Abstract
Coordination of mitosis and cytokinesis is crucial for ensuring proper chromosome segregation and genomic stability. In Schizosaccharomyces pombe, the sid genes (cdc7, cdc11, cdc14, spg1, sid1, sid2 and sid4) define a signaling pathway that regulates septation and cytokinesis. Here we describe the characterization of a novel protein kinase, Sid1p. Sid1p localizes asymmetrically to one spindle pole body (SPB) in anaphase. Sid1p localization is maintained during medial ring constriction and septum synthesis and disappears prior to cell separation. Additionally, we found that Cdc14p is in a complex with Sid1p. Epistasis analysis places Sid1p–Cdc14p downstream of Spg1p–Cdc7p but upstream of Sid2p. Finally, we show that cyclin proteolysis during mitosis is unaffected by inactivating the sid pathway; in fact, loss of Cdc2–cyclin activity promotes Sid1p–Cdc14p association with the SPB, possibly providing a mechanism that couples cytokinesis with mitotic exit.
Keywords: cdc7/cdc14/cytokinesis/Schizosaccharomyces pombe/sid1
Introduction
Cytokinesis occurs immediately following mitosis and results in irreversible cell cleavage. Mitosis and cytokinesis are tightly regulated both temporally and spatially so that cleavage occurs between separated chromosomes and not before mitotic spindle assembly and chromosome segregation. This ensures that each daughter cell receives only one copy of the genome. Cellular signals that regulate mitotic events have been studied extensively. Cellular mechanisms that coordinate mitosis and cytokinesis are less well defined.
The fission yeast Schizosaccharomyces pombe has proven to be an excellent model organism for studying cytokinesis (for reviews, see Gould and Simanis, 1997; Le Goff et al., 1999). Like animal cells, S.pombe divides through use of an actin–myosin-based contractile ring. During interphase, F-actin patches localize to the growing ends of cells. In early mitosis, the medial ring, which is composed of F-actin, myosin II and numerous other proteins, forms at the cell center. F-actin patches subsequently re-localize to the medial ring. Once anaphase is complete, the spindle disassembles, the medial ring constricts and synthesis of a septum is initiated. Follow ing septum formation, primary septal material is degraded and daughter cells physically separate.
At least eight genes are involved in triggering cytokinesis in S.pombe. They include the sid genes (septation initiation-deficient), cdc7, cdc11, cdc14, spg1, sid1, sid2 and sid4 (Nurse et al., 1976; Schmidt et al., 1997; Balasubramanian et al., 1998), as well as plo1 kinase (Ohkura et al., 1995). Under restrictive conditions, cells bearing temperature-sensitive alleles of the sid genes show normal medial ring assembly and redistribution of actin patches, but fail to initiate ring constriction and septum deposition. These cells proceed through additional nuclear division cycles without septation, becoming long and multinucleate before eventually lysing (Fankhauser et al., 1995; Balasubramanian et al., 1998). Plo1p seems to play a role in cytokinesis since it is required for septum formation and its overexpression in interphase results in septum formation (Ohkura et al., 1995).
Genetic and biochemical studies suggest that the sid gene products function in a novel signaling cascade regulating cytokinesis, yet their order of function is unclear (Marks et al., 1992; Balasubramanian et al., 1998; Sohrmann et al., 1998). Based on localization studies, at least part of this cascade appears to function by transducing septum-promoting signals from spindle pole bodies (SPBs) to the division site. Spg1p, a small GTPase of the ras superfamily, localizes to SPBs throughout interphase in an active GDP-bound form. Upon entry into mitosis, Spg1p is converted to an active GTP-bound state until anaphase, when conversion of Spg1p–GTP to Spg1p–GDP occurs at one SPB (Sohrmann et al., 1998). Bry4p and Cdc16p function together as the GTPase-activating protein (GAP) for Spg1p (Furge et al., 1998); however, the GDP/GTP exchange factor (GEF) has not yet been identified. Moreover, Byr4p/Cdc16p co-localizes with Spg1p–GDP but not Spg1p–GTP (Cerutti and Simanis, 1999). The key function of Spg1p–GTP appears to be in localizing Cdc7p kinase to the SPB during mitosis since Cdc7p kinase activity does not seem to be regulated (Sohrmann et al., 1998). Sid2p kinase is localized to the single SPB in interphase, both SPBs during mitosis and transiently to the cleavage site during medial ring constriction and septation. Sid2p kinase activity peaks during late anaphase and its localization and kinase activity require the other sid genes, suggesting that it functions late in the cascade (Sparks et al., 1999). Cdc14p has been cloned but does not display significant sequence similarity to any known proteins (Fankhauser and Simanis, 1993). Overproduction of Cdc14p results in a G2–M block; however, Cdc14p localization and where it functions in the sid pathway have not been described.
In this study, we investigated the role of S.pombe Sid1p and Cdc14p in the sid pathway. We show that Sid1p is a PAK/GC family protein kinase that functions in a complex with Cdc14p, downstream of Spg1p–Cdc7p and upstream of Sid2p. We also show that Sid1p localization may be regulated by Cdc2–cyclin activity, suggesting a mechanism that may couple cytokinesis with completion of mitosis.
Results
Cloning and disruption of sid1
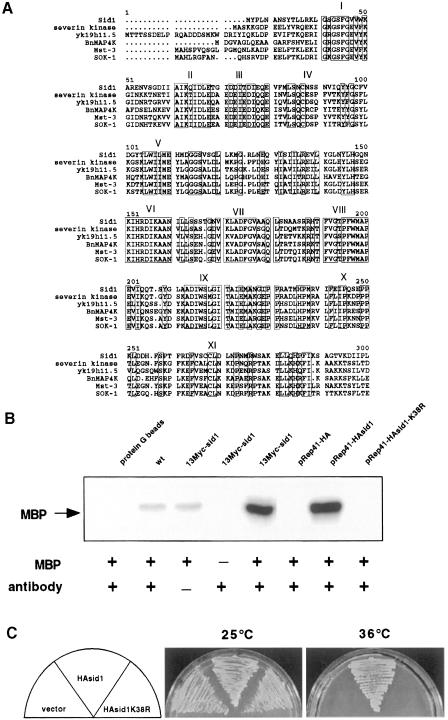
The sid1 gene was cloned by complementation of a sid1-239 temperature-sensitive mutant. sid1 corresponded to an open reading frame with GenBank accession No. CAB11493. sid1 encodes a 471 amino acid protein kinase of the PAK/GC kinase family (for a review, see Sells and Chernoff, 1997). A sequence comparison of the catalytic domain of Sid1p and catalytic domains from related proteins is shown in Figure 1A. Sid1p is most related to GC kinases that have an N-terminal kinase domain and a C-terminal half with little homology to known proteins. A null allele of sid1 was constructed using a one-step gene replacement strategy in a diploid strain, and sid1 was found to be essential for viability (see Materials and methods). sid1 null cells become long and multinucleate, identical to the phenotype of cells with sid1 temperature-sensitive alleles at restrictive temperature (data not shown).
Fig. 1. Kinase activity is essential. (A) Alignment of the catalytic domain of Sid1p with the catalytic domains of the most closely related PAK/GC family members as determined by the BLAST and MULTALIN programs. Relatives include severin kinase from Dictyostelium discoideum (AAC24522), Caenorhabditis elegans contig yk19h11.5 (AAC9038), BnMAPK4 from Brassica napus (CAA08757), human Ste20-like kinase Mst-3 (4505261) and human oxidant stress-activated Ste20-like kinase SOK-1 (5454174). Roman numerals indicate the 11 subdomains characteristic ofSer/Thr protein kinases (Hanks et al., 1988). Boxes enclose conserved residues. (B) Immune complex kinase assays were performed using Myc or HA antibodies as appropriate (see Materials and methods) on lysates prepared from wild-type cells, or cells expressing 13Myc-Sid1p, HA-Sid1p or HA-Sid1-K38R. Assays were carried out with (+) or without (–) antibody or the artificial substrate MBP. A control assay was also carried out in the absence of cell lysate (protein G beads). (C) sid1-125 cells (YDM435) transformed with vector alone or with plasmids expressing either HA-Sid1p or HA-Sid1-K38R were plated at 25 or 36°C.
