CSN complex controls the stability of selected synaptic proteins via a torsinA-dependent process
The synaptic chaperone torsin A is mutated in early-onset DYT1 dystonia. This study reveals a disease-relevant interaction between torsin A and subunit 4 of the CSN signalosome that regulates the turnover of proteins involved in the synaptic vesicle cycle.
Keywords: CSN4, dystonia, DYT1, snapin, stonin 2
Abstract
DYT1 dystonia is caused by an autosomal dominant mutation that leads to a glutamic acid deletion in torsinA (TA), a member of the AAA+ ATPase superfamily. In this study, we identified a novel-binding partner of TA, the subunit 4 (CSN4) of CSN signalosome. TA binds CSN4 and the synaptic regulator snapin in neuroblastoma cells and in brain synaptosomes. CSN4 and TA are required for the stability of both snapin and the synaptotagmin-specific endocytic adaptor stonin 2, as downregulation of CSN4 or TA reduces the levels of both proteins. Snapin is phosphorylated by the CSN-associated kinase protein kinase D (PKD) and its expression is decreased upon PKD inhibition. In contrast, the stability of stonin 2 is regulated by neddylation, another CSN-associated activity. Overexpression of the pathological TA mutant (ΔE-TA) reduces stonin 2 expression, causing the accumulation of the calcium sensor synaptotagmin 1 on the cell surface. Retrieval of surface-stranded synaptotagmin 1 is restored by overexpression of stonin 2 in ΔE-TA-expressing cells, suggesting that the DYT1 mutation compromises the role of TA in protein stabilisation and synaptic vesicle recycling.
Introduction
A three base pair deletion (ΔE302–303) in the DYT1gene leads to loss of a glutamate residue in the protein torsinA (TA), and causes the most common genetic form of dystonia, early-onset DYT1 dystonia (Ozelius et al, 1997). This is a severe movement disorder characterised by involuntary sustained muscle contractions that lead to twisting movements of the limbs and abnormal postures. DYT1 dystonia is characterised by the absence of neurodegeneration, implying that neuronal dysfunction rather than loss is the cause of dystonic movement.
TA belongs to the AAA+ superfamily of ATPases and shares homology with the heat shock protein, HSP 100/Clp (Neuwald et al, 1999). This similarity suggests a chaperone-like function for TA, which has not, however, been confirmed to date. TA has been found associated with several subcellular compartments, where it interacts with a number of proteins of diverse function, including cytoplasmic motor protein kinesin light chain and cytoskeletal-associated proteins, such as vimentin and actin (Kamm et al, 2004; Hewett et al, 2006). TA also interacts with nuclear envelope components, such as LAP1 and nesprin-3, and with endoplasmic reticulum resident proteins LULL1 and printor (Goodchild and Dauer, 2005; Nery et al, 2008; Giles et al, 2009). In addition, TA shows a vesicle-like distribution pattern and partially co-localises with synaptic vesicle (SV) markers (Hewett et al, 2000; Ferrari-Toninelli et al, 2004) and the SNARE-associated protein snapin (Granata et al, 2008).
Based on the homology with AAA+ ATPases, it is believed that the active form of TA is an oligomeric complex (Neuwald et al, 1999; Breakefield et al, 2001). The ΔE deletion could compromise the formation of this complex and consequently disrupt TA interaction with its multiple-binding partners, giving rise to a loss of function mutation, which in turn may lead to a dystonic phenotype. Alternatively, mutant TA carrying the ΔE deletion (ΔE-TA) might bind wild-type TA (wt-TA) and lead to the mislocalisation of TA and its interacting proteins to the nuclear envelope and the characteristic perinuclear inclusions (Hewett et al, 2000; Goodchild and Dauer, 2005; Goodchild et al, 2005; Misbahuddin et al, 2005), which may provide a molecular explanation for the dominant-negative character of the mutant protein.
Recent work suggests that TA regulates vesicular traffic, possibly affecting dopamine turnover. Imbalance of dopamine signalling has been observed in several DYT1 mouse models (Balcioglu et al, 2007; Zhao et al, 2008), and TA has been shown to modulate plasma membrane delivery of the dopamine transporter DAT-1 (Torres et al, 2004). Consistent with this hypothesis, our previous work revealed an association between TA and snapin, a SNARE-associated protein (Granata et al, 2008). Snapin was originally identified as a component of the exocytic machinery and was suggested to promote the interaction between the SNARE complex and synaptotagmin 1 (Syt1), a calcium-binding protein and putative SV calcium sensor (Ilardi et al, 1999). Although some aspects of this mechanism remain controversial (Vites et al, 2004), the contribution of snapin to docking of SVs was confirmed by studies in animal models (Tian et al, 2005; Pan et al, 2009). In human neuroblastoma cells stably expressing wt- and ΔE-TA, mutant TA altered SV exo-endocytosis and caused retention of Syt1 on the plasma membrane (Granata et al, 2008). Notably, knockdown of TA or overexpression of ΔE-TA gave rise to a similar phenotype, supporting the proposal that ΔE-TA is a loss of function mutation (Torres et al, 2004; Dang et al, 2005; Granata et al, 2008). These findings suggest that TA acts as a synaptic chaperone that controls transport and/or targeting of synaptic proteins.
In this study, we report a newly identified binding partner of TA, the subunit 4 (CSN4) of CSN signalosome, a complex highly conserved from yeast to human, which has been involved in a variety of biological processes, such as cell cycle regulation, DNA repair and apoptosis (Kato and Yoneda-Kato, 2009). CSN4 interacts exclusively with wt-TA and either snapin or the synaptotagmin-specific endocytic adaptor, stonin 2 (Martina et al, 2001; Walther et al, 2004). We showed that snapin and stonin 2 are targeted by CSN-dependent modification and we provide evidence for a novel mechanism, in which TA is required for the stability of snapin and stonin 2. Furthermore, overexpression of stonin 2 in neuroblastoma cells expressing ΔE-TA partially restores Syt1 internalisation, suggesting that the abnormal accumulation of Syt1 on the plasma membrane is caused by stonin 2 downregulation. Consistent with this observation, analysis of SV recycling in primary neurons expressing pH-dependent SytPhluorin in the presence of ΔE-TA showed a reduced efficiency of Syt1 retrieval after synaptic stimulation. Altogether, our data identify a key role of TA in the regulation of the turnover of specific synaptic proteins essential for SV exocytosis and recycling.
Results
The subunit 4 of the CSN complex (CSN4) is a novel-binding partner of TA
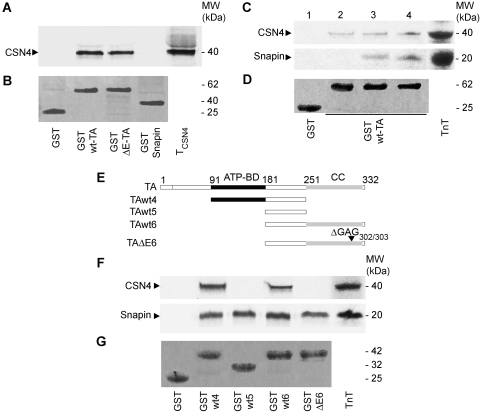
From a yeast two-hybrid screen targeted at the identification of proteins that bind wt-TA and ΔE-TA, we identified the subunit 4 (CSN4) of the CSN signalosome complex (Wei and Deng, 1999, 2003) as potential-interacting partner of TA. X-gal positive colonies were observed for both wt-TA and ΔE-TA co-transformed with CSN4 (data not shown). These interactions were confirmed by GST pull-down assays, which showed that both wt-TA and ΔE-TA expressed as GST-fusion proteins are able to bind in vitro translated CSN4 (Figure 1A). No interaction of CSN4 with snapin, another TA-interacting partner (Granata et al, 2008), was observed under these experimental conditions. We previously demonstrated that TA binds snapin via the region containing both the ATP-binding (ATP-BD) and the coiled-coil (CC) domains (Granata et al, 2008). To investigate whether CSN4 and snapin interact simultaneously with TA, a competition assay was performed by adding increasing quantities of snapin to the in vitro translation mix containing a constant amount of CSN4. As shown in Figure 1C, the binding of CSN4 to TA is not affected by snapin, suggesting that these two interactions are likely to be independent.
Figure 1.
CSN4 and snapin bind TA independently. (A) GST pull downs using in vitro transcribed-translated CSN4 (TCSN4) labelled with [35S]-methionine show that both wt-TA and ΔE-TA bind CSN4, while snapin does not interact directly with CSN4. (B) The corresponding Coomassie Blue stained gel shows GST-wt-TA, GST-ΔE-TA, GST-snapin and GST bands. (C) CSN4 and snapin do not compete for binding on GST-wt-TA. Increasing quantities of snapin (2, no snapin; 3, half snapin; 4, equimolar CSN4 and snapin) were added to a [35S]-labelled in vitro transcription translation (TnT) mix containing a constant quantity of CSN4. (D) The corresponding Coomassie Blue stained gel shows GST-wt-TA and GST bands. (E) Schematic representation of the full-length and truncated versions of wt and mutant TA; ATP-BD, ATP-binding domain; CC, coiled-coil domain. The filled region (91–181) marks the position of the ATPase domain. ΔGAG302/303 indicates the position of the three base pair deletion. (F) GST pull downs with the truncated mutants of TA show that CSN4 interacts with both wt4 and wt6 mutants while no signal was detected for ΔE6. In a parallel experiment, snapin binds to all TA fragments. (G) The corresponding Coomassie Blue stained gel shows GST-TA-wt4, -wt5, -wt6 and -ΔE6, and GST used as control.
