Abstract
The PmrA–PmrB two-component system of Salmonella enterica controls resistance to the peptide antibiotic polymyxin B and to several antimicrobial proteins from human neutrophils. Transcription of PmrA-activated genes is induced by high iron, but can also be promoted by growth in low magnesium in a process that requires another two-component system, PhoP–PhoQ. Here, we define the genetic basis for the interaction between the PhoP–PhoQ and PmrA–PmrB systems. We have identified pmrD as a PhoP-activated gene that mediates the transcriptional activation of PmrA-regulated genes during growth in low magnesium. When transcription of pmrD is driven from a heterologous promoter, expression of PmrA-activated genes occurs even at repressing magnesium concentrations and becomes independent of the phoP and phoQ genes. The PmrD effect is specific for PmrA-regulated genes and requires functional PmrA and PmrB proteins. A pmrD mutant is sensitive to polymyxin if grown in low magnesium, but resistant if grown in high iron. The PmrD protein controls the activity of the PmrA–PmrB system at a post-transcriptional level.
Keywords: magnesium/PhoP–PhoQ/PmrA–PmrB/signal transduction/transcription
Introduction
Two-component regulatory systems are signal transduction machineries employed by eubacteria, archaebacteria, and the cell-wall-containing eukaryotes Saccharomyces cerevisiae, Neurospora crassa and Arabidopsis thaliana (Loomis et al., 1997; Perraud et al., 1999). These systems use reversible phosphorylation between two proteins, a sensor kinase and a response regulator, to mediate the adaptation to changing environmental conditions. Sensors are usually integral membrane proteins that respond to particular chemical or physical signals by modulating the phosphorylated state of their cognate regulators, which are often transcription factors whose conformation or affinity for their promoter sequences is controlled by phosphorylation (Stock et al., 1995). Eubacterial species harbor many two-component systems, each responding to distinct cues and controlling the expression of discrete sets of genes.
The PmrA–PmrB two-component system of Salmonella enterica serovar typhimurium is required for resistance to polymyxin B and to other antimicrobial compounds (Mäkelä et al., 1978; Shafer et al., 1984a,b). The PmrA–PmrB system controls the transcription of several loci including: pbgP and ugd (designated pmrF and pmrE, respectively, by Gunn et al., 1998), which mediate the modification of the lipopolysaccharide and are necessary for polymyxin resistance (Groisman et al., 1997; Gunn et al., 1998); pmrG, which encodes a protein of unknown function (Gunn et al., 1998); and the pmrCAB operon (Gunn and Miller, 1996; Soncini and Groisman, 1996), indicative that the PmrA–PmrB system is autogenously regulated (Figure 1). In addition, it has been hypothesized that the pmrD gene might be PmrA regulated because, when present in a medium-copy-number plasmid, it confers polymyxin resistance in a PmrA-dependent manner (Roland et al., 1994). The PmrA protein has been shown to bind and footprint the promoter region of the pbgP, pmrC and pmrG genes (Wösten and Groisman, 1999).
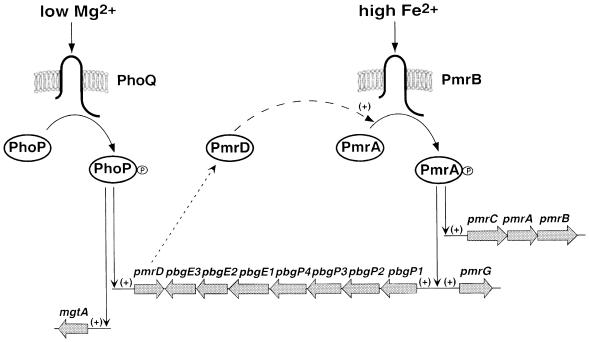
Fig. 1. Model for the activation of the PmrA–PmrB two-component system by the PhoP–PhoQ two-component system. Transcription of PmrA-activated genes can be induced by growth in high iron independently of the PhoP–PhoQ two-component system (right) or by growth in low magnesium in a PhoP–PhoQ-dependent manner (left). During growth in low magnesium, the PhoP–PhoQ system promotes expression of the pmrD gene. The PmrD protein controls the activity of the PmrA–PmrB system at a post-transcriptional level. The seven-gene pbgP/E operon has also been designated the pmrF locus (Gunn et al., 1998).
Transcription of PmrA-activated genes is promoted by either of two stimuli: (i) growth in low extracellular magnesium in a process that requires PhoP–PhoQ, a two-component system that responds to the magnesium levels in the environment (García Véscovi et al., 1996); and (ii) growth in the presence of high iron (M.M.S.M.Wösten and E.A.Groisman, unpublished results) or mild acid pH (Soncini and Groisman, 1996) in a process that does not require PhoP–PhoQ. The PmrB protein is necessary for iron sensing (M.M.S.M.Wösten and E.A.Groisman, unpublished results) and its cytoplasmic domain can phosphorylate the regulatory protein PmrA (Wösten and Groisman, 1999). The mechanism by which the PhoP–PhoQ system transduces the low magnesium signal to the PmrA–PmrB system remains unknown. However, it does not appear to involve cross-talk (i.e. phosphorylation of the regulatory protein PmrA by the sensor protein PhoQ) or a classical regulatory cascade (i.e. the PhoP–PhoQ system being solely responsible for transcription of the pmrAB genes) (Soncini and Groisman, 1996).
In this paper, we define the genetic basis for the interaction between the PhoP–PhoQ and PmrA–PmrB two-component systems. We show that pmrD is a PhoP-activated gene that mediates the transcriptional induction of PmrA-activated genes during growth in low magnesium, and demonstrate that pmrD expression does not depend on the PmrA–PmrB system. We establish that the PmrD protein acts specifically on PmrA-activated genes and that functional PmrA and PmrB proteins are required for the PmrD protein to exert its effect. Transcription of PmrA-activated genes during growth in high iron is independent of PmrD, which controls the activity of the PmrA–PmrB system at a post-transcriptional level.
Results
PmrD is required for transcription of PmrA-activated genes during growth in low magnesium
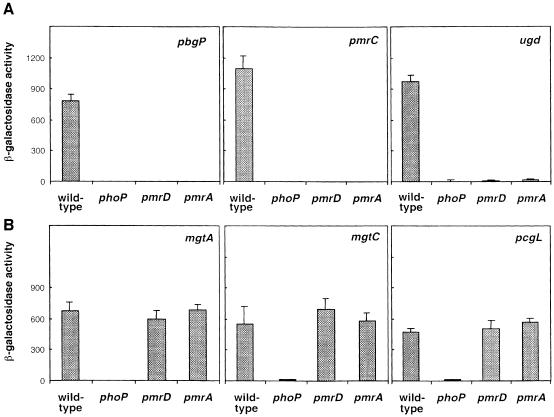
To address the role of the PmrD protein in the PhoP–PhoQ-mediated activation of the PmrA–PmrB system, we examined whether transcription of PmrA-regulated genes required pmrD. This gene is located downstream of the PmrA-activated pbgP/pmrF locus (Gunn et al., 1998) and has been implicated in polymyxin resistance (Roland et al., 1994). When bacteria were grown in low-magnesium media, a condition that promotes expression of PmrA-activated genes in a PhoP- and PhoQ-dependent manner (Soncini and Groisman, 1996), transcription of the pbgP, pmrC and ugd genes was abolished in the pmrD mutant (Figure 2A). This effect was specific for PhoP-activated genes that are PmrA dependent because the PmrA-independent PhoP-activated genes mgtA, mgtC and pcgL were expressed at wild-type levels in a pmrD mutant strain (Figure 2B). As expected, a phoP null mutation abolished transcription of all six PhoP-activated genes, whereas a pmrA null allele prevented expression of the three PmrA-dependent loci (Figure 2A). These results show that PmrD is essential for the transcriptional induction of PmrA-activated genes that occurs during growth in low magnesium.
