miR-301a as an NF-κB activator in pancreatic cancer cells
The authors identify miR-301a as functional regulator of NF-κB activation in pancreatic cancer. Molecularly, NKRF (negative regulator of NF-κB) is revealed as direct miR-301a target. Interestingly, miR-301a is itself induced by NF-κB, establishing a crucial feedback-forward cycle that seems operational also in tumour tissue samples.
Keywords: microRNA, miR-301a, NF-κB, NF-κB-repressing factor, pancreatic cancer
Abstract
NF-κB is constitutively activated in most human pancreatic adenocarcinoma, which is a deadly malignancy with a 5-year survival rate of about 5%. In this work, we investigate whether microRNAs (miRNAs) contribute to NF-κB activation in pancreatic cancer. We demonstrate that miR-301a down-regulates NF-κB-repressing factor (Nkrf) and elevates NF-κB activation. As NF-κB promotes the transcription of miR-301a, our results support a positive feedback loop as a mechanism for persistent NF-κB activation, in which miR-301a represses Nkrf to elevate NF-κB activity, which in turn promotes miR-301a transcription. Nkrf was found down-regulated and miR-301a up-regulated in human pancreatic adenocarcinoma tissues. Moreover, miR-301a inhibition or Nkrf up-regulation in pancreatic cancer cells led to reduced NF-κB target gene expression and attenuated xenograft tumour growth, indicating that miR-301a overexpression contributes to NF-κB activation. Revealing this novel mechanism of NF-κB activation by an miRNA offers new avenues for therapeutic interventions against pancreatic cancer.
Introduction
NF-κB is a transcription factor (TF) consisting of dimers of NF-κB1 (p50), NF-κB2, RelA, RelB, and c-Rel proteins with p50:RelA heterodimers generally as the dominating form. NF-κB is activated in eukaryotic cell systems in response to inflammation, carcinogens, and other stimuli (Pahl, 1999). NF-κB is constitutively activated in human pancreatic cancer, a deadly malignancy with a 5-year survival rate of ∼5% (Jemal et al, 2009). Somatic mutation has been implicated to have a critical function in NF-κB in some types of cancers as a large number of genetic abnormalities found in genes involved in the canonical or alternative NF-κB pathways. For example, the oncogenic CARD11 mutations were found in activated B-cell like diffuse large B-cell lymphoma (Lenz et al, 2008), as well as inactivating mutations of IκBα in Hodgkin's lymphoma (Jost and Ruland, 2007), and a dozen of NF-κB-relevant genes in multiple myeloma (Annunziata et al, 2007; Keats et al, 2007). A recent comprehensive genetic analysis of 24 pancreatic tumours revealed that there are plenty of mutations on genes in 12 cellular signalling pathways and processes, but little on genes in the NF-κB network (Jones et al, 2008), indicating that somatic mutations are unlikely the cause for NF-κB activation in pancreatic cancer.
MicroRNAs (miRNAs) are short 20–25 nucleotide RNA molecules that negatively regulate gene expression in animals and plants. Though miRNAs were first discovered to have crucial functions in Caenorhabditis elegans development, recent progress in cancer biology has shown that miRNAs are frequently dysregulated in human cancers. Moreover, miRNAs are acting as modulators or effectors of the NF-κB pathway. For example, miR-146a and miR-146b down-regulate IL-1 receptor-associated kinase 1 and TNF receptor-associated factor 6 protein levels, which demonstrate miRNA's regulatory roles in the NF-κB pathway (Taganov et al, 2006). Furthermore, NF-κB-responsive miR-155 and miR-125b have a function in innate immune response (Tili et al, 2007). Most recently, miR-199a is shown to regulate IKKβ, a known modulator of the tumour inflammatory microenvironment (Chen et al, 2008).
In this study, we first found that miR-301a, an miRNA that is specifically up-regulated in pancreatic cancer (Lee et al, 2007), activates NF-κB by negatively regulating the expression of the NF-κB-repressing factor (NKRF) gene. We also found that the expression of miR-301a is up-regulated when NF-κB is activated. Therefore, a positive feedback loop exists for persistent NF-κB activation, whereby miR-301a down-regulates Nkrf levels leading to elevated NF-κB activity, which in turn further promotes miR-301a transcription. Additionally, we demonstrated that Nkrf is down-regulated in pancreatic adenocarcinomas, and that miR-301a inhibition or Nkrf overexpression in pancreatic cancer cells led to reduced NF-κB activation and tumour growth, indicating that this NF-κB activation mechanism could be a target for therapeutic intervention against pancreatic adenocarcinoma.
Results
miR-301a as the most potent NF-κB activator
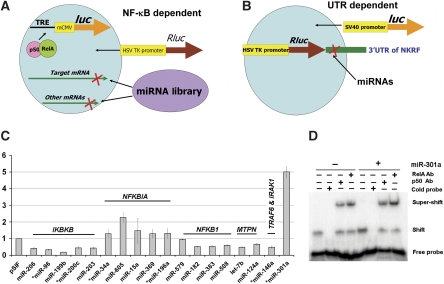
We constructed an miRNA genetic library that is based on a feline immunodeficiency virus vector (Supplementary Figure S1) and formulated a reporter strategy to screen miRNAs from this library that modulate NF-κB activation (Figure 1A). The first assay (NF-κB-dependent reporter) uses a cell line 293NF-κB (Supplementary data) that expresses a firefly luciferase gene (luc) under the control of a minimal CMV promoter and NF-κB transcription response elements. The miRNAs that up-regulate the NF-κB-dependent reporter are subjected to the second assay (UTR-dependent reporter) to test whether they suppress the expression of a Renilla luciferase gene (Rluc) with a chosen 3′UTR. Testing the first assay using known negative regulatory miRNAs in NF-κB signalling showed that miR-146a (Taganov et al, 2006), let-7b, and miR-124a (Gupta et al, 2002) down-regulated the NF-κB-dependent reporter (Figure 1C), suggesting that this assay was rather effective and sensitive. When all the miRNA constructs in the library were screened with the first assay, we found that miR-301a was the most potent activator that up-regulated NF-κB-dependent reporter activity, ∼5.0-fold, whereas miR-199b down-regulated reporter activity the most, ∼82% (Figure 1C). We also revealed a few miRNAs as potent NF-κB modulators that are dysregulated in pancreatic cancer and are predicted to target NFKB1, NFKBIA, or IKBKB and up- or down-regulate reporter activities accordingly (Figure 1C; Supplementary Table S1). In this study, we focused on miR-301a because (1) it was the most potent NF-κB activator from our reporter screening; (2) its NF-κB activation function was confirmed by an electrophoretic mobility shift assay (EMSA), in which the NF-κB DNA-binding ability was increased ∼2.8-fold in cells with miR-301a overexpression (Figure 1D); and (3) miR-301a was first demonstrated to be specifically up-regulated ∼34-fold in pancreatic tumours (Lee et al, 2007) and later in hepatocellular carcinoma, at a much lower level (Jiang et al, 2008), whereas other dysregulated miRNAs were not exclusive to pancreatic cancer.