CSN4 and snapin were tested in parallel pull-down assays with a series of truncation mutants of wt-TA and ΔE-TA to confirm that binding to CSN4 involves different domains of TA (Figure 1E and F). Four fragments were generated: TAwt4, which contains the ATPase-binding and intermediate domains, TAwt5, which contains the intermediate region only, and TAwt6 and TAΔE6 containing the intermediate fragment and the CC domain without or with the ΔE deletion, respectively (Figure 1E). CSN4 interacts with both TAwt4 and TAwt6, but not with TAwt5 (Figure 1F), suggesting that either CSN4 binds the intermediate domain of TA, but that this region requires the flanking domains to maintain correct folding or an open conformation competent for binding, or that two independent binding sites for CSN4 are present on the ATP-BD and CC domains. In contrast to the GST pull-down data described in Figure 1A, no binding of CSN4 was observed with the TAΔE6 fragment, suggesting that the dystonia-inducing mutation may affect the molecular interaction between TA and CSN (Figure 1F). As described previously (Granata et al, 2008), the binding of snapin to TA is not affected by the ΔE deletion (Figure 1F).
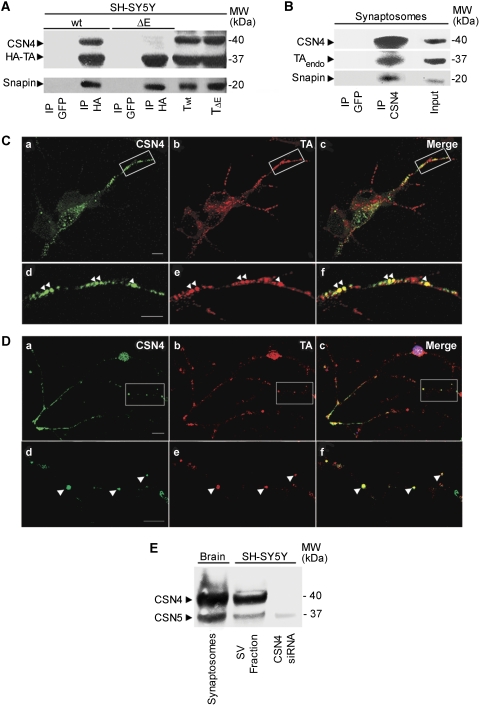
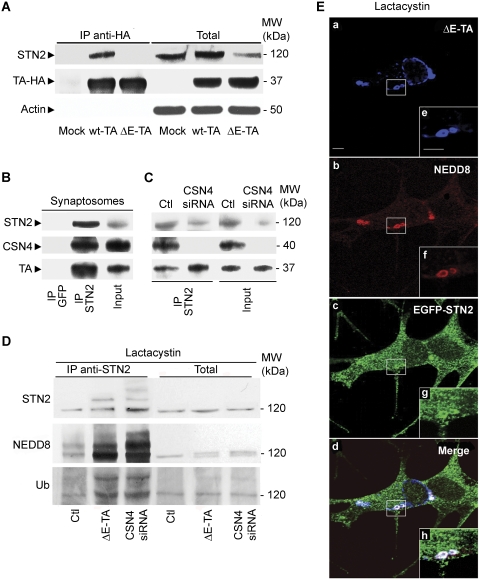
These conclusions are strengthened by immunoprecipitation experiments performed in SH-SY5Y neuroblastoma cells expressing HA-wt-TA and HA-ΔE-TA, which showed that endogenous CSN4 only interacts with wt-TA and not with ΔE-TA in cell extracts (Figure 2A). Under the same experimental conditions, both HA-wt-TA and HA-ΔE-TA efficiently bind snapin (Figure 2A). To confirm this finding, CSN4 was immunoprecipitated using an antibody recognising the native protein. As shown in Figure 2B, a triple complex, which includes endogenous CSN4, wt-TA and snapin, was detected in an extract from brain synaptosomes.
Figure 2.
Endogenous CSN4 forms a complex with wt-TA and snapin in SH-SY5Y cells and brain synaptosomes. (A) Lysates of SH-SY5Y cells overexpressing HA-wt-TA or HA-ΔE-TA were incubated with either anti-HA or anti-GFP antibodies. Immunoprecipitated complexes (IP) were then analysed by western blot with specific antibodies against CSN4, snapin and HA tag. A triple complex containing CSN4, snapin and TA was observed in lysates expressing wt-TA. Lanes Twt and TΔE show 1/10th of the starting material. (B) A complex of endogenous CSN4, TA and snapin was detected in rat brain synaptosomes using a specific anti-CSN4 antibody for immunoprecipitation. Total lysate (input) consists of 1/10th of the starting material. (C) Immunofluorescence analysis of endogenous CSN4 and TA in SH-SY5Y cells. Both CSN4 (green; a, d) and TA (red; b, e) show a punctate pattern. A partial overlapping of CSN4 and TA is observed along neurites (c, f). Scale bar=5 μm (a–c) and 2 μm (d–f). (D) Immunofluorescence of CSN4 and TA in primary cerebellar granule neurons. CSN4 (green; a, d) and TA (red; b, e) co-localise along neurites (c, f). Scale bar=10 μm (a–c) and 3 μm (d–f). (E) The subunit 5 (CSN5) of CSN was also detected in the synaptic vesicle fraction (SV) of SH-SY5Y cells and in brain synaptosomes. Downregulation of CSN4 by siRNA destabilises CSN5.
In agreement with this biochemical evidence, immunostaining with CSN4 and TA antibodies showed a partial co-localisation of both proteins on discrete puncta along neurites in SH-SY5Y cells (Figure 2C) and in primary cerebellar neurons (Figure 2D), which are reminiscent of synaptic boutons. Notably, CSN4 was seen to partially overlap with SV markers, such as Syt1 in SH-SY5Y cells (Supplementary Figure 1A–F) and in primary hippocampal neurons (Supplementary Figure 1G–L). CSN4 distribution was not affected by ΔE-TA expression or siRNA-mediated knockdown of TA (data not shown). Crucially, CSN4 and another CSN subunit, CSN5, which has been found to translocate between nucleus and cytosol (Tomoda et al, 2002), were found to associate with the SV-like compartment in SH-SY5Y cells and rat brain synaptosomes (Figure 2E), suggesting the possibility that the CSN complex is present at synaptic sites. Altogether, these findings indicate that in addition to snapin, TA binds to the CSN complex via its CSN4 subunit in neuronal cells and brain tissue.
CSN4 regulates the cellular levels of snapin
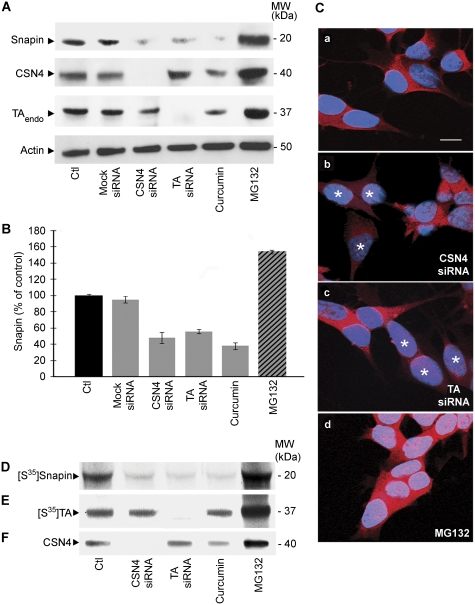
Based on the evidence that CSN subunits share structural similarities with the lid subcomplex of the proteasome and regulate the ubiquitin-dependent degradation of several target proteins (Schwechheimer, 2004), we investigated whether CSN4 affects the cellular levels of TA and its binding partner, snapin. To test this hypothesis, we used two independent approaches in SH-SY5Y cells: pre-treatment with curcumin, a well-recognised inhibitor of CSN-associated kinase activity (Bech-Otschir et al, 2001) and siRNA-mediated CSN4 knockdown (Figure 3A). The latter approach was very effective in reducing the expression level of CSN4 (Figure 3A; Supplementary Figure 2) and indirectly, of other CSN subunits, such as CSN5 in whole cell lysates (data not shown) and in the SV fraction (Figure 2E). This result is in agreement with previous studies, indicating that the whole CSN complex is depleted in cells treated with siRNAs directed against its individual subunits (Peth et al, 2007). No significant effect was observed on endogenous TA levels after CSN4 downregulation or incubation with curcumin (Figure 3A; Supplementary Figure 3). In contrast, both treatments caused a marked reduction in snapin levels (Figure 3A). In addition, treatment with siRNA against TA decreased snapin expression similarly to that observed for CSN4 knockdown (Figure 3A). Remarkably, CSN4 expression is reduced in response of curcumin (Figure 3A), which may be able to regulate the stability of CSN subunits, along with the organisation and dynamics of CSN as shown previously (Fukumoto et al, 2005; Tomoda et al, 2005). In addition, we considered if snapin expression levels could be downregulated in the presence of ΔE-TA in SH-SY5Y cells; however, no changes were observed (data not shown).
