Abstract
C1s is the highly specific modular serine protease that mediates the proteolytic activity of the C1 complex and thereby triggers activation of the complement cascade. The crystal structure of a catalytic fragment from human C1s comprising the second complement control protein (CCP2) module and the chymotrypsin-like serine protease (SP) domain has been determined and refined to 1.7 Å resolution. In the areas surrounding the active site, the SP structure reveals a restricted access to subsidiary substrate binding sites that could be responsible for the narrow specificity of C1s. The ellipsoidal CCP2 module is oriented perpendicularly to the surface of the SP domain. This arrangement is maintained through a rigid module–domain interface involving intertwined proline- and tyrosine-rich polypeptide segments. The relative orientation of SP and CCP2 is consistent with the fact that the latter provides additional substrate recognition sites for the C4 substrate. This structure provides a first example of a CCP–SP assembly that is conserved in diverse extracellular proteins. Its implications in the activation mechanism of C1 are discussed.
Keywords: complement/modular structure/molecular recognition/serine protease/substrate specificity
Introduction
The complement cascade is a major system of innate immunity against pathogenic micro-organisms in mammals and in other vertebrate species. Activation of the classical pathway of complement is mediated by C1, a 790 000 Da macromolecular complex comprising three components: C1q, C1r and C1s. C1q is a hexamer of collagen-rich heterotrimers that has the overall shape of a bunch of tulips and internally binds two modular proteases C1r and C1s. The binding of C1q to the target micro-organism leads to autolytic activation of C1r, which, in turn, activates C1s by cleaving the Arg426–Ile427 bond (Cooper, 1985; Arlaud et al., 1987, 1998). In contrast with the low specificity of the initial recognition step mediated by C1q, C1s restrictively cleaves at a single arginyl peptide bond in both complement C4 and C2 proteins, the natural substrates of C1. A cascade of subsequent proteolytic reactions results in various biological activities designed to provide a first line of defense against microbial infection. Another facet of the protective action of complement lies in its role in immune tolerance, as exemplified by its involvement in the clearance of apoptotic cells (Mevorach et al., 1998) and in graft rejection (Dalmasso, 1992). Uncontrolled side-effects of complement may also result in various autoimmune pathologies, including Alzheimer’s disease (Rogers et al., 1992).
C1s comprises, starting from the N-terminus, a CUB module (Bork and Beckmann, 1993), an epidermal growth factor (EGF)-like module, a second CUB module, two complement control protein (CCP, also known as sushi and SCR) modules (Mackinnon et al., 1987; Tosi et al., 1987) and a C-terminal chymotrypsin-like serine protease (SP) domain. This modular architecture is also found in C1r (Arlaud et al., 1998) and in the mannan-binding lectin-associated serine proteases (MASPs), a family of proteolytic enzymes involved in the activation of the recently discovered ‘lectin pathway’ of complement activation (Sato et al., 1994; Thiel et al., 1997; Endo et al., 1998).
Insights into the molecular architecture and functional role of the catalytic region of C1s have been obtained only in the past few years. Studies based on chemical cross-linking and homology modeling strongly suggested the occurrence of a close interaction between the second CCP module and the SP domain in both C1s and C1r (Rossi et al., 1995; Lacroix et al., 1997). A similar conclusion (Gaboriaud et al., 1998) was reached from a comparative analysis of amino acid sequence variability in the family of proteins containing the CCP–SP motif found in species ranging from invertebrates to mammals (Tosi et al., 1989; Muta et al., 1991; Matsushita et al., 1998). Studies carried out with recombinant modular fragments from the C1s catalytic region indicated that although C2 cleavage and reactivity towards C1 inhibitor are mediated mainly by the SP domain, efficient C4 cleavage requires substrate binding sites located in both CCP modules (Rossi et al., 1998).
CCP modules are known to be involved in many recognition processes, including the binding of several complement factors to fragments C3b and C4b (Reid and Day, 1989). Five CCP module structures have been determined by NMR spectroscopy: H5, the H15–H16 pair from human complement factor H (Barlow et al., 1992, 1993) and the VC3–VC4 module pair from the vaccinia virus complement control protein (VCP) (Wiles et al., 1997). More recently, the 3 Å resolution X-ray structure of the N-terminal pair of CCP modules from the complement regulatory protein CD46 has also been reported (Casasnovas et al., 1999). Structural studies performed on pairs of CCP modules indicate that these may exhibit quite different inter-modular orientations and show varying degrees of flexibility at the interface between the two modules (Barlow et al., 1993; Wiles et al., 1997; Casasnovas et al., 1999; Kirkitadze et al., 1999). As discussed below, this flexibility may be relevant for the activation and activity of the proteases of C1.
There is relatively little structural information concerning the proteases of the complement system. Pioneering studies in this field have been performed on factor D, providing a structural basis for the unique activation and control mechanisms of this chymotrypsin-like serine protease (Volanakis and Narayana, 1996; Jing et al., 1999). The present study describes the first three-dimensional (3D) structure of an activated catalytic fragment from a modular protease of the complement system and provides information that can be used as a basis for further studies of the structure–function relationships of C1s as well as other modular proteins of the CCP–SP family.
Results and discussion
We have determined the structure of a recombinant fragment from the catalytic domain of human C1s, consisting of the CCP2 module (residues 342–406) linked to the C-terminal chymotrypsin-like SP domain (residues 410–668). The structure was solved with the combined use of molecular replacement (MR) and automatic model building (WARP) and refined at 1.7 Å resolution (see Materials and methods). The final Rwork and Rfree factors are 0.185 and 0.220, respectively, and the stereochemistry of the model is of good quality (Table I).
Table I. Final refinement and model geometry statistics.
| Protein residues | 303 |
| Protein atoms | 2284 |
| Carbohydrate residues | 3 |
| Waters/ions/buffer | 320/2 sulfates/1TES |
| Resolution (Å) | 15–1.7 |
| Rwork/No. of observations | 0.185/36847 |
| Rfree/No. of observations | 0.220/1969 |
| Residues in dual conformations | 10a |
| Residues with disordered side chains after Cβ | 14b |
| R.m.s. bond lengths (Å) | 0.011 |
| R.m.s. bond angle associated distances (Å) | 0.024 |
| R.m.s. planarity (Å) | 0.016 |
aSer349, Ser365, Cys371, Val395, Cys410, Thr469, Ser474, Cys580, Cys613, Lys614.
bGlu351, Glu356, Asp357, Glu359, Glu373, Glu385, Glu397, Glu402, Arg414, Glu506, Asp541, Met545, Lys629, Gln665.
Structure of the SP domain
As in other trypsin-like serine proteinases, the core of the C-terminal domain of C1s folds into two six-stranded β-barrels connected by three trans-segments, several surface loops and one C-terminal α-helix (Figures 1A and 2A). The catalytic residues Ser617(195), His460(57) and Asp514(102) (C1s numbering, followed by chymotrypsinogen numbering in brackets) are located at the junction of both barrels in a manner virtually identical to the one observed in trypsin. C1s also presents all the other key structural features of the active conformation (for a review, see Perona and Craik, 1997). Asp611(189), the well-established determinant of the specificity of cleavage after Arg and/or Lys residues, lies at the bottom of the primary specificity pocket (S1). A putative sulfate ion is found at the active site, where three of its oxygen atoms form hydrogen bonds with the active Ser617(195) Oγ (2.68 Å), the His460(57) Nε2 (2.84 Å) and the Lys614(192) Nζ (2.9 Å), respectively.
