Introduction
The deployment of a cell’s genetic program in a multicellular organism must be tightly controlled for the sake of the organism as a whole. Over the past 20 years the transforming growth factor-β (TGF-β) family of secretory polypeptides has emerged as a major source of signals exerting this type of control. This family includes various forms of TGF-β, the bone morphogenetic proteins (BMPs), the Nodals, the Activins, the anti-Müllerian hormone, and many other structurally related factors in vertebrates, insects and nematodes (Massagué, 1998). Produced by diverse cell types, these factors regulate cell migration, adhesion, multiplication, differentiation and death throughout the life span of the organism. Many of these responses result from changes in the expression of key target genes. Hence, transcriptional control by the TGF-β family has become a subject of intense investigation in recent years. The present knowledge of these mechanisms is reviewed here.
Ask not what a signal can do with a cell, but what a cell can do with a signal
One basic concept concerning the role of the TGF-β family as hormonally active agents warrants mention at the outset. Unlike classical hormones, whose actions are few and concrete, the members of the TGF-β family have many different effects depending on the type and state of the cell. For example, in the same healing wound TGF-β may stimulate or inhibit cell proliferation depending on whether the target is a fibroblast or a keratinocyte (Ashcroft et al., 1999); in mammary epithelial cells TGF-β will cause growth arrest or metastatic behavior depending on the level of oncogenic Ras activity present in the cell (Oft et al., 1996); and human BMP4 and its Drosophila ortholog, DPP, can signal dorsalization in the fly (Padgett et al., 1993) yet bone formation in a vertebrate (Sampath et al., 1993). TGF-β family members are multifunctional hormones, the nature of their effects depending on what has been called ‘the cellular context’.
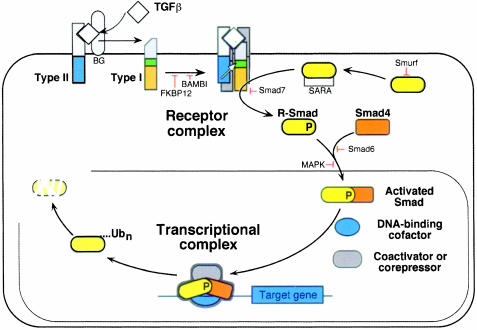
It was plausible that the TGF-β signal transduction pathways might have to be numerous and complex in order to account for this diversity of responses. On the contrary, a disarmingly simple system has been elucidated recently that mediates many diverse TGF-β responses. This system involves a family of membrane receptor protein kinases and a family of receptor substrates, the Smad proteins, that march into the nucleus where they act as transcription factors. The ligand TGF-β assembles a receptor complex that activates Smads, and the Smads assemble multisubunit complexes that regulate transcription (Figure 1) (reviewed in Massagué, 1998). Two general steps thus suffice to carry the hormonal stimulus to target genes.
Fig. 1. Schematic representation of the TGF-β/Smad signaling engine. This system involves a family of membrane receptor protein kinases and a family of receptor substrates (the Smad proteins) that march into the nucleus where they act as transcription factors. The ligand TGF-β assembles a receptor complex that activates Smads, and the Smads assemble multisubunit complexes that regulate transcription. Two general steps suffice to carry the hormonal stimulus to target genes. The central components of this signaling system are indicated along with the sites of action of various positive and negative regulators. See the text for further details.
How can such a simple system mediate a variety of cell-specific responses? An incoming Smad complex is met in the nucleus by a set of partner proteins that are specific to a particular cell type in each particular set of conditions. These partners determine the DNA sequences that the Smad complex will bind, the transcriptional co-activators or co-repressors it will recruit, the other transcription factors it will cooperate with, and how long all this will last. The mix of Smad partners and regulators present in a given cell at the time of TGF-β stimulation thus decides the outcome of the response, and defines, in molecular terms, the cellular context. Identifying these partners and regulators is, therefore, critical for understanding TGF-β action.
Smad activation
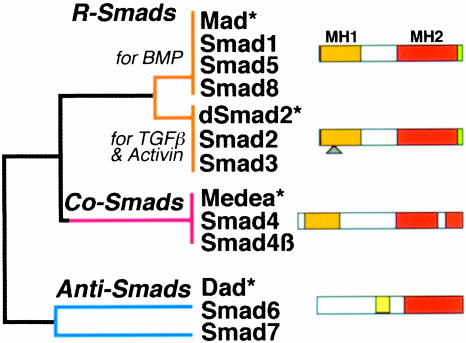
The Smad proteins are a family of transcription factors found in vertebrates, insects and nematodes (Figure 2) (Heldin et al., 1997; Massagué, 1998). To date, the Smads are the only TGF-β receptor substrates with a demonstrated ability to propagate signals. The mechanism of activation of the TGF-β receptor itself has been reviewed in detail elsewhere (Massagué, 1998). Briefly, two different transmembrane protein serine/threonine kinases, known as receptor types I and II, are brought together by the ligand, which acts as a receptor assembly factor (Figure 1). Before this occurs, receptor I is catalytically inactive because a wedge-shaped GS region is inserted into the kinase domain, dislocating the catalytic center (Huse et al., 1999). In the ligand-induced complex, receptor II phosphorylates the GS region, resulting in activation of the receptor I kinase.
Fig. 2. The Smad family. Simplified dendrogram of sequence similarity between the three Smad subfamilies. The receptor-regulated Smads (R-Smads) and their cooperating Smads (Co-Smads) contain conserved N-terminal (MH1) and C-terminal (MH2) domains separated by a divergent region. Only the MH2 domain is conserved in the inhibitory Smads (Anti-Smads). The green sliver represents the receptor phosphorylation sites at the extreme C-terminus of the R-Smads. The triangle represents the alternatively spliced insert in Smad2. Asterisks denote representative members from Drosophila.
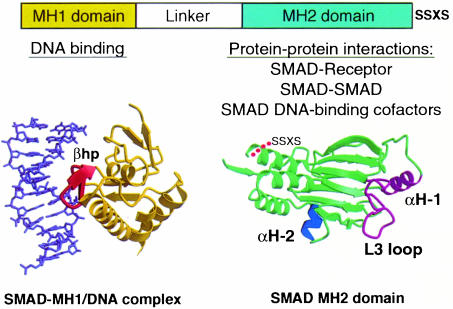
The type I receptors specifically recognize the Smad subgroup known as receptor-activated Smads (R-Smads) (Massagué, 1998 and references therein). These include Smad2 and Smad3, which are recognized by TGF-β and Activin receptors, and Smads 1, 5 and 8, recognized by BMP receptors (Figure 2). The R-Smads consist of two conserved domains that form globular structures separated by a linker region (Figures 2 and 3) (Shi et al., 1997, 1998). The N-terminal MH1 domain has DNA-binding activity whereas the C-terminal MH2 drives translocation into the nucleus and has transcription regulatory activity. Receptor-mediated phosphorylation of the C-terminal sequence SSxS appears to relieve these two domains from a mutually inhibitory interaction and leads to R-Smad activation and accumulation in the nucleus (Figure 1).
Fig. 3. Smad structural domains and their functions. Representation of the three-dimensional structures of the Smad3 MH1 domain bound to the AGAC sequence, and the Smad2 MH2 domain. The principal interactions of these two domains are listed. The structures involved in these interactions are shown in different colors: the β-hairpin (βhp) that mediated DNA binding, the L3 loop and α-helix 1 (αH-1) that specify Smad interactions with type I receptors, and the α-helix 2 (αH-2) that specifies Smad2 interaction with FAST. SSXS, receptor phosphorylation sites (adapted from Shi et al., 1997, 1998; Wu et al., 2000).
