Abstract
Losses of heterozygosity are the most common molecular genetic alteration observed in human cancers. However, there have been few systematic studies to understand the mechanism(s) responsible for losses of heterozygosity in such tumors. Here we report a detailed investigation of the five chromosomes lost most frequently in human colorectal cancers. A total of 10,216 determinations were made with 88 microsatellite markers, revealing 245 chromosomal loss events. The mechanisms of loss were remarkably chromosome-specific. Some chromosomes displayed complete loss such as that predicted to result from mitotic nondisjunction. However, more than half of the losses were associated with losses of only part of a chromosome rather than a whole chromosome. Surprisingly, these losses were due largely to structural alterations rather than to mitotic recombination, break-induced replication, or gene conversion, suggesting novel mechanisms for the generation of much of the aneuploidy in this common tumor type.
Aneuploidy is a nearly ubiquitous feature of both common epithelial cancers and experimentally transformed cells (1–5). It is thought that these chromosome changes may be required for malignancy for one of two reasons. First, tumor suppressor genes present anywhere within the lost regions of chromosomes will be deleted from the cell, leading to abnormal growth control if the first allele is mutationally inactivated or silenced (6–8). Second, losses and gains of chromosomal regions have the capacity to alter patterns of gene expression, precisely tailoring them to optimal growth in the abnormal microenvironment within neoplasms (9, 10). Chromosome losses and gains are driven by a chromosomal instability that persists throughout the lifetime of tumor cells (11). Only a few human cancers have been shown to lack aneuploidy, and these generally have a different form of instability that involves subtle sequence changes rather than gross chromosomal events (12).
At the molecular level, aneuploidy is reflected in allelic imbalance and often is associated with losses of heterozygosity (LOH), i.e., loss of one of the parental alleles present in the patient's normal cells (13). In the first study to use DNA polymorphisms to evaluate allelic losses in cancer, it was observed that several potential mechanisms might have contributed to the loss of chromosome 13q in retinoblastomas (13–16). Surprisingly, however, the mechanisms underlying the thousands of LOH that since have been reported in other cancers have not been a focus of study, with rare exceptions (17–20). Instead, most investigations have concentrated on defining the minimal regions of loss of specific chromosomes in various cancers in an effort to identify the putative tumor suppressor genes targeted by the losses. Because knowledge about the mechanisms involved in generating LOH may have significant implications for understanding the pathogenesis of cancer, we have performed detailed molecular genetic and cytogenetic studies of LOH in colorectal cancers. The results, described below, provide insights into the processes underlying aneuploidy in this common tumor type.
Materials and Methods
Colorectal Cancer Cell Lines.
Because primary tumors always contain nonneoplastic cells, a reduced but not absent signal from one allele can never be interpreted unambiguously by molecular techniques. To circumvent this difficulty, we analyzed early passage cell lines passaged in vitro or in nude mice, in which allele losses were unambiguous. It has been shown previously that the genetic alterations in such cell lines usually are indistinguishable from those found in the primary tumors (21–24). Surgically removed colorectal tumors were disaggregated and implanted into nude mice or into in vitro culture conditions as described previously (21, 25). Fifteen of the lines were passaged in vitro and an additional 47 lines were passaged in nude mice, and DNA was prepared at early times (<3 passages) after their establishment. DNA was purified by following standard SDS-proteinase K digestion and phenol-chloroform extraction. The lines studied represented all those available to us in which (i) normal tissue was also available for analysis and (ii) microsatellite instability was not observed in any of five mononucleotide and dinucleotide markers (26, 27). The latter requirement was made because such instability precludes analysis with the polymorphic markers used (28) and because it has been shown previously that tumors with microsatellite instability do not commonly acquire karyotypic changes or LOH events (11).
Allelic Loss Analysis.
