RITA, a novel modulator of Notch signalling, acts via nuclear export of RBP-J
This study demonstrates that Notch signalling can be inhibited by RITA, which binds the transcription factor RBP-J and promotes its nuclear export. Consequently, RITA knockdown disrupts Notch-regulated neurogenesis during Xenopus development.
Keywords: neurogenesis, notch, nucleo-cytoplasmic shuttling, RBP-J, Xenopus
Abstract
The evolutionarily conserved Notch signal transduction pathway regulates fundamental cellular processes during embryonic development and in the adult. Ligand binding induces presenilin-dependent cleavage of the receptor and a subsequent nuclear translocation of the Notch intracellular domain (NICD). In the nucleus, NICD binds to the recombination signal sequence-binding protein J (RBP-J)/CBF-1 transcription factor to induce expression of Notch target genes. Here, we report the identification and functional characterization of RBP-J interacting and tubulin associated (RITA) (C12ORF52) as a novel RBP-J/CBF-1-interacting protein. RITA is a highly conserved 36 kDa protein that, most interestingly, binds to tubulin in the cytoplasm and shuttles rapidly between cytoplasm and nucleus. This shuttling RITA exports RBP-J/CBF-1 from the nucleus. Functionally, we show that RITA can reverse a Notch-induced loss of primary neurogenesis in Xenopus laevis. Furthermore, RITA is able to downregulate Notch-mediated transcription. Thus, we propose that RITA acts as a negative modulator of the Notch signalling pathway, controlling the level of nuclear RBP-J/CBF-1, where its amounts are limiting.
Introduction
The Notch signalling pathway is a highly conserved key player in the regulation of fundamental cellular processes, including stem cell maintenance, control of cell differentiation, and proliferation (Artavanis-Tsakonas et al, 1999; Kopan and Ilagan, 2009). Aberrant Notch signalling occurs in a variety of human disorders (reviewed in Miele et al, 2006; Koch and Radtke, 2007; Roy et al, 2007). Notch signalling appears to be a short-range communication that is activated via direct cell-to-cell contacts. Membrane-associated ligands (Delta, Jagged (Serrate in Drosophila melanogaster)) have been identified that upon binding induce proteolytic cleavage, resulting in the release of the Notch intracellular domain (NICD). NICD subsequently translocates to the nucleus and activates transcription of Notch target genes. NICD does not bind to DNA by itself but interacts with the DNA-binding protein recombination signal sequence-binding protein Jκ (RBP-J), also called CSL (CBF-1, Su(H), LAG1)) (Tamura et al, 1995).
In the absence of NICD in the nucleus, RBP-J inhibits transcription of Notch target genes by recruiting repressor complexes. A number of components of the repressor complexes have been identified so far (Borggrefe and Oswald, 2009), including Mint/SHARP (Oswald et al, 2002; Kuroda et al, 2003), CtBP (Morel et al, 2001; Oswald et al, 2005), ETO (Salat et al, 2008), HDAC activity (Kao et al, 1998), and H3K4 demethylase activity (Moshkin et al, 2009; Liefke et al, 2010). When NICD is present in the nucleus, RBP-J-corepressor complexes disassemble. NICD and RBP-J form the core of a transcriptional activator complex that enables transcription of Notch target genes. The transcriptional activation includes amongst others the function of the Mastermind-like coactivator as well as HAT activity (Wu et al, 2000; Oswald et al, 2001; Wallberg et al, 2002; Wilson and Kovall, 2006). Subsequent NICD phosphorylation, ubiquitination and proteosomal degradation leads to a rapid termination of the Notch signal (Fryer et al, 2004).
Notch signalling can be modulated on different levels of this pathway. It mainly takes place at the level of the Notch receptor and its ligands. Mechanisms include temporally and spatially restricted expression of ligand and receptor, ligand endocytosis and trafficking, availability and state of the receptor on the cell surface, and proteolytic events that finally result in the release of NICD (Bray, 2006; Fortini, 2009). However, the transcription factor RBP-J may represent a second regulatory bottleneck of Notch signalling, as it is the crucial component for recognition of the DNA target sequences in both, the repressor complex and activator complexes.
Here, we report for the first time a potential mechanism modulating the Notch signalling pathway on the level of the RBP-J transcription factor. We identify and functionally characterize RBP-J interacting and tubulin associated (RITA) (C12ORF52) as a novel RBP-J-interacting protein. RITA is a highly conserved, 36 kDa protein that has no significant homologies to any other protein. We identify a tubulin-interaction domain, a functional nuclear localization signal (NLS), a nuclear export signal (NES), and the RBP-J-interaction domain. On a functional level, RITA interferes with Notch- and RBP-J-mediated transcription. It is subject to rapid nucleo-cytoplasmic shuttling and, most importantly, mediates the nuclear export of RBP-J to tubulin fibres. In Xenopus laevis RITA counteracts the transcriptional activation of Notch target genes and the resulting loss of primary neurogenesis induced by dominant active Notch-1. This observation points to a novel regulatory function on the Notch signalling pathway in vivo: RITA induces nuclear export of RBP-J and thereby may function as a negative modulator of an activated Notch signalling pathway via the regulated shuttling of RBP-J.
Results
In order to identify RBP-J-interacting proteins, we performed a yeast two-hybrid screen on a human embryonic brain library, using the human splice variant RBP-2N as a bait. We identified a 618-bp open reading frame fused to the Gal4 activation domain. BLAST searches revealed an identity to the not yet characterized hypothetical protein C12ORF52 (FLJ14827, NM_0328489). We named this novel 269 amino acid (aa) protein RITA (Supplementary Figure S1).
RITA physically interacts with RBP-J
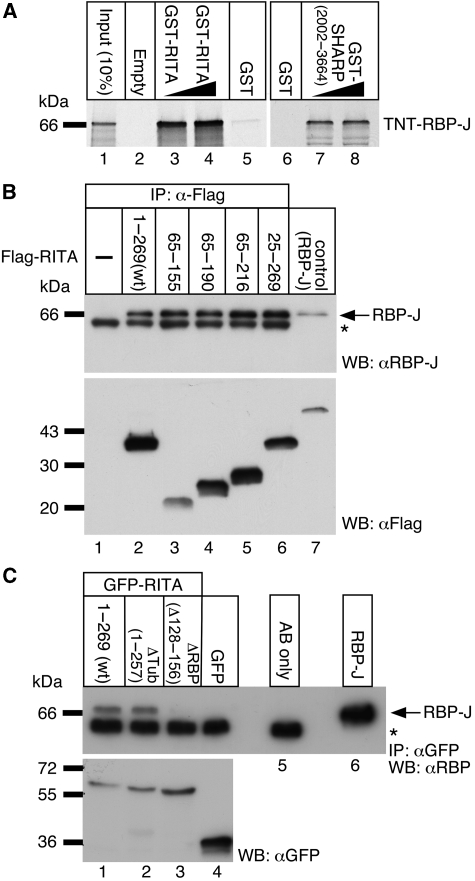
Interaction of RITA with RBP-J was first confirmed in vitro by using GST pull-down assays in a cell-free system (Figure 1A). The radiolabelled full-length RBP-J interacted specifically with a bacterially expressed GST-RITA fusion protein (lanes 3 and 4). GST-SHARP(2002–3664) (Oswald et al, 2002) served as a positive control for RBP-J interaction (lanes 7 and 8). No interaction was detectable with glutathione-sepharose beads (lane 2) or with GST alone (lanes 5 and 6).
Figure 1.