Forming a Smad complex
Little is known about the mechanism of phosphorylation-induced Smad nuclear accumulation. However, it is well established that en route to the nucleus, R-Smads associate with members of a second group known as the Co-Smads (Figure 1) (Massagué, 1998). Two highly related Co-Smads are known in vertebrates: Smad4 and Smad4β (the latter also known as Smad10 and to date identified only in Xenopus) (Figure 2) (Howell et al., 1999; Masuyama et al., 1999). The Co-Smads have an MH1–linker–MH2 domain structure, but lack the SSxS sequence and are not phosphorylated by the receptor. Their interaction with R-Smads is primarily mediated by MH2 domain contacts (Hata et al., 1997; Wu et al., 1997). The Co-Smads are shared by all R-Smads (Lagna et al., 1996; Masuyama et al., 1999) and are required not for nuclear accumulation but for the formation of functional transcriptional complexes (Liu et al., 1997). Both the R-Smad and the Co-Smad in this complex may participate in DNA binding and recruitment of transcriptional cofactors.
Smad4 forms homotrimers in solution, and inactivating mutation of key residues in the monomer interfaces occurs in cancer (Shi et al., 1997). Putative trimer interface residues are also mutated in inactive R-Smad alleles. A trimeric configuration has been suggested to be optimal for Smad binding to DNA (Johnson et al., 1999). However, Smads also exist as monomers (Kawabata et al., 1998). The Smad functions that require a monomeric or trimeric configuration, and the stoichiometry of Smad complexes in general, remain to be elucidated.
Control over the activation process
The two basic steps in TGF-β/Smad signaling, receptor activation and Smad activation, are controlled by a web of regulatory proteins that exert tight control over the activity of this system (reviewed in Massagué and Chen, 2000). Positive regulators include ligand accessory receptors and substrate anchoring factors (Figure 1). The membrane-anchored proteoglycan betaglycan (also known as the TGF-β type III receptor) binds TGF-β and increases its affinity for the signaling receptors (Massagué, 1998; Brown et al., 1999). The protein Smad anchor for receptor activation (SARA), which is thought to associate with endosomal membranes via lipid-binding FYVE domains, binds Smad2 and Smad3, facilitating their interaction with TGF-β receptors (Tsukazaki et al., 1998). SARA clamps onto the MH2 domain as an extended structure without occluding receptor recognition surfaces (Wu et al., 2000).
Other proteins exert negative control (Figure 1). Various proteins bind TGF-β, Activin, Nodal or BMP, inhibiting their interaction with receptors (Massagué and Chen, 2000). The immunophilin FKBP12 binds to the GS domain, occluding it from ligand-independent receptor phosphorylation (Huse et al., 1999). The pseudoreceptor BAMBI forms inactive dimers with type I receptors (Onichtchouk et al., 1999). Smad6 acts as a Smad4 decoy that blocks activated Smad1 (Hata et al., 1998). Another antagonistic Smad, Smad7, blocks activated receptors (Hayashi et al., 1997; Nakao et al., 1997). Erk kinases activated via the Ras pathway phosphorylate R-Smads in the linker region, inhibiting Smad nuclear accumulation (Kretzschmar et al., 1999). The Smad1 ubiquitin ligase Smurf1 regulates the basal levels of Smad1 (Zhu et al., 1999), whereas Smad2 in the nucleus is specifically targeted for ubiquitylation and degradation, effectively ending its function (Lo and Massagué, 1999). Many of these regulators participate in Smad feedback loops or as mechanisms for integration with other signaling pathways.
Receptor–Smad interaction: a first level of target gene selection
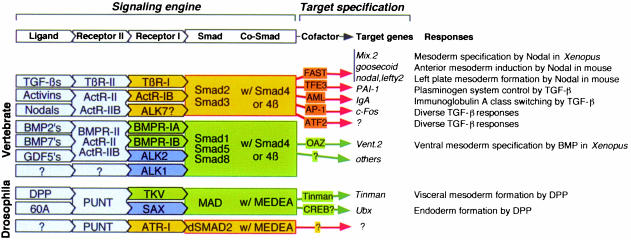
Based on structural and functional similarities, the type I receptors fall into distinct subtypes that specifically recognize one of two subtypes of R-Smads (Figure 4). These two sets of R-Smads differ in the genes that they control, and lead to radically different cellular responses (Heldin et al., 1997; Massagué, 1998). Mutations that switch the recognition of R-Smads by TGF-β and BMP receptors also switch the responses to these receptors (Chen et al., 1998). Therefore, the choice of R-Smad by a TGF-β family receptor provides a first level of target gene selection.
Fig. 4. Making choices through the Smad system.The combinatorial organization of this system as presently understood.
Small structural elements on the surface of the type I receptors and the R-Smads determine the specificity of a receptor–Smad interaction. These elements are the L45 loop in the small lobe of the type I receptor kinase (Feng and Derynck, 1997; Chen et al., 1998; Huse et al., 1999) and the L3 loop in the MH2 domain of R-Smads (Lo et al., 1998) (Figure 3). A few key amino acid residues in the L45 loop and the L3 loop are conserved only within a given receptor subtype or Smad subtype. Exchanging these residues between receptor or Smad subtypes is sufficient to switch the signaling specificity of the TGF-β and BMP pathways (Chen et al., 1998). ALK1 and ALK2 specifically recognize Smads 1, 5 and 8 even though the L45 loop sequence of these receptors is quite different from that of the other BMP receptors. In this case, R-Smad recognition requires the L3 loop as well as the adjacent α-helix 1 in the MH2 domain (Figure 3) (Chen and Massagué, 1999).
The DNA-binding function of Smads
The fusion of a Smad MH2 domain to a heterologous DNA-binding domain, such as that of Gal4p, demonstrated that, once recruited to DNA, a Smad complex is able to activate transcription (Liu et al., 1996). Smad recruitment to DNA is therefore a key step in determining which set of genes will be activated in response to a TGF-β stimulus. Both R-Smads and Co-Smads can bind to DNA via the MH1 domain (Kim et al., 1997; Shi et al., 1998). Optimal binding is achieved with the 5 bp sequence CAGAC, although AGAC suffices (Shi et al., 1998; Zawel et al., 1998). Such Smad-binding elements (SBEs) are often present in the responsive region of TGF-β, Activin or BMP target genes. [The original identification of the palindrome GTCTAGAC as an SBE may have resulted from dimerization of recombinant Smads used in oligonucleotide selection experiments (Zawel et al., 1998)]. The three-dimensional structure of the Smad3 MH1 domain bound to cognate DNA shows that the MH1 domain monomer binds precisely to either half of this sequence (Shi et al., 1998). DNA binding is mediated by a β-hairpin structure that protrudes from the surface of the MH1 domain and binds in the major groove (Figure 3).
In several genes, the ability to respond to TGF-β family signals requires the presence of one or more SBEs (Vindevoghel et al., 1998; Hua et al., 1999; Nagarajan et al., 1999; Yeo et al., 1999; Hata et al., 2000). This rule, however, may not apply to all Smad target genes, as suggested by the absence of a canonical SBE in mouse goosecoid (Kim et al., 1997; Labbé et al., 1998). Moreover, it is not essential that every Smad subunit in a transcriptional complex contact DNA (Yeo et al., 1999). In fact, the most common splice form of Smad2 lacks DNA-binding activity because of an insert located next to the β-hairpin (Shi et al., 1998).