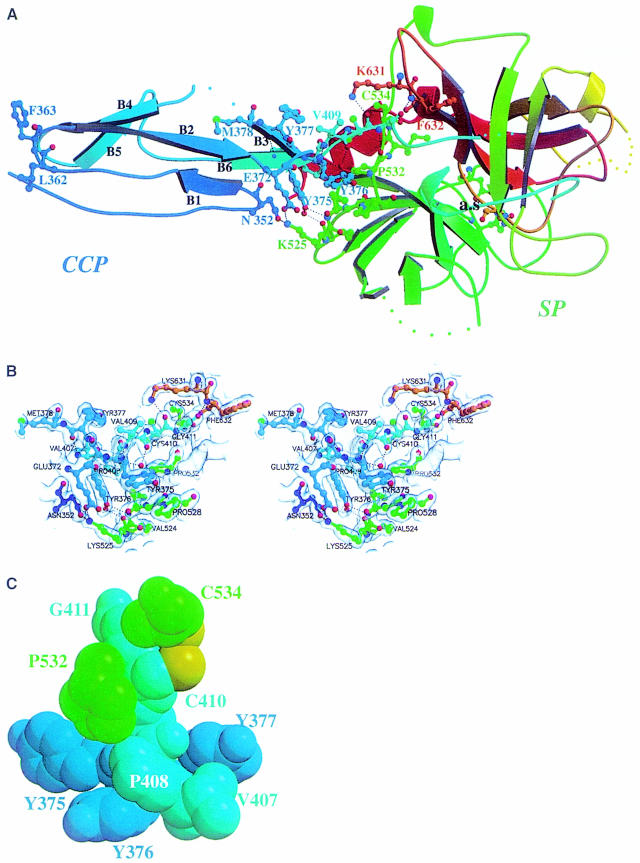
Fig. 1. Three-dimensional structure of the C1s CCP module–SP domain assembly. (A) Overall structure. The CCP2 module (residues 342–406, in blue) is held in a perpendicular position relative to the SP domain (residues 410–668) through a rigid interface. A sequence-ramping color scheme has been used, going from blue (N-terminus) to red (C-terminus). The short 407–410 CCP2–SP intersubunit segment lies on the back of the molecule (cyan). The catalytic triad at the active site (a.s.) is shown at the bottom of the molecule. The hydrogen bonding network at the interface is depicted by dotted lines. The exposed Leu362 and Phe363, located near the N-terminal end of the CCP2 module, are shown as ball-and-stick side chains. Colored dots represent residues not defined in the model. The numbering used is that of C1s. (B) Closer stereo view of the interface, with the corresponding (2Fo – Fc) electron density map contoured at the 1σ level. (C) CPK representation of the assembly of segments 375–377, 407–411 and residues 532 and 534, involved in the interface. The intersubunit segment 407–411 (cyan) is rigidly maintained from both sides. All but two of these residues are highly conserved in the CCP–SP family (Gaboriaud et al., 1998). The two variable positions (Y377 and V407 in C1s) face each other at the periphery of the interface. Tyr376, Pro532(120), Cys534(122), Pro408 (Ala in haptoglobin and related sequences), Val409 (Ile in the ascidian MASP sequences), Cys410(1) and Gly411(2) are fully conserved except for the cases mentioned in parentheses. The aromatic residue at position 375 is also well conserved, with nine Tyr, two Trp and three Phe found in homologous proteins (Gaboriaud et al., 1998). The interchain disulfide bridge Cys410(1)–Cys534(122) is not observed in the X-ray structure probably because of radiation damage; however, the cysteine side chains are displayed here in a conformation compatible with disulfide bridge formation (sulfur atoms are shown in yellow).
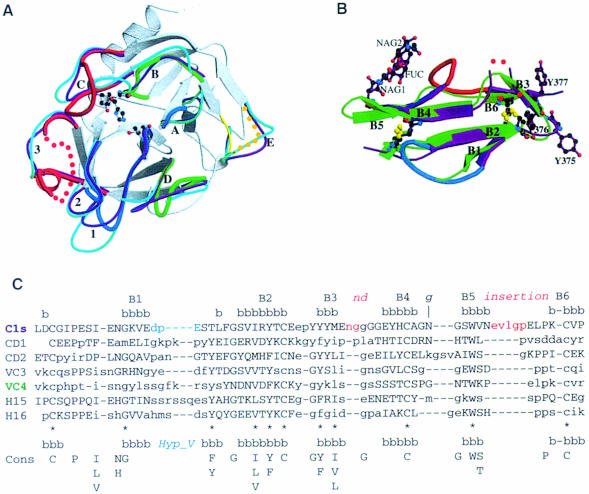
Fig. 2. Comparison of the C1s SP domain (A) and the C1s CCP2 module (B and C) with the 3D structures of homologous proteins. (A) Superimposition of the structures of the SP domains of C1s, human trypsin (magenta), and human thrombin (cyan). The structures are shown in the ‘standard’ orientation, i.e. with the active-site cleft facing the viewer and the substrate binding site running from left to right. The catalytic triad residues of C1s are shown with ball-and-sticks (distances: Ser Oγ–His Nε2 = 2.86 Å; His Nδ1–Asp Oδ2 = 2.74 Å). The common core is shown in white, and only the variable surface loops around the active site are highlighted. The C1s loops are thicker and color-coded red for major insertions, blue for deletions, and green for other significant modifications. Dots represent disordered segments. The loops are labeled as in Perona and Craik (1997): A, 34–41; B, 56–64; C, 97–103; D, 143–149; E, 74–86; 1, 185–188; 2, 217–225; 3, 169–175. (B) Superimposed structures of the CCP modules from C1s (magenta) and CD46-2 (green). The two insertions in the C1s structure are shown in red, the deletion in blue. Red dots represent the disordered Gly–Gly stretch. The three carbohydrate residues observed in the structure are also displayed. The disulfide bridges from the two CCPs are superimposed. (C) Structural alignment of the CCP modules. The secondary structure (bbb) and strand numbering in the C1s structure are shown above. Pairs of residues with Cα–Cα distances of <2.0 Å are shown in upper case, the other residues in lower case. The deletion–insertion color code is the one used in (B). The consensus sequence (Cons), and the positions of the eight commonly observed β-strands (bbb) depicted above it, are taken from Wiles et al. (1997). The 11 consensus residues used for the core superposition (see Table III) are indicated by *. The carbohydrate attachment site is indicated by g.
The surface segments of C1s that differ in conformation relative to the other SPs of known 3D structure are listed in Table II. In the mammalian SPs, fine substrate specificity is determined by surface loops 1–3 and A–E (Perona and Craik, 1997). C1s displays unique structural features in all of these loops (Table IIB and Figure 2A). The two major insertions in loops 3 and C are grouped on one side of the entrance of the active site, whereas the deletions in loops 1, 2 and A are on the opposite side (Figures 2A and 3). Disordered conformations are observed in loops 3 and E at both ends of the specific substrate binding region.