Sid1p is a protein kinase
To determine if Sid1p displayed kinase activity in vitro, we constructed a strain that expressed a 13Myc epitope-tagged version of Sid1p from the chromosomal sid1 locus. 13Myc-Sid1p immune complexes were prepared from cell lysates and kinase assays were performed using myelin basic protein (MBP) as a substrate. Immune complexes prepared from 13Myc-sid1 cell lysates displayed high kinase activity. To confirm that kinase activity is specific to Sid1p, a hemagglutinin (HA)-tagged kinase-dead version of Sid1p (HA-Sid1p-K38R) was constructed and expressed from the pREP41 vector under the control of the thiamine-repressible nmt1 promoter (see Materials and methods). Kinase activity was associated with HA-Sid1p immune complexes (Figure 1B) and was absent in preparations from cells expressing kinase-dead HA-Sid1-K38R or the empty plasmid vector (Figure 1B). We next tested the ability of the HA-Sid1-K38R mutant to rescue the sid1-125 temperature-sensitive defect. As shown in Figure 1C, only the wild-type version of Sid1p rescued the sid1-125 phenotype.
Localization of Sid1p
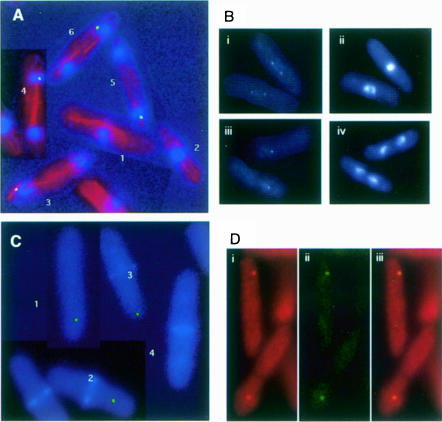
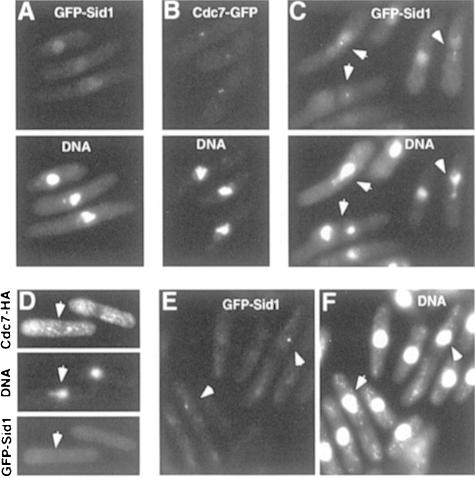
Cells expressing green fluorescent protein (GFP)–Sid1p from the chromosomal locus were stained with antibodies against tubulin to assess clearly the cell cycle-specific intracellular distribution of Sid1p. In cells with an inter phase array of microtubules (Figure 2A, cells 1 and 2), diffuse Sid1p signal was observed. In contrast, cells in anaphase (Figure 2A, cells 3–5) or telophase (Figure 2A, cell 6) exhibited strong GFP signal at one SPB. Because weak GFP signals are often difficult to see following immunofluorescence processing, we also examined cells just prior to processing. As before, interphase cells displayed diffuse GFP–Sid1p signal (data not shown), while anaphase or telophase cells showed strong GFP–Sid1p signal at one SPB (Figure 2B, iii and iv). Interestingly, GFP–Sid1p could be seen faintly at both SPBs in metaphase or early anaphase (Figure 2B, i and ii). To determine the temporal relationship between Sid1p appearance at the SPB and initiation of septation, we stained GFP–Sid1p-expressing cells with calcofluor, which preferentially stains the septum. Sid1p arrived at the SPB prior to initiation of septation since cells could be observed that had not yet initiated septation but had GFP–Sid1p signal at one SPB (Figure 2C, cell 1). Sid1p was always observed at one SPB in septating cells that had not completed septum formation (Figure 2C, cells 2 and 3). Once cells had completed septum formation but prior to cell separation, Sid1p disappeared from the SPB (Figure 2C, cell 4). Cdc7p, after localizing initially to both SPBs during mitosis, also localizes to only one SPB in late anaphase (Sohrmann et al., 1998). We tested whether Sid1p localized to the same SPB as Cdc7p. Cells expressing both Cdc7-HA (Figure 2D, i) and GFP–Sid1p (Figure 2D, ii) gave signals that overlapped (Figure 2D, iii), suggesting that Cdc7p and Sid1p localize to the same SPB during septum formation.
Fig. 2. Localization of GFP–Sid1p. (A) Cells expressing GFP–Sid1p (green) were processed for immunofluorescence and stained for tubulin (red) and DNA (blue). Numbers indicate cells at stages of the cell cycle described in the text. (B) GFP–Sid1p cells in early (i and ii) and late (iii and iv) anaphase were fixed and stained for DNA. Panels i and iii show GFP–Sid1p fluorescence, and the corresponding DNA fluorescence is displayed in panels ii and iv. (C) Cells were fixed and stained for septum material (blue) with calcofluor. (D) Cells expressing Cdc7p-HA and GFP–Sid1p were fixed and processed for immunofluorescence using HA antibodies. Cdc7-HA fluorescence (i) and GFP–Sid1p fluorescence (ii) were merged (iii) to show co-localization of the two proteins.
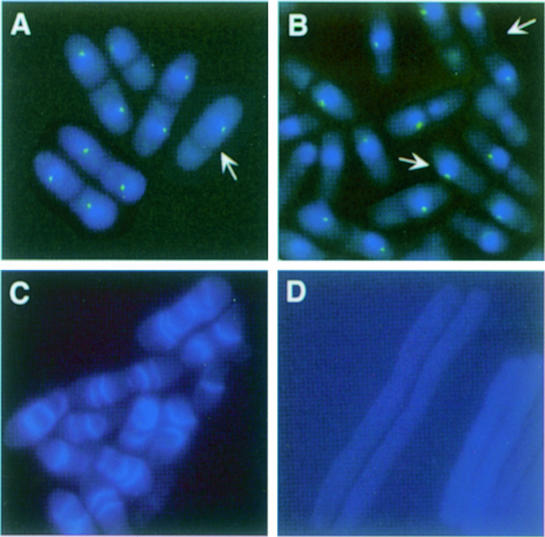
We next tested whether factors that affect Cdc7p localization also affect Sid1p localization. Inactivation of cdc16 results in localization of Cdc7p to both SPBs in late anaphase, an effect that is also observed upon overexpression of Spg1p (Sohrmann et al., 1998). Furthermore, overexpression of Spg1p results in inappropriate recruitment of Cdc7p to the SPB in interphase cells. Like Cdc7p, Sid1p localized to both SPBs in cdc16-116 cells (Figure 3A) and in cells overexpressing Spg1p (Figure 3B; see arrows), as well as to the SPB in interphase cells overexpressing Spg1p (Figure 3B). After shift to restrictive temperature, cdc16-116 cells display two phenotypes termed type I and type II cells (Minet et al., 1979; Cerutti and Simanis, 1999). Type I cells have two nuclei and make multiple septa. Type II cells have a single nucleus and a septa. It has been proposed that the type II cells immediately septate again after division because they inherit the SPB that contains active Spg1p (Cerutti and Simanis, 1999). In support of this hypothesis, Sid1p was present at the SPB in type II cells (Figure 3A; see arrow). These observations suggest that Sid1p may function downstream of Spg1p. Consistent with this model, Spg1p overexpression causes wild-type cells to form multiple septa (Schmidt et al., 1997) (Figure 3C); however, septa are not formed when Spg1p is overexpressed in a sid1-125 mutant (Figure 3D).
Fig. 3. spg1 overexpression or cdc16 inactivation cause Sid1p to localize to the SPB. (A) GFP–sid1 cdc16-116 cells (YDM560) were grown at 25°C, shifted to 36°C for 2 h and then fixed. Cells were stained for DNA (blue), and images of the DNA staining and the GFP–Sid1p signal (green) were captured. (B) spg1 expression was induced for 16 h in nmt1-spg1 GFP–sid1 cells (YDM564) and images of DNA (blue) and GFP–Sid1p (green) were captured. (C) nmt1-spg1 (YDM551) or (D) sid1-125 nmt1-spg1 cells (YDM566) were grown to mid-log phase at 25°C in medium containing thiamine. The culture was then shifted to medium lacking thiamine, grown at 25°C for an additional 23 h and subsequently placed at 36°C for 5 h. Cells were then fixed and stained with calcofluor.