Role of the SBE in Smad binding to DNA
The SBE CAGAC sequence is calculated to be present on average once every 1024 bp in the genome, or about once in the regulatory region of any average size gene. If binding to the SBE were sufficient for Smad-dependent transcriptional activation, an activated Smad protein would lead to the non-selective activation of massive numbers of genes. However, this is not the case. Activation of target genes solely via an SBE is not feasible for two reasons. First, the affinity of a Smad MH1 domain for the SBE is in the 10–7 M range (Shi et al., 1998), which is too weak for effective binding in vivo without the involvement of additional DNA contacts. In artificial concateners it takes many SBEs to achieve Smad activation of a reporter gene (Zawel et al., 1998). Secondly, Smad binding to the SBE lacks selectivity, as Smads 1, 3 and 4 have a similar affinity for the SBE. This is not surprising because the β-hairpin sequence is identical in all R-Smads and highly conserved in Smad4. Therefore, additional DNA contacts appear necessary for specific, high-affinity binding of a Smad complex to a target gene.
A regulatory region of junB containing multiple copies of the SBE has been shown, when tested as a dimer, to bind Smads and support activation of a reporter gene when co-transfected with Smads (Jonk et al., 1998). However, the ability of TGF-β to activate the natural junB promoter through this element remains to be confirmed and the involvement of other proteins as cofactors of endogenous Smads in this response has not been ruled out. The latter also applies to the TGF-β response element in Smad7, which includes an SBE palindrome GTCTAGAC (Nagarajan et al., 1999).
DNA-binding partners that determine the choice of target genes
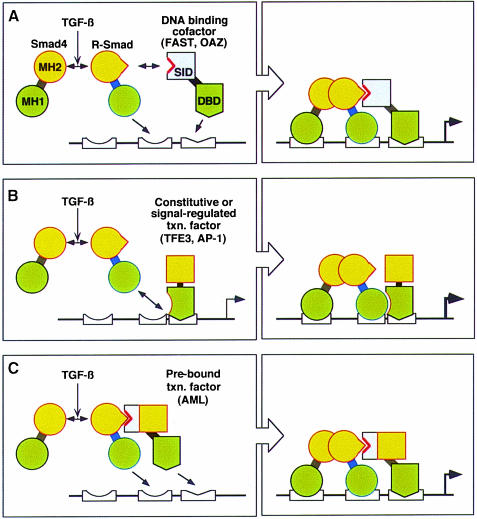
By associating with DNA-binding partners, forming complexes of specific composition and geometry, the Smads can achieve high-affinity, selective interactions with cognate DNA (Figure 5). The DNA-binding domains of Smads and their partners in the same complex will be able to act synergistically if their corresponding cognate sequences are present at the right distance from each other in a target promoter.
Fig. 5. Smad transcriptional partners. General models for the recognition and regulation of specific target genes by Smads in concert with DNA-binding adaptors such as FAST and OAZ (model A) and constitutive (e.g. TFE and CBF) or signal-regulated (e.g. AP-1) transcription factors that interact with the MH1 domain upon agonist activation (model B) or with the MH2 domain in the basal state (model C). MH1 and MH2, Smad domains; orange boxes, transactivator domains; SID, Smad interaction domain. Although two Smad DNA sites are depicted in each model, only one may be used in certain response elements.
FAST as a Smad partner in Nodal pathways
An interaction of Smad proteins with a DNA-binding factor, resulting in tight binding of this complex to DNA, was first described for the Xenopus FAST-1 protein (Chen et al., 1996). FAST-1 is a member of the winged-helix family of DNA-binding proteins. It was identified by its ability to bind to the ARE, an enhancer responsive to an Activin-like factor—most likely, Nodal—in the Xenopus Mix.2 homeobox gene. FAST-1 is now known to mediate activation of an entire panel of homeobox genes in the specification of mesoderm (Watanabe and Whitman, 1999). A mammalian homolog (alternatively named FAST-1 or FAST-2) may mediate activation of the homeobox gene goosecoid following gastrulation (Labbé et al., 1998; Zhou et al., 1998) and the TGF-β family members lefty-2 and nodal during establishment of the left-side lateral plate mesoderm in response to an initial pulse of Nodal (Saijoh et al., 2000). Other Smad-dependent TGF-β or Activin responses do not involve FAST, suggesting that different DNA-binding partners may mediate these responses.
Studies on Smads and FAST have revealed basic principles governing Smad interactions with DNA-binding cofactors (Chen et al., 1997; Liu et al., 1997; Yeo et al., 1999). FAST interacts with Smad2–Smad4 or Smad3–Smad4 complexes, but not with BMP-activated Smad complexes. A few subtype-specific residues in the α-helix 2 region of the Smad2/3 MH2 domain determine the specificity of this interaction (Figure 3) (Chen et al., 1998). Both FAST and the associated Smads are required for efficient binding to target enhancers. The Smad4 MH1 domain is not required for high-affinity binding of Smad3–Smad4–FAST complexes to DNA, but is required when the complex contains Smad2, which lacks intrinsic DNA-binding activity. The FAST-binding sequence in the Mix.2, goosecoid, nodal and lefty2 enhancers is similar, but the proposed Smad-binding sequences may differ. Smads appear to contact an inverted SBE (GTCT) downstream of the FAST site in Mix.2, long GC-rich sequences (that may represent degenerate SBEs) upstream of the FAST site in goosecoid, and SBEs in various orientations in nodal and lefty2 (Labbé et al., 1998; Yeo et al., 1999; Saijoh et al., 2000). Both Smad4 and Smad2 (or Smad3) are required for efficient transactivation from the ARE, suggesting that both types of Smads jointly recruit coactivators regardless of their individual contributions to DNA binding. Via a motif homologous to the Smad interacting domain of FAST, the homeodomain proteins Mixer and Milk recruit Smad2–Smad4 to the activin-responsive enhancer of goosecoid during Xenopus development (Germain et al., 2000).
OAZ as a bifunctional Smad partner in the BMP pathway
In the BMP pathway, OAZ has been identified as a DNA-binding cofactor that associates with an activated Smad1–Smad4 complex and allows recognition and activation of the homeobox gene Xvent-2 (Hata et al., 2000). Xvent-2, which controls mesoderm ventralization and suppresses neuralization in Xenopus, contains an enhancer, the BMP response element (BRE), which has a CAGAC SBE for Smad binding and a separate binding site for OAZ. Both OAZ and Smads are required for efficient binding to this element. Like the FAST proteins, OAZ appears to lack intrinsic transactivating activity; this activity is provided by the Smads in the complex. In spite of these similarities, OAZ and FAST are structurally unrelated. OAZ is a zinc-finger protein with 30 Krüppel-type fingers distributed in several groups. Distinct groups directly recognize the BRE on Vent.2 and the MH2 domain on Smad1. Together these two groups form a signaling module that is necessary and sufficient for activation of the Xvent-2 promoter in response to BMP2.
OAZ has also been implicated as a partner of the transcription factor Olf-1/EBF in the control of gene expression during the development of the olfactory epithelium and pre-B lymphocytes (Tsai and Reed, 1998). The BMP signaling module of OAZ is separate from the regions involved in the Olf-1/EBF interaction (Hata et al., 2000). The mutually exclusive use of OAZ by the BMP–Smad and Olf pathways suggests a dual role of this multi-zinc-finger protein in separate signal transduction pathways during development.