Table II. Structural comparisons of the C1s SP domain with homologous X-ray structures.
| A Overall structure | |||||
| Chymotrypsin | Thrombin | Factor Xa | Trypsin | Protein C | |
| Global r.m.s. (Å)/nb Ca | 1.0/194 | 0.8/182 | 1.0/188 | 0.8/183 | 1.0/194 |
| Sequence identity (%) | 37 | 38 | 35 | 36 | 37 |
| PDB code | 3gch | 1fpc | 1fax | 1trn | 1aut |
| B Surface segments of the C1s SP domain significantly different (d >1.5 Å) from the homologous segments of other SP domains (chymotrypsin,trypsin, thrombin, factor VII, factor D, factor Xa, and factor IXa) | |||||
| C1s numbering |
Chymotrypsin numbering |
Loop labela |
Type of modification |
| 430–431 | 23–24 | same length | |
| 441–443 | 34–40 | A | deletion |
| 466–467 | 61–62 | B | variable |
| 501–509 | 96–98 | C | major insertion |
| 538–543 | 126–129 | minor insertion | |
| 560–562 | 145–146 | D | variable |
| 577–596 | 165–175 | 3 | major insertion |
| 607–608 | 185–187 | 1 | deletion |
| 625–630 | 203–205 | minor insertion | |
| 646 | 221–224 | 2 | deletion |
aLoop labels as defined by Perona and Craik (1997) and detailed in Figure 2A. Loop E is not defined in C1s structure.
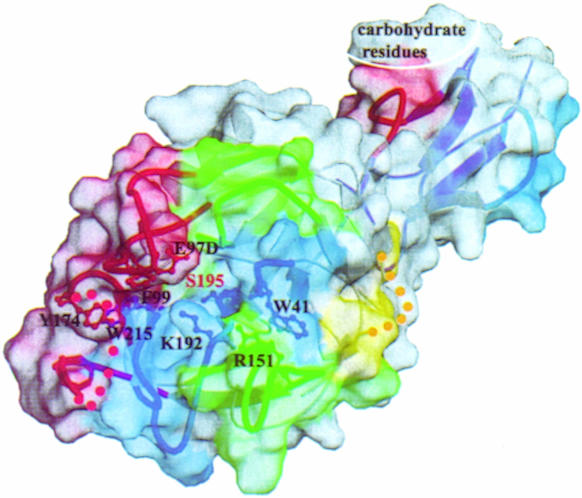
Fig. 3. Specific surface features around the catalytic site and substrate binding sub-sites of the C1s catalytic domain. The surface and the underlying loops have been colored using the same code. Also shown are the side chains of residues restricting the access of the substrate (see text) and of the active site Ser195. The CCP module constitutes an extension of the SP domain substrate binding site. The molecule is shown in the same orientation as in Figure 2A. The chymotrypsinogen residue numbering has been used throughout.
The C-terminal segment 669–673 is flexible, as observed previously in the NMR structural analysis of the C-terminal segments 653–673 and 656–673 of C1s (Gans et al., 1998). The disorder observed at the C-terminus of the cleaved activation peptide (418–422) and in the loop homologous to the trypsin calcium binding loop (478–486) are common features of SP domain structures. The salt bridges between Glu418 and Lys608, and between Glu672 and Lys405, previously identified by chemical cross-linking (Rossi et al., 1995), are not observed in the crystal structure. This is because these glutamic acid residues are located at the two disordered C-termini. This suggests that these segments have alternative configurations, one of which was trapped by the chemical reaction. Four cysteines (371, 410, 580, 613) are observed with dual conformations, and the inter-chain disulfide bridge (Cys410–Cys534) is not formed, most likely because of radiation damage (Burmeister, 2000). However, no conformational change is required to bridge cysteines 410 and 534.
Structure of the CCP2 module
As with other CCP modules, CCP2 folds into a small and compact hydrophobic core enveloped by six β-strands and stabilized by four conserved cysteines forming disulfide bridges in a 1–3, 2–4 pattern (Figures 1A, 2B and C). There are overall lower structural similarities among the CCP modules (Table III) than among the SP domains (Table II). Unexpectedly, the VCP CCP3 module has the least similar structure relative to C1s CCP2, despite it having the most homologous amino acid sequence (Table III). Structurally, the CD46 CCP2 module is the closest to C1s CCP2 (Table III; Figure 2B). As shown in Figure 2C, the relative structural orientation of the B2 and B4 strands is shared by all the CCP structures, whereas the topology of the other strands relative to this central conserved core is variable, especially at the regions that form the interfaces with the preceding and following module or domain.
Table III. Structural comparisons of the C1s CCP2 module with homologous structures.
| VC3a | VC4a | H15b | H16b | CD1c | CD2c | |
|---|---|---|---|---|---|---|
| PDB code | 1vvc | 1vvc | 1hfi | 1hcc | 1ckl | 1ckl |
| R.m.s on cysteines (Å) | 1.6 | 1.0 | 1.1 | 0.9 | 1.2 | 0.4 |
| Core r.m.sd (Å) | 1.6 | 1.3 | 1.0 | 1.3 | 1.8 | 0.7 |
| Global r.m.s. (Å)/nb Ca | 2.2/56 | 2.0/54 | 1.7/56 | 2.0/55 | 1.7/51 | 1.5/57 |
| Sequence identitye (%) | 30.3 | 24.1 | 28.6 | 27.3 | 23.5 | 24.5 |
aModules 3 and 4 from the vaccinia virus complement control protein (Wiles et al., 1997).
bModules 15 and 16 from the complement factor H (Barlow et al., 1993).
cModules 1 and 2 from the complement regulatory protein CD46 (Casasnovas et al., 1999).
dThe superposition was based on the Cα of the 11 consensus residues comprised in conserved strands, as indicated by * in Figure 2C.
eThe sequence identity was measured on the homologous residues defined by the global superposition.
The most prominent structural features specific to the C1s CCP module are: (i) the presence of tyrosines 375, 376 and 377, which form a surface patch near the C-terminal extremity of the module; and (ii) the insertion loops located between B3 and B4 and between B5 and B6 (Figure 2B and C). The large insertion between B5 and B6 is found in only 23 of the 1109 known CCP module amino acid sequences. The B1 to B2 loop, previously called the ‘hypervariable loop’ (Wiles et al., 1997), lies on the side opposite to these insertions (Figure 2B) and is shorter in C1s CCP2 than in most other CCP modules.
The glycosylation site of C1s is located in the loop connecting B4 to B5 (Figure 2B), on the side of the protein that contains the insertion loops. Two N-acetyl-glucosamines (NAG) and a fucose bound to the proximal NAG (Pétillot et al., 1995) (Figure 2B) were built into well-defined electron density (not shown).
Structure of the rigid CCP2 module–SP domain interface
The overall shape of the CCP2–SP assembly is that of a bludgeon with the ellipsoidal CCP2 module tightly anchored on the more globular SP domain, on the side opposite to the active site (Figures 1A and 4A). The perpendicular orientation of the long axis of CCP2 occurs in the absence of crystallographic packing constraints and results from a very rigid interface. This rigidity arises from interactions among a proline- and tyrosine-rich hydrophobic framework involving residues within the 372–377 stretch from CCP2, the 526(114)–534(122) region from SP and the short intermediary segment spanning residues 407–410 (Figure 1B and C).
