Summary
Variation in disease resistance is a widespread phenomenon in wild plant-pathogen associations. Here, we review current literature on natural plant-pathogen associations to determine how diversity in disease resistance is distributed at different hierarchical levels – within host individuals, within host populations, among host populations at the metapopulation scale and at larger regional scales.
We find diversity in resistance across all spatial scales examined. Furthermore, variability seems to be the best counter-defence of plants against their rapidly evolving pathogens. We find that higher diversity of resistance phenotypes also results in higher levels of resistance at the population level.
Overall, we find that wild plant populations are more likely to be susceptible than resistant to their pathogens. However, the degree of resistance differs strikingly depending on the origin of the pathogen strains used in experimental inoculation studies. Plant populations are on average 16% more resistant to allopatric pathogen strains than they are to strains that occur within the same population (48 % vs. 32 % respectively).
Pathogen dispersal mode affects levels of resistance in natural plant populations with lowest levels detected for hosts of airborne pathogens and highest for waterborne pathogens.
Detailed analysis of two model systems, Linum marginale infected by Melampsora lini, and Plantago lanceolata infected by Podosphaera plantaginis, show that the amount of variation in disease resistance declines towards higher spatial scales as we move from individual hosts to metapopulations, but evaluation of multiple spatial scales is needed to fully capture the structure of disease resistance.
Synthesis: Variation in disease resistance is ubiquitous in wild plant-pathogen associations. While the debate over whether the resistance structure of plant populations is determined by pathogen-imposed selection versus non-adaptive processes remains unresolved, we do report examples of pathogen-imposed selection on host resistance. Here we highlight the importance of measuring resistance across multiple spatial scales, and of using sympatric strains when looking for signs of coevolution in wild plant-pathogen interactions.
Keywords: Coevolution, host-parasite interactions, life-history, metapopulation dynamics, gene-for-gene, major resistance, sympatric, disease resistance, epidemiology
Introduction
How plant resistance to disease is distributed across space can affect fundamental components of disease biology and epidemiology. Ultimately the resistance structure of plant populations will determine where pathogens occur, as a pathogen can only infect hosts whose defence strategies it can overcome. Furthermore, host resistance may be considered the main driving force of pathogen evolution, with pathogens evolving to overcome host resistance strategies. This process is believed to be reciprocal so hosts and their pathogens are engaged in a co-evolutionary arms-race with hosts evolving new counter defences to escape attack, and pathogens in turn evolving to overcome these new forms of resistance (e.g. Clay & Kover 1996).
The interaction between hosts and pathogens is characterized by a great disparity in genome size, generation time and speed of adaptability which, on the surface, would generally appear to favour the pathogen. Indeed, there are numerous examples of pathogens adapting to their hosts, both from studies of local adaptation (Parker 1985; Thrall et al. 2002; Laine 2005; Niemi et al. 2006; Sicard et al. 2007; Springer 2007) (but see Kaltz et al. 1999 for an exception), and from classic boom-and-bust cycles, particularly in agricultural crops, where newly deployed resistance genes increase in frequency but then rapidly lose their effectiveness as pathogens adapt to that new variety (Browning & Frey 1969). However, there is notably little evidence of the opposite – hosts evolving resistance under the pressure of pathogen attack. At the same time, variation in disease resistance is widespread (Table 1; Salvaudon et al. 2008), which implies that natural plant populations have the capacity to undergo significant adaptive evolution in response to pathogen attack.
Table 1.
Summary information for studies used for assessing whether there is spatial variation in host resistance at different spatial scales. Studies with superscript ‘1’ in the Reference column were used for the analysis of levels of resistance presented in Fig. 1 and studies with superscript ‘2’ were used for the analysis of resistance diversity and average resistance presented in Fig. 2.
| Reference | Species | Nr host populations | Spatial scale (Min. and max. distance separating host populations, km) | How resistance measured | Qualitative or quantitave resistance | At what scale variation in resistance detected |
|---|---|---|---|---|---|---|
| Alexander 1989 1 | Silene alba (white campion) - Microbotryum violaceum (anther-smut fungus) | 1 | Single population | Common garden | Quantitative | Within populations |
| Alexander et al. 1993 | Silene alba (white campion) - Microbotryum violaceum (anther-smut) | 1 | Single population | Inoculation | Quantitative | Within populations |
| Bevan et al. 1993a 1, 2 | Senecio vulgaris (groundsel) - Golovinomyces cichoracearum var. fischeri (powdery mildew fungus) | 2 | 480 | Inoculations | Qualitative and quantitative | Within and between populations |
| Burdon 1980 | Trifolium repens (white clover)- Cymadothea trifolii, Pseudopeziza trifolii (black blotch and leaf spotting fungi) | 1 | Single population | Inoculations | Quantitative | Within populations |
| Burdon et al. 1983 | Avena barbata, A. fatua, A. ludoviciana (wild oat species) – P. coronata (rust fungus) | 21 | ~ 30 / 650 | Inoculations | Qualitative and quantitative | Within and between populations |
| Burdon & Jarosz 1991 1, 2 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 1 | Single population | Inoculations | Qualitative | Within populations |
| Burdon & Thompson 1995 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 1 | Single population | Inoculations | Qualitative | Within populations |
| Burdon et al. 1999 1, 2 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 11 | 0.2 / ~200 | Inoculations | Qualitative | Within and between populations |
| Burdon et al. 2002 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 42 | 56 / 3228 | Inoculations | Qualitative | Continental |
| Carlsson-Granér 1997 1 | Silene dioica (red campion) - Microbotryum violaceum (anther-smut fungus) | 3 | <10 km | Inoculations | Quantitative | Within and between populations |
| Carlsson-Granér et al. 19991 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 3 | 0.1 km | Inoculations | Qualitative | Within and between populations |
| Davelos et al. 1996 1 | Spartina pectinata - Puccinia seymouriana and Puccinia sparganioides (rust fungi) | 5 | 0.2 / 120 | Inoculations, Common garden | Quantitative | Between populations |
| de Meaux & Neema 2003 | Phaseolus vulgaris (Common bean) – Colletotrichum lindemuthianum (fungal pathogen) | 10 | NA / 5000 | Molecular markers (AFLPs), inoculations | Qualitative | Within and between populations also at regional scales |
| de Meaux et al. 2003 | Phaseolus vulgaris (Common bean) – Colletotrichum lindemuthianum (fungal pathogen) | 15 | ~ 20 / 500 | RGC polymorphism, inoculations | Qualitative | Within and between populations |
| De Nooij & van Damme 1988a | Plantago lanceolata (ribwort plantain)- Phomopsis subordinaria (fungal pathogen) | 3 | NA | Inoculations | Quantitative | Within population |
| Ericson & Burdon 2009 1, 2 | Betula pubescens (white birch) - Melampsoridium betulinum (rust fungus) | 1 | Single population | Inoculations | Qualitative and quantitative | Within population |
| Ericson et al. 2002 1 | Filipendula ulmaria (meadowsweet) - Triphragmium ulmariae (rust fungus) | 6 | ~2 / 363 | Inoculations | Qualitative | Wihin and between populations |
| Espiau et al. 1998 1, 2 | Chondrilla juncea (skeletonweed)- Puccinia chondrillina (rust fungus) | 1 | Single population | Inoculations | Qualitative | Within host, within population |
| Goss & Bergelson 2006 | Arabidopsis thaliana - Pseudomonas viridiflava (bacterial pathogen) | 6 | 3 / 48 | Inoculations | Quantitative | Within and between populations |
| Jarosz & Burdon 1990 | Glycine argyrea (wild relative of soybean) – Phakopsora pachyrhizi (rust fungus) | 1 | Single population | Inoculations | Quantitative | Within population |
| Jarosz & Burdon 1991 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 10 | ~2 / ~110 | Inoculations | Qualitative | Within and between populations |
| Jorgensen & Emerson 2008 1 | Arabidopsis thaliana - Golovinomyces orontii (powdery mildews) | 8 | 2 / 155 | Molecular diversity in RPW8, inoculations | Quantitative | Within and between populations |
| Kaltz et al. 1999 1 | Silene latifolia (white campion) – Microbotryum violaceum (anther-smut fungus) | 14 | 2 / 166 | Inoculations | Qualitative | Within and between populations |
| Kniskern & Rausher 2006 1 | Ipomoea purpurea (common morning glory) – Coleosporium ipomoeae (rust fungus) | 11 | ~1 / 622 | Inoculations | Qualitative | Within and between populations |
| Laine 2004 1, 2 | Plantago lanceolata (ribwort plantain) – Podosphaera plantaginis (powdery mildew fungus) | 8 | 0.2 / 10 | Inoculations | Qualitative | Within and between populations |
| Laine 2005 1 | Plantago lanceolata (ribwort plantain) – Podosphaera plantaginis (powdery mildew fungus) | 20 | 0.1 / 45 | Inoculations | Qualitative | Within populations, between populations and metapopulations |
| Laine 2006 1, 2 | Plantago lanceolata (ribwort plantain) – Podosphaera plantaginis (powdery mildew fungus) | 2 | 0.2 | Inoculations | Qualitative | Within and between populations |
| Lebeda et al. 2008 | Lactuca serriola (prickly lettuce)- Bremia lactucae (downy mildew fungus) | 16 | NA | Inoculations | Qualitative and quantitative | Within and between populations |
| Niemi et al. 2006 | Salix triandra (almond willow) - Melampsora amygdalinae (rust fungus) | 4 | 355 / 907 | Inoculations | Qualitative | Within and between populations |
| Parker 1985 1 | Amphicarpaea bracteata (hog peanut)- Synchytrium decipiens (fungal pathogen) | 3 | 1 / 100 | Inoculations, transplant | Quantitative | Within and between populations |
| Parker 1988a1 | Amphicarpaea bracteata (hog peanut)- Synchytrium decipiens (fungal pathogen) | 1 | Single population | Allozyme variation, inoculations | Quantitative | Within population |
| Parker 1988b1 | Amphicarpaea bracteata (hog peanut)- Synchytrium decipiens (fungal pathogen) | 1 | Single population | Inoculations | Quantitative | Within population |
| Petrzelova & Lebeda 2010 | Lactuca serriola (prickly lettuce)- Bremia lactucae (downy mildew fungus) | 50 | Continental | Inoculations | Qualitative and quantitative | Within and between populations |
| Springer 2007 | Hesperolinon californicum (California dwarf flax)- Melampsora lini (rust fungus) | 16 | 1 / 180 | Inoculations | Qualitative | Within and between populations |
| Thrall et al. 2001 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 16 | 0.1 / 10 | Allozyme variation, inoculations | Qualitative | Within and between populations |
| Thrall et al. 2002 1 | Linum marginale (wild flax) – Melampsora lini (rust fungus) | 6 | 0.2 / 10 | Inoculations | Qualitative | Within and between populations |
Theoretical models of coevolution imply indirect negative frequency-dependent selection (FDS) between hosts and parasites within local populations, in which the selection rate for resistance depends on the frequency of parasite avirulence alleles and vice versa (Jayakar 1970; Leonard 1977; Bergelson et al. 2001). In systems with such FDS, costs of resistance and virulence have traditionally been required to maintain polymorphism in these traits (e.g. Jayakar 1970; Leonard 1977; Frank 1993), but recent theoretical work has shown metapopulation dynamics and spatially heterogenous selection to be as important, or even a sufficient condition, for the maintenance of polymorphism (Gandon et al. 1996; Gomulkiewicz et al. 2000; Nuismer et al. 2000; Gandon & Michalakis 2002; Thrall & Burdon 2002; Nuismer & Gandon 2008). Indeed, host-pathogen coevolutionary processes are intrinsically spatial as well as temporal, and occur at many different scales (Burdon & Thrall 2001). These range from single populations dominated by demographic and genetic stochasticity (Parker 1985; Burdon & Jarosz 1991; Alexander et al. 1993; Bevan et al. 1993a; Espiau et al. 1998), to metapopulations in which extinction/colonization dynamics have a large influence (Thrall et al. 2001; Ericson et al. 2002; Thrall et al. 2002; Smith et al. 2003; Antonovics 2004; Laine & Hanski 2006; Soubeyrand et al. 2009), and larger geographic regions where phylogenetic patterns and historical events become more important (Burdon et al. 2002; de Meaux et al. 2003; de Meaux & Mitchell-Olds 2003). Coevolutionary interactions between hosts and pathogens may occur at a broad range of spatial scales, from those encompassed by a single individual through various intermediate levels to those of the species as a whole.