Primers for each of the five chromosomes analyzed were obtained from Research Genetics (Huntsville, AL). Mapping and sequence information for each of the markers can be obtained from the authors. A subset of the marker data on chromosome 18 has been published previously (21); all other results are reported here for the first time, to the best of our knowledge. To label primers, one primer of each pair was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase, and amplifications were carried out in 96-well plates in 10-μl reaction volumes (21). The reaction mixtures contained 67 mM Tris⋅HCl (pH 8.8), 16.6 mM ammonium sulfate, 6.7 mM magnesium chloride, 10 mM 2-mercaptoethanol, 6% DMSO, 100 μM each of dATP, dGTP, dCTP, and dTTP, 0.02 mM each of the primers, 10 ng of DNA template, and 0.5 μl of Platinum Taq (GIBCO/BRL). An initial denaturation at 95°C for 2 min was followed by 30 cycles, each carried out at 95°C for 30 sec, 55–60°C for 1 min, and 70°C for 1 min.
Fluorescence in Situ Hybridization (FISH).
Prometaphase chromosome spreads and interphase nuclei were fixed on slides and pretreated with RNase and pepsin as described previously (29). P1 clones were labeled with biotin-16-dUTP and/or digoxigenin-11-dUTP by nick translation. FISH was performed by following standard procedures (30). Biotinylated probes were detected with Texas Red Avidin-DCS. Digoxigenin-labeled probes were detected with FITC-conjugated sheep antidigoxigenin and donkey anti-sheep mAbs. Cells were counterstained with 0.05 μg/ml 4′,6-diamidino-2-phenylindole. Photographs were taken by using a charge-coupled device camera (Princeton) mounted on a Nikon E200 microscope (31).
Results
To understand the mechanisms through which losses of heterozygosity are generated in a common cancer, we studied a panel of colorectal cancers with a large number of highly polymorphic markers spanning the five chromosomes lost most frequently in this disease. We used only early passage cell lines and xenografts for this purpose, because only such pure populations of cells allow distinction between true losses of heterozygosity vs. allelic imbalance from gains of chromosomes. The lines studied represented all those available to us in which normal tissue also was available for analysis and in which microsatellite instability was not observed. A total of 10,216 assays were carried out, using 88 markers dispersed among the five chromosomes at an average distance of 8.8 cM. The assays revealed 245 separate loss events, with at least 29 loss events observed for each of the five chromosomes studied.
Chromosome Arm Specificity.
The first result apparent from these studies was a remarkable specificity with respect to chromosome arm loss. Thus, 32 of the tumors lost markers from chromosome 1p without losing any marker from chromosome 1q, whereas only one tumor lost markers from chromosome 1q without losing any part of chromosome 1p (Fig. 1). On chromosome 5, all 29 losses observed involved the q arm, and none involved the p arm alone. Similar arm-specific losses were observed on chromosomes 8, 17, and 18 (Table 1). These results strongly imply that LOH is due to highly specific events coupled with clonal selection for cells that have lost tumor suppressor genes on the short arms of chromosome 1, 8, and 17 and the long arms of chromosomes 5 and 18 (33, 34).
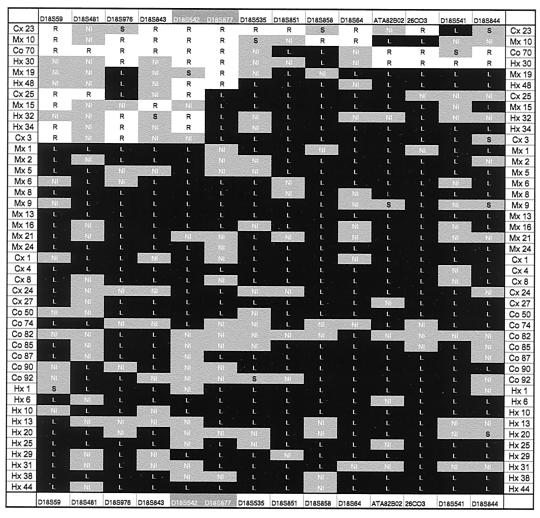
Figure 1.