Figure 1.
miRNAs modulate NF-κB signalling and miR-301a is the most potent activator. (A) The first assay to identify miRNAs modulating NF-κB-dependent reporter expression. (B) The second assay to determine whether an miRNA targets a specific 3′UTR. (C) Screening miRNA in NF-κB signalling. miRNAs labelled for NFKB1 (encoding p50), NFKBBIA (IκBα), and IKBKB (IKKβ) are based on computational predictions, whereas those for MTPN (Myotrophin), TRAF6 (TNF receptor-associated factor 6), and IRAK1 (interleukin-1 receptor-associated kinase 1) are experimentally tested by other groups. The y axis is the relative luminescence unit (RLU) of luc normalized to that of Rluc from pRL-TK compared with that of the vector control. ‘*' denotes miRNA that was dysregulated in pancreatic cancer. Error bars represent s.d. (D) EMSA of nuclear extracts of 293T cells with miR-301a (lanes 5–8) or the parental vector (lanes 1–4). Super-shift was performed using antibodies (Ab) against p50 or RelA. The densitometry of band intensity was calculated as (Shift/(Shift+Free Probe)) with the signal density of lane 5 ∼2.8 times to that of lane 1.
miR-301a targets NKRF
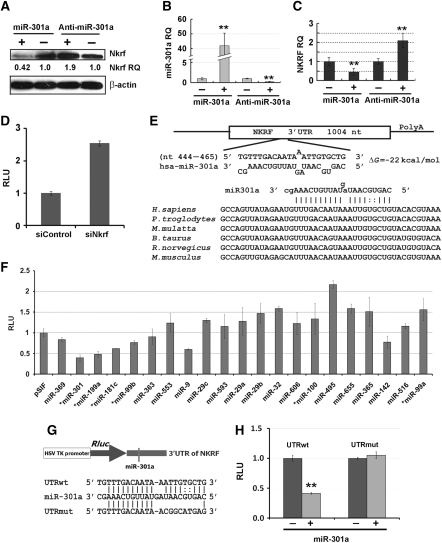
We investigated how miR-301a activates NF-κB as it is not predicted to target any Rel, IKB, or IKK gene by four widely used miRNA target prediction methods: Miranda (John et al, 2004), TargetScan (Lewis et al, 2005), PicTar (Krek et al, 2005), or RNA22 (Miranda et al, 2006). Using these four programmes with relaxed stringency, computational analyses were performed to predict target genes of miR-301a. Over 1000 genes were predicted targets of miR-301a after screening all human genes. We next examined proteins known to inhibit NF-κB activation, which include IKBs (Karin et al, 2002), Cyld (Trompouki et al, 2003), A20 (Krikos et al, 1992), Tax1bp1 (Shembade et al, 2008), Itch (Shembade et al, 2008), Tnip1 (Mauro et al, 2006), and Nkrf (Nourbakhsh and Hauser, 1999). Of these, only TAX1BP1 and NKRF were predicted targets of miR-301a by the Miranda algorithm (John et al, 2004). Thus, we transfected 293T cells with the miR-301a construct and extracted soluble proteins to probe the expression levels of Tax1bp1 and Nkrf. Nkrf protein was reduced ∼3-fold when miR-301a was overexpressed ∼42-fold in HEK-293T cells (Figure 2A and B), while Tax1bp1 levels did not change. We tested whether down-regulation of miR-301a in 293T cells affected Nkrf expression. As shown in Figure 2A and B, when miR-301a expression levels were decreased ∼80% using an antisense inhibitor (anti-miR-301a), Nkrf protein expression was increased approximately two-fold. A qRT–PCR reaction was performed to determine whether the mRNA level of NKRF changed when miR-301a expression was modulated. NKRF mRNA increased when miR-301a was down-regulated, and it decreased with miR-301a overexpression (Figure 2B and C). These data suggested that NKRF expression was negatively regulated by miR-301a via mRNA degradation. Additionally, when Nkrf expression level was reduced by the siRNA against Nkrf (siNkrf), NF-κB-dependent reporter expression was up-regulated ∼2.5-fold (Figure 2D), indicating that activation of NF-κB by miR-301a is likely through down-regulation of Nkrf. Furthermore, our in silico analyses revealed that the miR-301a:NKRF interaction is conserved in human, rat, and mouse (Lewis et al, 2005)—a finding indicative of regulatory importance (Figure 2E). It is noteworthy that there is a lack of a perfect seed match between miR-301a and the NKRF 3′UTR. Two G:U wobble-base pairs are present in the seed region, but there is a great deal of compensatory 3′ pairing (10 pairs) (Bartel, 2009). To further test the importance of the compensatory 3′ pairing, we introduced miR-130a that has the same seed sequence, but 9 out of the 10 compensatory base pairs are disrupted compared with miR-301a (Supplementary Figure S2). Neither Nkrf expression level (Supplementary Figure S2B) nor NF-κB reporter activity (Supplementary Figure S2C) was affected by miR-130a, indicating that the complementation of 3′ part of miR-301a to 3′UTR of NKRF is essential for Nkrf-negative regulation. Thus, we conclude that the interaction of miR-301a and NKRF represents a 3′ compensatory site (Bartel, 2009) and the first one with two G:U pairs.
Figure 2.
miR-301a targets NF-κB-repressing factor (NKRF). (A) miR-301a down-regulated Nkrf expression and anti-miR-301a up-regulated its expression in 293T cells. ‘Nkrf RQ', relative quantity of Nkrf protein as normalized to β-actin. (B) Expression of miR-301a in 293T cells from (A) measured by qPCR. (C) NKRF mRNA level by qPCR in 293T cells from (A). (D) Down-regulation of Nkrf by the siRNA against Nkrf (siNkrf) increased NF-κB-dependent reporter expression. (E) An miR-301a target site resides at nts 444–465 of the NKRF 3′UTR, and is highly conserved in six species. (F) Modulation of the expression of Rluc upstream of the 3′UTR of NKRF with miRNAs that are predicted to target NKRF. ‘*' denotes miRNA that was dysregulated in pancreatic cancer. (G) Diagram of NKRF 3′UTR downstream of a reporter. Wild-type 3′UTR (UTRwt) contains a native miR-301a-binding site, while a mutant 3′UTR (UTRmut) contains mutations that abolished the seed match with miR-301a. (H) The second assay (Figure 1B) with UTRwt or UTRmut. **P⩽0.001 with n=3–7 compared with respective controls. Error bars represent s.d.