Dependencies of localization of Sid1p, Cdc7p and Cdc14p
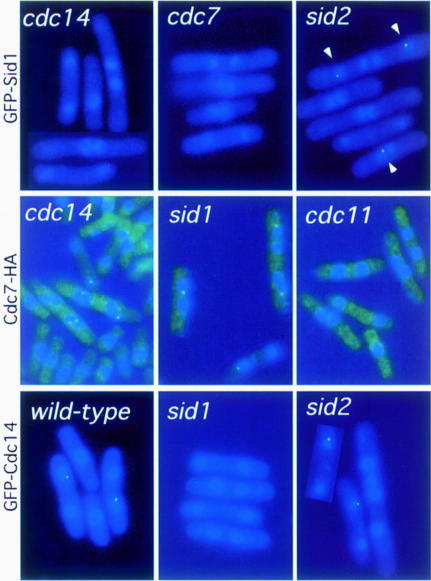
To determine where Sid1p functions with respect to the other sid gene products, we examined whether GFP–Sid1p localized to the SPB in other sid mutants. cdc7-24, cdc11-123, cdc14-118, sid2-250, spg1-106 and sid4-A1 cells expressing GFP–Sid1p were shifted to restrictive temperature and localization of GFP–Sid1p was assessed. Sid1p localized to the SPB in 12% (32/227) of wild-type cells and 22% (69/240) of sid2-250 mutants; however, examination of >200 cells showed that Sid1p was unable to localize in cdc7, cdc11, cdc14, spg1 or sid4 mutant cells (Figure 4; Table I; data not shown). This is consistent with Sid1p functioning upstream of Sid2p but downstream of the other sid proteins. Similar experiments examining Cdc7p localization in other mutant backgrounds showed that Cdc7p cannot localize to the SPB(s) in cdc11 (Figure 4; Table I) and spg1 (Sohrmann et al., 1998) mutant cells (>200 cells examined). In contrast, Cdc7 localized to the SPBs in 27% (47/124) of cdc14 mutant cells and in 27% (102/277) of sid1 mutant cells, which is similar to that seen in sid2 mutants at permissive and restrictive temperature (28 and 53%, respectively; Sparks et al., 1999; Figure 4; Table I). These results indicate that Cdc14p, Sid1p and Sid2p probably function downstream of Cdc7p, with Cdc14p either functioning upstream of or in conjunction with Sid1p.
Fig. 4. Dependencies for localization of Sid1p, Cdc14p and Cdc7p. The localizations of GFP–Sid1p, Cdc7-HA and GFP–Cdc14p were examined in wild-type and different sid mutant cells after incubation at 36°C for 2 h. All samples were fixed and stained for DNA (blue), GFP–Sid1p and GFP–Cdc14p signal, and Cdc7-HA staining is shown in green. Cdc7-HA cells have a bright green haze as a result of background staining from the secondary antibody.
Table I. Dependencies for localization of Cdc7p, Sid1p and Cdc14p.
| Cdc7p localization | Sid1p localization | Cdc14p localization | |
|---|---|---|---|
| sid2-250 | + | + | + |
| sid1-125 | + | – | |
| cdc14-118 | + | – | |
| cdc7-24 | – | – | |
| cdc11-123 | – | – | – |
| spg1-106 | n.d. | – | – |
| sid4-A1 | n.d. | – | – |
+ and – denote whether or not the protein was able to localize properly in the given mutant background. n.d. indicates that this particular strain was not assayed.
Cdc14p localizes to one spindle pole body during septum formation
Previous studies have shown synthetic lethal interactions between sid1-239 and cdc14-118 (Balasubramanian et al., 1998). Furthermore, our epistasis studies suggest that Cdc14p functions just upstream of or at the same point as Sid1p. Therefore, we further characterized the relationship between Sid1p and Cdc14p. We first examined the intracellular distribution of Cdc14p in a strain expressing an integrated copy of GFP–Cdc14p from the Cdc14 promoter. In wild-type cells, GFP–Cdc14p had an identical localization pattern to that of Sid1p. Cdc14p localized to a single spot on the nuclear periphery of cells at the end of anaphase or telophase (Figure 4). GFP–Cdc14p presumably localized to the same SPB as Sid1p and Cdc7p, but we were unable to confirm this since the signal did not survive immunofluorescence processing. To circumvent this problem, a strain was constructed that expressed a GFP-tagged version of both Sid1p and Cdc14p. Similarly to cells expressing GFP–Sid1p or GFP–Cdc14p alone, all late anaphase cells (30/30) expressing both GFP–Sid1p and GFP–Cdc14p showed GFP signal at just one SPB, suggesting that Sid1p and Cdc14p co-localize. Examination of Cdc14p localization in different mutant backgrounds revealed that Cdc14p localized normally to the SPB in sid2 mutant cells (30/139) compared with wild type (22/199), and was unable to localize in sid1, sid4, spg1, cdc7 and cdc11 mutant cells (Figure 4; Table I; data not shown). Thus it appears that Sid1p and Cdc14p may function together since they depend on each other for localization.
Cdc14p and Sid1p show physical interaction
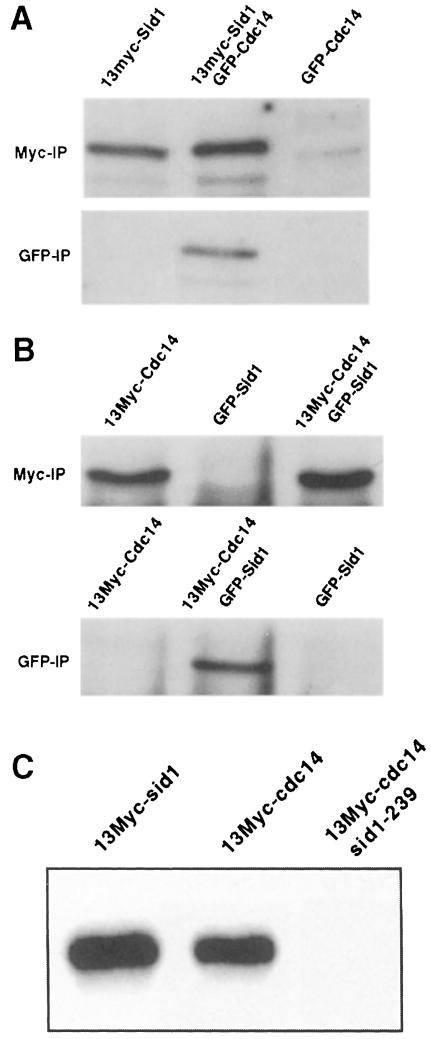
Since localization experiments suggested that Sid1p and Cdc14p may function together, we tested whether the two proteins formed a complex. To facilitate these studies, we constructed strains expressing epitope-tagged versions of both Sid1p and Cdc14p (see Materials and methods). Examination of lysates prepared from cells expressing 13Myc-Sid1p and GFP–Cdc14p showed that 13Myc-Sid1p could be co-immunoprecipitated with GFP–Cdc14p (Figure 5A). When immunoprecipitations were immunoblotted with anti-GFP antibodies, we did not detect GFP–Cdc14p in either 13Myc-Sid1p immune complexes or control GFP–Cdc14p immune complexes (data not shown). We presumed that this was because Western blotting using anti-GFP antibody was not as sensitive as when anti-Myc antibody was used against the 13Myc tag. To test this, we switched the tags and prepared lysates from cells expressing 13Myc-Cdc14p and GFP–Sid1p. Samples were analyzed as above and we found that 13Myc-Cdc14p co-immunoprecipitated with GFP–Sid1p (Figure 5B). Consistent with the observation that Cdc14p and Sid1p form a complex, we found that Cdc14p has Sid1-dependent associated kinase activity (Figure 5C).
Fig. 5. Sid1p and Cdc14p associate in vivo. (A) Lysates were prepared from cells expressing 13Myc-Sid1p (YDM617), GFP–Cdc14p (YDM627) or both (YDM629). (B) Lysates were prepared from cells expressing 13Myc-Cdc14p (YDM617), GFP–Sid1p (YDM558) or both (YDM626). Each lysate was split in two, and immunoprecipitations were performed using anti-Myc or anti-GFP antibodies. Immune complexes were analyzed by Western blotting using anti-Myc antibodies. (C) Wild-type cells expressing 13Myc-Sid1p (YDM587) or 13Myc-Cdc14p (YDM617), or sid1-239 (YDM637) cells expressing 13Myc-Cdc14p were started at 25°C, shifted to 36°C for 3 h, and kinase assays were performed on 13Myc-Cdc14 or 13Myc-Sid1 immune complexes from these cells.