Figure 3.
Downregulation of CSN4 and TA affect snapin levels in SH-SY5Y cells. (A) Equal amount of lysates from untreated SH-SY5Y cells (Ctl), cells treated with mock siRNA, with specific siRNA to knockdown CSN4 (CSN4 siRNA) and TA (TA siRNA), with curcumin, a specific inhibitor for CSN and with the proteasome inhibitor MG132 were analysed by western blot and detected with specific antibodies. β-actin was used as loading control. (B) The amount of snapin from three independent experiments is shown. Snapin levels are drastically reduced in cells treated with siRNA against CSN4 and TA, or curcumin, while treatment with MG132 increases the amount of endogenous snapin. Error bars represent s.e.m. (C) The immunofluorescence signal for snapin (red) is unaffected by transfection with a control siRNA alone (a), whereas is decreased in cells transfected with CSN4 siRNA (b) and TA siRNA (c). Snapin is accumulated in cells after MG132 treatment (d). Stars indicate transfected cells. Scale bar=10 μm. (D–F) Metabolic labelling of snapin in SH-SY5Y cells. Untreated cells (Ctl) and cells treated with siRNA against CSN4 or TA, curcumin and MG132 were incubated overnight with [35S]-methionine. After 2 h incubation with cycloheximide, cells were analysed by immunoprecipitation using a specific anti-snapin (D) or anti-TA antibody (E) and radioactive bands revealed by autoradiography. (F) CSN4 levels were monitored by western blot.
Quantitative analysis from three independent experiments (Figure 3B) confirmed that snapin expression is negatively affected by CSN4 or TA downregulation, or by curcumin treatment, whereas adding MG132, a proteasome inhibitor, increased the cellular levels of snapin. Immunofluorescence analysis confirmed the reduced levels of snapin in SH-SY5Y cells treated with CSN4 or TA siRNAs (Figure 3C).
This observation was confirmed by metabolic labelling of snapin in SH-SY5Y cells (Figure 3D–F). [35S]-methionine was added to samples incubated with CSN4 or TA siRNAs, or treated with curcumin or MG132 overnight before addition of cyclohexamide to block protein synthesis. Snapin was immunoprecipitated and detected by autoradiography. We observed a marked decrease in total snapin levels in cells treated with siRNAs directed against CSN4 and TA, or treated with curcumin (Figure 3D), suggesting that CSN and TA have a function in the mechanism, that regulates snapin stability.
Knockdown of CSN4 or ΔE-TA overexpression reduce the cellular levels of stonin 2
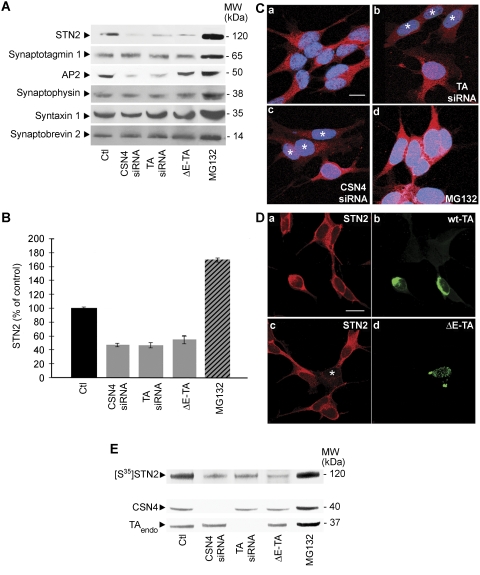
The above findings opened the possibility that, in addition to snapin, CSN complex controls the stability of other synaptic proteins in a TA-dependent manner, a function that may be compromised by ΔE-TA. To address this hypothesis, we performed quantitative western blot analyses comparing the total amount of synaptic markers in control and ΔE-TA SH-SY5Y cells with those found upon CSN4 or TA knockdown (Figure 4A). Among the tested proteins, stonin 2 (STN2), a synaptotagmin-specific sorting adaptor (Maritzen et al, 2010) and the AP2 adaptor complex (Jung and Haucke, 2007; Dittman and Ryan, 2009) were negatively affected by siRNA knockdowns and showed a reduction quantitatively similar to that observed for snapin (compare Figures 4A with 3A). Strikingly, the expression level of stonin 2, but not that of AP2 is also markedly reduced in cells expressing the ΔE-TA mutant (Figure 4A; Supplementary Figure 4). These observations were confirmed by measuring the expression level of stonin 2 in three independent experiments (Figure 4B), as well as by analysing the distribution and intensity of the immunofluorescence signal for stonin 2 in SH-SY5Y cells following CSN4 or TA downregulation (Figure 4C). Consistent with these biochemical data, stonin 2 expression also appeared to be reduced at the light microscopic level in cells overexpressing HA-ΔE-TA (Figure 4D), while the intensity of the staining for snapin, observed in a parallel experiment, remains the same (data not shown). To confirm the effect of CSN4 and TA knockdown on stonin 2 protein levels, metabolic labelling of stonin 2 was performed as previously described for snapin (Figure 4E). After incubation with [35S]-methionine and treatment with cycloheximide, SH-SY5Y cells were analysed by immunoprecipitation with anti-stonin 2 antibodies. As predicted, the expression level of stonin 2 was significantly reduced in cells transfected with CSN4 and TA siRNA in comparison with control cells (Figure 4E). These results suggest that both CSN4 and TA control the stability of stonin 2. As stonin 2 was the only protein that appeared to be affected by the ΔE-TA mutant, we decided to further investigate the interaction between stonin 2, TA and ΔE-TA. To explore the possibility of a direct interaction between stonin 2 and the TA–CSN complex, we performed co-immunoprecipitation experiments in SH-SY5Y cell lines overexpressing HA-wt-TA or HA-ΔE-TA (Figure 5A). Stonin 2 was found to co-immunoprecipitate with HA-wt-TA, but not with HA-ΔE-TA, suggesting that ΔE mutation affects stonin 2 binding similarly to that seen for CSN4. Alternatively, the reduction in stonin 2 levels observed in HA-ΔE-TA-expressing cells (Figure 5A) may also influence its efficient detection in the immunoprecipitate by immunoblotting. Complex formation between stonin 2, TA and CSN4 was verified by immunoprecipitation from brain synaptosomes using specific anti-stonin 2 antibodies (Figure 5B). Furthermore, the interaction between stonin 2 and TA was also observed in SH-SY5Y cells treated with CSN4 siRNA, indicating that the binding of stonin 2 to TA is direct and independent of CSN4 (Figure 5C).
Figure 4.
Stonin 2 levels are affected by ΔE-TA overexpression, and CSN4 and TA knockdown. (A) The amount of the synaptic markers stonin 2 (STN2), synaptotagmin 1, AP2, synaptophysin, syntaxin 1 and VAMP2/synaptobrevin 2 was analysed by western blot in control SH-SY5Y cells, cells treated with CSN4 or TA siRNAs and cells expressing ΔE-TA. Endogenous stonin 2 and AP2 levels were reduced after cells treatment with CSN4 and TA siRNAs. Lower levels of stonin 2 were also seen in ΔE-TA-expressing cells. The total amount of synaptic proteins was increased as a consequence of MG132 treatment. (B) Total expression level of stonin 2 from three independent experiments is shown. Stonin 2 is reduced in cells treated with siRNA against CSN4 and TA and in cells expressing ΔE-TA, whereas it is increased in cells treated with MG132. Error bars represent s.e.m. (C) Immunofluorescence analysis of stonin 2 (red) showed reduced signal in cells transfected with siRNAs directed against TA (b) and CSN4 (c) compared with those transfected with control siRNA (a). A higher stonin 2 signal was seen after MG132 treatment (d). Stars indicate transfected cells. Scale bar=10 μm. (D) Immunofluorescence analysis shows reduced signal for stonin 2 (c; red) in cell overexpressing ΔE-TA (d; green). No changes in stonin 2 levels (a) were seen in cells expressing wt-TA (b). Scale bar=10 μm. (E) Metabolic labelling of stonin 2 in SH-SY5Y cells. Untreated cells (Ctl), cells treated with siRNA against CSN4 (CSN4 siRNA), TA (TA siRNA), ΔE-TA cell line and cells treated with MG132 were incubated overnight with [35S]-methionine. After 2 h treatment with cycloheximide, cells were analysed by immunoprecipitation using an anti-stonin 2 antibody and radioactive bands were revealed by autoradiography. The quantification of the efficiency of CSN4 and TA knockdown is shown in Supplementary Figure 2.
Figure 5.
