). Reconstructions were performed using a Fourier-Bessel algorithm, as implemented in the program EM3DR (24).
Using these programs, a model-based icosahedrally symmetric reconstruction was calculated and refined by successively increasing the higher-resolution limit of the calculation until no further improvement in the FSC was obtained. In each such iteration, improved alignment parameters were obtained for each boxed image, and thereby, a more accurate density model was obtained from their average.
We calculated an asymmetrical map by assigning to each boxed image 1 of the 60 orientations that were symmetry equivalent to its previously refined icosahedral orientation. These assignments were refined over 100 successive cycles, using the cryo-ET map as an initial reference, and then iteratively, using the previous cycle's average as a reference for each subsequent cycle. The resolution was estimated to be 18 Å for the symmetrized map and 45 Å for the asymmetric map. All image manipulations were carried out using the BSOFT program package (29).
RESULTS AND DISCUSSION
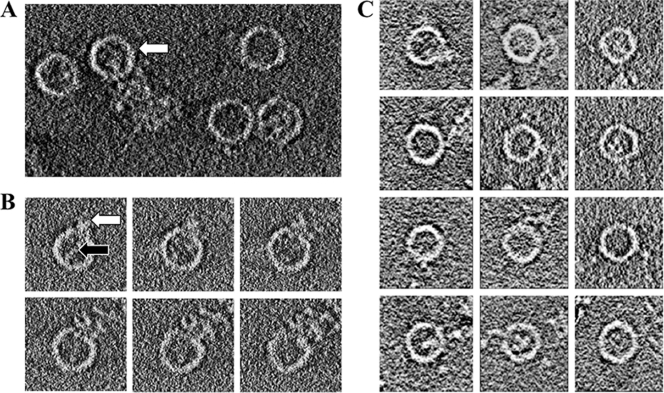
Receptor-induced viral uncoating can be recapitulated in vitro by heating purified virus at 50 to 56°C (37, 40). By all criteria evaluated to date, including proteolysis, antigenicity, and sedimentation properties, the 80S particles produced by heating in this way are identical to the ones that are produced at 37°C during the infection of cells (6, 16, 53). While studying the structure of the 80S end-state empty capsids prepared in this manner by cryo-electron tomography (here) and by cryo-electron microscopy (here and previously [7, 38]), we discovered that our procedure produces at least three different classes of structures, including a class (accounting for as many as 20% of the particles) in which the virions appear to have been caught in the act of RNA release (Fig. 1 A). Thus, several individual particles each clearly show a large string of RNA traversing the capsid shell, while a small amount of RNA density continues to be present inside the capsid (Fig. 1B and C). We should note that the greater proportion of reliably caught-in-the-act particles in the cryo-electron tomography study (20%, versus only 5% in the single-particle cryo-EM projections) is primarily due to our ability to find favorable orientations for viewing three-dimensional tomograms and to section them computationally (as in Fig. 1B). Both samples were prepared in similar ways, so that the higher 20% estimate of caught-in-the-act particles is probably the more reliable estimate of their prevalence in the population.
FIG. 1.
Cryo-electron tomographic data showing the release of RNA from poliovirus. The outer diameter of each particle is about 33 nm. (A) A 10-nm slice through a reconstructed tomogram of frozen hydrated poliovirus 80S particles. Density for RNA is visible on the inside and/or outside of many of the virions. The white arrow indicates a particle associated with RNA at both the capsid interior and exterior and caught in the act of RNA release. (B) A series of horizontal 3-nm-thick slices through a subtomogram of an individual 80S virus particle caught in the act of releasing RNA. Note that the RNA density that is apparent in the particle interior (and indicated by the black arrow in the frame at the upper left) can be traced continuously through successive sections to connect with RNA density on the exterior of the particle (indicated by the white arrow). (C) A gallery of horizontal slices, each obtained from a different subtomogram, showing empty 80S virus particles releasing their RNA.
