Recently, a five-center collaborative study1 reported that genetic variations of glypican 5 (GPC5) may significantly contribute to an increased risk of lung cancer in never smokers. GPC5 gene-expression levels in normal lung tissues were found significantly lower in individuals who carry high-risk alleles, and the GPC5 expression level in adenocarcinoma tissue was significantly lower than in matched normal lung tissue. Reduction of expression of GPC5 may lead to the development of lung cancer, suggesting that this gene normally functions as a tumor suppressor.
GPC5 is a member of the glypican gene family. Glypicans are a family of heparan sulphate proteoglycans (HSPGs) that are linked to the exocytoplasmic surface of the plasma membrane by a glycosyl-phosphatidylinositol (GPI) anchor. There are six glypican family members in the Human Genome (GPC1 to GPC6).2 GPC1 is overexpressed in human pancreatic 3 and breast cancers.4 GPC3, the family member that shows highest homology to GPC5, is overexpressed in neuroblastoma, Wilm’s tumors,5 and melanoma.6 GPC3 was also shown to be overexpressed in hepatocellular carcinoma,7 and engineered GPC3 overexpression in hepatocellular carcinoma cell lines was associated with modulated proliferation.8 Missense mutations in GPC3 are found in Simpson-Golabi-Behemel syndrome, which is associated with overgrowth and a reported predisposition to develop pediatric tumors.9
GPC5 gene has eight exons encoding 572 amino acids and spans a large genomic region of 1.47 Mb at chromosome 13q31.3.10 Alterations at the GPC5 locus are a common event in various human tumors. Amplifications at 13q31–32 are frequently seen across several tumor types, including lymphomas, breast cancers, and neurological tumors.11–13 For lung cancer, an array CGH-based study reported a homozygous deletion at 13q31.3 in a non-small-cell lung cancer cell line.14 Another array CGH based study recently analyzed a series of 14 patients with 13q partial deletion syndrome and noted lung hypoplasia as one of the common phenotypes. Among the 14 patients, two had lung hypoplasia.15 Amplification and overexpression of the miR-17-92 miRNA cluster at 13q31.3 was recently reported in lung cancers from one study;16 another study showed that inhibition of miR-17-5p and miR-20a with antisense oligonucleotides can induce apoptosis selectively in lung cancer cells overexpressing miR-17-92.17 When using TargetScan (http://www.targetscan. org/vert_50/) to predict biological targets of miRNAs, miR-17, miR-20a, miR-20b, miR-23a, and miR-23b were found to target GPC5 3’UTR. These results suggested that miRNA regulating GPC5 expression may be important in the development of lung cancers and warrant in-depth investigation. Moreover, epigenetic silencing by hypermethylation of the CpG-rich region of GPC5 leads to loss of GPC5 function, which may in turn lead to carcinogenesis. Additional studies will be required to unravel the mechanisms of silencing or mutations of 13q31.3 region and its role in lung cancer.
Different SNPs in GPC5 have also been implicated in susceptibility to multiple sclerosis (MS).18 A recent meta-analysis of cancer risk in MS has shown that the risk of lung cancer is significantly decreased in individuals with MS.19 One study showed a particularly strongly decreased risk for cancers of the respiratory tract in MS patients.20 HSPGs were found to be often expressed in reduced amounts in non-small-cell lung carcinomas, particularly poorly differentiated tumors, compared to normal epithelia.21 Our results also showed that GPC5 expression is significantly lower in adenocarcinoma than in matched normal lung tissue.1 In the Oncomine microarray databases, there are nine studies on lung cancer,22–30 seven 22–28 of which included lung adenocarcinoma. Two datasets 25, 28 showed significant downregulation of GPC5 in adenocarcinoma tumors compared with normal lung tissue. Importantly, two studies 25, 26 included smoking status information, and both showed lower expression in never smokers than in smokers. Four studies 23, 24, 29, 30 reported the GPC5 expression information from other histological types, including carcinoid, squamous, small-cell carcinoma, and large-cell carcinoma, and showed no significant differences in GPC5 expression between tumor and normal tissues. Thus, reduced GPC5 expression could be specific to adenocarcinoma in never smokers. However, owing to sample sizes and the different characteristics of study samples available in Oncomine, this conclusion needs to be further validated. All of the current evidence appears to support GPC5 to be a protective factor from developing lung cancer; mechanisms are open for further investigation.
