Abstract
The plastid of Plasmodium falciparum (or ‘apicoplast’) is the evolutionary homolog of the plant chloroplast and represents a vestige of a photosynthetic past. Apicoplast indispensability indicates that it still provides essential functions to parasites. Similar to plant chloroplasts, the apicoplast is dependent on many nucleus-encoded genes to provide these functions. The apicoplast is surrounded by four membranes, two more than plant chloroplasts. Thus, protein targeting to the apicoplast must overcome additional membrane barriers. In P.falciparum we have analyzed apicoplast targeting using green fluorescent protein (GFP). We demonstrate that protein targeting is at least a two-step process mediated by bipartite N-terminal pre-sequences that consist of a signal peptide for entry into the secretory pathway and a plant-like transit peptide for subsequent import into the apicoplast. The P.falciparum transit peptide is exceptional compared with other known plastid transit peptides in not requiring serine or threonine residues. The pre-sequence components are removed stepwise during apicoplast targeting. Targeting GFP to the apicoplast has also provided the first opportunity to examine apicoplast morphology in live P.falciparum.
Keywords: green fluorescent protein/malaria/plastid targeting/secretory pathway/transfection
Introduction
The discovery of a remnant plastid in apicomplexan parasites (including the malarial causative agents Plasmodium spp. and the opportunistic human pathogen Toxoplasma gondii) (McFadden et al., 1996; Wilson et al., 1996; Köhler et al., 1997) caused an immediate revision of our understanding of the evolutionary origin of this highly specialized and successful assemblage of endoparasites. However, in addition to suggesting that these parasites once lived an autotrophic lifestyle based on photosynthesis, the indispensability (Fichera and Roos, 1997) of the apicomplexan plastid (apicoplast) shows that it remains an integral organelle within parasite cell biology and metabolism. All plastids ultimately derive from a cyanobacterial endosymbiont and, therefore, as for the plastids of plants and algae the apicoplast is likely to contain many prokaryote-type metabolic pathways. This offers great promise for combating malaria and other human and animal diseases caused by apicomplexan parasites because such pathways potentially offer many specific drug targets (McFadden and Waller, 1997; McFadden et al., 1997). Indeed, the apicoplast has already provided a rationale for the anti-parasite activity of many antibiotics (including doxycycline, lincosamides and macrolides) (McFadden and Roos, 1999) as well as the identification of several new antimalarials (Clough et al., 1997, 1999; Waller et al., 1998; Jomaa et al., 1999). This provides a strong impetus to learn more of apicoplast metabolism.
We previously identified nucleus-encoded genes from Plasmodium falciparum and T.gondii whose products are targeted to the apicoplast (Waller et al., 1998). This offers a powerful approach to increasing our knowledge of apicoplast metabolism because the apicoplast genome contains few genes other than those necessary for organelle transcription and translation (Wilson et al., 1996; Köhler et al., 1997). In plants it is estimated that between 1000 and 5000 chloroplast proteins are encoded by nuclear genes (Martin and Herrmann, 1998) and, by analogy, in Apicomplexa most of the protein content of the apicoplast will most likely also be nucleus encoded. If we take the more conservative estimate of plant chloroplast-targeted genes, and subtract from this the genes for functions not implicated in apicoplast function (photosynthesis and the shikimate pathway for aromatic amino acid synthesis; Keeling et al., 1998), we estimate that at least 800 nucleus-encoded proteins may be targeted to the apicoplast. Hence, the imminent completion of the P.falciparum genome sequencing project provides an excellent opportunity to apply a genomics-based approach to studying apicoplast function. Such an approach is facilitated by the presence of conspicuous targeting information, in the form of a bipartite N-terminal pre-sequence on apicoplast-targeted proteins (Waller et al., 1998). Understanding the nature and routing of protein targeting to the apicoplast is therefore essential for implementing a predictive tool for genes involved in apicoplast function.
In this study we have characterized protein targeting to the apicoplast of P.falciparum using green fluorescent protein (GFP) as a reporter molecule. We have tested that the N-terminal pre-sequences are sufficient and necessary to target proteins to the P.falciparum apicoplast. Furthermore, by testing altered forms of the pre-sequences, by mutations and truncations, we have identified functional domains that define protein routing to the apicoplast. This is the first reported use of a transgenic reporter protein being used for protein trafficking studies in P.falciparum. The evolutionary significance of the routing of proteins to the apicoplast is discussed.
Results
Bipartite pre-sequences target GFP to the apicoplast in P.falciparum
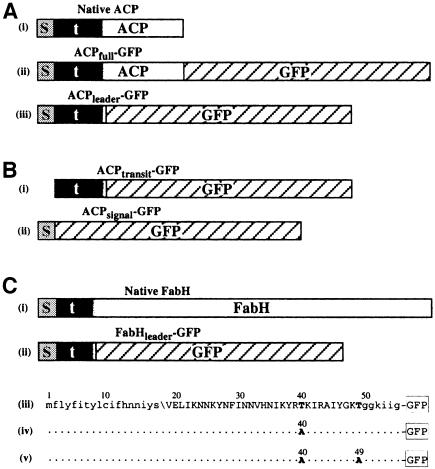
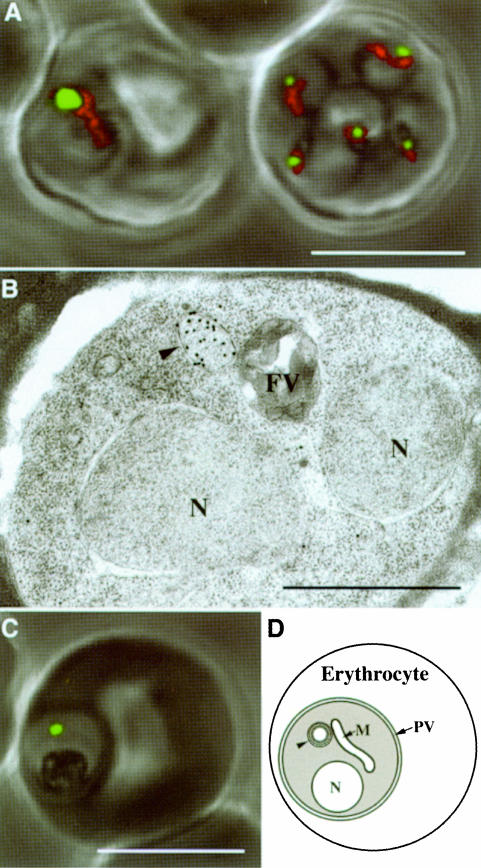
To verify that P.falciparum gene products previously identified as possessing bipartite pre-sequences (Waller et al., 1998) were indeed apicoplast targeted in P.falciparum we generated C-terminal fusions with the reporter molecule GFP. The gene for the fatty acid synthase protein acyl carrier protein (ACP) was ligated upstream of the GFP coding sequence and the resultant GFP fusion protein (ACPfull–GFP; Figure 1A, ii) was expressed under the control of the calmodulin regulatory sequences from stably maintained episomes within transformed blood-stage parasites. Expression of this fusion resulted in green fluorescence localized to a well defined region within the parasites. A similar fusion was constructed lacking the coding sequence for the mature ACP such that only the bipartite leader (first 60 amino acids) was fused with GFP (ACPleader–GFP; Figure 1A, iii). An identical pattern of fluorescence was observed when this fusion was expressed (Figure 2A). This region of fluorescence was in close association but distinct from the mitochondrion as seen by co-staining with the mitochondrial-specific live stain MitoTracker® Red (Figure 2A). Immunoelectron microscopy against GFP shows that targeting was confined to the lumen of a multi-membrane-bound compartment consistent with the apicoplast (Figure 2B). No fusion protein was detectable in other regions of the cell.