Fig. 2. The pmrD gene is necessary for transcription of PhoP-activated PmrA-dependent genes, but does not affect transcription of PhoP-activated PmrA-independent genes. β-galactosidase activities (Miller units) expressed by strains grown in N-minimal medium pH 7.7 with 10 µM Mg2+ were determined for mutants harboring a lac transcriptional fusion to the PhoP-activated, PmrA-dependent genes pbgP, pmrC and ugd (A) and the PhoP-activated, PmrA-independent genes mgtA, mgtC and pcgL (B). The transcriptional activity was investigated in four genetic backgrounds: wild type, phoP, pmrD and pmrA. Data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if greater than the resolution of the figure).
Transcription of the pmrD gene is PhoP dependent but PmrA independent
It has been hypothesized that pmrD might be a PmrA-regulated gene because a pmrD-containing plasmid conferred polymyxin resistance only in strains harboring a functional pmrA gene (Roland et al., 1994). To explore the possibility that PmrA might control transcription of the pmrD gene, we carried out S1 nuclease experiments to map the pmrD promoter in wild-type, pmrA and phoP strains.
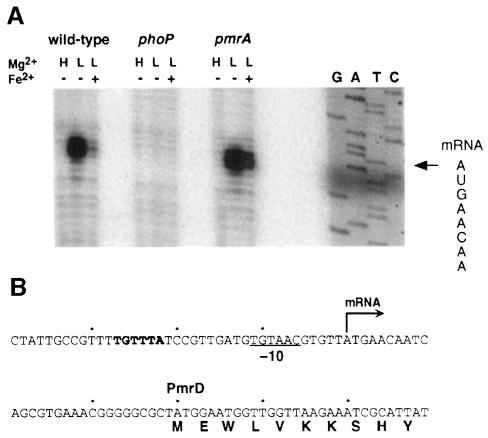
A transcription start site was identified 29 bp from the pmrD start codon with RNA harvested from wild-type bacteria grown in low magnesium (Figure 3A). Transcription of pmrD was dependent on a functional phoP gene, but did not require pmrA (Figure 3A). (Similar results were obtained in primer extension experiments; data not shown.) Consistent with these results, pmrD transcription was not detected in wild-type bacteria grown in high-magnesium media (Figure 3A), a condition that represses transcription of PhoP-activated genes (García Véscovi et al., 1996). Furthermore, the pmrD promoter harbors a –10 region typical of σ70 promoters and a relatively weak –35 consensus, but also contains the sequence TGTTTA 23 bp upstream of the pmrD transcription start site (Figure 3B). This sequence matches the consensus (T/G)GTTTA, which is found as a direct repeat some 25 bp upstream of the transcription start sites of several PhoP-dependent promoters in Escherichia coli (Kato et al., 1999). That pmrD transcription is not regulated by the PmrA protein is supported by the absence of a PmrA binding site (Wösten and Groisman, 1999) in the promoter region of pmrD (Figure 3B). Furthermore, growth of wild-type Salmonella in low magnesium with 100 µM iron, a condition that promotes expression of PmrA-activated genes in a PhoP- and PhoQ-independent manner (M.M.S.M.Wösten and E.A.Groisman, unpublished results), did not promote transcription of pmrD (actually, lower levels of pmrD transcription were observed; Figure 3A). These results demonstrate that pmrD is a PhoP-activated gene and that PmrA is not necessary for pmrD transcription.
Fig. 3. Transcription of pmrD requires PhoP, but is PmrA independent. (A) S1 mapping of pmrD transcripts produced in wild-type (14028s), phoP (MS7953s) and pmrA (EG7139) bacteria grown in N-minimal medium pH 7.7, containing 10 mM Mg2+, 10 µM Mg2+ or 10 µM Mg2+ and 100 µM Fe2+ and harvested during the logarithmic phase. The S1 protection assay was performed as described in Materials and methods. Lanes G, A, T and C correspond to dideoxy chain termination sequence reactions corresponding to this region. The sequence spanning the transcription start site is shown, and the transcription start site is marked with an arrow. (B) DNA sequence of the promoter region of the pmrD gene. The arrow corresponds to the +1 transcription start site, the underlined sequence is the predicted –10 region for the pmrD promoter and the sequence in bold matches the consensus found as a direct repeat in the promoter region of PhoP-activated genes in E.coli K-12 (Kato et al., 1999).
Role of the PhoP–PhoQ and PmrA–PmrB systems in transcription of the PmrA-activated gene pbgP
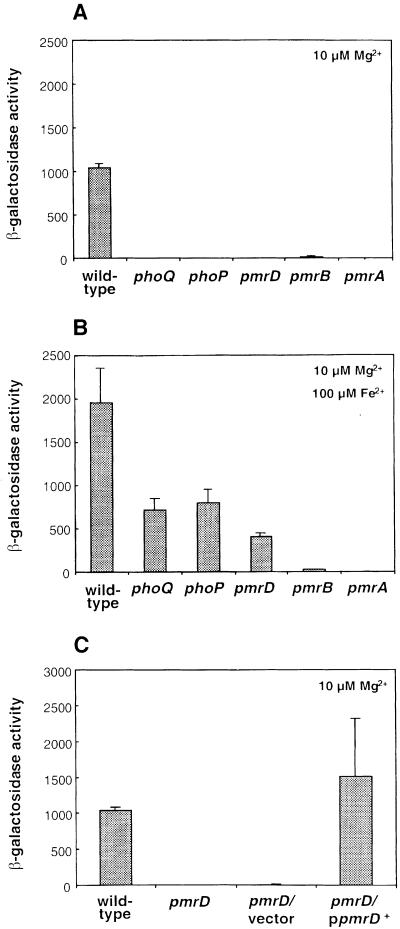
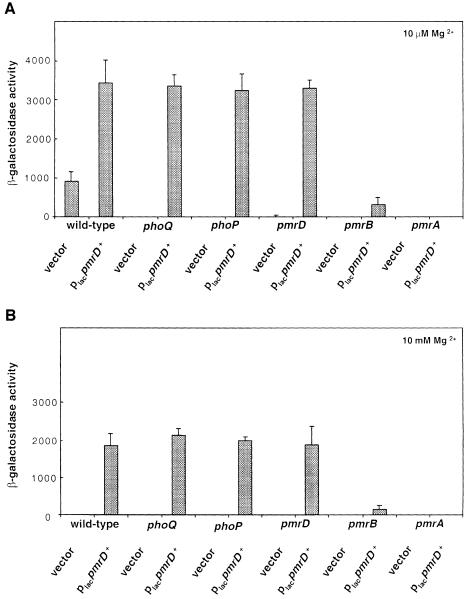
We examined the role of the PhoP–PhoQ and PmrA–PmrB two-component systems in transcription of pbgP, which was chosen as a prototypical PmrA-activated gene because the PmrA protein binds to the pbgP promoter (Wösten and Groisman, 1999). When bacteria were grown in low magnesium, pbgP transcription required functional phoQ, phoP, pmrA and pmrB genes (Figure 4A). On the other hand, when bacteria were grown in low-magnesium media containing 100 µM iron, pbgP transcription was observed in the phoQ and phoP mutants, but not in the pmrA or pmrB mutant strains (Figure 4B). The levels of pbgP transcription in the phoP and phoQ mutants grown in the presence of 100 µM iron were slightly reduced to 40–45% of those observed in the isogenic wild-type strain (Figure 4B).