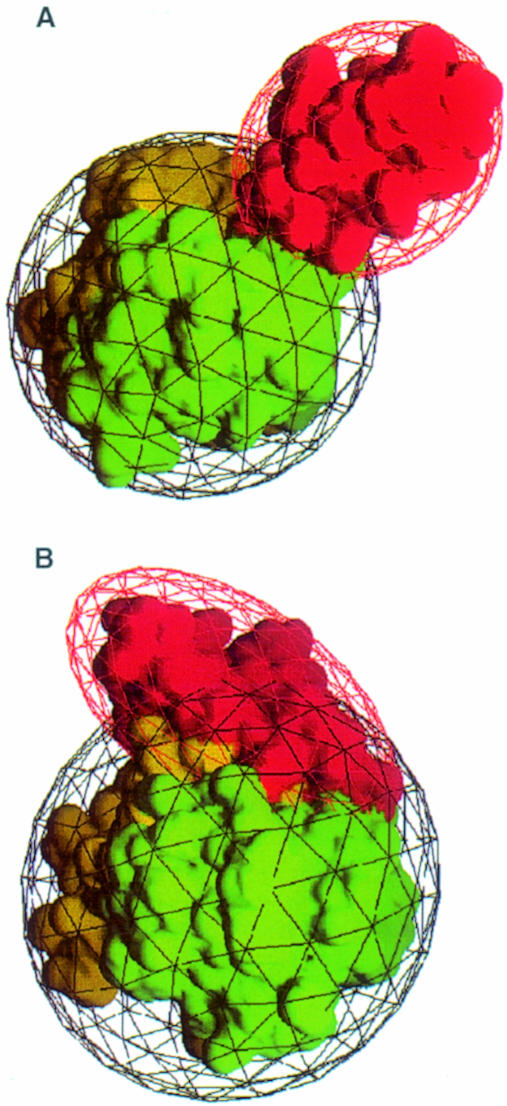
Fig. 4. The C1s and the vitamin K-dependent blood-clotting enzyme SP domains bear rigid modular N-terminal extensions, but they have strikingly different orientations. (A) The CCP modular extension of C1s (depicted in red) sits perpendicular to the SP domain surface (towards the viewer). The two SP subdomains are colored yellow and green. The molecule is seen in a perpendicular top view relative to Figures 3 and 2A, the binding site lying at the bottom of the figure. (B) The EGF modular extension (red) of blood coagulation factor VIIa (PDB code 1dan) lies tangentially to the surface of the SP domain. The SP domain in viewed in the same orientation as in (A) and the same color code has been used.
The connecting 407–410 segment is clamped on one side by the CCP module β-stranded tyrosine 375–377 patch and, on the opposite side, by residues Pro532(120) to Cys534(122) of the SP domain (Figure 1C). This clamp is strengthened on both sides by other direct interactions between the SP domain and the CCP module: Lys525(113) with Asn352 and Glu372, and the 526(114)–528(116) segment with Tyr375 and Tyr376 (Figure 1B).
In the crystal structure, there are no covalent bonds connecting the regions of SP and CCP involved in the interface. On the one hand, cleavage of the Arg426(15)–Ile427(16) bond has occurred at the C-terminal end of the chymotrypsinogen-like activation peptide [residues 410(1)–426(15)], generating two polypeptide chains. On the other hand, as indicated above, the inter-subunit Cys410(1)–Cys534(122) disulfide bridge is not formed. The following non-covalent interactions are observed: 23 van der Waals contacts, seven direct and four water-mediated hydrogen bonds and one salt bridge.
A template structure for other CCP module–SP domain assemblies involved in several humoral defense systems
In addition to the C1r and MASPs complement proteases, which share the modular architecture of C1s, the CCP–SP family includes several proteins belonging to humoral defense systems: Limulus clotting factor C, which initiates a small defensive coagulation cascade in the hemolymph of the primitive horseshoe crab in the presence of traces of endotoxin lipopolysaccharides (Muta et al., 1991); haptoglobin (Hp), which is an acute-phase plasma glycoprotein (Dobryszycka, 1997) that binds free hemoglobin with extremely high affinity in order to remove it from plasma (Hwang and Greer, 1980); and the haptoglobin-related protein (Hpr), which is the toxic component of the trypanosome lytic factors directed against the African cattle parasite Trypanosoma brucei brucei (Smith et al., 1995). As detailed in the legend to Figure 1C, the residues maintaining the interface framework, which are clustered around the connecting segment, are highly conserved in the various sequences of the CCP–SP family. This strongly suggests that the rigid CCP2–SP assembly observed in C1s is also present in these proteins (Gaboriaud et al., 1998). Consequently, the C1s structure constitutes a reliable template for the study of proteins of this family.
Obstruction of C1s binding sub-sites: a striking similarity with blood clotting enzymes
In the absence of inhibitor in the CCP2–SP structure, the substrate binding sub-sites are unoccupied and cannot be unambiguously identified. However, canonical ‘substrate binding-like’ conformations of the P4 to P3′ binding sites for the inhibitors have been observed in numerous serine protease–inhibitor complexes (Bode and Huber, 1992). We have superimposed such complexes onto the structure of the C1s SP domain in order to identify S4 to S2 and S1′ to S3′ C1s sub-sites (P1, P2, etc. and P1′, P2′, etc. refer to residue positions on the N- and C-terminal sides of the substrate scissile peptide bond, respectively, whereas S1, S2, etc. and S1′, S2′, etc. represent the corresponding binding sites on the protease). It appears from this analysis that the access to the C1s binding sub-sites is severely restricted, especially when compared with the open canyon found in digestive enzymes such as trypsin. This situation is reminiscent of the occlusion of the binding sites observed in some highly specific proteases of the coagulation cascade (Bode et al., 1997).
The major structural features of the C1s sub-sites (Figure 3) may be summarized as follows. (i) Access to S1 may be affected by the partially disordered Lys614(192) that lies immediately above its entrance. (ii) The major insertion loop C is found just above the entrance to the active site [see Glu506(97D) in Figure 3], in a position similar to that of loop B in thrombin where it restricts the access to S2 (Figure 2A). (iii) The cluster formed by Phe511(99), Tyr595(174)and Trp640(215) is reminiscent of the hydrophobic box of coagulation factors IXa and Xa (Bode et al., 1997), although the first two residues are significantly displaced in C1s: Phe511(99) fills up part of the space normally assigned to S2 and Tyr595(174) occupies S4. (iv) On the opposite side of the active site, the accessibility of S2′ is restricted by Trp444(41) and Arg563(151), which are slightly displaced relative to the coagulation factors because of the conformation of loops A and D (Table II).
Implications for the specific inhibition and substrate recognition of C1s
The steric constraints observed in the sub-sites of C1s SP are likely to contribute to its high substrate specificity. They may explain (i) why there is only one plasma inhibitor of C1s and (ii) why C1s is not inhibited by the archetypal bovine pancreatic trypsin inhibitor (BPTI) (Arlaud and Thielens, 1993). In the case of thrombin, the poor inhibition by BPTI has been shown to be due to both the insertion at loop B and the presence of Glu(192) (instead of Gln) at the entrance of the active site (van de Locht et al., 1997). By analogy, in C1s the presence of both the loop C insertion and Lys614(192) above the active site entrance may explain the lack of inhibition by BPTI (Figure 3). The restricted access to S2 may also explain why either a Ala→Val or a Ala→Asp change at the P2 position of the human C1 inhibitor leads to a dysfunctional molecule with diminished inhibitory activity towards both C1s and C1r [a condition resulting in systemic lupus erythematosus (Zahedi et al., 1997)]. It should be noted, however, that although in C2 the P2 sub-site is occupied by Gly, it corresponds to a Gln in C4. The fitting of the bulky Gln to a not very accessible S2 sub-site may be compensated by additional binding sub-sites. Indeed, the presence of both C1s CCP modules is required for efficient proteolytic cleavage of C4, indicating an extended recognition surface (Rossi et al., 1998). In addition, the occurrence in C1s of a cluster containing Phe511(99), Tyr595(174) and Trp640(215) (Figure 3) may explain its affinity for hydrophobic P3 residues (Leu, Val) and small P4 residues (Gly, Ser), as found in C4, C2 and C1 inhibitor.