Fig. 1. Schematics of GFP fusions expressed in P.falciparum. (A) Plasmodium falciparum ACP with (i) bipartite leader, and GFP fusions (ii) ACPfull–GPF and (iii) ACPleader–GFP (s, signal peptide; t, transit peptide; shaded transition between the transit peptide and ACP indicates uncertainty of the precise boundary). (B) Signal and transit peptide deletion GFP fusions (i) ACPtransit–GFP and (ii) ACPsignal–GFP. (C) Plasmodium falciparum FabH protein with (i) bipartite leader and (ii) GFP fusion FabHleader–GFP represented schematically and (iii) in sequence form. Mutations engineered into fusions FabHleader[T40A]–GFP and FabHleader[T40/49A]–GFP are indicated in (iv) and (v), respectively. N-terminal lower case sequence represents the signal peptide with the predicted cleavage point indicated (\). Upper case sequence indicates transit peptide domain and the C-terminal lower case sequence indicates the first conserved residues shared amongst FabH sequences in other organisms.
Fig. 2. Plasmodium falciparum expressing apicoplast-targeted GFP in human erythrocytes. (A) ACPleader–GFP expressing parasites co-stained for mitochondria (red) [note, the right erythrocyte contains multiple (5) infections]. (B) Detection of GFP in the apicoplast (arrowhead) by immunogold labeling of ultrathin sections of ACPleader–GFP-expressing parasite. (C) Green fluorescence of FabHleader–GFP-expressing parasite. (D) Schematic of P.falciparum with major organelles. The parasite is contained within the parasitophorous vacuole (PV) inside the erythrocyte. Arrowhead, apicoplast; N, nucleus; FV, food vacuole; M, mitochondrion; PV, parasitophorous vacuole. White scale bars, 5 µm; black scale bar, 1 µm.
An equivalent GFP fusion protein was also made with the bipartite leader of the FabH (β-ketoacyl-ACP synthase III, also a protein of fatty acid synthase) protein (first 55 amino acids) (FabHleader–GFP; Figure 1C). Expression of this fusion resulted in an identical pattern of fluorescence to that of the ACP-based fusions (Figure 2C). In addition, we tested whether the T.gondii ACP pre-sequence (first 104 amino acids), which had previously been demonstrated to be sufficient to target a protein to the T.gondii apicoplast (Waller et al., 1998), could target GFP to the P.falciparum apicoplast. This gene fusion construct (ACPToxo–GFP) also resulted in the pattern of plastid fluorescence (not shown). These data demonstrate that the bipartite pre-sequence is sufficient to target a protein to the apicoplast of P.falciparum.
Signal peptide and transit peptide domains are essential for apicoplast targeting
The bipartite pre-sequence of proteins targeted to the apicoplast appears to contain two functional domains: a signal peptide and a transit peptide (Waller et al., 1998). The signal peptide consists of a short hydrophobic domain followed by a von Heijne-type cleavage motif. Prediction programs SignalP and PSORT (Nakai and Kanehisa, 1992; Nielsen et al., 1997) assign high confidence values to the presence of these signal peptides and their cleavage sites in P.falciparum. Following the signal peptide is an additional pre-sequence component. Similar to plant plastid transit peptides this domain carries a net positive charge (Waller et al., 1998). We sought to test this model of bipartite pre-sequences by individually testing the targeting properties of each domain.
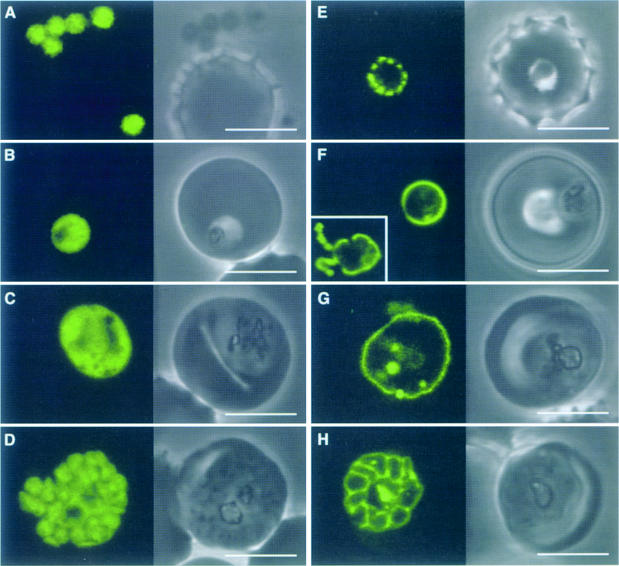
To test the role of the predicted signal peptide a gene construct was made to create a fusion of GFP with the P.falciparum ACP pre-sequence minus the signal peptide domain (using amino acids 14–60) (ACPtransit–GFP; Figure 1B, i). In these transfectants GFP accumulated in the parasite cytoplasm (Figure 3A–D). This result indicates that entry into the secretory pathway is a necessary step for apicoplast targeting, and that a transit peptide alone was not sufficient.
Fig. 3. Plasmodium falciparum expressing ACPtransit–GFP (A–D) and ACPsignal–GFP (E–H). ACPtransit–GFP expression results in cytoplasmic GFP in merozoites (A) through to schizonts (D). GFP is apparently excluded from the food vacuole (B and C) and other organelles (C). ACPsignal–GFP expression results in GFP export to the parasitophorous vacuolar space (in which the parasite resides within the erythrocyte, compare with Figure 2D). Newly invaded ring parasites (E) show punctate fluorescence in the parasitophorous vacuole, which may indicate some early formative stage of this specialized structure. Subsequent stages show continuous fluorescence surrounding the parasite (F and G), which ultimately delineates the products of schizogony (H). GFP is also seen in the tubulovesicular network [(F), inset, fluorescence only]. Some internalized GFP probably represents protein that re-enters the parasite in food vacuoles. Scale bars, 5 µm.