Although there is no direct evident for GPC5, HSPGs as a group are known to interact with many proteins including growth factors, chemokines, and structural proteins of the extracellular matrix to influence cell growth, differentiation, and the cellular response to the environment.31 The main function of the membrane-attached glypicans is to regulate the signaling pathways of Wnt, hedgehog (Hh), and fibroblast growth factors (FGF).32 A recent study has shown that GPC5 increases proliferation in rhabdomyosarcoma through potentiating the effects of FGF2 and Wnt1; GPC5 enhanced the intracellular signaling of FGF2 and altered the cellular distribution of FGF2.33 Previous and recent studies have highlighted potentially significant roles for Wnt, Hh, and FGF signaling pathways in lung cancer development.
Dysregulated Wnt signaling has been found in lung cancer, in particular, non-small cell lung cancer (NSCLC).34, 35 Several Wnt proteins are differentially expressed in NSCLC specimens, including Wnt1, -2, and -7a. Wnt1 and Wnt2 are overexpressed in NSCLC samples, and cancer cells expressing Wnt1 are resistant to apoptotic therapies.36, 37 Conversely, inhibition of Wnt1 and Wnt2 led to apoptosis in human cancer cells and reduced tumor growth in vivo and in vitro.36, 38 Wnt7a is decreased in NSCLC; its re-expression leads to growth inhibition of NSCLC cell lines.39
Persistent Hh pathway activation is seen in small cell lung cancer (SCLC), manifested by a high level of expression of sonic hedgehog (Shh), Patched, and GLi1.40 Treatment of SCLC cell lines with a specific inhibitor of the Hh pathway (cyclopamine) produced tumour growth arrest. Cell lines were protected from cyclopamine inhibition by constitutive overexpression of the Hh pathway transcription factor GLi1.41 Studies on cell lines demonstrated that 5 of 7 SCLC cell lines expressed both Shh and Gli1 in contrast to NSCLC, which expressed only Shh but not Gli1. Analysis of clinical samples of human lung cancer tissue demonstrated 50% (five of 10) of SCLC expressed both Shh and Gli1 compared to only 10% (four of 40) of NSCLC.42 Another study reported that 85% (34 of 40) of SCLC expressed Gli1 and more than 60% have a medium to strong expression correlating with increased Hh signaling.43 Thus, it appears that the degree of dependence on Hh signaling varies among the subtypes of lung cancer.
Fibroblast growth factor (FGF) belongs to a family of ubiquitously expressed ligands, which bind to the extracellular domain of fibroblast growth factor receptors (FGFRs), initiating a signal transduction cascade, which promotes cell proliferation, motility, and angiogenesis. Dysregulation of the FGF signaling pathway has been associated with cancer development.44–46 The FGF pathway has been shown to be activated in lung cancer.47–51 Elevated levels of FGF and FGFR proteins have been detected in NSCLC cell lines and in human lung cancers.49, 52, 53 Behrens et al 54 described high levels of immunohistochemical expression of basic FGF, FGFR1, and FGFR2 in a large series of NSCLC specimens, including the two most frequent histologic types, squamous cell carcinoma (SCC) and adenocarcinoma. Behrens et al found higher levels of basic FGF and FGFRs expression in tumor cells than in adjacent normal bronchial epithelia at cytoplasmic localization in both SCC and adenocarcinoma. Additionally, in adenocarcinoma specimens, Behrens et al detected differences in the expression of the three markers and patients' smoking status, with cytoplasmic FGFR1 expression being significantly higher in smokers and nuclear FGFR1 and FGFR2 significantly higher in never smokers. These differences highlight the potential differential role of these proteins in the pathogenesis of both smoking and non-smoking-related lung cancers. Another recent study supports the previous studies and also provides molecular evidence for an active FGF autocrine signaling pathway in a subset of NSCLC cell lines.55
In summary, with all the information of GPC5 and the glypican family, we hypothesize that GPC5 regulates lung cancer development through a complex pathway network, particularly through Wnt, Hh, and FGF signaling pathways and their interactions. As shown in Figure 1, depending on the context, GPC5 may have a stimulatory or inhibitory activity on these pathways, which are important in regulating cell proliferation, division, and survival. The challenge in the future will be to elucidate the precise regulation mechanisms of the GPC5 signaling pathway in lung tumorigenesis and in lung cancer of never smokers and smokers. A better understanding of GPC5’s role in signaling pathways, particularly the upstream event that suppresses GPC5 expression and reduces functional GPC5, or the downstream event that unleashes the oncogenic processes may provide a unique opportunity for developing novel and effective strategies for early detection, targeted chemoprevention, and treatment of lung cancer.