OAZ is expressed in BMP-sensitive adult tissues, suggesting that it may be involved in other BMP responses. However, OAZ is specific for genes that have a Vent2-like BRE. It does not mediate the activation of BMP2-responsive genes that have a different type of BRE, such as the homeobox gene Tlx2. In fact, OAZ overexpression inhibits BMP2-induced activation of the Tlx.2 promoter, suggesting that OAZ and a Tlx2-specific activator may compete for BMP-activated Smads (Hata et al., 2000).
In Drosophila, the homeobox protein Tinman cooperates with Mad and Medea (the orthologs of Smad1 and Smad4, respectively) in the induction of tinman itself by DPP during formation of the visceral mesoderm (Xu et al., 1998).
Three levels of specificity
DNA-binding Smad cofactors like FAST and OAZ provide three levels of specificity to a Smad-dependent gene response. By cooperating only with Activin-activated Smads or BMP-activated Smads, respectively, FAST and OAZ ensure pathway specificity. By recognizing target genes that contain the appropriate ARE (such as Mix.2) or BRE (such as Xvent-2), but not other Activin–TGF-β or BMP target genes, FAST and OAZ provide target gene specificity within their respective Activin and BMP pathways; and, by being expressed in some cell types but not others, FAST and OAZ provide cell-type specificity to the Activin or BMP response.
Smad interactions with other transcription factors
The FAST and OAZ proteins cannot activate transcription on their own because they apparently lack a transactivation domain. These proteins may therefore be considered as DNA-binding adaptors for the Smads. However, various proteins that act as transcription factors on their own have recently been shown to recruit Smads to certain promoters. These include constitutive as well as signal-activated transcription factors that were previously known to act in other contexts. Such interactions may be mediated either by the MH1 or MH2 domain of Smads. Like FAST and OAZ, the transcription factors may mediate Smad binding to SBEs located at the appropriate distance from their own binding sites (Figure 5). In this case, however, Smad recruitment may serve to augment or modify the activity of an existing transcriptional complex, rather than creating a transcriptional complex de novo as occurs with FAST or OAZ.
TFE3
The transcription factor TFE3 was originally identified as a regulator of the immunoglobulin µ heavy chain enhancer (Beckmann et al., 1990). TFE3 is a basic helix–loop–helix leucine-zipper transcription factor that binds the coactivator p300/CBP and the DNA sequence CACGTG (known as the E-box) as a homodimer. On the µ heavy chain enhancer, TFE3 can stimulate binding of another transcription factor, Ets-1 (Tian et al., 1999). TFE3 binds to an E-box in one of the TGF-β response elements of plasminogen activator inhibitor-1 (PAI-1) (Hua et al., 1999). TFE3 binding to this site allows recruitment of a Smad3–Smad4 complex to two adjacent SBEs. In response to TGF-β, the Smads increase the basal activity observed with TFE3 alone. This effect requires a distance of 3 bp between the E-box and the closest SBE, and may involve direct contacts between TFE3 and the MH1 domain of one of the Smads in the complex. Of note, this is one of four distinct Smad-interacting regions identified in PAI-1, the other regions apparently not involving TFE3 (Stroschein et al., 1999a).
Contacts with AP-1 and ATF-2
Various gene responses to TGF-β appear to require the presence of Fos–Jun (AP-1) transcriptional activity in the cell (Zhang and Derynck, 1999). Smads and AP-1 have been shown to synergize in transcriptional activation from artificial promoters (Zhang et al., 1998; Wong et al., 1999) and to form complexes in vitro and under conditions of overexpression (Zhang et al., 1998; Liberati et al., 1999). However, based on the three-dimensional structure of an AP-1-bound Fos–Jun complex and an SBE-bound Smad complex, it is not apparent how these two protein complexes could simultaneously contact the DNA site (Shi et al., 1998). It has been suggested that a Smad3–Smad4 complex and an AP-1 complex synergize in the transcriptional activation from the c-Jun promoter by binding to separate sites located 120 bp apart from each other (Wong et al., 1999). Adding further complexity to the involvement of AP-1 factors in TGF-β signaling, the expression of c-fos, c-jun and junB is rapidly increased by TGF-β in various cell types. Furthermore, a growing body of evidence indicates that TGF-β and BMP can activate the protein kinases Erk, JNK and p38, which in turn regulate the activity of Fos, Jun and ATF-2 and related transcription factors (reviewed in Zhang and Derynck, 1999; Massagué and Chen, 2000). An ability of overexpressed Smads to associate with ATF-2 has been reported (Hanafusa et al., 1999; Sano et al., 1999). The related factor CREB may be involved in Ubx induction by DPP during endoderm formation in Drosophila (Eresh et al., 1997). Because they are also responsive to many other signals, these factors could have diverse effects on TGF-β target genes that contain AP-1 or ATF-2 sites.
The acute myelogenous leukemia (AML) family
The family of proteins variously known as AMLs, core binding factors (CBFs) or polyoma enhancer binding protein (PEPB2s) exist in three heterodimeric forms each consisting of a different α-subunit (αA, αB and αC) and a common β-subunit (Werner et al., 1999). The α-subunits contain a DNA-binding Runt domain and a transactivation domain. The β-subunit enhances the DNA-binding activity of the α-subunits. Molecular, genetic and biochemical studies have shown that AML1 (αB) is required for normal hematopoiesis, and is disrupted by inherited mutations or somatic chromosomal translocations associated with myelodysplasia and leukemia, whereas AML3 (αA) is essential for osteoblast differentiation and bone formation (Werner et al., 1999). AML2 (αC), and also AML1, control immunoglobulin A (IgA) class switching by activating the germline IgA1 and IgA2 promoters via an Iα enhancer element, favoring IgA rearrangement (Pardali et al., 2000 and references therein). TGF-β activates this promoter and induces IgA class switching in splenic B cells (Lin and Stavnezer, 1992).
The Iα element contains SBE and AML sites that bind Smad3, Smad4 and AML1 or AML2 in response to TGF-β (Hanai et al., 1999; Pardali et al., 2000). Under conditions of protein overexpression, as well as in vitro, all three AML α-subunits can form complexes with TGF-β-activated Smads as well as BMP-activated Smads. These interactions involve the MH2 domain of Smads and multiple regions in AML (Pardali et al., 2000). The AML1–Smad complex appears to be formed constitutively in the cytoplasm, becoming active by association with additional factors in the nucleus in response to TGF-β (Pardali et al., 2000). Although the significance of all these interactions needs to be established, they raise the possibility that AML–Smad complexes may have targets other than IgA, and may function in other TGF-β family pathways. Like TFE3, the AMLs interact with Ets-1 and bind cooperatively to DNA (Wotton et al., 1994). The relationship between this phenomenon and the Smad interaction is unknown.