Functional implications of SP rigid modular extensions in C1s and blood-clotting enzymes
As the proteases of the complement cascade, the blood-clotting enzymes are under tight regulation to prevent potentially harmful effects. These highly selective serum enzymes all bear a modular N-terminal extension and act in the context of macromolecular complexes, attached to their site of action. Their activity is limited in time and space by inhibitors and modulated by several effectors.
A major difference between C1s and the vitamin K-dependent blood-clotting enzymes is the orientation of the module at the N-terminal side of the SP domain. In the X-ray structures of coagulation factors Xa (Padmanabhan et al., 1993), IXa (Brandstetter et al., 1995), VIIa (Banner et al., 1996) and of protein C (Mather et al., 1996), the epidermal growth factor-like module (EGF2) lies tangent and binds extensively to the chymotrypsin-like SP domain (Figure 4B). The mean surface buried at the EGF module–SP domain interface in the vitamin K-dependent enzymes goes from 562 Å2 in factor IXa [code 1pfx in the Protein Data Bank (PDB; Bernstein et al., 1977)] to 742 Å2 in factor VIIa (PDB code 1dan). Furthermore, EGF2 interacts with the SP domain on the side opposite to the substrate binding site, whereas C1s CCP2 extends in the same direction as this site (Figures 3 and 4A). As in C1s, only two residues link the C-terminal end of the EGF2 to the N-terminus of the SP domain [the conserved inter-subunit Cys(1)–Cys(122) disulfide bond]. Because of the perpendicular orientation of C1s CCP2, the surface buried by the module/domain assembly represents only 8.3% of the total surface of the CCP module and 3.6% of the total surface of the SP domain (378 Å2 out of 4528 Å2 and 10526 Å2, respectively). Thus, the buried surface in the coagulation factors represents a 150–200% increase relative to C1s.
Although both the EGF and CCP modules are involved in the control of protease activity, the different module–SP domain interaction implies a different kind of modulation. In the case of coagulation factors VIIa, IXa and Xa, the pair of EGF modules and the preceding GLA domain are known to bind the respective cofactors (Banner et al., 1996; Bode et al., 1997). In C1s, the CCP modules probably provide additional binding sites for C4 (Rossi et al., 1998). The catalytic domain of C1s represents the first example of a rigid modular extension of the substrate binding site of an SP domain. An extension of the recognition site of an SP has also been reported in the microplasmin–staphylokinase–microplasmin ternary complex (Parry et al., 1998). However, in that case the interaction takes place between a bacterial cofactor and a plasma SP.
A trypsin-like, closed S1 pocket
A further difference between C1s and the coagulation enzymes lies in the Na+-induced allosteric regulation of activity observed in the latter. This regulation has been structurally correlated to the architecture of a water channel where Na+ binds, which connects the bottom of the S1 binding site to an aperture at the bottom surface of the molecule (Dang and Dicera, 1996; Guinto et al., 1999). Although it was postulated that this channel should be common to all proteases exhibiting either Tyr or Phe at position 225 (Guinto et al., 1999), the position of Tyr(225) in C1s is closer to Pro225 of trypsin (Ca–Ca distance = 0.4 Å) than to Tyr225 of thrombin (distance = 1.4 Å), casting doubts about the generality of the above proposition. This significant difference between C1s and the coagulation enzymes may result from the three-residue deletion in loop 2, which appears to be a unique feature of C1s. This deletion induces a narrowing of the bottom of the S1 specificity pocket, which is closed as in the case of trypsin.
The CCP module extension may act as a spacer and a handle
Current C1 models (Schumaker et al., 1986; Weiss et al., 1986; Arlaud et al., 1987) propose that the SP domain of C1s, once activated by C1r (an event expected to take place inside the C1 complex), becomes exposed to the solvent in order to be able to cleave C4 and C2. Both the orientation of CCP2 and its very rigid interaction with SP suggest that it could be involved, as both a spacer and a handle, in the movement required to expose the active site of C1s. Figure 5 depicts a very simple model that illustrates this hypothesis. In the initial state, the SP domains of C1r and C1s are buried inside C1 and oriented in a position compatible with the activating cleavage of the Arg446–Ile447 bond of C1s by C1r. This position would be similar to the one observed in the microplasmin–staphylokinase–microplasmin ternary complex (Parry et al., 1998). In this configuration, the C1s catalytic domain cannot cleave its protein substrates, because the essential S′ binding sub-sites are buried and face the C1r catalytic domain (Figure 5).
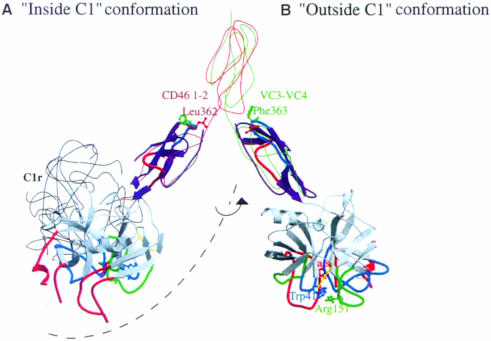
Fig. 5. Illustration of the potential role of the C1s CCP2 module handle in the putative displacement of the SP active site. The activation of C1s by C1r inside the C1 complex (A) requires an orientation of the C1s SP that is not compatible with its ability to cleave C4 and C2: the side chains of Trp41 (blue), Arg151 (green) and the active site Ser195 (red) face the C1r molecule inside the C1 complex. The thin black line indicates the position of the homologous C1r catalytic domain as required for C1s activation. In this orientation, the homologous CCP2 modular extension of C1r lies behind the SP domain. Correct positioning of C1s for proteolysis would require a conformational change in the CCP1–CCP2 hinge region (B). C1s colors are coded as in Figure 3. The thin red and green lines show the conformation of the CCP module pairs (CD46 CCP 1–2 and VCP CCP 3–4, respectively) used as references in the construction of this model, as detailed in the text. The exposed hydrophobic side chains of Leu362 (red) and Phe363 (green) observed in the structure are proposed to stabilize each of the two alternative conformations of the C1s CCP1–CCP2 module pair.
In the next step, we propose that the region connecting the CCP1 and CCP2 modules acts as a hinge allowing the SP domain to move to a position outside the C1 complex, a likely requirement for the cleavage of C4 and C2 (Figure 5). The proposed initial and final positions of SP are based on the comparison between C1s CCP2 and the structures of the CCP1–CCP2 module pair of CD46 (Casasnovas et al., 1999) and the CCP3–CCP4 module pair of VCP (Wiles et al., 1997). In the buried conformation, Leu362 (Figures 1A and 5), could help stabilize the CCP1–CCP2 interactions as observed for the corresponding modules of CD46 (Casasnovas et al., 1999; Figure 5). In the exposed orientation, Phe363 would stabilize the C1s CCP1–CCP2 interface as does the homologous Tyr81 in VCP CCP4 (Wiles et al., 1997). A similar movement may take place in the homologous C1r catalytic domains, which are expected to switch from a position suitable for their own activation to a position appropriate for C1s activation.