Fig. 4. PmrD is essential for transcription of the PmrA-activated gene pbgP during growth in low magnesium. Bacteria were grown in N-minimal medium pH 7.7 with 10 µM Mg2+ (A and C) or 10 µM Mg2+ and 100 µM Fe2+ (B). β-galactosidase activities (Miller units) were determined for mutants harboring a lac transcriptional fusion to pbgP. The transcriptional activity was determined in six genetic backgrounds: wild type, phoQ, phoP, pmrD, pmrB and pmrA (A and B) or wild-type and pmrD backgrounds for the complementation assay (C). The pmrD– strains used for this assay were transformed with a plasmid harboring the pmrD gene expressed from its own promoter (pLK23) or the cloning vector (pUC19). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if greater than the resolution of the figure).
In a pmrD mutant, transcription of pbgP was similar to that observed in the phoP or phoQ mutants: it was absent in bacteria grown in low magnesium (Figure 4A), but present in bacteria grown in the presence of 100 µM iron (Figure 4B). Plasmid pLK23, harboring a wild-type copy of the pmrD gene expressed from its own promoter, restored wild-type levels of pbgP expression to the pmrD mutant (Figure 4C), demonstrating that the lack of pbgP transcription is solely due to inactivation of pmrD. Cumulatively, these results establish that the phoQ, phoP and pmrD genes are essential for pbgP transcription during growth in low magnesium (mediating >100-fold activation in pbgP expression), but unnecessary (<5-fold effect) during growth in high iron.
PmrD acts downstream of PhoP–PhoQ
The results described above suggest the following model for the transcription of PmrA-activated genes during growth in low magnesium: the PhoQ protein activates the PhoP protein (presumably by promoting its phosphorylated state), which induces transcription of PhoP-activated genes including pmrD. The PmrD protein then specifically promotes transcription of PmrA-activated genes. We tested this model (i.e. that PmrD acts downstream of PhoP–PhoQ) by examining whether ectopic expression of pmrD could bypass the requirement for phoP, phoQ and growth in low magnesium. Plasmid pLK24, in which the pmrD gene is transcribed from the lac promoter in pUC19, restored pbgP transcription to phoP and phoQ mutants as well as to wild-type and pmrD cells, whether bacteria were grown under inducing (i.e. low magnesium) or repressing (i.e. high magnesium) conditions (Figure 5). In contrast, plasmid pLK23, in which the pmrD gene is expressed from its own promoter, could not restore pbgP expression to a phoP mutant (data not shown). These results suggest that the role of the PhoP–PhoQ system in activation of PmrA-activated genes is to promote expression of the pmrD gene.
Fig. 5. PmrD acts downstream of the PhoP–PhoQ system and upstream of the PmrA–PmrB system. β-galactosidase activities (Miller units) expressed by transformants grown in N-minimal medium pH 7.7 with 10 µM Mg2+ (A) or 10 mM Mg2+ (B) were determined for mutants harboring a lac transcriptional fusion to pbgP. The transcriptional activity was determined in six genetic backgrounds: wild type, phoQ, phoP, pmrD, pmrB and pmrA. The strains were transformed with a plasmid harboring the pmrD gene expressed from the lac promoter (pLK24) or the cloning vector (pUC19). The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if greater than the resolution of the figure).
PmrD acts upstream of PmrA–PmrB
We conducted epistasis experiments to examine whether the PmrD-mediated transcription of PmrA-activated loci required the pmrA and/or pmrB genes. Consistent with the notion that PmrD acts upstream or at the same level as PmrA, plasmid pLK24 (in which the pmrD gene is transcribed from the lac promoter in pUC19) could not restore pbgP expression to the pmrA null mutant: the levels of pbgP transcription were similar to those obtained in a strain harboring the pUC19 vector (Figure 5). Likewise, expression of pmrD from plasmid pLK24 in the pmrB mutant restored only 6% of the pbgP transcription levels found in the wild-type strain. These results establish that the PmrD protein requires a functional PmrA–PmrB system to exert its effect.
A pmrA constitutive mutant bypasses the requirement for pmrD in the low-magnesium activation of pbgP
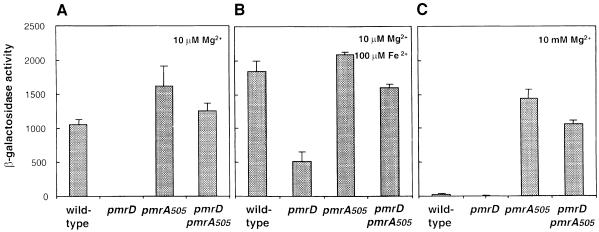
Roland et al. (1993) described an allele of the pmrA gene, pmrA505, which confers heightened levels of polymyxin resistance to wild-type Salmonella. The pmrA505 allele has a single nucleotide substitution, which results in an arginine to histidine substitution at amino acid 81 in the N-terminal response regulator domain of the PmrA protein (Roland et al., 1993).
We established that pmrA505 is a constitutive allele of pmrA: transcription of the pbgP gene was reduced <20% under repressing magnesium and iron concentrations (Figure 6), whereas identical growth conditions decreased pbgP expression >100-fold in an isogenic pmrA+ strain (Figure 6). If the pmrD gene functions upstream of pmrA, pbgP transcription should be pmrD independent in a pmrA505 mutant. Consistent with this notion, the pmrD::cat mutation hardly affected pbgP transcription in the pmrA505 mutant regardless of the magnesium and iron concentrations (Figure 6).
Fig. 6. Transcripton of pbgP in a strain grown in low magnesium expressing the constitutive pmrA505 allele is independent of pmrD. β-galactosidase activities (Miller units) expressed by transformants grown in N-minimal medium pH 7.7 with 10 µM Mg2+ (A), 10 µM Mg2+ and 100 µM Fe2+ (B) or 10 mM Mg2+ (C) were determined for a mutant harboring a lac transcriptional fusion to pbgP. The transcriptional activity was investigated in four genetic backgrounds: wild type, pmrD, pmrA505 and pmrD pmrA505. The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if greater than the resolution of the figure).
PmrD controls the activity of the PmrA–PmrB system at a post-transcriptional level
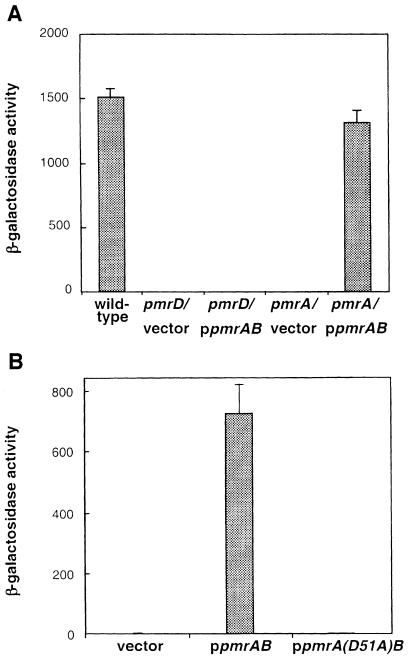
The PmrD protein may function by regulating the transcription, stability and/or the activity of the PmrA and PmrB proteins. If the PmrD protein acts at a post-transcriptional level, a pmrD mutation should abolish transcription of PmrA-activated genes even if the pmrAB genes are transcribed from a heterologous promoter. We tested this hypothesis by examining pbgP transcription in a strain carrying a plasmid with the pmrAB genes under the control of a derivative of the lac promoter (this strain also harbored wild-type copies of the pmrA and pmrB genes at their normal chromosomal location). When grown in low magnesium, the pmrAB-expressing plasmid could not rescue the pmrD mutant: pbgP transcription was abolished as observed in the vector-carrying strain (Figure 7A). Control experiments demonstrated that the pmrAB-expressing plasmid could rescue the pmrA mutant, restoring wild-type levels of pbgP transcription (Figure 7A).