To test whether miR-301a targets the 3′UTR of the NKRF gene directly, we cloned the 3′UTR of NKRF downstream of the Rluc gene of pRL-TK and performed the second assay with miR-301a and 21 other miRNAs, all of which are predicted to target NKRF. We found that the reporter was down-regulated the most when miR-301a was introduced (Figure 2F). We subsequently mutated the 3′UTR to disrupt its interaction with the seed sequence of miR-301a and found that reporter down-regulation by miR-301a was largely abolished (Figure 2G and H). These results indicate that overexpression of miR-301a down-regulates Nkrf protein levels and that down-regulation of Nkrf by miR-301a is dependent on the NKRF 3′UTR. To further demonstrate that the interaction of miR-301a and Nkrf is specific, we performed reporter assays and qPCR with three controls: (1) miR-130a, which has a seed sequence as miR-301a, but has no 10-bp 3′ compensatory site; (2) miR-223 has a 10-bp (one G:U) pairing, but no seed sequence match with the UTR; and (3) miR-301a mutant (scrambled miR-301a), in which the seed sequence was disrupted (Supplementary Figure S2A). miR-301a, but not these controls, up-regulated NF-κB-dependent reporter expression (Supplementary Figure S2C), down-regulated NKRF 3′UTR-dependent reporter activity (Supplementary Figure S2D), down-regulated Nkrf mRNA levels, and increased the expression of two NF-κB-transactivation targets (Supplementary Figure S2E).
miR-301a expression is regulated by NF-κB
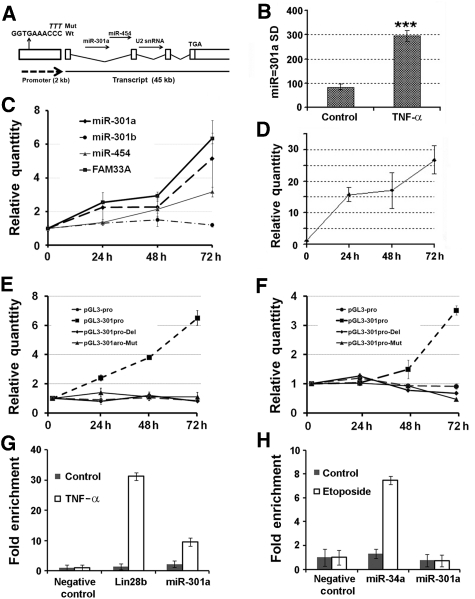
The miR-301a gene is located on chromosome 17: 54 583 279–54 583 364 [−], within the first intron of a ∼45 kb mRNA transcript FAM33A (Figure 3A). Interestingly, there are two more non-coding RNAs in this transcript: miR-454 in the same intron and a predicted U2 snRNA homologue in the second intron, although we have not examined the functions of the other non-coding RNAs or FAM33A in this work. We profiled the expression of all miRNAs in 293T cells using miRNA microarray. miR-301a was up-regulated in cells treated with TNF-α (approximately three-fold; Figure 3B). To further test whether miR-301a is induced by TNF-α, we treated 293NF-κB cells with TNF-α for 24, 48, and 72 h. The transcription of endogenous miR-301a and miR-454 was induced as measured by qRT–PCR (Figure 3C) when NF-κB was activated by TNF-α as monitored by the reporter assay (Figure 3D). In addition, the expression of FAM33A mRNA was also induced using β-actin mRNA as a reference. We noted that there are two copies of the miR-301 gene in the human genome: miR-301a and miR-301b with the latter located in an intergenic region on Chromosome 22 (Griffiths-Jones et al, 2006); and the expression of miR-301b was not induced by TNF-α treatment (Figure 3C). Using the computer program MAPPER (Marinescu et al, 2005), a search engine that identifies TF-binding sites, we examined whether there is a κB site on the 1.8 kb DNA sequence upstream of the FAM33A start codon (Hanisch et al, 2006). We found a putative p50-RelA-binding site (Figure 3A). To determine whether this 1.8 kb DNA fragment is an active promoter and this κB site is authentic, we cloned this DNA fragment upstream of the ATG codon of FAM33A and placed it upstream of luc. The DNA segment was able to drive the expression of luc, indicating that this 1.8 kb DNA fragment is functional as a promoter. Furthermore, the luc activity was increased by the addition of TNF-α for 24, 48, and 72 h, while mutations either altering or deleting the κB site abolished the response to TNF-α treatment, but not basal luc expression in 293T cells (Figure 3E). Similar results were observed in a pancreatic cancer cell line PANC-1 (Figure 3F).
Figure 3.
NF-κB activation promotes miR-301a expression. (A) The gene structure of miR-301a/FAM33A. The sequence ‘GGTGAAACCC' is a predicted p50-RelA-binding site in the miR-301a promoter (301aPro) and the upper italic letters ‘TTT' replaces ‘CCC' in the miR-301a promoter mutation construct (301aPro-Mut). (B) miR-301a up-regulation in a microarray assay for 293T cells treated with TNF-α. The y axis denotes the relative signal density (SD) for miR-301a. ***P⩽0.0001 with n=7. (C) Expression of the transcriptional unit of miR-301a and FAM33A in 293NF-κB cell lines with TNF-α treatment. ‘Relative Quantity' in (C) is the relative expression level of genes determined by qPCR when referenced by that of β-actin or U6 snRNA. (D) Luciferase reporter assays with TNF-α treatment in 293NF-κB cell line. TNF-α induced NF-κB activity in a time-dependent manner. (E) The promoter of miR-301a (pGL3-301aPro) was active and responsive to TNF-α treatment in 293T cells. The mutant (pGL3-301aPro-Mut and pGL-301aPro-Del) and pGL3-pro (SV40-promoter) were active, but unresponsive to TNF-α. (F) Similar to (E), but in PANC-1 cells. ‘Relative Quantity' in (D–F) is the ratio of the firefly luciferase activity driven by various promoters normalized to that of Renilla luciferase from a co-transfected construct (pRL-TK) before referenced to that from cells without TNF-α stimulation. (G) NF-κB (RelA) occupancy (fold enrichment) at the miR-301a locus. ChIP assays were performed on lysates prepared from 293T cells with or without TNF-α treatment using antibodies against RelA. (H) p53 occupancy assay. Similar assay was performed as in (G), but with p53 antibodies and etoposide treatment. ‘Fold enrichment' in (G) and (H) was represented by two to the power of Ct (input) subtracted by Ct (elute) before normalized to that of the negative control. Error bars represent s.d.
To further demonstrate that miR-301a was transcriptionally controlled by NF-κB, we performed a chromatin immunoprecipitation (ChIP) assay using lysates from 293T cells with or without TNF-α stimulation. We found that RelA (NF-κB) bound the miR-301a promoter strongly upon TNF-α stimulation compared with a negative control (Figure 3G). A similar result was observed with a positive control, that is NF-κB-regulated lin28b (Iliopoulos et al, 2009). In contrast, p53, another TF, bound the miR-34a promoter, but not the miR-301a promoter (Figure 3H) as the miR-301a promoter appears to contain no p53 transcription response elements. These data suggest that the promoter of miR-301a contains an authentic NF-κB-binding site. Taken together, our results support that miR-301a is both an NF-κB activator and an NF-κB-regulated gene.