Endogenous stonin 2 interacts with wt-TA and CSN4 and it is a potential target for CSN-mediated deneddylation. (A) Immunoprecipitation using an anti-HA antibody against HA-wt-TA and ΔE-TA overexpressed in SH-SY5Y cells shows that stonin 2 binds specifically wt-TA. Cells transfected with an empty vector were used as control (mock). Stonin 2 levels are reduced in cells overexpressing HA-ΔE-TA. β-actin was used as loading control. Lanes on the right (total) represent 1/10th of the starting material. (B, C) Stonin 2 forms a complex together with CSN4 and TA. Immunoprecipitation with a specific antibody against stonin 2 shows that it exists in a complex with TA and CSN4 in rat brain synaptosomes (B) and in SH-SY5Y cell lysates (C; Ctl). In cells in which CSN4 has been downregulated (CSN4 siRNA), stonin 2 still binds TA. Total lysates consist of 1/10th of the starting material. (D) Extracts from control (Ctl), ΔE-TA expressing and CSN4 knockdown (CSN4 siRNA) SH-SY5Y cells previously treated with lactacystin, a proteasome inhibitor, were immunoprecipitated with an anti-stonin 2 antibody and then blotted for stonin 2, NEDD8 and ubiquitin. Higher amount of neddylated and poly-ubiquitinated stonin 2 levels were recovered in ΔE-TA and CSN4 siRNA samples. The panel on the right shows 1/10th of the starting material. (E) Immunostaining with anti-NEDD8 (b; red) and anti-HA (a; blue) antibodies performed in ΔE-TA SH-SY5Y cells transfected with EGFP-STN2 (c; green) and pre-treated with lactacystin showed accumulation of EGFP-STN2 and NEDD8 in perinuclear ΔE-TA-positive inclusions (d; merge).
CSN complex affects the post-translational modifications of stonin 2 and snapin
Independent lines of evidence suggest that the CSN complex regulates protein stability via an isopeptidase activity that removes the ubiquitin-like protein NEDD8 from its substrates (Rabut and Peter, 2008). NEDD8 becomes covalently conjugated to a subset of cellular proteins, which includes the cullin subunit of the SCF ubiquitin E3 ligase, in a manner similar to ubiquitination (Hotton and Callis, 2008). We, therefore, examined whether stonin 2 or snapin could be targets for neddylation by performing immunoprecipitation assays from SH-SY5Y cells using antibodies against endogenous stonin 2 and snapin. As shown in Figure 5D, multiple bands recognised by the stonin 2-specific antibody were detected in immmunoprecipitates from cells treated with lactacystin, a proteasome inhibitor. The intensity of these bands increased in ΔE-TA overexpressing cells and in cells in which CSN4 was downregulated (Figure 5D). Blotting with specific antibodies against NEDD8 and ubiquitin showed that the upper bands recovered in anti-stonin 2 immunoprecipitation samples correspond to neddylated and polyubiquitinated form of stonin 2 (Figure 5D). These results suggest that stonin 2 may be a potential target for neddylation and consequent ubiquitination and that the neddylated form of stonin 2 accumulates in cells expressing ΔE-TA as well as in cells after treatment with siRNA against CSN4. Similarly, immunoprecipitation analysis with anti-NEDD8 antibody showed multiple bands for stonin 2, where intensity increased in ΔE-TA cells (Supplementary Figure 5). Consistent with this, immunofluorescence analysis for NEDD8 in SH-SY5Y cells expressing HA-ΔE-TA and transfected with EGFP-stonin 2 (EGFP-STN2) revealed an increased signal for NEDD8 and STN2 in the typical membrane inclusions induced by the ΔE-TA mutant upon treatment with lactacystin (Figure 5E). This observation, together with the presence of a strong NEDD8 signal in ΔE-TA positive membrane whorls (Supplementary Figure 6A), strongly suggests that neddylated proteins, including stonin 2, accumulate at the level of these perinuclear inclusions. In contrast, in control cells, NEDD8 displayed a more homogeneous distribution across the cytoplasm, with some accumulation at the tip of neurites (Supplementary Figure 6B). NEDD8 expression level and distribution are unaffected in cells overexpressing wt-TA (Supplementary Figure 6C).
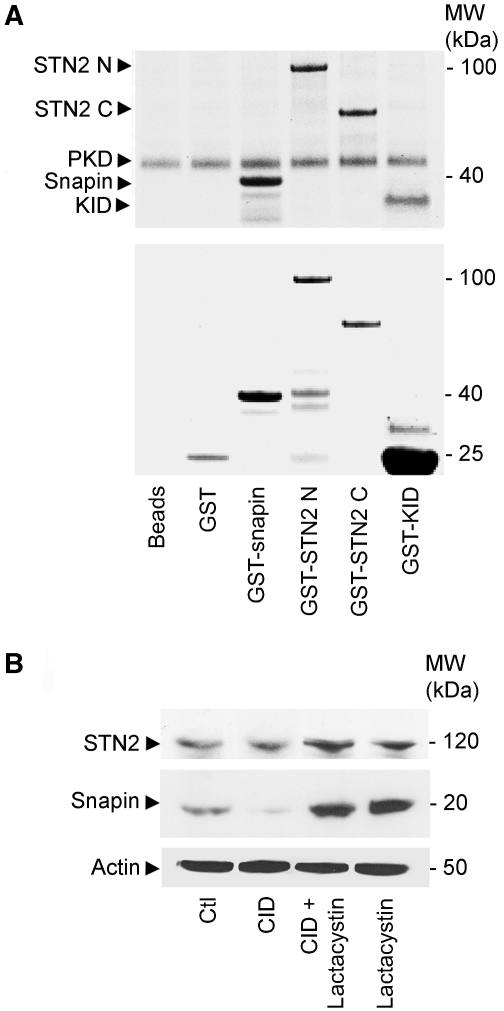
Strikingly, no signal for snapin was detectable in NEDD8 immmunoprecipitates (data not shown), suggesting that the mechanism of protein stabilisation mediated by the TA–CSN complex may involve other pathways. This view is supported by our in vitro phosphorylation experiments, showing that GST-snapin, but not GST alone, is phosphorylated in vitro by protein kinase D (PKD) (Figure 6A), a well-known CSN-associated kinase activity (Uhle et al, 2003). Interestingly, both the N- and C-terminal stonin 2 GST-fusion proteins are also efficiently phosphorylated by recombinant PKD in vitro. We further explored the possibility that the stability of snapin and stonin 2 could be affected by CSN-associated kinases by using a specific PKD inhibitor (CID; Figure 6B). We observed reduced snapin levels in cells treated with CID, which could be rescued by treatment with lactacystin, while stonin 2 stability was unaffected (Figure 6B). Consistent with this result, we found that curcumin, an inhibitor of CSN-associated kinase activity, altered the steady-state levels of snapin, (Figure 3A), without affecting stonin 2 (data not shown). This indicates that the mechanisms of stabilisation of stonin 2 and snapin are different and are likely to involve distinct post-translational modifications.
Figure 6.
Snapin levels are affected by PKD inhibition. (A) Recombinant GST-snapin, GST-N-terminal (1–555 aa; GST-STN2 N) and C-terminal (557–898 aa; GST-STN2 C) domains of stonin 2 are phosphorylated in vitro by recombinant PKD. A fragment of Kidins220/ARMS (KID; 871–961 aa) containing the phosphorylation site for PKD (Ser919) was used as positive control. (B) SH-SY5Y cells were treated with a specific inhibitor for PKD (CID) alone or in combination with lactacystin, PKD inhibition decreases the cellular levels of snapin compared with untreated cells (Ctl), while the level of stonin 2 appeared unaffected. β-actin was used as loading control.