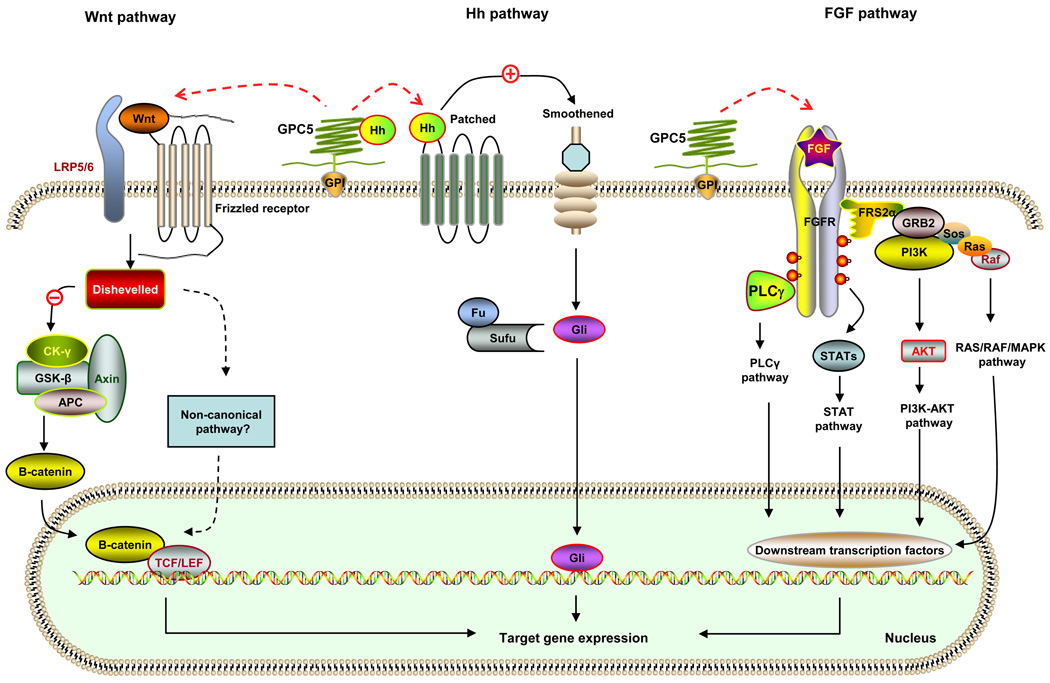
Figure 1. Possible signal network of GPC5 in lung cancer development.
The main function of membrane-attached GPC5 is thought to regulate the pathway signaling of Wnt, Hedgehogs (Hh), and fibroblast growth factors (FGFs). Depending on the biological context, GPC5 can either stimulate or inhibit cell signaling activity. GPC5 may bind to Wnt and to the Frizzled receptor in association with the co-receptors LRP5/6. Activation of the Frizzled receptor results in subsequent activation of Dishevelled. Through an unknown mechanism, Dishevelled inhibits the function of a multi-protein complex including Axin, APC, CK-γ, and GSK-β, which blocks phosphorylation of β-catenin and prevents its degradation. Increasing levels of free β-catenin translocate to the nucleus, complex with the TCF/LEF cofactors and activate target genes. In the case of the Hh pathway, it is proposed that GPC5 may inhibit the signaling by competing with Patched (the Hh receptor) for Hh binding. Binding Hh with the Patched receptor alters the interaction of Patched with Smoothened, a G protein-coupled receptor, resulting in the activation of Smoothened. This initiates a cascade of events resulting in the Gli entering the nucleus and acting as transcriptional activators. When the Hh signaling is lacking, Gli is bound to a multiprotein complex consisting of Fused (Fu) and the suppressor of Fu (SuFu) and will not enter the nucleus. GPC5 may activate the FGF pathway through unknown mechanisms. Following FGF ligand binding and receptor dimerization, the kinase domains transphosphorylate each other, leading to the docking of adaptor proteins and the activation of four key downstream pathways: RAS–RAF–MAPK, PI3K–AKT, signal transducer and activator of transcription (STAT) and phospholipase Cγ (PLCγ). The biological outcomes of FGF/FGFR activation are dependent on a complex network of signaling and transcriptional events regulated by multiple factors. CK-γ, casein kinase γ; GSK-β, glycogen synthase kinase β; APC, adenomatous polyposis coli; TCF, T cell–specific transcription factor; LEF, lymphoid enhancer–binding factor; Hh, hedgehog; Fu, fused; SuFu, suppressor of Fu; GPI, glycosylphosphatidylinositol; FGF, fibroblast growth factor; FGFR, fibroblast growth factor receptor; FRS2α, FGFR substrate 2α; GRB2, growth factor receptor-bound 2; Sos, son of sevenless; PLC γ, phospholipase C γ; STAT, signal-transducer and activator of transcription protein; MAPK, mitogen activated kinase-like protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li Y, Sheu CC, Ye Y, et al. Genetic variants and risk of lung cancer in never smokers: a genome-wide association study. Lancet Oncol. 2010;11(4):321–330. doi: 10.1016/S1470-2045(10)70042-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Filmus J, Capurro M, Rast J. Glypicans. Genome Biol. 2008;9(5):224. doi: 10.1186/gb-2008-9-5-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleeff J, Ishiwata T, Kumbasar A, et al. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J Clin Invest. 1998;102(9):1662–1673. doi: 10.1172/JCI4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matsuda K, Maruyama H, Guo F, et al. Glypican-1 is overexpressed in human breast cancer and modulates the mitogenic effects of multiple heparin-binding growth factors in breast cancer cells. Cancer Res. 2001;61(14):5562–5569. [PubMed] [Google Scholar]
- 5.Saikali Z, Sinnett D. Expression of glypican 3 (GPC3) in embryonal tumors. Int J Cancer. 2000;89(5):418–422. [PubMed] [Google Scholar]
- 6.Nakatsura T, Kageshita T, Ito S, et al. Identification of glypican-3 as a novel tumor marker for melanoma. Clin Cancer Res. 2004;10(19):6612–6621. doi: 10.1158/1078-0432.CCR-04-0348. [DOI] [PubMed] [Google Scholar]