Fig. 7. PmrD activates the PmrA–PmrB system by a post-transcriptional mechanism. (A) Expression of the pmrAB genes from a heterologous promoter does not rescue pbgP transcripton in a pmrD mutant. β-galactosidase activities (Miller units) expressed by bacteria harboring a lac transcriptional fusion to the pbgP gene grown in N-minimal medium pH 7.7 with 10 µM Mg2+. The transcriptional activity was investigated in three genetic backgrounds: wild type, pmrD and pmrA, the latter carrying either a plasmid with the pmrAB genes under the control of a derivative of the lac promoter or the plasmid vector. The data correspond to mean values of four independent experiments performed in duplicate. (B) pmrD-mediated activation is dependent on the putative site of PmrA phosphorylation.β-galactosidase activities (Miller units) expressed by bacteria harboring a lac transcriptional fusion to the pbgP gene grown in N-minimal medium pH 7.7 with 10 µM Mg2+. The transcriptional activity was investigated in three genetic backgrounds: a pmrA null mutant harboring plasmid pEG9102 with the pmrAB genes under the control of a derivative of the lac promoter, plasmid pLK39 with the pmrA(D51A)B genes under the control of a derivative of the lac promoter or the plasmid vector. Similar results were obtained with plamids harboring only the pmrA gene under the control of a derivative of the lac promoter: the D51A mutation abolished pbgP transcription. The data correspond to mean values of four independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if greater than the resolution of the figure).
Mutation of the putative site of PmrA phosphorylation abolishes PmrD-mediated activation of pbgP
Response regulators are normally activated by phosphorylation, typically from their cognate sensor kinases. The PmrA protein is phosphorylated by the cytoplasmic domain of the PmrB protein (Wösten and Groisman, 1999) and mutation of the putative site of PmrA phosphorylation abolished the high-iron (PmrD-independent) activation of the pbgP gene (data not shown). Consistent with the notion that the low-magnesium (i.e. PmrD-mediated) activation of PmrA-regulated genes is a phosphorylation-dependent mechanism, a strain with the chromosomal copy of the pmrA gene inactivated and harboring a plasmid expressing a PmrA protein mutated in the putative phosphorylation site (i.e. D51A) failed to activate pbgP transcription whether bacteria were grown in low-magnesium (Figure 7B), high-magnesium or high-iron conditions (data not shown). In contrast, the isogenic pmrA+-plasmid-containing strain exhibited a normal profile of pbgP transcription: high expression during growth in low magnesium (Figure 7B) or high iron and no expression in high-magnesium media (data not shown).
The pmrD gene is necessary for polymyxin resistance during growth in low magnesium but expendable during growth in high iron
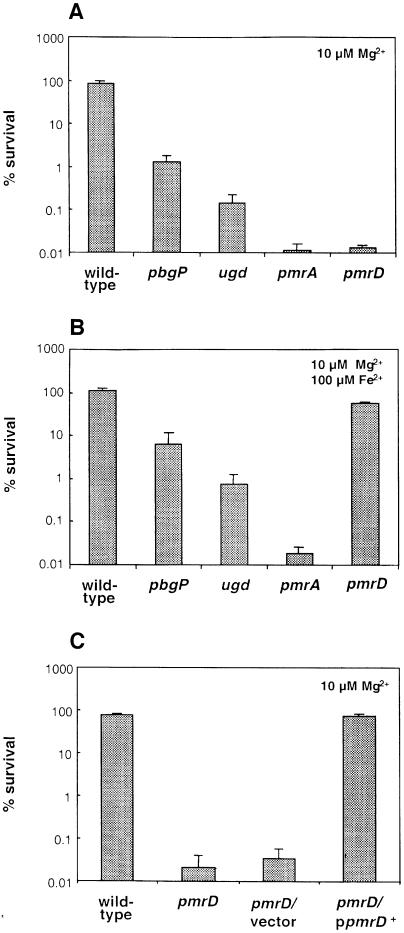
It was originally reported that the pmrD gene confers polymyxin resistance when present in a medium-copy plasmid, but that a strain with a tet resistance cassette in the chromosomal copy of the pmrD gene displays wild-type levels of polymyxin resistance (Roland et al., 1994). We hypothesized that the inability to detect differences between wild-type and pmrD Salmonella might have been due to not growing the microorganism under conditions promoting expression of PmrA-activated genes (which were not known at that time). Thus, we performed polymyxin sensitivity assays with bacteria grown in defined media with pH 5.8 and 10 µM Mg2+, a condition that promotes transcription of PmrA-activated genes and the phenotypic display of polymyxin resistance in wild-type Salmonella (Groisman et al., 1997). The pmrD mutant was >5000-fold more sensitive to polymyxin than the wild-type strain; this level of sensitivity is similar to that displayed by a pmrA null mutant and much higher than that exhibited by strains harboring mutations in the PmrA-activated genes pbgP and ugd (Figure 8A). The polymyxin sensitivity of the pmrD mutant was solely due to inactivation of pmrD because wild-type levels of polymyxin resistance could be restored to the pmrD mutant by plasmid pLK23, which harbors a wild-type copy of the pmrD gene (Figure 8C). These data demonstrate that pmrD is necessary for polymyxin resistance during growth in low magnesium.
Fig. 8. The pmrD gene is required for polymyxin resistance during growth in low magnesium. Wild-type (14028s), pbgP (EG9241), ugd (EG9524), pmrA (EG7139) and pmrD (EG11491) bacteria were grown to logarithmic phase in N-minimal medium pH 5.8 with 10 µM Mg2+ (A and C) or 10 µM Mg2+ and 100 µM Fe2+ (B), washed, and incubated in the presence of polymyxin (a final concentration of 2.5 µg/ml) for 1 h at 37°C. Samples were diluted in phosphate-buffered saline and plated on LB-agar plates to assess bacterial viability. Survival values are relative to the original inoculum. Complementation experiments were carried out with the pmrD mutant transformed with a plasmid harboring the pmrD gene expressed from its own promoter (pLK23) or the cloning vector (pUC19) (C). Data correspond to mean values from three independent experiments performed in duplicate. Error bars correspond to the standard deviation (and are only shown if bigger than the resolution of the figure).
Because growth in 100 µM iron restored pbgP transcription to the pmrD mutant (Figure 4B), we hypothesized that the pmrD mutant might become resistant to polymyxin if grown in the presence of 100 µM iron. Consistent with this notion, the pmrD mutant exhibited wild-type levels of polymyxin resistance when grown in N-minimal medium pH 5.8 and 10 µM Mg2+ supplemented with 100 µM iron (Figure 8B). In contrast, the pmrA mutant remained >5000-fold more sensitive to polymyxin than the wild-type strain (Figure 8B). Likewise, growth in 100 µM iron could not rescue polymyxin resistance in the pbgP and ugd mutants (Figure 8B). These results argue that the PmrD protein mediates polymyxin resistance only indirectly, by promoting transcription of PmrA-regulated genes during growth in low magnesium.