Sid1p cell cycle regulation
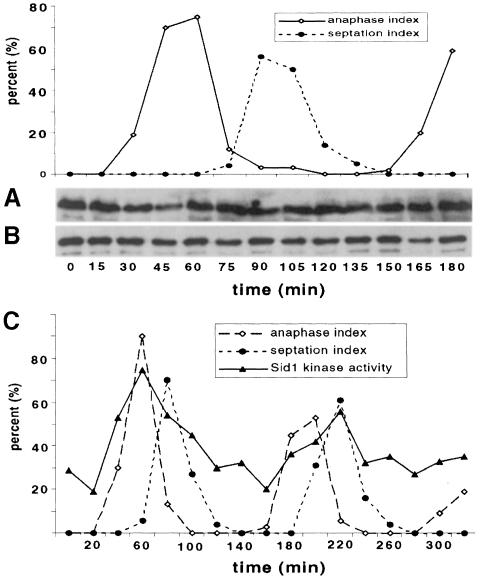
We next tested if Sid1p and Cdc14p protein levels or their ability to form a complex fluctuated throughout the cell cycle. cdc25-22 13Myc-sid1 GFP–cdc14 cells were synchronized by shifting them to 36°C to block them in G2, then they were released by returning them to permissive temperature (25°C). Samples were collected every 15 min and lysates were prepared and analyzed directly for Sid1p protein (Figure 6A) or subjected to co-immunoprecipitation experiments (Figure 6B). Western analysis of lysates revealed that Sid1p (Figure 6A) and Cdc14p (data not shown) protein levels remain unchanged throughout the cell cycle. Moreover, Western analysis of GFP–Cdc14p immune complexes with Myc antibody revealed that 13Myc-Sid1p remained in a complex with GFP–Cdc14 throughout the cell cycle (Figure 6B). These results suggest that neither the levels of Sid1p nor its association with Cdc14p are regulated in a cell cycle-dependent manner.
Fig. 6. Sid1p kinase activity, but not protein level or ability to co-immunoprecipitate with Cdc14p, is cell cycle regulated. (A and B) cdc25-22 13myc-sid1 GFP–cdc14 cells were released in synchrony from a Cdc25-dependent G2 block. Cells were collected at 15 min intervals and scored for the percentage of total cells in anaphase or septation. (A) Western analysis was performed directly on lysates with Myc antibody to detect Sid1p. (B) GFP–Cdc14 immune complexes were prepared from lysates using GFP antibody and subjected to Western analysis with Myc antibody. (C) cdc25-22 13Myc-sid1 cells were released from a G2, samples were collected at 20 min intervals and kinase assays were performed on 13Myc-Sid1p immune complexes. Kinase activity is displayed in arbitrary units.
Cdc7p kinase remains active throughout the cell cycle (Sohrmann et al., 1998) while Sid2p kinase activity peaks at the end of anaphase (Sparks et al., 1999). Therefore, we wondered if Sid1p kinase activity was cell cycle regulated. cdc25-22 13Myc-sid1 cells were released from a G2 block as described above, and samples were collected at 20 min intervals through two rounds of the cell cycle. Kinase assays were then performed on 13Myc-Sid1p immune complexes. Sid1p kinase activity peaked at anaphase/telophase (Figure 6C), indicating that Sid1p becomes more active at around the same time that it localizes to the SPB (Figure 2). This suggests that Sid1p kinase activity is cell cycle regulated.
Sid1p recruitment to the SPB may be triggered by cyclin destruction
Our localization studies suggest that the Sid1p–Cdc14p complex functions immediately downstream of Cdc7p and requires Cdc7p to localize to the SPB. However, the mere presence of Cdc7p at the SPB is not sufficient to recruit Sid1p–Cdc14p since Cdc7p appears at the SPB(s) at the beginning of mitosis and Sid1p–Cdc14p does not localize to the SPB until anaphase. We reasoned that some other signal, perhaps spindle elongation or anaphase proteolysis events, might contribute to Sid1p–Cdc14p recruitment. To address this issue, we looked at Sid1p and Cdc7p localization in the tubulin mutant nda3-km311, which arrests in a pre-metaphase state, unable to form a mitotic spindle. After 4 h at restrictive temperature, 70% of nda3-km311 cells become arrested in mitosis as judged by the presence of condensed chromatin and absence of septa (Table II). GFP–Cdc7p signal was observed at the SPB(s) in 69% of cells, consistent with GFP–Cdc7p localizing to the SPB(s) in cells blocked in mitosis (Table II; Figure 7B). In contrast, GFP–Sid1p signal was not observed at SPBs in these cells although faint nuclear signal was observed (Figure 7A). This suggests that some aspect of completion of mitosis is required in order for Sid1p to localize. Mitotic progression in nda3-km311 mutants is prevented by the spindle assembly checkpoint. We therefore addressed whether GFP–Sid1p localized in septating nda3-km311 cells under conditions in which the spindle assembly checkpoint was rendered inactive by introduction of a mad2 mutation in the nda3-km311 GFP–sid1 strain. Unlike nda3-km311 mutants alone, double mutant cells enter mitosis but are unable to inhibit anaphase proteolysis events. This triggers mitotic exit and cytokinesis, often resulting in cleavage of the undivided nucleus (He et al., 1997). We found that after 4 h at restrictive temperature, only 6% of these cells were in mitosis as judged by condensed chromatin, 46% had septated without undergoing chromosome separation, and GFP–Sid1p could be observed in 50% of the aberrantly septating cells (Table II; Figure 7C, see arowheads). GFP–Sid1p was not observed in every aberrantly septated cell since many cells had presumably finished septation and Sid1p normally disappears from the SPB once the septum has formed. We conclude that Sid1p localization to the SPB requires anaphase-induced proteolysis events but not chromosome segregation and spindle elongation.
Table II. Cdc7p–GFP and GFP–Sid1p localization in an nda3 block.
| Strain | % Condensed chromatin | % Pole Staining | % Septa |
|---|---|---|---|
| cdc7-GFP nda3-km311 | 70 | 69 | 1 |
| GFP–sid1 nda3-km311 | 71 | 0 | 0 |
| GFP–sid1 nda3-km311 mad2Δ | 6 | 23 | 46 |
For all data points, at least 200 cells were scored.
Fig. 7. Cdc2p inactivation triggers localization of Sid1p to the SPB. The nda3-km311 GFP–sid1 (A), nda3-km31 GFP–cdc7 (YDM667) (B) and nda3-km31 GFP–sid1 mad2Δ (YDM555) (C) strains were grown at 30°C and then shifted to 19°C for 4 h, fixed, stained for DNA and the GFP signals were visualized. (D–F) cdc2-33 GFP–sid1 cdc7-HA cells (YDM651) containing a nmt1-mad2 plasmid were grown at 25°C in the presence of thiamine, then thiamine was removed to induce mad2 expression and cells were allowed to continue growth at 25°C for 20 h. At this time, a portion of the culture was fixed and stained for DNA, Cdc7-HA and GFP–Sid1p. The rest of the culture was shifted to 36°C for 50 min, fixed and stained for DNA (F) and the GFP–Sid1p signal was visualized (E). Arrowheads indicate cells described in the text.
Multiple proteins, such as Cut2p (Funabiki et al., 1996) and the B-type cyclin Cdc13p (Yamano et al., 1996), are ubiquitylated and degraded in anaphase; therefore, we attempted to determine which proteins need to be destroyed for Sid1p to localize to the SPB. Previous experiments suggested that inactivation of Cdc2p kinase is essential for cytokinesis (He et al., 1997). He et al. showed that overexpression of mad2 resulted in a metaphase block. If mad2 was overexpressed in a temperature-sensitive cdc2 or cdc13 mutant strain at permissive temperature (25°C), cells arrested in metaphase. When arrested cells were shifted to restrictive temperature (36°C), they exited mitosis without executing anaphase and underwent cytokinesis. These observations suggest that loss of Cdc2p kinase activity, presumably brought about by cyclin degradation, is essential for triggering cytokinesis. We asked if GFP–Sid1p could be targeted to the SPB by inactivating cdc2 in Mad2p-overproducing cells. mad2 was overexpressed in cdc2-33 cells that also expressed Cdc7p-HA and GFP–Sid1p. Cells were stained with anti-tubulin or anti-HA monoclonal antibodies. It was reported previously that mad2 overexpression in cdc2-33 cells at permissive temperature resulted in 50% of cells becoming blocked in metaphase (He et al., 1997). In our strains, ∼20% of cells became blocked with metaphase spindles. However, this percentage is still substantially elevated over the 2% that are observed in asynchronous wild-type cells (Hagan and Hyams, 1988). Twenty-one percent of cells displayed a metaphase Cdc7p staining pattern, consistent with Cdc7p being present at the SPBs of metaphase-arrested cells (Figure 7D, see arrow). Sid1p was never observed at the SPBs in these cells (Figure 7D, see arrow). All cells that displayed Sid1p SPB signal (87/87) were in late anaphase and presumably not yet arrested. After a shift to restrictive temperature for 50 min to inactivate Cdc2p, cells could now be observed that were septating without undergoing anaphase (Figure 7E and F, see arrows). At this point, 96% (68/71) of cells displaying Sid1p signal at the SPB were septating without having undergone anaphase (Figure 7E and F, see arrows). Thus, it appears that loss of Cdc2p activity in metaphase is sufficient to allow Sid1p to localize to the SPB and for initiation of cytokinesis.