It has already been noted by several authors that variation in disease resistance is widespread (e.g. Parker 1985; Springer 2007; Jorgensen & Emerson 2008) yet, to date, we know little about how this variation is structured across space, and what are the processes driving host resistance structure at different spatial scales. This is despite the general recognition that variation in host resistance (and pathogen infectivity and aggressiveness) is of central importance to understanding patterns of infection (Hill 1998; Lockett et al. 2001). Indeed, study of the genetic components of host-pathogen interactions has lagged far behind work documenting the demographic impacts of disease. Thus, while some work has shown negative relationships between the overall diversity of hosts and parasitism (Coltman et al. 1999; Meagher 1999), in only a very few cases has host genetic variation been implicated in rates of epidemic spread of disease or patterns of disease prevalence (e.g. Thrall & Burdon 2000; Laine 2004).
Here we review studies on how resistance is distributed in wild plant-pathogen interactions at different spatial scales: within individual hosts, within host populations, among populations at the metapopulation scale, and at regional scales. Understanding this hierarchical spatial structure has the potential to provide valuable insights into the influence of pathogens on host evolution, and spatial variation in the magnitude of their effect. Specifically, here we ask: 1) What are the kinds of resistance responses we find in wild plant populations (i.e. qualitative or quantitative)? 2) Given that there is polymorphism in resistance, are wild plant populations more likely to be resistant or susceptible to their pathogens? 3) How do host and pathogen life-history and origin of pathogen strain affect resistance of plant populations? 4) How is resistance diversity structured at these different spatial levels? We include here an analysis of two plant-pathogen interactions that have been studied in detail across several different spatial scales. One of these, the interaction between the wild flax, Linum marginale, and its obligate rust fungus Melampsora lini, is a native host-pathogen association which occurs across southern Australia. The other is the long-standing interaction between Plantago lanceolata and its obligate powdery mildew fungus, Podosphaera plantaginis in the southwest archipelago of Finland. Knowledge of these systems allows us to explicitly estimate how variation in disease resistance is hierarchically distributed at the different spatial scales. Finally, we conclude with a consideration of empirical and conceptual future directions for the study of plant-pathogen interactions.
Literature search
Studies were compiled for the review by searching the literature in the ISI Web of Science database using a combination of search terms such as ‘plant’, ‘pathogen’, ‘resistance’, ‘spatial scale’ or ‘spatial structure’. Additional studies were gathered by scouring cross-citations from reviews and plant coevolutionary studies, as well as from colleagues. Studies were discarded if they were purely theoretical or review articles, or if the study was focused on agricultural populations (where reciprocal evolutionary change was prevented by the host presumably evolving under human imposed selection). Altogether, this resulted in the identification of 36 studies which included explicit information on plant resistance to its pathogens within a spatial framework. The following measurements were extracted from these studies: 1) the type of interaction (e.g. plant-fungus, plant-bacteria); 2) the study species; 3) how resistance was measured (e.g. inoculation or sequence diversity); 4) type of resistance (qualitative vs. quantitative); 5) number of populations included in the study and the study design; 6) the spatial scale of the study - the shortest and longest distances separating the study populations; 7) whether the study detected resistance variation and at what spatial scale; 8) average resistance (reported only when resistance data was in the form of infected/not, percent of infected individuals etc.), and whether resistance was measured with sympatric pathogen strains (sampled from the same population), nearby allopatric strains (collected from less than 2 km away) or allopatric strains collected from 2 km or further away (in three cases the origin of the pathogen strains was not specified); and 9) the number of resistance phenotypes that were detected in inoculations. These data are summarized in Table 1, and in Figures 1a and 2. Additional information was gathered on the life-history and dispersal characteristics of both host and pathogen when available. These data are presented in Supporting Information in Table S1 and in Fig. 1b.
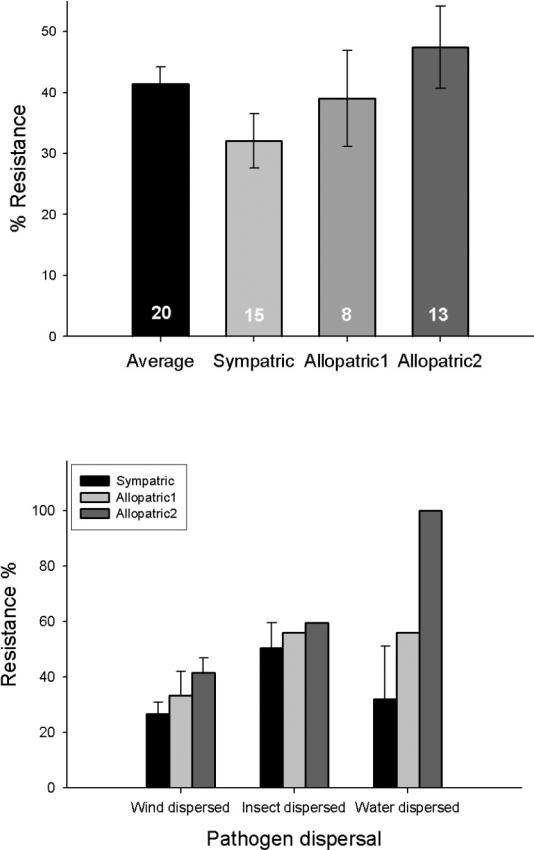
FIGURE 1.
a) Population level qualitative resistance (%) averaged for all inoculations regardless of pathogen origin (black bar), and divided into categories: resistance against pathogen strains collected from the same population (Sympatric; light grey bar), resistance against strains from neighbouring populations (Allopatric1; medium grey bar) and resistance against strains from faraway populations (Allopatric2; dark grey bar). Error bars are based on standard errors of the mean. The overall trend was not statistically significant (P = 0.157), while the comparison between Sympatric and Allopatric2 was marginally significant (P = 0.061). Numbers in the bars show the number of studies used for each estimate. b) Qualitative disease resistance levels differed depending on the mode of pathogen dispersal (P = 0.009), and the origin of strains used to measure resistance also had a significant effect on the level of resistance measured (P = 0.021). Studies used for these analyses are depicted in Table 1 with superscript ‘1’ and presented in Table S1.
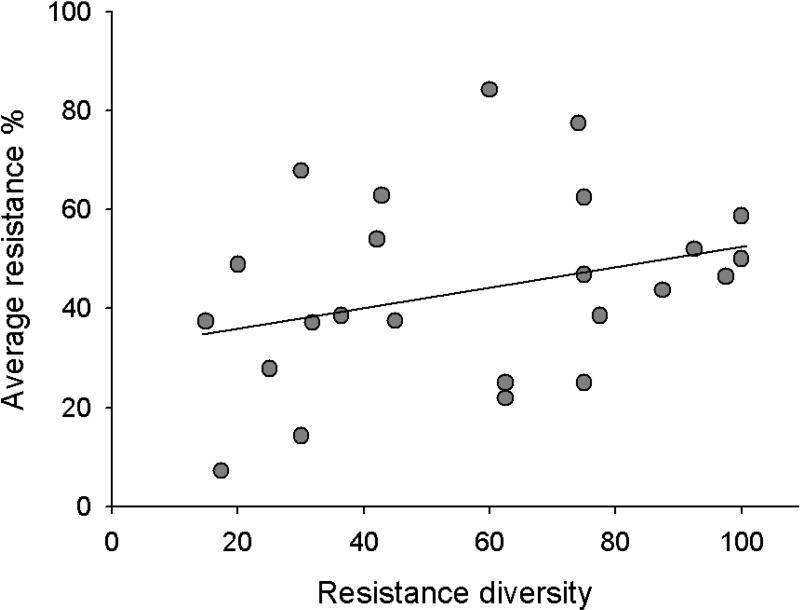
FIGURE 2.
Average population level qualitative resistance (%) plotted and the diversity of resistance phenotypes detected in the same population are positively associated (P= 0.0152; R2 = 0.49). Studies used for the estimation are depicted in Table 1 with superscript ‘2’.
Although these studies cover a wide range of unmanaged plant-pathogen interactions, notably 35 of these studies were on plant-fungus interactions and there was only a single study on a plant-bacterial association (Table 1; Goss & Bergelson 2006). While pathogenic fungi of plants are highly prevalent and diverse, this result is also likely to reflect the ease of detection of biotrophic fungi such as rusts and powdery mildews, compared to, for example, many viruses that require immunological assays for their detection. Only a small subset of the studies would have met the formal requirements of a meta-analysis (Rosenberg et al. 2000). We did not want to exclude a large number of studies providing valuable information on host resistance in wild host-pathogen interactions and hence, we decided to forgo a meta-analysis approach. Instead, we summarized the data in a more qualitative manner (but including quantitative data analyses where possible) to provide as comprehensive a view as possible on what we know to date on how host resistance to pathogens is spatially structured.
The genetic basis of resistance
The gene-for-gene (GFG) interaction is probably the best characterized genetic system of interaction for plant-pathogen populations (Thompson & Burdon 1992). The central assumption is that each resistance (R) gene in the host interacts specifically with a corresponding avirulence (AVR) gene in the pathogen (Flor 1956). An elicitor allele carried by an AVR-pathogen is recognized by the host R-allele, triggering local and systemic defence responses in the host (Hammond-Kosack & Jones 1997; Dangl & Jones 2001). Plants that lack the R allele are called susceptible, and parasites with a modified AVR factor (i.e. one not able to be recognised by the plant) are able to infect the host. Despite early scepticism (Barrett 1985; Frank 1993), it is now clear from hundreds of genetically and molecularly characterised examples that GFG is a common mechanism of interactions between wild and cultivated plant species and their parasites (bacteria, viruses, fungi, nematodes, insects and Oomycetes; (Jones & Dangl 2006). This genetic interaction is now understood in molecular terms as resulting from recognition between highly polymorphic host immune receptors (R proteins) and pathogen effector (Avr) proteins (Dodds & Rathjen 2010).
While the underlying genetic control of host resistance has only been verified for a few of the host-pathogen interactions that have been studied in wild populations (Burdon 1988; Burdon 1994), the strain-specific response typical of many plant-pathogen interactions is usually taken as evidence that resistance is controlled by major genes. Indeed, the majority of the studies reviewed here demonstrate or postulate resistance that is determined by genes with major effects (Table 1). Hence, to date, much more is known about the levels of diversity that may be found in interacting species whose interactions is governed by major genes, than is known about spatial variation in resistance that is quantitative in character (i.e. controlled by many genes; de Nooij & van Damme 1988a; Alexander 1989; Goss & Bergelson 2006).