Discussion
Two-component systems are signal transduction machineries that promote changes in gene expression or bacterial behavior in response to distinct physical or chemical cues. Eubacterial species typically harbor several dozen two-component systems that exhibit exquisite specificity so that a sensor kinase only modulates the activity of its cognate response regulator. Yet, certain two-component systems appear to communicate with one another, allowing their regulated genes to respond to more than one cue (Wanner, 1992; Birkey et al., 1998). In this paper, we have described a novel type of interaction between a pair of two-component systems, PhoP–PhoQ and PmrA–PmrB, which allows Salmonella to regulate expression of antimicrobial resistance determinants in response to two different conditions: high iron and low magnesium.
Our results suggest the following model (Figure 1) for the transcription of PmrA-regulated genes: high extracellular iron is detected by the sensor protein PmrB (M.M.S.M.Wösten and E.A.Groisman, unpublished results), which promotes phosphorylation of the regulatory protein PmrA; phosphorylated PmrA binds to the promoter region of the pbgP and pmrC genes (Wösten and Groisman, 1999), thereby activating their transcription and resulting in the phenotypic display of polymyxin resistance. Mutants lacking functional pmrA or pmrB genes fail to express PmrA-activated genes regardless of the iron or magnesium concentrations (Figure 4) and are hypersensitive to polymyxin (Figure 8). The PhoP–PhoQ system is not necessary for the iron-mediated activation of the pbgP and pmrC genes (Figure 4), allowing for transcription of PmrA-regulated genes to occur independently of the PhoP–PhoQ system.
The pbgP and ugd genes are essential for growth in low-magnesium solid media (Groisman et al., 1997) and, not surprisingly, PmrA-regulated genes are transcriptionally induced also during growth in low magnesium in a PhoP–PhoQ-dependent manner (Figure 2). We suggest that this occurs by the PhoQ protein serving as a magnesium sensor (Waldburger and Sauer, 1996; García Véscovi et al., 1997) that modulates the ability of the PhoP protein to activate transcription of the pmrD gene (Figure 3), which harbors a sequence matching the consensus found as a direct repeat in the promoter region of several PhoP-activated genes in E.coli K-12 (Kato et al., 1999). Consistent with this notion (Figure 1), transcription of the pmrD gene from a heterologous promoter bypassed the requirement for low magnesium and functional phoP and phoQ genes in transcription of the pbgP gene (Figure 5). Moreover, it ruled out models involving activation via the PhoP protein binding to the pbgP promoter, the PhoQ protein serving as a phosphate donor for the PmrA protein, or the PhoP protein serving as a phosphate donor for the PmrB protein.
On the other hand, transcription of the pmrD gene from a heterologous promoter did not rescue pbgP transcription in pmrA or pmrB mutants (Figure 5), which argues against the PmrD protein being a transcription factor acting on the pbgP promoter independently of the PmrA protein. Moreover, cells harboring a constitutive allele of the pmrA gene (Roland et al., 1993) allowed pbgP transcription in a pmrD-independent manner (Figure 6), indicating that pmrD functions upstream of pmrA. This raised the possibility of the PhoP–PhoQ system controlling transcription of the pmrAB genes via the PmrD protein. For example, when phosphate levels are limiting, the PhoP–PhoR two-component system of Bacillus subtilis (which is unrelated to the Salmonella PhoP–PhoQ system) activates the ResD–ResE two-component system by the PhoP protein binding to the promoter region and activating transcription of the resABCDE operon (Birkey et al., 1998). While a role for PmrD in controlling pmrAB transcription can not be ruled out, our data show that the PmrD protein does have an effect at the post-transcriptional level: inactivation of pmrD prevented the low-magnesium activation of pbgP when the pmrAB genes were transcribed from a heterologous promoter (Figure 7A).
PmrD-mediated activation appears to be a phosphorylation-dependent mechanism because mutation of the putative site of PmrA phosphorylation abolished pbgP transcription (Figure 7B). Thus, the PmrD protein may function by increasing the levels of phosphorylated PmrA protein, which binds the promoters of the pmrC and pbgP genes with higher affinity than unphosphorylated PmrA (Wösten and Groisman, 1999). This could be achieved by promoting the phosphorylation of the PmrA protein and/or by inhibiting its dephosphorylation. Increased PmrA phosphorylation may result from the PmrB protein adopting an active conformation during growth in high iron or in the presence of the PmrD protein, conditions that promote pbgP expression in a PmrB-dependent manner (Figure 4; M.M.S.M.Wösten and E.A.Groisman, unpublished results). An active PmrB conformation may have higher autokinase activity and/or be a better phosphate donor for the PmrA protein. Alternatively, or in addition, PmrD might inhibit an autophosphatase activity of PmrA or a PmrA phosphatase activity of PmrB. For example, certain phosphorylated response regulators exhibit autophosphatase activity (e.g. P-CheY, P-CheB, P-NRI, P-PhoB; Stock et al., 1989), whereas others are dephosphorylated by their cognate sensor kinases. For example, the sensor EnvZ of E.coli catalyzes both phosphorylation of the response regulator OmpR and dephosphorylation of phosphorylated OmpR (Igo et al., 1989). The PmrD protein could also antagonize the activity of a yet to be described PmrA phosphatase, such as PrpA, which in vivo exhibits histidine and/or aspartyl phosphatase activity towards the CpxA–CpxR two-component system of E.coli (Missiakas and Raina, 1997), or SixA, which in vitro displays histidine phosphatase activity towards the histidine-containing phosphotransfer domain of the ArcB sensor of E.coli (Ogino et al., 1998).
The 85 amino acid PmrD protein has no homologs in the sequence databases, and thus it does not resemble the kinases or the phosphatase inhibitors controlling the two-component phosphorelay that governs sporulation in B.subtilis (Fabret et al., 1999). Yet, the phosphatase inhibitor NH4VO3 could rescue pbgP transcription in phoP, phoQ and pmrD mutants (to 25% of wild-type levels), but not in pmrA or pmrB mutants (our unpublished results). This effect appears to be specific to PmrA-activated genes because NH4VO3 could not rescue transcription of PhoP-activated genes, which are PmrA independent. While this result is consistent with PmrD being a phosphatase inhibitor, we can not exclude other possible roles for NH4VO3, such as affecting the cellular ATP pools, thereby impacting on the ability of the PmrB protein to become phosphorylated.
PmrD is the first example of a protein mediating the activation of a two-component system by another two-component system at a post-transcriptional level. Thus, PmrD-mediated activation is different from classical cascades in which a two-component system activates a second two-component system by the response regulator of the first system binding to the promoter and promoting transcription of the genes encoding the second two-component system (Birkey et al., 1998; Lee et al., 2000). Moreover, unlike the well characterized kinases and phosphatases controlling the onset of sporulation in B.subtilis, which channel different inputs into a single phosphorelay system (Perego and Hoch, 1996), PmrD connects two distinct two-component systems (PhoP–PhoQ and PmrA–PmrB) that can be activated independently of one another.
It was originally reported that a medium-copy-number plasmid harboring the pmrD gene conferred polymyxin resistance in a pmrA-dependent manner, but that a Salmonella pmrD mutant retained wild-type levels of polymyxin resistance (Roland et al., 1994). Our experiments provide a molecular interpretation for these seemingly contradictory results: when bacteria are grown in low-magnesium media (to promote expression of PmrA-activated genes) the pmrD mutant is >5000-fold more sensitive to polymyxin than wild-type Salmonella (Figure 8A). Roland et al. (1994) grew the bacteria under non-inducing conditions, and thus could only detect differences in polymyxin resistance when the pmrD gene was expressed from a plasmid vector. Likewise, the pmrA dependence of pmrD-mediated polymyxin resistance (Roland et al., 1994) reflects the fact that the PmrD protein acts upstream of the PmrA–PmrB system (Figures 5 and 6). Finally, our data show that the pmrD mutant exhibits wild-type levels of polymyxin resistance when grown in the presence of iron (Figure 8B). This in dicates that the PmrD protein mediates polymyxin resistance only indirectly, by promoting transcription of PmrA-regulated genes during growth in low magnesium.
