Molecular mechanism by which the nucleoid occlusion factor, SlmA, keeps cytokinesis in check
Nucleoid occlusion (NO) restricts bacterial cell division to prevent chromosome guillotining in the cell midzone when replication or segregation is delayed. Structural work suggests that the NO factor SlmA (synthetic lethal with a defective Min system) interferes with formation of the cytokinetic Z-ring by altering associations between FtsZ protofilaments.
Keywords: bacterial cell division, chromosome segregation, FtsZ Z-ring formation, nucleoid occlusion, SlmA
Abstract
In Escherichia coli, cytokinesis is orchestrated by FtsZ, which forms a Z-ring to drive septation. Spatial and temporal control of Z-ring formation is achieved by the Min and nucleoid occlusion (NO) systems. Unlike the well-studied Min system, less is known about the anti-DNA guillotining NO process. Here, we describe studies addressing the molecular mechanism of SlmA (synthetic lethal with a defective Min system)-mediated NO. SlmA contains a TetR-like DNA-binding fold, and chromatin immunoprecipitation analyses show that SlmA-binding sites are dispersed on the chromosome except the Ter region, which segregates immediately before septation. SlmA binds DNA and FtsZ simultaneously, and the SlmA–FtsZ structure reveals that two FtsZ molecules sandwich a SlmA dimer. In this complex, FtsZ can still bind GTP and form protofilaments, but the separated protofilaments are forced into an anti-parallel arrangement. This suggests that SlmA may alter FtsZ polymer assembly. Indeed, electron microscopy data, showing that SlmA–DNA disrupts the formation of normal FtsZ polymers and induces distinct spiral structures, supports this. Thus, the combined data reveal how SlmA derails Z-ring formation at the correct place and time to effect NO.
Introduction
Cell division or cytokinesis is one of the most fundamental processes in biology and is essential for the propagation of all living species. In Escherichia coli, cell division occurs by in-growth of the membrane envelope at the cell centre and is orchestrated by the FtsZ protein (Margolin, 2005; Osawa et al, 2008; Adams and Errington, 2009). FtsZ has a tubulin-like fold and self-assembles into linear protofilaments in a GTP-dependent manner by the interaction of the plus end of one subunit with the minus end of another subunit, resulting in a head-to-tail geometry. While FtsZ and tubulin protofilaments are similar, the higher order polymers they form are notably different. Specifically, tubulin protofilaments interact to produce microtubules, while FtsZ protofilaments combine to form a cytoskeletal scaffold called the Z-ring (Erickson et al, 1996; Nogales, 2000). The Z-ring provides the framework for the assembly of the division apparatus and determines the site of cytokinesis (Erickson et al, 1996; Adams and Errington, 2009). Several studies have suggested that the functional unit of FtsZ used in Z-ring formation consists of parallel interacting FtsZ protofilaments, which have been termed ‘thick filaments' (Löwe and Amos, 1999; Oliva et al, 2003). However, the precise arrangement of FtsZ protofilaments within the Z-ring is currently unknown. The total amount of FtsZ molecules in a cell significantly exceeds the concentration required for Z-ring formation, and this concentration remains constant during the cell cycle. Hence, Z-ring formation must be highly regulated, both temporally and spatially. In particular, the assembly of Z-rings at the cell poles and over chromosomal DNA must be prevented. These inhibitory roles are played by two key regulatory systems called the Min system and the nucleoid occlusion (NO) system (Yu and Margolin, 1999).
The Min system has been extensively studied and, in E. coli, is comprised of the FtsZ inhibitor, MinC, membrane-associated ATPase called MinD and MinE, a factor that binds and spatially organizes the MinCD complex (Hu and Lutkenhaus, 2001; Hu et al, 2002; Shih et al, 2003). MinC, which interacts with MinD, inhibits FtsZ polymerization by preventing lateral interactions required for Z-ring formation. MinE binds MinCD and oscillates from pole to pole (Raskin and de Boer, 1999a, 1999b). The net result of this oscillatory process is the formation of a zone of FtsZ inhibition at the cell poles. However, the replicated nucleoid DNA near the midcell must also be protected from bisection by the Z-ring and this is ensured by NO. In contrast to the Min system, the mechanisms responsible for NO have been unclear. Indeed, although the process of NO was proposed over 20 years ago by Woldringh et al. (1990, 1991), it took until 2004 for Wu and Errington to identify a factor, Noc, that is responsible for NO in Bacillus subtilis (Wu and Errington, 2004). Cells lacking Noc had no obvious cell division phenotype, but inhibiting DNA replication, in a Min-mutant background, resulted in aberrant formation of cell division septa over unpartitioned nucleoids and subsequent nucleoid guillotining. Furthermore, Noc localized to nucleoids and excess Noc inhibited division. These findings established Noc as a bone fide NO factor. Subsequently, it was shown that Noc binds to specific DNA sites with the consensus, 5′-ATTTCCCGGGAAAT-3′ in the B. subtilis chromosome (Wu et al, 2009). However, the mechanism by which Noc prevents Z-ring formation over the nucleoid is still unclear, as it does not appear to bind FtsZ or any regulator of cell division.
Following the discovery of Noc, a 198 residue protein called SlmA (synthetic lethal with a defective Min system) was shown to be the effector of NO in E. coli (Bernhardt and de Boer, 2005). SlmA was identified similarly to Noc, in a screen designed to isolate mutations that were lethal in the absence of Min, hence the name SlmA. Like Noc, SlmA was shown to bind DNA and localized to the nucleoid fraction of the cell. However, SlmA and Noc show no sequence homology and belong to different families of DNA-binding proteins. While Noc is a ParB family member, SlmA contains a putative N-terminal helix-turn-helix (HTH) motif and a predicted C-terminal coiled coil (Bernhardt and de Boer, 2005; Schumacher, 2008). Light scattering experiments suggested that SlmA interacts with FtsZ–GTP and alters its polymerization properties. However, this interaction appeared to enhance rather than disrupt polymer formation, leaving in question how it could be involved in NO. Here, we describe studies that reveal the molecular mechanism by which SlmA mediates NO in E. coli. Specifically, we determined the crystal structure of SlmA, identified its DNA-binding site specificity and mapped its binding sites on the E. coli chromosome by chromatin immunoprecipitation (ChIP) experiments. We went on to determine the SlmA–FtsZ structure by small-angle X-ray scattering (SAXS) and examined the affect of SlmA–DNA on FtsZ polymerization by electron microscopy (EM). Our combined data show how SlmA is able to disrupt Z-ring formation through its interaction with FtsZ in a specific temporal and spatial manner and hence prevent nucleoid guillotining during cell division.