To test the role of the predicted transit peptide domain a gene construct was made where the ACP signal peptide, but no transit peptide, was fused to GFP (amino acids 1–20) (ACPsignal–GFP; Figure 1B, ii). Upon expression in parasites, GFP accumulated outside the parasite within the parasitophorous vacuole (see Figure 2D for diagram of parasitophorous vacuole). This secreted form of GFP was evident as punctate green fluorescence surrounding ring stage parasites (Figure 3E), continuous fluorescence surrounding the parasites in trophozoite stages (Figure 3F and G) and cobblestone-patterned fluorescence surrounding the developing merozoites of schizont stages (Figure 3H). GFP fluorescence was also seen within the tubulovesicular network in the erythrocyte cytoplasm (Figure 3F, inset), but was not associated with merozoites (data not shown), consistent with GFP being secreted. Some GFP appears to re-enter the parasites with erythrocyte cytoplasm ingestion and was associated with the food vacuoles and hemozoin. Export of GFP out of the parasite cell confirms that the signal peptide domain mediates protein entry into the secretory pathway. Furthermore, it shows that once within the secretory pathway the transit peptide domain is essential for targeting into the apicoplast. Failing this, the protein appears to follow a default route of secretion and exits the cell.
Together these results demonstrate that protein targeting to the apicoplast in P.falciparum is an ordered two-step process. Proteins must first enter the secretory pathway. Once there, the transit peptide then mediates final transfer into the lumen of the apicoplast.
Serine and threonine residues are not essential components of P.falciparum transit peptides
Plastid-targeting transit peptides have long defied researchers’ efforts to identify an underlying structure, as seen with mitochondrial transit peptides (von Heijne and Nishikawa, 1991). However, enrichment for hydroxylated residues (serines and threonines) has been noted as a consistent feature of plant transit peptides (Keegstra and Olsen, 1989; von Heijne and Nishikawa, 1991; Cline and Henry, 1996), and this feature is conserved in transit peptides of algal proteins (Liaud et al., 1997; Sulli et al., 1999). Plasmodium falciparum presents an exception to this rule as its transit peptides show no bias for such residues (Waller et al., 1998). In fact, the transit peptide domain of the FabH pre-sequence contains only two threonines and no serines. This presented an opportunity to test whether serine or threonine residues are an essential component of P.falciparum transit peptides. Based on the GFP fusion construct FabHleader–GFP (Figure 1C), two constructs bearing mutations within the FabH transit peptide were made with either one or both threonines replaced with an alanine (FabHleader[T40A]–GFP and FabHleader[T40/49A]–GFP, respectively) (Figure 1C, iv and v). Expression of these in P.falciparum was examined for perturbation of GFP targeting to the apicoplast. In neither was mis-targeting seen. Both mutant forms displayed fluorescence patterns indistinguishable from those of the non-mutated form of the FabH pre-sequence (e.g. Figure 5F) and, indeed, of all pre-sequences tested. Furthermore, these fusion proteins were processed identically to the non-mutated FabH (see below and Figure 4C), which further implies correct targeting.
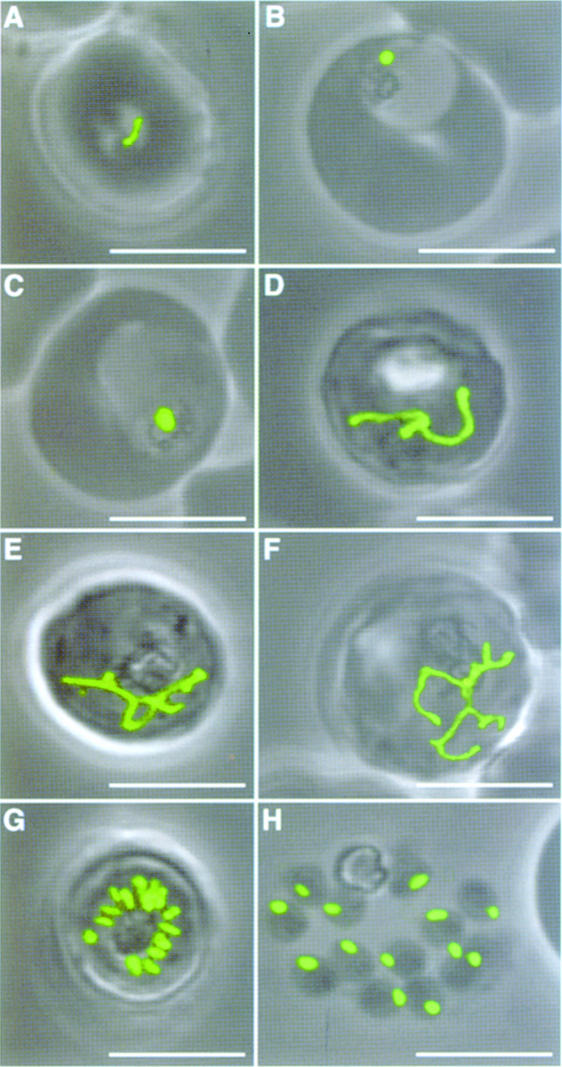
Fig. 5. Apicoplast morphology throughout the asexual life cycle of blood stage parasites expressing targeted GFP. Stages represent ring form (A), small trophozoite (B), large trophozoite (C), three stages of early schizonts (D–F), late schizont (G) and free merozoites (H). Single apicoplasts enlarge slowly, elongate to branched forms during schizogony and then divide and segregate prior to merozoite egression. These stages were seen for all transfectants with apicoplast-targeted GFP. (A–E) and (G–H) represent ACPleader–GFP-expressing parasites and (F) represents a FabHleader[T40/49A]–GFP-expressing parasite. Scale bars, 5 µm.
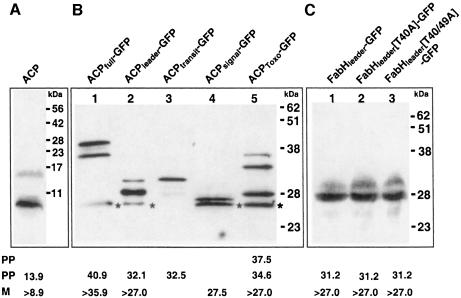
Fig. 4. Western blot analyses of apicoplast-targeted proteins using ACP antiserum to detect endogenous ACP (A) or GFP antiserum to detect ACP–GFP-based fusion proteins (B) and FabH–GFP-based fusion proteins (C). Bands marked with ‘*’ indicate GFP degradation products. Predicted sizes of mature (‘M’) and pre-processed (‘PP’) forms of each protein as discussed in the text are shown below each lane in kDa. Sizes marked with ‘>’ indicate minimal estimates due to uncertainty of the precise processing point.