RITA interacts with RBP-2N in vitro (A) and in cellular extracts (B, C). Cell-free synthesized 35S-labelled RBP-2N (RBP-J) interacts with GST-tagged RITA immobilized on glutathione-sepharose beads (A, lanes 3 and 4), but not with empty beads (lane 2) or GST alone (lanes 5 and 6). GST-SHARP(2002–3664) served as a positive control for RBP-J interaction (lanes 7 and 8). (B) HEK293 cells were transfected with expression constructs for the indicated Flag-tagged RITA proteins. Expression of RITA(wt) and specific truncations were verified by western blotting (lower) using an anti-Flag antibody. Endogenous RBP-J protein was coimmunoprecipitated with RITA proteins using the anti-Flag antibody and detected by an anti-RBP antibody (lanes 2–6). In HEK293 extracts transfected with an empty vector, no RBP-J protein was coimmunoprecipitated (lane 1). Flag-tagged RBP-2N (lane 7, lower) or purified RBP-J (lane 7, upper) served as control. (C) Mapping of the RBP-J-interaction domain within RITA. Endogenous RBP-J proteins were coimmunoprecipitated from HEK293 cellular lysates stably expressing GFP-RITA(wt) (lane 1) and GFP-RITA (aa 1–257, ΔTub) (lane 2) but not in lysates from HEK293 cells stably expressing an in frame deletion (aa 128–156) of RITA, RITA-ΔRBP (lane 3) or GFP alone (lane 4). Asterisks denote the heavy chain of the antibodies used for immunoprecipitation.
To examine the interaction between RITA and RBP-J in a cellular context, we performed immunoprecipitation experiments on a cellular background. HEK293 cells were transiently transfected with various Flag-tagged RITA expression plasmids (Figure 1B). Expression of Flag-RITA proteins (aa 1–269, 65–155, 65–190, 65–216, 25–269) was tested by western blotting (Figure 1B, lower). After incubation of lysates with an anti-Flag antibody, coimmunoprecipitated endogenous RBP-J proteins were detected using an anti-RBP-J antibody (Figure 1B, lanes 2–6, upper). RBP-J was coimmunoprecipitated even with the smallest RITA fragment employed in this experiment (aa 65–155). By contrast, RBP-J was not coimmunoprecipitated from untransfected HEK293 lysates (Figure 1B, lane 1) or from HEK293 lysates transfected with an empty Flag construct (not shown).
To locate and characterize the RBP-J-binding domain within the RITA sequence, HEK293 cell lines were established, stably expressing RITA proteins and fragments fused to GFP. Coimmunoprecipitation experiments were performed using an anti-GFP antibody. Again, endogenous RBP-J protein was coimmunoprecipitated from HEK293 lysates expressing either GFP-RITA(wt) or GFP-RITA (aa 1–257=ΔTub) (Figure 1C, lanes 1 and 2). However, RBP-J was not coimmunoprecipitated from lysates expressing a GFP-RITA protein, where aa 128–156 are deleted, GFP-RITA-ΔRBP (Figure 1C, lane 3) or from lysates expressing GFP alone (Figure 1C, lane 4). Therefore, aa 128–156 appear to be involved in the RBP-J interaction.
To identify the domains of RBP-J that are interacting with RITA, RBP-J fragments (aa 1–315, aa 166–487, and aa 166–334) and full-length RBP-J (aa 1–487) were synthesized in a cell-free system and examined in GST pull-down assays (Supplementary Figure S2) using GST-mNICD and GST-RITA as baits. Both, NICD and RITA strongly interact with RBP-J (aa 166–487; Supplementary Figure S2C and D, lane 3) and RBP-J (aa 166–334; Supplementary Figure S2C and D, lane 4). The later fragment represents the Beta-Trefoil-Domain (BTD) of RBP-J (Kovall, 2008). We conclude that NICD and RITA bind to the BTD of RBP-J.
RITA interferes with Notch-1 and RBP-VP16-mediated transcription
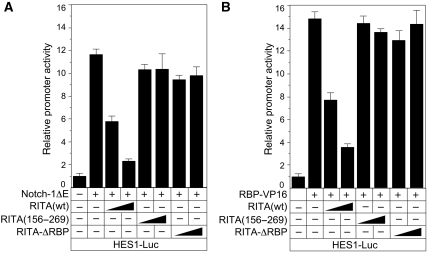
To study the effects of RITA on the transcriptional regulation of a Notch target gene, luciferase experiments were performed in HeLa cells using an HES1-specific reporter construct together with expression plasmids for mNotch-1ΔE, RBP-VP16, RITA, and RITA proteins defective in RBP-J interaction (Figure 2). As shown previously (Oswald et al, 2005; Salat et al, 2008), expression of the dominant active form of mNotch-1 (Notch-1ΔE) strongly activated the HES1 luciferase reporter. Additional expression of increasing amounts of RITA gradually impaired transcriptional activity from the reporter. This appeared to be specific, as expression of RITA proteins lacking the RBP-J-interaction domain (RITA 156–269 and RITA-ΔRBP) had no effect on Notch-1ΔE-mediated transcriptional activation (Figure 2A). Similar effects on HES1 promoter activity were observed with a transcriptional activator RBP-J protein (RBP-VP16). Again, transcriptional activation was lost after coexpression of RITA, but not after coexpression of interaction-defective RITA proteins (Figure 2B). Similar results were obtained, when a luciferase construct with 12 RBP-J-binding sites in front of a minimal promoter (pGa981/6) was used (data not shown).
Figure 2.
RITA interferes with Notch- (A) and RBP-VP16- (B) mediated transcriptional activation of the HES1 promoter. The reporter construct HES1-Luc was transfected into HeLa cells alone (1 μg) or together with 50 ng mNotch-1ΔE expression plasmid (A) or 50 ng RBP-VP16 expression plasmid (B) and increasing amounts of expression plasmid (50 and 100 ng) for RITA(wt) or for the RBP-J-interaction-defective RITA (156–269) and RITA-ΔRBP. Mean values and s.d. (error bars) based on at least four independent experiments are shown. − denotes absence of; + denotes presence of.
RITA is a tubulin-binding protein
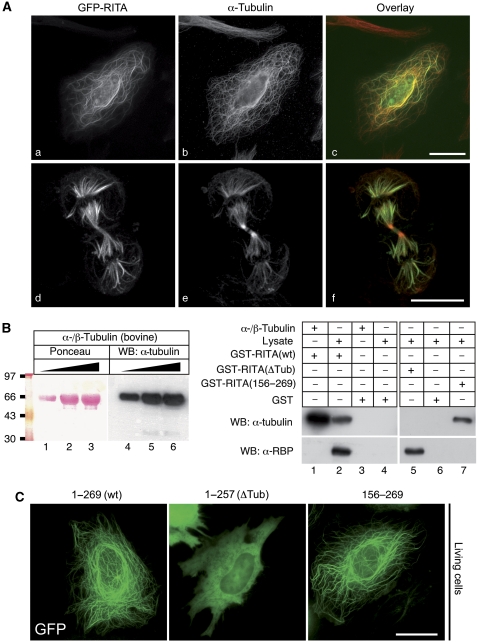
We studied the subcellular localization of RITA by fluorescence microscopy on HeLa cells transfected with a construct coding for RITA fused to GFP. Surprisingly, the RITA-GFP fusion protein showed a fibrillar localization in interphase cells (Figure 3Aa). Costaining of GFP-RITA-transfected cells with an anti-α-tubulin antibody showed clear regions of colocalization in interphase cells (Figure 3Ab and c) as well as in dividing cells (Figure 3Ad, e, and f), suggesting that RITA associates with tubulin. Similar structures were detected with immunofluorescence assays using Flag-tagged RITA constructs (data not shown). Upon treatment of transfected cells with nocodazole, which interferes with the polymerization of microtubules, the fibrillar staining disappeared within 2 h (Supplementary Figure S3A; Supplementary Movie S1). During mitosis, GFP-RITA highlighted the spindle apparatus (Figure 3Ad). In dividing HEK293 cells stably expressing GFP-RITA, labelling of the microtubule organizing centre and the mitotic spindle apparatus was observed (Supplementary Figure S3B; Supplementary Movie S2). A similar localization of GFP-RITA was found in dissociated embryonic cells from X. laevis (Supplementary Figure S3C). GST pull-down assays were performed using purified bovine α-/β-tubulin as well as lysates from HEK293 cells (Figure 3B). GST-RITA interacted with bovine tubulin as confirmed by western blotting using the anti-α-tubulin antibody (Figure 3B, right, lane 1). In lysates from HEK293 cells, both endogenous α-tubulin and endogenous RBP-J protein were pulled down with GST-RITA(wt) (Figure 3B, right, lane 2). RBP-J and tubulin interact with different domains of RITA, as the GST-RITA(ΔTub) lost tubulin-binding capacity, but still interacted with RBP-J (Figure 3B, right, lane 5). In contrast GST-RITA(156–269) failed to bind RBP-J, but still interacted with tubulin (Figure 3B, right, lane 7). Neither α-tubulin nor RBP-J interacted with GST alone (Figure 3B, right, lanes 3, 4, and 6).