Results and discussion
Crystal structure of E. coli SlmA
To gain insight into the function of SlmA, we first determined its crystal structure to 2.50 Å resolution by multiple wavelength anomalous diffraction (MAD; Supplementary Table SI). The final SlmA structure consists of residues 9–25, 32–113, 120–148, 150–198, contains 14 solvent molecules and has Rwork/Rfree values of 22.4%/26.5%. The structure shows that SlmA is comprised of nine helices (α1–α9) and can be divided into two domains, a small N-terminal domain (residues 1–53) and a C-terminal domain (residues 54–198) (Figure 1A). The N-terminal domain is formed by the first three helices (α1–α3). Helices 2 and 3 form a canonical HTH motif, suggesting that this domain functions in DNA binding. Helices α4–α9 form the C-terminal domain, and crystal packing analyses reveal that this region mediates dimerization. The dimer interface buries an extensive 2640 Å2 of protein surface from solvent. Dimerization or protein–protein binding energy has been shown to be directly related to the buried hydrophobic surface area (Janin et al, 1988). The SlmA dimer interface is notable in this regard as it is almost entirely hydrophobic. Residues that are involved in dimerization include Leu171, Gln175, Ala178, Phe179 and Met183 on the internal face of helix α8, and Ala202 and Ala209 from helix α9 (Figures 1B). Mutation of three hydrophobic residues, Leu171Arg, Gln175Arg and Phe179Arg, resulted in insoluble protein that was found in inclusion bodies, underscoring the important role these residues have in dimerization and hence proper protein folding (Supplementary Figure S1A–B). Size-exclusion chromatography analyses, which resulted in a calculated mass of 48 kDa, support that SlmA is dimeric (Figure 1C).
Figure 1.
Overall structure of SlmA and analysis of SlmA dimerization domain. (A) Ribbon diagram of the SlmA dimer. In the left subunit, the helices are coloured differently and each helix is labelled. (B) SlmA dimer interface. Residues on α8 and α9 that mediate dimerization are shown as green sticks and labelled. (C) SlmA and the SlmA–DNA stoichiometries as determined by size-exclusion chromatography. (D) Structural comparison of the SlmA (yellow) and QacR (cyan) DNA-binding domains (helices α1 to α3) and dimerization domains (helices α4 to α6).
Database searches using the Dali server (Holm et al, 2008) revealed that the SlmA structure is most similar to that of the QacR protein, thus establishing SlmA as a new member of the TetR family. The DNA-binding domains of all TetR proteins show sequence homology; however, their C-terminal domains do not. Despite this, all TetR members whose structures have been solved possess C-terminal domains that are similar structurally. All TetR proteins are dimers and their C-terminal domains mediate dimerization (Ramos et al, 2005). A multiple sequence alignment of SlmA with TetR members that have been structurally characterized showed that the most conserved region between the proteins lies within the HTH, which overall shows 23% sequence similarity compared with the 6% sequence correspondence found in the comparison of their C-terminal domains (Supplementary Figure S2). Despite the lack of sequence similarity, structural superimpositions of SlmA with TetR members QacR and TetR, reveal that SlmA has the same structural topology as these TetR proteins. In particular, comparison of the DNA binding and dimerization domains of SlmA with QacR yielded a root mean squared deviation (RMSD) of 1.9 Å and 3.9 Å, respectively (Figure 1D; Hinrichs et al, 1994; Orth et al, 2000; Schumacher et al, 2001; Schumacher and Brennan, 2002).
The biological functions of 85 TetR members have been elucidated (Ramos et al, 2005). Notably, all these proteins function as transcriptional regulators. The genes they regulate encode products involved in diverse pathways such as multidrug resistance, catabolism, antibiotic biosynthesis, osmotic stress and the pathogenicity of Gram-negative and Gram-positive bacteria. To carry out their regulatory functions, TetR proteins respond to small-molecule ligand sensors (Hinrichs et al, 1994; Orth et al, 2000; Schumacher et al, 2001; Frénois et al, 2004; Ramos et al, 2005). The lack of sequence homology within the C-terminal domains of TetR proteins reflects the fact that, in addition to dimerization, this domain also functions as a ligand-binding domain. Ligand binding leads to structural changes that cause the proteins to dissociate from their DNA sites, allowing transcription. Although SlmA is clearly a member of this family of transcriptional regulators, it has a very different function, which is NO (Bernhardt and de Boer, 2005). Consistent with this distinction, analysis of the SlmA structure reveals that unlike canonical TetR proteins, the SlmA C-terminal domain contains only a small cavity with a volume of ∼360 Å3. Moreover, there is also no clear entrance to this potential pocket, as it is occluded by helix α8′ from the other subunit in the dimer (Supplementary Figure S3). Thus, although SlmA is a structural member of the TetR family of regulators, it is unique among these proteins in that it does not function in transcription and it also lacks an obvious ligand-binding site within its C-terminal domain.
Identification of the SlmA–DNA-binding sequence
Previous studies showed that the ability of SlmA to associate with the nucleoid is essential for its NO function. In particular, cells containing an N-terminal truncation of residues 1–64 were not functional in NO (Bernhardt and de Boer, 2005). Our SlmA structure, showing that it contains a N-terminal HTH and is a TetR member, suggested that it may bind a palindromic DNA site as a homodimer, in a manner similar to other TetR proteins (Orth et al, 2000; Schumacher et al, 2002). With this a priori assumption, we went on to determine whether SlmA displays DNA-binding specificity by conducting a restriction endonuclease protection, selection and amplification (REPSA) experiment (Van Dyke et al, 2007). The 43 unique possible binding sequences identified via REPSA were analysed with the sequence motif discovery program, Multiple Expectation Maximum for Motif Elicitation (MEME) (Supplementary Figure S4) (Bailey et al, 2006). The results indicated that SlmA binds in a specific manner to DNA duplexes containing a 12-bp palindromic site with the consensus, 5′-GTGAGTACTCAC-3′, herein called the SlmA–DNA-binding sequence (SBS).