Bipartite pre-sequences are removed during targeting to the apicoplast
In plant cells, following plastid targeting, transit peptides are cleaved off by a plastid peptidase to create a mature form of the protein (Cline and Henry, 1996). We have previously shown that this is also the case for proteins targeted to the apicoplast in T.gondii (Waller et al., 1998). In this study we have performed Western analyses of native P.falciparum apicoplast-targeted ACP as well as GFP fusion proteins. Figure 4A shows detection of endogenous ACP in untransfected cells (parasite cloned line D10). The precise cleavage site of the transit peptide from the remainder of ACP is currently unknown. However, based on alignments with other ACP proteins we estimate that the mature form of the P.falciparum ACP will be ∼8.9 kDa (denoted ‘>8.9’ in Figure 4 to indicate that the cleavage site may be slightly upstream of the limits of N-terminal sequence conservation). Figure 4A shows a protein band at ∼10.2 kDa, consistent with ACP having its pre-sequence removed. In addition to this processed band was a second larger band of size equivalent to a pre-processed form of ACP still possessing a transit peptide (Figure 4A). This is consistent with previous observations of processing intermediates in T.gondii (Waller et al., 1998).
Western analysis of the various ACP–GFP fusion proteins expressed in transfected parasites further supports that targeting pre-sequences are removed (Figure 4B). In each case the larger pre-processed forms can also be detected. Fusion proteins ACPfull–GFP and ACPleader–GFP are detected as two bands (Figure 4B, lanes 1 and 2) of predicted sizes for the pre-processed and mature forms [a third, smaller band appears to represent a GFP degradation product due to its size being smaller than GFP alone, and was present in only ACP–GFP fusions that enter the secretion system (lanes 1, 2, 4 and 5) and not in untransfected parasites (not shown)]. Fusion protein ACPtransit–GFP did not target to the apicoplast, and lane 3 (Figure 4B) reveals a band of expected size for the unprocessed fusion protein with transit peptide still attached. This result indicates that transit peptide removal is dependent upon targeting to the apicoplast. Furthermore, this unprocessed band was equivalent in size to the putative pre-processed form of ACPleader–GFP, which supports our argument that this band in lane 2 (Figure 4B) lacks the signal peptide. This is consistent with the signal peptide being removed during co-translational import into the endoplasmic reticulum (ER). Fusion protein ACPsignal–GFP (Figure 4B, lane 4) was secreted out of the cell and was represented by a band of the size of GFP alone, again lacking the signal peptide. This band is marginally smaller than the mature form of ACPleader–GFP as this latter fusion includes a small part of the mature ACP protein in front of GFP, whereas ACPsignal–GFP does not.
The GFP fusion protein made with the T.gondii ACP pre-sequence, ACPToxo–GFP, demonstrates the tolerance of the apicoplast-targeting system as this fusion was correctly targeted. Furthermore, lane 5 (Figure 4B) indicates that it was correctly processed at the predicted transit peptide–ACP boundary. Hence, the T.gondii transit peptide cleavage site was recognized in P.falciparum. A pre-processed band with the transit peptide attached was again evident. However, so too was an additional larger band of size indicating that the signal peptide was also attached in this form. This may indicate that the T.gondii signal peptide cleavage site was less efficiently recognized in P.falciparum.
Western analyses were also performed on the cells expressing the FabHleader–GFP fusion and the two mutant forms (Figure 4C). For FabHleader–GFP an equivalent pattern of two bands, with sizes corresponding to a mature and a pre-processed form, was observed (Figure 4C, lane 1). Identical Western blot profiles were seen for the two mutants (FabHleader[T40A]–GFP and FabHleader[T40/49A]–GFP) (Figure 4C, lanes 2 and 3), further supporting that the ablation of serine and threonine residues from the transit peptide does not affect protein targeting to, or processing within, the P.falciparum apicoplast.
Apicoplast life cycle and division in P.falciparum
Because the apicoplast is unpigmented, targeted GFP has provided our first opportunity to visualize the apicoplast, and track its development, throughout the asexual propagation of parasites in human erythrocytes. Each parasite contains only one apicoplast and Figure 5 shows its progress from young parasites (ring stage) through to cell division (schizogony) and egression of invasive merozoites. In ring stages the apicoplast is crescent shaped, very thin compared with subsequent forms, and typically at the periphery of the cell (Figure 5A). As the trophozoite grows, the apicoplast rounds up to form a sphere (Figure 5B and C). When the parasite has enlarged to fill most of the erythrocyte, a dramatic change in apicoplast morphology takes place. The apicoplast elongates and develops into a complex, multiply branched form (Figure 5D–F). The most highly branched state persists until the beginning of cytokinesis. The next apicoplast form, within mature schizonts immediately prior to merozoite release, consists of multiple, discrete, slightly rod-shaped apicoplasts (Figure 5G). Upon parasite release, each merozoite inherits only one apicoplast, and there is no apparent wastage left with parent cell debris (Figure 5H).
Discussion
An overwhelming trend in plastid evolution is the transfer of molecular genetic responsibility from the endosymbiont to the host cell (Martin and Herrmann, 1998; Martin et al., 1998; Palmer and Delwiche, 1998). No known plastid has completely lost its prokaryote-type organelle genome, but all show considerable genome reduction and dependency on the nucleus to fulfill plastid functions (Martin and Herrmann, 1998). Most of the nucleus-encoded genes that service the plastid appear to be derived from the endosymbiont genome, but the process of gene relocation is poorly understood (Martin et al., 1998). Regardless of origin, any nuclear gene whose protein product is required within the plastid demands some mechanism of protein trafficking.
Trafficking routes to plastids fall into two groups reflecting the two processes through which eukaryotic cells have acquired their plastids. The theory of primary endosymbiosis describes one of these processes and proposes that a cyanobacterium was directly engulfed and retained by a eukaryote (Cavalier-Smith, 1982; Delwiche, 1999). Such plastids (seen in plants, green algae, red algae and glaucophytes) are typically surrounded by two membranes and protein targeting to them is facilitated by an N-terminal transit peptide that is subsequently removed by a plastid-located peptidase (Cline and Henry, 1996; Schwartzbach et al., 1998). This scenario is analogous to that of mitochondria.