Figure 3.
RITA is a tubulin-binding protein. (A) RITA fused to GFP colocalizes with endogenous tubulin in interphase cells (a–c) and highlights the spindle apparatus during mitosis (d–f). HeLa cells were transiently transfected with an expression plasmid for GFP-RITA. At 24 h after transfection, cells were fixed and immunostained using an anti-tubulin antibody. The subcellular localization of GFP-RITA and tubulin was determined by fluorescence microscopy. Magnification × 630. (B) RITA interacts with tubulin and with RBP-J with different binding domains. (Left panel) The anti-tubulin antibody used in GST pull-down experiments recognizes the bovine α-/β-tubulin as shown by western blotting. (Right panel) Bovine α-tubulin interacts with GST-RITA (lane 1) but not with GST alone (lane 3). Endogenous α-tubulin together with endogenous RBP-J from HEK293 cell lysates is pulled down with GST-RITA (lane 2) but not with GST alone (lanes 4 and 6). GST-RITA(ΔTub, aa 1–257) is defective in tubulin binding (lane 5) and GST-RITA (aa 156–269) is defective in RBP-J binding (lane 7). (C) The C-terminus of RITA (aa 156–269) is necessary for tubulin interaction. HeLa cells were transiently transfected with expression plasmids for the indicated RITA proteins fused to GFP. At 24 h after transfection, the living cells were analysed by fluorescence microscopy; scale bars, 5 μm; α, anti; − denotes absence of; + denotes presence of.
The region of RITA that confers the tubulin interaction was confirmed by fluorescence microscopy after transfection of GFP-RITA deletions into HeLa cells. The C-terminal truncated RITA (1–257, ΔTub) (Figure 3C, middle panel) showed diffuse localization mainly in the cytoplasm, whereas an N-terminal truncated RITA (156–269; Figure 3C, right panel) showed a tubulin-like localization similar to full-length RITA (1–269; Figure 3C, left panel). Colocalization of tubulin with either GFP-RITA(wt) or GFP-RITA (156–269) were shown using an anti-tubulin antibody (Supplementary Figure S3D). These results demonstrate that (i) RITA localizes at tubulin structures, (ii) it interacts directly with tubulin, and (iii) tubulin and RBP-J interaction occur at separate domains of RITA.
RITA localizes at centrosomes
In fluorescence microscopy images of RITA, we noticed specific punctate structures in the cells, possibly centrosomes. Thus, we probed for colocalization of RITA with different centrosomal markers (Figure 4). Indeed, the GFP-RITA spots (Figure 4A, upper, left) colocalized with antibody-labelled pericentrin (Figure 4A, lower, right). Whereas the pericentrin antibody labelled an extended region around the centrosomes (Figure 4B, middle), RITA could only be detected in two clearly distinct substructures (Figure 4B, left), suggesting that RITA localizes directly at the centrioles. A 3D reconstruction of pericentrin/RITA localization at the centrosomal region is shown in Supplementary Movie S3. Centrosomal localization of RITA was also shown using an antibody against the NIMA-related protein kinase Nek2, which is known to localize at centrosomes (Fry et al, 1998) (Figure 4C), and in live HeLa cells transfected with GFP-RITA and γ-tubulin fused to the red fluorescent protein mRuby (Figure 4D).
Figure 4.
RITA localizes at centrosomes. (A, B) Subcellular localization of RITA and pericentrin (arrows) analysed by epifluorescence (A) or confocal microscopy (B). (C) Subcellular localization of RITA and Nek2 (arrows). HeLa cells were transiently transfected with an expression plasmid for GFP-RITA. At 24 h after transfection, cells were fixed and immunostained. Cells were counterstained with DAPI (A, C) to show the nucleus. (D) Colocalization of RITA and γ-tubulin. HeLa cells were transiently cotransfected with GFP-RITA and mRuby-γ-tubulin. After 24 h, living cells were analysed by fluorescence microscopy; scale bars, 5 μm.
RITA is subject of rapid nucleo-cytoplasmic shuttling
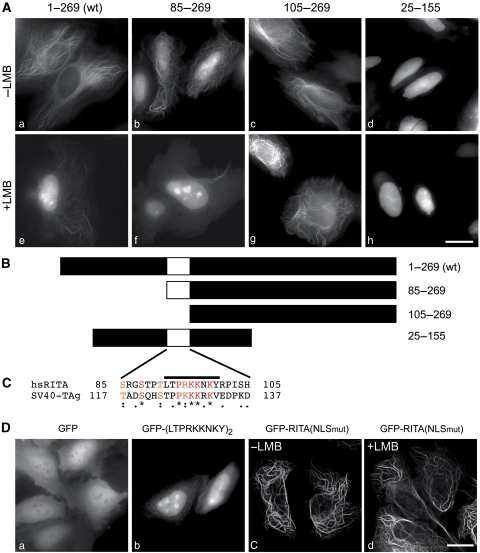
We observed that some of the RITA deletion mutants localized in the nucleus, suggesting that RITA may undergo nuclear import and export processes. Indeed, GFP-RITA expressing cells treated with the nuclear export inhibitor leptomycin B (LMB), displayed GFP-RITA localization in the nucleus within 60 min (Figure 5Ae). We obtained similar results using HEK293 cells stably expressing GFP-RITA (Supplementary Movie S4). Nuclear accumulation was independent of tubulin binding, because RITA(ΔTub), which only shows diffuse localization in the cytoplasm (compare Figure 3C, middle panel, and Supplementary Figure S3D, middle panel), also accumulated in the nucleus after LMB treatment (Supplementary Movie S5). In contrast, an N-terminally truncated form of RITA (aa 25–269) binds to microtubules but shows enhanced nuclear localization even without LMB treatment (Supplementary Movie S6). We searched for an NLS within RITA, using the four deletion constructs shown in Figure 5B. RITA (aa 85–269) shows nuclear accumulation (Figure 5Ab), which is enhanced only marginally after LMB treatment (Figure 5Af). In contrast, RITA (aa 105–269) shows tubulin association (Figure 5Ac) and, remarkably, fails to accumulate in the nucleus after LMB treatment (Figure 5Ag). An N-terminally and C-terminally truncated RITA (aa 25–155) fails to bind to tubulin and locates to the nucleus even without LMB treatment (Figure 5Ad and h). From these experiments, we conclude that the region between aa 85 and 105 may contain a putative NLS. Alignment with a canonical NLS from SV40 large T-antigen (TAg) revealed a similarity of basic amino acids within this region (Figure 5C). Cells transfected with a C-terminal fusion of the identified peptide sequence (L T P R K K N K Y) in tandem to GFP showed nuclear staining (Figure 5Db). In addition, the RKK to AAA substitution in GFP-RITA(NLSmut) did not show nuclear accumulation after LMB treatment (compare Figure 5Dc and d). These data clearly prove the presence of a functional NLS.
Figure 5.
RITA undergoes rapid nucleo-cytoplasmic shuttling. (A) Subcellular localization of the indicated GFP-RITA proteins without (upper panel) and 1 h after Leptomycin B (LMB; 2.5 ng/ml) treatment (lower panel). (B) Schematic representation of RITA proteins used in (A) and localization of the deduced putative NLS within RITA. (C) Alignment of the NLS within RITA and a functionally characterized NLS from SV40 large T-antigen. (*) identical residue, (:) conserved substitution, (.) semiconserved substitution according to clustalW. (D) Subcellular localization of GFP (a) and GFP fused to a tandem of the indicated RITA NLS peptide (b). GFP-RITA(NLSmut) does not accumulate in the nucleus after LMB treatment (c, d). HeLa cells were transiently transfected with expression plasmids for GFP or the indicated GFP-RITA proteins. After 24 h, the living cells were analysed by fluorescence microscopy; scale bars, 5 μm.