However, the distinction between qualitative and quantitative resistance is not as generally conveyed. While most plant resistance responses may be categorized as resistant or susceptible and are governed by major genes, partial resistance phenotypes are also common and may come about through different mechanisms. Firstly, even in GFG systems there can still be varying degrees of expression of resistance (Table 1; Islam & Mayo 1990). It was demonstrated with M. lini and its wild host, L. marginale, that, like their fully resistant and susceptible counterparts, partially resistant phenotypes are controlled by single dominant genes (Burdon 1994). Similar control of partially resistant phenotypes has been demonstrated on many occasions in the interaction between wheat and its rust pathogens Puccinia graminis, P. triticina and P. striiformis (McIntosh et al. 1995). Secondly, non GFG resistance mechanisms, where multiple genes with additive effects contribute to non-race specific resistance, can give rise to partial resistance phenotypes. A mixture of quantitative and qualitative traits is suggested to govern resistance in several wild host-pathogen interactions (Bevan et al. 1993a; Ericson & Burdon 2009). In some cases, non-race specific resistance mechanisms can be ascribed to single major genes. For instance the RPW8 gene (Recognition of Powdery Mildew) has been identified as the major source of powdery mildew resistance in Arabidopsis thaliana (Xiao et al. 1997; Schiff et al. 2001; Wilson et al. 2001; Xiao et al. 2001), but differs from most known R genes by conferring broad spectrum resistance to a number of different powdery mildew species (Xiao et al. 2001; Jorgensen & Emerson 2008).
Environmental heterogeneity may further diversify resistance expression. For example, in the interaction between groundsel and its powdery mildew, incubation temperature and plant age influenced the infection types of some isolate/plant line combinations (Bevan et al. 1993a). An age-related, developmentally regulated general form of resistance was also suggested to explain variation in resistance in the interaction between A. thaliana and its bacterial pathogen Pseudomonas viridiflava (Goss & Bergelson 2006). In the interaction between Pl. lanceolata and its powdery mildew, Po. plantaginis, it has been demonstrated that the initial recognition steps remain robust over temperature and nutrient gradients, while further infection development is strongly regulated directly by environmental variation as well as indirectly through genotype specific (both host and pathogen) responses to that variation (Laine 2007b). Indeed, a general conclusion emerging from these studies is that resistance expression is far from the stable phenotype assumed in epidemiological models and models of host-parasite coevolution.
Additional predictors of disease levels in the field include phenological traits affecting transmission (e.g. timing of leaf flushing; Desprez-Loustau et al. 2010), timing of flowering and flower production for pollinator-transmitted diseases such as anther smuts; (Thrall & Jarosz 1994), implying genetic differences among plants that affect their exposure to inoculum (Alexander 1989; Alexander et al. 1996). These passive resistance mechanisms, in which plants may be physiologically susceptible but avoid infection due to genetic traits that reduce the probability of contact between host and pathogen, are likely to be an important component of resistance in some systems (Burdon 1987).
How is resistance measured?
Inoculations and transplant experiments
Experimental inoculations continue to be the most common way of classifying whether or not host plants harbour resistance to particular pathogen lines (Table 1). The development of high throughput genetic technologies such as next generation sequencing, and the increasing profile of ecogenomic studies that explicitly integrate molecular and ecological approaches, suggests that we will likely see an increase in work documenting sequence variation in particular host resistance genes; this could usefully expand our understanding, which is currently based on model species such as Arabidopsis, to a diversity of wild plant-pathogen associations (cf. Vera et al. 2007). While molecular methods provide exciting new opportunities for the study of host-pathogen interactions, inoculating a given host genotype with a given pathogen genotype will nevertheless remain a powerful way of directly documenting infection outcomes for a given genotype-by-genotype interaction. However, it could be argued that many inoculation studies have yielded little information about the actual structure of plant-pathogen interactions in natural environments, since plants have been tested with arbitrary pathogen genotypes not necessarily representative of those found in their native habitats. The origin of the strains used to score host responses has a tremendous impact on the levels of resistance likely to be identified (see ‘Resistant or not’ below). Hence, while the use of sympatric, allopatric or standard sets of pathogen strains all yield valuable information on host resistance, study designs need to match the questions being asked.
Transplant experiments where plants are introduced back into their native environment and their infection status monitored, have several benefits over inoculation studies carried out in controlled environments. First, as discussed above, infection outcomes may be mediated by locally varying environmental conditions generating variation in coevolutionary trajectories (Thompson 1999; Thompson 2005; Ridenhour & Nuismer 2007; Laine 2008). Second, transplant studies capture the natural range of pathogen variability, in contrast to laboratory experiments that typically only use a subsample of the local pathogen population (Laine 2007a). Third, for plant-pathogen interactions where passive mechanisms such as disease avoidance are an important component of resistance (Alexander 1989), transplant experiments are more likely to capture the full resistance profile of the individual (Ridenhour & Nuismer 2007). However, a major downside of transplant experiments is that it may be difficult to uncover the underlying variability that is characteristic of strain specific resistance under field conditions, where a few highly infective clones may cause the majority of infections (Laine 2007a).
Linking molecular and phenotypic resistance diversity
Linking phenotypic and molecular data on disease resistance has not been done frequently, although it has been strongly argued that, to further our understanding of coevolutionary processes, traditional phenomenological approaches need to be complemented by studies that focus on the molecular genetics of host-pathogen interactions (Woolhouse et al. 2002). There are as yet few examples where this type of analysis has been attempted, but these are valuable first steps to the wider application of molecular tools to studies of host-pathogen interactions. Jorgensen and Emerson (2008) studied the molecular diversity of the RPW8 gene (Recognition of Powdery Mildew) in A. thaliana. They did not find a tight correlation between disease resistance phenotype and RPW8 haplotype but, notably, resistant individuals consistently possessed at least one copy of RPW8.1 or RPW8.2, or of both of them. Previous QTL (Quantitative Trait Loci) analyses identified the regions on chromosome 3 containing RPW8 as the major source of resistance to powdery mildew pathogens, but the finding of other minor QTLs in these analyses suggest that resistance is a polygenic trait (Jorgensen & Emerson 2008), which explains why a tight association is not expected between the phenotype and the RPW8 haplotype. Barrett et al. (2009) found that isolates of M. lini from L. Marginale populations show extensive sequence variation and a signature of strong positive selection at the AvrP4 and AvrP123 loci. Transient expression of these AvrP123 and AvrP4 variants in L. marginale plants triggered hyper-sensitive responses in some host genotypes, suggesting that there is differential recognition of these genes by R genes in the host population, and indeed there is variation in the frequency of AvrP123 and AvrP4 variants between populations (Barrett et al. 2009). The Arabidopsis-downy mildew (Hyaloperonospora arabidopsidis) interaction offers a powerful set of molecular tools for population studies. Both genomes have now been sequenced, and at least nine RPP resistance genes in Arabidopsis and two corresponding Avr genes from downy mildew have been cloned. Hall et al. (2009) showed extensive variation in the RPP13 and corresponding ATR13 loci amongst host and pathogen isolates from wild populations. In the interaction between wild bean (Phaseolus vulgaris) and a deuteromycete fungus Colletotrichum lindemuthianum, populations (separated by some tens of kilometres) were differentiated for both neutral RAPD (Random Amplified Polymorphic DNA) markers as well as for phenotypic resistance and RGC (Resistance Gene Candidate family) markers. However, at a larger spatial scale, regions separated by some hundreds of kilometres were differentiated only for the two RGC molecular markers, suggesting that resistance diversity may not be driven by the same selective forces as at the molecular and phenotypic levels (de Meaux et al. 2003). These findings highlight the importance of taking complementary approaches at both the molecular and phenotypic level when studying patterns of disease resistance.
Resistant or not
We used studies that reported qualitative measures of resistance (% of hosts infected or % of resistant responses) to first measure the frequency of resistance in natural plant populations. We then tested how origin of the pathogen strains affects the level of resistance measured in inoculations. The origin of strains was categorised as being ‘Sympatric’ (from the same population), ‘Allopatric1’ (originating from populations ≤ 2 km away), and ‘Allopatric2’ (originating from populations > 2 km away). The classification of the two allopatry categories was justified by studies that show the probability of contact between host and pathogen populations separated by more than 1-2 kilometres drops drastically (Thrall et al. 2002; Laine 2005; Niemi et al. 2006; Roslin et al. 2007). Data on resistance levels were normally distributed and hence we used ANOVAs to determine whether levels of resistance depend on the origin of the strains.
We find that on average, wild plant populations are more likely to be susceptible than resistant to their pathogens (58.6% susceptibility vs. 41.4% resistance, Fig. 1a). Interestingly, we also find that the choice of strains used to classify host resistance is a major determinant of the levels of resistance observed, and thus is an important consideration. When hosts are scored with sympatric strains, studies typically measure much lower levels of resistance (average 32.1%, Fig. 1a). When plants are scored with pathogens from neighbouring populations (< 2 km away), resistance levels are somewhat higher (39%). Plant populations harbour the highest levels of resistance to allopatric pathogen strains originating typically some tens to hundreds of kilometres away (Table 1; 47.5%, Fig. 1a). The overall trend of an increase in resistance with greater distance between the host population and pathogen source population was not statistically significant (F2, 35 = 1.95, P = 0.157). However, the comparison between resistance measured against sympatric strains and strains originating from furthest away (Allopatric2) was marginally significant (F1, 27 = 3.82, P = 0.061; Fig. 1a).
Finding susceptibility to sympatric pathogen strains is in itself not surprising, since the collection of pathogen samples from plants within a particular host population inevitably involves a bias as only strains capable of attacking at least some of these hosts will be found. Finding that resistance increases as distance between host and pathogen origin increases can certainly be taken as evidence of pathogen local adaptation. Indeed, seven of the 36 studies reviewed were specifically designed to test whether pathogens are adapted to their local hosts, and six studies found evidence to support this (Parker 1985; Thrall et al. 2002; Laine 2005; Niemi et al. 2006; Sicard et al. 2007; Springer 2007). However, our results show an increase in host resistance to pathogen strains the host is likely to never or rarely encounter. This may seem counter-intuitive given that some studies suggest that resistance is a costly trait for the host (Bergelson & Purrington 1996; Tian et al. 2003), although this may not be a general feature (Brown 2003b; Brown 2003c; Brown 2003a; Burdon & Thrall 2003). In this respect, one would expect ‘unnecessary’ resistance to be selected against, although the rate at which this occurs across metapopulations will at least partly depend on the degree of among-population isolation as well as colonisation-extinction processes; even in situations where disease is absent, the time to lose even a costly R gene may be many generations (Thrall & Antonovics 1995).
However, it is also possible that in natural populations resistance is not always as costly as traditionally has been assumed (Laine & Tellier 2008). At least one plant protein belonging to the major class of R proteins has a developmental role (Faigon-Soverna et al. 2006), so it is possible that some R proteins may contribute to host fitness in other ways than the resistance they confer to their hosts. One mechanism that can explain some instances of apparently high resistance costs is hybrid incompatibility. In several cases the underlying basis of this phenomenon has been characterised as an incompatibility between different members of an R protein complex that leads to inappropriate activation of defence response (Bomblies & Weigel 2007; Yamamoto et al. 2010) . However, such interactions are also important for maintaining reproductive barriers, and hence there may be underlying benefits as well. Furthermore, resistance genes that are ineffective against all current pathogen races have been reported to have some residual effects on pathogen development (Pedersen & Leath 1988). Alternatively, these ‘unnecessary’ resistance genes may in fact serve an important function at the population level by reducing the effectiveness of incoming inoculum, even for relatively infrequent colonization attempts by the pathogen. This may delay disease development in such host stands by weeks, months or even entire growing seasons (Burdon & Jarosz 1991; Burdon et al. 1996).