Down-regulation of miR-301a in pancreatic cancer cells reduces NF-κB activation
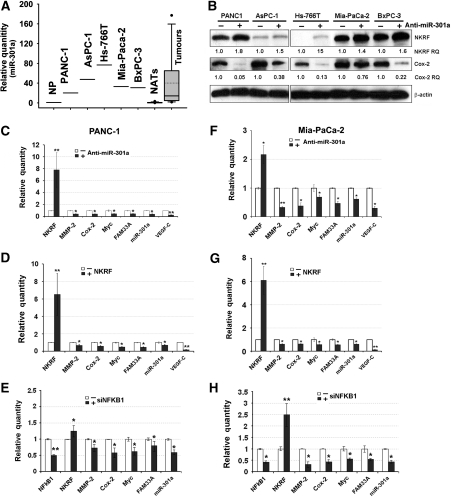
We profiled miR-301a expression levels in five pancreatic cancer cell lines (PANC-1, AsPC-1, Hs-766T, Mia-PaCa-2, and BxPC-3) and found it was up-regulated compared with total RNAs extracted from a pool of normal pancreases (Figure 4A), and miR-301a expression levels also were elevated in pancreatic adenocarcinoma tissues compared with paired normal adjacent tissues as normalized by U6 snRNA (P=0.001; n=24) (Figure 4A; Supplementary Figure S3). When miR-301a expression was inhibited, Nkrf level in each of these cell lines was increased (Figure 4B). Moreover, the protein level of an NF-κB target gene COX2, expressed in invasive pancreatic ductal carcinoma (Aoki et al, 2002), was significantly down-regulated (Figure 4B). We tested whether down-regulation of Cox2 is NF-κB/Nkrf dependent in pancreatic cancer cells. We transfected PANC-1 cells with anti-miR-301a or an Nkrf overexpression construct and the Cox2 promoter upstream of a luc reporter (Yamamoto et al, 1995). When either anti-miR-301a (Supplementary Figure S4A) or an Nkrf expression construct (Supplementary Figure S4B) was introduced, the activities of the native Cox2 promoter, but not the mutant with an altered NF-κB-binding site, were inhibited. This result was in agreement with the Cox2 mRNA levels in PANC-1 cells (Supplementary Figure S4C). In addition, we tested whether a modest (two- to three-fold) down-regulation of the Cox2 mRNA led to an ∼10-fold protein level change using an siRNA against Cox2. As shown in Supplementary Figure S4D and E, when Cox2 mRNA was down-regulated ∼2.5-fold, its protein level was robustly reduced in PANC-1 cells. These results indicate that anti-miR-301a-mediated down-regulation of Cox2, a known NF-κB-transactivational target, in pancreatic cancer cells is dependent on Nkrf.
Figure 4.
A positive feedback loop for NF-κB activation in pancreatic cancer cells. (A) miR-301a expression levels in normal human pancreases (NP, a pool of ten), five pancreatic cancer cell lines, twenty-four pancreatic tumour tissues and their paired adjacent non-cancerous tissues (NATs). The relative quantities of miR-301a normalized to that of NP are presented by a Whisker-box plot with the boxes indicating 25th and 75th percentile, solid lines in the boxes indicating 50th percentile, dotted lines denoting the mean values, Whisker caps indicating 5th and 95th percentile, and filled circles indicating outliers. (B) Western blotting analysis in pancreatic cancer cell lines treated with anti-miR-301a or its negative control. ‘Nkrf RQ' or ‘Cox-2 RQ', relative quantity of Nkrf or Cox-2 proteins normalized to β-actin. (C) Inhibiting miR-301a expression up-regulated Nkrf expression and reduced the expression of NF-κB target genes in PANC-1 cells. (D) Overexpression of Nkrf reduced NF-κB activation and miR-301a expression in PANC-1 cells. (E) Inhibition of NFKB1 expression by an siRNA (siNFKB1) reduced miR-301a expression and up-regulated Nkrf in PANC-1 cells. (F–H) are similar to (C–E), but in Mia-PaCa-2 cells. ‘Relative Quantity' in (C–H) is the relative expression level of genes determined by qPCR when referenced by that of β-actin or U6 snRNA; *P⩽0.05; **P⩽0.01; with n=3–6 compared with respective controls. Error bars represent s.d.
We used two cell lines (PANC-1 and Mia-PaCa-2) to further test the miRNA-mediated NF-κB activation in pancreatic cancer cells. Anti-miR-301a was introduced into PANC-1 and Mia-PaCa-2 cells to inhibit miR-301a expression. As expected, Nkrf expression was up-regulated and five NF-κB target genes (MMP2, COX2, MYC, VEGFC, and FAM33A) were all down-regulated (Figure 4C and F). Next, we introduced an Nkrf overexpression construct into these two cell lines and found that Nkrf mRNA levels were significantly increased (Figure 4D and G). Five NF-κB target genes and miR-301a, however, were down-regulated (Figure 4D and G). We also determined whether down-regulation of NFKB1 gene expression impacted the expression of Nkrf and miR-301a. As shown in Figure 4E and H, when NFKB1 expression was inhibited by an siRNA, the expression of four NF-κB target genes was inhibited. Furthermore, miR-301a expression was decreased, accompanied by increased expression of Nkrf (Figure 4E and H). We next tested whether blocking other genes in the NF-κB pathway also reduced miR-301a expression. Down-regulation of RELA or IKBKB in PANC-1 cells led to lower expression of miR-301a and other NF-κB-transactivational targets (Supplementary Figure S5A and B). Finally, we tested whether reintroduction of miR-301a inhibited down-regulation of NF-κB target genes in pancreatic cancer cells caused by ectopic expression of Nkrf. We found that the down-regulation of MMP2, COX2, MYC, and FAM33A/miR-301a and the up-regulation of Nkrf in PANC-1 and Mia-PaCa-2 cells were abolished when both miR-301a and Nkrf overexpression constructs were introduced (Supplementary Figure S5C). These results support the miR-301a-mediated NF-κB activation in pancreatic cancer cells.
Beyond pancreatic cancer cells, we also tested miR-301a-mediated NF-κB activation in 293T and HeLa cells by modulating miR-301a or Nkrf or using TNF-α stimulation. When miR-301a was overexpressed, Nkrf was down-regulated by siRNA (siNkrf), or NF-κB was activated by TNF-α, the expression of NF-κB target genes such as NOS2A, IL8, IFNB, and FAM33A was up-regulated (Supplementary Figure S6A, C, and E–G). In contrast, these genes were down-regulated when miR-301a was inhibited (Supplementary Figure S6B). We also found that NF-κB activation as measured by EMSA was reduced by miR-301a inhibition both in the presence and absence of TNF-α (Supplementary Figure S6D). Moreover, miR-301a expression was induced either by siNkrf (Supplementary Figure S6C and G) or by TNF-α treatment (Supplementary Figure S6F). These results support a feedback loop for NF-κB activation: miR-301a negatively regulates NKRF and lower levels of Nkrf elevate NF-κB activity, which in turn, promotes miR-301a transcription. It is noteworthy that miR-301a overexpression led to ∼50% down-regulation of Nkrf in HeLa cells, which is similar to the level achieved by TNF-α or siNkrf treatment, yet the expression of FAM33A and MMP2 (another NF-κB target gene) was increased significantly higher by miR-301a than that of TNF-α or siNkrf in HeLa cells (Supplementary Figure S6E–G). This result indicated that miR-301a may target other genes beyond Nkrf to regulate NF-κB activation. Given that recent proteomic studies have revealed that one miRNA is expected to repress the expression of a large number of genes (Baek et al, 2008; Selbach et al, 2008), Nkrf is likely one of the targets rather than the target of miR-301a in NF-κB signalling.