Partial loss of chromosome 1. Allelic loss analysis of chromosome 1 revealed that 32 of the tumors lost markers from the short arm of chromosome 1 without losing any marker from chromosome 1q, whereas only one tumor (Mx1) lost markers from the long arm of chromosome 1 without losing any part of chromosome 1p. The two outermost vertical columns indicate the tumors analyzed. The outermost horizontal columns list the markers used. The highlighted markers (white lettering) straddle the centromere. A black box containing a “L” indicates that an allele of the indicated marker was lost; a white box containing an “R” indicates that both alleles were retained. Gray boxes containing “NI” indicate that the marker was not informative because the patient's normal cells were not heterozygous for the marker. A gray box marked “S” indicates that there was a new band (“shift”) in the tumor that was not present in normal cells; occasional length changes of microsatellite sequences in tumors are well known (32).
Table 1.
Whole vs. partial chromosome loss in colorectal cancers
| Chromosome
|
|||||
|---|---|---|---|---|---|
| 1 | 5 | 8 | 17 | 18 | |
| % with any LOH | 53.2 | 46.8 | 55.8 | 72.5 | 78.1 |
| % with p arm LOH only | 51.6 | 0 | 48.1 | 41.9 | 0 |
| % with q arm LOH only | 1.6 | 35.5 | 0 | 0 | 14.5 |
| % with whole chromosome loss | 0 | 11.3 | 7.7 | 30.6 | 63.6 |
Whole Chromosome vs. Partial Chromosome Loss.
Each of the chromosomes was affected by LOH in a high frequency of cases, ranging from 47% (chromosome 5) to 78% (chromosome 18). However, the mechanisms underlying these losses were strikingly chromosome-specific. For example, 33 cases of chromosome 1 LOH were noted, and, in each case, only part of chromosome 1, rather than the whole chromosome 1, was lost. In marked contrast, the majority (80%) of losses of chromosome 18 involved the whole chromosome (Figs. 1 and 2 and Table 1). On chromosomes 5 and 8, partial losses were predominant, whereas on chromosome 17, there was a roughly equal mixture of partial and whole chromosome losses (Table 1). Of the 245 losses of chromosome arms, 139 (57%) involved part of a chromosome and the remaining 106 (43%) appeared to involve the whole chromosome.
Figure 2.
Loss of whole chromosome 18. In 32 of 40 tumors, allelic losses of chromosome 18 involved the whole chromosome. Labeling of boxes is the same as in Fig. 1.
In view of this dichotomy, it was of interest to determine whether the patterns of chromosome loss were specific to individual tumors rather than to individual chromosomes. Of the 62 cancers tested, 58 (94%) exhibited LOH of at least one of the five chromosomes analyzed. Of these 58, 42 (78%) exhibited partial loss of one chromosome and complete loss of another. Thus, both processes occurred in most cancers analyzed.
Mechanisms Underlying LOH.
Several basic mechanisms have been considered to account for LOH: mitotic nondisjunction, loss of a segment of chromosome resulting from a deletion event, mitotic recombination between two homologous chromosomes, break-induced replication, recombination between two nonhomologous chromosomes (translocation), and gene conversion (13, 35–38). Some of these patterns can be distinguished by the patterns of marker loss. In addition, we used FISH to help elucidate the mechanism in selected cases. Although we could perform FISH only on a subset of the total cases analyzed (xenografts and cell lines that did not passage well in vitro could not be used), these examinations proved informative.