- 7.Capurro M, Wanless IR, Sherman M, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/s0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 8.Midorikawa Y, Ishikawa S, Iwanari H, et al. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer. 2003;103(4):455–465. doi: 10.1002/ijc.10856. [DOI] [PubMed] [Google Scholar]
- 9.Cano-Gauci DF, Song HH, Yang H, et al. Glypican-3-deficient mice exhibit developmental overgrowth and some of the abnormalities typical of Simpson-Golabi-Behmel syndrome. J Cell Biol. 1999;146(1):255–264. doi: 10.1083/jcb.146.1.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Veugelers M, Vermeesch J, Reekmans G, et al. Characterization of glypican-5 and chromosomal localization of human GPC5, a new member of the glypican gene family. Genomics. 1997;40(1):24–30. doi: 10.1006/geno.1996.4518. [DOI] [PubMed] [Google Scholar]
- 11.Neat MJ, Foot N, Jenner M, et al. Localisation of a novel region of recurrent amplification in follicular lymphoma to an approximately 6.8 Mb region of 13q32–33. Genes Chromosomes Cancer. 2001;32(3):236–243. doi: 10.1002/gcc.1187. [DOI] [PubMed] [Google Scholar]
- 12.Ojopi EP, Rogatto SR, Caldeira JR, et al. Comparative genomic hybridization detects novel amplifications in fibroadenomas of the breast. Genes Chromosomes Cancer. 2001;30(1):25–31. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1057>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 13.Reardon DA, Jenkins JJ, Sublett JE, et al. Multiple genomic alterations including N-myc amplification in a primary large cell medulloblastoma. Pediatr Neurosurg. 2000;32(4):187–191. doi: 10.1159/000028932. [DOI] [PubMed] [Google Scholar]
- 14.Imoto I, Izumi H, Yokoi S, et al. Frequent silencing of the candidate tumor suppressor PCDH20 by epigenetic mechanism in non-small-cell lung cancers. Cancer Res. 2006;66(9):4617–4626. doi: 10.1158/0008-5472.CAN-05-4437. [DOI] [PubMed] [Google Scholar]
- 15.Kirchhoff M, Bisgaard AM, Stoeva R, et al. Phenotype and 244k array-CGH characterization of chromosome 13q deletions: an update of the phenotypic map of 13q21.1-qter. Am J Med Genet A. 2009;149A(5):894–905. doi: 10.1002/ajmg.a.32814. [DOI] [PubMed] [Google Scholar]
- 16.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65(21):9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara H, Takeuchi T, Nishikawa E, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene. 2007;26(41):6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 18.Baranzini SE, Wang J, Gibson RA, et al. Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet. 2009;18(4):767–778. doi: 10.1093/hmg/ddn388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handel AE, Joseph A, Ramagopalan SV. Multiple sclerosis and lung cancer: an unexpected inverse association. QJM. 2010;103(8):625–626. doi: 10.1093/qjmed/hcq071. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen NM, Rostgaard K, Rasmussen S, et al. Cancer risk among patients with multiple sclerosis: a population-based register study. Int J Cancer. 2006;118(4):979–984. doi: 10.1002/ijc.21437. [DOI] [PubMed] [Google Scholar]
- 21.Nackaerts K, Verbeken E, Deneffe G, et al. Heparan sulfate proteoglycan expression in human lung-cancer cells. Int J Cancer. 1997;74(3):335–345. doi: 10.1002/(sici)1097-0215(19970620)74:3<335::aid-ijc18>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 22.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8(8):816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 23.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98(24):13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98(24):13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Landi MT, Dracheva T, Rotunno M, et al. Gene expression signature of cigarette smoking and its role in lung adenocarcinoma development and survival. PLoS One. 2008;3(2):e1651. doi: 10.1371/journal.pone.0001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Powell CA, Spira A, Derti A, et al. Gene expression in lung adenocarcinomas of smokers and nonsmokers. Am J Respir Cell Mol Biol. 2003;29(2):157–162. doi: 10.1165/rcmb.2002-0183RC. [DOI] [PubMed] [Google Scholar]