Overexpression of EGFP-stonin 2 in ΔE-TA cells restores Syt1 recycling
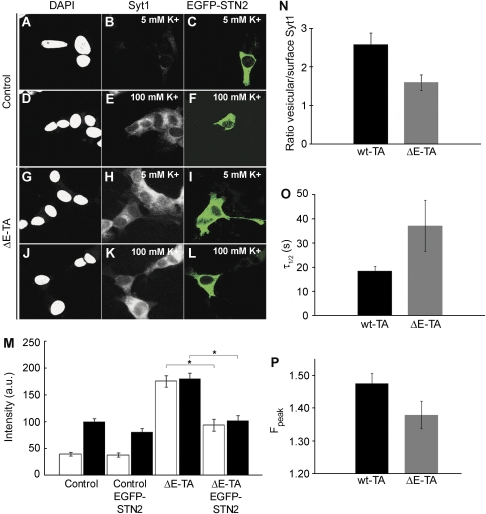
Our previous studies have demonstrated a role for TA in regulating SV trafficking. We found that endocytic recycling of Syt1, the main calcium sensor of exocytosis and an important factor in exo-endocytic coupling of SVs, is impaired in neuronal cells expressing ΔE-TA, resulting in Syt1 accumulation on the neuronal surface in both resting and depolarising conditions (Granata et al, 2008). This phenotype is reminiscent to that observed following manipulation of stonin 2 in primary neurons (Diril et al, 2006; Jung et al, 2007). Interestingly, while the total amount of Syt1 in ΔE-TA-expressing cells is unaffected, the level of expression of stonin 2, a main adaptor for Syt1 endocytosis, is negatively affected by ΔE-TA overexpression (Figure 4A). We, therefore, hypothesised that reduced levels of stonin 2 at the plasma membrane in ΔE-TA-expressing cells may cause defective Syt1 recycling. If this is the case, this defect might be partially restored by overexpression of stonin 2 in ΔE-TA cells. To test this hypothesis, we performed rescue experiments by overexpressing EGFP-stonin 2 in SH-SY5Y cells expressing ΔE-TA. After 24 h of transfection, quantification of Syt1 levels on the plasma membrane was carried out in both control and ΔE-TA cells using antibodies recognising the luminal domain of Syt1 (Granata et al, 2008) under resting (5 mM KCl; Figure 7A–C and G–I) or depolarising conditions (100 mM KCl; Figure 7D–F and J–L). Cells were then washed, fixed and stained with a fluorescently labelled secondary antibody. Mean fluorescence intensity of cells expressing EGFP-stonin 2 from three independent experiments was quantified and compared with control or ΔE-TA-expressing cells (Figure 7M). As expected, depolarisation led to increased surface expression of Syt1 on the plasma membrane of control cells (Figure 7E) when compared with resting conditions (Figure 7B). This was also observed in cells transfected with EGFP-stonin 2 (Figure 7C and F). As previously shown, neuroblastoma cells expressing mutant ΔE-TA displayed increased surface levels of Syt1 in both resting (Figure 7H) and depolarising conditions (Figure 7K). Strikingly, overexpression of EGFP-stonin 2 in ΔE-TA cells reduced the amount of surface-stranded Syt1 on the plasma membrane in resting conditions (Figure 7I and M) and completely rescued the Syt1 phenotype under depolarising conditions (Figure 7L and M). No significant changes in the total amount of Syt1 were seen in control, ΔE-TA cells untransfected or transfected with EGFP-stonin 2 in both resting and depolarising conditions (Supplementary Figure 7).
Figure 7.
The accumulation of Syt1 on the surface of ΔE-TA-expressing cells is rescued by EGFP-stonin 2 overexpression. Control and ΔE-TA-expressing SH-SY5Y cells were transfected with EGFP-stonin 2 (EGFP-STN2; in green) and analysed for Syt1 recycling in resting (5 mM K+) and depolarising conditions (100 mM K+). In control cells expressing EGFP-stonin 2 (A–F), the surface labelling for Syt1 increases in response to depolarisation (E, F) compared with resting conditions (B, C), similarly to untransfected control cells (EGFP negative; A–F). In contrast, in cells co-expressing ΔE-TA and EGFP-stonin 2 (G–L), the level of Syt1 on the cell surface is significantly reduced in resting cells (H, I) and depolarising conditions (K, L) compared with ΔE-TA-expressing cells. (M) Quantification of the intensity of anti-Syt1 antibody staining on the plasma membrane. Empty columns refer to resting condition (5 mM K+), whereas filled columns show the extent of Syt1 staining in depolarising condition (100 mM K+) (n=3). Error bars represent s.e. The asterisks indicate P<0.05 in Student's t-test. Scale bar=5 μm. (N) The vesicular-to-surface-stranded pool ratio of Syt1 was quantified in control wt-TA and ΔE-TA SH-SY5Y cells. Overexpression of ΔE-TA causes a decrease in the vesicular-to-surface pool ratio. Error bars represent s.e.m. (n=3; P<0.05 by Student's t-test). (O) wt-TA overexpressing neurons display faster retrieval of SytpHluorin when compared with ΔE-TA. Hippocampal neurons (DIV15) co-expressing SytpHluorin with wt- or ΔE-TA were stimulated with 200 APs at 20 Hz. Data are derived from the time course of normalised SytpHluorin fluorescence traces [(F−F0)/(Fpeak−F0)]. Time constants (τ1/2±s.e.m.) for retrieval were 18.37 s±2.06 for wt-TA and 36.92 s±10.85 for ΔE-TA (n=2; 565 boutons for w-TA and 567 boutons for ΔE-TA; P<0.06 by Student's t-test). (P) Effects of TA expression on SytpHluorin exo-endocytosis. Hippocampal neurons (DIV 15) expressing wt-TA or ΔE-TA were stimulated with 200 APs (20 Hz) and exo-endocytosis was monitored by following the time course of SytpHluorin fluorescence. Peak values of SytpHluorin relative fluorescence are plotted. Peak values±s.e.m. for wt-TA and ΔE-TA were 1.48±0.03 and 1.38±0.04, respectively; P<0.04 by Student's t-test).
To confirm these observations, we used pH-sensitive pHluorin GFP fused to the lumenal domain of Syt1 (SytpHluorin) as a reporter for Syt1 recycling (Diril et al, 2006) and to assess the distribution of Syt1 between SVs and the neuronal surface. SytpHluorin distribution was monitored in SH-SY5Y cells (Figure 7N) and in hippocampal neurons (Figure 7O and P). SH-SY5Y cells expressing wt- or ΔE-TA were transfected with SytpHluorin; 24 h post-transfection cells were stimulated with high potassium. Image analysis was performed in standard media and upon alkalinisation of the vesicular SV lumen using NH4Cl to maximise the fluorescence intensity of the vesicular SytpHluorin pool (quenching–dequenching conditions; Supplementary Figure 8). Fluorescence analysis after quenching–dequenching showed that under resting conditions, expression of ΔE-TA caused a partial retention of Syt1 on the plasma membrane as indicated by a decrease of the vesicular-to-surface-stranded pool ratio (Figure 7N). This effect resembles the phenotype observed upon overexpression of a dominant-negative stonin 2 mutant in neurons (Diril et al, 2006). These data were confirmed by kinetic analysis of SytpHluorin retrieval in primary hippocampal neurons co-expressing wt or mutant TA. Retrieval kinetics of SytpHluorin were assessed by measuring the time constant (τ1/2) required to retrieve half of all surface-stranded sytpHluorin molecules exocytosed during stimulation. Endocytosis kinetics were significantly faster in wt-TA compared with ΔE-TA-expressing neurons, suggesting that TA indeed facilitates sytpHluorin retrieval from the neuronal surface (Figure 7O). Conversely, fluorescence peak amplitudes were significantly smaller in ΔE-TA-expressing neurons (Figure 7P), indicating that enhanced exocytosis is not the cause for the kinetic differences in retrieval efficiency. Collectively, our data establish a molecular mechanism underlying the observed increase in surface-stranded Syt1 in neurons expressing the pathological TA mutant.
Discussion
In this study, we provide evidence for a novel regulatory mechanism of synaptic function via CSN, which controls the stability of specific SV proteins through interaction with TA. Previous studies have shown the association of TA with vesicles, axons and synaptic terminals in human and animal brains (Granata et al, 2009). In this study, we show that TA binds CSN4, the subunit 4 of the CSN complex, and modulates the CSN-dependent degradation of specific synaptic components. The core of CSN is composed of eight subunits (CSN1–8), which are related structurally to subunits of the lid of the 26S proteasome. Accordingly, CSN has been shown to control protein degradation through the ubiquitin–proteasome system (Schwechheimer and Deng, 2001; Bech-Otschir et al, 2002; von Arnim, 2003) and regulate a wide range of biological functions including embryonic development, cell cycle, checkpoint control, signal transduction, autophagy and circadian rhythm (Wei and Deng, 2003).
TA affects SV protein stability via CSN-associated activities
We have demonstrated that TA forms a complex with CSN4, snapin and stonin 2 in SH-SY5Y cells and synaptosomal fractions. Depletion of CSN4 in SH-SY5Y cells does not affect the binding of TA to snapin and stonin 2, indicating that these are direct interactions. However, downregulation of CSN4 leads to a significant reduction in the protein levels of snapin and stonin 2, as well as of the clathrin adaptor complex AP2. This result is consistent with the idea that CSN has an important function in synaptic protein turnover. Although the molecular mechanisms of CSN-mediated protein degradation remain to be clarified, recent studies have shown that CSN regulates the stability of target proteins by modifying their neddylation levels, as shown by its ability to deneddylate the cullin-RING-E3 ubiquitin ligase (Bosu and Kipreos, 2008). Conjugation to NEDD8 is a widespread post-translational modification and several substrates besides cullins have been reported, including p53 (Xirodimas et al, 2004). Among CSN subunits, CSN5, known also as Jun-activation-domain-binding protein 1 (Jab1) (Claret et al, 1996) has the unique ability to translocate from the nucleus to the cytoplasm and to bind various intracellular regulators, such as Cdk inhibitor p27, p53 and various transcriptional factors (Tomoda et al, 2005; Zhang et al, 2008). CSN5 has an important function in deneddylation (Cope et al, 2002), being the only subunit of CSN with a metalloenzyme domain, which is required to remove NEDD8. However, recent studies showed that other CSN subunits are required for this deneddylating activity (Sharon et al, 2009). We found that CSN5 is present in synaptosomal fractions and its stability is affected by CSN4 knockdown, which is likely to alter the overall neddylation activity of the complex. Our work demonstrates that stonin 2 is a potential target for deneddylation and CSN-mediated regulation of protein stability. Indeed, stonin 2 appeared preferentially neddylated in cells treated with siRNA against CSN4 or overexpressing the ΔE-TA mutant, and its protein levels were significantly reduced in these conditions. Such effect may be explained by the ΔE-TA-induced redistribution of wt-TA from its usual cellular localisation to perinuclear inclusions and the nuclear envelope. This possibility is supported by the finding that the interaction between wt-TA and mutant ΔE-TA is preserved (Supplementary Figure 9). In turn, the redistribution and/or sequestration of TA may affect the stability of stonin 2 by altering its degradation rate.