Conclusions and perspectives
Here we report the first 3D structure of a modular complement protease. The extreme specificity of C1s seems to arise from a combination of restriction elements in the vicinity of the active site, and additional specific recognition elements on the CCP modules for C4. Analogous exclusion mechanisms have been reported in the case of the blood-clotting enzymes. However, the module–SP interactions are very different in the two cases. The rigid perpendicular orientation of CCP2 relative to the SP domain in C1s is very likely to exist in other members of the CCP–SP family. In addition to their postulated substrate recognition function, the CCP2 modules of both C1s and C1r could play the role of rigid spacers and handles helping to position the SP domain so that it can either be activated or exert its catalytic function. This certainly represents a key feature of C1r and C1s, as these proteases are held together through their N-terminal ends (Thielens et al., 1990) as parts of a macromolecular complex, instead of acting freely in solution. Similar conclusions may also be extrapolated to the homologous MASPs proteases, which also participate in multimolecular complexes (Sato et al., 1994; Thiel et al., 1997; Endo et al., 1998).
We plan to use site-directed mutagenesis in order to further increase our understanding of the role played by the different structural determinants on tuning the proteolytic activity of C1s.
Materials and methods
Protein expression and purification
A recombinant fragment corresponding to the second CCP module and the SP domain of C1s was expressed and purified as described previously (Rossi et al., 1998). The protein comprised the human C1s segment Asp343–Asp673 preceded by an Asp–Leu sequence added at the N-terminal end due to the introduction of a restriction site at the 5′ end of the cDNA. Briefly, recombinant baculoviruses containing the plasmid pNT-Bac/CCP2-ap-SP were generated by using the Bac-to-Bac system (Life Technologies, Inc.) and used to infect Sf21 insect cells in serum-free medium. The recombinant protein was isolated from the cell culture supernatant by anion-exchange chromatography on a Mono Q column followed by hydrophobic interaction chromatography on a TSK-Phenyl column.
Crystallization and data collection
Pooled fractions were concentrated to ∼6 mg/ml in 50 mM triethanolamine–hydrochloride, 145 mM NaCl pH 7.4. Crystals were obtained by mixing 2 µl of protein solution with 2 µl of reservoir solution on a coverslip and then sealing over the reservoir using vacuum grease. The reservoirs contained 1 ml of 32–26% PEG 4000 at pH 5.6, 7.4 or 8.4, with or without 100 mM ammonium sulfate. Crystals were allowed to grow in the cold room (4°C) for periods of 1 to 3 months. Crystals were initially characterized in the laboratory (space group P21, cell parameters listed in Table IV) and data were collected subsequently on the BM02 and ID14-EH4 synchrotron beamlines at the European Synchrotron Radiation Facility (ESRF), Grenoble. Crystals were cooled at cryo-temperature and stored in solid propane as described by Vernède and Fontecilla-Camps (1999). The first native dataset was measured to a resolution of 2.2 Å. An additional 180° of data to 1.7 Å resolution were recorded later from a different cryo-cooled crystal on the beamline ID14-EH4. All data were processed, reduced and scaled using the XDS package (Kabsch, 1993). Details are given in Table IV.
Table IV. Data collection and processing statistics relevant to the two distinct steps of refinement of the model.
| Native 1 | Native 2 | |
|---|---|---|
| Crystallization parameters | pH 5.7 | pH 7.4 |
| [PEG4K] 32% | [PEG4K] 34% | |
| [(NH4)2SO4] = 100 mM | ||
| Unit cell parameters | a = 39.08 Å | a = 39.54 Å |
| b = 79.65 Å | b = 80.39 Å | |
| c = 60.21 Å | c = 61.31 Å | |
| β = 91.24° | β = 91.21° | |
| Wavelength (Å) | 0.98 | 0.93 |
| Resolution (Å) | 2.2 | 1.7 |
| No. of measurements | 64464 | 115710 |
| No. of unique reflections | 18464 | 39489 |
| Wilson B factor (Å2) | 20.8 | 21.7 |
| Coverage overall (%) | 98.0 (95.3)a | 96.7 (75.8)a |
| I/Sigma(I) all data | 11.6 (3.5)a | 16.9 (3.4)a |
| Rsym overall (%)b | 6.7 (14.8)a | 3.7 (24.7)a |
aNumbers in parentheses correspond to the highest resolution shell: 2.4–2.2 Å for native 1, 1.75–1.7 Å for native 2.
bRsym = Σ[I – <I>]/ΣI, where the summation is over all symmetry equivalent reflections.
Structure determination, refinement and analysis
The structure was solved in two steps. First, molecular replacement was applied using the native 1 dataset (Table IV). Rotational and translational searches were performed using the AMoRe package (Navaza, 1994) with data from 15 to 3 Å resolution and the bovine thrombin structure (1fpc from the PDB; Bernstein et al., 1977) as a search model. The rotational search showed a unique solution with a correlation value on intensities of 0.21. Translational search and rigid body fitting resulted in a contrasted solution with a correlation value of 0.28 and an R value of 50.7% between 15 and 3 Å resolution. Similar results were obtained with other SP models [chymotrypsin (2cga), protein C (1aut)]. Correction and refinement of the model were carried out in the areas where electron density maps clearly showed up relevant information. Rounds of refinement using X-PLOR (Brünger, 1992) were interspersed with interactive computer graphics model building using O (Jones et al., 1991). At this stage, the Rwork and Rfree were 0.413 and 0.516, respectively, using data between 15 and 2.2 Å resolution. However, at this stage it was not possible to build unambiguously the surface insertion loops, the activation peptide and the CCP module.
The high-resolution native 2 dataset (Table IV) combined with the use of the powerful WARP procedure (Perrakis et al., 1999) allowed the complete structure of the C1s CCP2–SP to be solved in a second step. The main chain was automatically built for 279 residues whereas the side chains were constructed using O (Jones et al., 1991) into a good electron density map. This latter step was restricted in most cases to the manual choice of the correct rotamer. Twenty additional residues were built manually, along with three carbohydrate residues and two sulfate ions that were clearly defined in the electron density map. Refinement was then carried out using Refmac (Murshudov et al., 1997), starting from an Rwork of 0.296 and an Rfree of 0.309 for data between 15 and 1.7 Å resolution. ARP (Lamzin and Wilson, 1993) was used for solvent building. A number of residues displayed alternative conformations and were modeled as such. Some residues poorly defined in the electron density maps were truncated to alanines (Table I). The stereochemistry of the structure was assessed with PROCHECK (Laskowski et al., 1993). Details and statistics of the final model are presented in Table I. The structure factors of the high-resolution dataset 2 and the atomic coordinates have been deposited in the Protein Data Bank with accession code 1elv.
The detailed analysis of the contacts between the CCP module and the SP domain was carried out using Hbplus (McDonald and Thornton, 1994) and LIGPLOT (Wallace et al., 1995). The surface areas were calculated with NACCESS (Hubbard and Thornton, 1993). Figures were generated with several combined uses of MOLSCRIPT (Kraulis, 1991), BOBSCRIPT, GRASP (Nicholls et al., 1991) and Raster3D (Merritt and Bacon, 1997). The sequence alignments were the same as described previously (Gaboriaud et al., 1998) or extracted from the recent version of the pfam database (Bateman et al., 1999).