Transcriptional activation
Smad coactivators
Both receptor-activated Smads and Smad4 are able to activate transcription, and this function resides primarily within the MH2 domain (Massagué, 1998). Transcrip tional activation by R-Smads has been shown to occur, in part at least, by their ability to recruit the general coactivators p300 and CBP (Feng et al., 1998; Janknecht et al., 1998; Pouponnot et al., 1998; Shen et al., 1998). p300 and CBP have histone acetyl transferase (HAT) activity, suggesting that their recruitment by a Smad complex may increase transcription of target genes by altering nucleosome structure and thereby remodeling the chromatin template. This interaction is directly mediated by the MH2 domain on R-Smads. An interaction between p300 and Smad4 has been mapped to the N-terminal end of the MH2 domain (de Caestecker et al., 2000). MSG1, a protein that binds to this region, might be involved in the formation of this complex (Shioda et al., 1998).
Some of the observed cooperative effects of Smads and other DNA-binding proteins may be at the level of recruitment of coactivators. p300 and CBP are large proteins with separate regions for interaction with different transcription factors. It has been suggested that the cooperative signaling of BMP2 and the cytokine LIF in astrocyte formation is mediated by a complex between Smad1 and Stat3 bridged by contacts with separate regions of p300 (Nakashima et al., 1999). The vitamin D receptor (VDR) has been shown to physically and functionally interact with Smad3 (but not Smad2) in transcriptional assays (Yanagi et al., 1999). This interaction could involve the steroid receptor coactivator-1 (SRC-1), another protein with associated HAT activity. The significance of this interaction can be assessed by inspecting VDR-null mice and Smad3-null mice for possible similarities in their phenotypes.
De-repression
Smad proteins have also been proposed to activate transcription by relieving the action of transcriptional repressors. Members of the Hox family of homeodomain proteins repress transcription when bound to their cognate DNA sites. Recent evidence suggests that a BMP-activated Smad complex is able to relieve repression of osteopontin by direct interaction with Hoxc-8 (Shi et al., 1999). Interestingly, this derepression appears to occur by the Smad1 protein binding to and dislodging Hoxc-8 from DNA. Another repressor, SIP1, was identified by its ability to interact with the Smad1 MH2 domain (Verschueren et al., 1999). SIP1 binds DNA through two separate sets of zinc fingers, and contains a homeodomain and a Smad-interacting region (Remacle et al., 1999). Smad binding could block the ability of SIP-1 to repress transcription or remove SIP-1 from DNA, thus relieving repression. The zinc-finger transcriptional repressor protein Evi1, which also exists as a truncated version generated by a chromosomal rearrangement of MDS1/Evi1 in leukemia, has been reported to antagonize Smad signaling by binding to Smad3 and interfering with its binding to DNA (Kurokawa et al., 1998). However, this mechanism has been called into question (Sood et al., 1999). The Drosophila Brinker protein is likely to be a transcriptional repressor, the action of which is prevented by Smad signaling (Campbell and Tomlinson, 1999; Jazwinska et al., 1999; Minami et al., 1999). As yet, it is not clear whether this is via direct interaction or by inhibion of Brinker expression by Smad signals.
Smad corepressors
TGIF
A Smad complex bound to DNA has the option to recruit not only coactivators but also corepressors. The homeodomain protein TGIF represses Smad-dependent transcription in part by recruiting histone deacetylases (HDACs) (Wotton et al., 1999a). TGIF interacts with Smad2/3 in a TGF-β-inducible manner, resulting in the recruitment of TGIF to Smad-responsive DNA elements. The recruitment of TGIF and associated HDAC results in the repression of Smad-activated transcription. This can affect genes that are normally activated by TGF-β signaling. For such genes, the expression level of TGIF appears to set a maximum transcriptional response to TGF-β, in part by competing with p300 for binding to the Smad complex. The TGIF homeodomain on its own binds to the RXR response element (Bertolino et al., 1995), and can repress transcription from this element independently of Smads (Wotton et al., 1999b). It is unclear whether the DNA-binding function of TGIF is involved in the context of a Smad transcriptional complex.
Ski and SnoN
Ski was originally discovered as the product of a retroviral oncogene (v-ski) that causes transformation in chick embryo fibroblasts and muscle hypertrophy in mice, and its cellular counterpart c-Ski and the related SnoN protein were later found to be corepressors that recruit HDAC via the adaptor protein N-CoR (Luo et al., 1999 and references therein). Recently, c-Ski and SnoN have been identified as Smad3- and Smad4-interacting proteins, and both have been shown to act as Smad2/3 corepressors (Akiyoshi et al., 1999; Luo et al., 1999; Sun et al., 1999a). In contrast to the interaction of Smads with TGIF, which is induced by TGF-β stimulation, the interaction of Smads with Ski and SnoN is observed under basal conditions and disappears during the first hours of TGF-β stimulation (Stroschein et al., 1999b; Sun et al., 1999b). This effect appears to be cell type dependent and may be mediated by TGF-β-induced, proteasome-mediated degradation of these proteins.
Two possible functions of Smad corepressors
The regulation of transcription by TGF-β therefore depends on the ability of Smads to recruit proteins with different chromatin-remodeling activities. TGIF may function as a negative regulator of agonist-activated Smad complexes whereas Ski and SnoN may serve to protect against agonist-independent transactivation by Smads in the basal state. A wave of SnoN expression several hours after TGF-β addition may contribute to terminating the TGF-β response (Stroschein et al., 1999b). TGIF levels may also change dependent on the conditions (Wotton et al., 1999a). Therefore, the relative levels of coactivators and corepressors that interact with Smads can be modulated by the same and other signaling pathways, resulting in negative regulation of the transcriptional response to TGF-β.
These corepressors should not, however, be simply viewed as inhibitors of Smad signaling. Certain Smad complexes might have a preference for corepressors over coactivators and primarily mediate gene downregulation. TGIF was recently identified as a holoprosencephaly gene in humans (our unpublished work). Loss-of-function mutations in the repressor or Smad-binding regions of TGIF are associated with devastating effects in brain and craniofacial development, and the resulting phenotypes are reminiscent of the phenotype observed in nodal-deficient mice. It could be that TGIF mediates Nodal-induced transcriptional repression responses, with a loss of function in either nodal or TGIF leading to a loss of such responses.
A dynamic process
Clearly, the regulation of transcription by TGF-β family signaling is a dynamic process. At its simplest, it is the transition from inactive, or perhaps actively repressed, to activated transcription of a target gene via the recruitment of a Smad complex. Competition for Smad interaction between coactivators and corepressors may determine the outcome of signaling events, and this balance may shift depending on the relative levels of these proteins and on signaling inputs that affect their activities. In this context, it should be noted that many of the known Smad cofactors, coactivators and corepressors have roles in other pathways as well. The activity of these other pathways may also influence the outcome of a TGF-β response in a given cell at a given time.
Important variations in the current scheme of Smads as transcriptional regulators are to be expected. For example, it is possible that R-Smads mediate gene responses in concert with Co-Smads other than Smad4 (Hocevar et al., 1999), or no Co-Smad at all. If the role of Smad4 is to help R-Smads bind to DNA and recruit coactivators, other proteins could substitute for Smad4 in fulfilling these functions. Furthermore, although Smad function has been shown to be essential for many of the classical physiological effects of TGF-β and related factors, it is conceivable that some transcriptional responses to these factors may not involve Smads. Considering that TGF-β factors signal through receptor protein kinases, and considering the bewildering array of pathways that are coupled to the other major class of receptor kinases—the tyrosine kinases—it would be peculiar if a single family of substrates mediated all the effects of the TGF-β receptors. Thus, our understanding of transcriptional control by TGF-β is still at a minimalist stage. To elaborate on the idea that this process is likely to be more complex would be indulging in the obvious. To discern and exploit the principles that govern this complexity is the real challenge for the future.