Both snapin and stonin 2 undergo phosphorylation by PKD, a kinase associated with CSN, but only snapin is selectively regulated by PKD inhibition. Several other proteins, such as c-Jun, NF-κB, IκBα and p53 have been identified as targets of CSN-associated kinases (Harari-Steinberg and Chamovitz, 2004). Interestingly, CSN-dependent phosphorylation has been shown to influence the stability of p53 (Bech-Otschir et al, 2001). CSN4 silencing may, therefore, reduce the phosphorylation levels of snapin and consequently promote its degradation. This is supported by the observation that snapin levels are reduced upon treatment with curcumin, which is known to inhibit CSN-associated kinases (Henke et al, 1999).
ΔE-TA inhibits Syt1 recycling by promoting stonin 2 degradation
Our previous work demonstrated that Syt1 accumulated on the plasma membrane as a consequence of mutant ΔE-TA expression. We have now shown that this Syt1 recycling defect is caused by degradation of stonin 2 triggered by ΔE-TA. Stonin 2 is a specific sorting adaptor mediating the interaction between the SV calcium sensor Syt1 and components of the endocytic machinery, such as AP2, both of which are required for Syt1 internalisation and recycling (Diril et al, 2006). Interestingly, we showed that AP2 levels are also decreased upon CSN4 and TA downregulation, suggesting that TA and CSN4 could be part of a general mechanism regulating SV protein stability. Previous studies have shown that stonin 2 co-localises with Syt1 at presynaptic sites, in which it appears to affect the partitioning of Syt1 between the plasma membrane and SVs (Diril et al, 2006). Interestingly, cells expressing a dominant-negative stonin 2 mutant displayed a reduced ability to internalise Syt1 from the plasma membrane, generating a phenotype that closely resembles the Syt1 recycling deficit found in SH-SY5Y cells overexpressing ΔE-TA (Diril et al, 2006; Granata et al, 2008). On this basis, we investigated whether the accumulation of Syt1 on the plasma membrane of ΔE-TA-expressing cells is caused by a depletion of stonin 2. Overexpression of stonin 2 successfully rescued Syt1 retrieval in ΔE-TA-expressing cells after depolarisation. However, a substantial amount of Syt1 is present on the plasma membrane of cells co-expressing ΔE-TA and exogenous stonin 2 in resting conditions, suggesting that the machinery coupling calcium sensing with SV exo-endocytosis at the synapse is still impaired. AP2 and other still uncharacterised CSN substrates may also have a function in this pathway. Consistent with these observations, SytpHluorin recycling assays in SH-SY5Y and primary neurons revealed increased surface levels of this reporter, implicating reduced retrieval efficiency of Syt1 in ΔE-TA overexpressing cells compared with wt-TA controls.
CSN regulates the degradation machinery at the synapse
CSN is essential for life as the knockout of any of its subunits in mice leads to early lethality (Lykke-Andersen et al, 2003). A similar phenotype is observed for NEDD8 knockout (Tateishi et al, 2001). In addition, deregulation of neddylation appears to be involved in human diseases, such as neurodegeneration (Mori et al, 2005) and cancer (Chairatvit and Ngamkitidechakul, 2007). Notably, NEDD8 localises at the tips of the neurites in neuroblastoma cells, suggesting that it may be involved in neurite outgrowth and/or SV function. In contrast, in cells expressing ΔE-TA, NEDD8 was found to accumulate in ΔE-TA positive membrane-rich inclusions. This may imply that the NEDD8 pathway could be altered or disrupted by mutant TA.
TA appears to be important in ensuring the stability of its interacting partners, such as snapin and stonin 2, as TA knockdown mimics the effect of CSN4 silencing. TA may be able to regulate the correct folding of these proteins, which in turn influences their stability. To date, the only evidence that TA may have a role in regulating protein stability comes from the analysis of sarcoglycan traffic (Esapa et al, 2007), in which TA has been suggested to co-operate with the proteasome to enhance the clearance of misfolded ɛ-sarcoglycan.
Here, we propose that TA may have an important function in regulating the stability of the specific synaptic proteins snapin and stonin 2, and possibly others, including AP2, which are rescued from proteasomal degradation via CSN-associated activities. Dysregulation of this process in the presence of ΔE-TA may lead to increased degradation, reducing the availability of snapin and stonin 2, which in turn can compromise the recycling of Syt1 at the surface and affect neurotransmitter release by altering SV exo-endocytosis.
Materials and methods
Chemicals and antibodies
Reagents were from Sigma-Aldrich (St Louis, MO), unless otherwise specified. Primary antibodies are listed in Supplementary Table 1. AlexaFluor488-, AlexaFluor555- and AlexFluor633-conjugated secondary antibodies were from Invitrogen (Paisley, UK). Horseradish peroxidase (HRP)-conjugated secondary antibodies were from DAKO (Ely, UK) and ECL was from GE Healthcare (Little Chalfont, UK). Curcumin was from BIOMOL International (Exeter, UK) and MG132 from Calbiochem (Darmstadt, DE). Recombinant PKD catalytic domain was a kind gift from H Jefferies and PJ Parker (Cancer Research UK London Research Institute, UK). SytpHluorin, EGFP-stonin 2 and GST N- and C-terminal stonin 2 constructs were previously described (Walther et al, 2004).
Cell culture
SH-SY5Y cell lines stably transfected with pcDNA3.1, containing either wild-type DYT1 (wt-TA), GAG-deleted DYT1 (ΔE-TA) or no insert were grown in of DMEM/F12 Nutrient 1:1 mixture (Invitrogen) and 10% foetal calf serum at 37°C and 5% CO2 under selective conditions (0.4 mg/ml G418; Invitrogen).
Yeast two-hybrid screen
The Matchmaker yeast two-hybrid system 3 (Clontech, Mountain View, CA) was used according to manufacturer's instructions. The generation of the baits and the screening procedure have been previously described (Granata et al, 2008). Full-length CSN4 was found as a hit for both wt-TA and ΔE-TA baits, when co-transformed in AH109 yeast cells.
Pull-down assays
A total of 1 μg of CSN4 or snapin cDNAs were used to generate [35S]-labelled proteins using TnT Quick coupled transcription/translation system (Promega, Wisconsin, WI). In case of the competition pull down, both CSN4 and snapin cDNAs were added to the reaction mix, which was pre-cleared on glutathione-Sepharose for 1 h at 4°C and then incubated with either pre-bound GST-fusion proteins or GST alone for 1 h at 4°C in Hank's-BSA (20 mM Hepes-NaOH pH 7.4, 0.44 mM KH2PO4, 0.42 mM NaH2PO4, 5.36 mM KCl, 136 mM NaCl, 0.81 mM MgSO4, 1.26 mM CaCl2, 6.1 mM glucose, 0.1% BSA). Beads were then washed with ice-cold Hank's-BSA containing 250 mM NaCl, 1% Triton X-100, resuspended in loading buffer and after SDS–PAGE, analysed by autoradiography.
Immunoprecipitation and western blot
Synaptosomes and SH-SY5Y cells expressing HA-wt-TA and HA-ΔE-TA were lysed in lysis buffer (50 mM Hepes-NaOH pH 7.4, 0.1 mM EDTA and protease inhibitors) containing 0.5% CHAPS (Calbiochem) and 1% Triton X-100 for 20 min at 4°C under constant agitation. Cell extracts and synaptosomes (20 μg protein/lane) were incubated with anti-HA or anti-CSN4 or anti-stonin 2 antibodies (10 μg antibody/sample) at 4°C overnight. As a negative control, lysates were mixed with an irrelevant antibody (anti-GFP). Anti-NEDD8 antibody was added to SH-SY5Y cells previously lysed in a buffer containing 0.5% NP40 (50 mM Tris–HCl, pH 8.0; 120 mM NaCl). Anti-GFP antibody was used to immunoprecipitate EGFP-wt-TA, generated by subcloning full-length wt DYT1 into EGFP-N2 vector and transfected in HA-ΔE-TA cells. Protein G or A-Sepharose beads (GE Healthcare) were then added to each samples and incubated for 1 h at 4°C under constant stirring. After extensive washes with lysis buffer containing 0.5% CHAPS and 1% Triton X-100 or 0.5% NP40 for NEDD8-bound beads, samples were resuspended in loading buffer, boiled and analysed in SDS–PAGE followed by western blot. PVDF membranes were incubated with anti-HA (1:1000), anti-CSN4 (1:250), anti-snapin (1:500), anti-stonin 2 (1:250), anti-NEDD8 (1:500), anti-poly-ubiquitinylated proteins (1:200) and anti-GFP (1:1000) antibodies, followed by HRP-conjugated secondary antibodies and revealed by ECL.