Resistance and life-history
Life-history is considered to play a major role in determining the genetic structure and evolutionary potential of host-pathogen interactions (for reviews, please see Barrett et al. 2008; Burdon & Thrall 2009). Essentially all key stages of the interaction are determined by life-history characteristics, starting from how frequent contact is between host and pathogen, to which life history stages of the host are affected by infection. We extracted information on both host and pathogen life history features, as summarised in Supporting Information, Table S1 for the same set of studies reporting qualitative measures of resistance as used for the analysis presented in Figure 1a. We used ANOVAs to test how life history, the origin of pathogen strains, and their interaction, affect levels of host resistance. The results of all ANOVAs are presented in Table S2. Hosts were least resistant to pathogens that are wind dispersed, with higher resistance detected for pathogens dispersed by insect vectors and highest resistance observed for water dispersed pathogen (F2, 28 = 5.56, P = 0.009; Fig. 1b). As found earlier, highest levels of resistance were found against ‘Allopatric2’ strains (F2, 28 = 4.43, P = 0.021; Fig. 1b), and this trend did not vary significantly for the different pathogen dispersal modes (F4, 28 = 1.43, P = 0. 251; Fig. 1b). These differences in overall resistance may be related to asymmetries in fitness effects of infection caused by pathogens with qualitatively different life histories. For example, many airborne pathogen have general debilitative effects while vector-borne smuts frequently castrate their hosts (Burdon & Thrall 2009).
None of the other life-history characteristics that we tested (host mating system, mode of pollen dispersal, host dispersal model, or pathogen specificity) had significant effects on resistance (Table S2). However, the data suggest interesting differences for many of these classifications that we were simply not able to pick up because of a lack of statistical power.
Resistance variation
While mutational change at individual resistance or avirulence loci may not be grossly dissimilar, the major differences that occur in propagule production between hosts and pathogens (typically many orders of magnitude) strongly support the contention that pathogens would, through faster evolution (Parker 1985; Clay & Kover 1996; Thrall et al. 2002; Laine 2005; Niemi et al. 2006; Sicard et al. 2007; Springer 2007), leave their hosts defenseless, were it not for the basic feature of the host's counter strategy – variability. Here we find that a higher level of diversity in resistance phenotypes within populations does indeed provide greater protection against pathogens in wild plant-pathogen interactions (Fig. 2). For studies reporting host population phenotypic diversity and average resistance (qualitative measured resistance; studies are identified in Table 1), we tested whether higher levels of phenotypic resistance diversity in fact provide better protection against pathogens. For this purpose we used ANCOVA analysis (implemented in SAS 9.1) with the number of resistance phenotypes divided by the number of host genotypes tested as a covariate, and study system (i.e. which host and pathogen species) as a fixed factor to explain average resistance (with a normal distribution of errors). There was a significant positive association between the diversity of resistance phenotypes and average population level of resistance (F1, 24= 7.28, P = 0.0152), As expected, the average level of resistance differed between the study systems, although not significantly (F6, 24 = 2.25, P = 0.0879). The association between resistance phenotype diversity and average level of resistance did not vary between the different study systems, thus, the statistically non-significant study system × phenotypic diversity interaction was not included in the final model. There is considerable variation in this trend of higher diversity conferring higher resistance levels (R2 = 0.49), which in part may be explained by the fact that the origin of the pathogen isolates used to score resistance varies between the different studies (Table 1, see above ‘Resistant or not’). Very few studies had explicitly explored this relationship between resistance diversity and level of resistance (See below ‘Host population differentiation: Implications for pathogen epidemiology and evolution’).
Patterns of resistance at different spatial scales
Essentially all the studies reviewed here report variation in disease resistance, regardless of the spatial scale at which resistance was studied (Table 1). Below we briefly review the results from a range of different studies on levels of resistance and resistance diversity.
Within host variation
While several studies identified host genotypes that were monomorphic in their resistance response (fully susceptible or fully resistant; e.g. Bevan et al. 1993a; Burdon et al. 1999; Laine 2004; Lebeda et al. 2008), individual level variation was detected in all studies where it was explicitly looked for. For resistance controlled by major genes the genes underlying defence-traits are highly variable, resulting in the occurrence of genotypes with varying degrees of susceptibility to pathogens (Bergelson et al. 2001). A striking feature of plant R genes is that they can be highly variable in copy number, gene structure and allelic composition within species, resulting in the occurrence of genotypes that vary in their degree of susceptibility to pathogens (Bergelson et al. 2001). Over the last fifteen years, the identification and cloning of over 100 resistance genes has revealed that they share sequence similarities and can be categorized into several R-gene classes. Intragenic recombination and point mutations appear to be the major evolutionary mechanisms generating novelty in R genes (de Meaux et al. 2003). The most common functional domain found in R genes is the C-terminal Leucine Rich Repeat (LRR) domain. The LRR domain has been shown to be highly variable, with the hallmark of diversifying selection detected on the solvent exposed residues of many R genes. Thus, LRR diversity may be primarily shaped by pathogen diversity (Michelmore & Meyers 1998).
To a great extent, what then determines variability within host individuals depends on the number of loci involved in R gene mediated defence. The estimates from wild plant species vary. For the interaction between Chondrilla juncea and its obligate rust pathogen, Puccinia chondrillina, it was necessary to postulate a minimum of 11 resistance genes to explain the observed patterns of resistance diversity (Espiau et al. 1998). In two populations of Senecio vulgaris six and ten genes for resistance to Erysiphe fischeri were suggested to occur (Bevan et al. 1993a; Bevan et al. 1993b). A minimum of six resistance genes was needed to explain the observed patterns of resistance within a single L. marginale population attacked by the rust pathogen M. lini (Burdon & Jarosz 1991). Overall, the existence of multiple resistance loci appears to be common in natural plant populations (Thrall & Burdon 1997).
Within and among population variation
Despite indications that R gene diversity may generally be high, for many wild plant-pathogen associations, some populations are found in which variation for disease resistance does not occur or is limited (Jarosz & Burdon 1991; de Meaux et al. 2003; Kniskern & Rausher 2006); while in others considerable intra-population diversity for resistance has been detected (Espiau et al. 1998; Thrall et al. 2001; de Meaux et al. 2003; Laine 2004; Niemi et al. 2006). Many studies report high numbers of resistance phenotypes occurring over very small spatial scales (measured over only a few metres within populations; e.g Bevan et al. 1993a; Burdon & Thompson 1995; Laine 2006). Within host populations no strong spatial structuring of resistance phenotypes (i.e. similar phenotypes occurring more closely together than would be expected by chance) has been detected; (Burdon & Jarosz 1991; Laine 2006). However, it is possible that spatial structuring occurs over smaller spatial scales than have been examined (A. Nemri et al. unpublished data).
As expected, there is variation in host resistance among populations of the same plant-pathogen interaction. The studies we have considered cover very different spatial scales, ranging from neighbouring populations which are separated by some hundreds of metres to populations separated by several hundreds of kilometres (Table 1). While some studies find significant differences among populations in their average level of resistance (Thrall et al. 2002; Niemi et al. 2006), other systems show relatively similar overall levels of resistance (Carlsson-Granér 1997; Thrall et al. 2001). In the Plantago-Podosphaera interaction populations with a history of infection showed more similar levels of resistance than did host populations that were known to have been uninfected for several successive years (Laine 2004; Laine 2005).
How evenly resistance diversity is distributed among populations varies between study systems. Parker (Parker 1985) found little variation in the observed phenotypic distribution among plant families within sites and large differences among sites, which is not surprising for isolated, inbred subpopulations undergoing genetic drift such as those of Amphicarpaea bracteata. In the Pl. lanceolata – Po. plantaginis interaction no evidence was found for greater similarity in the resistance phenotypic compositions of neighbouring than far-away populations (Laine 2004). A similar, seemingly random distribution was observed by Davelos et al. (1996) in resistance to rust infection of the clonal plant, Spartina pectinata. In contrast, Thrall and colleagues (2001) found evidence for greater similarity in the resistance phenotypic compositions of neighbouring than far-away populations in the L. marginale – M. lini interaction for the most common resistance phenotypes. These common resistance phenotypes also tended to show a higher degree of susceptibility than the rarer forms to 12 standard pathogen isolates (each with a unique infectivity phenotype). Interestingly, the number of resistant responses shown by individual hosts was negatively correlated with their prevalence across the metapopulation (Thrall et al. 2001), which could suggest negative frequency dependent selection.
Recent studies have demonstrated the fundamental role that biophysical variation in the surrounding environment may play in determining the outcome of coevolutionary interactions between hosts and parasites (Thomas & Blanford 2003; Price et al. 2004; Mitchell et al. 2005; Fels & Kaltz 2006; Lambrechts et al. 2006; Laine 2007b; Laine 2008; Vale et al. 2008). Thus, it is not surprising that a number of studies have found strong differentiation in resistance that correlates with different habitat types. In the interaction between A. thaliana and its bacterial pathogen P. viridiflava, whether plants originated from a fallow or agricultural field had a significant effect on measures of resistance with average resistance being significantly greater for plants from cultivated sites (Goss & Bergelson 2006). In the interaction between L. marginale and M. lini, there was a significant effect of ecotype due to the overall lower resistance of bog populations as compared to the more resistant hill populations (Thrall et al.2001). What maintains this type of resistance structure difference among ecotypes is currently being investigated.
Regional patterns of resistance
To date there are few data available on the resistance structure of wild host populations over large regional spatial scales. In the interaction between L. marginale and M. lini, populations of Linum located in the mountains and plains regions of New South Wales of Australia are separated by several hundred kilometres. Detailed studies of host resistance structure within and between these regions showed large differences in average resistance, both among populations within the mountains and plains areas, as well as between regions (Burdon et al. 1999). Levels of within-family heterogeneity in disease resistance in the mountain populations were low while heterogeneity within family lines was much more frequent in the plains. This may be explained by differences in the mating system of the host plant between these regions; L. marginale is predominantly selfing in the mountains while significant outcrossing has been detected in the plains. Despite the higher levels of heterozygosity and outcrossing in the plains populations, gene diversity (h) was actually higher in the mountain populations. These populations are more polymorphic although with a greater proportion of variation occurring in multilocus homozygous genotypes, whereas in the plains metapopulation there is considerable evidence for continuing recombination through outcrossing (Burdon et al. 1999). A continental analysis of host resistance in L. marginale to M. lini demonstrated that three host lines were susceptible to all 37 isolates of M. lini while the remaining 39 showed resistance to various combinations of the M. lini isolates. None were resistant to all isolates (Burdon et al. 2002). In contrast with pathogen infectivity data, which indicated clustering of strains according to their area of origin, there was no evidence for regional groupings of different multi-pathogen resistance phenotypes. Host lines from Tasmania, New South Wales, and South Australia were all apparently randomly distributed throughout the cluster tree (Burdon et al. 2002).
In the Lactuca serriola - Bremia lactucae interaction, resistance of the host was screened along an east-to-west transect across the Czech Republic, Germany, Netherlands and the United Kingdom. The results show large variation both among and within individual populations and countries. A clear gradient of increasing uniformity of race-specificity was detected when moving from central to western Europe, as well as a slight decrease in the diversity of resistance phenotypes (Petrzelova & Lebeda 2010). Forty-five resistance phenotypes against B. lactucae were found in 16 L. serriola populations across the Czech Republic. Eight common resistance phenotypes represented 80% of the studied plants, and the remaining genotypes were generally rare (Lebeda et al.2008). In contrast to the high levels of diversity detected in other systems, the interaction between wild bean and C. lindemuthianum was characterised by three areas, separated by some hundreds of kilometres, which were not differentiated with respect to host resistance phenotypes, and where overall levels of diversity were similar (de Meaux et al. 2003).