References
- Akiyoshi S., Inoue,H., Hanai,J., Kusanagi,K., Nemoto,N., Miyazono,K. and Kawabata,M. (1999) c-Ski acts as a transcriptional co-repressor in transforming growth factor-β signaling through interaction with smads. J. Biol. Chem., 274, 35269–35277. [DOI] [PubMed] [Google Scholar]
- Ashcroft G.S. et al. (1999) Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nature Cell Biol., 1, 260–266. [DOI] [PubMed] [Google Scholar]
- Beckmann H., Su,L.K. and Kadesch,T. (1990) TFE3: a helix–loop–helix protein that activates transcription through the immunoglobulin enhancer muE3 motif. Genes Dev., 4, 167–179. [DOI] [PubMed] [Google Scholar]
- Bertolino E., Reimund,B., Wildt-Perinic,D. and Clerc,R. (1995) A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J. Biol. Chem., 270, 31178–31188. [DOI] [PubMed] [Google Scholar]
- Brown C.B., Boyer,A.S., Runyan,R.B. and Barnett,J.V. (1999) Requirement of type III TGF-β receptor for endocardial cell transformation in the heart. Science, 283, 2080–2082. [DOI] [PubMed] [Google Scholar]
- Campbell G. and Tomlinson,A. (1999) Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell, 96, 553–562. [DOI] [PubMed] [Google Scholar]
- Chen X., Rubock,M.J. and Whitman,M. (1996) A transcriptional partner of MAD proteins in TGF-β signalling. Nature, 383, 691–696. [DOI] [PubMed] [Google Scholar]
- Chen X., Weisberg,E., Fridmacher,V., Watanabe,M., Naco,G. and Whitman,M. (1997) Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature, 389, 85–89. [DOI] [PubMed] [Google Scholar]
- Chen Y.G. and Massagué,J. (1999) Smad1 recognition and activation by the ALK1 group of transforming growth factor-β family receptors. J. Biol. Chem., 274, 3672–3677. [DOI] [PubMed] [Google Scholar]
- Chen Y.G., Hata,A., Lo,R.S., Wotton,D., Shi,Y., Pavletich,N. and Massagué,J. (1998) Determinants of specificity in TGF-β signal transduction. Genes Dev., 12, 2144–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Caestecker M.P., Yahata,T., Wang,D., Parks,W.T., Huang,S., Hill,C.S., Shioda,T., Roberts,A.B. and Lechleider,R.J. (2000) The smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J. Biol. Chem., 275, 2115–2122. [DOI] [PubMed] [Google Scholar]
- Eresh S., Riese,J., Jackson,D.B., Bohman,D. and Bienz,M. (1997) A CREB-binding site as a target for decapentaplegic signalling during Drosophila endoderm induction. EMBO J., 16, 2014–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.H. and Derynck,R. (1997) A kinase subdomain of transforming growth factor-β (TGF-β) type I receptor determines the TGF-β intracellular signaling activity. EMBO J., 16, 3912–3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng X.-H., Zhang,Y., Wu,R.-Y. and Derynck,R. (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for Smad3 in TGF-β-induced transcriptional activation. Genes Dev., 12, 2153–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain S., Howell,M., Esslemont,G.M. and Hill,C. (2000) Homeo domain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev., 14, 435–451. [PMC free article] [PubMed] [Google Scholar]
- Hanafusa H., Ninomiya-Tsuji,J., Masuyama,N., Nishita,M., Fujisawa,J., Shibuya,H., Matsumoto,K. and Nishida,E. (1999) Involvement of the p38 mitogen-activated protein kinase pathway in transforming growth factor-β-induced gene expression. J. Biol. Chem., 274, 27161–27167. [DOI] [PubMed] [Google Scholar]
- Hanai J. et al. (1999) Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Cα promoter. J. Biol. Chem., 274, 31577–31582. [DOI] [PubMed] [Google Scholar]
- Hata A., Lo,R., Wotton,D., Lagna,M. and Massagué,J. (1997) Mutations increasing autoinhibition inactivate the tumour suppressors Smad2 and Smad4. Nature, 388, 82–86. [DOI] [PubMed] [Google Scholar]
- Hata A., Lagna,G., Massagué,J. and Hemmati-Brivanlou,A. (1998) Smad6 inhibits BMP/Smad1 signaling by specifically competing with the Smad4 tumor suppressor. Genes Dev., 12, 186–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A., Seoane,J., Lagna,G., Montalvo,E., Hemmati-Brivanlou,A. and Massagué,J. (2000) OAZ uses distinct DNA- and protein-binding zinc fingers in separate BMP-Smad and Olf signaling pathways. Cell, 100, 229–240. [DOI] [PubMed] [Google Scholar]
- Hayashi H. et al. (1997) The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell, 89, 1165–1173. [DOI] [PubMed] [Google Scholar]
- Heldin C.-H., Miyazono,K. and ten Dijke,P. (1997) TGF-β signalling from cell membrane to nucleus through SMAD proteins. Nature, 390, 465–471. [DOI] [PubMed] [Google Scholar]
- Hocevar B.A., Brown,T.L. and Howe,P.H. (1999) TGF-β induces fibronectin synthesis through a c-Jun N-terminal kinase-dependent, Smad4-independent pathway. EMBO J., 18, 1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell M., Itoh,F., Pierreux,C.E., Valgeirsdottir,S., Itoh,S., ten Dijke,P. and Hill,C.S. (1999) Xenopus Smad4β is the Co-Smad component of developmentally regulated transcription factor complexes responsible for induction of early mesodermal genes. Dev. Biol., 214, 354–369. [DOI] [PubMed] [Google Scholar]
- Hua X., Miller,Z.A., Wu,G., Shi,Y. and Lodish,H.F. (1999) Specificity in transforming growth factor β-induced transcription of the plasminogen activator inhibitor-1 gene: interactions of promoter DNA, transcription factor µE3 and Smad proteins. Proc. Natl Acad. Sci. USA, 96, 13130–13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huse M., Chen,Y.G., Massagué,J. and Kuriyan,J. (1999) Crystal structure of the cytoplasmic domain of the type I TGF β receptor in complex with FKBP12. Cell, 96, 425–436. [DOI] [PubMed] [Google Scholar]
- Janknecht R., Wells,N.J. and Hunter,T. (1998) TGF-β-stipulated cooperation of Smad proteins with the coactivators CBP/p300. Genes Dev., 12, 2114–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazwinska A., Kirov,N., Wieschaus,E., Roth,S. and Rushlow,C. (1999) The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell, 96, 563–573. [DOI] [PubMed] [Google Scholar]
- Johnson K., Kirkpatrick,H., Comer,A., Hoffmann,F.M. and Laughon,A. (1999) Interaction of Smad complexes with tripartite DNA-binding sites. J. Biol. Chem., 274, 20709–20716. [DOI] [PubMed] [Google Scholar]
- Jonk L.J., Itoh,S., Heldin,C.H., ten Dijke,P. and Kruijer,W. (1998) Identification and functional characterization of a Smad binding element (SBE) in the JunB promoter that acts as a transforming growth factor-β, activin and bone morphogenetic protein-inducible enhancer. J. Biol. Chem., 273, 21145–21152. [DOI] [PubMed] [Google Scholar]