A second method of plastid acquisition is termed secondary endosymbiosis. In this process a eukaryotic cell engulfs and retains another eukaryote that already possesses a plastid (McFadden and Gilson, 1997; Delwiche, 1999). Whilst this internalized eukaryote is typically stripped of many of its redundant features, its plastid with its two-membrane envelope is retained by the new host. However, this plastid is contained within additional membranes believed to derive from the endosymbiont’s plasma membrane and the host’s food vacuolar membrane, and is thus termed a complex plastid. Such plastids are observed in many algal groups as well as in apicomplexan parasites (Delwiche, 1999; McFadden and Roos, 1999). The extra membranes of complex plastids present an additional obstacle for protein targeting to overcome. An accepted model of routing of proteins to such plastids is through the secretory pathway (Gibbs, 1979; Schwartzbach et al., 1998; Bodyl, 1999; Kroth and Strotmann, 1999). Targeted proteins must thus possess an N-terminal extension that contains a signal peptide, for entry into the secretory pathway, in front of a transit peptide for final targeting and import into the plastid. Evidence for this model comes from studies of algae where direct and indirect pieces of evidence support this protein routing to complex plastids (Bhaya and Grossman, 1991; Osafune et al., 1991; Sulli and Schwartzbach, 1995; Liaud et al., 1997; Lang et al., 1998). Our study further substantiates this model by demonstrating that P.falciparum also targets proteins to its complex plastid via the secretory pathway, and that this is mediated by N-terminal pre-sequences that consist of a signal and transit peptide.
The ability of the P.falciparum bipartite pre-sequences to target proteins to the apicoplast is consistent with previous studies in T.gondii where the equivalent bipartite pre-sequence composition is seen (Waller et al., 1998). Indeed, the capacity of the T.gondii leader to target proteins correctly to the P.falciparum apicoplast is strong evidence for a common apicoplast protein routing system in the two parasites. The reciprocal case has also been demonstrated, where P.falciparum pre-sequences will target proteins to the T.gondii apicoplast (Jomaa et al., 1999). In this study we have further dissected apicoplast targeting by examining the individual properties of the P.falciparum leader domains. A signal peptide is necessary to mediate protein entry into the secretory pathway. Without a signal peptide the protein accumulates in the cytoplasm and, presumably, is unable to reach the apicoplast. Once a protein enters the secretory pathway a transit peptide directs the protein into the apicoplast. Without a transit peptide proteins cannot enter the apicoplast and are secreted from the cell. Hence, neither component can successfully affect apicoplast targeting alone. Targeting to the apicoplast of P.falciparum, therefore, is at least a two-step process. Similar studies in T.gondii also reveal that a signal peptide and transit peptide mediate protein targeting via the secretory pathway to the apicoplast (D.Roos, personal communication), and indicate that this sequence of protein trafficking events to apicoplasts is probably common to all apicomplexan parasites.
Western blot analyses indicate that the components of the bipartite leader are removed in a similar two-step, sequential fashion. The signal peptide is removed first, most likely during co-translational import into the secretory system. Following this, the transit peptide is removed. Consistent with previous findings for T.gondii (Waller et al., 1998), pools of pre-processed apicoplast-targeted proteins (with transit peptides still attached) can be detected by Western blot analysis in P.falciparum. The lack of detectable GFP in the secretory system (by either fluorescence or immunoelectron microscopy) suggests that these stocks of pre-processed proteins are chiefly within the apicoplast and await final processing there. This is consistent with plant transit peptidases being located within chloroplasts (Richter and Lamppa, 1998). The exact cleavage sites for the transit peptides remain unknown. In plants and algae, where large numbers of plastid-targeted proteins have been defined, cleavage motifs have been identified (Emanuelsson et al., 1999). No such motif is yet evident in P.falciparum. However, given that P.falciparum is able to cleave the T.gondii ACP transit peptide appropriately it seems likely that a site with some conserved features will present itself, either as more apicoplast proteins are assembled or through N-terminal sequencing.
In P.falciparum, protein targeting to the apicoplast from within the endomembrane network is apparently highly efficient given the lack of any observed GFP in either the ER or other non-apicoplast regions. This tells us that the transit peptide is a potent sorting cue because proteins are directed to many specific destinations from within the secretion system of P.falciparum. These include intracellular destinations such as invasion-related organelles (rhoptries and micronemes) (Howard and Schmidt, 1995) and several extracellular locations, either in the parasitophorous membrane or within the host erythrocyte cytoplasm (Foley and Tilley, 1998). Targeting information for the apicoplast must discriminate between these options. At present the precise route through the endomembrane network to the apicoplast is unknown. No direct continuity is apparent between the apicoplast and the ER in P.falciparum nor are ribosomes evident on the outermost membrane of the apicoplast. Hence, immediate exposure of the apicoplast to proteins translationally imported into the ER probably does not occur (as is the case in several algal groups). Two possibilities exist: that apicoplast-targeted proteins utilize a specific component of the secretory pathway, with the transit peptide being instrumental in this discrimination; or, that the apicoplast is located within a more general secretory flow, perhaps through the Golgi, and that transit peptide-bearing proteins are trapped by the apicoplast as they wash past. From this study it is clear, however, that if a protein fails to be imported into the apicoplast from within the secretory pathway (in the case of the construct lacking a transit peptide) it joins the bulk flow out of the cell.
A caveat to the model of protein routing to complex plastids via the secretory pathway is its inability to explain trafficking across all four membranes of the complex plastid. The signal peptide is predicted to overcome the outermost membrane. The transit peptide, by analogy with plants, is expected to mediate transport across the two innermost membranes, and homologous components of this import machinery (Keegstra and Cline, 1999) are found in P.falciparum (our unpublished data). However, this leaves one membrane passage (the second in from the outside) unaccounted for. How much of an obstacle this extra membrane presents to the apicoplast and the other four membrane-bound plastids, and how it is overcome, remains unknown. We note that one study suggests that the P.falciparum apicoplast may only be surrounded by three membranes (Hopkins et al., 1999) as is also seen in select algae that possess complex plastids. This would make the above, bipartite model complete. However, our ultrastructural studies suggest that the P.falciparum apicoplast has four membranes as in T.gondii (Köhler et al., 1997; McFadden and Roos, 1999) and the interchangeability of pre-sequences is consistent with both apicoplasts having the same number of membranes.
In P.falciparum the paucity of serines and threonines in the transit peptides is in contrast to all other plastid-containing cells (von Heijne and Nishikawa, 1991; Liaud et al., 1997; Sulli et al., 1999). Although the role of such residues is uncertain, they have been shown in plants to be essential for transit peptide phosphorylation–dephosphorylation cycles that precede import of some chloroplast proteins, and may form a regulatory function in plastid protein import (Waegemann and Soll, 1996). These residues have also been implicated in amphipathic helix formation upon contact with plastid envelope lipids and hence affect transit peptide presentation to the import machinery (Wienk et al., 1999). The low number of serines and threonines in P.falciparum transit peptides (which extends to numerous unpublished sequences) may reflect the high AT codon bias of the P.falciparum genome (both residues require G- or C-containing codons). Given their putative roles in plants we considered that P.falciparum may represent the minimal requirement. However, the ablation of all such residues from the FabH pre-sequence had no observable consequence for protein targeting (as observed by both fluorescence microscopy and Western blot analysis). This may indicate a divergence of function of P.falciparum transit peptides, or that other residues fulfill the function of serine and threonines in P.falciparum.