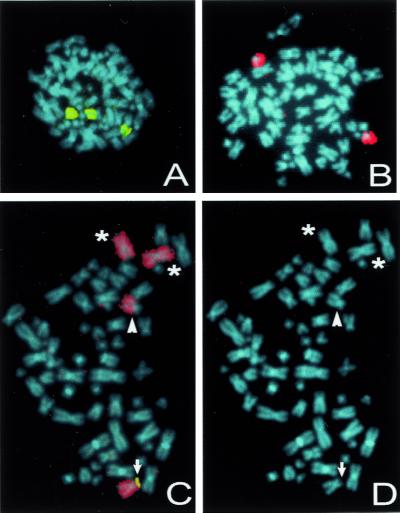
Mitotic nondisjunction likely was responsible for those cases undergoing loss of an entire chromosome as assessed by LOH analysis. This conclusion was buttressed by FISH analyses of five cases by using whole chromosome-specific paint probes that hybridized along the length of chromosomes 17 or 18 (examples in Fig. 3 A and B). Interestingly, in each of these five cases, there were two, three, or four apparently intact copies of the tested chromosome. These cases therefore were homozygous rather than hemizygous for all loci on chromosomes 17 and 18. Both the allelotyping and FISH data pointed to an abnormal mitotic division in which the two chromatids of one chromosome segregated to a single-daughter cell, whereas neither chromatid of the sister chromosome segregated to this daughter cell. In some cases, such an abnormal mitotic division was followed by a reduplication of the chromosome.
Figure 3.
Examples of FISH analysis of colorectal cancers with LOH. (A and B) Although LOH analyses showed that this cancer lost both chromosomes 17 and 18, FISH showed three intact copies of chromosome 17 (painted yellow in A) and two intact copies of chromosome 18 (painted red in B). (C and D) Another colorectal cancer suffered loss of the short arm of chromosome 8 as assessed by molecular LOH analyses. Multicolor FISH analysis with a chromosome 8-specific paint probe (red) and a P1 clone mapping to 8p22 (yellow) revealed translocation of the involved chromosome 8 to another chromosome coinciding with the loss of its short arm (arrowhead). In addition, one intact chromosome 8 (arrow) and two isochromosomes 8q (asterisks) could be observed. (D) Same chromosome spread counterstained with 4′,6-diamidino-2-phenylindole (blue).
In those cases undergoing loss of only part of a chromosome, we expected the major mechanism to be mitotic recombination (13, 35–39). However, FISH analysis revealed that in each of nine cases analyzed, there was a gross structural alteration of the chromosome containing the loss (examples in Fig. 3 C and D). In each of these cases (four on chromosome 5, three on chromosome 8, one on chromosome 17, and one on chromosome 1), the involved chromosomes could be observed to be fused with other chromosomes, and complex intrachromosomal recombinations could be excluded. Thus, the losses of heterozygosity observed in these cases must have been derived by interchromosomal recombinations and deletions associated with DNA double-strand breaks rather than through simple, homologous mitotic recombination, break-induced replication, or gene conversion.
Discussion
Our studies confirm the high prevalence of losses of specific chromosomal regions in epithelial cancers. The arm specificity component of the data suggests further that most LOH events are selected for during tumor development and are not simply “passenger” changes. The data implicating chromosome-specific mechanisms for loss are consistent with previous karyotypic data (3, 40–42). For example, karyotypes of colorectal cancers often reveal loss of an entire chromosome 18, consistent with a mitotic nondisjunction event (34, 41). However, our FISH data on the cases exhibiting only subchromosomal losses were unexpected. Based on previous studies of human cancers, we anticipated that these losses would be the result of processes that did not involve structural rearrangements of chromosomes, such as mitotic recombination (13, 35, 37, 38). Recent mechanistic studies in murine cells also have suggested that LOH in tumors is likely to result from mitotic recombination (39). Indeed, had we not performed FISH, our allelotyping studies and standard karyotypes of these tumors (data not shown) also would have suggested mitotic recombination to be the major mechanism involved. The differences between our study and previous ones may be due to the different cell types analyzed. There have been very few detailed studies of the mechanisms underlying LOH in common forms of human cancer, and current views are based largely on classic studies of retinoblastomas (13, 36, 38). Although our conclusions are based on a relatively small number of cases, they are unbiased in that we examined every cancer cell line in the cohort in which high-quality mitoses could be obtained. In every case analyzed, the losses of whole chromosomes detected by allelotyping were found to be associated with duplications or triplications of the remaining chromosome. In addition, in every case analyzed, the partial losses of chromosomes detected by allelotyping were found to be associated with gross structural changes associated with interchromosomal recombinations. Our data are consistent with recent spectral karyotype and Multiplex-FISH data indicating a far higher prevalence of structural chromosomal abnormalities in cancers than indicated by classic cytogenetics or suspected from LOH analyses (43, 44). Based on this information, it can be predicted that much of the LOH observed in other common human cancers will prove to be due to structural changes rather than mitotic recombination.