Probing SlmA–DNA-binding specificity
To determine the affinity of SlmA for the SBS and further dissect its DNA-binding preferences, we performed a series of fluorescence polarization (FP) assays (Lundblad et al, 1996; Materials and methods). These analyses showed that SlmA binds the SBS with a Kd of ∼50 nM. By contrast, SlmA showed no detectable binding to DNA containing randomized sequences (Supplementary Figure S5). Next, each of the six corresponding positions of the palindromic 12-bp SBS were systematically mutated and their binding affinities for SlmA determined (Figure 2A). These results showed that there is a strong preference for a G, T, A and G at positions 1, 2, 4 and 5, respectively. Mutation of these bases significantly impaired SlmA binding. However, SBS position 3 is able to accommodate any purine nucleotide, as mutation of the guanine at this position to an adenine yielded a Kd similar to the consensus SBS of ∼60 nM. Lastly, position 6 is the most flexible in terms of nucleotide specificity. Any pyrimidine in this position allowed high-affinity binding to SlmA, and mutation to guanine allowed binding but with reduced affinity. The DNA sequence preferences for SlmA revealed by these studies are summarized as a sequence logo in Figure 2B.
Figure 2.
SlmA–DNA binding preference. (A) FP results summarizing the affect of SBS double mutations on SlmA binding. *Indicates the nucleotides that were not changed from the top strand and the nucleotides that were mutated are shown as letters. (B) Sequence logo summarizing the preference of SlmA for DNA based on the FP studies in A.
Distribution of SlmA-binding sites on the E. coli chromosome
We hypothesized that the sequence-specific yet relaxed DNA-binding capability of SlmA likely has a role in its NO function. Thus, to efficiently identify all possible SlmA-binding sites, we performed a ChIP followed by DNA sequencing (ChIP–Seq) (Materials and methods; Supplementary Materials and methods). After mapping the tag sequences onto the E. coli chromosome, 52 peaks were identified to be statistically significant (Supplementary Figure S6A–B; Kent et al, 2002). ChIP followed by PCR (ChIP–PCR) experiments conducted on these sites confirmed the positive signals (Supplementary Figure S7A–B). Moreover, the Motif Alignment and Search Tool (MAST) revealed that 50 of the 52 sites conform to the SBS motif shown in Figure 2B, indicating that the SBS identified by REPSA is the specific sequence recognized by SlmA in vivo (Bailey and Gribskov, 1998). Examination of the location of the SBS sites revealed that they are primarily clustered in specific defined regions of the chromosome called macrodomains (MDs). Studies have demonstrated that the bacterial chromosome is organized into four ordered MDs, the Ori, Ter, Right and Left MDs and two less structured regions (termed non-structured regions; Valens et al, 2004; Boccard et al, 2005; Espeli et al, 2008). These parts of the chromosome form compact regions and are concentrated in the same cellular space. The Ori MD contains the origin of replication and is located opposite the Ter MD, which contains the replication terminus site. On either side of the Ter domain are the Left and Right MD, while the Ori MD is flanked by the two non-structured regions. The SBS sites cluster within the Ori MD and non-structured regions, and, notably, none of these sites are located in promoter regions, consistent with previous data indicating that SlmA does not exert its NO function via transcription regulation (Figure 3A; Bernhardt and de Boer, 2005). In addition, we see no evidence of spreading of SlmA along the DNA from its target sites, as has been observed for Noc and other ParB proteins (Wu and Errington, 2004). Perhaps, the most significant finding, however, was that SBS sites are essentially absent in the Ter MD and largely absent from the MDs that surround the Ter, most notably the Right MD (Figure 3A).
Figure 3.
Mapping of SlmA-binding sites on the E. coli chromosome. (A) SlmA-binding sites as determined by ChIP-Seq are represented as red triangular ticks. The four E. coli chromosomal macrodomains, Ori (red), Right (pink), Ter (blue) and Left (yellow), are shown as blocks. (B) The predicted locations of SlmA-binding sites on the chromosome of three E. coli strains, uropathogenic E. coli strain 536 (GB: CP000247), enterotoxigenic E. coli strain E24377A (GB: CP000800) and avian pathogenic E. coli strain APEC O1 (GB: CP000468). The putative sites are represented as black triangular ticks and the MD are represented as in A.
Multiple sequence alignments of SlmA proteins show that the region corresponding to the recognition helix is conserved among these proteins in Gram-negative bacteria and γ-proteobacteria (Supplementary Figure S8A–B). This suggests that these proteins likely bind DNA sites with the same or similar sequences. Hence, we used the Find Individual Motif Occurrence program to map the putative SBS sites on the chromosomes of the uropathogenic E. coli strain 536 (GB: CP000247), enterotoxigenic E. coli strain E24377A (GB: CP000800), avian pathogenic E. coli strain APEC O1 (GB: CP000468), Salmonella typhimurium (GB: AE006468), Klebsiella pneumoniae (GB: CP000647) and Enterobacter (GB: CP000653) (Bailey and Gribskov, 1998). The main finding from this analysis is that the predicted (putative) SBS sites are dispersed over the chromosomes, with the exception of the Ter MD (Figure 3B, Supplementary Figure S8B).
The finding that SlmA sites are clustered in specific regions of the chromosome was intriguing, as it has been speculated that the formation of specific MDs may have roles in certain cellular processes, key among them being cell division (Valens et al, 2004; Boccard et al, 2005; Espeli et al, 2008). In particular, previous studies have indicated that Z-ring assembly appears to be coordinated with chromosome segregation (Den Blaauwen et al, 1999). These studies showed that after replication, the Ori MD migrates towards the cell poles, with the other MDs following. The Ter MD is the last to migrate and its segregation coincides with the onset of cell division (Espeli et al, 2008). SlmA binding to non-Ter DNA to prevent Z-ring formation at these regions and not the Ter region is consistent with these events. The relative lack of SBS sites in the Right and, to a lesser extent Left, MDs may serve as a buffer to ensure that septation does not occur at the Ter MD. This mechanism may work in concert with FtsK, which pumps DNA to the correct cellular compartments (Bigot et al, 2007). It appears that in B. subtilis and Caulobacter crescentus, DNA segregation with cell division are similarly coordinated. Specifically, the B. subtilis chromosome was also shown to lack binding sites in its Ter MD for its NO factor, Noc. However, it is still unknown how Noc interacts with the division machinery to inhibit cell division (Wu et al, 2009). In the case of C. crescentus, the MipZ protein interacts with ParB, localizing it near the Ori region, and interferes with Z-ring assembly to restrict its formation to the midcell (Thanbichler and Shapiro, 2006).