While the experiments described in this paper were carried out in Salmonella, the PmrD activation mechanism is likely to operate in other species because homologs of the phoP, phoQ, pmrA, pmrB and pmrD genes have been detected in at least five Gram-negative species (Groisman et al., 1989; our unpublished results). Moreover, the pmrD gene of E.coli K-12 fully rescued a pmrD mutant of Salmonella, indicating that the E.coli pmrD gene is functional and has a similar role in both enteric species (our unpublished results).
What is the physiological significance of our findings? When enteric bacteria experience low-magnesium environments, the PhoP–PhoQ system becomes activated, resulting in the expression of several proteins including the MgtA and MgtB Mg2+ transporters (Soncini et al., 1996). PhoP also promotes PmrD expression, which activates the PmrA–PmrB system and results in the synthesis of PmrA-regulated genes, some of which are necessary for growth in low-magnesium media (Groisman et al., 1997). However, the PmrA–PmrB system can also be activated by high iron in a mechanism that is PhoP-, PhoQ- and PmrD-independent (M.M.S.M.Wösten and E.A.Groisman, unpublished results), consistent with PmrA-activated genes being required for growth in high iron (M.M.S.M.Wösten and E.A.Groisman, unpublished results). This physiological plasticity allows the independent activation of two distinct two-component regulatory systems in response to a broader spectrum of environmental cues.
Materials and methods
Bacterial strains, plasmids, recombinant molecular techniques and growth conditions
Bacterial strains and plasmids used in this study are listed in Table I. Mutants were constructed by phage P22-mediated transductions as described elsewhere (Davis et al., 1980). Construction of the ΔpmrB::cat mutant strain will be described elsewhere. Recombinant DNA techniques were performed according to standard protocols (Sambrook et al., 1989). Bacteria were grown at 37°C in Luria broth (LB) or in a modified N-minimal medium containing 0.1% casamino acids and 38 mM glycerol (Snavely et al., 1991) in which the 100 mM Tris–HCl was replaced by a mixture of 50 mM bis-Tris and 50 mM Tris adjusted to pH 7.7 or 5.8 with HCl. MgCl2 was added to a final concentration of 10 µM or 10 mM. FeSO4 was used at a final concentration of 0.1 mM from a freshly prepared 0.1 M stock solution. Kanamycin was used at a final concentration of 50 µg/ml, chloramphenicol at 25 µg/ml, tetracycline at 10 µg/ml, ampicillin at 50 µg/ml and polymyxin B (6000 U/mg) (Sigma) at 2.5 µg/ml.
Table I. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| S.enterica serovar typhimurium | ||
| 14028s | wild type | Fields et al. (1986) |
| AA3007 | polA2 ara-9 | Whitfield and Levine (1973) |
| MS7953s | phoP7953::Tn10 | Fields et al. (1989) |
| MS5996s | phoQ5996::Tn10 | Fields et al. (1989) |
| EG11491 | pmrD1::cat | this work |
| EG7139 | pmrAl ::cat | Soncini and Groisman (1996) |
| EG10056 | ΔpmrB::cat | this work |
| EG9241 | pbgP1::MudJ | Soncini et al. (1996) |
| EG9168 | pbgP1::MudJ phoP7953::Tn10 | Soncini et al. (1996) |
| EG10236 | pbgP1::MudJ phoQ5996::Tn10 | this work |
| EG11775 | pbgP1::MudJ pmrD::cat | this work |
| EG9813 | pbgP1::MudJ pmrA1::cat | this work |
| EG10065 | pbgP1::MudJ ΔpmrB::cat | this work |
| EG9524 | ugd (pbgC)9228::MudJ | García Véscovi et al. (1996) |
| EG9526 | ugd9228::MudJ phoP7953::Tn10 | García Véscovi et al. (1996) |
| EG11773 | ugd9228::MudJ pmrD1::cat | this work |
| EG9811 | ugd9228::MudJ pmrA1::cat | this work |
| EG9521 | mgtA9226::MudJ | García Véscovi et al. (1996) |
| EG9523 | mgtA9226::MudJ phoP7953::Tn10 | García Véscovi et al. (1996) |
| EG11777 | mgtA9226::MudJ pmrD1::cat | this work |
| EG11779 | mgtA9226::MudJ pmrA1::cat | this work |
| EG9527 | mgtC9232::MudJ | García Véscovi et al. (1996) |
| EG9529 | mgtC9232::MudJ phoP7953::Tn10 | García Véscovi et al. (1996) |
| EG11781 | mgtC9232::MudJ pmrD1::cat | this work |
| EG11783 | mgtC9232::MudJ pmrA1::cat | this work |
| EG10627 | pcgL9331::MudJ | Soncini et al. (1996) |
| EG10745 | pcgL9331::MudJ phoP7953::Tn10 | Soncini et al. (1996) |
| EG11789 | pcgL9331::MudJ pmrD1::cat | this work |
| EG11791 | pcgL9331::MudJ pmrA1::cat | this work |
| EG9460 | psiD9065(pmrC)::MudJ | García Véscovi et al. (1996) |
| EG9280 | psiD9065(pmrC)::MudJ phoP7953::Tn10 | García Véscovi et al. (1996) |
| EG11785 | psiD9065(pmrC)::MudJ pmrD1::cat | this work |
| EG11787 | psiD9065(pmrC)::MudJ pmrA1::cat | this work |
| EG9888 | pbgP1::MudJ zjd::Tn10d-Cam | Groisman et al. (1997) |
| EG9868 | pbgP1::MudJ pmrA505 zjd::Tn10d-Cam | Groisman et al. (1997) |
| EG12115 | pbgP1::MudJ pmrD::cat zjd::Tn10d-Cam | this work |
| EG12117 | pbgP1::MudJ pmrA505 pmrD::cat zjd::Tn10d-Cam | this work |
| E.coli | ||
| DH5α | supE44 ΔlacU169 (φ80 lacZΔM15) hsdR17 | Hanahan (1983) |
| recA1 endA1 gyrA96 thi-1 relA1 | ||
| Plasmids | ||
| pKRP10 | reppMB1 Apr Cmr | Reece and Phillips (1995) |
| pUC19 | reppMB1 lacZa ApR lacI q | Yanisch-Perron et al. (1985) |
| pLK4 | reppMB1 Apr pbgP/E pmrD | this work |
| PLK7 | reppMB1 Apr pbgP/EΔpbgP3 pmrD | this work |
| PLK10 | reppMB1 Apr pbgP/EΔpbgP3 pmrD::cat | this work |
| pLK38 | reppMB1 lacZa- ApR pmrA(D51A)B | this work |
| pLK23 | reppMB1 Apr pmrD | this work |
| pLK24 | reppMB1 lacZa Apr laqI q pmrD | this work |
| pUHE21-2laqIq | reppMB1 Apr lacI q | Soncini et al. (1995) |
| pEG9102 | reppMB1 Apr lacI q pmrAB | Soncini and Groisman (1996) |
| pLK39 | reppMB1 ApR lacI q pmrA(D51A)B | this work |
aGene designations are as described (Sanderson et al., 1995).