Nkrf is expressed at a lower level in pancreatic tumours than normal pancreas tissues
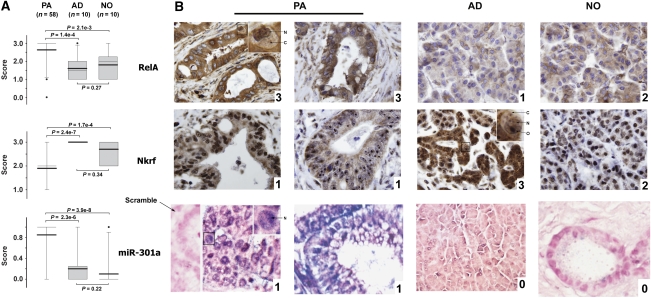
As miR-301a is up-regulated (Lee et al, 2007) and NF-κB is activated in pancreatic cancer (Wang et al, 1999; Weichert et al, 2007), Nkrf protein levels are expected to be down-regulated in pancreatic tumours if miR-301a overexpression causes constitutive NF-κB activation by repressing Nkrf. We examined the expression levels of Nkrf in human pancreatic adenocarcinomas using a tumour microarray containing 58 adenocarcinoma tissues and 20 non-cancerous tissues (i.e. 10 non-cancerous adjacent tissues and normal pancreas tissues). Immunohistochemistry (IHC) of RelA and Nkrf and in situ hybridization of miR-301a were performed to determine the expression levels of these proteins and the miRNA, respectively. As shown in Figure 5B, miR-301a was shown in perinuclear regions of tumour cells from cancer specimens, but not in non-cancerous tissues, including tissues adjacent to pancreatic adenocarcinoma and normal pancreas tissues. There is sporadic nuclear RelA staining in pancreatic tumour sections, but not in sections from normal adjacent tissue or normal pancreas, consistent with previous reports (Wang et al, 1999; Weichert et al, 2007). In contrast, Nkrf was expressed at a significantly lower level in tumour tissues than in non-cancerous tissues (P<0.001). Similar to a previous report (Niedick et al, 2004), Nkrf was predominantly localized in the nucleolus, as well as in the cytoplasm and nucleoplasm (Figure 5B). There was no statistical difference between the expression levels of these proteins in adjacent non-cancerous tissues and in normal pancreas tissues (Figure 5A). This result indicates that decreased Nkrf protein expression coincides with miR-301a overexpression and NF-κB activation in pancreatic adenocarcinomas.
Figure 5.
Decreased Nkrf expression in pancreatic adenocarcinomas (PA). (A) Expression levels of Nkrf, RelA, and miR-301a in PA, adjacent non-cancerous tissues (AD), and normal pancreas tissues (NO). Whisker-box plot with the boxes indicate 25th and 75th percentile; thin lines in the boxes indicate 50th percentile and thick lines denote the mean values (50th percentile and mean value for Nkrf overlap); Whisker caps indicate 5th and 95th percentile; filled circles indicate outliers. P-values of two-tailed Student's t-test are provided. (B) Representative photos taken at × 20 magnification. Arrows point to cytoplasm (C), nucleoplasm (N), and nucleolus (O). A scrambled probe for miR-301a was used as a negative control that did not stain any tissue sections (one image shown in the leftmost). PA, pancreatic adenocarcinomas; AD, adjacent non-cancerous tissues; and NO, normal pancreas tissues. Numbers on the right-bottom corner of each panel denote histological scores.
miR-301a inhibition reduces xenograft tumour growth
Finally, we tested whether down-regulation of miR-301a reduced pancreatic cancer growth using a mouse xenograft model. We first used oligonucleotide-based antisense to miR-301a (anti-miR-301a) to transfect PANC-1 cells and then injected these transfected tumour cells subcutaneously into nude mice. Eight weeks post-injection, mice were euthanized and tumours were dissected. We did not observe significant tumour reduction in mice injected with PANC-1 cells transiently transfected with anti-miR-301a compared with the negative control injected cells. Ensuing examination found limited presence of anti-miR-301a (determined by qRT–PCR) in xenograft tumours after 8 weeks, indicating that transient introduction of miR-301a inhibitor was insufficient to inhibit in vivo xenograft growth. We next tested a newly developed tough decoy RNA (TuD) for stable suppression of miR-301a function in pancreatic cancer cells using a lentiviral vector. These RNAs contain two copies of antisense RNA against miR-301a embedded in two stems with 18 and 8 bp in length, respectively (Figure 6A) (Haraguchi et al, 2009). The vector contained a GFP marker so that cell sorting by flow cytometry can be used to isolate infected cells. TuD:anti-miR-301a was stably incorporated into PANC-1 cells via lentiviral infection and GFP-positive tumour cells were sorted and expanded in culture. The introduction of TuD:anti-miR-301a into PANC-1 cells increased miR-301a target Nkrf expression approximately two-fold (Supplementary Figure S7B). We then injected tumour cells subcutaneously into nude mice and examined tumour growth for 8 weeks. Tumour volume from mice with TuD:anti-miR-301a was significantly less than the TuD control (Figure 6B and C). Furthermore, anti-miR-301a-containing tumours had less dilated pancreatic ducts and less intraductal mucin (Figure 6D), indicating that the malignancy of these tumours was reduced (Kern et al, 2002). We next probed the expression levels of RelA and Nkrf and found that the RelA staining was significantly reduced and Nkrf expression was up-regulated in tumours with anti-miR-301a treatment (Figure 6E; Supplementary Figure S7B). Interestingly, the RelA mRNA levels were not significantly altered, suggesting that Nkrf may be involved in the posttranslational regulation of RelA as it binds RelA (Nourbakhsh and Hauser, 1999). Nonetheless, these results indicate that stable inhibition of miR-301a reduced NF-κB activation and pancreatic cancer xenograft growth in mice.