HIERARCHICAL RESISTANCE STRUCTURE
Long-term studies of the L. marginale – M. lini and Pl. lanceolata – Po. plantaginis interactions provide an opportunity to examine how resistance diversity is hierarchically distributed across different spatial scales within the same pathosystem. We have comparable datasets from both of these interactions where sympatric and allopatric pathogen strains were used to score resistance within host populations and in neighbouring host populations separated by some hundreds of metres up to two kilometres (called here the ‘within metapopulation’ scale), as well as between metapopulations separated by some tens of kilometres. For details on study designs and experimental protocols, please see Thrall et al. (2002) and Laine (2005). Data from previous studies have shown that these arerelevant scales of examination for these interactions: pathogen populations show adaptation at thismetapopulation scale and have frequent gene flow among host populations within the metapopulations (Thrall et al. 2002; Laine 2005). We use generalised linear mixed models (GLMM; Breslow & Clayton 1993) to estimate variance components for random effects which were: individual plants within populations, populations within metapopulations and among metapopulations(hierarchical levels of variation, as described in Roslin et al.(2006)). Analyses were carried out in SAS System for Windows 9.2, using the GLIMMIX macro (Littell et al. 1996). The response variable in both analyses is host resistance/susceptibility (0/1) against a given pathogen strain, and hence the model assumes binomially distributed errors and a logit link function.
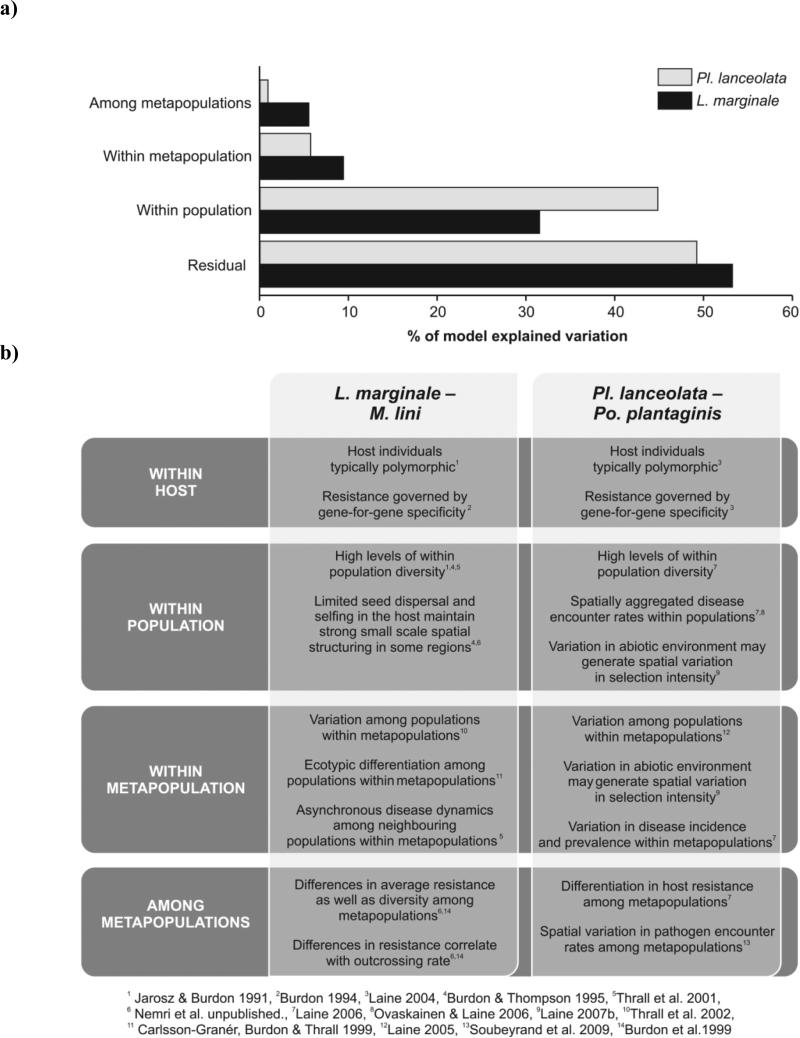
For both associations we find that the level of variation explained decreases at higher spatial scales (Fig. 3). The amount of variation not explained (‘Residual’ in Fig. 3) by the model variables – spatial scales – are 53% and 49% for L. marginale and Pl. lanceolata, respectively. For both L.marginale and Pl. lanceolata most of the variation in resistance diversity can be found within host populations (32% in L. marginale and 45% in Pl. lanceolata; Fig. 3). Highly asymmetric disease encounter rates coupled with microclimatic variation are considered to generate such high levels of within population variation in Pl. lanceolata (Laine 2004; Fig. 3; Laine 2006; Laine 2007b)(Fig 3). For L. marginale, higher levels of variation in resistance were detected among neighbouring populations than in Pl. lanceolata (10% vs. 6%; Fig. 3). This could be explained by ecotypic differentiation among neighbouring L. marginale populations that occur in hill and bog habitats (Thrall et al. 2001). In Pl. lanceolata negligible variation in disease resistance could be attributed to differences among metapopulations (~1%), while in L. marginale 6% of the variation explained by the model was found among metapopulations. In Figure 3 we summarize the different processes that have been identified to drive resistance structure at different spatial scales for these interactions in long-term studies.
Figure 3.
a) Variance components for resistance diversity at the different spatial scales in the interaction between L. marginale – M. lini (black bars) and Pl. lanceolata - Po. plantaginis (grey bars); and b) Summary of processes affecting resistance diversity at different spatial scales of the same interactions.
What generates variation in disease resistance?
There is sufficient evidence to conclude that disease resistance in wild plant populations has a genetic basis, is heritable, and is a trait containing high levels of variability in space and time (See review by (Salvaudon et al.2008), providing ample opportunities for selection to act. However, assessing the relative importance of adaptive vs. non-adaptive processes (e.g. random drift, bottlenecks, and founder effects) in driving the resistance structure of wild plant populations has remained difficult.
While pathogens are widely recognized as one of the most potent ecological and evolutionary forces driving the dynamics of their hosts, we have little direct evidence of adaptive changes in the resistance structure of wild plant populations (e.g. Burdon & Thompson 1995) in response to pathogen attack. Aside from selection that may be imposed by costs of resistance (Bergelson & Purrington 1996; Tian et al. 2003), non-adaptive evolution has been suggested to be a strong force shaping plant populations. The sessile growth form of plants, together with restricted recombination due to high levels of self-fertilization and clonal reproduction, increases the chance of nonadaptive, or even maladaptive, evolution. Furthermore, extinctions and rapid population expansions following colonizations from a few founder individuals are frequent, and are associated with small effective population size and random genetic drift (Silvertown & Charlesworth 2001). It has also been suggested that genetic hitchhiking resulting from linkage between resistance genes and other traits under even more intense selection may affect the genetic composition of plant populations, where selection on resistance is swamped by these processes (Burdon & Thompson 1995). Alternatively, disease could impose selection on traits other than resistance, for example on demography or phenology, when larger or earlier emerging plants are more likely to become infectedand may support higher levels of infection,and hence may suffer higher mortality (Burdon & Thompson 1995; Desprez-Loustau et al. 2010).
Detecting signs of adaptive responses to pathogens in nature may be further complicated by the fact that selection intensity imposed by pathogens is characterized by the temporal and spatial asynchrony characteristic of pathogen epidemics, variation in epidemic amplitudes between host populations, and distance-dependent migration and gene-flow (Burdon et al. 1995; Antonovics et al. 1998; Ericson et al. 1999; Smith et al. 2003; Antonovics 2004; Laine & Hanski 2006; Soubeyrand et al. 2009) (Smith et al. unpublished data). In addition to the spatially and temporally heterogeneous nature of selection pressure of pathogens, patterns of selection are also likely to be influenced by a range of factors including the life-history and dispersal mechanisms of both host and pathogen (Barrett et al. 2008). For example, a comparison of the reaction of a Trifolium repens population to two different pathogens, Cymadothea trifolii and Pseudopeziza trifolii, revealed marked differences in the frequency distribution of disease resistance (Burdon 1980). Resistance to C. trifolii was normally distributed while resistance to P. trifolii was strongly skewed, with the majority of the population showing a high degree of resistance. The higher severity of past epidemics in the study population, and putative higher fitness costs associated with infection by P. trifolii are likely causes of the difference in resistance against these two pathogens in the host population (Burdon 1980). Studies measuring host resistance profiles within the same populations, butagainst different pathogens, especially those which differ in their effects on host fitness, could yield valuable tests of the potential pathogens have as selective forces driving the resistance of their hosts.
Potentially, spatial pattern could arise in a host-pathogen association through an interaction between microclimate and pathogen development rates, leading to marked differences in selection pressures over very short distances (i.e. within local populations). To date one of the few studies reporting pathogen imposed selection on host resistance at this scale comes from the Pl. lanceolata-Po. plantaginis interaction (Laine 2006). Detailed epidemiological data collected within host populations over several consecutive years demonstrated encounter rates between host and pathogen to be highly divergent at a scale of a few metres, due to a highly predictable seasonal spread of local epidemics. Resistance was higher in areas within host populations where disease encounter rates have been systematically high than in areas where they have been low. Furthermore, resistance was higher toward sympatric than allopatric pathogen strains, further suggesting a response to selection caused by the local pathogen population. The fine scale selection mosaic may have formed through an interaction with the physical environment as the study coincided with severe drought,with the highest levels of mortality in areas of the populations where disease had been most prevalent (Laine 2006). In the interaction between groundsel and its powdery mildew, it was suggested that that climatic conditions were most favourable to mildew at the site where resistance was found to be highest, having resulted from greater selection intensity (Bevan et al. 1993a).
Suggestions of pathogen-mediated selection also come from the pollinator-transmitted anther smut infecting Silene dioica. Rapid rates of disease spread in some newly infected populations, as compared to sites with a long history of disease, could be explained by a higher frequency of resistant genotypes accumulating following exposure to the fungus (Alexander et al. 1996). Elmqvist et al. (1993) further found that S. dioica plants derived from a completely healthy site had larger flowers and longer styles, and also received more spores, than plants derived from a site with high disease, suggesting pathogen selection (mediated by the vector) for floral traits conferring resistance (Alexander et al. 1996). In the interaction between Ipomoea purpurea and its rust pathogen Coleosporium ipomoea, differences among populations in the frequencies of resistant plants are postulated to be caused by an underlying divergence in allele frequencies at Rci1 (Kniskern & Rausher 2006). Although direct information on the cause of this divergence is not available, it is unlikely to be entirely due to genetic drift. Coleosporium ipomoea is a potent pathogen that substantially reduces plant fitness. Moreover, resistant genotypes exhibit a substantial cost of resistance that can reach 15.5%; this cost generates a fitness differential at the Rci1 locus at low pathogen densities. It thus seems unlikely that variation at this locus is neutral, allowing for divergence by drift (Kniskern & Rausher 2006).
Host population differentiation: Implications for pathogen epidemiology and evolution
It is commonly argued that the greater genetic diversity of host plants in natural communities may give rise to more stable associations with pathogens than the ‘boom-and-bust’ cycles seen in many agricultural crops (Burdon et al. 2006). While many factors will influence the prevalence and severity of disease in host populations, both resistance diversity and average level of resistance are likely to have an impact on both within-season and between-season dynamics of pathogen populations (Thrall & Burdon 2000; Laine 2004). In agricultural systems it has become evident that increasing resistance diversity within populations often leads to reduced pathogen prevalence, at least temporarily (Browning & Frey 1969; Wolfe 1985; DiLeone & Mundt 1994; Zhu et al. 2000). Most of what is known about natural systems comes from studies that have taken an experimental approach. Using a series of experimental host populations with dissimilar genetic compositions, several studies have shown that disease levels are higher in less diverse host stands (Schmid 1994; Thrall & Jarosz 1994), and that disease declines in resistant populations while the host and the pathogen may be more likely to coexist in susceptible populations (Alexander et al. 1996).