Construction of plasmids
Plasmid pLK4, containing the complete pbgP/E and pmrD loci, was constructed by simultaneous ligation of a 4.5 kb SphI–KpnI fragment from pbgP/E plasmid pML3 and a 4.2 kb KpnI–PvuII fragment from pbgP/E plasmid pML1 into the PvuII-digested plasmid pUC19 (Yanisch-Perron et al., 1985). Plasmids pML1 and pML3 were identified by screening a Mud5005-generated library of Salmonella for clones hybridizing to PCR-generated probes corresponding to sequences immediately adjacent to the MudJ insertions in the pbgP1::MudJ and pbgE1::MudJ mutants (Soncini et al., 1996). Plasmid pLK7 was constructed by deleting the third open reading frame in the pbgP/E operon (pbgP3) in plasmid pLK4 by digesting with AgeI and HindIII, filling in the 5′ overhanging ends with T4 DNA polymerase (Gibco-BRL) and ligating to the pUC19 vector. This plasmid was designated pLK7. The nucleotide sequence of the AgeI–HindIII junction was checked by sequence analysis on a ABI Prism® 310 Genetic Analyzer (Perkin Elmer) using the ABI Prism® Big Dye®Terminator Cycle Sequencing Ready Reaction Kit. Plasmid pLK10 was constructed by inserting the 0.8 kb cat EcoRI fragment from plasmid pKRP10 (Reece and Phillips, 1995) into the unique EcoRI site of plasmid pLK7 within the pmrD open reading frame. Digestion of the plasmid with ScaI showed that the cat gene was present in the opposite orientation of the pmrD gene.
Plasmid pLK23 contains the pmrD gene expressed from its own promoter and was constructed by digesting plasmid pLK15 DNA with NdeI and BamHI, repairing the overhanging ends with T4 DNA polymerase and ligation. Plasmid pLK15 is a derivative of plasmid pLK7 in which a BamHI site was created between pmrD and the seventh open reading frame of the pbgP/E operon (pbgE3). To construct pLK15, we simultaneously ligated four fragments: the 0.8 kb cat fragment from plasmid pKRP10, a PCR fragment generated with primers 647 (5′-TGG CTGGGGTTACTGCCTGG-3′) and 1028 (5′-CGCGGATCCTCATGA TCTGGCGGGCAG-3′) and plasmid pLK4 DNA as template and digested with ApaI and BamHI, a second PCR product generated with primers 1029 (5′-CCCGGATCCTCATGATGGCTTCGCCGTCA-3′) and 641 and plasmid pLK4 DNA as template and digested with EcoRI and BamHI, and plasmid pLK7 digested with ApaI–EcoRI. Digestion of pLK15 with ScaI showed that the cat gene is inserted with the same orientation as the pmrD gene. Nucleotide sequence analysis showed that the PCR-generated DNAs in pLK15 had a wild-type sequence.
Plasmid pLK24 was constructed by cloning between the XbaI and KpnI sites of pUC19 a PCR fragment containing the pmrD-coding region generated with primers 1090 (5′-GCTCTAGAAACGGGGGCGC TATGG-3′) and 1089 (5′-GGGGTACCAGATCATGATGGCTTGC-3′) and Salmonella chromosomal DNA as template, and digested with XbaI and KpnI. Nucleotide sequence analysis showed that the PCR-generated DNAs in pLK24 had a wild-type sequence.
Plasmid pLK39, a pEG9102 derivative containing a mutation in the putative phosphorylation site of PmrA (D51A), was constructed as follows. First, we performed an inverse PCR with primers 1185 (5′-CTAGCTAGCTTTAGGGCTGCCCGATGAG-3′) and 1186 (5′-CTAGCTAGCACCATCAGACTGTAATG-3′) and a pUC19 derivative harboring the pmrAB genes as template. After digestion of the PCR product with NheI and self-ligation, the ligation mixture was used to transform E.coli DH5α cells. Ampicillin-resistant transformants were screened by colony PCR with primers 351 (5′-AAGGATCCA GGAGACTAAGCG-3′) and 352 (5′-GGCAAGCTTAGCTTTCCT CAG-3′) and digestion of the generated PCR product with NheI. One of the plasmids that could be digested with NheI was designated pLK38. Nucleotide sequencing confirmed the presence of wild-type pmrAB sequence except in the predicted mutated codon. A BamHI–Bsu36I fragment from plasmid pLK38 harboring the mutant portion of the pmrA gene was ligated to pEG9102 DNA that had been digested with BamHI and Bsu36I. After transformation of E.coli DH5α cells, transformants were screened for the presence of the mutation by colony PCR with primers 351 and 352. One of the plasmids that could be digested with NheI was designated pLK39. Western blot analysis with anti-PmrA antibodies showed similar steady-state levels of PmrA+ and PmrA(D51A) proteins.
Construction of chromosomal pmrD mutant
The pmrD::cat mutation in pLK10 was transferred to the chromosome of S.enterica serovar typhimurium strain 14028s as described previously (Groisman et al., 1993). One of eight transductants analyzed harbored a wild-type pbgP3 gene as tested by colony PCR using primers 655 (5′-GCCGTGCGCCGCTAAACTGG-3′) and 587 (5′-CCGCGATAT CGACGCTACGC-3′). The structure of the disrupted pmrD gene in this transductant, designated EG11491, was confirmed by colony PCR using primers 1001 (5′-CTGTCTAGACTGCCCGCCAGATCATGA-3′) and 641, and further verified by Southern hybridization of chromosomal DNA digested with NsiI, HphI and HphI–ApaI double digests using pmrD- and cat-specific probes. The pmrD probe was the KpnI–XbaI fragment from plasmid pLK24. The cat probe corresponds to the 0.8 kb EcoRI fragment from pKRP10.
Polymyxin resistance and β-galactosidase assays
One-hour polymyxin sensitivity assays were performed as described previously (Groisman et al., 1997). β-galactosidase activity was determined as described elsewhere (Miller, 1972).
S1 nuclease assay
The S1 nuclease protection assay was performed as described previously (García Véscovi et al., 1996) with RNA harvested from mid-exponential phase cultures (OD600 0.4–0.6) grown in 50 ml N-minimal medium pH 7.7, containing either 10 mM MgCl2, 10 µM MgCl2 or 10 µM MgCl2 and 0.1 mM FeSO4. Total RNA was isolated with Trizol (Gibco-BRL) according to the manufacturer’s specifications. A PCR product generated with primers 1064 (5′-GCCCTCTTTTTGACATAATG-3′) and 641 (5′-CGTGCCGGTAGAAGATAAAG-3′) and Salmonella chromosomal DNA as template was used as probe. This probe was labeled at the 5′ end by phosphorylation with [γ-32P]ATP using T4 polynucleotide kinase (Gibco-BRL). In brief, total RNA (50 µg) and the labeled DNA probe were mixed in 50 µl of hybridization buffer (80% formamide, 20 mM HEPES pH 6.5, 0.4 M NaCl). The mixture was incubated at 75°C for 10 min and then left to cool down in an incubator at 37°C overnight. After adding 220 µl of H2O, 30 µl of 10× S1 nuclease buffer (0.3 M sodium acetate pH 4.5, 0.5 M NaCl, 10 mM ZnSO4, 50% glycerol), the mixture was treated with 10 U of S1 nuclease (Promega) for 30 min. The reaction was stopped by the addition of 300 µl of phenol–chloroform, and the aqueous phase was precipitated with ethanol. The precipitate was dissolved in sequence loading buffer and electrophoresed on a 6% acrylamide–7 M urea gel together with a sequence ladder initiated with primer 1064 using the T7 Sequenase version 2.0 DNA sequencing kit (Amersham).