Figure 6.
miR-301a inhibition or Nkrf up-regulation in PANC-1 cells reduces tumour formation in mice. (A) Schematic representation of the tough decoy (TuD) RNA that inhibits miR-301a function. (B) Tumour volumes of mouse xenografts with TuD:anti-miR-301a were smaller than that of the control (empty vector pSIF-H1-copGFP). P⩽0.05 from Weeks 4 to 9 and P=0.05 at Week 10. (C) A representative photo of tumours from mice injected with PANC-1 cells with TuD:anti-miR-301a (right) or its control (left). (D) Haematoxylin and eosin staining of tissue sections from mouse xenografts. (E) Expression levels of RelA and Nkrf by IHC (right and middle) or qPCR (right) in tumour tissues from PANC-1 cells with TuD:anti-miR-301a or its control. (F) PANC-1 cells with Nkrf constructs carrying a wild-type coding sequence (CDS) and a wild-type (UTRwt) or a mutant 3′UTR (UTRmut) (Figure 2G). (G) Tumour volumes of mouse xenografts with cells expressing Nkrf from (F). P⩽0.05 from Week 7 and P=0.03 at Week 8 between UTRwt and UTRmut. ‘Control' denotes PANC-1 cells. (H) mRNA levels of Nkrf and VEGF-C by qPCR from tumour sections from (G). (I) PANC-1 cells stably expressing TuD:anti-miR-301a were infected by viruses carrying siNkrf or a control (siControl). (J) Tumour volumes of mouse xenografts with resultant cells from (I). PANC-1 cells expressing TuD:anti-miR-301a from (I) were infected with viruses carrying siControl or siNkrf. ‘Control' denotes PANC-1 cells with TuD:anti-miR-301a only as in (A). P⩽0.05 from Weeks 6 to 7 and P=0.04 at Week 8 between siControl and siNkrf. (K) mRNA levels of Nkrf and VEGF-C by qPCR from tumour sections from (J). For (B, G, J), five animals per group were used. There is no statistical difference in tumour volumes between ‘Control' in (B) and ‘Control' in (G) or between ‘Control' and ‘siControl' in (J). For (E, H, K), one section from each tumour (five total per group) was extracted for total RNAs for qPCR or was used for IHC. Error bars represent s.d.
We investigated the mechanism of PANC-1 xenograft tumour growth inhibition by TuD:anti-miR-301a. In petri dishes, there was no difference in cell proliferation, apoptosis, and/or cell cycle between PANC-1 cells with anti-miR-301a and the TuD control (Supplementary Figure S7A). An in vitro transwell assay indicated that the cell migration of PANC-1 or Mia-PaCa-2 cells with anti-miR-301a was reduced (Supplementary Figure S7C). However, this effect likely did not contribute to tumour growth inhibition in vivo as there was no metastasis in our subcutaneous xenograft inoculated with PANC-1 with anti-miR-301a, the TuD control, or with naive PANC-1 cells. Yet in xenografts, cell proliferation was certainly reduced in tumours from PANC-1 cells with anti-miR-301a as judged by tumour volume (Figure 6) and proliferating cell nuclear antigen staining (PCNA, a hallmark of DNA synthesis and cell proliferation) (Supplementary Figure S8). We next examined the expression of another NF-κB-transactivation target, vascular endothelial growth factor C (VEGF-C) (Chilov et al, 1997). VEGF-C, an angiogenesis factor, was up-regulated in pancreatic cancer tissues compared with normal pancreas and is abundant in PANC-1 cells (Tang et al, 2001). We found that VEGF-C was expressed at a significantly lower level in PANC-1 cells with anti-miR-301a than the control in either petri dishes (Figure 4C) or xenografts (Supplementary Figure S8). As angiogenesis is required by xenograft tumour growth, but not for PANC-1 cells in petri dishes, it is likely that reduced NF-κB activation, subsequent down-regulation of VEGF-C, and decreased blood vessel formation as judged by reduced CD31 levels (Supplementary Figure S8) attenuated tumour growth in vivo.
Next, we determined whether anti-miR-301a-mediated tumour inhibition was dependent on Nkrf. We first made two constructs (Figure 6F) in which the Nkrf coding sequence was placed upstream either of a native 3′UTR (NKRF-UTRwt) or a mutant construct that miR-301a cannot target (NKRF-UTRmut; Figure 2C). We packaged these two constructs into lentivirus and infected PANC-1 cells, which endogenously express high levels of miR-301a. Similar to anti-miR-301a, cell proliferation in vitro (petri dishes) was not changed in cells with NKRF-UTRwt compared with NKRF-UTRmut cells. When cells were inoculated into nude mice, however, tumours xenografted with cells carrying NKRF-UTRmut were significantly smaller than those with NKRF-UTRwt. Nkrf expression was significantly greater in xenograft tumour sections with NKRF-UTRmut, whereas VEGF-C expression was reduced (Figure 6G and H; Supplementary Figure S8). Given that the only difference between NKRF-UTRwt and NKRF-UTRmut is the miR-301a-binding site and that miR-301a is highly expressed in PANC-1 cells, these results suggest that the interaction between miR-301a and Nkrf 3′UTR is causal in Nkrf repression and xenograft tumour growth. Furthermore, we introduced siNkrf into PANC-1 cells stably expressing TuD:anti-miR-301a (Figure 6I) and inoculated subcutaneously the resulting cells into nude mice. We found that siNkrf reversed tumour growth reduction mediated by TuD:anti-miR-301a (Figure 6J). It coincided with down-regulation of Nkrf and up-regulation of VEGF-C (Figure 6K; Supplementary Figure S8). However, there was a lack of statistical significance in CD31 staining in xenografts with PANC-1 cells carrying anti-miR-301a treated with either siNkrf or a control (Supplementary Figure S8D), suggesting that modulation of miR-301a:Nkrf expression may attenuate tumour growth through mechanisms other than angiogenesis, or in addition to angiogenesis. This is also an indication that miR-301a has other functions than repressing Nkrf as observed in Supplementary Figure S6.
Discussion
To explore the possibility that miRNAs are involved in NF-κB activation in pancreatic cancer, we formulated a strategy to screen for miRNAs that modulate NF-κB signalling and found that miR-301a is the most potent NF-κB activator. We then identified the NKRF gene as an authentic target of miR-301a. Nkrf was first found to interact with specific negative regulatory elements (NREs) to mediate NF-κB's transcriptional activity, which regulates the expression of some NF-κB-responsive genes, including IL8, IFNB, and NOS2A (Nourbakhsh and Hauser, 1999; Nourbakhsh et al, 2001; Feng et al, 2002). However, several lines of evidence suggest that Nkrf-mediated inhibition of NF-κB transactivation is not dependent on Nkrf's DNA-binding ability, that is NREs are not required for Nkrf function. First, it was reported that a mutant Nkrf lacking the DNA-binding domain also inhibits NF-κB-dependent gene expression (Nourbakhsh and Hauser, 1999). Second, our data show that the expression of NF-κB target genes without NREs (MMP and MYC) and three with NREs (NOS2A, IFNB, and IL8) were all up-regulated when NKRF was negatively regulated by miR-301a; and they were down-regulated with higher expression levels of Nkrf. Third, the luc screening reporter construct we used in this study contains no NRE sequences, yet down-regulation of Nkrf caused luc up-regulation. Fourth, the EMSA demonstrated that miR-301a promotes NF-κB activation (Figure 1D), in which the DNA probe contains no NREs. Finally, there are no NRE sequences in the promoters for Cox2 and miR-301a, which are modulated by both Nkrf and NF-κB, and their flanking 10 kb genomic sequences, suggesting that Nkrf and NF-κB regulation of Cox2 and miR-301a is independent of NREs. Collectively, these data support that Nkrf, like IκBα, broadly inhibits NF-κB-transactivational activities. Nkrf is abundant in many human cell lines and adult tissues (Nourbakhsh and Hauser, 1999) with localization in cytoplasm, nucleoplasm, and dominantly nucleoli (Niedick et al, 2004), supporting its availability to participate in the regulation of its target genes. The DNA-independent function is likely from cytoplasmic Nkrf proteins that inhibit the expression of MMP2, COX2, MYC, and FAM33A∼miR-301a by binding NF-κB (Nourbakhsh and Hauser, 1999), that is in the cytoplasm, Nkrf acts to hold NF-κB in an inactive state. This is in contrast to the DNA-dependent regulation of NOS2A, IFNB, and IL in nucleoplasm, where Nkrf interacts with NREs to mediate transcriptional repression by inhibition of the NF-κB activity at their promoters. The biological activity of nucleolar Nkrf, however, remains unknown with a speculation that the Nkrf-regulated genes are kept inactive in nucleoli, but are released to the nucleoplasm upon transcriptional activation (Niedick et al, 2004).