SlmA binds simultaneously to DNA and FtsZ
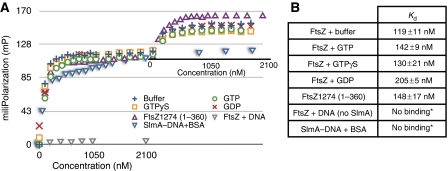
Our data show that the location of SBS sites on the chromosome optimally positions SlmA to act as a negative regulator of cell division. One way in which SlmA could effect such inhibition is via interactions with proteins involved in divisome assembly. Perhaps, most effective would be an interaction with FtsZ, as it initiates cell division. In fact, previous data suggested that SlmA and FtsZ may interact. However, these studies, based on light scattering, implied that polymerization is not inhibited by SlmA, which appeared to be contrary to the mechanism of NO (Bernhardt and de Boer, 2005). Thus, to investigate whether SlmA interacts with FtsZ and, importantly, whether SlmA can interact with DNA and FtsZ simultaneously, we used FP (Lundblad et al, 1996). Similar to previous FP studies, SlmA was titrated into SBS mixtures until saturation was reached. Then, increasing concentrations of FtsZ were added to the same reaction mixture. A clear second binding event was observed on FtsZ addition (Figure 4A). As a control for molecular crowding, BSA was titrated in the place of FtsZ and revealed no second binding event. In addition, when FtsZ was titrated into a reaction tube with only labelled SBS, there was no appreciable change in polarization, showing that FtsZ alone does not bind the SBS (Figure 4A). The titration curve for the second binding event of FtsZ to the SlmA–DNA complex was used to calculate an apparent Kd of ∼120 nM. The interaction of FtsZ with SlmA–DNA did not require GTP, nor was it affected by guanine nucleotides; binding assays performed in the presence of GTP, GTPγS, GDP and buffer alone yielded apparent affinities of 142±9 nM, 130±21 nM, 205±5 nM and 119±11 nM, respectively (Figure 4B). For assays measuring FtsZ binding to SlmA–DNA, the presence of an N-terminal His-tag on FtsZ did not affect binding, as the His-tagged protein bound to SlmA–DNA with essentially the same apparent affinity as the non-tagged FtsZ.
Figure 4.
Analyses of FtsZ binding to SlmA–DNA. (A) FtsZ binding to SlmA–DNA as measured by FP. SlmA was initially titrated into DNA until saturation (first plot) and then FtsZ (or BSA) was added to assess binding to the SlmA–DNA complex (second binding curve). These assays were carried out in the same buffer conditions with or without guanine nucleotides as follows: FtsZ + buffer (+), FtsZ+1 mM GTP (○), FtsZ +1 mM GTPγS (□), FtsZ + 1 mM GDP (×), FtsZ(1–360) + buffer (△) and BSA with no FtsZ (blue ▽). Additionally, FtsZ was titrated into SBS alone to control for the possible binding of FtsZ to the DNA (grey ▽). (B) Table of apparent binding affinities of FtsZ, FtsZ(1–360) and BSA to SlmA–DNA. The second binding curve from FP assays in A was used to calculate the apparent Kd. *No appreciable change in polarization was detected, thus further analysis of binding affinity could not be conducted.
FtsZ interacts with a number of proteins involved in cell division or its regulation. Most of these interactions have been shown to be mediated by the extended C-terminal tail of FtsZ, including its binding to FtsA and ZipA (Liu et al, 1999; Ma and Margolin, 1999; Mosyak et al, 2000; Haney et al, 2001). Interestingly, the TetR protein EthR binds extended ligands such as ethionamide and TetR itself can bind peptides, which act as tetracycline agonists (Frénois et al, 2004; Luckner et al, 2007). Both proteins bind these ligands in the pockets located in their C-terminal domains. This suggested that the SlmA C-terminal domain might similarly bind the FtsZ C-tail, as although its C-domain pocket appears inaccessible, structural alterations may allow entrance and binding of the FtsZ tail. To test this possibility, a FtsZ truncation mutant, FtsZ(1–360), was used in binding assays with SlmA–DNA. The FP analyses revealed that FtsZ(1–360) bound SlmA–DNA with an apparent affinity that was essentially equal to wild-type FtsZ (148±17 nM compared with ∼120 nM) (Figure 4B). Thus, these combined data show that SlmA can bind DNA and FtsZ simultaneously, and that guanine nucleotides are not required for this interaction. Moreover, SlmA does not interact with the FtsZ C-terminal tail.
Small-Angle X-Ray analyses on the SlmA–FtsZ complex
The finding that SlmA–DNA interacts with FtsZ provides a direct link between an NO factor and the key cell division protein. However, to ascertain how this interaction might lead to NO necessitates a molecular understanding of the SlmA–FtsZ complex and how it may impinge on Z-ring formation. Thus, to gain insight into the molecular interactions between FtsZ with SlmA, we employed SAXS (Putnam et al, 2007). SAXS analyses were carried out on SlmA, FtsZ and the SlmA–FtsZ complex. The SAXS profile and RG (radius of gyration) of SlmA alone over a concentration range of 1–6 mg/ml indicated that the protein is homogeneous (Supplementary Figure S9A–E). The experimental RG of SlmA was 31.4±0.01 Å, which agrees well with the value of 28.8 Å derived from our crystal structure. By contrast, the SAXS profile of FtsZ in the presence or absence of guanine nucleotides shows that it is prone to aggregation, which is expected as FtsZ is known to form protofilaments and other polymer structures (Erickson et al, 1996; Adams and Errington, 2009). Guinier analysis of FtsZ samples at low concentrations of 1–2 mg/ml, yielded a RG of 75.9±0.97 Å, and a rod analysis yielded an RG of 26.3±0.10 Å for the cross-section. The RG estimated for the cross-section agrees well with the calculated RG of 26.2 Å for a FtsZ monomer, suggesting that, at this concentration, FtsZ exists largely as protofilament-like structure (Supplementary Figure S9B).