A putative NES was identified within the N-terminal region of RITA proteins from different species (including X. laevis and human) by the netNES 1.1 software (la Cour et al, 2004) (Figure 6A). The presence of the NES was verified by site-directed mutagenesis of RITA from both species. Substitution of critical amino acids (asterisks in Figure 6A, lower) to alanine resulted in nuclear accumulation of GFP-xlRITA(NESmut) (Figure 6B, middle panel) and GFP-hsRITA(NESmut) (Figure 6B, right panel) without LMB treatment. Consequently, (i) RITA is a nucleo-cytoplasmic shuttle protein, (ii) nuclear import of RITA is mediated by an NLS consisting of aa 92–100, and (iii) a functional NES at the N-terminus mediates nuclear export.
Figure 6.
Identification of a nuclear export signal (NES) within RITA. (A, upper) Putative NES sequences within xlRITA (left panel) and hsRITA (right panel) were identified by the netNES 1.1 software. (A, lower) Putative NES sequences (red) and positions within the RITA proteins. The marked amino acids (asterisks) are mutated to alanine in the RITA(NESmut) constructs. (B) Mutation of NES sequences within RITA proteins from Xenopus laevis (middle panel) and human (right panel) leads to nuclear accumulation without LMB treatment. Wild-type RITA from Xenopus laevis (left panel) served as a control. HeLa cells were transiently transfected with the indicated GFP-RITA constructs. After 24 h, living cells were analysed by fluorescence microscopy; scale bar, 5 μm.
RITA mediates nuclear export of RBP-J, but not of NICD
We showed that RITA interacts with RBP-J and is involved in rapid nucleo-cytoplasmic shuttling, which raises the question, if RITA colocalizes with RBP-J in the nucleus. RBP-J, fused to the red fluorescent protein mRuby (Kredel et al, 2009), showed nuclear localization (Figure 7Aa, e, and i). Surprisingly, when GFP-RITA(wt) was coexpressed with mRuby-RBP-J, both proteins were observed in the cytoplasm at tubulin fibres (Figure 7Ab, f, and j). After coexpression with RITA-ΔRBP, RBP-J was found in the nucleus, whereas RITA still localized at the tubulin fibres (Figure 7Ac, g, and k). And finally, GFP-RITA(wt) and mRuby-RBP-J colocalized in the nucleus after LMB treatment (Figure 7Ad, h, and l). Endogenous RBP-J was assayed in nuclear and cytoplasmic fractions of HEK293 cells after RITA overexpression (Figure 7B and C) or knockdown (Figure 7D and E). Overexpression of RITA(wt) and RITA(ΔTub), but not of GFP alone, resulted in an increase of cytoplasmic RBP-J and a decrease of nuclear RBP-J (Figure 7B, lanes 4 and 6, densitometric measurements in 7C). Conversely, knockdown of endogenous RITA by two siRNAs (Figure 7D, left) resulted in decrease of cytoplasmic and increase of nuclear RBP-J proteins (Figure 7D, right, lanes 5 and 6, densitometric measurements in 7E). Interestingly, RITA expression did not change the subcellular localization of NICD (Supplementary Figure S4A), indicating the absence of a RITA–RBP-J–NICD complex. In addition, we find that RITA prevents formation of a RBP-J/NICD complex (Supplementary Figure S4B). Cell-free synthesized RBP-J and NICD were used in a modified GST pull-down-Co-IP assay. RBP-J alone or combined with NICD was added to GST (Supplementary Figure S4B, lanes 1–3) or GST-RITA (Supplementary Figure S4B, lanes 4–6). RBP-J was only coprecipitated with NICD in the absence of GST-RITA (Supplementary Figure S4B, lanes 3 and 7). These results clearly indicate that RITA modulates the nuclear export of RBP-J and protects RBP-J from NICD loading.
Figure 7.
Nucleo-cytoplasmic shuttling of RITA together with RBP-J. (A) Colocalization of RITA and RBP-J. RBP-J-mRuby is found in the nucleus (a, e, i). Coexpression of RITA directs RBP-J out of the nucleus to tubulin fibres (b, f, j). After coexpression of RITA-ΔRBP (defective for RBP-J interaction), RBP-J remains in the nucleus (c, g, k). LMB treatment leads to colocalization of RITA and RBP-J in the nucleus (d, h, l). HeLa cells were transiently cotransfected with GFP-RITA or GFP-RITA-ΔRBP together with RBP-J-mRuby. After 24 h, living cells were analysed by fluorescence microscopy. Where indicated, cells were treated with leptomycin B (LMB; 2.5 ng/ml) 1 h prior to imaging, scale bar=5 μm. (B–E) Changes of nuclear versus cytoplasmic RBP-J after overexpression (B, C) or knockdown (D, E) of RITA. HEK293 cells were transiently transfected with the indicated GFP-RITA constructs (B, lanes 1, 3, 4, and 6) or GFP alone (B, lanes 2 and 5). At 24 h after transfection, cellular lysates were fractionated. (D, E) HEK293 cells were transiently transfected with RITA siRNAs (D, right panel, lanes 2, 3, 5, and 6). At 48 h after transfection, cellular lysates were fractionated. Cytoplasmic and nuclear fractions were analysed by western blotting. Knockdown of RITA was evaluated on mRNA level (D, left panel) by qPCR. Densitometric analysis in (C, E) was performed with the ImageJ software. Signal intensity in the control experiment was set to 1. Mean and standard error from three experiments are shown.
RITA expression reverses Notch-induced loss of primary neurogenesis in Xenopus laevis
A search for homologous sequences in other species yielded RITA proteins in many different organisms, representing a broad range of the metazoan phylum and including the placozoan Trichoplax adhaerens (Supplementary Figure S5A). Tubulin association as well as physical interaction of Trichoplax RITA (taRITA) and RBP-J (taRBP) are conserved (Supplementary Figure S5B and C). We did not find RITA protein sequences in D. melanogaster by BLAST searches. However, Drosophila Su(H), the homologue of RBP-J, interacts with human RITA in GST pull-down assays (Supplementary Figure S5D), suggesting that a functional homologue of RITA also exists in flies. Expression analysis in mouse embryos and adult tissue indicates a ubiquitous expression of RITA, although the levels vary in different tissues (Supplementary Figure S6A and B). In embryos of the amphibian X. laevis, maternal expression of RITA is detected. Zygotic expression levels are low during gastrula stages and then increase during neurula stages (Supplementary Figure S6C). Analysis of the spatial expression pattern by quantitative PCR on embryonic fragments (Supplementary Figure S6D) and by whole-mount in situ hybridization at neurula and tadpole stages (Supplementary Figure S6E) indicate a ubiquitous spatial expression. The temporal expression pattern suggests that RITA may be a component of the Notch signalling pathway during neurogenesis. To test this hypothesis, we first verified that the RBP-J/RITA interaction is conserved for the X. laevis proteins (Supplementary Figure S7A and B). Subsequently, we analysed how manipulations of RITA, as a putative novel modulator of the Notch signal transduction pathway, affects neurogenesis and organogenesis in Xenopus embryos. Numbers for the following experiments are listed in Supplementary Table S3.
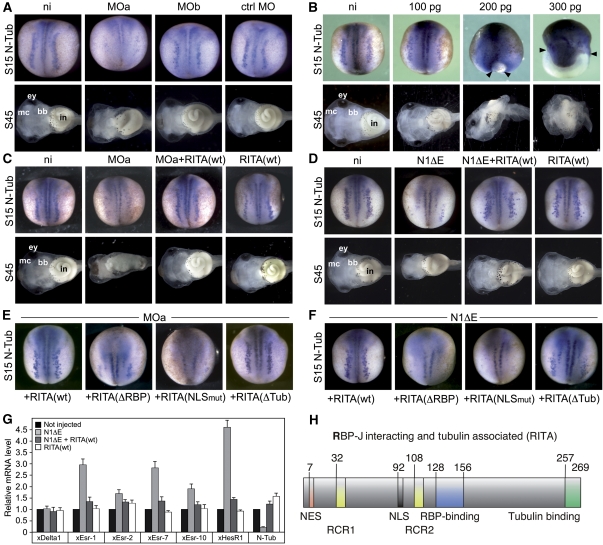
We pursued a loss-of-function approach. Antisense morpholino oligonucleotides against the start of the RITA transcript (MOa) efficiently inhibited translation of RITA(wt), but not of a Flag-tagged RITA(wt) in vitro (Supplementary Figure S7C). In vivo, a clear reduction of the neuronal marker N-tubulin was observed at neurula stage 15 after morpholino injection, but not in controls without morpholinos (Figure 8A, upper). Changing the morpholino sequence in five positions (MOb), thereby reducing its affinity to RITA mRNA, abolished this effect. Moreover, a standard control morpholino (ctMO) had no influence on the N-tubulin expression (Figure 8, upper). None of these injections resulted in gastrulation defects (data not shown). At the late tadpole stage 45, several malformations were noticed in MOa-injected embryos. The most obvious effects were smaller eyes, reduced elements of the jaw (Meckel's cartilage, palatoquadrate, ceratohyale), reduced branchial arches, and a deformation of the intestine (Figure 8A, lower). Oedemas were observed regularly, axial structures were not significantly affected (not shown). Effects of MOb injections were much milder, showing an almost normal phenotype, ctMO-injected embryos looked like not injected controls (Figure 8A).