Immunofluorescence
SH-SY5Y cells were plated onto glass coverslips and grown overnight. Cells were fixed with 4% paraformaldehyde (PFA) in PBS for 20 min at room temperature, washed and incubated with PBS containing 10% goat serum for 1 h. Primary antibodies were diluted (anti-TA 1:500, anti-CSN4 1:200, anti-NEDD8 1:500 and anti-STN2 1:100) in PBS containing 0.1% Triton X-100 and 10% goat serum, and incubated for 1 h. After rinsing with PBS three times for 10 min, secondary antibodies diluted in PBS (1:1000) were applied for 1 h. Coverslips were then washed and mounted with Mowiol 4–88 (EMD Bioscience, La Jolla, CA), containing Hoechst 33258 (1:1000; Invitrogen). Images were acquired by confocal microscopy (Zeiss LSM510; Carl Zeiss, Jena, DE) with a 63x Plan-Apochromat oil immersion objective.
siRNA and inhibitors
On-target plus Smartpool of siRNAs for human TA (60 nM) and human CSN4 (40 nM, Dharmacon, Chicago, IL) were used to knockdown gene expression. In total, 20 nM siGLO control fluorescent siRNA (Dharmacon) was used alone as control or combined with TA or CSN4 siRNA to visualise transfected cells. siRNAs were transfected into SHSY-5Y cells using the DharmaFECT transfection protocol (Dharmacon) and cells were harvested at 72 h after transfections. Knockdown of TA and CSN4 was verified by western blot using anti-TA and rabbit anti-CSN4 antibodies. When necessary, cells were incubated with 50 μM of curcumin or 10 μM of MG132 in medium overnight or lactacystin (50 μM) for 4 h at 37°C, harvested and lysed as described. Protein concentration was measured by Bradford (Bio-Rad Laboratories, Hercules, CA); 20 μg of protein were loaded and analysed for snapin and stonin 2 by western blot. Bands were quantified (n=3) using NIH Image 1.61 (http://rsb.info.nih.gov/nih-image/). For immunofluorescence, SH-SY5Y cells were plated on coverslips, transfected with siRNA for TA or CSN4 for 72 h or treated with MG132 overnight, then fixed with 4% PFA and permeabilised with 0.05% saponin. After blocking with 10% goat serum, cells were stained with anti-snapin (1:500) or anti-stonin 2 (1:200) and Hoechst 33342, followed by AlexaFluor555 secondary antibody.
Cells transfection and Syt1 recycling assay
Electroporation was used to transfect EGFP-stonin 2 (Walther et al, 2004) into wt-TA and ΔE-TA-expressing SH-SY5Y cells. Cells were detached with EDTA/trypsin, washed with PBS and resuspended with 10 μl of resuspension buffer at 0.5 × 105 cell/ml. Resuspended cells were mixed with 0.5 μg/μl of EGPF-stonin 2 and transfected in a pipette-type electroporator (MicroPorator MP-100, Digital Bio, Huston, TX) using the following conditions: 1100 V for 30 ms; n=2. After 24 h transfection, SV recycling was monitored using a polyclonal antibody (Syt-136) against the intraluminal domain of Syt1 as described previously (Granata et al, 2008). Cells were then fixed and stained with AlexaFluor555-conjugated secondary antibody. Images were acquired by confocal microscopy using a 63x Plan-Apochromat oil immersion objective with the pinhole fully open. The experiment was repeated three times, 10 pictures for each sample, including control, ΔE-TA, control positive for EGFP-stonin 2 and ΔE-TA positive for stonin 2, were taken and the mean fluorescence intensity of individual cells was measured using Zeiss LSM 510 software version 3.2.
Supplementary Material
Acknowledgments
We thank H Jefferies and PJ Parker (Cancer Research UK London Research Institute, UK) for the catalytic domain of recombinant PKD. The microscopy analysis was supported by the Confocal Microscopy Unit managed by Dr Shane Minogue. This work was supported by grants from The Brain Research Trust, The Dystonia Society (AG and TTW), Cancer Research UK (GS), The Wellcome Trust (AG, GS and TTW), the Deutsche Forschungsgemeinschaft (Exc-257; HA2686/6-1; VH), the Schram Foundation (VH) and the Helmholtz International Research School for Molecular Neurobiology (SJK).
Footnotes
The authors declare that they have no conflict of interest.
References
- Balcioglu A, Kim MO, Sharma N, Cha JH, Breakefield XO, Standaert DG (2007) Dopamine release is impaired in a mouse model of DYT1 dystonia. J Neurochem 102: 783–788 [DOI] [PubMed] [Google Scholar]
- Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, Dubiel W (2001) COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J 20: 1630–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D, Seeger M, Dubiel W (2002) The COP9 signalosome: at the interface between signal transduction and ubiquitin-dependent proteolysis. J Cell Sci 115: 467–473 [DOI] [PubMed] [Google Scholar]
- Bosu DR, Kipreos ET (2008) Cullin-RING ubiquitin ligases: global regulation and activation cycles. Cell Div 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breakefield XO, Kamm C, Hanson PI (2001) TorsinA: movement at many levels. Neuron 31: 9–12 [DOI] [PubMed] [Google Scholar]
- Chairatvit K, Ngamkitidechakul C (2007) Control of cell proliferation via elevated NEDD8 conjugation in oral squamous cell carcinoma. Mol Cell Biochem 306: 163–169 [DOI] [PubMed] [Google Scholar]
- Claret FX, Hibi M, Dhut S, Toda T, Karin M (1996) A new group of conserved coactivators that increase the specificity of AP-1 transcription factors. Nature 383: 453–457 [DOI] [PubMed] [Google Scholar]
- Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298: 608–611 [DOI] [PubMed] [Google Scholar]
- Dang MT, Yokoi F, McNaught KS, Jengelley TA, Jackson T, Li J, Li Y (2005) Generation and characterization of Dyt1 DeltaGAG knock-in mouse as a model for early-onset dystonia. Exp Neurol 196: 452–463 [DOI] [PubMed] [Google Scholar]
- Diril MK, Wienisch M, Jung N, Klingauf J, Haucke V (2006) Stonin 2 is an AP-2-dependent endocytic sorting adaptor for synaptotagmin internalization and recycling. Dev Cell 10: 233–244 [DOI] [PubMed] [Google Scholar]
- Dittman J, Ryan TA (2009) Molecular circuitry of endocytosis at nerve terminals. Annu Rev Cell Dev Biol 25: 133–160 [DOI] [PubMed] [Google Scholar]
- Esapa CT, Waite A, Locke M, Benson MA, Kraus M, McIlhinney RA, Sillitoe RV, Beesley PW, Blake DJ (2007) SGCE missense mutations that cause myoclonus-dystonia syndrome impair epsilon-sarcoglycan trafficking to the plasma membrane: modulation by ubiquitination and torsinA. Hum Mol Genet 16: 327–342 [DOI] [PubMed] [Google Scholar]
- Ferrari-Toninelli G, Paccioretti S, Francisconi S, Uberti D, Memo M (2004) TorsinA negatively controls neurite outgrowth of SH-SY5Y human neuronal cell line. Brain Res 1012: 75–81 [DOI] [PubMed] [Google Scholar]
- Fukumoto A, Tomoda K, Kubota M, Kato JY, Yoneda-Kato N (2005) Small Jab1-containing subcomplex is regulated in an anchorage- and cell cycle-dependent manner, which is abrogated by ras transformation. FEBS Lett 579: 1047–1054 [DOI] [PubMed] [Google Scholar]
- Giles LM, Li L, Chin LS (2009) Printor, a novel torsinA-interacting protein implicated in dystonia pathogenesis. J Biol Chem 284: 21765–21775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Dauer WT (2005) The AAA+ protein torsinA interacts with a conserved domain present in LAP1 and a novel ER protein. J Cell Biol 168: 855–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodchild RE, Kim CE, Dauer WT (2005) Loss of the dystonia-associated protein torsinA selectively disrupts the neuronal nuclear envelope. Neuron 48: 923–932 [DOI] [PubMed] [Google Scholar]
- Granata A, Schiavo G, Warner TT (2009) TorsinA and dystonia: from nuclear envelope to synapse. J Neurochem 109: 1596–1609 [DOI] [PubMed] [Google Scholar]
- Granata A, Watson R, Collinson LM, Schiavo G, Warner TT (2008) The dystonia-associated protein torsinA modulates synaptic vesicle recycling. J Biol Chem 283: 7568–7579 [DOI] [PubMed] [Google Scholar]
- Harari-Steinberg O, Chamovitz DA (2004) The COP9 signalosome: mediating between kinase signaling and protein degradation. Curr Protein Pept Sci 5: 185–189 [DOI] [PubMed] [Google Scholar]