Interestingly, compared with the behaviour of FtsZ alone, SAXS profiles of the SlmA–FtsZ complex, at a concentration range of 1–5 mg/ml, revealed it to be aggregation free (Supplementary Figure S9C). Thus, these data were used to calculate ab initio SAXS envelopes for the SlmA–FtsZ complex. Multiple calculations of independent models with the ab initio shape determination programs, DAMMIN and GASBOR, yielded consistent SAXS envelopes with only small variations between runs (Svergun, 1999; Svergun et al, 2001) (Supplementary Figure S10A–B). The overall shape of the envelope can be described as a symmetric ellipsoid. A homology model of the E. coli FtsZ protein along with our atomic model of SlmA were used in the protein–protein docking servers, ClusPro and PatchDock (Comeau et al, 2004; Schneidman-Duhovny et al, 2005; Arnold et al, 2006). These predictions were then used as inputs for the multidomain modelling program BUNCH (Petoukhov and Svergun, 2005). The best-fit model from BUNCH (Supplementary Figure S11) is a structure with a 1:1 SlmA:FtsZ ratio with one SlmA dimer sandwiched between two FtsZ subunits (Figure 5A). The overall fit of the model was quite good, except for the presence of unaccounted for density near the FtsZ molecules. However, this unaccounted portion of the envelope could be explained by the large number of missing residues (residues 317–383) from the FtsZ structure that was used to model the SlmA–FtsZ complex, which contains the full-length FtsZ protein. Examination of the best-fit model shows that the last visible C-terminal residue of FtsZ lies next to this extra density, suggesting how the C-terminal residues may extend into the envelope (Figure 5A). Hence, the model is consistent with our biochemical data showing that the FtsZ C-tail does not bind SlmA. In addition, consistent with the model is the fact that the calculated RG of the model, 45.6 Å, compares remarkably well with the experimentally calculated RG for the complex of 46.0 Å (Supplementary Figure S9E). While the SlmA dimer can be docked in the envelope, the precise orientation of the FtsZ proteins was more ambiguous because of its spherical shape. Thus, to obtain additional constraints on the FtsZ orientation in the envelope, SAXS analyses were carried out on a SlmA–FtsZ complex containing a FtsZ fusion protein in which GFP was attached after FtsZ residue 316. The presence of the GFP protein was evident from the calculated SAXS envelope of the complex and confirmed the previously obtained orientation (Figure 5B). The structure indicates that SlmA helices α4 and α7, which contain several basic residues, from each subunit interact with helices on the surface exposed face of each FtsZ C-terminal domain, which contain multiple glutamate residues.
Figure 5.
SAXS envelopes and models of SlmA-FtsZ complexes. (A) The average SlmA–FtsZ envelope, as determined by DAMAVER, is displayed as grey spheres (Svergun, 1999; Svergun et al, 2001). The model of the SlmA–FtsZ complex was calculated by BUNCH (Petoukhov and Svergun, 2005). In this model, a SlmA dimer (yellow) is flanked by two FtsZ molecules (turquoise and magenta). (B) The SAXS envelope of the SlmA–(FtsZ–GFP) complex was calculated as in A. Compared with the SlmA–FtsZ envelope (A) additional density was clearly observed, which corresponded to the GFP (green) fused at the C terminus of FtsZ(1–316). (C) SAXS structure of SlmA–FtsZ showing that when bound to SlmA, FtsZ protofilaments can form, but emanate in opposite directions relative to each other. The two FtsZ oligomers (cyan and magenta) in the structure flank the SlmA dimer (yellow).
EM studies on SlmA–DNA–FtsZ: SlmA–DNA alters higher order polymer assembly by FtsZ
The SlmA–FtsZ structure suggests that SlmA may function in NO by inserting between FtsZ molecules and perturbing the assembly of FtsZ polymers. In fact, studies indicate that even subtle changes in FtsZ polymer assembly can result in large changes in Z-ring formation (Romberg and Levin, 2003). In the SlmA–FtsZ structure, the SlmA dimer interacts with helices on the surface exposed face of each FtsZ C-terminal domain and not the GTP-binding domain. As a result, the FtsZ–GTP-binding pockets and T7 loops, which are required for protofilament formation, remain exposed in the SlmA–FtsZ complex (Supplementary Figure S12A–B). This suggests that SlmA binding would not prevent the linear polymerization of FtsZ. Indeed, modelling indicates that FtsZ protofilament formation would still be possible when bound to SlmA (Figure 5C). Strikingly, examination of the model of SlmA–DNA bound to FtsZ protofilaments shows that when bound to the SlmA–DNA, FtsZ protofilaments would be forced to grow in anti-parallel directions relative to each other (Supplementary Figure S12A–B). This would prevent the formation of parallel thick filaments, which have been proposed to be involved in FtsZ Z-ring formation (Figure 5C; Löwe and Amos, 1999; Oliva et al, 2003). However, to further address the affect of SlmA on FtsZ protofilament interactions, we performed negative stain EM experiments on SlmA and its complexes with DNA and FtsZ (Materials and methods). As previously observed by others, our EM images show that FtsZ forms filament bundles in the presence of GTP/Mg2+ (Erickson et al, 1996; Figure 6A). The addition of SBS DNA, SlmA or SlmA with non-SBS DNA had no affect on the appearance of these bundles (Figure 6B and Supplementary Figure S13A–B). In contrast, addition of SlmA and SBS DNA prevented FtsZ–GTP/Mg2+ from forming long bundles and instead led to the creation of ordered helical-like structures, of a fairly uniform size (typical lengths of approximately 150–200 nm; Figure 6C and D). The filamentous structures within the spirals resemble FtsZ protofilament bundles but are packed in a side by side orientation (Figure 6C and D). Although the resolution prevents a detailed description of the EM structures, the close packing of the two filamentous structures is consistent with the idea that SlmA–DNA enforces an anti-parallel arrangement of FtsZ polymers.
Figure 6.
SlmA–DNA complex induces formation of spiral FtsZ bundles. (A) Negative stain EM of 3 μM FtsZ under polymerizing condition (Materials and methods); negative stain causes FtsZ filaments to bundle into higher ordered structures. (B) Negative stain EM of 3 μM FtsZ and 0.6 μM SlmA. Bundling of FtsZ protofilaments as in A can be observed. (C) Negative stain EM of 3 μM FtsZ, 0.6 μM SlmA and 1 μM SBS DNA. Discrete and uniform helical structures are observed throughout the sample. (D) Higher magnification of helical-like structures seen in C. These structures range from containing a single spiral to multiple spirals with closed loops. Scale bar represents 500 nm in A and C, and 100 nm in B and D.