Figure 8.
Functional characterization of RITA during development of Xenopus laevis. (A) Loss-of-function experiments. Injection of an antisense Morpholino oligonucleotide (MOa) results in reduction of N-tubulin expression at stage 15 (S15 N-Tub), compared to not injected embryos (ni). Injection of either the mutated Morpholino (MOb) or the standard control Morpholino (ctrl MO) did not result in this reduction. At stage 45 (S45), MOa-injected embryos show reduction of the jaw (mc, Meckel's cartilage), reduced branchial baskets (bb), smaller eyes (ey), and a deformed intestine (in). MOb and ctrl MO-injected embryos develop as not injected controls. (B) Gain-of-function experiments. Increasing doses of RITA(wt) mRNA (100, 200, 300 pg per embryo) resulted in gastrulation defects as indicated by the open blastopore (arrowheads) and in ectopic activation of N-tubulin at stage 15. Phenotypes at stage 45 show extensive malformations at higher doses, correlated to the gastrulation defects. (C) Rescue of the MO phenotype with low doses of MO-insensitive Flag-RITA(wt) mRNA. The knockdown of N-tubulin activation by the MO approach at stage 15 (MOa, compared to not injected control ni) is compensated by coinjection with the Flag-RITA(wt) mRNA (MOa+RITA(wt)), even at mRNA doses that solely do not have effects on N-tubulin expression (RITA(wt)). Correspondingly, the MOa effect on stage 45 embryos is restored by coinjection of Flag-RITA(wt) mRNA. (D) Rescue of a Notch-induced neurogenesis phenotype by RITA. A knockdown of N-tubulin at stage 15 by injection of the mNotch-1ΔE mRNA (N1ΔE), compared to not injected control (ni), is restored by coinjection of RITA mRNA (N1ΔE+RITA(wt)) at mRNA doses that solely do not have a significant effect on N-tubulin expression (RITA(wt)). Stage 45 phenotypes are rescued as well. (E) Influence of different MO-insensitive RITA constructs on the effects of MOa injections. The inhibition of N-tubulin expression at stage 15 by the MOa is compensated by coinjection with RITA(wt). This effect is absent in both, the construct deficient of RBP-J binding (+RITA(ΔRBP)) and the NLS-mutated construct (+RITA(NLSmut)), but it is mimicked by the tubulin binding deficient RITA(ΔTub). (F) Influence of different RITA constructs on the effects of Notch-1ΔE. The inhibition of N-tubulin expression by Notch-1ΔE (N1DE) is antagonized by coinjection with RITA(wt). Again, this is mimicked by RITA(ΔTub), but neither by RITA(ΔRBP) nor by RITA(NLSmut). (G) RITA(wt) restores missexpression of Notch target genes after injection of the constitutive active mNotch-1ΔE. Upregulation of xEsr-1, xEsr-2, xEsr-7, xEsr-10, and downregulation of N-tubulin after Notch-1ΔE injection (N1ΔE) was rescued by coinjection with a low dose of RITA(wt) (N1ΔE+RITA(wt)) that solely did not show effects (RITA(wt)). Mean values and standard error of the mean (error bars) based on three experiments are shown. Numbers of injected embryos and phenotypes for (A–D) are listed in Supplementary Table S3. (H) Schematic drawing of identified domains within RITA. Numbers (of amino acids) indicate positions in respect of human RITA. NES nuclear export signal, RCR1 and RCR2 RITA conserved repeats, NLS nuclear localization signal, RBP-binding domain, Tubulin-binding domain.
A gain-of-function approach by injecting RITA(wt) mRNA showed dose-dependent responses (Figure 8B). RNA doses of <100 pg per embryo resulted in normal N-tubulin patterns at stage 15 and a normal phenotype at stage 45. Doses >100 pg induced gastrulation defects and an apparent increase of N-tubulin expression (Figure 8B, upper). Identical results were obtained with mRNA encoding the Flag-tagged form of RITA(wt) (not shown). Stage 45 tadpoles assembled defects of many organ systems, including eyes, CNS, head and axial skeleton, and intestine (Figure 8B, lower). A dose >300 pg RITA mRNA per embryo was lethal before the end of neurulation (not shown).
The specificity of the RITA knockdown phenotypes was further supported by an experiment attempting to override the MOa effect by injection of an MO-insensitive mRNA, using a Flag-tagged RITA(wt) construct. As the MOa sequence is not close to the start of translation of this construct, the inhibition is inefficient (Supplementary Figure S7C). A low dose of Flag-tagged RITA mRNA (40 pg per embryo) did not have any visible effect, if injected solely. However, this dose was entirely sufficient to compensate the effects of MOa injections, as shown by a normal N-tubulin expression at stage 15 and normal phenotypes at stage 45 (Figure 8C).
Activation of the Notch signalling pathway by the Delta ligand results in a process called lateral inhibition during neurogenesis (Chitnis, 1995; Chitnis et al, 1995). In line with this process, injection of mRNA encoding a dominant active form of Notch-1 (Notch-1ΔE) resulted in a reduced number of primary neurons (Oswald et al, 2002). We tested the functional role of RITA as a negative modulator of Notch signalling by performing coinjection experiments using Notch-1ΔE together with RITA(wt). Importantly, coinjections with fairly low doses of RITA(wt) mRNA (100 pg per embryo) were indeed efficient to antagonize the effects of the constitutive active Notch-1ΔE on N-tubulin expression at stage 15 and phenotypes at stage 45 (Figure 8D). To further investigate the function of RITA as a negative modulator of the Notch signalling pathway, mRNA synthesized from different MO-insensitive flag-tagged constructs was injected into Xenopus embryos in combination with either MOa or in combination with the constitutive active Notch-1ΔE (N1ΔE). As shown above, RITA(ΔTub) is defective in tubulin binding, RITA(ΔRBP) is defective in RBP-J binding, and in RITA(NLSmut) nuclear localization is disabled. Whereas RITA(ΔRBP) and the RITA(NLSmut) mRNAs lost the ability to restore the MOa effects on N-tubulin expression, RITA(ΔTub) mRNA mimicked the rescue effect of RITA(wt) (Figure 8E). Therefore, restoration depends on nuclear localization and on RBP-J binding of RITA, but not on its ability to interact with tubulin. Consistently, in coinjection experiments with Notch-1ΔE, antagonistic effects were obtained from RITA(ΔTub) and from RITA(wt). RITA(ΔRBP) and the RITA(NLSmut) failed to restore the N-tubulin expression, confirming the necessity of both, nuclear localization and RBP-J interaction of RITA (Figure 8F). In addition, quantitative PCR analysis revealed that the transcriptional activation of several Notch targets (xEsr-1, xEsr-2, xEsr-7, xEsr-10, xHesR1) by dominant-activated Notch-1ΔE as well as the resulting downregulation of N-tubulin was reversed after coinjection of RITA(wt) (Figure 8G).
In summary, the RITA-specific gain-of-function and loss-of-function experiments in X. laevis show strong effects on Notch targets during embryogenesis. Most importantly, in agreement with reporter-gene assays in cell culture, RITA acts as a negative modulator of the Notch signalling pathway. This function of RITA depends on both, nuclear localization and RBP-J-interaction domains, but not on the ability to bind to tubulin.