We therefore propose a model for aneuploidy in human cancer cells that involves two separate defects. The first to occur, perhaps at the earliest stage of tumorigenesis (45), involves defects that lead to abnormalities in chromosome number in the absence of chromosome structural changes. Such defects may include those affecting cell cycle checkpoints and centrosome number (46). These defects would explain the aneuploidy present in benign tumors (45, 47, 48) and would account for those cases in our study involving whole chromosome loss. The second proposed defect, however, cannot be accounted for by such well studied mechanisms. This defect presumably results in a higher frequency of structural changes in chromosomes associated with interchromosomal recombination, perhaps resulting from chromosome breakage and fusion events. More than half of the loss events observed in colorectal cancers was attributable to this second type of defect. The mechanisms responsible for such alterations are mostly obscure. Because such events likely begin with double-strand breaks, however, it is possible that they arise from defects in the double-strand repair–recombination machinery (e.g., ref. 49). Accordingly, recently developed techniques for evaluating yeast chromosomal breakpoints at the molecular level could prove valuable (50, 51), pointing to human genes that lead to high rates of structural alterations when mutated.
Acknowledgments
We thank L. Meszler from the Cell Imaging Core Facility of the Johns Hopkins Oncology Center for technical assistance. This work was supported by the V Foundation (C.L.), the Clayton Fund, and by grants from the National Institutes of Health.
Abbreviations
- LOH
loss(es) of heterozygosity
- FISH
fluorescence in situ hybridization
References
- 1.Boveri T. Zur Frage der Enstehung maligner Tumoren. Vol. 1. Jena, Germany: Fischer; 1914. [Google Scholar]
- 2.Rowley J D. Nature (London) 1983;301:290–291. doi: 10.1038/301290a0. [DOI] [PubMed] [Google Scholar]
- 3.Mitelman F, Johansson B, Mertens F. Catalog of Chromosome Aberrations in Cancer. New York: Wiley; 1994. [Google Scholar]
- 4.Devilee P, Cornelisse C J. Biochim Biophys Acta. 1994;1198:113–130. doi: 10.1016/0304-419x(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 5.Sandberg A A, Chen Z. In Vivo. 1994;8:807–818. [PubMed] [Google Scholar]
- 6.Knudson A G., Jr Semin Oncol. 1978;5:57–60. [PubMed] [Google Scholar]
- 7.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 8.Herman J G, Baylin S B. Curr Top Microbiol Immunol. 2000;249:35–54. doi: 10.1007/978-3-642-59696-4_3. [DOI] [PubMed] [Google Scholar]
- 9.Duesberg P, Rausch C, Rasnick D, Hehlmann R. Proc Natl Acad Sci USA. 1998;95:13692–13697. doi: 10.1073/pnas.95.23.13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lucito R, West J, Reiner A, Alexander J, Esposito D, Mishra B, Powers S, Norton L, Wigler M. Genome Res. 2000;10:1726–1736. doi: 10.1101/gr.138300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 12.Lengauer C, Kinzler K W, Vogelstein B. Nature (London) 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 13.Cavenee W K, Dryja T P, Phillips R A, Benedict W F, Godbout R, Gallie B L, Murphree A L, Strong L C, White R L. Nature (London) 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert F. Nature (London) 1983;305:761–762. doi: 10.1038/305761a0. [DOI] [PubMed] [Google Scholar]