Thrall & Burdon (2000) demonstrated a negative relationship between resistance diversity and disease prevalence in the natural populations of L. marginale infected by M. lini. Furthermore, host resistance varied as a function of population connectedness, with average population resistance increasing in connected populations, compared to more isolated host populations. This could explain why disease prevalence decreased in regions where host populations were more continuous (Carlsson-Granér & Thrall 2002). Antonovics (2004) proposed that increased disease resistance in the metapopulation as a whole could partly explain why rates of disease colonization have declined in the Silene – Microbotryum pathosystem in SW Virginia, USA. Overall, the diversity of disease resistance that we find in wild plant populations is likely to partly explain why damaging regional epidemics are so rare in natural situations. In Pl. lanceolata, resistance diversity and mean resistance both have potentially strong effects on pathogen epidemiology – populations with higher resistance diversity and mean level of resistance are less likely to be colonized by the pathogen than more susceptible and less diverse host populations (Laine 2004).
Much less is known about how resistance diversity may affect pathogen evolution. There is some evidence that high levels of resistance select for high levels of infectivity in the associated pathogen population (Thrall & Burdon 2003; Niemi et al. 2006). However, while these findings appear to support the ‘naïve’ coevolutionary expectation for a host-pathogen arms race, highly virulent pathogen strains do not seem to dominate natural pathosystems (de Nooij & van Damme 1988b; Bevan et al. 1993c; Thrall et al. 2002; Laine 2005). This suggests that pathogen evolution may be constrained by trade-offs between transmission and infectivity (Bull 1994; Thrall & Burdon 2003), where pathogens balance reductions in the quality and quantity of host resources against the benefits of their own rapid growth and reproduction.
Conclusions and future research
In conclusion, diversity in disease resistance in natural host plant populations is ubiquitous, with resistance diversity being found across all spatial scales, ranging from within-individual polymorphism to continental scales. While any given plant population is more likely to be susceptible than resistant to the pathogens attacking it, diversity in resistance appears to be the best counterstrategy for plants against their pathogens. Much of the variation seen in disease resistance is found within host populations, but this scale alone does not explain the patterns of diversity in natural plant-pathogen interactions, as demonstrated by the hierarchical analysis of the M. lini and Pl. lanceolata resistance structures in Figure 3. We have some convincing examples of pathogen-imposed selection on host population resistance structure, yet metapopulation dynamics and heterogeneous selection processes must be taken into account if we want to understand how resistance diversity is structured and maintained in plant populations.
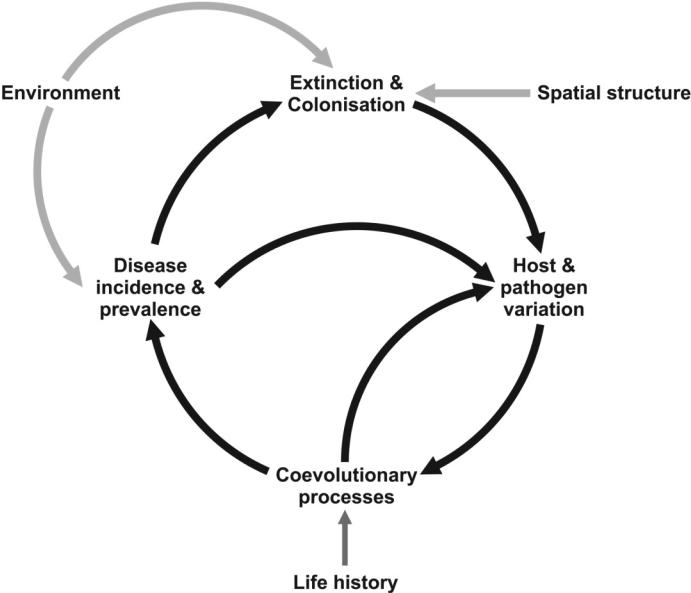
Figure 4 summarizes the interplay between genetic diversity, metapopulation dynamics, the environment and spatial structure in driving host-pathogen coevolution. Host and pathogen life-history (e.g. dispersal, mating system, transmission mode) is an important determinant of the outcomeht of the coevolving interaction. The relative importance of these different components on host-pathogen coevolution in natural systems is likely to vary spatially and temporally. Thus, we argue that a key aim for host-pathogen research is to generate a consensus of how life-histories, spatial structure and population dynamics interact to jointly determine patterns of genetic and phenotypic variation in resistance and virulence. Specifically, under which conditions are we to expect high levels of diversity and what situations may favour a decrease or maintenance of low diversity? What conditions favour the emergence and spread of novel pathogen and host genotypes? Furthermore, we need to better understand how disease resistance impacts disease dynamics and pathogen evolution. Simplified assumptions about links between host resistance and disease underlie much of modern crop protection strategies, yet there is little data to validate these ideas.
Figure 4.
A schematic representation of the biotic and abiotic processes that drive the coevolutionary cycle generating host resistance diversity across space and time. The relative importance of the different processes may vary both spatially and temporally.
Hence, we identify five areas where future research could assist with estimation of the relative importance of processes outlined in Figure 4 in driving coevolutionary dynamics in plant-pathogen interactions:
Studies of the same interactions across multiple spatial scales will allow us to more accurately determine the relative importance of selection, and how it varies across space and time, metapopulation dynamics, and environmental heterogeneity, in driving host population resistance structure.
Increasing the number of studies that span a greater range of host-pathogen interactions will enable us to better estimate how coevolutionary trajectories are determined by the life-histories of the host and its pathogen. Our preliminary analyses of a limited number studies suggest that both levels of resistance and their spatial structure may be driven by life history features.
In the last decade it has become increasingly clear that both resistance and infectivity may be profoundly affected by environmental conditions through G (× G) × E interactions. Identification of key environmental variables driving the coevolutionary cycle is required to understand patterns of disease and how polymorphism is maintained in the interaction traits of both hosts and pathogens.
While the influence of spatial structure of coevolutionary dynamics has long been recognized, we are still far from a predictive understanding of how connectivity at different spatial scales generates spatial variation in disease resistance. Mapping of resistance across different spatial settings coupled with studies of experimental evolution will be most useful for this purpose. Species distributions are rapidly changing under human imposed fragmentation and climate change and we need to understand how this may impact on the resistance structure of natural plant populations and disease dynamics.
We are still a long way from linking resistance phenotypes with molecular diversity, and the structure and function of particular proteins but, given that resistance may be a much more complex trait than traditionally assumed, an integrated ecogenomic approach is needed to fully understand variation in disease resistance.
Supplementary Material
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1. Summary of studies that were used for analysis of how host and pathogen life history features may affect host resistance.
Table S2. ANOVA results for how life history and origin of pathogen affect host population resistance.
Acknowledgements
This work was supported by NIH grant 5RO1 GMO74265-01A2.
References
- Alexander HM. An experimental field study of anther-smut disease of Silene alba caused by Ustilago violacea: genotypic variation and disease incidence. Evolution. 1989;43:835–847. doi: 10.1111/j.1558-5646.1989.tb05181.x. [DOI] [PubMed] [Google Scholar]
- Alexander HM, Antonovics J, Kelly AW. Genotypic variation in plant disease resistance - physiological resistance in relation to field disease transmission. Journal of Ecology. 1993;81:325–333. [Google Scholar]
- Alexander HM, Thrall PH, Jarosz AM, Oudemans PV. Population dynamics and genetics of plant disease: a case study of anther-smut disease. Ecology. 1996;77:990–996. [Google Scholar]
- Antonovics J. Long-term study of a plant-pathogen metapopulation. In: Hanski I, Gaggiotti OE, editors. Metapopulation Ecology, Genetics, and Evolution. Academic Press; Amsterdam: 2004. pp. 471–488. [Google Scholar]
- Antonovics J, Thrall PH, Jarosz AM. Genetics and the spatial ecology of species interactions: the Silene-Ustilago system. In: Tilman D, Kareiva P, editors. Spatial ecology: the role of space in population dynamics and interspecific interactions. Princeton University Press; 1998. pp. 158–180. [Google Scholar]
- Barrett JA. The genetics of host-pathogen interaction. In: Rollinson D, Anderson RM, editors. Ecology and genetics of host-parasite interactions. Academic Press; London: 1985. [Google Scholar]
- Barrett LG, Thrall PH, Burdon JJ, Linde CC. Life history determines genetic structure and evolutionary potential of host-parasite interactions. Trends in Ecology and Evolution. 2008;23:678–685. doi: 10.1016/j.tree.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett LG, Thrall PH, Dodds PN, van der Merwe M, Linde CC, Lawrence GJ, Burdon JJ. Diversity and Evolution of Effector Loci in Natural Populations of the Plant Pathogen Melampsora lini. Molecular Biology and Evolution. 2009;26:2499–2513. doi: 10.1093/molbev/msp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergelson J, Kreitman M, Stahl EA, Tian D. Evolutionary dynamics of plant R-genes. Science. 2001;292:2281–2285. doi: 10.1126/science.1061337. [DOI] [PubMed] [Google Scholar]
- Bergelson J, Purrington CB. Surveying patterns in the costs of resistance in plants. American Naturalist. 1996;148:536–558. [Google Scholar]
- Bevan JR, Clarke DD, Crute IR. Resistance to Erysiphe fischeri in two populations of Senecio vulgaris. Plant Pathology. 1993a;42:636–646. [Google Scholar]
- Bevan JR, Crute IR, Clarke DD. Diversity and variation in expression of resistance to Erysiphe fischeri in Senecio vulgaris. Plant Pathology. 1993b;42:647–653. [Google Scholar]
- Bevan JR, Crute IR, Clarke DD. Variation for virulence in Erysiphe fischeri from Senecio vulgaris. Plant Pathology. 1993c;42:622–635. [Google Scholar]
- Bomblies K, Weigel D. Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nature Reviews Genetics. 2007;8:382–393. doi: 10.1038/nrg2082. [DOI] [PubMed] [Google Scholar]
- Breslow NE, Clayton DG. Approximate inference in generalized linear models. Journal of American Statistical Association. 1993;88:9–25. [Google Scholar]
- Brown JKM. A cost of disease resistance: paradigm or peculiarity? Trends in Genetics. 2003a;19:667–671. doi: 10.1016/j.tig.2003.10.008. [DOI] [PubMed] [Google Scholar]
- Brown JKM. Little else but parasites. Science. 2003b;299:1680–1681. doi: 10.1126/science.1083033. [DOI] [PubMed] [Google Scholar]
- Brown JKM. Yield penalties of disease resistance in crops. Current Opinion in Plant Biology. 2003c;5:339–344. doi: 10.1016/s1369-5266(02)00270-4. [DOI] [PubMed] [Google Scholar]
- Browning JA, Frey KJ. Multiline cultivars as means of disease control. Annual Review of Phytopathology. 1969;7:355–382. [Google Scholar]
- Bull JJ. Virulence. Evolution. 1994;48:1423–1435. doi: 10.1111/j.1558-5646.1994.tb02185.x. [DOI] [PubMed] [Google Scholar]
- Burdon JJ. Variation in disease resistance within a population of Trifolium repens. Journal of Ecology. 1980;68:737–744. [Google Scholar]
- Burdon JJ. Diseases and plant population biology. Cambridge University Press; Cambridge, UK: 1987. [Google Scholar]