The amount of RNA density on the inside of the capsids appears to be variable (Fig. 1C) (7, 38), which suggests that RNA release is gradual and can be trapped at several different stages. Indeed, when the amount of interior density was analyzed and plotted for some 10,000 80S virions in the 80Se and 80Sl classes (7, 38), an apparent continuum of interior density values was seen, with no obvious preference for any particular density value. On the interior of the virion, and especially on the exterior, the RNA density appears to be cohesive, knobby, and highly branched (Fig. 1A, B, and C), suggesting the presence of significant secondary structure. We postulate that the need to unwind secondary structure as the RNA traverses the capsid (and subsequent rewinding, once the RNA is on the outside) is responsible for the apparent slowness of the RNA release process and for our ability to trap the RNA in various stages of release, giving rise to various amounts of residual density on the interior. The hypothesis that RNA needs to be unwound while exiting (but refolds once outside, as seen here) is also supported by the double-label fluorescence microscopy studies of Brandenburg et al. (5), tracking viruses in real time in infected cells, showing that Syto 82 dye is lost when RNA is released from the capsid. Importantly, Syto 82 dye is known to bind strongly only to the duplex form of RNA and remains stably bound to isolated viral RNA labeled in vitro, even after extensive dialysis in buffer that lacks the dye. Finally, it should be noted that the in vitro trigger for RNA release (raising the temperature of the sample to 56°C) brings the sample very close to the melting temperature of RNA.
Causing a similar unwinding to occur at physiological temperatures would presumably require a catalyst, and some (as yet unknown) transient specific interaction between the RNA and the capsid protein would be an obvious candidate for providing such a catalyst. From the knobby and cohesive appearance of the RNA density on the interior and exterior (Fig. 1), we surmise that the unwinding process might be most efficient within the stretch of RNA that is currently traversing the capsid and involves an (as yet unidentified) portion of the capsid protein that is kept busy doing something else in the 160S and 135S forms of the virus, prior to the time that RNA release would be favorable for the virus.
Clearly, it would be desirable to better characterize the details of RNA interaction with the capsid protein, particularly concerning the location of the RNA exit site, identification of the aminoacyl residues involved, and ultimately, visualization of the details of the interaction at high resolution. Thus, both to improve the resolution of our images and to determine an icosahedral frame of reference for interpreting them, we apply two different kinds of averaging to our tomographic images: one that can average icosahedrally related densities within the capsid (mostly done for determining an orientation for each tomogram) and another that can average asymmetrically between particles, so as to preserve information about whatever structurally distinguishes the RNA exit site from the remainder of the capsid but is common to the many particles that were caught releasing RNA. Note that determining icosahedral orientations for a subtomogram and subsequently breaking that symmetry by applying a lower-symmetry subset of the symmetry operators is a well-established procedure that we and others have used successfully (4, 8-11, 21, 35, 48).
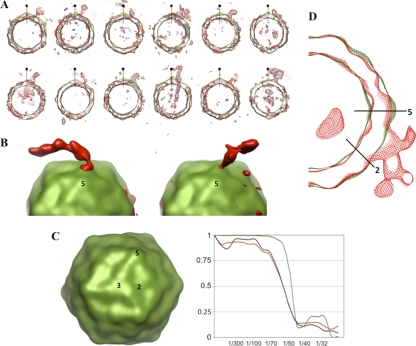
An icosahedrally averaged tomographic reconstruction at approximately 45-Å resolution (Fig. 2 C) was calculated by averaging 200 individual subtomograms selected from 10 complete tilt series. Even at this resolution, the major surface features of the virus, including peaks at the 3-fold and 5-fold axes, are evident. Observe that the averaging process does much to overcome the distortions present in individual subtomograms, including breaks in the capsid density (Fig. 2A) caused by a low signal-to-noise ratio and by the missing wedge of Fourier terms. To visualize the RNA density exiting from the capsid, we selected subtomograms in which the putative RNA features extended from the particle in a direction that was roughly perpendicular to the electron beam, in order to minimize shape distortions caused by the missing Fourier terms and in order to make it less likely that the features were artifacts of the distortion. Several individual tomograms give clear indications of the location of the RNA exit site (Fig. 2A and B). Because those exit positions appear to be consistent with one another from one subtomogram to the next, we believe that they are unlikely to be artifacts of the missing wedge.
FIG. 2.