- 15.Benedict W F, Srivatsan E S, Mark C, Banerjee A, Sparkes R S, Murphree A L. Cancer Res. 1987;47:4189–4191. [PubMed] [Google Scholar]
- 16.Zhu X, Dunn J M, Goddard A D, Squire J A, Becker A, Phillips R A, Gallie B L. Cytogenet Cell Genet. 1992;59:248–252. doi: 10.1159/000133261. [DOI] [PubMed] [Google Scholar]
- 17.Yandell D W, Dryja T P, Little J B. Mutat Res. 1990;229:89–102. doi: 10.1016/0027-5107(90)90011-r. [DOI] [PubMed] [Google Scholar]
- 18.Mertens F, Johansson B, Mitelman F. Genes Chromosomes Cancer. 1994;10:221–230. doi: 10.1002/gcc.2870100402. [DOI] [PubMed] [Google Scholar]
- 19.de Nooij-van Dalen A G, van Buuren-van Seggelen V H, Lohman P H, Giphart-Gassler M. Genes Chromosomes Cancer. 1998;21:30–38. doi: 10.1002/(sici)1098-2264(199801)21:1<30::aid-gcc5>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 20.Bovee J V, Cleton-Jansen A M, Kuipers-Dijkshoorn N J, van den Broek L J, Taminiau A H, Cornelisse C J, Hogendoorn P C. Genes Chromosomes Cancer. 1999;26:237–246. [PubMed] [Google Scholar]
- 21.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Wilson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 22.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K, Markowitz S, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 23.Cahill D P, Lengauer C, Yu J, Riggins G J, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nature (London) 1998;392:300–303. doi: 10.1038/32688. [DOI] [PubMed] [Google Scholar]
- 24.Thiagalingam S. In: Methods in Molecular Medicine. Powell S M, editor. Vol. 50. Totowa, NJ: Humana; 2001. pp. 149–165. [DOI] [PubMed] [Google Scholar]
- 25.Willson J K, Bittner G N, Oberley T D, Meisner L F, Weese J L. Cancer Res. 1987;47:2704–2713. [PubMed] [Google Scholar]
- 26.Parsons R, Myeroff L L, Liu B, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Cancer Res. 1995;55:5548–5550. [PubMed] [Google Scholar]
- 27.Boland C R, Thibodeau S N, Hamilton S R, Sidransky D, Eshleman J R, Burt R W, Meltzer S J, Rodriguez-Bigas M A, Fodde R, Ranzani G N, et al. Cancer Res. 1998;58:5248–5257. [PubMed] [Google Scholar]
- 28.Aaltonen L A, Peltomaki P, Leach F S, Sistonen P, Pylkkanen L, Mecklin J P, Jarvinen H, Powell S M, Jen J, Hamilton S R, et al. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 29.Ried T, Lengauer C, Cremer T, Wiegant J, Raap A K, van der Ploeg M, Groitl P, Lipp M. Genes Chromosomes Cancer. 1992;4:69–74. doi: 10.1002/gcc.2870040109. [DOI] [PubMed] [Google Scholar]
- 30.Lichter P, Boyle A L, Cremer T, Ward D C. Genet Anal Tech Appl. 1991;8:24–35. doi: 10.1016/1050-3862(91)90005-c. [DOI] [PubMed] [Google Scholar]
- 31.Lengauer C, Speicher M R, Popp S, Jauch A, Taniwaki M, Nagaraja R, Riethman H C, Donis-Keller H, D'Urso M, Schlessinger D, et al. Hum Mol Genet. 1993;2:505–512. doi: 10.1093/hmg/2.5.505. [DOI] [PubMed] [Google Scholar]