Acknowledgments
Acknowledgements
We thank F.Solomon and T.Latiffi for technical assistance, G.Phillips for plasmid pKRP10 and R.Kranz for critical reading of the manuscript. This work was supported by grant AI42236 from the NIH to E.A.G., who is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Birkey S.M., Liu,W., Zhang,X., Duggan,M.F. and Hulett,F.M. (1998) Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol. Microbiol., 30, 943–953. [DOI] [PubMed] [Google Scholar]
- Davis R.W., Bolstein,D. and Roth,J.R. (1980) Advanced Bacterial Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Fabret C., Feher,V.A. and Hoch,J.A. (1999) Two-component signal transduction in Bacillus subtilis: how one organism sees its world. J. Bacteriol., 181, 1975–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields P.I., Swanson,R.V., Haidaris,C.G. and Heffron,F. (1986) Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl Acad. Sci. USA, 83, 5189–5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields P.I., Groisman,E.A. and Heffron,F. (1989) A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science, 243, 1059–1062. [DOI] [PubMed] [Google Scholar]
- García Véscovi E., Soncini,F.C. and Groisman,E.A. (1996) Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell, 84, 165–174. [DOI] [PubMed] [Google Scholar]
- García Véscovi E., Ayala,M., Di Cera,E. and Groisman,E.A. (1997) Characterization of the bacterial sensor protein PhoQ. J. Biol. Chem., 272, 1440–1443. [DOI] [PubMed] [Google Scholar]
- Groisman E.A., Chiao,E., Lipps,C.J. and Heffron,F. (1989) Salmonella typhimurium phoP virulence gene is a transcriptional regulator. Proc. Natl Acad. Sci. USA, 86, 7077–7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E.A., Sturmoski,M.A., Solomon,F., Lin,R. and Ochman,H. (1993) Molecular, functional and evolutionary analysis of sequences specific to Salmonella. Proc. Natl Acad. Sci. USA, 90, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E.A., Kayser,J. and Soncini,F.C. (1997) Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol., 179, 7040–7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J.S. and Miller,S.I. (1996) PhoP–PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol., 178, 6857–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J.S., Lim,K.B., Krueger,J., Kim,K., Guo,L., Hackett,M. and Miller,S.I. (1998) PmrA–PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol., 27, 1171–1182. [DOI] [PubMed] [Google Scholar]
- Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- Igo M.M., Ninfa,A.J., Stock,J.B. and Silhavy,T.J. (1989) Phosphorylation and dephosphorylation of a bacterial activator by a transmembrane receptor. Genes Dev., 3, 1725–1734. [DOI] [PubMed] [Google Scholar]
- Kato A., Tanabe,H. and Utsumi,R. (1999) Molecular characterization of the PhoP–PhoQ two-component system in Escherichia coli K12: Identification of extracellular Mg2+-responsive promoters. J. Bacteriol., 181, 5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee A.K., Detweiler,C.S. and Falkow,S. (2000) OmpR regulates the two-component system SsrA–SsrB in Salmonella pathogenicity island 2. J. Bacteriol., 182, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomis W.F., Shaulski,G. and Wang,N. (1997) Histidine kinases in signal transduction pathways of eukaryotes. J. Cell Sci., 110, 1141–1145. [DOI] [PubMed] [Google Scholar]
- Mäkelä P.H., Sarvas,M., Calcagno,S. and Lounatmaa,K. (1978) Isolation and genetic characterization of polymyxin-resistant mutants of Salmonella. FEMS Microbiol. Lett., 3, 323–326. [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Missiakas D. and Raina,S. (1997) Signal transduction pathways in response to protein misfolding in the extracytoplasmic compartments of E.coli: role of new phosphoprotein phosphatases PrpA and PrpB. EMBO J., 16, 1670–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T., Matsubara,M., Kato,N., Nakamura,Y. and Mizuno,T. (1998) An Escherichia coli protein that exhibits phosphohistidine phosphatase activity towards the HPt domain of the ArcB sensor involved in the multistep His-Asp phosphorelay. Mol. Microbiol., 27, 573–585. [DOI] [PubMed] [Google Scholar]
- Perego M. and Hoch,J.A. (1996) Protein aspartate phosphatases control the output of two-component signal transduction systems. Trends Genet., 12, 97–101. [DOI] [PubMed] [Google Scholar]
- Perraud A.L., Weiss,V. and Gross,R. (1999) Signalling pathways in two-component phosphorelay systems. Trends Microbiol., 7, 115–120. [DOI] [PubMed] [Google Scholar]
- Reece K.S. and Phillips,G.J. (1995) New plasmids carrying antibiotic-resistance cassettes. Gene, 165, 141–142. [DOI] [PubMed] [Google Scholar]
- Roland K.L., Martin,L.E., Esther,C.R. and Spitznagel,J.K. (1993) Spontaneous pmrA mutants of Salmonella typhimurium LT2 define a new two-component regulatory system with possible role in virulence. J. Bacteriol., 175, 4154–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K.L., Esther,C.R. and Spitznagel,J.K. (1994) Isolation and characterization of a gene, pmrD, from Salmonella typhimurium that confers resistance to polymyxin when expressed in multiple copies. J. Bacteriol., 176, 3589–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sanderson K.E., Hessel,A. and Rudd,K.E. (1995) Genetic map of Salmonella typhimurium, edition VIII. Microbiol. Rev., 59, 241–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer W.M., Casey,S.G. and Spitznagel,J.K. (1984a) Lipid A and resistance of Salmonella typhimurium to antimicrobial granule proteins of human neutrophils. Infect. Immun., 43, 834–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer W.M., Martin,L.E. and Spitznagel,J.K. (1984b) Cationic antimicrobial proteins isolated from human neutrophil granulocytes in the presence of diisopropyl fluorophosphate. Infect. Immun., 53, 651–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely M.D., Miller,C.G. and Maguire,M.E. (1991) The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem., 266, 815–823. [PubMed] [Google Scholar]
- Soncini F.C. and Groisman,E.A. (1996) Two-component regulatory systems can interact to process multiple environmental signals. J. Bacteriol., 178, 6796–6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini F.C., García Véscovi,E. and Groisman,E.A. (1995) Transcrip tional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol., 177, 4364–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini F.C., García Véscovi,E., Solomon,F. and Groisman,E.A. (1996) Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol., 178, 5092–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J.B., Ninfa,A.J. and Stock,A.M. (1989) Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev., 53, 450–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J.B., Surette,M.G., Levit,M. and Park,P. (1995) Two component signal transduction systems: structure–function relationships and mechanisms of catalysis. In Hoch,J.A. and Silhavy,T.J. (eds), Two-Component Signal Transduction. ASM Press, Washington, DC, pp. 25–51. [Google Scholar]
- Waldburger C.D. and Sauer,R.T. (1996) Signal detection by PhoQ: characterization of the sensor domain and a response-impaired mutant that identifies ligand-binding determinants. J. Biol. Chem., 271, 26630–26636. [DOI] [PubMed] [Google Scholar]
- Wanner B.L. (1992) Is cross regulation by phosphorylation of two-component response regulator proteins important in bacteria? J. Bacteriol., 174, 2053–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield H.J. and Levine,G. (1973) Isolation and characterization of a mutant of Salmonella typhimurium deficient in a major deoxyribonucleic acid polymerase activity. J. Bacteriol., 116, 54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wösten M.M.S.M. and Groisman,E.A.G. (1999) Molecular characterization of the PmrA regulon. J. Biol. Chem., 274, 27185–27190. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]