- Kawabata M., Inoue,H., Hanyu,A., Imamura,T. and Miyazono,K. (1998) Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J., 17, 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Johnson,K., Chen,H.J., Carroll,S. and Laughon,A. (1997) Drosophila Mad binds to DNA and directly mediates activation of vestigial by Decapentaplegic. Nature, 388, 304–308. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M., Doody,J., Timokhina,I. and Massagué,J. (1999) A mechanism of repression of TGFb/Smad signaling by ongenic ras. Genes Dev., 13, 804–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M., Mitani,K., Irie,K., Matsuyama,T., Takahashi,T., Chiba,S., Yazaki,Y., Matsumoto,K. and Hirai,H. (1998) The oncoprotein Evi-1 represses TGFβ signalling by inhibiting Smad3. Nature, 394, 92–96. [DOI] [PubMed] [Google Scholar]
- Labbé E., Silvestri,C., Hoodless,P.A., Wrana,J.L. and Attisano,L. (1998) Smad2 and Smad3 positively and negatively regulate TGFβ-dependent transcription through the forkhead DNA-binding protein FAST2. Mol. Cell, 2, 109–120. [DOI] [PubMed] [Google Scholar]
- Lagna G., Hata,A., Hemmati-Brivanlou,A. and Massagué,J. (1996) Partnership between DPC4 and SMAD proteins in TGFβ signalling pathways. Nature, 383, 832–836. [DOI] [PubMed] [Google Scholar]
- Liberati N.T., Datto,M.B., Frederick,J.P., Shen,X., Wong,C., Rougier-Chapman,E.M. and Wang,X.F. (1999) Smads bind directly to the Jun family of AP-1 transcription factors. Proc. Natl Acad. Sci. USA, 96, 4844–4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.C. and Stavnezer,J. (1992) Regulation of transcription of the germ-line Ig α constant region gene by an ATF element and by novel transforming growth factor-β1-responsive elements. J. Immunol., 149, 2914–2925. [PubMed] [Google Scholar]
- Liu F., Hata,A., Baker,J., Doody,J., Cárcamo,J., Harland,R. and Massagué,J. (1996) A human Mad protein acting as a BMP-regulated transcriptional activator. Nature, 381, 620–623. [DOI] [PubMed] [Google Scholar]
- Liu F., Pouponnot,C. and Massagué,J. (1997) Dual role of the Smad4/DPC4 tumor suppressor in TGFβ-inducible transcriptional responses. Genes Dev., 11, 3157–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo R.S. and Massagué,J. (1999) Ubiquitin-dependent degradation of TGF-β-activated Smad2. Nature Cell Biol., 1, 472–478. [DOI] [PubMed] [Google Scholar]
- Lo R.S., Chen,Y.G., Shi,Y.G., Pavletich,N. and Massagué,J. (1998) The L3 loop: a structural motif determining specific interactions between SMAD proteins and TGF-β receptors. EMBO J., 17, 996–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K., Stroschein,S.L., Wang,W., Chen,D., Martens,E., Zhou,S. and Zhou,Q. (1999) The ski oncoprotein interacts with the smad proteins to repress TGFβ signaling. Genes Dev., 13, 2196–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J. (1998) TGFβ signal transduction. Annu. Rev. Biochem., 67, 753–791. [DOI] [PubMed] [Google Scholar]
- Massagué J. and Chen,Y.G. (2000) Controlling TGFβ signaling. Genes Dev., 14, 627–644. [PubMed] [Google Scholar]
- Masuyama N., Hanafusa,H., Kusakabe,M., Shibuya,H. and Nishida,E. (1999) Identification of two Smad4 proteins in Xenopus. Their common and distinct properties. J. Biol. Chem., 274, 12163–12170. [DOI] [PubMed] [Google Scholar]
- Minami M., Kinoshita,N., Kamoshida,Y., Tanimoto,H. and Tabata,T. (1999) brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature, 398, 242–246. [DOI] [PubMed] [Google Scholar]
- Nagarajan R.P., Zhang,J., Li,W. and Chen,Y. (1999) Regulation of Smad7 promoter by direct association with Smad3 and Smad4. J. Biol. Chem., 274, 33412–33418. [DOI] [PubMed] [Google Scholar]
- Nakao A. et al. (1997) Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signaling. Nature, 389, 631–635. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Yanagisawa,M., Arakawa,H., Kimura,N., Hisatsune,T., Kawabata,M., Miyazono,K. and Taga,T. (1999) Synergistic signaling in fetal brain by STAT3–Smad1 complex bridged by p300. Science, 284, 479–482. [DOI] [PubMed] [Google Scholar]
- Oft M., Peli,J., Rudaz,C., Schwarz,H., Beug,H. and Reichmann,E. (1996) TGF-β1 and Ha-Ras collaborate in modulating the phenotypic plasticity and invasiveness of epithelial tumor cells. Genes Dev., 10, 2462–2477. [DOI] [PubMed] [Google Scholar]
- Onichtchouk D., Chen,Y.G., Dosch,R., Gawantka,V., Delius,H., Massagué,J. and Niehrs,C. (1999) Silencing of TGF-β signalling by the pseudoreceptor BAMBI. Nature, 401, 480–485. [DOI] [PubMed] [Google Scholar]
- Padgett R.W., Wozney,J.M. and Gelbart,W.M. (1993) Human BMP sequences can confer normal dorsal-ventral patterning in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 90, 2905–2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali E., Xie,X.Q., Tsapogas,P., Itoh,S., Arvanitidis,K., Heldin,C.H., ten Dijke,P., Grundström,T. and Sideras,P. (2000) Smad and AML proteins synergistically confer transforming growth factor β1 responsiveness to human germ-line IgA genes. J. Biol. Chem., 275, 3552–3560. [DOI] [PubMed] [Google Scholar]
- Pouponnot C., Jayaraman,L. and Massagué,J. (1998) Physical and functional interactions of Smads and p300/CBP. J. Biol. Chem., 273, 22865–22868. [DOI] [PubMed] [Google Scholar]
- Remacle J.E., Kraft,H., Lerchner,W., Wuytens,G., Collart,C., Verschueren,K., Smith,J.C. and Huylebroeck,D. (1999) New mode of DNA binding of multi-zinc finger transcription factors: δEF1 family members bind with two hands to two target sites. EMBO J., 18, 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijoh Y. et al. (2000) Left–right assymetric expression of lefty2 and nodal is induced by a signaling pathway that includes the transcription factor FAST2. Mol. Cell, 5, 35–47. [DOI] [PubMed] [Google Scholar]
- Sampath T.K., Rashka,K.E., Doctor,J.S., Tucker,R.F. and Hoffman,F.M. (1993) Drosophila transforming growth factor β superfamily proteins induce endochondral bone formation in mammals. Proc. Natl Acad. Sci. USA, 90, 6004–6008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano Y., Harada,J., Tashiro,S., Gotoh-Mandeville,R., Maekawa,T. and Ishii,S. (1999) ATF-2 is a common nuclear target of Smad and TAK1 pathways in transforming growth factor-β signaling. J. Biol. Chem., 274, 8949–8957. [DOI] [PubMed] [Google Scholar]
- Shen X., Hu,P.P., Liberati,N.T., Datto,M.B., Frederick,J.P. and Wang,X.F. (1998) TGF-β-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol. Biol. Cell, 9, 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Yang,X., Chen,D., Chang,Z. and Cao,X. (1999) Smad1 interacts with homeobox DNA-binding proteins in bone morphogenetic protein signaling. J. Biol. Chem., 274, 13711–13717. [DOI] [PubMed] [Google Scholar]