- 32.Mao L, Schoenberg M P, Scicchitano M, Erozan Y S, Merlo A, Schwab D, Sidransky D. Science. 1996;271:659–662. doi: 10.1126/science.271.5249.659. [DOI] [PubMed] [Google Scholar]
- 33.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 34.Bardi G, Sukhikh T, Pandis N, Fenger C, Kronborg O, Heim S. Genes Chromosomes Cancer. 1995;12:97–109. doi: 10.1002/gcc.2870120204. [DOI] [PubMed] [Google Scholar]
- 35.Meuth M. Biochim Biophys Acta. 1990;1032:1–17. doi: 10.1016/0304-419x(90)90009-p. [DOI] [PubMed] [Google Scholar]
- 36.Stanbridge E J. Annu Rev Genet. 1990;24:615–657. doi: 10.1146/annurev.ge.24.120190.003151. [DOI] [PubMed] [Google Scholar]
- 37.Haber J E. Trends Biochem Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 38.Cavenee W K, Hansen M F, Nordenskjold M, Kock E, Maumenee I, Squire J A, Phillips R A, Gallie B L. Science. 1985;228:501–503. doi: 10.1126/science.3983638. [DOI] [PubMed] [Google Scholar]
- 39.Luo G, Santoro I M, McDaniel L D, Nishijima I, Mills M, Youssoufian H, Vogel H, Schultz R A, Bradley A. Nat Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- 40.Ried T, Knutzen R, Steinbeck R, Blegen H, Schrock E, Heselmeyer K, du Manoir S, Auer G. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 41.Remvikos Y, Muleris M, Salmon R J, Dutrillaux B. Cancer Genet Cytogenet. 1997;93:63–73. doi: 10.1016/s0165-4608(96)00305-6. [DOI] [PubMed] [Google Scholar]
- 42.Nakao K, Shibusawa M, Tsunoda A, Yoshizawa H, Murakami M, Kusano M, Uesugi N, Sasaki K. Surg Today. 1998;28:567–569. doi: 10.1007/s005950050185. [DOI] [PubMed] [Google Scholar]
- 43.Speicher M R, Petersen S, Uhrig S, Jentsch I, Fauth C, Eils R, Petersen I. Lab Invest. 2000;80:1031–1041. doi: 10.1038/labinvest.3780108. [DOI] [PubMed] [Google Scholar]
- 44.Melcher R, Steinlein C, Feichtinger W, Muller C R, Menzel T, Luhrs H, Scheppach W, Schmid M. Cytogenet Cell Genet. 2000;88:145–152. doi: 10.1159/000015508. [DOI] [PubMed] [Google Scholar]
- 45.Shih, I. M., Zhou, W., Goodman, S. N., Lengauer, C., Kinzler, K. W. & Vogelstein, B. (2001) Cancer Res., in press. [PubMed]
- 46.Doxsey S. Nat Genet. 1998;20:104–106. doi: 10.1038/2392. [DOI] [PubMed] [Google Scholar]
- 47.Hemmer S, Wasenius V M, Knuutila S, Joensuu H, Franssila K. Br J Cancer. 1998;78:1012–1017. doi: 10.1038/bjc.1998.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dohna M, Reincke M, Mincheva A, Allolio B, Solinas-Toldo S, Lichter P. Genes Chromosomes Cancer. 2000;28:145–152. [PubMed] [Google Scholar]
- 49.Ferguson D O, Sekiguchi J M, Chang S, Frank K M, Gao Y, DePinho R A, Alt F W. Proc Natl Acad Sci USA. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. . (First Published May 23, 2000; 10.1073/pnas.110152897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chen C, Kolodner R D. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- 51.Hiraoka M, Watanabe K, Umezu K, Maki H. Genetics. 2000;156:1531–1548. doi: 10.1093/genetics/156.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]