Targeted GFP to the P.falciparum apicoplast provides additional insights into apicoplast biology by being a live marker of the apicoplast throughout the in vitro life cycle. The close association of the apicoplast with the mitochondrion has also been noted in electron microscopy studies (Hopkins et al., 1999). More intriguing is the nature and timing of division of the single apicoplast during parasite division (schizogony). The number of daughter cells produced per P.falciparum schizont is variable, generally between 8 and 24 (Kudo, 1971). This number must be tied to the final number of nuclei after a schizont completes successive and non-synchronous rounds of DNA replication and nuclear division (Read et al., 1993). The process of schizogony is unusual in that multiple nuclei are produced prior to any cytokinesis. For the apicoplast to segregate such that each daughter cell receives one apicoplast, apicoplast division needs to receive some cue as to the number of nuclei and divide accordingly. Delaying apicoplast division until the completion of nuclear division may thus be necessary. Elaborate branched forms of the apicoplast, which persist up until cytokinesis begins, may represent such a delay to achieve correct apicoplast number for equal segregation. Immunoelectron microscopy confirms that these branched forms are present in multi-nucleate parasites (not shown). Furthermore, in other apicomplexan parasites an association of the apicoplast with centrioles is seen, supporting a link between nuclear division and apicoplast segregation (Speer and Dubey, 1999). No sequentially dividing branched forms of the apicoplast were observed, suggesting that division may be simultaneous and is perhaps closely tied to segregation into the daughter cells as occurs with the two daughter apicoplasts of T.gondii (McFadden et al., 1997).
Materials and methods
GFP expression constructs
The GFP coding sequence was excised from the plasmid pCAT-GFP/HXGPRT (Striepen et al., 1998) using restriction endonucleases AvrII and PstI and ligated into pLITMUS 28 (New England BioLabs). To generate C-terminal GFP fusions with P.falciparum targeting sequences a primary GFP fusion was made with the P.falciparum ACP gene (DDBJ/EMBL/GenBank accession No. AF038928) by PCR amplification of P.falciparum cDNA using 5′ primer 1 (containing SacI–XhoI–BglII restriction sites) and 3′ primer 2 (with AvrII restriction site replacing the endogenous stop codon of the ACP gene). The amplicon was then restricted with SacI and AvrII and ligated to SacI–AvrII-restricted GFP-pLITMUS. The ACP–GFP fusion gene was then excised and inserted as a XhoI fragment into the P.falciparum expression vector pHC1 (Crabb et al., 1997) to create the construct ACPfull–GFP. Subsequent constructs were made by excising ACP from the primary GFP-pLITMUS fusion using SacI and AvrII, and replacing it with new amplicons. As XhoI fragments these GFP gene fusions were similarly inserted into pHC1. The constructs generated in this manner, and primers and DDBJ/EMBL/GenBank accession Nos of templates used to generate the 5′ fusion element are as follows: ACPleader–GFP, primers 1/3, AF038928; ACPtransit–GFP, primers 4/3, AF038928; ACPToxo–GFP, primers 5/6, AF038922; FabHleader–GFP, primers7/8, AF038929; FabHleader[T40A]–GFP, primers7/9, AF038929; and FabHleader[T40/49A]–GFP, primers7/10, AF038929.
A subsequent P.falciparum expression vector, pHH1 (Reed et al., 2000), was developed based on the existing expression vector pHC4 (Crabb et al., 1997). In this vector the T.gondii dhfr-ts region was excised with BamHI and KpnI and replaced with human dhfr mutated to encode resistance to WR99210 (Fidock and Wellems, 1997). This pHH1 vector was then digested with SacII and HinCII to release a 512 base pair fragment from the hsp86 upstream regulatory sequence. The SacI end was subsequently ‘filled in’ and vector religated to create the expression vector pHH2. GFP construct ACPsignal–GFP was based on pHH2. The P.falciparum ACP signal peptide region was amplified with primers 11/12, restricted and ligated between BglII and AvrII sites in the GFP-pLITMUS primary vector, and the ACPsignal–GFP fusion gene ligated as a XhoI fragment into pHH2.
Oligonucleotide primer numbers for PCR are listed below in 5′ to 3′ orientation. Upper case denotes complementary sequence to template DNA. Lower case denotes adapter sequences for cloning with restriction sites shown in brackets. Primers 9 and 10 created mutations in the FabH pre-sequence (shown underlined) as described in Results.
1. (SacI–XhoI–BglII) taggagctcgagatcTTAGAATGAAGATCTT ATTACTTTG
2. (AvrII) tggacctaggTTGCTTATTATTTTTTTCTATATAATCTAT AGC
3. (AvrII) tggacctaggTTTTAAAGAGCTAGATGGG
4. (SacI–XhoI) taggagctcgagaaaatgGTTAACGCTTTTAAAAATAC ACAAAAAGA
5. (SacI–XhoI) tacgagctcgagATGGAGATGCATCCCCG
6. (AvrII) catcctaggATCAGAACTCGCCTCGTC
7. (SacI–XhoI) taggagctcGAGAAAATGTTTCTATATTTTATTACATA TCTGTG
8. (AvrII) tggacctagGTCCTATTATTTTACCTCCAGTTTTCCC
9. (AvrII) tggacctagGTCCTATTATTTTACCTCCAGTTTTCCCATAA ATAGCACGTATTTTAGC
10. (AvrII) tggacctagGTCCTATTATTTTACCTCCAGCTTTCCCATA AATAGCACGTATTTTAGC
11. (BglII) ggaagatcTAGAATGAAGATATTATTACTTTGTAT
12. (AvrII) tggacctagGTGTATTTTTAAAAGCGTTAACATAATATAG
13. (BamHI) cgcggatccAGTACTTTTGATGATATTAAAAAAATT
14. (XhoI) tacctcgagTTATTGCTTATTATTTTTTTCTATATAATC
Parasite transformation
Plasmodium falciparum (parasite cloned line D10) parasites were cultured in vitro as previously described (Trager and Jensen, 1976). Transfection of GFP plasmid constructs was carried out by electroporation (Fidock and Wellems, 1997) and drug selection using either 0.1 µM pyrimethamine for HCl-based plasmids (Triglia et al., 1998), or 0.25 µM WR99210 (kindly supplied by D.Jacobus) for human dhfr-based plasmids. Transfectant lines were maintained on antibiotic selection.