- Burdon JJ. Major gene resistance to Phakopsora pachyrhizi in Glycine canescens, a wild relative of soybean. Theoretical and Applied Genetics. 1988;75:923–928. [Google Scholar]
- Burdon JJ. The distribution and origin of genes for race-specific resistance to Melampsora lini in Linum marginale. Evolution. 1994;48:1564–1575. doi: 10.1111/j.1558-5646.1994.tb02196.x. [DOI] [PubMed] [Google Scholar]
- Burdon JJ, Ericson L, Müller WJ. Temporal and spatial changes in a metapopulation of the rust pathogen Triphragmium ulmariae and its host, Filipendula ulmaria. Journal of Ecology. 1995;83:979–989. [Google Scholar]
- Burdon JJ, Jarosz AM. Host-pathogen interactions in natural populations of Linum marginale and Melampsora lini: I Patterns of resistance and racial variation in a large host population. Evolution. 1991;45:205–217. doi: 10.1111/j.1558-5646.1991.tb05278.x. [DOI] [PubMed] [Google Scholar]
- Burdon JJ, Thompson JN. Changed patterns of resistance in a population of Linum marginale attacked by the rust pathogen Melampsora lini. Journal of Ecology. 1995;83:199–206. [Google Scholar]
- Burdon JJ, Thrall PH. The fitness costs to plants of resistance to pathogens. Genome Biology. 2003;4:227. doi: 10.1186/gb-2003-4-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon JJ, Thrall PH. Coevolution at multiple scales: Linum marginale - Melampsora lini - from the individual to the species. Evolutionary Ecology. 2001;14:261–281. [Google Scholar]
- Burdon JJ, Thrall PH. Coevolution of plants and their pathogens in natural habitats. Science. 2009;324:755–756. doi: 10.1126/science.1171663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdon JJ, Thrall PH, Brown AHD. Resistance and virulence structure in two Linum marginale -Melampsora lini host-pathogen metapopulations with different mating systems. Evolution. 1999;53:704–716. doi: 10.1111/j.1558-5646.1999.tb05365.x. [DOI] [PubMed] [Google Scholar]
- Burdon JJ, Thrall PH, Ericson L. The current and future dynamics of disease in plant communities. Annual Review of Phytopathology. 2006;44:19–39. doi: 10.1146/annurev.phyto.43.040204.140238. [DOI] [PubMed] [Google Scholar]
- Burdon JJ, Thrall PH, Lawrence GJ. Coevolutionary patterns in the Linum marginale -Melampsora lini association at the continental scale. Canadian Journal of Botany. 2002;80:288–296. [Google Scholar]
- Burdon JJ, Wennström A, Elmqvist E, Kirby GC. The role of race specific resistance in natural plant populations. Oikos. 1996;76:411–416. [Google Scholar]
- Carlsson-Granér U. Anther-smut disease in Silene dioica: variation in susceptibility among genotypes and populations, and patterns of disease within populations. Evolution. 1997;5:1416–1426. doi: 10.1111/j.1558-5646.1997.tb01465.x. [DOI] [PubMed] [Google Scholar]
- Carlsson-Granér U, Thrall PH. The spatial distribution of plant populations, disease dynamics and evolution of resistance. Oikos. 2002;97:97–110. [Google Scholar]
- Clay K, Kover PX. The Red Queen hypothesis and plant/pathogen interactions. Annual Review of Phytopathology. 1996;34:29–50. doi: 10.1146/annurev.phyto.34.1.29. [DOI] [PubMed] [Google Scholar]
- Coltman DW, Pilkington JG, Smith JA, Pemberton JM. Parasite-mediated selection against inbred Soay sheep in a free-living island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- Davelos AL, Alexander HM, Slade NA. Ecological genetic interactions between a clonal host plant (Spartina pectinata) and associated rust fungi (Puccinia seymouriana and Puccinia sparganioides). Oecologia. 1996;105:205–213. doi: 10.1007/BF00328548. [DOI] [PubMed] [Google Scholar]
- de Meaux J, Cattan-Toupance I, Lavigne C, Langin T, Neema C. Polymorphism of a complex resistance gene candidate family in wild populations of common bean (Phaseolus vulgaris) in Argentina: comparison with phenotypic resistance polymorphism. Molecular Ecology. 2003;12:263–273. doi: 10.1046/j.1365-294x.2003.01718.x. [DOI] [PubMed] [Google Scholar]
- de Meaux J, Mitchell-Olds T. Evolution of plant resistance at the molecular level: ecological context of species interactions. Heredity. 2003;91:345–152. doi: 10.1038/sj.hdy.6800342. [DOI] [PubMed] [Google Scholar]
- de Nooij M, van Damme JMM. Variation in host susceptibility among and within populations of Plantago lanceolata infected by the fungus Phomopsis subordinaria (Desm.) Trav. Oecologia. 1988a;75:535–538. doi: 10.1007/BF00776417. [DOI] [PubMed] [Google Scholar]
- de Nooij M, van Damme JMM. Variation in the pathogenicity among and within populations of the fungus Phomopsis subordinaria infecting Plantago lanceolata. Evolution. 1988b;42:1166–1171. doi: 10.1111/j.1558-5646.1988.tb04177.x. [DOI] [PubMed] [Google Scholar]
- Desprez-Loustau M-L, Vitasse Y, Delzon S, Capdevielle X, Marcais B, Kremer A. Are plant pathogen populations adapted for encounter with their host? A case study of phenological synchrony between oak and an obligate fungal parasite along an altitudinal gradient. Journal of Evolutionary Biology. 2010;23:87–97. doi: 10.1111/j.1420-9101.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- DiLeone JA, Mundt CC. Effect of wheat cultivar mixtures on populations of Puccinia striiformis races. Plant Pathology. 1994;43:917–930. [Google Scholar]
- Dodds PN, Rathjen JP. Pathogen perception and responses in plant immunity. Nature Reviews Genetics. 2010 doi: 10.1038/nrg2812. in press. [DOI] [PubMed] [Google Scholar]
- Elmqvist T, Liu D, Carlsson U, Giles BE. Anther-smut infection in Silene dioica: variation in floral morphology and patterns of spore deposition. Oikos. 1993;68:207–216. [Google Scholar]
- Ericson L, Burdon JJ. Linking field epidemiological and individual plant resistance patterns in the Betula pubescens-Melampsoridium betulinum host-pathogen interaction. Oikos. 2009;118:225–232. [Google Scholar]
- Ericson L, Burdon JJ, Müller J. The rust pathogen Triphragmium ulmariae as a selective force affecting its host, Filipendula ulmaria. Journal of Ecology. 2002;90:167–178. [Google Scholar]
- Ericson L, Burdon JJ, Müller WJ. Spatial and temporal dynamics of epidemics of the rust fungus Uromyces valerianae on populations of its host Valeriana salina. Journal of Ecology. 1999;87:649–658. [Google Scholar]
- Espiau C, Riviere D, Burdon JJ, Gartner S, Daclinat B, Hasan S, Chaboudez P. Host-pathogen diversity in a wild system: Chondrilla juncea - Puccinia chondrillina. Oecologia. 1998;113:133–139. doi: 10.1007/s004420050361. [DOI] [PubMed] [Google Scholar]
- Faigon-Soverna A, Harmon FG, Storani L, Karayekov E, Staneloni RJ, Gassmann W, Mas P, Casal JJ, Kay SA, Yanovsky MJ. A constitutive shade-avoidance mutant implicates TIR-NBS-LRR proteins in Arabidopsis photomorphogenic development. Plant Cell. 2006;18:2919–2928. doi: 10.1105/tpc.105.038810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fels D, Kaltz O. Temperature dependent transmission and latency of Holospora undulata, a micronucleus-specific parasite of the ciliate Paramecium caudatum. Proceedings of the Royal Society of London Series B. 2006;273:1031–1038. doi: 10.1098/rspb.2005.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor HH. The complementary genic systems in flax and flax rust. Advances in Genetics. 1956;8:29–54. [Google Scholar]
- Frank SA. Coevolutionary genetics of plants and pathogens. Evolutionary Ecology. 1993;7:45–75. [Google Scholar]
- Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proceedings of the Royal Society of London Series B. 1996;263:1003–1009. [Google Scholar]
- Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, population size and generation time. Journal of Evolutionary Biology. 2002;15:451–462. [Google Scholar]
- Gomulkiewicz R, Thompson JN, Holt RD, Nuismer SL, Hochberg ME. Hot spots, cold spots, and the geographic mosaic theory of coevolution. American Naturalist. 2000;156:156–174. doi: 10.1086/303382. [DOI] [PubMed] [Google Scholar]
- Goss EM, Bergelson J. Variation in resistance and virulence in the interaction between Arabidopsis thaliana and a bacterial pathogen. Evolution. 2006;60:1562–1573. [PubMed] [Google Scholar]
- Hall SA, Allen RL, Baumber RE, Baxter LA, Fisher K, Bittner-Eddy PD, Rose LE, Holub EB, Beynon JL. Maintenance of genetic variation in plants and pathogens involves complex networks of gene-for-gene interactions. Molecular Plant Pathology. 2009;10:449–457. doi: 10.1111/j.1364-3703.2009.00544.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annual Review of Plant Physiology and Plant Molecular Biology. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hill AV. The immunogenetics of human infectious diseases. Annual Review of Immunology. 1998;16:593–617. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- Islam MR, Mayo GME. A compendium of host genes in flax conferring resistance to flax rust. Plant Breeding. 1990;104:89–100. [Google Scholar]
- Jarosz AM, Burdon JJ. Host-pathogen interactions in natural populations of Linum marginale and Melampsora lini: II. Local and regional variation in patterns of resistance and racial structure. Evolution. 1991;45:1618–1627. doi: 10.1111/j.1558-5646.1991.tb02667.x. [DOI] [PubMed] [Google Scholar]
- Jayakar SD. A mathematical model for interactions of gene frequencies in a parasite and its host. Theoretical Population Biology. 1970;1:140–164. doi: 10.1016/0040-5809(70)90032-8. [DOI] [PubMed] [Google Scholar]
- Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Jorgensen TH, Emerson BC. Functional variation in a disease resistance gene in populations of Arabidopsis thaliana. Molecular Ecology. 2008;17:4912–4923. doi: 10.1111/j.1365-294X.2008.03960.x. [DOI] [PubMed] [Google Scholar]
- Kaltz O, Gandon S, Michalakis Y, Shykoff JA. Local maladaptation in the anther-smut fungus Microbotryum violaceum to its host plant Silene latifolia: evidence from a cross-inoculation experiment. Evolution. 1999;53:395–407. doi: 10.1111/j.1558-5646.1999.tb03775.x. [DOI] [PubMed] [Google Scholar]
- Kniskern JM, Rausher MD. Major-gene resistance to the rust pathogen Coleosporium ipomoeae is common in natural populations of Ipomoea purpurea. New Phytologist. 2006;171:137–144. doi: 10.1111/j.1469-8137.2006.01729.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Resistance variation within and among host populations in a plant-pathogen metapopulation - implications for regional pathogen dynamics. Journal of Ecology. 2004;92:990–1000. [Google Scholar]
- Laine A-L. Spatial scale of local adaptation in a plant-pathogen metapopulation. Journal of Evolutionary Biology. 2005;18:930–938. doi: 10.1111/j.1420-9101.2005.00933.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Evolution of host resistance: looking for coevolutionary hotspots at small spatial scales. Proceedings of the Royal Society of London Series B. 2006;273:267–273. doi: 10.1098/rspb.2005.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laine A-L. Detecting local adaptation in a natural plant-pathogen metapopulation – laboratory vs. field transplant approach. Journal of Evolutionary Biology. 2007a;20:1665–1673. doi: 10.1111/j.1420-9101.2007.01359.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Pathogen fitness components and genotypes differ in their sensitivity to nutrient and temperature variation in a wild plant-pathogen association. Journal of Evolutionary Biology. 2007b;20:2371–2378. doi: 10.1111/j.1420-9101.2007.01406.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L. Temperature-mediated patterns of local adaptation in a natural plant-pathogen metapopulation. Ecology Letters. 2008;11:327–337. doi: 10.1111/j.1461-0248.2007.01146.x. [DOI] [PubMed] [Google Scholar]