Discussion
In this study we present the novel, highly conserved RBP-J-interacting protein RITA. Identified domains are depicted in Figure 8H. We show that besides interaction with RBP-J, RITA interacts with microtubules and locates at the centrosomes. On a molecular level, RITA undergoes a rapid nucleo-cytoplasmic shuttle process and transports RBP-J out of the nucleus to tubulin fibres. We propose that it thereby limits the amount of nuclear RBP-J. On a functional level, RITA affects Notch-mediated transcription and primary neurogenesis. From our data, we postulate that RITA acts as a negative modulator of Notch signalling, representing a novel regulatory mechanism on the level of DNA binding.
RITA–RBP-J interaction
Experiments with deletion constructs demonstrate that the central part of RITA is necessary for RBP-J interaction (Figure 1B). RITA interacts with the BTD of RBP-J (Supplementary Figure S2). We do not have evidence that RITA forms a ternary complex with RBP-J and NICD. In contrast to the colocalization of RITA and RBP-J (Figure 7), NICD remains localized in the nucleus, when RITA is accumulated at the tubulin fibres (Supplementary Figure S4A). Notch proteins form a high-affinity RBP-J-interaction module with their RBP-J association module (RAM) domain. A highly conserved W-X-P motif within the RAM domain contributes to the interaction with the RBP-J BTD (Tamura et al, 1995). Interestingly, we also find a W-X-P motif within the RBP-J-interaction domain of RITA proteins from various species (Supplementary Figure S5A). This W-X-P motif within RITA may contribute to RBP-J interaction and thereby compete against the NICD binding to RBP-J (Supplementary Figure S4B).
Identification of specific domains within RITA
Initially, we could not identify conserved domains within RITA using bioinformatics tools. Microscopic and biochemical analysis revealed that RITA interacts with tubulin. The C-terminus turned out to be essential for tubulin binding, as a deletion of the last 12 aa resulted in a diffuse cytoplasmic localization (Figure 3). This is further supported by the fact that a RITA protein N-terminally fused to EGFP lost its tubulin-binding capability (not shown), and the high conservation of the last amino acids within different species (see Supplementary Figure S5A). A carboxy-terminal fusion of the last 14 aa of human RITA to EGFP did not bind to tubulin (not shown), suggesting that the C-terminus of RITA is necessary, but not sufficient for the tubulin interaction. However, aa 156–269 of human RITA are sufficient for tubulin binding (see Figure 3C), indicating that additional sequences mediate tubulin binding. RITA also localizes at the centrosomes (Figure 4). The region responsible for this interaction appears to differ from the one that confers binding to α-/β-tubulin, as a carboxy-terminally truncated RITA protein (aa 1–257) still localizes at the centrosomes (Supplementary Movie S1). The biological function of RITA at the centrosomes is not yet clarified. A recent study has shown a role of centrosome inheritance during asymmetric cell divisions in the developing brain, maintaining neural progenitor cells in the neocortex (Wang et al, 2009). One could speculate, if RITA has a role in this context.
RITA rapidly accumulates in the nucleus after inhibition of nuclear export by LMB. This behaviour indicates that the subcellular localization of RITA depends on nuclear import and export processes. We identified a functional NLS together with a functional NES within RITA, mediating nucleo-cytoplasmic shuttling (Figures 5 and 6). In addition, nuclear transport of RITA is an essential prerequisite for its function in X. laevis primary neurogenesis (Figure 8E and F). Similarly to human RITA, the RITA proteins from X. laevis and T. adhaerens accumulate in the nucleus after LMB treatment of transfected cells (not shown).
How is the nuclear–cytoplasmic shuttle process of RITA regulated?
It has been shown previously, that RBP-J/CSL proteins can be detected in the nucleus as well as in the cytoplasm and their subcellular distribution changes in defined physiological contexts (Sakai et al, 1995; Zhou and Hayward, 2001; Krejci and Bray, 2007). Here, we provide clear evidence that RITA mediates this redistribution. Most signalling pathways make use of limiting their central transcription factors in the nucleus to activate, modulate, or fine-tune their transcriptional output. Regulation of subcellular localization of RBP-J by RITA may depend on phosphorylation/dephosphorylation of RITA. Besides the C-terminal sequences, the NLS, and the W-X-P motif, two highly conserved, almost identical amino acid stretches (RITA Conserved Repeat 1 and 2 (RCR1 and RCR2 in Figure 8H and Supplementary Figure S5A)) can be exclusively identified within RITA proteins. RCR1 and RCR2 show the consensus motif S-Y-X-D-E-(S/T)-L-F-G, which could represent specific protein–protein interaction motifs or target sites for posttranslational modifications. Indeed, RITA is highly phosphorylated (not shown). A database search for putative kinases, using the Minimotif Miner (MnM 2.0) software (Balla et al, 2006), revealed target sites for several serine/threonine kinases. Therefore, yet not specified kinases and phosphatases may mediate RITA action in regulating the subcellular localization of RBP-J.
Consequences of limiting the amount of nuclear RBP-J by RITA
RBP-J is a key element in the regulation of transcription by Notch signalling. After nuclear translocation, NICD interacts with RBP-J and recruits a coactivator complex to activate transcription of Notch target genes. The transcriptional response to NICD may be controlled by the amount of RBP-J in the nucleus. In addition, NICD was reported to interact with nuclear components of additional signal transduction pathways. Especially crosstalk between Notch signalling, NF-κB signalling, and TGFβ signalling have been described (Kluppel and Wrana, 2005; Osipo et al, 2008; Poellinger and Lendahl, 2008). Given the fact, that after signal activation NICD is found at very low concentrations in the nucleus, the abundance of its major interaction partner RBP-J might be critical for functional association of NICD with additional nuclear factors. Therefore, regulation of the subcellular distribution of RBP-J by RITA and, as a consequence, its concentration in the nucleus, might be critical for the crosstalk of Notch signalling with other pathways. In this regard, it will be interesting to see differential expression of RITA during development and disease and to find correlations to possible changes in Notch target gene expression.
RITA modulates early organogenesis in Xenopus laevis
The Notch signalling pathway affects many different developmental processes, most prominently the effect on primary neurogenesis (Chitnis et al, 1995; Wettstein et al, 1997). Moreover, effects on germ layer formation (Abe et al, 2005; Contakos et al, 2005), somitogenesis (Jen et al, 1997; Davis et al, 2001), development of heart (Rones et al, 2000; Miazga and McLaughlin, 2009), pronephros (McLaughlin et al, 2000), head structures and eyes (Onuma et al, 2002; Ogino et al, 2008), and axial structures (Lopez et al, 2003) have been described in X. laevis and other organisms. A loss-of-function approach, using a morpholino against RITA, and RITA gain-of-function experiments resulted in defects that are in correspondence with Notch overexpression phenotypes. An initial analysis in mouse shows highest levels of RITA expression in several of the organs that are affected by Notch pathway manipulations, for example brain, heart, muscle, kidney, and intestine (Supplementary Figure S6A and B). In accordance with a function of RITA as regulator of Notch signalling are the rescue experiments. A RITA mRNA coinjection in Xenopus embryos restored all observed effects of a constitutive active Notch construct Notch-1ΔE, including restoration of the N-tubulin expression, phenotypical effects, and changes in the expression levels of known downstream targets of the Notch signalling pathway.
These results and overlapping expression patterns of RITA and Notch strengthen the paradigm that RITA acts as a general negative modulator of an activated Notch signalling pathway. RITA may do so by interacting with RBP-J, regulating its cytoplasmic/nuclear localization and consequently modulating the Notch-dependent transcriptional response. This general molecular mechanism, that is the regulated targeting of a transcription factor in or out of the nucleus by posttranslational modifications or a specific inhibitory protein, is now for the first time indicated within the Notch signalling pathway.
Materials and methods
Oligonucleotides
A list of all oligonucleotides used for plasmid construction and real-time PCR and the morpholino sequences, used for a specific knockdown in X. laevis, are included in Supplementary data.
Yeast two-hybrid screening
Yeast two-hybrid screening was carried out with the MATCHMAKER system (Clontech) according to the manufacturer's instructions as described previously (Oswald et al, 2002).