Quality of the model
Most of the structure was well-defined in a good high resolution electron density map. The final refined model comprises residues 342–379, 382–417, 423–477, 487–582 and 595–668. Of the 303 amino acid residues defined in the structure, 99.6% are in the favorable or additionally allowed regions in the Ramachandran plot. Only Thr646 is in the generously allowed region. Five segments (380–381, 418–422, 478–486, 583–594 and 669–673) display high flexibility or disordered conformations.
Acknowledgments
Acknowledgements
The authors are grateful to M.Roth and W.Burmeister as local contacts at the ESRF. The quality of the beam-lines they developed was critical for this study. The authors are grateful to D.Housset for the collection of the high resolution dataset and for numerous stimulating discussions. We thank A.Perrakis for his advice and help in the installation of the WARP software. The work of M.Qian and C.Olivier at early stages of the C1s project is also acknowledged. This work was supported by the Commissariat à l’Energie Atomique and the Centre National de la Recherche Scientifique.
References
- Arlaud G.J. and Thielens,N. (1993) Human complement serine proteases C1r and C1s and their proenzymes. Methods Enzymol., 223, 61–82. [DOI] [PubMed] [Google Scholar]
- Arlaud G.J., Colomb,M.G. and Gagnon,J. (1987) A functional model of the human C1 complex. Immunol. Today, 8, 106–111. [DOI] [PubMed] [Google Scholar]
- Arlaud G.J., Volanakis,J.E., Thielens,N.M., Narayana,S.V.L., Rossi,V. and Xu,Y. (1998) The atypical serine proteases of the complement system. Adv. Immunol., 69, 249–307. [PubMed] [Google Scholar]
- Banner D.W., D’Arcy,A., Chene,C., Winkler,F.K., Guha,A., Konigsberg,W.H., Nemerson,Y. and Kirchhofer,D. (1996) The crystal structure of the complex of blood coagulation factor VIIa with soluble tissue factor. Nature, 380, 41–46. [DOI] [PubMed] [Google Scholar]
- Barlow P.N., Norman,D.G., Steinkasserer,A., Horne,T.J., Pearce,J., Driscoll,P.C., Sim,R.B. and Campbell,I.D. (1992) Solution structure of the fifth repeat of factor H: a second example of the complement control protein module. Biochemistry, 31, 3626–3634. [DOI] [PubMed] [Google Scholar]
- Barlow P.N., Steinkasserer,A., Norman,D.G., Kieffer,B., Wiles,A.P., Sim,R.B. and Campbell,I.D. (1993) Solution structure of a pair of complement modules by nuclear magnetic resonance. J. Mol. Biol., 232, 268–284. [DOI] [PubMed] [Google Scholar]
- Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L. (1999) Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res., 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein F.C., Koetzle,T.F., Williams,G.J., Meyer,E.E.,Jr, Brice,M.D., Rodgers,J.R., Kennard,O., Shimanouchi,T. and Tasumi,M. (1977) The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol., 112, 535–542. [DOI] [PubMed] [Google Scholar]
- Bode W. and Huber,R. (1992) Natural protein proteinase inhibitors and their interaction with proteinases. Eur. J. Biochem., 204, 433–451. [DOI] [PubMed] [Google Scholar]
- Bode W., Brandstetter,H., Mather,T. and Stubbs,M.T. (1997) Comparative analysis of haemostatic proteinases: structural aspects of thrombin, factor Xa, factor IXa and protein C. Thromb. Haemost., 78, 501–511. [PubMed] [Google Scholar]
- Bork P. and Beckmann,G. (1993) The CUB domain. A widespread module in developmentally regulated proteins. J. Mol. Biol., 231, 539–545. [DOI] [PubMed] [Google Scholar]
- Brandstetter H., Bauer,M., Huber,R., Lollar,P. and Bode,W. (1995) X-ray structure of clotting factor IXa: active site and module structure related to Xase activity and hemophilia B. Proc. Natl Acad. Sci. USA, 92, 9796–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR version 3.1, a System for X-ray Crystallography and NMR. Yale University Press, New Haven, CT. [Google Scholar]
- Burmeister W.P. (2000) Structural changes in a cryo-cooled protein crystal due to radiation damage. Acta Crystallogr. D, 56, 328–341. [DOI] [PubMed] [Google Scholar]
- Casasnovas J.M., Larvie,M. and Stehle,T. (1999) Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J., 18, 2911–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N.R. (1985) The classical complement pathway: activation and regulation of the first complement component. Adv. Immunol., 37, 151–216. [DOI] [PubMed] [Google Scholar]
- Dalmasso A.P. (1992) The complement system in xenotransplantation. Immunopharmacology, 24, 149–160. [DOI] [PubMed] [Google Scholar]
- Dang Q.D. and Di Cera,E. (1996) Residue 225 determines the Na+-induced allosteric regulation of catalytic activity in serine proteases. Proc. Natl Acad. Sci. USA, 93, 10653–10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobryszycka W. (1997) Biological functions of haptoglobin—new pieces to an old puzzle. Eur. J. Clin. Chem. Clin. Biochem., 35, 647–654. [PubMed] [Google Scholar]
- Endo Y., Takahashi,M., Nakao,M., Saiga,H., Sekine,H., Matsushita,M., Nonaka,M. and Fujita,T. (1998) Two lineages of mannose-binding lectin-associated serine protease (MASP) in vertebrates. J. Immunol., 161, 4924–4930. [PubMed] [Google Scholar]
- Gaboriaud C., Rossi,V., Fontecilla-Camps,J.C. and Arlaud,G.J. (1998) Evolutionary conserved rigid module–domain interactions can be detected at the sequence level: the examples of complement and blood coagulation proteases. J. Mol. Biol., 282, 459–470. [DOI] [PubMed] [Google Scholar]
- Gans P., Rossi,V., Gaboriaud,C., Bally,I., Hernandez,J.F., Blackledge,M.J. and Arlaud,G.J. (1998) NMR structures of the C-terminal end of human complement serine protease C1s. Cell. Mol. Life Sci., 54, 171–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinto E.R., Caccia,S., Rose,T., Fütterer,K., Waksman,G. and Di Cera,E. (1999) Unexpected crucial role of residue 225 in serine proteases. Proc. Natl Acad. Sci. USA, 96, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.J. and Thornton,J.M. (1993) ‘NACCESS’, Computer Program, Department of Biochemistry and Molecular Biology, University College London, UK. [Google Scholar]
- Hwang P.K. and Greer,J. (1980) Interaction between hemoglobin subunits in the hemoglobin.haptoglobin complex. J. Biol. Chem., 255, 3038–3041. [PubMed] [Google Scholar]
- Jing H., Macon,K., Moore,D., DeLucas,L.J., Volanakis,J.E. and Narayana,S.V.L. (1999) Structural basis of profactor D activation: from a highly flexible zymogen to a novel self-inhibited serine protease, complement factor D. EMBO J., 18, 804–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A, Zou,J.-Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kirkitadze M.D. et al. (1999) Independently melting modules and highly structured intermodular junctions within complement receptor type 1. Biochemistry, 38, 7019–7031. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lacroix M., Rossi,V., Gaboriaud,C., Chevallier,S., Jaquinod,M., Thielens,N.M., Gagnon,J. and Arlaud,G.J. (1997) Structure and assembly of the catalytic region of human complement protease C1r: a three-dimensional model based on chemical cross-linking and homology modeling. Biochemistry, 36, 6270–6282. [DOI] [PubMed] [Google Scholar]