Superposition of cryo-electron tomographic data on icosahedrally symmetrized capsids. (A) A gallery of tomogram central slices, 5 nm thick, of capsids releasing their RNA (red), each shown superimposed on the corresponding slice of the icosahedrally symmetrized and averaged reconstruction of the capsid (green). Particles were chosen in which the area of RNA exit is not strongly distorted by the missing wedge of Fourier terms. A 5-fold-symmetry axis lying in the plane of the section is indicated in each panel. (B) A surface representation of two virions caught in the process of genome release: icosahedral averaging was applied only to the capsid region (green). An isocontour surface of the RNA density is rendered in red. (C) The icosahedrally symmetrized average of subtomograms, viewed along a 3-fold axis, shows improved resolution. Axes of symmetry are indicated by numbers. Unsurprisingly, its Fourier shell correlation curve (plotted in green at the right) extends to a higher resolution than the FSC curve for the unsymmetrized average of aligned subtomograms (red) or for the icosahedrally symmetrized version of a single subtomogram (black). (D) Central slice through the asymmetric average of multiple subtomograms, showing 80S virions releasing their genomes (red). The icosahedrally symmetrized average of the reconstructions is superimposed (in green). Twofold and 5-fold axes of symmetry are indicated by black lines. FSC curves for these two density maps are plotted in red and green, respectively, in panel C.
Our best assessment of the location of the RNA footprint on the virus surface comes from calculating an asymmetric average of subtomograms (Fig. 2D). The calculation proceeded in two phases. In the first step, icosahedral orientations were determined for each subtomogram, using only the Fourier terms unaffected by the missing wedge to assess alignment, following the procedure described elsewhere (4). The 24 subtomograms with the best icosahedral agreement statistics (and thus the most reliable alignments) were selected and then combined in a second step by choosing the 1 alignment (out of 60 possible symmetry equivalents) for each subtomogram that would optimally superimpose RNA densities. Fourier shell correlation comparisons (Fig. 2C) show that the resolution of the resulting asymmetric average of 24 reconstructions is similar to what is obtained from the 60-fold icosahedral average of an individual subtomogram. The resolution is modest, partly resulting from our need for relatively high defocus in order to visualize RNA unambiguously. Interestingly, at the unique site of RNA release on the capsid (Fig. 2B, left panel, and Fig. 2D), there is some suggestion, from the shape of the density in many particles, that a thick outward projection may be present where the RNA contacts the outer surface of the capsid. Possibly either the shift of some of the capsid proteins outward or the addition or reordering of certain polypeptides, or both, might occur at this site. In the alternative, the RNA itself might have a tendency to form such a thickened structure in the vicinity of the capsid. Higher-resolution experiments will be needed to determine whether the projection is real and, if so, what causes it.
Using similarly prepared samples, micrographs were collected for a single-particle cryo-EM study (38). Out of roughly 10,000 individual boxed particle projections, 540 projections were identified that showed good evidence for having RNA on both the inside and outside of the capsid at the same time. As noted above, the proportion of particles for which the RNA was easily visible was smaller than in the cryo-ET data, even though the method of preparation was the same. Several reasons can explain this observation. In the higher-resolution cryo-EM experiment, both the lower defocus and lower electron dose led to reduced contrast, making reliable identification of RNA more difficult. Additionally, in a single projected image, the structure is visible from only a single viewpoint and the direction of RNA exit cannot be established unambiguously. In the end, we used a set of 540 boxed projections of virus particles that appeared to be in the process of releasing their RNA.