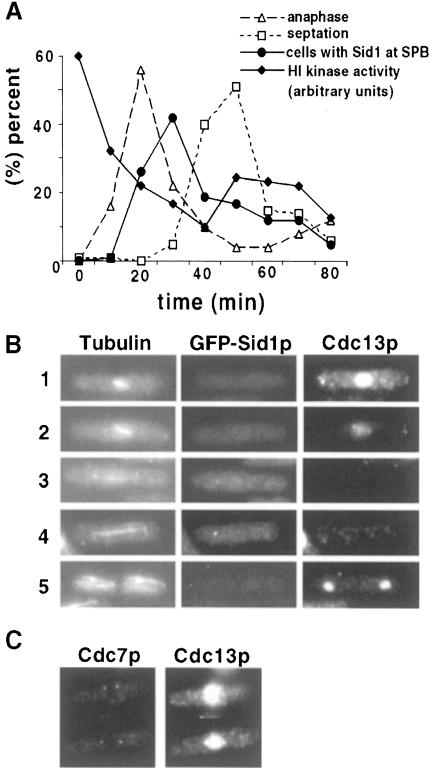
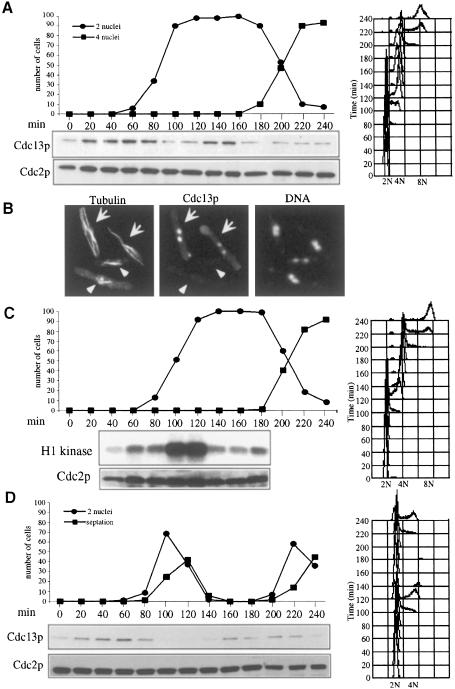
If loss of Cdc2p activity triggers Sid1p localization to the SPB, one would expect Sid1p localization to follow Cdc2p kinase inactivation. To test this directly, H1 kinase activity and GFP–Sid1p localization were monitored as cells progressed through mitosis. As expected, Sid1p localization peaked as cells completed anaphase, coincident with loss of Cdc2 kinase activity, but prior to septation (Figure 8A). APC/C-dependent degradation of Cdc13p upon anaphase onset results in decreased Cdc2 kinase activity. It has been shown previously using indirect immunofluorescence that Cdc13p accumulates in the nucleus until anaphase onset, when it becomes degraded (Alfa et al., 1990). Consistent with loss of Cdc2p kinase activity triggering Sid1p localization, cells in early mitosis that have not degraded Cdc13p do not display GFP–Sid1p at the SPB (Figure 8B, cells 1 and 2), but do show Cdc7p SPB signal (Figure 8C). GFP–Sid1p does not become localized to the SPB until cells degrade Cdc13p (Figure 8B, cells 3 and 4). Interestingly, post-mitotic cells that finished sepation and no longer show GFP–Sid1p SPB signal have begun to accumulate cyclin (Figure 8B, cell 5). These experiments are consistent with the notion that Sid1p does not localize to the SPB until anaphase proteolysis of cyclin (and inactivation of Cdc2p kinase) occurs.
Fig. 8. Sid1p SPB localization coincides with Cdk inactivation and cyclin destruction. (A) nda3-km311 GFP–sid1 cells were blocked in early mitosis by incubation at 19°C for 6 h, released from the block by shifting to 30°C and samples were collected every 10 min and scored for percentage anaphase, septation, GFP–Sid1p localization and H1 kinase activity. (B) GFP–sid1 cells were fixed and stained for tubulin and cyclin (Cdc13p). Numbers indicate the cell cycle stages described in the text. (C) cdc7-HA cells were fixed and stained for Cdc7-HA and cyclin.
The sid mutants are not defective in Cdc13p cyclin proteolysis
Our results suggest that the sid pathway in S.pombe, while clearly functioning in cytokinesis, may be regulated by cyclin proteolysis. The observation that sid mutants continue to undergo multiple nuclear division cycles also suggests that the sid pathway does not play a major role in regulating cyclin proteolysis in S.pombe; however, sid mutants have not been examined previously for fluctuations in cyclin levels. We therefore investigated whether the S.pombe sid genes were required for cyclin proteolysis. Synchronous populations of cdc7-24 (Figure 9A) and wild-type (Figure 9D) cells were analyzed for Cdc13p levels, DNA content and number of nuclei (Figure 9A). This analysis showed that in cdc7-24 cells, like wild-type cells, Cdc13p proteolysis proceeded normally as cells entered anaphase. The loss of Cdc13p in cdc7-24 cells could also be observed by immunofluorescence. Cdc13p was observed in the nucleus of interphase cells as judged by the presence of interphase microtubule arrays (Figure 9B, see arrows). However, in cells that displayed anaphase spindles, Cdc13p had disappeared from the nucleus (Figure 9B, see arrowheads). Similar results were obtained when all of the other sid mutants were examined (data not shown). To determine whether Cdc2p kinase activity dropped as cells entered anaphase, Cdc2p kinase activity was also analyzed (Figure 9C). It was only feasible to synchronize sufficient cells for analysis of Cdc2p kinase activity through the first mitosis; however, we analyzed the number of nuclei and DNA content through another mitosis to demonstrate that cells were undergoing a first cell cycle block. This analysis showed that like Cdc13p levels, Cdc2p kinase activity dropped as cells entered anaphase. Thus, it seems clear that in S.pombe the sid pathway is not essential for cyclin destruction and mitotic exit; however, it remains possible that the sid pathway could play a minor role in cyclin destruction.
Fig. 9. The sid genes are not required for Cdc13p–cyclin destruction in mitosis. (A) cdc7-24 (YDM168) cells, grown at 25°C, were synchronized in early G2 phase by centrifugal elutriation and then shifted to 36°C. Every 20 min, samples were removed and examined for number of nuclei, Cdc13p and Cdc2p levels by Western blotting, and DNA content by FACS. (B) Asynchronous cdc7-24 cells were grown at 25°C, shifted to 36°C for 2 h, fixed and stained for tubulin, Cdc13p and DNA. Arrows indicate interphase cells, and arrowheads indicate post-metaphase cells. (C) cdc7-24 cells were grown as in (A) and H1 kinase assays were performed on samples through the first mitosis. (D) Wild-type cells were analyzed as in (A).