- Henke W, Ferrell K, Bech-Otschir D, Seeger M, Schade R, Jungblut P, Naumann M, Dubiel W (1999) Comparison of human COP9 signalsome and 26S proteasome lid. Mol Biol Rep 26: 29–34 [DOI] [PubMed] [Google Scholar]
- Hewett J, Gonzalez-Agosti C, Slater D, Ziefer P, Li S, Bergeron D, Jacoby DJ, Ozelius LJ, Ramesh V, Breakefield XO (2000) Mutant torsinA, responsible for early-onset torsion dystonia, forms membrane inclusions in cultured neural cells. Hum Mol Genet 9: 1403–1413 [DOI] [PubMed] [Google Scholar]
- Hewett JW, Zeng J, Niland BP, Bragg DC, Breakefield XO (2006) Dystonia-causing mutant torsinA inhibits cell adhesion and neurite extension through interference with cytoskeletal dynamics. Neurobiol Dis 22: 98–111 [DOI] [PubMed] [Google Scholar]
- Hotton SK, Callis J (2008) Regulation of cullin RING ligases. Annu Rev Plant Biol 59: 467–489 [DOI] [PubMed] [Google Scholar]
- Ilardi JM, Mochida S, Sheng ZH (1999) Snapin: a SNARE-associated protein implicated in synaptic transmission. Nat Neurosci 2: 119–124 [DOI] [PubMed] [Google Scholar]
- Jung N, Haucke V (2007) Clathrin-mediated endocytosis at synapses. Traffic 8: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Jung N, Wienisch M, Gu M, Rand JB, Muller SL, Krause G, Jorgensen EM, Klingauf J, Haucke V (2007) Molecular basis of synaptic vesicle cargo recognition by the endocytic sorting adaptor stonin 2. J Cell Biol 179: 1497–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamm C, Boston H, Hewett J, Wilbur J, Corey DP, Hanson PI, Ramesh V, Breakefield XO (2004) The early onset dystonia protein torsinA interacts with kinesin light chain 1. J Biol Chem 279: 19882–19892 [DOI] [PubMed] [Google Scholar]
- Kato JY, Yoneda-Kato N (2009) Mammalian COP9 signalosome. Genes Cells 14: 1209–1225 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen K, Schaefer L, Menon S, Deng XW, Miller JB, Wei N (2003) Disruption of the COP9 signalosome Csn2 subunit in mice causes deficient cell proliferation, accumulation of p53 and cyclin E, and early embryonic death. Mol Cell Biol 23: 6790–6797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maritzen T, Podufall J, Haucke V (2010) Stonins—specialized adaptors for synaptic vesicle recycling and beyond? Traffic 11: 8–15 [DOI] [PubMed] [Google Scholar]
- Martina JA, Bonangelino CJ, Aguilar RC, Bonifacino JS (2001) Stonin 2: an adaptor-like protein that interacts with components of the endocytic machinery. J Cell Biol 153: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misbahuddin A, Placzek MR, Taanman JW, Gschmeissner S, Schiavo G, Cooper JM, Warner TT (2005) Mutant torsinA, which causes early-onset primary torsion dystonia, is redistributed to membranous structures enriched in vesicular monoamine transporter in cultured human SH-SY5Y cells. Mov Disord 20: 432–440 [DOI] [PubMed] [Google Scholar]
- Mori F, Nishie M, Piao YS, Kito K, Kamitani T, Takahashi H, Wakabayashi K (2005) Accumulation of NEDD8 in neuronal and glial inclusions of neurodegenerative disorders. Neuropathol Appl Neurobiol 31: 53–61 [DOI] [PubMed] [Google Scholar]
- Nery FC, Zeng J, Niland BP, Hewett J, Farley J, Irimia D, Li Y, Wiche G, Sonnenberg A, Breakefield XO (2008) TorsinA binds the KASH domain of nesprins and participates in linkage between nuclear envelope and cytoskeleton. J Cell Sci 121: 3476–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV (1999) AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res 9: 27–43 [PubMed] [Google Scholar]
- Ozelius LJ, Hewett JW, Page CE, Bressman SB, Kramer PL, Shalish C, de LD, Brin MF, Raymond D, Corey DP, Fahn S, Risch NJ, Buckler AJ, Gusella JF, Breakefield XO (1997) The early-onset torsion dystonia gene (DYT1) encodes an ATP-binding protein. Nat Genet 17: 40–48 [DOI] [PubMed] [Google Scholar]
- Pan PY, Tian JH, Sheng ZH (2009) Snapin facilitates the synchronization of synaptic vesicle fusion. Neuron 61: 412–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peth A, Berndt C, Henke W, Dubiel W (2007) Downregulation of COP9 signalosome subunits differentially affects the CSN complex and target protein stability. BMC Biochem 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Peter M (2008) Function and regulation of protein neddylation. ‘Protein modifications: beyond the usual suspects'. EMBO Rep 9: 969–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C (2004) The COP9 signalosome (CSN): an evolutionary conserved proteolysis regulator in eukaryotic development. Biochim Biophys Acta 1695: 45–54 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Deng XW (2001) COP9 signalosome revisited: a novel mediator of protein degradation. Trends Cell Biol 11: 420–426 [DOI] [PubMed] [Google Scholar]
- Sharon M, Mao H, Boeri EE, Stephens E, Zheng N, Robinson CV (2009) Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure 17: 31–40 [DOI] [PubMed] [Google Scholar]
- Tateishi K, Omata M, Tanaka K, Chiba T (2001) The NEDD8 system is essential for cell cycle progression and morphogenetic pathway in mice. J Cell Biol 155: 571–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian JH, Wu ZX, Unzicker M, Lu L, Cai Q, Li C, Schirra C, Matti U, Stevens D, Deng C, Rettig J, Sheng ZH (2005) The role of Snapin in neurosecretion: snapin knock-out mice exhibit impaired calcium-dependent exocytosis of large dense-core vesicles in chromaffin cells. J Neurosci 25: 10546–10555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoda K, Kato JY, Tatsumi E, Takahashi T, Matsuo Y, Yoneda-Kato N (2005) The Jab1/COP9 signalosome subcomplex is a downstream mediator of Bcr-Abl kinase activity and facilitates cell-cycle progression. Blood 105: 775–783 [DOI] [PubMed] [Google Scholar]
- Tomoda K, Kubota Y, Arata Y, Mori S, Maeda M, Tanaka T, Yoshida M, Yoneda-Kato N, Kato JY (2002) The cytoplasmic shuttling and subsequent degradation of p27Kip1 mediated by Jab1/CSN5 and the COP9 signalosome complex. J Biol Chem 277: 2302–2310 [DOI] [PubMed] [Google Scholar]
- Torres GE, Sweeney AL, Beaulieu JM, Shashidharan P, Caron MG (2004) Effect of torsinA on membrane proteins reveals a loss of function and a dominant-negative phenotype of the dystonia-associated DeltaE-torsinA mutant. Proc Natl Acad Sci USA 101: 15650–15655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhle S, Medalia O, Waldron R, Dumdey R, Henklein P, Bech-Otschir D, Huang X, Berse M, Sperling J, Schade R, Dubiel W (2003) Protein kinase CK2 and protein kinase D are associated with the COP9 signalosome. EMBO J 22: 1302–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vites O, Rhee JS, Schwarz M, Rosenmund C, Jahn R (2004) Reinvestigation of the role of snapin in neurotransmitter release. J Biol Chem 279: 26251–26256 [DOI] [PubMed] [Google Scholar]
- von Arnim AG (2003) On again-off again: COP9 signalosome turns the key on protein degradation. Curr Opin Plant Biol 6: 520–529 [DOI] [PubMed] [Google Scholar]
- Walther K, Diril MK, Jung N, Haucke V (2004) Functional dissection of the interactions of stonin 2 with the adaptor complex AP-2 and synaptotagmin. Proc Natl Acad Sci USA 101: 964–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei N, Deng XW (1999) Making sense of the COP9 signalosome. A regulatory protein complex conserved from Arabidopsis to human. Trends Genet 15: 98–103 [DOI] [PubMed] [Google Scholar]
- Wei N, Deng XW (2003) The COP9 signalosome. Annu Rev Cell Dev Biol 19: 261–286 [DOI] [PubMed] [Google Scholar]
- Xirodimas DP, Saville MK, Bourdon JC, Hay RT, Lane DP (2004) Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell 118: 83–97 [DOI] [PubMed] [Google Scholar]
- Zhang XC, Chen J, Su CH, Yang HY, Lee MH (2008) Roles for CSN5 in control of p53/MDM2 activities. J Cell Biochem 103: 1219–1230 [DOI] [PubMed] [Google Scholar]
- Zhao Y, DeCuypere M, LeDoux MS (2008) Abnormal motor function and dopamine neurotransmission in DYT1 DeltaGAG transgenic mice. Exp Neurol 210: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.