In the experiments, the ratio of FtsZ to SlmA used was 5:1, in an effort to establish conditions close to the physiological state. The typical filament bundles formed by FtsZ–GTP (Figure 6A) were never observed in SlmA–DNA–FtsZ samples. Indeed, these samples consistently showed only the uniform structures shown in Figure 6C and D. This suggests that a small amount of SlmA–DNA is sufficient to inhibit the formation of functional FtsZ bundles and further indicates that SlmA–DNA act as nucleation sites to promote the growth of non-functional FtsZ spirals, which can propagate up to several hundred nanometers. Interestingly, SlmA must be bound to SBS DNA to impart this effect, as EM samples with FtsZ and SlmA alone or SlmA and non-SBS DNA failed to affect FtsZ polymer assembly. Because our SAXS structure was obtained using a 1:1 ratio of SlmA to FtsZ, it cannot address how the SlmA–DNA-binding domain may impact the polymerization properties of a growing FtsZ protofilament attached to SlmA–DNA. Like other TetR proteins, the SlmA–DNA-binding domains are flexible and likely only become fixed on cognate DNA binding. It seems probable that the precise orientation of the DNA bound form of the SlmA–DNA-binding domains and the DNA itself may be necessary in steering the growing FtsZ protofilaments into the specific helical-like structures that we observe. The inability of SlmA alone to affect FtsZ polymer assembly could also function as a failsafe measure to prevent unwanted perturbation of cytosolic FtsZ polymers where Z-ring assembly is desired. However, it is likely that there is little SlmA present in the cytosol. In fact, previous studies showed that SlmA is localized within the nucleoid fraction of the cell (Bernhardt and de Boer, 2005). Moreover, data suggest that DNA-binding proteins that are not bound to their cognate sites interact nonspecifically and slide along the DNA or are engaged in rapid dissociation/reassociation from/onto DNA (Dowd and Lloyd, 1990; van Noort et al, 1998). Thus, the DNA bound form of SlmA is the physiologically relevant form.
Molecular model for SlmA-mediated NO
Our combined data suggest a molecular mechanism for SlmA-mediated NO (Figure 7). First, ChIP analyses revealed that the SlmA-binding sites are dispersed on non-Ter regions of the chromosome. The fact that the Ter MD regions are not bound by SlmA and therefore do not exhibit NO is consistent with the finding that the Z-ring formation occurs nearly concomitantly with replication of the Ter region (Espeli et al, 2008). In this regard, the ability of SlmA to bind DNA and FtsZ simultaneously is crucial for NO, as it localizes SlmA to the non-Ter MD. Our finding that SlmA does not prevent FtsZ polymerization is also consistent with and suggests an explanation for previous data showing that FtsZ can form larger polymers when bound to SlmA (Bernhardt and de Boer, 2005). Our EM experiments demonstrate that SlmA–DNA does not prevent FtsZ filament formation but severely affects the higher order assembly of FtsZ filaments, leading to unique higher order spiral-like structures. The sequestration of FtsZ molecules by SlmA could also have a role in Z-ring inhibition, and when combined, these mechanisms would provide multiple levels of protection against nucleoid bisection. Given the high conservation of SlmA in Gram-negative bacteria, we propose that this NO mechanism is likely used by all bacteria that harbour a SlmA protein.
Figure 7.
Model for SlmA-mediated NO. Schematic representation of SlmA-mediated NO. OriC sites, opposite the Ter region, are shown as red circles. FtsZ molecules are green arrows with yellow outline. Left: Without functional SlmA, FtsZ polymerizes to form a premature Z-ring, and in the presence of cellular stress caused by the inhibition of chromosome partition, DNA replication or a non-functional Min system, this leads to nucleoid guillotining and DNA fragmentation. Right: In the presence of SlmA (blue), the Z-ring cannot form over the DNA until replication of the Ter MD because SlmA does not bind the Ter MD. Cell division is thus coordinated with DNA segregation as the cell division machinery assembles right before the commencement of DNA segregation. Close up of the SlmA–FtsZ interaction, illustrating that SlmA disrupts Z-ring formation by inserting between FtsZ molecules and ultimately perturbing FtsZ higher order polymerization.
Materials and methods
Crystallization and structure determination of SlmA
The slmA gene was purchased from Genscript Corporation (Piscataway, NJ, USA; http://www.genscript.com). The gene was subcloned into pET15b such that an N-terminal hexa-histidine tag was expressed and the protein was purified using Ni-NTA chromatography. SlmA protein was concentrated to 6 mg/ml and crystallized in 100 mM Tris (pH 8.5), 10% PEG 400 and 58 mM LiSO4 by hanging-drop vapour diffusion. Data were collected at the Advanced Light Source (ALS) beamline 8.2.1 and processed with MOSFLM and SCALA (Supplementary Table SI). The SlmA structure was solved by MAD using crystals grown with selenomethionine-substituted protein. MAD data were collected and the selenium sites were located using SOLVE (Terwilliger and Berendzen, 1999). Model building was carried out using Coot (Emsley and Cowtan, 2004) and refinement with CNS (Brünger et al, 1998). The SlmA structure contains one molecule per asymmetric unit, and has Rwork/Rfree values of 22.4%/26.5% to 2.5 Å resolution (Supplementary Table S1). The oligomeric states of SlmA and SlmA–DNA were determined by size-exclusion chromatography on a Superdex 200 26/60 column, using appropriate standards.
FP assay
FP is a technique that can be used to obtain binding constants for macromolecular interactions. A powerful use of the method is the measurement of protein binding to a fluorescein-tagged oligonucleotide. In such measurements, the rotational motion of the fluorescein-tagged oligonucleotide is slowed by protein binding, increasing the fluorescence emission anisotropy value for the tagged DNA. Proteins may also be fluorescently tagged but DNA is easier to label in a manner that does not interfere with binding. Additionally, because of the rod-like geometry of DNA, protein binding to an oligonucleotide generally has a greater effect on rotational motion, leading to larger changes in fluorescence anisotropy. FP assays were performed with a PanVera Beacon 2000 fluorescence polarization system. Samples were excited at 490 nm, and fluorescence emission was measured at 520 nm. All oligonucleotides used in these assays contain a 5′ fluorescein tag. Each assay was carried out with 1 nM oligonucleotide in the binding buffer (200 mM NaCl, 25 mM Tris–HCl (pH 7.5), 2.5 mM MgCl2). Either SlmA or FtsZ was titrated into the reaction mixture. The polarization data were analysed with KaleidaGraph and fitted to a simple bimolecular binding model by nonlinear regression (Lundblad et al, 1996).