Plasmids
The vectors pcDNA3-Flag-RBP-2N, CMV-RBP-VP16, HES1-LUC, pcDNA3-Flag-1-SHARP(RBPID), pcDNA3mNotch-1ΔE, plasmid for expression of GST-mNotch-1-IC (GST-mNICD) were described in Oswald et al (2002). The expression plasmids for mNotch-1-IC mRuby and mNotch-1-IC(ΔRBP/ΔEP)-mRuby were made by exchanging the fluorescent tag in the corresponding GFP constructs (Oswald et al, 2001). Constructs for bacterial expression of SHARP-specific GST-fusion proteins (SHARP-RBPID, SHARP-2002–3663) were described in Oswald et al (2005). The expression construct pcDNA3-RBP-2N-mRuby was described in Salat et al (2008). The expression plasmid for X. laevis RBP-J, pCS2-xlSu(H), was a kind gift from Ying Cao, University of Ulm. The expression plasmid for γ-Tubulin fused to mRuby, was kindly provided by MW Davidson (The Florida State University, Tallahassee, FL). Expression plasmids for the RITA proteins (human, X. laevis, and T. adhaerens) were made by PCR. For further details see Supplementary data.
Cell culture and preparation of cell extracts
HEK293 cells were transfected with DNA using the calcium phosphate coprecipitation method (Promega). HEK293 were transfected with siRNA using the GeneEraser™ siRNA transfection reagent (Stratagene). HeLa cells were transfected using the Nanofectine (PAA) transfection reagent. All transfections were performed according to the manufacturer's instructions. The cell lines HEK293 (ATCC CRL 1573) and HeLa (ATCC CCL 2) were grown in Dulbecco's modified Eagle's medium (DMEM, Gibco) supplemented with 10% fetal calf serum, penicillin, and streptomycin. The HEK293 cell lines stably expressing GFP and GFP-RITA fusion proteins (HEK293RITA 1–269(wt), HEK293RITA 1–257, HEK293RITA ΔRBP, HEK293RITA 25–269, HEK293EGFP) were propagated with 300 μg/ml Neomycin (G418, PAA) within the medium.
Coimmunoprecipitation and western blotting
Whole-cell lysates for western blotting and immunoprecipitation experiments were prepared as described previously (Salat et al, 2008). Nuclear and cytoplasmic fractions were prepared using standard procedures as described in Supplementary data. IP experiments were carried out using whole-cell extracts either from HEK293 cells stably expressing GFP-RITA fusion proteins or HEK293 cells 24 h after transfection with Flag-RITA expressing constructs as described in Supplementary data.
In vitro protein translation
Proteins were synthesized in the presence of [35S] methionine using a reticulocyte lysate-coupled transcription/translation system (Promega). Translation and labelling quality were monitored by SDS–PAGE.
GST pull-down assay
GST-protein and the GST-fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene) and stored as whole bacterial lysates at −80°C. Approximately 1 μg of GST-protein and GST-fusion protein were immobilized with glutathione-sepharose beads (Amersham) and incubated together with the in vitro translated proteins under rotation at 4°C for 1 h. Beads were washed four times with 600 μl buffer A (40 mM HEPES (pH 7.5), 5 mM MgCl2, 0.2 mM EDTA, 0.5% Nonidet P40 (NP-40), and 100 mM KCl) and four times with 600 μl buffer B (equivalent to buffer A, but containing 300 mM KCl). After the washing steps, the beads were resuspended in SDS–PAGE loading buffer and the proteins were separated by SDS–PAGE. The gels were dried and exposed to X-ray films.
Luciferase assay
HeLa cells (5 × 104) were transfected in 24-well plates with 1 μg of reporter plasmid DNA together with various amounts of expression plasmid. Luciferase activity was determined from at least three independent transfections with 20 μl of cleared lysate in an LB 9501 luminometer (Berthold) using the luciferase assay system from Promega.
Quantitative RNA analysis
Total RNA was isolated from different mouse tissues, ES cells and X. laevis embryos as previously described (Oswald et al, 2002). Half Xenopus embryos were cut with eyebrow knives after removal of the perivitelline membrane using forceps. Expression levels of mRNA were quantified using real-time PCR (TaqMan, PE Applied Biosystems). For PCR, cDNAs were reverse transcribed from 2 μg of total RNA. The PCR reaction (denaturation 95°C for 2 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min) was performed using Sybr Green PCR Core Reagents (PE Applied Biosystems) and primer combinations are listed in Supplementary data.
Injection of mRNA and morpholinos
Embryos were staged according to Nieuwkoop and Faber (1956). In vitro fertilization, embryo culture, and microinjections were carried out as previously described (Wacker et al, 2000). For details about injection of mRNAs and MOs see Supplementary data. Numbers for the injection experiments and distribution of attained phenotypes are listed in Supplementary Table S3.
Detection of gene expression by in situ hybridization
Whole-mount in situ hybridization was performed as previously described (Harland, 1991), except that the RNAse step was omitted. RITA digoxigenin-labelled probes were generated from pcDNA3-RITA(xl) as a template (sense T7/Xba, antisense SP6/EcoRI). An antisense digoxigenin-labelled probe was used for N-tubulin (Marcus et al, 1998).
Fluorescence microscopy
For immunofluorescence imaging, HeLa cells were prepared as described in Supplementary data. The following antibodies and antisera were used: anti-α-Tubulin, mouse monoclonal IgG, (T9026, Sigma), secondary antibody, Cy3™-coupled sheep anti-mouse IgG (C2181, Sigma), anti-Pericentrin (Martin-Subero et al, 2003), secondary antibody, Alexa-Fluor-568-coupled goat anti-rabbit IgG (A11011, Invitrogen). Production of the anti-NEK-2 antiserum is described in Supplementary data. The secondary antibody was A11011 (Invitrogen).
Supplementary Material
Acknowledgments
We thank Y Cao and B Schierwater for providing us with reagents. We thank B Korte, N Heymann, R Rittelman, and S Schirmer for excellent technical assistance. This study was supported by the Deutsche Forschungsgemeinschaft (DFG, SFB 497/B9 and SFB 518/A18 to FO, SFB 497/A1 to WK, Wi1990/2-1 to JW). GUN acknowledges support by the State of Baden-Württemberg and the DFG (CFN, Ni 291/9 and SFB 497/C10).
Authors' contributions: FO and SW wrote the manuscript; FO and WK conceived and designed the experiments; FO, SW, CA, GW, UK, KC, HH, and BB performed the experiments; and FO, SW, CA, JW, GUN, and TB analysed the data.
Footnotes
The authors declare that they have no conflict of interest.