- Lamzin V. and Wilson,K. (1993) Automated refinement of protein model. Acta Crystallogr. D, 49, 129–147. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Mackinnon C.M., Carter,P.E., Smyth,S.J., Dunbar,B. and Fothergill,J.E. (1987) Molecular cloning of cDNA for human complement component C1s. The complete amino acid sequence. Eur. J. Biochem., 169, 547–553. [DOI] [PubMed] [Google Scholar]
- Mather T., Oganessyan,V., Hof,P., Huber,R., Foundling,S., Esmon,C. and Bode,W. (1996) The 2.8 Å crystal structure of Gla-domainless activated protein C. EMBO J., 15, 6822–6831. [PMC free article] [PubMed] [Google Scholar]
- Matsushita M., Endo,Y., Nonaka,M. and Fujita,T. (1998) Complement-related serine proteases in tunicates and vertebrates. Curr. Opin. Immunol., 10, 29–35. [DOI] [PubMed] [Google Scholar]
- McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mevorach D., Mascarenhas,J.O., Gershov,D. and Elkon,K.B. (1998) Complement-dependent clearance of apoptotic cells by human macrophages. J. Exp. Med., 188, 2313–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Muta T., Miyata,T., Misumi,Y., Tokunaga,F., Nakamura,T., Toh,Y., Ikehara,Y. and Iwanaga,S. (1991) Limulus factor C. An endotoxin-sensitive serine protease zymogen with a mosaic structure of complement-like, epidermal growth factor-like and lectin-like domains. J. Biol. Chem., 26, 6554–6561. [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Padmanabhan K., Padmanabhan,K.P., Tulinsky,A., Park,C.H., Bode,W., Huber,R., Blankenship,D.T., Cardin,A.D. and Kisiel,W. (1993) Structure of human Des (1–45) factor Xa at 2.2 Å resolution. J. Mol. Biol., 232, 947–966. [DOI] [PubMed] [Google Scholar]
- Parry M.A, Fernandez-Catalan,C., Bergner,A., Huber,R., Hopfner,K.P., Schlott,B., Guhrs,K.H. and Bode,W. (1998) The ternary microplasmin–staphylokinase–microplasmin complex is a proteinase–cofactor–substrate complex in action. Nature Struct. Biol., 10, 917–923. [DOI] [PubMed] [Google Scholar]
- Perona J.J. and Craik,C.S. (1997) Evolutionary divergence of substrate specificity within the chymotrypsin-like serine protease fold. J. Biol. Chem., 272, 29987–29990. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Pétillot Y. et al. (1995) Analysis of the N-linked oligosaccharides of human C1s using electrospray ionisation mass spectrometry. FEBS Lett., 358, 323–328. [DOI] [PubMed] [Google Scholar]
- Reid K.B.M. and Day,A.J. (1989) Structure–function relationships of the complement components. Immunol. Today, 10, 177–180. [DOI] [PubMed] [Google Scholar]
- Rogers J. et al. (1992) Complement activation by β-amyloid in Alzheimer disease. Proc. Natl Acad. Sci. USA, 89, 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi V., Gaboriaud,C., Lacroix,M., Ulrich,J., Fontecilla-Camps,J.C., Gagnon,J. and Arlaud,G.J. (1995) Structure of the catalytic region of human complement protease C1s: study by chemical cross-linking and three-dimensional homology modeling. Biochemistry, 34, 7311–7321. [DOI] [PubMed] [Google Scholar]
- Rossi V., Bally,I., Thielens,N.M., Esser,A.F. and Arlaud,G.J. (1998) Baculovirus-mediated expression of truncated modular fragments from the catalytic region of human complement serine protease C1s. Evidence for the involvement of both complement control protein modules in the recognition of the C4 protein substrate. J. Biol. Chem., 273, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Sato T., Endo,Y., Matsushita,M. and Fujita,T. (1994) Molecular characterization of a novel serine protease involved in activation of the complement system by mannose-binding protein. Int. Immunol., 6, 665–669. [DOI] [PubMed] [Google Scholar]
- Schumaker V.N., Hanson,D.C., Kilchherr,E., Phillips,M.L. and Poon,P.H. (1986) A molecular mechanism for the activation of the first component of complement by immune complexes. Mol. Immunol., 23, 557–565. [DOI] [PubMed] [Google Scholar]
- Smith A.B., Esko,J.D. and Hajduk,S.L. (1995) Killing of trypanosomes by the human haptoglobin related protein. Science, 268, 284–286. [DOI] [PubMed] [Google Scholar]
- Thiel S. et al. (1997) A second serine protease associated with mannan-binding lectin that activates complement. Nature, 386, 506–510. [DOI] [PubMed] [Google Scholar]
- Thielens N.M., Aude,C.A., Lacroix,M.B., Gagnon,J. and Arlaud,G.J. (1990) Ca2+ binding properties and Ca2+-dependent interactions of the isolated NH2-terminal α fragments of human complement proteases C1r and C1s. J. Biol. Chem., 265, 14469–14475. [PubMed] [Google Scholar]
- Tosi M., Duponchel,C., Meo,T. and Julier,C. (1987) Complete cDNA sequence of human complement C1s and close physical linkage of the homologous genes C1s and C1r. Biochemistry, 26, 8516–8524. [DOI] [PubMed] [Google Scholar]
- Tosi M., Duponchel,C., Meo,T. and Couture-Tosi,E. (1989) Complement genes C1r and C1s feature an intronless serine protein domain closely related to haptoglobin. J. Mol. Biol., 208, 709–714. [DOI] [PubMed] [Google Scholar]
- van de Locht A., Bode,W., Huber,R., Le Bonniec,B.F., Stone,S.R., Esmon,C.T. and Stubbs,M.T. (1997) The thrombin E192Q–BPTI complex reveals gross structural rearrangements: implications for the interaction with antithrombin and thrombomodulin. EMBO J., 16, 2977–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernède X. and Fontecilla-Camps,J.C. (1999) A method to stabilize reduced and/or gas-treated protein crystals by flash-cooling under a controlled atmosphere. J. Appl. Crystallogr., 32, 505–509. [Google Scholar]
- Volanakis J.E. and Narayana,S.V.L. (1996) Complement factor D, a novel serine protease. Protein Sci., 5, 553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Weiss V., Fauser,C. and Engel,J. (1986) Functional model of sub component C1 of human complement. J. Mol. Biol., 189, 573–581. [DOI] [PubMed] [Google Scholar]
- Wiles A.P., Shaw,G., Bright,J., Perczel,A., Campbell,I.D. and Barlow,P.N. (1997) NMR studies of a viral protein that mimics the regulators of complement activation. J. Mol. Biol., 272, 253–265. [DOI] [PubMed] [Google Scholar]
- Zahedi R., Wisnieski,J. and Davis,A.E.,III (1997) Role of the P2 residue of complement 1 inhibitor (Ala443) in determination of target protease specificity: inhibition of complement and contact system proteases. J. Immunol., 159, 983–988. [PubMed] [Google Scholar]