Identification of the SlmA–DNA-binding sequence
The REPSA experiment was conducted as previously described (Van Dyke et al, 2007). Briefly, 40 μM SlmA was bound to 4 ng of REPSA selection template in binding buffer (10 mM Tris–HCl (pH 7.9), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT) for 30 min at 37°C. The cleavage reaction was then performed with either 0.5 U of FokI or BpmI restriction enzyme for 5 min at 37°C. Products bound by SlmA and consequently protected from endonuclease digestion were amplified by PCR. Resulting PCR products were subjected to additional rounds of selection until convergence, as detected by DNA sequencing. The resulting DNA sequences were analysed by the MEME program (Bailey et al, 2006). Default parameters were used to search for palindromic motifs. The position-specific scoring matrix from the MEME analysis was input into Find Individual Motif Occurrence program (Bailey and Gribskov, 1998) with default parameters. The E. coli strain K-12 sub-strain MG1655 (GenBank ID: U00096) was used as the sequence input.
ChIP-Seq/PCR analysis
ChIP-Seq experiments were carried out similar to a previously described method (Grainger et al, 2004). E. coli cells were treated with formaldehyde to induce DNA crosslinking, and FLAG-tagged SlmA was then immunoprecipitated. The SlmA–DNA complexes were un-crosslinked and the purified DNA analysed via sequencing with an Illumina Solexa Genome Analyzer II. The sequence data were analysed using a modified method developed by Zhang et al (2008). Details of these experiments and the adapted methods of analyses that were used are included in Supplementary Materials and methods.
Expression and purification of FtsZ
Full-length FtsZ and C-terminal truncated FtsZ, FtsZ(1–360), from E. coli were produced as previously described with minor modifications (Romberg et al, 2001). Specifically, an extra 25% ammonium sulphate precipitation was performed and the precipitant was solubilized in storage buffer (50 mM Tris (pH 8), 100 mM NaCl, 1 mM MgSO4). Proteins were polymerized with 10 mM MgSO4, 1 M monosodium glutamate and 1 mM GTP at 37°C for 30 min. The resultant pellets were then redissolved in storage buffer. The FtsZ(1–316)–GFP fusion protein was generated by cloning a C-terminal truncated version of E. coli FtsZ (residues 1–316) along with GFP in the pET15b vector. The protein was expressed in BL21(DE3) cells and purified in one step using Ni-NTA chromatography.
SAXS data collection and evaluation
SAXS data were collected at the ALS beamline 12.3.1 at a wavelength of 1 Å and a temperature of 10°C (Lawrence Berkeley National Laboratory, Berkeley, CA; Hura et al, 2009). A detailed description of the SAXS sample preparation is given in Supplementary Materials and methods. SAXS data were collected for protein samples over a range of concentration, and the profiles were evaluated for aggregation using Guinier analyses (Koch et al, 2003). The radius of gyration (RG) was derived by the Guinier approximation I(q)=I(0) exp(−q2RG2/3), with the limits qRG<1.3. The program GNOM (Svergun, 1992) was used to compute the pair distance distribution functions, P(r). The overall shapes were calculated from the experimental data using the program DAMMIN (Svergun, 1999) or GASBOR (Svergun et al, 2001). The models generated by BUNCH (Petoukhov and Svergun, 2005) were evaluated for q of ranges (0.020–0.40/Å).
Negative stain EM
All samples (FtsZ, SlmA and their complexes with and without DNA) were taken in a buffer consisting of 25 mM HEPES (pH 7.4), 100 mM potassium glutamate, 300 mM potassium acetate, 5 mM magnesium acetate and 2 mM GTP. The concentration of FtsZ was 3 μM and that of SlmA was 0.6 μM. The concentration of DNA (5′-GCAGTGAGTACTCACTGC-3′; top strand) was 1 μM. Samples were placed on 100-mesh formvar-coated copper grids treated with poly-L-lysine for 1 h. Excess samples were blotted with filter paper and then stained with filtered 2% uranyl acetate for 1 min. Stain was blotted dry from the grids with filter paper and samples were allowed to dry. Samples were then examined in a JEM 1010 transmission electron microscope (JEOL USA Inc., Peabody, MA) at an accelerating voltage of 80 Kv. Digital images were obtained using the AMT Imaging System (Advanced Microscopy Techniques Corp., Danvers, MA).
Accession number
Coordinates and structure factor amplitudes for the SlmA structure have been deposited with the Protein Data Bank under the accession code 3NXC.
Supplementary Material
Acknowledgments
This work was supported by an MD Anderson Trust Fellowship (to MAS), the National Institutes of Health (GM068453 to MAS and GM61074 to WM) and the National Institutes of Health Molecular Biophysics Training grant (T32 GM008280 to NKT). We thank the Advanced Light Source (ALS) and their support staff. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences and Material Science Division of the US Department of Energy at the Lawrence Berkeley National Laboratory. We also thank the SIBYLS beamline staff at 12.3.1 for aiding solution scattering data collection and the MDA Institutional Core Grant no. CA16672 High Resolution Electron Microscopy Facility for help with EM experiments.