References
- Abe T, Furue M, Kondow A, Matsuzaki K, Asashima M (2005) Notch signaling modulates the nuclear localization of carboxy-terminal-phosphorylated smad2 and controls the competence of ectodermal cells for activin A. Mech Dev 122: 671–680 [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 [DOI] [PubMed] [Google Scholar]
- Balla S, Thapar V, Verma S, Luong T, Faghri T, Huang CH, Rajasekaran S, del Campo JJ, Shinn JH, Mohler WA, Maciejewski MW, Gryk MR, Piccirillo B, Schiller SR, Schiller MR (2006) Minimotif Miner: a tool for investigating protein function. Nat Methods 3: 175–177 [DOI] [PubMed] [Google Scholar]
- Borggrefe T, Oswald F (2009) The Notch signaling pathway: transcriptional regulation at Notch target genes. Cell Mol Life Sci 66: 1631–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ (2006) Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C (1995) Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 375: 761–766 [DOI] [PubMed] [Google Scholar]
- Chitnis AB (1995) The role of Notch in lateral inhibition and cell fate specification. Mol Cell Neurosci 6: 311–321 [PubMed] [Google Scholar]
- Contakos SP, Gaydos CM, Pfeil EC, McLaughlin KA (2005) Subdividing the embryo: a role for Notch signaling during germ layer patterning in Xenopus laevis. Dev Biol 288: 294–307 [DOI] [PubMed] [Google Scholar]
- Davis RL, Turner DL, Evans LM, Kirschner MW (2001) Molecular targets of vertebrate segmentation: two mechanisms control segmental expression of Xenopus hairy2 during somite formation. Dev Cell 1: 553–565 [DOI] [PubMed] [Google Scholar]
- Fortini ME (2009) Notch signaling: the core pathway and its posttranslational regulation. Dev Cell 16: 633–647 [DOI] [PubMed] [Google Scholar]
- Fry AM, Meraldi P, Nigg EA (1998) A centrosomal function for the human Nek2 protein kinase, a member of the NIMA family of cell cycle regulators. EMBO J 17: 470–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer CJ, White JB, Jones KA (2004) Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol Cell 16: 509–520 [DOI] [PubMed] [Google Scholar]
- Harland RM (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol 36: 685–695 [DOI] [PubMed] [Google Scholar]
- Jen WC, Wettstein D, Turner D, Chitnis A, Kintner C (1997) The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development 124: 1169–1178 [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T (1998) A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev 12: 2269–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluppel M, Wrana JL (2005) Turning it up a Notch: cross-talk between TGF beta and Notch signaling. Bioessays 27: 115–118 [DOI] [PubMed] [Google Scholar]
- Koch U, Radtke F (2007) Notch and cancer: a double-edged sword. Cell Mol Life Sci 64: 2746–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX (2009) The canonical Notch signaling pathway: unfolding the activation mechanism. Cell 137: 216–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovall RA (2008) More complicated than it looks: assembly of Notch pathway transcription complexes. Oncogene 27: 5099–5109 [DOI] [PubMed] [Google Scholar]
- Kredel S, Oswald F, Nienhaus K, Deuschle K, Rocker C, Wolff M, Heilker R, Nienhaus GU, Wiedenmann J (2009) mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One 4: e4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S (2007) Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev 21: 1322–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda K, Han H, Tani S, Tanigaki K, Tun T, Furukawa T, Taniguchi Y, Kurooka H, Hamada Y, Toyokuni S, Honjo T (2003) Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18: 301–312 [DOI] [PubMed] [Google Scholar]
- la Cour T, Kiemer L, Molgaard A, Gupta R, Skriver K, Brunak S (2004) Analysis and prediction of leucine-rich nuclear export signals. Protein Eng Des Sel 17: 527–536 [DOI] [PubMed] [Google Scholar]
- Liefke R, Oswald F, Alvarado C, Ferres-Marco D, Mittler G, Rodriguez P, Dominguez M, Borggrefe T (2010) Histone demethylase KDM5A is an integral part of the core Notch-RBP-J repressor complex. Genes Dev 24: 590–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez SL, Paganelli AR, Siri MV, Ocana OH, Franco PG, Carrasco AE (2003) Notch activates sonic hedgehog and both are involved in the specification of dorsal midline cell-fates in Xenopus. Development 130: 2225–2238 [DOI] [PubMed] [Google Scholar]
- Marcus EA, Kintner C, Harris W (1998) The role of GSK3beta in regulating neuronal differentiation in Xenopus laevis. Mol Cell Neurosci 12: 269–280 [DOI] [PubMed] [Google Scholar]
- Martin-Subero JI, Knippschild U, Harder L, Barth TF, Riemke J, Grohmann S, Gesk S, Hoppner J, Moller P, Parwaresch RM, Siebert R (2003) Segmental chromosomal aberrations and centrosome amplifications: pathogenetic mechanisms in Hodgkin and Reed-Sternberg cells of classical Hodgkin's lymphoma? Leukemia 17: 2214–2219 [DOI] [PubMed] [Google Scholar]
- McLaughlin KA, Rones MS, Mercola M (2000) Notch regulates cell fate in the developing pronephros. Dev Biol 227: 567–580 [DOI] [PubMed] [Google Scholar]
- Miazga CM, McLaughlin KA (2009) Coordinating the timing of cardiac precursor development during gastrulation: a new role for Notch signaling. Dev Biol 333: 285–296 [DOI] [PubMed] [Google Scholar]
- Miele L, Golde T, Osborne B (2006) Notch signaling in cancer. Curr Mol Med 6: 905–918 [DOI] [PubMed] [Google Scholar]
- Morel V, Lecourtois M, Massiani O, Maier D, Preiss A, Schweisguth F (2001) Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr Biol 11: 789–792 [DOI] [PubMed] [Google Scholar]
- Moshkin YM, Kan TW, Goodfellow H, Bezstarosti K, Maeda RK, Pilyugin M, Karch F, Bray SJ, Demmers JA, Verrijzer CP (2009) Histone chaperones ASF1 and NAP1 differentially modulate removal of active histone marks by LID-RPD3 complexes during NOTCH silencing. Mol Cell 35: 782–793 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J (1956) Normal Table of Xenopus laevis (Daudin). Amsterdam: North Holland Publishing Company [Google Scholar]
- Ogino H, Fisher M, Grainger RM (2008) Convergence of a head-field selector Otx2 and Notch signaling: a mechanism for lens specification. Development 135: 249–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onuma Y, Takahashi S, Asashima M, Kurata S, Gehring WJ (2002) Conservation of Pax 6 function and upstream activation by Notch signaling in eye development of frogs and flies. Proc Natl Acad Sci USA 99: 2020–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osipo C, Golde TE, Osborne BA, Miele LA (2008) Off the beaten pathway: the complex cross talk between Notch and NF-kappaB. Lab Invest 88: 11–17 [DOI] [PubMed] [Google Scholar]
- Oswald F, Kostezka U, Astrahantseff K, Bourteele S, Dillinger K, Zechner U, Ludwig L, Wilda M, Hameister H, Knöchel W, Liptay S, Schmid RM (2002) SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J 21: 5417–5426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Tauber B, Dobner T, Bourteele S, Kostezka U, Adler G, Liptay S, Schmid RM (2001) p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol Cell Biol 21: 7761–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knöchel W, Borggrefe T (2005) RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol 25: 10379–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poellinger L, Lendahl U (2008) Modulating Notch signaling by pathway-intrinsic and pathway-extrinsic mechanisms. Curr Opin Genet Dev 18: 449–454 [DOI] [PubMed] [Google Scholar]
- Rones MS, McLaughlin KA, Raffin M, Mercola M (2000) Serrate and Notch specify cell fates in the heart field by suppressing cardiomyogenesis. Development 127: 3865–3876 [DOI] [PubMed] [Google Scholar]
- Roy M, Pear WS, Aster JC (2007) The multifaceted role of Notch in cancer. Curr Opin Genet Dev 17: 52–59 [DOI] [PubMed] [Google Scholar]
- Sakai T, Furukawa T, Iwanari H, Oka C, Nakano T, Kawaichi M, Honjo T (1995) Loss of immunostaining of the RBP-J kappa transcription factor upon F9 cell differentiation induced by retinoic acid. J Biochem 118: 621–628 [DOI] [PubMed] [Google Scholar]
- Salat D, Liefke R, Wiedenmann J, Borggrefe T, Oswald F (2008) ETO, but not leukemogenic fusion protein AML1/ETO, augments RBP-Jkappa/SHARP-mediated repression of notch target genes. Mol Cell Biol 28: 3502–3512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Taniguchi Y, Minoguchi S, Sakai T, Tun T, Furukawa T, Honjo T (1995) Physical interaction between a novel domain of the receptor Notch and the transcription factor RBP-J kappa/Su(H). Curr Biol 5: 1416–1423 [DOI] [PubMed] [Google Scholar]
- Wacker S, Grimm K, Joos T, Winklbauer R (2000) Development and control of tissue separation at gastrulation in Xenopus. Dev Biol 224: 428–439 [DOI] [PubMed] [Google Scholar]
- Wallberg AE, Pedersen K, Lendahl U, Roeder RG (2002) p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol Cell Biol 22: 7812–7819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wettstein DA, Turner DL, Kintner C (1997) The Xenopus homolog of Drosophila suppressor of hairless mediates Notch signaling during primary neurogenesis. Development 124: 693–702 [DOI] [PubMed] [Google Scholar]
- Wilson JJ, Kovall RA (2006) Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124: 985–996 [DOI] [PubMed] [Google Scholar]
- Wu L, Aster JC, Blacklow SC, Lake R, Artavanis-Tsakonas S, Griffin JD (2000) MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat Genet 26: 484–489 [DOI] [PubMed] [Google Scholar]
- Zhou S, Hayward SD (2001) Nuclear localization of CBF1 is regulated by interactions with the SMRT corepressor complex. Mol Cell Biol 21: 6222–6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.