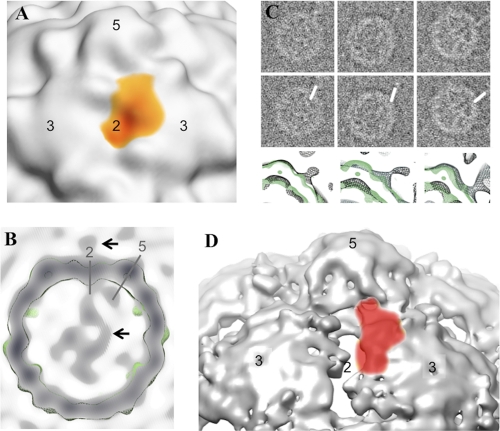
As in the cryo-ET reconstruction, image averaging from the untilted cryo-EM data proceeded in two stages. In the first step, an 18-Å-resolution icosahedrally symmetrized map was calculated from 540 boxed particles. During that process, alignment parameters were obtained for each particle image, expressed in an icosahedral frame of reference. In the second step, an iterative process, we broke the icosahedral symmetry by choosing the best-scoring orientation among the 60 symmetry-equivalent choices, as judged by their correlations with earlier and more approximate versions of the same asymmetric map. To make the automatic determination of orientation parameters more robust, the relatively weak density signal from external RNA could be enhanced artificially (Fig. 3C), following the procedure described elsewhere (9). The resulting cryo-EM map, at 45-Å resolution and calculated from unmodified images, confirmed the features observed in the cryo-ET study, including the site of RNA egress (Fig. 3B and Fig. 3C, bottom row of panels).
FIG. 3.
RNA release by 80S poliovirus particles, as determined by single-particle cryo-electron microscopy. (A) Cryo-EM reconstruction of 80S poliovirus showing the footprint of the density that corresponds to RNA: red indicates a higher contour level and yellow indicates a lower contour level. Axes of symmetry are indicated by numbers. (B) A central slice from an asymmetrical cryo-EM reconstruction, which was calculated from 540 boxed single-particle projections of poliovirus 80S empty capsids during RNA release. An isocontour level of the icosahedrally symmetrized reconstruction is shown superimposed in green. The 2-fold and 5-fold axes of symmetry are indicated by gray lines. Black arrows indicate RNA associated with the capsid exterior (upper arrow) and with the capsid interior (lower arrow). (C) Asymmetric alignment of particles was accomplished in stages. First, the 540 boxed projections were selected because they appeared to show RNA on both the interior and exterior of the capsid (panel C, top row). Second, unbiased alignment parameters were obtained for those projections, as usual, in an icosahedral frame of reference. Third, following the method of Bubeck et al. (9), putative RNA density was reinforced artificially (middle row). By applying the unbiased orientation parameters to the modified images, it was then possible to calculate an icosahedrally symmetrized reconstruction showing the location of the RNA footprint on the outer surface of the virus. Finally, the artificial reinforcement of RNA density makes it easy to determine which 1 of 60 symmetry-equivalent orientations to assign to each unmodified image, when reconstructing their asymmetric average (bottom row, black contour). The icosahedrally symmetrized capsid (green contour) is superimposed for comparison. (D) For comparison, the footprint of the putative RNA density from the asymmetrically averaged cryo-tomographic reconstruction is shown as a red semitransparent surface, superimposed on a portion of the icosahedrally symmetrized late 80S capsid at 10-Å resolution, as determined previously (38). Axes of symmetry are numbered. At the chosen contour, holes near the 2-fold axis are evident and partly covered by the putative RNA footprint. Note that the footprint is large, relative to the width of unfolded RNA.
Contrary to numerous speculations in the literature (2, 3, 7, 8, 25, 27, 28, 30, 42, 45), the RNA does not exit the capsid from a channel at the 5-fold axis. Instead, the footprint of the RNA density on the outer surface of the capsid is situated above a point near where VP1 and VP2 from one protomer interact with VP3 of a neighboring protomer (Fig. 3A and D). This point lies roughly along a line joining a particle 2-fold axis to a nearby particle 5-fold axis, closer to the 2-fold axis by a ratio of about 1:3 (Fig. 2B). This places the center of the RNA density footprint about 20 Å away from the 2-fold axis, immediately above an opening in the capsid (i.e., a low-density region) seen in the icosahedrally averaged reconstructions of the 80S particles at ∼10-Å resolution (38) (Fig. 3D).