Microscopy
Green fluorescence of GFP-expressing transfectant parasites was observed and captured in live cells using a Leica TCS 4D confocal microscope. Live mitochondrial staining was done using MitoTracker® Red CMXRos (Molecular Probes). Immunolabeling for electron microscopy was performed on ultrathin sections of cryopreserved parasites. Parasitized erythrocytes were drawn into dialysis tubing that was then immersed in hexadecene (Hohenberg et al., 1994) and rapidly frozen in a Leica EM HPF high pressure freezer. Cells were freeze substituted with acetone and embedded in Lowicryl HM20 resin using a Leica AFS automatic freeze substitution machine. Antibody incubations were performed at room temperature in phosphate-buffered saline (PBS) supplemented with bovine serum albumin (BSA) (0.8%) and Tween 80 (0.01%). After blocking in the above solution, incubations with GFP-antiserum (kindly provided by P.Silver) were followed by secondary antibodies conjugated to 20 nm gold particles (BioCell). Standard staining with uranium and lead preceded observation with a Philips CM120 BioTWIN transmission electron microscope at 120 kV.
Western blot analysis
Antiserum against the mature portion of P.falciparum ACP was generated in mice. The portion of the ACP gene encoding this region was amplified with PCR primers 13/14, and cloned in-frame into the pGEX 4T2 expression vector (Pharmacia). Glutathione S-transferase (GST)–ACP fusion protein was expressed in Escherichia coli and affinity-purified fusion protein used to immunize mice. Western blot analyses using ACP and GFP antisera were performed on lysates prepared from transfectant lines and untransfected blood stage parasites. Parasitized erythrocytes were saponin lysed and then boiled in reducing sample buffer. Secondary antibodies conjugated to horseradish peroxidase were used with the SuperSignal® (Pierce) chemiluminescent reagent for signal detection.
Acknowledgments
Acknowledgements
We are grateful to B.Striepen and D.Roos for providing the GFP clone and helpful guidance, D.Jacobus for providing WR99210, P.Silver for providing GFP-antiserum and T.Spurck for assistance with confocal microscopy. R.F.W. was supported by an Australian Postgraduate Research Award, M.B.R. by an NHMRC post-doctoral fellowship, and G.I.M and A.F.C. by Australian Research Council Awards. We thank the Red Cross Blood Service (Melbourne) for supply of blood products.
References
- Bhaya D. and Grossman,A. (1991) Targeting proteins to diatom plastids involves transport through an endoplasmic reticulum. Mol. Gen. Genet., 229, 400–404. [DOI] [PubMed] [Google Scholar]
- Bodyl A. (1999) How are plastid proteins of the apicomplexan parasites imported? A hypothesis. Acta Protozool., 38, 31–37. [Google Scholar]
- Cavalier-Smith T. (1982) The origins of plastids. Biol. J. Linn. Soc., 17, 289–306. [Google Scholar]
- Cline K. and Henry,R. (1996) Import and routing of nucleus-encoded chloroplast proteins. Annu. Rev. Cell. Dev. Biol., 12, 1–26. [DOI] [PubMed] [Google Scholar]
- Clough B., Strath,M., Preiser,P., Denny,P. and Wilson,R. (1997) Thiostrepton binds to malarial plastid rRNA. FEBS Lett., 406, 123–125. [DOI] [PubMed] [Google Scholar]
- Clough B., Rangachari,K., Strath,M., Preiser,P.R. and Wilson,R. (1999) Antibiotic inhibitors of organellar protein synthesis in Plasmodium falciparum. Protist, 150, 189–195. [DOI] [PubMed] [Google Scholar]
- Crabb B.S., Triglia,T., Waterkeyn,J.G. and Cowman,A.F. (1997) Stable transgene expression in Plasmodium falciparum. Mol. Biochem. Parasitol., 90, 131–144. [DOI] [PubMed] [Google Scholar]
- Delwiche C. (1999) Tracing the tread of plastid diversity through the tapestry of life. Am. Nat., 154, S164–S177. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Nielsen,H. and von Heijne,G. (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci., 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichera M.E. and Roos,D.S. (1997) A plastid organelle as a drug target in apicomplexan parasites. Nature, 390, 407–409. [DOI] [PubMed] [Google Scholar]
- Fidock D.A. and Wellems,T.E. (1997) Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl Acad. Sci. USA, 94, 10931–10936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley M. and Tilley,L. (1998) Protein trafficking in malaria-infected erythrocytes. Int. J. Parasitol., 28, 1671–1680. [DOI] [PubMed] [Google Scholar]
- Gibbs S.P. (1979) The route of entry of cytoplasmically synthesized proteins into chloroplasts of algae possessing chloroplast ER. J. Cell Sci., 35, 253–266. [DOI] [PubMed] [Google Scholar]
- Hohenberg H., Mannweiler,K. and Muller,M. (1994) High-pressure freezing of cell suspensions in cellulose capillary tubes. J. Microsc., 175, 34–43. [DOI] [PubMed] [Google Scholar]
- Hopkins J., Fowler,R., Krishna,S., Wilson,I., Mitchell,G. and Bannister,L. (1999) The plastid in Plasmodium falciparum asexual blood stages: a three-dimensional ultrastructural analysis. Protist, 150, 283–295. [DOI] [PubMed] [Google Scholar]
- Howard R.F. and Schmidt,C.M. (1995) The secretory pathway of Plasmodium falciparum regulates transport of p82/rap-1 to the rhoptries. Mol. Biochem. Parasitol., 74, 43–54. [DOI] [PubMed] [Google Scholar]
- Jomaa H. et al. (1999) Inhibitors of the nonmevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science, 285, 1573–1576. [DOI] [PubMed] [Google Scholar]
- Keegstra K. and Cline,K. (1999) Protein import and routing systems of chloroplasts. Plant Cell, 11, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra K. and Olsen,L. (1989) Chloroplastic precursors and their transport across the envelope membranes. Annu. Rev. Plant Physiol. Plant Mol. Biol., 40, 471–501. [Google Scholar]
- Keeling P.J., Palmer,J.D., Donald,R.G.K., Roos,D.S., Waller,R.F. and McFadden,G.I. (1998) Shikimate pathway in apicomplexan parasites. Nature, 397, 219–220. [DOI] [PubMed] [Google Scholar]