- Shi Y., Hata,A., Lo,R.S., Massagué,J. and Pavletich,N.P. (1997) A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature, 388, 87–93. [DOI] [PubMed] [Google Scholar]
- Shi Y., Wang,Y.-F., Jayaraman,L., Yang,H., Massagué,J. and Pavletich,N. (1998) Crystal structure of a Smad MH1 domain bound to DNA: Insights on DNA-binding in TGF-β signaling. Cell, 94, 585–594. [DOI] [PubMed] [Google Scholar]
- Shioda T., Lechleider,R.J., Dunwoodie,S.L., Li,H., Yahata,T., de Caestecker,M.P., Fenner,M.H., Roberts,A.B. and Isselbacher,K.J. (1998) Transcriptional activating activity of Smad4: roles of SMAD hetero-oligomerization and enhancement by an associating transactivator. Proc. Natl Acad. Sci. USA, 95, 9785–9790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R., Talwar-Trikha,A., Chakrabarti,S.R. and Nucifora,G. (1999) MDS1/EVI1 enhances TGF-β1 signaling and strengthens its growth-inhibitory effect but the leukemia-associated fusion protein AML1/MDS1/EVI1, product of the t(3;21), abrogates growth-inhibition in response to TGF-β1. Leukemia, 13, 348–357. [DOI] [PubMed] [Google Scholar]
- Stroschein S.L., Wang,W. and Luo,K. (1999a) Cooperative binding of Smad proteins to two adjacent DNA elements in the plasminogen activator inhibitor-1 promoter mediates transforming growth factor β-induced smad-dependent transcriptional activation. J. Biol. Chem., 274, 9431–9441. [DOI] [PubMed] [Google Scholar]
- Stroschein S.L., Wang,W., Zhou,S., Zhou,Q. and Luo,K. (1999b) Negative feedback regulation of TGF-β signaling by the SnoN oncoprotein. Science, 286, 771–774. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu,X., Eaton,E.N., Lane,W.S., Lodish,H.F. and Weinberg,R.A. (1999a) Interaction of the Ski oncoprotein with Smad3 regulates TGF-β signaling. Mol. Cell, 4, 499–509. [DOI] [PubMed] [Google Scholar]
- Sun Y., Liu,X., Ng-Eaton,E., Lodish,H.F. and Weinberg,R.A. (1999b) SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor β signaling. Proc. Natl Acad. Sci. USA, 96, 12442–12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian G., Erman,B., Ishii,H., Gangopadhyay,S.S. and Sen,R. (1999) Transcriptional activation by ETS and leucine zipper-containing basic helix–loop–helix proteins. Mol. Cell. Biol., 19, 2946–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai R.Y. and Reed,R.R. (1998) Identification of DNA recognition sequences and protein interaction domains of the multiple-Zn-finger protein Roaz. Mol. Cell. Biol., 18, 6447–6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T., Chiang,T.A., Davison,A.F., Attisano,L. and Wrana,J.L. (1998) SARA, a FYVE domain protein that recruits Smad2 to the TGFβ receptor. Cell, 95, 779–791. [DOI] [PubMed] [Google Scholar]
- Verschueren K. et al. (1999) SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J. Biol. Chem., 274, 20489–20498. [DOI] [PubMed] [Google Scholar]
- Vindevoghel L., Kon,A., Lechleider,R.J., Uitto,J., Roberts,A.B. and Mauviel,A. (1998) Smad-dependent transcriptional activation of human type VII collagen gene (COL7A1) promoter by transforming growth factor-β. J. Biol. Chem., 273, 13053–13057. [DOI] [PubMed] [Google Scholar]
- Watanabe M. and Whitman,M. (1999) FAST-1 is a key maternal effector of mesoderm inducers in the early Xenopus embryo. Development, 126, 5621–5634. [DOI] [PubMed] [Google Scholar]
- Werner M.H., Shigesada,K. and Ito,Y. (1999) Runt domains take the lead in hematopoiesis and osteogenesis. Nature Med., 5, 1356–1357. [DOI] [PubMed] [Google Scholar]
- Wong C., Rougier-Chapman,E.M., Frederick,J.P., Datto,M.B., Liberati,N.T., Li,J.M. and Wang,X.F. (1999) Smad3–Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor β. Mol. Cell. Biol., 19, 1821–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Ghysdael,J., Wang,S., Speck,N.A. and Owen,M.J. (1994) Cooperative binding of Ets-1 and core binding factor to DNA. Mol. Cell. Biol., 14, 840–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Lo,R.S., Lee,S. and Massagué,J. (1999a) A Smad transcriptional corepressor. Cell, 97, 29–39. [DOI] [PubMed] [Google Scholar]
- Wotton D., Lo,R.S., Swaby,L.A. and Massagué,J. (1999b) Multiple modes of repression by the smad transcriptional corepressor TGIF. J. Biol. Chem., 274, 37105–37110. [DOI] [PubMed] [Google Scholar]
- Wu G., Chen,Y.G., Ozdamar,B., Gyuricza,C.A., Chong,P.A., Wrana,J.L., Massagu,J. and Shi,Y. (2000) Structural basis of Smad2 recognition by the Smad anchor for receptor activation. Science, 287, 92–97. [DOI] [PubMed] [Google Scholar]
- Wu R.-Y., Zhang,Y., Feng,X.-H. and Derynck,R. (1997) Heteromeric and homomeric interactions correlate with signaling activity and functional cooperativity of Smad3 and Smad4/DPC4. Mol. Cell. Biol., 17, 2521–2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yin,Z., Hudson,J.B., Ferguson,E.L. and Frasch,M. (1998) Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev., 12, 2354–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagi Y., Suzawa,M., Kawabata,M., Miyazono,K., Yanagisawa,J. and Kato,S. (1999) Positive and negative modulation of vitamin D receptor function by transforming growth factor-β signaling through smad proteins. J. Biol. Chem., 274, 12971–12974. [DOI] [PubMed] [Google Scholar]
- Yeo C.Y., Chen,X. and Whitman,M. (1999) The role of FAST-1 and Smads in transcriptional regulation by activin during early Xenopus embryogenesis. J. Biol. Chem., 274, 26584–26590. [DOI] [PubMed] [Google Scholar]
- Zawel L., Dai,J.L., Buckhaults,P., Zhou,S., Kinzler,K.W., Vogelstein,B. and Kern,S.E. (1998) Human Smad3 and Smad4 are sequence-specific transcription activators. Mol. Cell, 1, 611–617. [DOI] [PubMed] [Google Scholar]
- Zhang Y. and Derynck,R. (1999) Regulation of Smad signaling by protein associations and signaling crosstalk. Trends Cell Biol., 9, 274–279. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Feng,X.H. and Derynck,R. (1998) Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature, 394, 909–913. [DOI] [PubMed] [Google Scholar]
- Zhou S., Zawel,L., Lengauer,C., Kinzler,K.W. and Vogelstein,B. (1998) Characterization of human FAST-1, a TGFβ and activin signal transducer. Mol. Cell, 2, 121–127. [DOI] [PubMed] [Google Scholar]
- Zhu H., Kavsak,P., Abdollah,S., Wrana,J.L. and Thomsen,G.H. (1999) A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature, 400, 687–693. [DOI] [PubMed] [Google Scholar]