The high-resolution icosahedrally symmetrized cryo-EM reconstructions identified two classes of 80S structures, one corresponding to an earlier-occurring intermediate and the other to a later-occurring one. In the earlier-occurring 80Se class, capsid protein rearrangements (relative to the 160S and 135S forms of the virus) produce significant thinning of the shell (at the site of the RNA footprint seen here), including small holes in the vicinity of the 2-fold axis and between 5-fold related protomers. In the later-occurring 80Sl class, the holes in the atomic model are larger (15 Å across at the 2-fold axis) and are thus easily large enough to permit single-stranded RNA (ssRNA) to pass through the capsid, provided that it is unwound (38). Importantly, our current asymmetric reconstructions show that these holes lie within the unique RNA footprint on the surface. Appropriately oriented slices through the asymmetric reconstruction also show a surprising amount of connected branched density for the RNA on the inside of the virion immediately below the contact point for the externalized RNA (Fig. 1 and 3B), which suggests that the RNA remaining inside the capsid has a significant level of structure that occurs consistently among the particles contributing to the reconstruction. The proximity of the contact points on the inner surface and the outer surface of the capsid would suggest that the RNA is passing directly through holes located at the site.
Although the resolution of the current asymmetric reconstructions is relatively low, RNA exiting from the capsid of a nonenveloped virus has never been visualized before, and the reconstructions permit us to come to some tentative conclusions about RNA egress. RNA egress takes place near one of the viral 2-fold axes, via a weak spot in the capsid (Fig. 3D), and probably involves a local asymmetric rearrangement of the capsid protein that is different from what occurs elsewhere in the capsid. Due to the low resolution of the reconstructions, this information about the location of the RNA footprint on the capsid surface (Fig. 3A and D) is much more reliable than any structural details seen at the exit site. There appears to be enlargement of the RNA-related density near the outer surface of the capsid, as seen in the cryo-ET and cryo-EM reconstructions, viewed either in profile or as a density footprint, but at the current resolution, we have to regard this finding as suggestive but not proven. Previous evidence on dye release from the double-labeling light microscopy experiments (5) indicates that the RNA is folded when inside the particle, exits the capsid gradually as ssRNA, and refolds after being released. From those results, we infer that the interaction between RNA and capsid protein sequences is likely to be structurally specific, as the localized unwinding of RNA at physiological temperatures probably needs to be catalyzed. Further, the variable levels of RNA density seen inside and out are consistent with a model in which RNA release is initiated and then pauses repeatedly at a number of points where unfolding of the secondary and/or tertiary structure is more difficult. Although it is still unclear how the process is initiated during normal infection, the unfolding of the RNA as it is externalized, followed by refolding once it is outside the capsid, provides a ratchet-like mechanism that could drive externalization and perhaps even translocation of the genome across a cellular membrane (most likely the membrane of an endocytic vesicle [5]).
It should also be recognized that the picture of RNA release seen here is only half complete, since these isolated 80S particles release their RNA into solution, rather than helping the RNA to traverse the endosomal membrane, as they would in a natural infection. Indeed, experiments using either equine rhinitis A virus or poliovirus have shown that the presence of RNase in endosomes does not impair infectivity (E. Groppeli and D. Rowlands, personal communication). That result indicates that RNA, leaving the virus, usually proceeds directly across either a pore or a breach in the membrane, without having first been dumped into a closed endosomal cavity. Additionally, fluorescence microscopy experiments showed that once poliovirions have been internalized in the endosomes of infected cells (a process that requires a conformational change to occur in the capsid), the process of RNA transfer to the cytoplasm is efficient, with virtually every internalized virus particle losing its noncovalent RNA label (5). Together, those two results imply that RNA release does not occur at physiological temperatures until after a continuous channel has been created that leads from the interior of the virion, through the membrane, and into the cytoplasm. Hypothetically, if externalized viral peptides do form a distinct pore structure in the membrane, then the RNA exit complex (visualized here as an asymmetric structural feature) would have to dock with the pore prior to RNA release. Alternatively, the asymmetric RNA exit structure could, itself, constitute the membrane-breaching or membrane-spanning structure.