- Laine A-L, Hanski I. Large-scale spatial dynamics of a specialist plant pathogen in a fragmented landscape. Journal of Ecology. 2006;94:217–226. [Google Scholar]
- Laine A-L, Tellier A. Heterogeneous selection promotes maintenance of polymorphism in host-parasite interactions. Oikos. 2008;117:1281–1288. [Google Scholar]
- Lambrechts L, Chavatte J-M, Snounou G, Koella JC. Environmental influence on the genetic basis of mosquito resistance to malaria parasites. Proceedings of the Royal Society of London Series B. 2006;273:1501–1506. doi: 10.1098/rspb.2006.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeda A, Petrzelova I, Maryska S. Structure and variation in the wild-plant pathosystem: Lactuca serriola-Bremia lactucae. European Journal of Plant Pathology. 2008;122:127–146. [Google Scholar]
- Leonard KJ. Selection pressures and plant pathogens. Annals of the New York Academy of Science. 1977;287:207–222. [Google Scholar]
- Littell RC, Milliken GA, Stroup WW, Wolfinger RD. SAS® system for mixed models. SAS Institute Inc; Cary, N. C.: 1996. [Google Scholar]
- Lockett SF, Robertson JR, Brettle RP, Yap PL, Middleton D, Leigh Brown AJ. Mismatched human leucocyte antigen alleles protect against heterosexual HIV transmission. Journal of Acquired Immune Deficiency Syndromes. 2001;27:227–280. doi: 10.1097/00126334-200107010-00010. [DOI] [PubMed] [Google Scholar]
- McIntosh RA, Wellings CR, Park RF. Wheat rusts: an atlas of resistance genes. CSIRO; Melbourne: 1995. [Google Scholar]
- Meagher S. Genetic diversity and Capillaria hepatica (Nematoda) prevalence in Michigan deer mouse populations. Evolution. 1999;53:1318–1324. doi: 10.1111/j.1558-5646.1999.tb04547.x. [DOI] [PubMed] [Google Scholar]
- Michelmore RW, Meyers BC. Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Research. 1998;8:1113–1130. doi: 10.1101/gr.8.11.1113. [DOI] [PubMed] [Google Scholar]
- Mitchell SE, Rogers ES, Little TJ, Read AF. Host-parasite and genotype-by-environment interactions: Temperature modifies potential for selection by a sterilizing pathogen. Evolution. 2005;59:70–80. [PubMed] [Google Scholar]
- Niemi L, Wennstrom A, Hjalten J, Waldmann P, Ericson L. Spatial variation in resistance and virulence in the host-pathogen system Salix triandra-Melampsora amygdalinae. Journal of Ecology. 2006;94:915–921. [Google Scholar]
- Nuismer SL, Gandon S. Moving beyond common-garden and transplant designs: Insight into the causes of local adaptation in species interactions. American Naturalist. 2008;171:658–668. doi: 10.1086/587077. [DOI] [PubMed] [Google Scholar]
- Nuismer SL, Thompson JN, Gomulkiewicz R. Coevolutionary clines across selection mosaics. Evolution. 2000;54:1102–1115. doi: 10.1111/j.0014-3820.2000.tb00546.x. [DOI] [PubMed] [Google Scholar]
- Parker MA. Local population differentiation for compatibility in an annual legume and its host-specific fungal pathogen. Evolution. 1985;39:713–723. doi: 10.1111/j.1558-5646.1985.tb00414.x. [DOI] [PubMed] [Google Scholar]
- Pedersen WL, Leath S. Pyramiding major genes for resistance to maintain residual effects. Annual Review of Phytopathology. 1988;26:369–378. [Google Scholar]
- Petrzelova I, Lebeda A. Distribution of race-specific resistance against Bremia lactucae in natural populations of Lactuca serriola. European Journal of Plant Pathology. 2010 In press. [Google Scholar]
- Price JS, Bever JD, Clay K. Genotype, environment and genotype by environment interactions determine quantitative resistance to leaf rust (Coleosporium asterum) in Euthamia graminifolia (Asteraceae). New Phytologist. 2004;162:729–743. doi: 10.1111/j.1469-8137.2004.01082.x. [DOI] [PubMed] [Google Scholar]
- Ridenhour BJ, Nuismer SL. Polygenic traits and parasite local adaptation. Evolution. 2007;61:368–376. doi: 10.1111/j.1558-5646.2007.00029.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg MS, Adams DC, Gurevitch J. Metawin, version 2.0: statistical software for meta-analysis. Sinauer; Sunderland, MA: 2000. [Google Scholar]
- Roslin T, Gripenberg S, Salminen J-P, Karonen M, O'Hara RB, Pihlaja K, Pulkkinen P. Seeing the trees for the leaves - oaks as mosaics for a host-specific moth. Oikos. 2006;113:106–120. [Google Scholar]
- Roslin T, Laine A-L, Gripenberg S. Spatial population structure in an obligate plant pathogen colonizing oak (Quercus robur). Functional Ecology. 2007;21:1168–1177. [Google Scholar]
- Salvaudon L, Giraud T, Shykoff JA. Genetic diversity in natural populations: a fundamental component of plant–microbe interactions. Current Opinion in Plant Biology. 2008;11:135–143. doi: 10.1016/j.pbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Schiff CL, Wilson IW, Somerville SC. Polygenic powdery mildew disease resistance in Arabidopsis thaliana: quantitative trait analysis of the accession Warschau-1. Plant Pathology. 2001;50:690–701. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid B. Effects of genetic diversity in experimental stands of Solidago altissima - evidence for the potential role of pathogens as selective agents in plant populations. Journal of Ecology. 1994;82:165–175. [Google Scholar]
- Sicard D, Pennings PS, Grandclément C, Acosta J, Kaltz O, Shykoff JA. Specialization and local adaptation of a fungal parasite on two host plant species as revealed by two fitness traits. Evolution. 2007;61:27–41. doi: 10.1111/j.1558-5646.2007.00003.x. [DOI] [PubMed] [Google Scholar]
- Silvertown JW, Charlesworth D. Introduction to plant population biology. Oxford: 2001. [Google Scholar]
- Smith DL, Ericson L, Burdon JJ. Epidemiological patterns at multiple spatial scales: an 11-year old study of a Triphragmium ulmariae-Filipendula ulmaria metapopulation. Journal of Ecology. 2003;91:890–903. [Google Scholar]
- Soubeyrand S, Laine A-L, Hanski I, Penttinen A. Spatio-temporal structure of interactions in a host-pathogen metapopulation. American Naturalist. 2009;174:308–320. doi: 10.1086/603624. [DOI] [PubMed] [Google Scholar]
- Springer Y. Clinal resistance structure and pathogen local adaptation in a serpentine flax-flax rust interaction. Evolution. 2007;61:1812–1822. doi: 10.1111/j.1558-5646.2007.00156.x. [DOI] [PubMed] [Google Scholar]
- Thomas MB, Blanford S. Thermal biology in insect-parasite interactions. Trends in Ecology and Evolution. 2003;18:344–350. [Google Scholar]
- Thompson JN. Specific hypothesis on the geographic mosaic of coevolution. American Naturalist. 1999;153:S1–S14. [Google Scholar]
- Thompson JN. The Geographic mosaic of coevolution. The University of Chicago Press; Chicago: 2005. [Google Scholar]
- Thompson JN, Burdon JJ. Gene-for-gene coevolution between plants and parasites. Nature. 1992;360:121–125. [Google Scholar]
- Thrall PH, Antonovics J. Theoretical and empirical studies of metapopulations: population and genetic dynamics of the Silene-Ustilago system. Canadian Journal of Botany. 1995;73:S1249–S1258. [Google Scholar]
- Thrall PH, Burdon JJ. Host-pathogen dynamics in a metapopulation context: the ecological and evolutionary consequences of being spatial. Journal of Ecology. 1997;85:743–753. [Google Scholar]
- Thrall PH, Burdon JJ. Effect of resistance variation in a natural plant host-pathogen metapopulation on disease dynamics. Plant Pathology. 2000;49:767–773. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of gene-for-gene systems in metapopulations: the effect of spatial scale of host and pathogen dispersal. Plant Pathology. 2002;51:169–184. [Google Scholar]
- Thrall PH, Burdon JJ. Evolution of virulence in a plant host-pathogen metapopulation. Science. 2003;299:1735–1737. doi: 10.1126/science.1080070. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ, Bever JD. Local adaptation in the Linum marginale-Melampsora lini host-pathogen interaction. Evolution. 2002;56:1340–1351. doi: 10.1111/j.0014-3820.2002.tb01448.x. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Burdon JJ, Young A. Variation in resistance and virulence among demes of a plant host-pathogen metapopulation. Journal of Ecology. 2001;89:736–748. [Google Scholar]
- Thrall PH, Jarosz AM. Host-pathogen dynamics in experimental populations of Silene alba and Ustilago violacea. II. Experimental tests of theoretical models. Journal of Ecology. 1994;82:561–570. [Google Scholar]
- Tian D, Traw MB, Chen JQ, Kreitman M, Bergelson J. Fitness costs of R-gene mediated resistance in Arabidopsis thaliana. Nature. 2003;423:74–77. doi: 10.1038/nature01588. [DOI] [PubMed] [Google Scholar]
- Vale PF, Stjernman M, Little TJ. Temperature-dependent costs of parasitism and maintenance of polymorphism under genotype-by-environment interactions. Journal of Evolutionary Biology. 2008;21:1318–1427. doi: 10.1111/j.1420-9101.2008.01555.x. [DOI] [PubMed] [Google Scholar]
- Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH. Rapid transcriptome characterization for a non-model organism using massively parallel 454 pyrosequencing. Molecular Ecology. 2007;17:1636–1647. doi: 10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- Wilson IW, Schiff CL, Hughes DE, Somerville SC. Quantitative trait loci analysis of powdery mildew disease resistance in the Arabidopsis thaliana accession Kashmir-1. Genetics. 2001;158:1301–1309. doi: 10.1093/genetics/158.3.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe MS. The current status and prospects of multiline cultivars and variety mixtures for disease resistance. Annual Review of Phytopathology. 1985;23:251–273. [Google Scholar]
- Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nature Genetics. 2002;32:569–557. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- Xiao SY, Ellwood S, Calis O, Patrick E, Li T, Coleman M, Turner JG. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science. 2001;291:118–120. doi: 10.1126/science.291.5501.118. [DOI] [PubMed] [Google Scholar]
- Xiao SY, Ellwood S, Findlay K, Oliver RP, Turner JG. Characterization of three loci controlling resistance of Arabidopsis thaliana accessions Ms-0 to two powdery mildew diseases. Plant Journal. 1997;12:757–768. doi: 10.1046/j.1365-313x.1997.12040757.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto E, Takashi T, Morinaka Y, Lin S, Wu J, Matsumoto T, Kitano H, Matsuoka M, Ashikari M. Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Molecular Genetics and Genomics. 2010;283:305–315. doi: 10.1007/s00438-010-0514-y. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Chen H, Fan J, Wang Y, Li Y, Chen J, Fan J, Yang S, Hus l., Leung H, Mew TW, Teng PS, Wang Z, Mundt CC. Genetic diversity and disease control in rice. Nature. 2000;406:718–722. doi: 10.1038/35021046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Additional supporting information may be found in the online version of this article:
Table S1. Summary of studies that were used for analysis of how host and pathogen life history features may affect host resistance.
Table S2. ANOVA results for how life history and origin of pathogen affect host population resistance.