- Köhler S., Delwiche,C.F., Denny,P.W., Tilney,L.G., Webster,P., Wilson,R.J.M., Palmer,J.D. and Roos,D.S. (1997) A plastid of probable green algal origin in apicomplexan parasites. Science, 275, 1485–1488. [DOI] [PubMed] [Google Scholar]
- Kroth P. and Strotmann,H. (1999) Diatom plastids: secondary endocytobiosis, plastid genome and protein import. Physiol. Plant., 107, 136–141. [Google Scholar]
- Kudo R. (1971) Protozoology. Charles C.Thomas, Springfield, IL. [Google Scholar]
- Lang M., Apt,K.E. and Kroth,P.G. (1998) Protein transport into complex diatom plastids utilizes two different targeting signals. J. Biol. Chem., 273, 30973–30978. [DOI] [PubMed] [Google Scholar]
- Liaud M.-F., Brandt,U., Scherzinger,M. and Cerff,R. (1997) Evolutionary origin of cryptomonad microalgae: two novel chloroplast/cytosol-specific GAPDH genes as potential markers of ancestral endosymbiont and host cell components. J. Mol. Evol. (Suppl. 1), 44, S28–S37. [DOI] [PubMed] [Google Scholar]
- Martin W. and Herrmann,R.G. (1998) Gene transfer from organelles to the nucleus: how much, what happens and why? Plant Physiol., 118, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W., Stoebe,B., Goremykin,V., Hansmann,S., Hasegawa,M. and Kowallik,K. (1998) Gene transfer to the nucleus and the evolution of chloroplasts. Nature, 393, 162–165. [DOI] [PubMed] [Google Scholar]
- McFadden G.I. and Gilson,P.R. (1997) What’s eating Eu? The role of eukaryote/eukaryote endosymbioses in plastid origins. In Schenk,H.E.A., Herrmann,R., Jeon,K.W., Müller,N.E. and Schwemmler,W. (eds), Eukaryotism and Symbiosis. Springer-Verlag, Heidelberg, Germany, pp. 24–39. [Google Scholar]
- McFadden G.I. and Roos,D.S. (1999) Apicomplexan plastids as drug targets. Trends Microbiol., 6, 328–333. [DOI] [PubMed] [Google Scholar]
- McFadden G.I. and Waller,R.F. (1997) Plastids in parasites of humans. BioEssays, 19, 1033–1040. [DOI] [PubMed] [Google Scholar]
- McFadden G.I., Reith,M., Munholland,J. and Lang-Unnasch,N. (1996) Plastid in human parasites. Nature, 381, 482. [DOI] [PubMed] [Google Scholar]
- McFadden G.I., Waller,R.F., Reith,M., Munholland,J. and Lang-Unnasch,N. (1997) Plastids in apicomplexan parasites. Pl. Syst. Evol. (Suppl.), 11, 261–287. [Google Scholar]
- Nakai K. and Kanehisa,M. (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics, 14, 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- Osafune T., Schiff,J. and Hase,E. (1991) Stage-dependent localization of LHCP II apoprotein in the Golgi of synchronized cells of Euglena gracilis by immunogold electron microscopy. Exp. Cell Res., 193, 320–330. [DOI] [PubMed] [Google Scholar]
- Palmer J.D. and Delwiche,C.F. (1998) The origin and evolution of plastids and their genomes. In Soltis,D.E., Soltis,P.S. and Doyle,J.J. (eds), Molecular Systematics of Plants II. Chapman Hall, New York, NY, pp. 375–409. [Google Scholar]
- Read M., Sherwin,T., Holloway,S., Gull,K. and Hyde,J. (1993) Microtubular organization visualized by immunofluorescence microscopy during erythrocytic schizogony in Plasmodium falciparum and investigation of post-translational modifications of parasite tubulin. Parasitology, 106, 223–232. [DOI] [PubMed] [Google Scholar]
- Reed M.B., Saliba,K.J., Caruana,S.R., Kirk,K. and Cowman,A.F. (2000) Pgh1 modulates sensitivity and resistance to multiple antimalarials in Plasmodium falciparum. Nature, 403, 906–909. [DOI] [PubMed] [Google Scholar]
- Richter S. and Lamppa,G.K. (1998) A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc. Natl Acad. Sci. USA, 95, 7463–7468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartzbach S.D., Osafune,T. and Loffelhardt,W. (1998) Protein import into cyanelles and complex chloroplasts. Plant Mol. Biol., 38, 247–263. [PubMed] [Google Scholar]
- Speer C.A. and Dubey,J.P. (1999) Ultrastructure of schizonts and merozoites of Sarcocystis falcatula in the lungs of budgerigars (Melopsittacus undulatus). J. Parasitol., 85, 630–637. [PubMed] [Google Scholar]
- Striepen B., He,C.Y., Matrajt,M., Soldati,D. and Roos,D.S. (1998) Expression, selection and organellar targeting of the green fluorescent protein in Toxoplasma gondii. Mol. Biochem. Parasitol., 92, 328–338. [DOI] [PubMed] [Google Scholar]
- Sulli C. and Schwartzbach,S.D. (1995) The polyprotein precursor of the Euglena light-harvesting chlorophyll a/b binding protein is transported to the Golgi apparatus prior to chloroplast import and polyprotein processing. J. Biol. Chem., 270, 13084–13090. [DOI] [PubMed] [Google Scholar]
- Sulli C., Fang,Z.W., Muchal,U. and Schwartzbach,S.D. (1999) Topology of Euglena chloroplast protein precursors within the endoplasmic reticulum to Golgi to chloroplast transport vesicles. J. Biol. Chem., 274, 457–463. [DOI] [PubMed] [Google Scholar]
- Trager W. and Jensen,J. (1976) Human malaria parasites in continuous culture. Science, 193, 673–675. [DOI] [PubMed] [Google Scholar]
- Triglia T., Wang,P., Sims,P.F.G., Hyde,J.E. and Cowman,A.F. (1998) Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J., 17, 3807–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. and Nishikawa,K. (1991) Chloroplast transit peptides: the perfect random coil? FEBS Lett., 278, 1–3. [DOI] [PubMed] [Google Scholar]
- Waegemann K. and Soll,J. (1996) Phosphorylation of the transit sequence of chloroplast precursor proteins. J. Biol. Chem., 271, 6545–6554. [DOI] [PubMed] [Google Scholar]
- Waller R.F., Keeling,P.J., Donald,R.G.K., Striepen,B., Handman,E., Lang-Unnasch,N., Cowman,A.F., Besra,G.S., Roos,D.S. and McFadden,G.I. (1998) Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc. Natl Acad. Sci. USA, 95, 12352–12357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienk H.L.J., Czisch,M. and de Kruijff,B. (1999) The structural flexibility of the preferredoxin transit peptide. FEBS Lett., 453, 318–326. [DOI] [PubMed] [Google Scholar]
- Wilson R.J.M. et al. (1996) Complete gene map of the plastid-like DNA of the malaria parasite Plasmodium falciparum. J. Mol. Biol., 261, 155–172. [DOI] [PubMed] [Google Scholar]