It is unclear how one end of the ssRNA genome picks a spot on the inner surface of the capsid to use as an exit portal (out of the 60 symmetry equivalent sites), assuming that there are no as-yet-unknown distinctions between the proteins. It is also unclear how the virus is smart enough to consistently pick an exit direction that points toward the membrane. Admittedly, interaction of part of the capsid with a component of the membrane could predispose that capsid site to becoming the RNA exit portal. But clearly, the presence of membrane is not essential, as heating the virus is sufficient to induce RNA release. The current experiment, visualizing RNA release, does present an alternative hypothesis that is worth considering. As we noted, electron density for a thickened projection from the outer viral surface was repeatedly seen near the point where RNA emerges from the capsid and (assuming that the projection is real) obviously forms in the absence of cellular membranes. It would be interesting if this projecting structure happens to surround the RNA, requires the presence of the free end of the RNA for its formation, includes sequences from VP4 and/or the amino terminus of VP1, and is directly involved in the breaching of the membrane. If this speculation is true, it would provide an attractively simple explanation for why RNA never comes out on the wrong (membrane-distal) side of the virion.
Acknowledgments
We thank Sriram Subramaniam and Alex Chou for assistance with the cryo-ET data collection on the Polara microscope at NCI/NIH. We also thank Niko Grigorieff and Chen Xu at Brandeis University for access to their F30 microscope and for assistance with the cryo-EM data acquisition.
This work was supported by NIH grants RO1 AI20566 (to J.M.H.) and F32 AI081427 (to H.L.).
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 2.Belnap, D. M., D. J. Filman, B. L. Trus, N. Cheng, F. P. Booy, J. F. Conway, S. Curry, C. N. Hiremath, S. K. Tsang, A. C. Steven, and J. M. Hogle. 2000. Molecular tectonic model of virus structural transitions: the putative cell entry states of poliovirus. J. Virol. 74:1342-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. U. S. A. 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bostina, M., D. Bubeck, C. Schwartz, D. Nicastro, D. J. Filman, and J. M. Hogle. 2007. Single particle cryoelectron tomography characterization of the structure and structural variability of poliovirus-receptor-membrane complex at 30 Å resolution. J. Struct. Biol. 160:200-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandenburg, B., L. Y. Lee, M. Lakadamyali, M. J. Rust, X. Zhuang, and J. M. Hogle. 2007. Imaging poliovirus entry in live cells. PLoS Biol. 5:e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breindl, M. 1971. The structure of heated poliovirus particles. J. Gen. Virol. 11:147-156. [DOI] [PubMed] [Google Scholar]
- 7.Bubeck, D., D. J. Filman, N. Cheng, A. C. Steven, J. M. Hogle, and D. M. Belnap. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745-7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bubeck, D., D. J. Filman, and J. M. Hogle. 2005. Cryo-electron microscopy reconstruction of a poliovirus-receptor-membrane complex. Nat. Struct. Mol. Biol. 12:615-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bubeck, D., D. J. Filman, M. Kuzmin, S. D. Fuller, and J. M. Hogle. 2008. Post-imaging fiducial markers aid in the orientation determination of complexes with mixed or unknown symmetry. J. Struct. Biol. 162:480-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardone, G., D. C. Winkler, B. L. Trus, N. Cheng, J. E. Heuser, W. W. Newcomb, J. C. Brown, and A. C. Steven. 2007. Visualization of the herpes simplex virus portal in situ by cryo-electron tomography. Virology 361:426-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, J. T., M. F. Schmid, F. J. Rixon, and W. Chiu. 2007. Electron cryotomography reveals the portal in the herpesvirus capsid. J. Virol. 81:2065-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J. Z., and N. Grigorieff. 2007. SIGNATURE: a single-particle selection system for molecular electron microscopy. J. Struct. Biol. 157:168-173. [DOI] [PubMed] [Google Scholar]
- 13.Chow, M., J. F. Newman, D. Filman, J. M. Hogle, D. J. Rowlands, and F. Brown. 1987. Myristylation of picornavirus capsid protein VP4 and its structural significance. Nature 327:482-486. [DOI] [PubMed] [Google Scholar]