- 27.Stearman RS, Dwyer-Nield L, Zerbe L, et al. Analysis of orthologous gene expression between human pulmonary adenocarcinoma and a carcinogen-induced murine model. Am J Pathol. 2005;167(6):1763–1775. doi: 10.1016/S0002-9440(10)61257-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su LJ, Chang CW, Wu YC, et al. Selection of DDX5 as a novel internal control for Q-RT-PCR from microarray data using a block bootstrap re-sampling scheme. BMC Genomics. 2007;8:140. doi: 10.1186/1471-2164-8-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talbot SG, Estilo C, Maghami E, et al. Gene expression profiling allows distinction between primary and metastatic squamous cell carcinomas in the lung. Cancer Res. 2005;65(8):3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 30.Wachi S, Yoneda K, Wu R. Interactome-transcriptome analysis reveals the high centrality of genes differentially expressed in lung cancer tissues. Bioinformatics. 2005;21(23):4205–4208. doi: 10.1093/bioinformatics/bti688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blackhall FH, Merry CL, Davies EJ, et al. Heparan sulfate proteoglycans and cancer. Br J Cancer. 2001;85(8):1094–1098. doi: 10.1054/bjoc.2001.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Cat B, David G. Developmental roles of the glypicans. Semin Cell Dev Biol. 2001;12(2):117–125. doi: 10.1006/scdb.2000.0240. [DOI] [PubMed] [Google Scholar]
- 33.Williamson D, Selfe J, Gordon T, et al. Role for amplification and expression of glypican-5 in rhabdomyosarcoma. Cancer Res. 2007;67(1):57–65. doi: 10.1158/0008-5472.CAN-06-1650. [DOI] [PubMed] [Google Scholar]
- 34.He B, Jablons DM. Wnt signaling in stem cells and lung cancer. Ernst Schering Found Symp Proc. 2006 May;:27–58. doi: 10.1007/2789_2007_043. [DOI] [PubMed] [Google Scholar]
- 35.Van Scoyk M, Randall J, Sergew A, et al. Wnt signaling pathway and lung disease. Transl Res. 2008;151(4):175–180. doi: 10.1016/j.trsl.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 36.He B, You L, Uematsu K, et al. A monoclonal antibody against Wnt-1 induces apoptosis in human cancer cells. Neoplasia. 2004;6(1):7–14. doi: 10.1016/s1476-5586(04)80048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen S, Guttridge DC, You Z, et al. Wnt-1 signaling inhibits apoptosis by activating beta-catenin/T cell factor-mediated transcription. J Cell Biol. 2001;152(1):87–96. doi: 10.1083/jcb.152.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.You L, He B, Xu Z, et al. Inhibition of Wnt-2-mediated signaling induces programmed cell death in non-small-cell lung cancer cells. Oncogene. 2004;23(36):6170–6174. doi: 10.1038/sj.onc.1207844. [DOI] [PubMed] [Google Scholar]
- 39.Winn RA, Marek L, Han SY, et al. Restoration of Wnt-7a expression reverses non-small cell lung cancer cellular transformation through frizzled-9-mediated growth inhibition and promotion of cell differentiation. J Biol Chem. 2005;280(20):19625–19634. doi: 10.1074/jbc.M409392200. [DOI] [PubMed] [Google Scholar]
- 40.Huang S, Yang L, An Y, et al. Expression of hedgehog signaling molecules in lung cancer. Acta Histochem. 2010 Jul 23; doi: 10.1016/j.acthis.2010.06.003. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Xie G, Fan Q, et al. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29(4):469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 42.Watkins DN, Berman DM, Burkholder SG, et al. Hedgehog signalling within airway epithelial progenitors and in small-cell lung cancer. Nature. 2003;422(6929):313–317. doi: 10.1038/nature01493. [DOI] [PubMed] [Google Scholar]