We found that the promoter for miR-301a contains an authentic κB site and miR-301a expression is regulated by NF-κB. This suggests a positive feedback loop: miR-301a down-regulates NKRF to elevate NF-κB activation, which in turn promotes miR-301a transcription. We validated this feedback loop by modulating the expression of Nkrf and miR-301a in 293T, HeLa, PANC-1, and Mia-PaCa-2 cells, respectively. This feedback loop helps to explain why miR-301a is the most potent NF-κB activator among all miRNAs tested. The ∼40-fold miR-301a overexpression in 293T cells resulting from a transient transfection of the miR-301a construct most likely comes from two sources: (1) from introduced ectopic plasmids that are not regulated by NF-κB (no κB sites in the vector) and (2) from the endogenous miR-301 gene (and FAM33A) that is regulated by NF-κB, that is the overexpression of miR-301a is amplified by NF-κB activation, which drives endogenous miR-301a expression.
While miRNA expression profiling in various cancers shares common features, some cancers have specific dysregulated miRNAs. For instance, miR-21 is overexpressed in nine types of solid tumours (lung, breast, stomach, prostate, colon, brain, head and neck, oesophagus, and pancreas), as well as in blood cancers, for example diffuse large B-cell lymphoma, leukaemia and so on (Liu et al, 2010). In contrast, miR-301a was first reported to be only specifically overexpressed in pancreatic adenocarcinoma tumours and tumour cell lines compared with normal pancreas and pancreatitis, but not in other types of cancers (Lee et al, 2007). Later, it was found to be overexpressed in hepatocellular carcinoma, yet at a much lower level and less significantly (Jiang et al, 2008) (2.19-fold with a P-value=0.0213), compared with that in pancreatic adenocarcinoma (Lee et al, 2007) (34.2-fold with a P-value=1.11E−05). Our results support the notion that miR-301a overexpression contributes to NF-κB activation in pancreatic cancer. Such a mechanism could render NF-κB activation in pancreatic cancer different from that in other cancers such as lymphoma and myeloma. As a multifaceted regulator, NF-κB controls a network of important genes including VEGF-C, which is critical to angiogenesis and tumour growth. Unveiling the NF-κB activation regulatory mechanism by miRNAs in pancreatic cancer cells supports the use of miRNA inhibitors to target NF-κB signalling for pancreatic cancer therapeutic intervention. Feasibility for this approach is shown when inhibiting miR-301a in pancreatic cancer cells reduces tumour formation in a mouse subcutaneous xenograft model, in which attenuated tumour growth is at least partially due to reduced expression of NF-κB-regulated VEGF-C. VEGF inhibition using monoclonal antibodies such as Bevacizumab (a.k.a. A4.6.1, RhuMAb, or Avastin® targeting all VEGF isoforms, including VEGF-C) has been reported to reduce xenograft pancreatic tumour growth and angiogenesis (Hotz et al, 2003).
It is intriguing that what causes miR-301a overexpression specifically in pancreatic cancer. There are a few possibilities. First, KRAS mutations are frequent in pancreatic adenocarcinomas and are detected in ∼30% of early neoplasms with the frequency rising to nearly 100% at advanced stages (Hezel et al, 2006), higher than any other types of cancer. One of the downstream effectors of Ras activation is NF-κB (Hezel et al, 2006). Activated NF-κB, in turn, could promote the transcription of miR-301a. To test this possibility, we introduce HRas V12 (activated Ras) (Serrano et al, 1997) into 293T cells and determine the expression of levels of miR-301a. miR-301a levels were not significantly changed with Ras activation, indicating that Ras activation alone cannot trigger miR-301a overexpression. We then down-regulated KRas using siRNAs in PANC-1 and Mia-PaCa-2 cells and found that the expression of miR-301a was not inhibited by KRas down-regulation. Collectively, these results suggest that Ras activation is unlikely responsible for miR-301a overexpression. Second, miR-301a is located in the human Chromosome 17q22–17q23.1, a region that has been reported to be amplified in 46% of 13 primary tissues and 24 cell lines of pancreatic adenocarcinomas (Aguirre et al, 2004). Other possibilities include epigenetic regulation of miR-301a expression and/or other trans or cis regulatory factors. All of these warrant extensive investigations beyond the scope of this work.
To summarize, in this study, we report that a large number of miRNAs may be involved in NF-κB signalling with the confirmation of miR-301a as an NF-κB activator through its ability to down-regulate an NF-κB inhibitor, Nkrf. The expression of miR-301a is regulated by NF-κB, rendering a novel miRNA-mediated positive feedback of NF-κB activation in which miR-301a down-regulates NKRF, and thus, elevates NF-κB activity, which in turn promotes miR-301a transcription. This mechanism is likely to be a driving force for constitutive NF-κB activation in pancreatic cancer cells as both lower Nkrf expression and miR-301a overexpression are observed in pancreatic tumour tissues compared with non-cancerous tissues, whereas inactivation of miR-301a results in higher Nkrf expression and decreased NF-κB activation in pancreatic cancer cells. Given that NF-κB activation is reported to have a causative function in cell transformation (Iliopoulos et al, 2009) and that stable expression of antisense RNAs against miR-301a in pancreatic cancer cells reduces tumour growth, modulation of miR-301a thus provides a potentially viable therapeutic option against this devastating cancer.