- 14.Colston, E., and V. R. Racaniello. 1994. Soluble receptor-resistant poliovirus mutants identify surface and internal capsid residues that control interaction with the cell receptor. EMBO J. 13:5855-5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coyne, C. B., K. S. Kim, and J. M. Bergelson. 2007. Poliovirus entry into human brain microvascular cells requires receptor-induced activation of SHP-2. EMBO J. 26:4016-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curry, S., M. Chow, and J. M. Hogle. 1996. The poliovirus 135S particle is infectious. J. Virol. 70:7125-7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danthi, P., M. Tosteson, Q. H. Li, and M. Chow. 2003. Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J. Virol. 77:5266-5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sena, J., and B. Mandel. 1977. Studies on the in vitro uncoating of poliovirus. II. Characteristics of the membrane-modified particle. Virology 78:554-566. [DOI] [PubMed] [Google Scholar]
- 19.Fenwick, M. L., and P. D. Cooper. 1962. Early interactions between poliovirus and ERK cells: some observations on the nature and significance of the rejected particles. Virology 18:212-223. [DOI] [PubMed] [Google Scholar]
- 20.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fokine, A., V. A. Kostyuchenko, A. V. Efimov, L. P. Kurochkina, N. N. Sykilinda, J. Robben, G. Volckaert, A. Hoenger, P. R. Chipman, A. J. Battisti, M. G. Rossmann, and V. V. Mesyanzhinov. 2005. A three-dimensional cryo-electron microscopy structure of the bacteriophage phiKZ head. J. Mol. Biol. 352:117-124. [DOI] [PubMed] [Google Scholar]
- 22.Frank, J., M. Radermacher, P. Penczek, J. Zhu, Y. Li, M. Ladjadj, and A. Leith. 1996. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116:190-199. [DOI] [PubMed] [Google Scholar]
- 23.Fricks, C. E., and J. M. Hogle. 1990. Cell-induced conformational change in poliovirus: externalization of the amino terminus of VP1 is responsible for liposome binding. J. Virol. 64:1934-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuller, S. D., S. J. Butcher, R. H. Cheng, and T. S. Baker. 1996. Three-dimensional reconstruction of icosahedral particles—the uncommon line. J. Struct. Biol. 116:48-55. [DOI] [PubMed] [Google Scholar]
- 25.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 26.Harrison, S. C. 1980. Protein interfaces and intersubunit bonding. The case of tomato bushy stunt virus. Biophys. J. 32:139-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewat, E. A., and D. Blaas. 2004. Cryoelectron microscopy analysis of the structural changes associated with human rhinovirus type 14 uncoating. J. Virol. 78:2935-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hewat, E. A., E. Neumann, and D. Blaas. 2002. The concerted conformational changes during human rhinovirus 2 uncoating. Mol. Cell 10:317-326. [DOI] [PubMed] [Google Scholar]
- 29.Heymann, J. B., and D. M. Belnap. 2007. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157:3-18. [DOI] [PubMed] [Google Scholar]
- 30.Hogle, J. M. 2002. Poliovirus cell entry: common structural themes in viral cell entry pathways. Annu. Rev. Microbiol. 56:677-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogle, J. M., M. Chow, and D. J. Filman. 1985. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science 229:1358-1365. [DOI] [PubMed] [Google Scholar]
- 32.Hogle, J. M., A. Maeda, and S. C. Harrison. 1986. Structure and assembly of turnip crinkle virus. I. X-ray crystallographic structure analysis at 3.2 Å resolution. J. Mol. Biol. 191:625-638. [DOI] [PubMed] [Google Scholar]
- 33.Huang, Y., J. M. Hogle, and M. Chow. 2000. Is the 135S poliovirus particle an intermediate during cell entry? J. Virol. 74:8757-8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobson, M. F., and D. Baltimore. 1968. Polypeptide cleavages in the formation of poliovirus proteins. Proc. Natl. Acad. Sci. U. S. A. 61:77-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang, W., J. Chang, J. Jakana, P. Weigele, J. King, and W. Chiu. 2006. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature 439:612-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kremer, J. R., D. N. Mastronarde, and J. R. McIntosh. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71-76. [DOI] [PubMed] [Google Scholar]