- 43.Vestergaard J, Pedersen MW, Pedersen N, et al. Hedgehog signaling in small-cell lung cancer: frequent in vivo but a rare event in vitro. Lung Cancer. 2006;52(3):281–290. doi: 10.1016/j.lungcan.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 44.Dailey L, Ambrosetti D, Mansukhani A, et al. Mechanisms underlying differential responses to FGF signaling. Cytokine Growth Factor Rev. 2005;16(2):233–247. doi: 10.1016/j.cytogfr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 45.Mohammadi M, Olsen SK, Ibrahimi OA. Structural basis for fibroblast growth factor receptor activation. Cytokine Growth Factor Rev. 2005;16(2):107–137. doi: 10.1016/j.cytogfr.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Powers CJ, McLeskey SW, Wellstein A. Fibroblast growth factors, their receptors and signaling. Endocr Relat Cancer. 2000;7(3):165–197. doi: 10.1677/erc.0.0070165. [DOI] [PubMed] [Google Scholar]
- 47.Bremnes RM, Camps C, Sirera R. Angiogenesis in non-small cell lung cancer: the prognostic impact of neoangiogenesis and the cytokines VEGF and bFGF in tumours and blood. Lung Cancer. 2006;51(2):143–158. doi: 10.1016/j.lungcan.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 48.Guddo F, Fontanini G, Reina C, et al. The expression of basic fibroblast growth factor (bFGF) in tumor-associated stromal cells and vessels is inversely correlated with non-small cell lung cancer progression. Hum Pathol. 1999;30(7):788–794. doi: 10.1016/s0046-8177(99)90139-9. [DOI] [PubMed] [Google Scholar]
- 49.Kuhn H, Kopff C, Konrad J, et al. Influence of basic fibroblast growth factor on the proliferation of non-small cell lung cancer cell lines. Lung Cancer. 2004;44(2):167–174. doi: 10.1016/j.lungcan.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 50.Takanami I, Tanaka F, Hashizume T, et al. The basic fibroblast growth factor and its receptor in pulmonary adenocarcinomas: an investigation of their expression as prognostic markers. Eur J Cancer. 1996;32A(9):1504–1509. doi: 10.1016/0959-8049(95)00620-6. [DOI] [PubMed] [Google Scholar]
- 51.Volm M, Koomagi R, Mattern J, et al. Prognostic value of basic fibroblast growth factor and its receptor (FGFR-1) in patients with non-small cell lung carcinomas. Eur J Cancer. 1997;33(4):691–693. doi: 10.1016/s0959-8049(96)00411-x. [DOI] [PubMed] [Google Scholar]
- 52.Berger W, Setinek U, Mohr T, et al. Evidence for a role of FGF-2 and FGF receptors in the proliferation of non-small cell lung cancer cells. Int J Cancer. 1999;83(3):415–423. doi: 10.1002/(sici)1097-0215(19991029)83:3<415::aid-ijc19>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 53.Chandler LA, Sosnowski BA, Greenlees L, et al. Prevalent expression of fibroblast growth factor (FGF) receptors and FGF2 in human tumor cell lines. Int J Cancer. 1999;81(3):451–458. doi: 10.1002/(sici)1097-0215(19990505)81:3<451::aid-ijc20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 54.Behrens C, Lin HY, Lee JJ, et al. Immunohistochemical expression of basic fibroblast growth factor and fibroblast growth factor receptors 1 and 2 in the pathogenesis of lung cancer. Clin Cancer Res. 2008;14(19):6014–6022. doi: 10.1158/1078-0432.CCR-08-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Marek L, Ware KE, Fritzsche A, et al. Fibroblast growth factor (FGF) and FGF receptor-mediated autocrine signaling in non-small-cell lung cancer cells. Mol Pharmacol. 2009;75(1):196–207. doi: 10.1124/mol.108.049544. [DOI] [PMC free article] [PubMed] [Google Scholar]