Discussion
A signaling pathway to initiate septation and cytokinesis
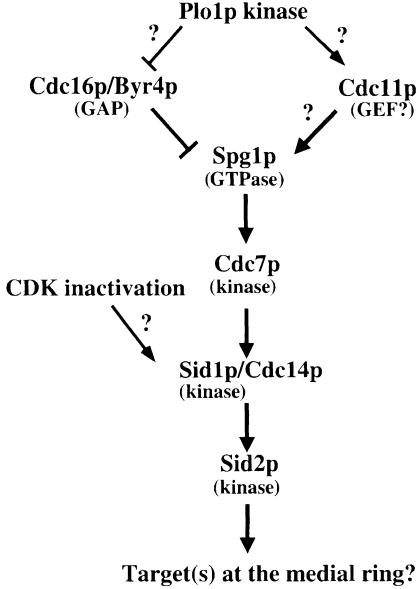
Several studies suggest that the sid gene products function in a signal transduction pathway (Marks et al., 1992; Schmidt et al., 1997; Balasubramanian et al., 1998; Sohrmann et al., 1998; Sparks et al., 1999). Overexpres sion of Cdc7p protein kinase rescues the cytokinesis defects of cdc11 and spg1 mutants (Fankhauser and Simanis, 1994; Schmidt et al., 1997), whereas overexpression of Spg1p rescues cdc11 mutants (Schmidt et al., 1997), suggesting that Cdc7p functions downstream of Spg1p, which, in turn, functions downstream of cdc11. Interestingly, Spg1p overexpression can induce septum formation during interphase in a Cdc7p-, Cdc14p-, Sid4p-, Sid1p- and Sid2p-dependent manner, consistent with Spg1p functioning early in the cascade (Schmidt et al., 1997; Balasubramanian et al., 1998; Sparks et al., 1999). Cdc16p and Byr4p have been identified as a two-component GAP for Spg1 (Furge et al., 1998). Because cdc11 mutants can be rescued by loss-of-function mutations in cdc16 (Marks et al., 1992), it has been proposed that Cdc11p may act as the GEF for Spg1p, although proof awaits the molecular cloning and characterization of cdc11 (Schmidt et al., 1997). Since localization of Cdc7p to the SPB has been shown to depend on Spg1p-GTP, the fact that Cdc7p does not localize in cdc11 mutants coincides with the proposed function of Cdc11p as an activator of Spg1p. Sid2p kinase localizes to the SPB throughout the cell cycle and transiently to the cleavage furrow in late anaphase. Sid2p kinase activity and localization require the function of the other sid gene products, suggesting that it functions late in the pathway (Sparks et al., 1999). In the present study, we found that Sid1p–Cdc14p localizes to one SPB after Cdc7p, just before Sid2p localizes to the cleavage furrow. Our localization studies suggest that Sid1p–Cdc14p functions at an intermediate step in the pathway, downstream of Cdc7p and upstream of Sid2p. We propose a model in which Spg1p, regulated by a two-component GAP comprised of Cdc16p/Byr4p and potential activator Cdc11p, functions upstream of Cdc7p kinase, while Sid1p kinase–Cdc14p functions downstream of Cdc7p and upstream of Sid2p kinase (Figure 10). While this model is consistent with genetic and localization studies presented here and elsewhere, further biochemical studies will be necessary to determine if the model is correct.
Fig. 10. A model for regulation of septation initiation in S.pombe (described in the text).
The role of Sid4p and Plo1p kinase in the sid pathway is unclear. Plo1p may function early in the pathway since its overexpression can induce septum formation in interphase, suggesting that Plo1p drives both actin ring formation and activation of the sid pathway (Ohkura et al., 1995). Plo1p overexpression also results in recruitment of Cdc7p to the SPB, presumably by activating Spg1p (Mulvihill et al., 1999). One prediction based on these data is that Plo1p might function to activate the GEF or inactivate the GAP for Spg1p.
The target of Sid1p phosphorylation in vivo is unknown; however, since Sid2p seems to function immediately downstream of Sid1p–Cdc14p, one model is that Sid2p or an associated protein could be substrates for Sid1p. Consistent with this idea, Sid1p peak kinase activity and Sid1p–Cdc14p SPB localization coincide with activation of Sid2p kinase activity and localization to the cleavage site. At present, it is unclear whether Sid1p acts directly on Sid2p or through another molecule(s). The function of Cdc14p is also unclear. Determination of how the sid proteins regulate each other will require studies using purified components in vitro.
Recruitment of Sid1p–Cdc14p to the SPB may be regulated by Cdc2–cyclin proteolysis
Studies involving expression of non-degradable forms of cyclin in S.pombe (Yamano et al., 1996), Saccharomyces cerevisiae (Surana et al., 1993) and mammalian cells (Wheatley et al., 1997) suggest that APC/C-dependent cyclin proteolysis and subsequent loss of Cdk activity is required to initiate cytokinesis. Surprisingly, inactivation of some APC/C subunits in S.pombe, which are required for cyclin ubiquitylation in mitosis, does not block cyto kinesis (Hirano et al., 1988; Samejima and Yanagida, 1994; Berry et al., 1999). It is possible that Cdc2p activity is inhibited in these APC mutants through tyrosine phosphorylation or the action of the inhibitor Rum1p (Moreno and Nurse, 1994), thus allowing cells to undergo cytokinesis. Activation of the sid pathway by inactivation of cdc16 can cause interphase cells to septate, but only if Cdc2p activity is low (Cerutti and Simanis, 1999). Similarly, Spg1p overproduction will not cause nda3 cells, which are blocked in early mitosis with high Cdc2p activity, to septate (Schmidt et al., 1997). Finally, overexpression of the spindle checkpoint protein Mad2p induces metaphase arrest by hyperactivating the spindle checkpoint (He et al., 1997). These cells maintain high Cdc2p kinase activity and do not septate. If the mitotic kinase (cdc2) or the associated mitotic cyclin (cdc13) is inactivated in cells blocked in metaphase by mad2 overexpression, cells form a division septum without undergoing anaphase. These data are consistent with the idea that high Cdc2p activity acts to inhibit initiation of septation. In this study, we show that activation of the spindle checkpoint by two different methods blocks Sid1p localization to the SPB, whereas Cdc7p can clearly be seen on spindle poles under these conditions. When we inactivated Cdc2p or the spindle checkpoint, septum formation was induced in the absence of anaphase and Sid1p localized to the SPB. It is unclear whether loss of Cdc2p activity directly regulates Sid1p–Cdc14p localization or functions through some intermediary; however, it is clear from our studies that cyclin degradation and Cdc2p inactivation occur prior to Sid1p–Cdc14p SPB localization. Both Sid1p and Cdc14p contain potential Cdc2p phosphorylation sites, but their functional significance is not known. In any case, although the data presented here are correlative, they suggest that Cdc2p inactivation, perhaps triggered by cyclin proteolysis, regulates Sid1p–Cdc14p localization to the SPB, which could provide a key control mechanism to link completion of mitosis with cytokinesis in S.pombe.
Evolutionary differences in sid pathway function
The S.pombe genes plo1, cdc16, byr4, spg1, cdc7 and sid2 all have counterparts in S.cerevisiae (CDC5, BUB2, BFA1, TEM1, CDC15 and DBF2, respectively). As in S.pombe, the products of these genes appear to define a signal transduction pathway required for late mitotic events and have been termed the mitotic exit network (Shou et al., 1999). Evidence from budding yeast suggests that this pathway is required for inactivation of cyclin-dependent kinase (Cdk) activity at the end of anaphase and acts through Cdc14 phosphatase to signal cyclin proteolysis and stabilization of the Cdk inhibitor, Sic1; however, a definitive link to cytokinesis has yet to be identified (Visintin et al., 1998; Alexandru et al., 1999; Shou et al., 1999; Visintin et al., 1999). The spindle checkpoint pathway, acting through Bub2, can block cyclin proteolysis and mitotic exit by inhibiting the mitotic exit network genes (Alexandru et al., 1999; Fesquet et al., 1999; Fraschini et al., 1999; Li, 1999). It has been unclear whether the sid pathway plays a role in cyclin proteolysis in fission yeast. Our results showing that Cdc13p (cyclin) destruction is unaffected in sid mutants are consistent with the sid pathway not contributing to cyclin proteolysis during anaphase. In fact, this study suggests that in S.pombe, the sid pathway itself may be regulated by Cdk inactivation. Some evidence suggests that the spindle checkpoint may inhibit the sid pathway directly in S.pombe through the Cdc16p GAP (the S.cerevisiae BUB2 homolog), which is required for the spindle checkpoint pathway (Fankhauser et al., 1993). Given what is known about the sid pathway, this result implies that response to activation of the spindle checkpoint may cause Cdc16p to inactivate Spg1p, in turn blocking Cdc7p recruitment to the SPB. However, we found that Cdc7p is present at the SPB in cells with the spindle checkpoint activated. Thus, if Cdc16p does function in the spindle checkpoint pathway in S.pombe, it seems unlikely that it functions by inactivating the sid pathway through Spg1p. Perhaps the different roles of the sid pathway in S.pombe and S.cerevisiae reflect the evolutionary divergence between the two organisms. It will be important in future studies to identify components of the sid pathway in higher eukaryotes and determine how they function.
Materials and methods
Yeast methods and strains
The S.pombe strains used in this study are listed in Table III. Fission yeast media and genetic manipulations were as previously described (Moreno et al., 1991). DNA sequencing was performed at the UMass Medical School Nucleic Acid Facility (Worcester, MA). Oligonucleotide primers were obtained from Integrated DNA Technologies (Coralville, IA) or from Operon Inc. (Alameda, CA).