Author contributions: NKT and MAS designed the research; NKT, STA, BP, MWVD and TKB performed the experimental research; SL and YL performed statistical analyses; MAS and NKT wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Adams DW, Errington J (2009) Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat Rev Microbiol 7: 642–653 [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22: 195–201 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Gribskov M (1998) Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14: 48–54 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34 (Web Server issue): W369–W373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG, de Boer PAJ (2005) SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol Cell 18: 555–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigot S, Sivanathan V, Possoz C, Barre FX, Cornet F (2007) FtsK, a literate chromosome segregation machine. Mol Microbiol 64: 1434–1441 [DOI] [PubMed] [Google Scholar]
- Boccard F, Esnault E, Valens M (2005) Spatial arrangement and macrodomain organization of bacterial chromosomes. Mol Microbiol 57: 9–16 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ (2004) ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res 32 (Web Server issue): W96–W99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Den Blaauwen T, Buddelmeijer N, Aarsman ME, Hameete CM, Nanninga N (1999) Timing of FtsZ assembly in Escherichia coli. J Bacteriol 181: 5167–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowd DR, Lloyd RS (1990) Biological significance of facilitated diffusion in protein-DNA interactions. Applications to T4 endonuclease V-initiated DNA repair. J Biol Chem 265: 3424–3431 [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA 93: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Mercier R, Boccard F (2008) DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol 68: 1418–1427 [DOI] [PubMed] [Google Scholar]
- Frénois F, Engohang-Ndong J, Locht C, Baulard AR, Villeret V (2004) Structure of EthR in a ligand bound conformation reveals therapeutic perspectives against tuberculosis. Mol Cell 16: 301–307 [DOI] [PubMed] [Google Scholar]
- Grainger DC, Overton TW, Reppas N, Wade JT, Tamai E, Hobman JL, Constantinidou C, Struhl K, Church G, Busby SJW (2004) Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J Bacteriol 186: 6938–6943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haney SA, Glasfeld E, Hale C, Keeney D, He Z, de Boer P (2001) Genetic analysis of the Escherichia coli FtsZ.ZipA interaction in the yeast two-hybrid system. Characterization of FtsZ residues essential for the interactions with ZipA and with FtsA. J Biol Chem 276: 11980–11987 [DOI] [PubMed] [Google Scholar]
- Hinrichs W, Kisker C, Düvel M, Müller A, Tovar K, Hillen W, Saenger W (1994) Structure of the Tet repressor-tetracycline complex and regulation of antibiotic resistance. Science 264: 418–420 [DOI] [PubMed] [Google Scholar]
- Holm L, Kääriäinen S, Rosenström P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Gogol EP, Lutkenhaus J (2002) Dynamic assembly of MinD on phospholipid vesicles regulated by ATP and MinE. Proc Natl Acad Sci USA 99: 6761–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2001) Topological regulation of cell division in E. coli. spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, Yang S-J, Scott JW, Dillard BD, Adams MWW, Tainer JA (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat Methods 6: 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J, Miller S, Chothia C. (1988) Surface, subunit interfaces and interior of oligomers. J Mol Biol 204: 155–164 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch MH, Vachette P, Svergun DI (2003) Small-angle scattering: a view on the properties, structures and structural changes of biological macromolecules in solution. Q Rev Biophys 36: 147–227 [DOI] [PubMed] [Google Scholar]
- Liu Z, Mukherjee A, Lutkenhaus J (1999) Recruitment of ZipA to the division site by interaction with FtsZ. Mol Microbiol 31: 1853–1861 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1999) Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J 18: 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luckner SR, Klotzsche M, Berens C, Hillen W, Muller YA (2007) How an agonist peptide mimics the antibiotic tetracycline to induce Tet-repressor. J Mol Biol 368: 780–790 [DOI] [PubMed] [Google Scholar]
- Lundblad JR, Laurance M, Goodman RH (1996) Fluorescence polarization analysis of protein-DNA and protein-protein interactions. Mol Endocrinol 10: 607–612 [DOI] [PubMed] [Google Scholar]
- Ma X, Margolin W (1999) Genetic and functional analyses of the conserved C-terminal core domain of Escherichia coli FtsZ. J Bacteriol 181: 7531–7544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W (2005) FtsZ and the division of prokaryotic cells and organelles. Nat Rev Mol Cell Biol 6: 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosyak L, Zhang Y, Glasfeld E, Haney S, Stahl M, Seehra J, Somers WS (2000) The bacterial cell-division protein ZipA and its interaction with an FtsZ fragment revealed by X-ray crystallography. EMBO J 19: 3179–3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogales E (2000) Structural insights into microtubule function. Annu Rev Biochem 69: 277–302 [DOI] [PubMed] [Google Scholar]
- Oliva MA, Huecas S, Palacios JM, Martín-Benito J, Valpuesta JM, Andreu JM (2003) Assembly of archaeal cell division protein FtsZ and a GTPase-inactive mutant into double-stranded filaments. J Biol Chem 278: 33562–33570 [DOI] [PubMed] [Google Scholar]
- Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W (2000) Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat Struct Biol 7: 215–219 [DOI] [PubMed] [Google Scholar]
- Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of contractile FtsZ rings in liposomes. Science 320: 792–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007) X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys 40: 191–285 [DOI] [PubMed] [Google Scholar]
- Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R (2005) The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69: 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999a) MinDE-dependent pole-to-pole oscillation of division inhibitor MinC in Escherichia coli. J Bacteriol 181: 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999b) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Levin PA (2003) Assembly dynamics of the bacterial cell division protein FtsZ: poised at the edge of stability. Annu Rev Microbiol 57: 125–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romberg L, Simon M, Erickson HP (2001) Polymerization of Ftsz, a bacterial homolog of tubulin. is assembly cooperative? J Biol Chem 276: 11743–11753 [DOI] [PubMed] [Google Scholar]
- Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ (2005) PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res 33 (Web Server issue): W363–W367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher MA (2008) Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem J 412: 1–18 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Brennan RG (2002) Structural mechanisms of multidrug recognition and regulation by bacterial multidrug transcription factors. Mol Microbiol 45: 885–893 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG (2001) Structural mechanisms of QacR induction and multidrug recognition. Science 294: 2158–2163 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG (2002) Structural basis for cooperative DNA binding by two dimers of the multidrug-binding protein QacR. EMBO J 21: 1210–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih Y-L, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI (1992) Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Cryst 25: 495–503 [Google Scholar]
- Svergun DI (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J 76: 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svergun DI, Petoukhov MV, Koch MH (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys J 80: 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanbichler M, Shapiro L (2006) MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126: 147–162 [DOI] [PubMed] [Google Scholar]
- Valens M, Penaud S, Rossignol M, Cornet F, Boccard F (2004) Macrodomain organization of the Escherichia coli chromosome. EMBO J 23: 4330–4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke MW, Van Dyke N, Sunavala-Dossabhoy G (2007) REPSA: general combinatorial approach for identifying preferred ligand-DNA binding sequences. Methods 42: 118–127 [DOI] [PubMed] [Google Scholar]
- Van Noort SJ, van der Werf KO, Eker AP, Wyman C, de Grooth BG, van Hilst NF, Greve J (1998) Direct visualization of dynamic protein-DNA interactions with a dedicated atomic force microscope. Biophys J 74: 2743–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142: 309–320 [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Valkenburg JA, Wientjes FB, Zaritsky A, Nanninga N (1990) Role of the nucleoid in the toporegulation of division. Res Microbiol 141: 39–49 [DOI] [PubMed] [Google Scholar]
- Wu L, Ishikawa S, Kawai Y, Oshima T, Ogasawara N, Errington J (2009) Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J 28: 1940–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LJ, Errington J (2004) Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117: 915–925 [DOI] [PubMed] [Google Scholar]
- Yu XC, Margolin W (1999) FtsZ ring clusters in min and partition mutants: role of both the Min system and the nucleoid in regulating FtsZ ring localization. Mol Microbiol 32: 315–326 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.