- 37.Le Bouvier, G. L. 1955. The modification of poliovirus antigens by heat and ultraviolet light. Lancet 269:1013-1016. [DOI] [PubMed] [Google Scholar]
- 38.Levy, H. C., M. Bostina, D. J. Filman, and J. M. Hogle. 2010. Catching a virus in the act of RNA release: a novel poliovirus uncoating intermediate characterized by cryo-electron microscopy. J. Virol. 84:4426-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Macadam, A. J., G. Ferguson, C. Arnold, and P. D. Minor. 1991. An assembly defect as a result of an attenuating mutation in the capsid proteins of the poliovirus type 3 vaccine strain. J. Virol. 65:5225-5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer, M. M., H. J. Rapp, B. Roizman, S. W. Klein, K. M. Cowan, and D. Lukens. 1957. The purification of poliomyelitis virus as studied by complement fixation. J. Immunol. 78:435-455. [PubMed] [Google Scholar]
- 41.Mendelsohn, C. L., E. Wimmer, and V. R. Racaniello. 1989. Cellular receptor for poliovirus: molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 56:855-865. [DOI] [PubMed] [Google Scholar]
- 42.Racaniello, V. R. 2007. Picornaviridae: the viruses and their replication, p. 795-838. In D. M. Knipe, B. Roizman, P. B. Howley, S. E. Strauss, and D. E. Griffin (ed.), Field's virology, vol. 1, 5th ed. Lippincott, Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 43.Robinson, I. K., and S. C. Harrison. 1982. Structure of the expanded state of tomato bushy stunt virus. Nature 297:563-568. [Google Scholar]
- 44.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, et al. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature 317:145-153. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G., J. Bella, P. R. Kolatkar, Y. He, E. Wimmer, R. J. Kuhn, and T. S. Baker. 2000. Cell recognition and entry by rhino- and enteroviruses. Virology 269:239-247. [DOI] [PubMed] [Google Scholar]
- 46.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of encephalomyocarditis (EMC) virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 47.Speir, J. A., S. Munshi, G. Wang, T. S. Baker, and J. E. Johnson. 1995. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure 3:63-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang, J., N. Olson, P. J. Jardine, S. Grimes, D. L. Anderson, and T. S. Baker. 2008. DNA poised for release in bacteriophage phi29. Structure 16:935-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tosteson, M. T., and M. Chow. 1997. Characterization of the ion channels formed by poliovirus in planar lipid membranes. J. Virol. 71:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tosteson, M. T., H. Wang, A. Naumov, and M. Chow. 2004. Poliovirus binding to its receptor in lipid bilayers results in particle-specific, temperature-sensitive channels. J. Gen. Virol. 85:1581-1589. [DOI] [PubMed] [Google Scholar]
- 51.Tuthill, T. J., D. Bubeck, D. J. Rowlands, and J. M. Hogle. 2006. Characterization of early steps in the poliovirus infection process: receptor-decorated liposomes induce conversion of the virus to membrane-anchored entry-intermediate particles. J. Virol. 80:172-180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tuthill, T. J., E. Groppelli, J. M. Hogle, and D. J. Rowlands. Picornaviruses. Curr. Top. Microbiol. Immunol. 2100:43-89. [DOI] [PMC free article] [PubMed]
- 53.Wetz, K., and T. Kucinski. 1991. Influence of different ionic and pH environments on structural alterations of poliovirus and their possible relation to virus uncoating. J. Gen. Virol. 72(Pt 10):2541-2544. [DOI] [PubMed] [Google Scholar]
- 54.Wien, M. W., S. Curry, D. J. Filman, and J. M. Hogle. 1997. Structural studies of poliovirus mutants that overcome receptor defects. Nat. Struct. Biol. 4:666-674. [DOI] [PubMed] [Google Scholar]
- 55.Xing, L., K. Tjarnlund, B. Lindqvist, G. G. Kaplan, D. Feigelstock, R. H. Cheng, and J. M. Casasnovas. 2000. Distinct cellular receptor interactions in poliovirus and rhinoviruses. EMBO J. 19:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, P., S. Mueller, M. C. Morais, C. M. Bator, V. D. Bowman, S. Hafenstein, E. Wimmer, and M. G. Rossmann. 2008. Crystal structure of CD155 and electron microscopic studies of its complexes with polioviruses. Proc. Natl. Acad. Sci. U. S. A. 105:18284-18289. [DOI] [PMC free article] [PubMed] [Google Scholar]