Table III. Strains used.
| Strain | Genotype | Source |
|---|---|---|
| YDM435 | sid1-125 ade6-210 ura4-D18 leu1-32 h+ | laboratory stock |
| YDM75 | sid1-239 ade6-210 ura4-D18 leu1-32 h+ | laboratory stock |
| YDM105 | leu1-32 ura4-D18 ade6-210 h– | laboratory stock |
| YDM108 | leu1-32 ura4-D18 ade6-216 h+ | laboratory stock |
| YDM579 | sid1::ura4+/sid1+ ura4-D18/ura4-D18 leu1-32/leu1-32 ade6-210/ade6-216 h+/h– | this study |
| YDM558 | GFP–sid1::ura4+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM587 | 13Myc-sid1::ura4+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM627 | GFP–cdc14::leu1+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM617 | 13Myc-cdc14::leu1+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM553 | GFP–sid1::ura+ cdc7-HA::ura+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM560 | GFP–sid1::ura4 cdc16-116 ura4-D18 leu1-32 h– | this study |
| YDM564 | GFP–sid1::ura4+ nmt1-spg1::leu1+ ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM551 | nmt1-spg1::leu1+ ura4-D18 leu1-32 ade6-210 h+ | K.Gould |
| YDM566 | sid1-125 nmt1-spg1::leu1+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM561 | GFP–sid1::ura4+ sid4-Al ura4-D18 leu1-32 ade6-210h– | this study |
| YDM562 | GFP–sid1::ura4 cdc11-123 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM568 | GFP–sid1::ura4+ cdc14-118 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM569 | GFP–sid1::ura4 cdc7-24 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM567 | GFP–sid1::ura4+ sid2-250 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM563 | GFP–sid1::ura4+ spg1-106 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM572 | cdc7-HA::ura4+ cdc11-123 ura4-D18 leu1-32 ade6-210h– | this study |
| YDM571 | cdc7-HA::ura4+ cdc14-118 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM542 | cdc7-HA::ura4+ sid1-125 ura4-D18 leu1-32h– | this study |
| YDM646 | GFP–cdc14::leu1+ cdc11-123 ura4-D18 leu1-32h– | this study |
| YDM647 | GFP–cdc14::leu1+ spg1-106 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM633 | GFP–cdc14::leu1+ sid1-125 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM632 | GFP–cdc14::leu1+ sid4-Al ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM631 | GFP–cdc14::leu1+ sid2-250 ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM636 | GFP–cdc14::leu1+ cdc7-24 ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM629 | GFP–cdc14::leu1+ 13Myc-sid1::ura4+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM626 | GFP–sid1::ura4+ 13Myc-cdc14::leu1+ ura4-D18 leu1-32 ade6-210 h– | this study |
| YDM637 | 13Myc-cdc14::leu1+ sid1-239 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM666 | GFP–sid1::ura4+ nda3-km311 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM555 | GFP–sid1::ura4+ nda3-km311 mad2Δ ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM667 | cdc7–GFP::ura4+ nda3 km311 ura4-D18 leu1-32 ade6-210 h+ | this study |
| YDM651 | cdc7-HA::ura4+ GFP–sid1::ura4+ cdc2-33 leu1-32h– | this study |
| YDM168 | cdc7-24 h– | P.Nurse |
Cloning of the sid1 gene
sid1-239 (YDM75) was transformed with a genomic library constructed in pUR19 (Barbet et al., 1992). Transformants were selected at 25°C, and then replica plated to 36°C. Four plasmids containing overlapping inserts were identified that were capable of rescuing the sid1 temperature-sensitive growth defect at 36°C. Integration mapping showed that sid1 mapped to the same region of the genome as the cloned gene, demonstrating that the cloned gene was sid1 and not a multicopy suppressor. The ends of the genomic inserts of two of the rescuing plasmids were sequenced. A search of the Sanger Center S.pombe genome sequencing project database revealed that these genomic clones corresponded to sequenced regions of the S.pombe genome.
Construction of sid1 deletion and epitope-tagged strains
A sid1 deletion strain was constructed using the one-step gene replacement strategy (Grimm et al., 1988). The knockout construct was made by first generating by PCR an 1100 bp SacI–NotI fragment containing the 5′-flanking region of sid1 that ends at the sid1 start codon. A sid1 3′-flanking region that contained the 1000 bp immediately downstream of the sid1 stop codon was also amplified by PCR as a NotI–SalI fragment and the two fragments were cloned in a three piece ligation into SacI–SalI-cut pSK to generate plasmid pDM356. A fragment containing the S.pombe ura4+ gene as a NotI fragment was then cloned into the NotI site of this plasmid to generate the sid1 deletion construct, plasmid pDM357. The SacI–SalI fragment of this plasmid was purified and transformed into a diploid strain (YDM105 crossed with YDM108) using the lithium acetate method. PCR analysis of DNA prepared from individual transformants identified a strain bearing the sid1 deletion (YDM579). The sid1 deletion strain was induced to sporulate, and tetrads were dissected, which resulted in only two viable ura4– progeny per tetrad. The viability of the sid1 deletion strain was rescued by a plasmid containing the sid1 coding region expressed from the nmt1 promoter (pDM321).
Single copy gene replacement strains expressing GFP and 13Myc-tagged versions of sid1 were constructed using the following strategy. PCR was used to amplify the first 660 bp of the sid1 coding region and to add a NotI site immediately after the start ATG. This fragment was cut with NotI–EcoRI and ligated, along with the SacI–NotI-cut sid1 promoter fragment from pDM356, into the integrating vector pJK210 that had been cut with SacI–EcoRI, generating plasmid pDM336. Then either the 13Myc or GFP tags bearing NotI sites at their ends were cloned into pDM336 to generate plasmids pDM360 and pDM339, respectively. Each of these plasmids was linearized at the NdeI site in the coding region and transformed into strain YDM105 to generate strains expressing a single copy of GFP-tagged (YDM558) and 13Myc-tagged (YDM587) sid1 expressed from its own promoter, as well as a truncated (non-functional) copy of sid1. Correct integration of each construct was confirmed by PCR.
Strains expressing a single copy of GFP- and 13Myc-tagged versions of cdc14 from the cdc14 promoter were constructed using a strategy similar to that described above for sid1. PCR was used to generate a 960 bp SacI–NotI fragment containing the 5′-flanking region of cdc14, as well as the entire cdc14 coding region as a NotI–BamHI fragment. These two fragments were cloned into the integrating vector pJK148 that had been cut with SacI–BamHI to generate plasmid pDM369. Then either the 13Myc or GFP tags bearing NotI sites at their ends were cloned into pDM369 to generate plasmids pDM374 and pDM370, respectively. Each of these plasmids was linearized at the NdeI site in the leu1+ gene and transformed into strain YDM105 to generate strains expressing a single copy of GFP-tagged (YDM627) and 13Myc-tagged (YDM617) cdc14 expressed from its own promoter, as well as a second, untagged copy of cdc14. Correct integration of each construct was confirmed by PCR.
Construction of sid1 kinase-dead plasmids
Triple HA-tagged (pRep41HA-Sid1) sid1 was constructed by adding BamHI sites to the ends of the sid1 coding region by PCR followed by cloning into the BamHI site of vector pRep41HA (Craven et al., 1998). Kinase-dead sid1 was created by site-directed mutagenesis with a QuikChange kit (Stratagene) using pRep41HA-Sid1 directly as a template. A single nucleotide change generated by the following oligonucleotides was sufficient to create the conservative lysine to arginine mutation at residue 38: 5′-GTGTCTGGCGACATTATCGCT ATTAGACAGGTAAGACTAATTAATTTTTAC-3′ and 5′-GTAAAA ATTAATTAGTCTTACCTGTCTTACCTGTCTAATAGCGATAATG TCGCCAGACAC-3′. Changes were confirmed by sequencing.
Microscopy
Indirect immmunofluorescence staining was performed as previously described (Balasubramanian et al., 1997). The tubulin antibody TAT-1 (Woods et al., 1989) was a generous gift from K.Gull (University of Manchester, UK). Cdc13 and HA antibodies (BABCO, Richmond, CA) were diluted 1:200. Primary antibodies were detected with anti-rabbit or anti-mouse Texas red or Alexa 594–IgG (Molecular Probes). DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI; Sigma) at 20 µg/ml. Images were captured using a Nikon Eclipse E600 microscope with a cooled CCD camera (Dage 300, MVI, Avon, MA) and IPlab Spectrum software (Signal Analytics Corporation, Vienna, VA).
Protein methods and kinase assays
Sid1 kinase assays were performed as previously described (Sparks et al., 1999) except that a different kinase assay buffer was used (10 mM MgCl2, 10 mM Tris pH 7.4). For co-immunoprecipitation studies, lysates were incubated with anti-GFP antibody, immune complexes were recovered with protein G–Sepharose beads and resolved on 7% polyacrylamide gels, and Western blots were probed with Myc antibody.
For block and release experiments, cells were incubated at restrictive temperatures (6 h at 19°C and 3.5 h at 36°C for nda3-km311 and cdc25-22 strains, respectively), then released by shifting to permissive temperature. For other synchronous cell experiments, cells were grown to mid-log phase at 25°C in YE medium, and G2 cells were isolated by centrifugal elutriation. Cells were collected on a filter, resuspended in pre-warmed YE medium, incubated for 30 min at 36.5°C, and then samples were collected every 20 min for fluorescence-activated cell sorting (FACS) analysis, nuclei count, septation count and cell lysates. H1 kinase assays were performed as previously described (Gould et al., 1991).
Acknowledgments
Acknowledgements
We are grateful to Hayes McDonald, Shelley Sazer, Viesturs Simanis, Paul Nurse, Pam Silver and Keith Gull for providing strains, antibodies and plasmids. We thank Mohan Balasubramanian for critical reading of the manuscript. This work was supported by National Institutes of Health (NIH) grant GM58406 to D.M., and by the Howard Hughes Medical Institute, of which K.L.G. is an Associate Investigator.
References
- Alexandru G., Zachariae,W., Schleiffer,A. and Nasmyth,K. (1999) Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J., 18, 2707–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfa C.E., Ducommun,B., Beach,D. and Hyams,J.S. (1990) Distinct nuclear and spindle pole body population of cyclin–cdc2 in fission yeast. Nature, 347, 680–682. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum,D. and Gould,K.L. (1997) Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol., 283, 494–506. [DOI] [PubMed] [Google Scholar]
- Balasubramanian M.K., McCollum,D., Chang,L., Wong,K.C., Naqvi,N.I., He,X., Sazer,S. and Gould,K.L. (1998) Isolation and characterization of new fission yeast cytokinesis mutants. Genetics, 149, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet N., Muriel,W.J. and Carr,A.M. (1992) Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene, 114, 59–66. [DOI] [PubMed] [Google Scholar]
- Berry L.D., Feoktistova,A., Wright,M.D. and Gould,K.L. (1999) The Schizosaccharomyces pombe dim1+ gene interacts with the anaphase-promoting complex or cyclosome (APC/C) component lid1+ and is required for APC/C function. Mol. Cell. Biol., 19, 2535–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L. and Simanis,V. (1999) Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci., 112, 2313–2321. [DOI] [PubMed] [Google Scholar]
- Craven R.A., Griffiths,D.J., Sheldrick,K.S., Randall,R.E., Hagan,I.M. and Carr,A.M. (1998) Vectors for the expression of tagged proteins in Schizosaccharomyces pombe. Gene, 221, 59–68. [DOI] [PubMed] [Google Scholar]
- Fankhauser C. and Simanis,V. (1993) The Schizosaccharomyces pombe cdc14 gene is required for septum formation and can also inhibit nuclear division. Mol. Biol. Cell, 4, 531–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C. and Simanis,V. (1994) The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J., 13, 3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Marks,J., Reymond,A. and Simanis,V. (1993) The S.pombe cdc16 gene is required both for maintenance of p34 Cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J., 12, 2697–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C., Reymond,A., Cerutti,L., Utzig,S., Hoffmann,K. and Simanis,V. (1995) The S.pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell, 82, 435–444. [DOI] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick,P.J., Johnson,A.L., Kramer,K.M., Toyn,J.H. and Johnston,L.H. (1999) A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J., 18, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Formenti,E., Lucchini,G. and Piatti,S. (1999) Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J. Cell Biol., 145, 979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H., Yamano,H., Kumada,K., Nagao,K., Hunt,T. and Yanagida,M. (1996) Cut2 proteolysis required for sister-chromatid separation in fission yeast. Nature, 381, 438–441. [DOI] [PubMed] [Google Scholar]
- Furge K.A., Wong,K., Armstrong,J., Balasubramanian,M. and Albright,C.F. (1998) Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol., 8, 947–954. [DOI] [PubMed] [Google Scholar]
- Gould K. and Simanis,V. (1997) The control of septum formation in fission yeast. Genes Dev., 11, 2939–2951. [DOI] [PubMed] [Google Scholar]
- Gould K.L., Moreno,S., Owen,D.J., Sazer,S. and Nurse,P. (1991) Phosphorylation at Thre167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J., 10, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Kohli,J., Murray,J. and Maundrell,K. (1988) Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4 gene as a selectable marker. Mol. Gen. Genet., 215, 81–86. [DOI] [PubMed] [Google Scholar]
- Hagan I.M. and Hyams,J.S. (1988) The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J. Cell Sci., 89, 343–357. [DOI] [PubMed] [Google Scholar]
- Hanks S.K., Quinn,M.A. and Hunter,T. (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science, 241, 42–52. [DOI] [PubMed] [Google Scholar]
- He X., Patterson,T.E. and Sazer,S. (1997) The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc. Natl Acad. Sci. USA, 94, 7965–7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T., Hiraoka,Y. and Yanagida,M. (1988) A temperature sensitive mutation of the Schizosaccharomyces pombe gene nuc2+ that encodes a nuclear scaffold-like protein blocks spindle elongation in mitotic anaphase. J. Cell Biol., 106, 1171–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Goff X., Utzig,S. and Simanis,V. (1999) Controlling septation in fission yeast: finding the middle and timing it right. Curr. Genet., 35, 571–584. [DOI] [PubMed] [Google Scholar]
- Li R. (1999) Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc. Natl Acad. Sci. USA, 96, 4989–4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F.C. and Winey,M. (1998) MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol. Biol. Cell, 9, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks J., Fankhauser,C. and Simanis,V. (1992) Genetic interactions in the control of septation in Schizosaccharomyces pombe. J. Cell Sci., 101, 801–808. [DOI] [PubMed] [Google Scholar]
- Minet M., Nurse,P., Thuriaux,P. and Mitchison,J.M. (1979) Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol., 137, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S. and Nurse,P. (1994) Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature, 367, 236–242. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast, Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- Mulvihill D.P., Petersen,J., Ohkura,H., Glover,D.M. and Hagan,I.M. (1999) Plo1 kinase recruitment to the spindle pole body and its role in cell division in Schizosaccharomyces pombe. Mol. Biol. Cell, 10, 2771–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P., Thuriaux,P. and Nasmyth,K. (1976) Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol. Gen. Genet., 146, 167–178. [DOI] [PubMed] [Google Scholar]
- Ohkura H., Hagan,I.M. and Glover,D.M. (1995) The conserved Schizosaccharomyces pombe kinase plo1, required to form a bipolar spindle, the actin ring and septum, can drive septum formation in G1 and G2 cells. Genes Dev., 9, 1059–1073. [DOI] [PubMed] [Google Scholar]
- Samejima I. and Yanagida,M. (1994) Bypassing anaphase by fission yeast cut9 mutation: requirement of cut9+ to initiate anaphase. J. Cell Biol., 6, 1655–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S., Sohrmann,M., Hofmann,K., Woollard,A. and Simanis,V. (1997) The Spg1p GTPase is an essential dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev., 11, 1519–1534. [DOI] [PubMed] [Google Scholar]
- Sells M.A. and Chernoff,J. (1997) Emerging from the Pak: the p21-activated protein kinase family. Trends Cell Biol., 7, 162–167. [DOI] [PubMed] [Google Scholar]
- Shou W., Seol,J.H., Shevchenko,A., Baskerville,C., Moazed,D., Chen,Z.W., Jang,J., Charbonneau,H. and Deshaies,R.J. (1999) Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell, 97, 233–244. [DOI] [PubMed] [Google Scholar]
- Sohrmann M., Schmidt,S., Hagan,I. and Simanis,V. (1998) Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev., 12, 84–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks C.A., Morphew,M. and McCollum,D. (1999) Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol., 146, 777–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Amon,A., Dowzer,C., McGrew,J., Byers,B. and Nasmyth,K. (1993) Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J., 12, 1969–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig,K., Hwang,E.S., Prinz,S., Tyers,M. and Amon,A. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell, 2, 709–718. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang,E.S. and Amon,A. (1999) Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature, 398, 818–823. [DOI] [PubMed] [Google Scholar]
- Wheatley S.P., Hinchcliffe,E.H., Glotzer,M., Hyman,A.A., Sluder,G. and Wang,Y. (1997) CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J. Cell Biol., 138, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods A., Sherwin,T., Sasse,R., MacRae,T.H., Baines,A.J. and Gull,K. (1989) Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci., 93, 491–500. [DOI] [PubMed] [Google Scholar]
- Yamano H., Gannon,J. and Hunt,T. (1996) The role of proteolysis in cell cycle progression in Schizosaccharomyces pombe. EMBO J., 15, 5268–5279. [PMC free article] [PubMed] [Google Scholar]