Materials and methods
Recombinant DNAs
The 3′UTRs of NKRF were PCR amplified from genomic DNA and cloned into pRL-TK (Promega) downstream of the Renilla luciferase gene. The mutations in pRL-TK-NKRF-3′UTRmut were cloned by PCR to contain the following sequences from the original 3′UTR from nts 440 to 481: AGA ATG TTT GAC AAT AAC GGC ATG AGT ACA CGT AAA TGT CCT. Nucleotides corresponding to the miRNA seed sequence are shown in bold. The promoter of miR-301a was amplified from genomic DNA using AGT CTT TTG CAG GAG TTG GC (ProFor) and GCC TCC ATG TTG AAT AGT TGA C (ProRev) and cloned into pGL3-Basic (Promega) upstream of the firefly luciferase gene. The mutations in pGL3-301aPro-Mut were cloned by PCR to contain the following sequences from the original miR-301a promoter from nts 160 to 196: GCC AAG AGG GTG AAA TTT GTC TCT ACT AAA AAT AC. The NF-κB-binding site is shown in bold and mutation nucleotides in italics. Promoter deletion was carried out using PCR with ProRev and ATA AGC TTC AAA AAT TAG CCG GGC GTA GC (ProDel). All constructs were confirmed by DNA sequencing. All siRNAs were short-hairpin mimicking a precursor to an miR-30 made by the RNAi Consortium (siNkrf, NM_029891.2-1030s1c1; siControl, NM_003998.2-2853s1c1; Sigma, St Louis, MO); details of other siRNAs are provided in Supplementary data. The miRNA library was constructed as described in Supplementary data and is available at GeneCopoeia, Inc. (Germantown, MD).
Animal experiments
For miR-301a inhibition, PANC-1 cells were infected with viruses carrying TuD:anti-miR-301a (sequence in Supplementary Figure S7) before subjected to GFP cell sorting. GFP-positive cells were used for xenografts. For Nkrf overexpression, Nkrf-UTRmut was first inserted into pCMV-SPORT6-NKRF via Bgl II and Not I sites. The native CDS-UTRwt or the mutant CDS-UTRmut was inserted into pGC188 (Addgene plasmid 19619; Addgene.org, Cambridge, MA) through Gateway® cloning (Invitrogen). The resulting plasmids were then inserted into pLenti CMV Hygro DEST (w117-1) (Addgene plasmid 17454) through Gateway cloning. Lentivirus with either NKRF-UTRwt or NKRF-UTRmut was then prepared to infect PANC-1 cells and hygromycin-resistant cells were used for xenografts. For siNkrf, lentivirus with siNkrf or siControl was used to infect PANC-1 cells stably expressing TuD:anti-miR-301a. Puromycin-resistant cells were then used for xenografts. Athymic female nude (Balb/c) mice (8 weeks old) were purchased from The Jackson Laboratory (Bar Harbor, ME) and were maintained in the University of Louisville's AAALAC-accredited animal facility. All animal studies were conducted in accordance with National Institutes of Health animal use guidelines and a protocol approved by the University of Louisville's Institutional Animal Care and Use Committee. Exponentially growing cells were harvested and injected subcutaneously (5.0 million cells/animal) into nude mice (five per group). Tumour size was monitored once per week for 8 or 10 weeks before killing and Student's t-test was used to analyse the tumour volumes. Tumours were harvested, weighed, and frozen immediately (−80°C). All other experimental details are provided in Supplementary data.
Supplementary Material
Acknowledgments
This project is supported by Clinical & Translational Science Pilot Grant Program Innovative Award and the Diabetes and Obesity Center at University of Louisville funded by NCRR/NIH (P20 RR024489). YL is also supported by the Center for Environmental Genomics and Integrated Biology at University of Louisville funded by NIEHS/NIH (P30 ES014443) and a Scientist Development Grant from American Heart Association. We are indebted to the critical reviews of Drs Nancy Martin and Robert Mitchell and Ms Juanita R Barker and to Cox2 promoter constructs from Drs Shozo Yamamoto and Tomoko Takano. YL is extremely grateful to Drs Aruni Bhatnagar and Kenneth S Ramos for their mentoring and support.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aguirre AJ, Brennan C, Bailey G, Sinha R, Feng B, Leo C, Zhang Y, Zhang J, Gans JD, Bardeesy N, Cauwels C, Cordon-Cardo C, Redston MS, DePinho RA, Chin L (2004) High-resolution characterization of the pancreatic adenocarcinoma genome. Proc Natl Acad Sci USA 101: 9067–9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, Dave S, Hurt EM, Tan B, Zhao H, Stephens O, Santra M, Williams DR, Dang L, Barlogie B, Shaughnessy JD Jr et al. (2007) Frequent engagement of the classical and alternative NF-κB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12: 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoki T, Nagakawa Y, Tsuchida A, Kasuya K, Kitamura K, Inoue K, Ozawa T, Koyanagi Y, Itoi T (2002) Expression of cyclooxygenase-2 and vascular endothelial growth factor in pancreatic tumors. Oncol Rep 9: 761–765 [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455: 64–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Alvero AB, Silasi DA, Kelly MG, Fest S, Visintin I, Leiser A, Schwartz PE, Rutherford T, Mor G (2008) Regulation of IKKβ by miR-199a affects NF-κB activity in ovarian cancer cells. Oncogene 27: 4712–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilov D, Kukk E, Taira S, Jeltsch M, Kaukonen J, Palotie A, Joukov V, Alitalo K (1997) Genomic organization of human and mouse genes for vascular endothelial growth factor C. J Biol Chem 272: 25176–25183 [DOI] [PubMed] [Google Scholar]
- Feng X, Guo Z, Nourbakhsh M, Hauser H, Ganster R, Shao L, Geller DA (2002) Identification of a negative response element in the human inducible nitric-oxide synthase (hiNOS) promoter: the role of NF-κB-repressing factor (NRF) in basal repression of the hiNOS gene. Proc Natl Acad Sci USA 99: 14212–14217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D140–D144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S, Purcell NH, Lin A, Sen S (2002) Activation of nuclear factor-κB is necessary for myotrophin-induced cardiac hypertrophy. J Cell Biol 159: 1019–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanisch A, Sillje HH, Nigg EA (2006) Timely anaphase onset requires a novel spindle and kinetochore complex comprising Ska1 and Ska2. EMBO J 25: 5504–5515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi T, Ozaki Y, Iba H (2009) Vectors expressing efficient RNA decoys achieve the long-term suppression of specific microRNA activity in mammalian cells. Nucleic Acids Res 37: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hezel AF, Kimmelman AC, Stanger BZ, Bardeesy N, DePinho RA (2006) Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev 20: 1218–1249 [DOI] [PubMed] [Google Scholar]
- Hotz HG, Hines OJ, Hotz B, Foitzik T, Buhr HJ, Reber HA (2003) Evaluation of vascular endothelial growth factor blockade and matrix metalloproteinase inhibition as a combination therapy for experimental human pancreatic cancer. J Gastrointest Surg 7: 220–228 [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K (2009) An epigenetic switch involving NF-κB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell 139: 693–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics, 2009. CA Cancer J Clin 59: 225–249 [DOI] [PubMed] [Google Scholar]
- Jiang J, Gusev Y, Aderca I, Mettler TA, Nagorney DM, Brackett DJ, Roberts LR, Schmittgen TD (2008) Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res 14: 419–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human MicroRNA targets. PLoS Biol 2: e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Zhang X, Parsons DW, Lin JC-H, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong S-M, Fu B, Lin M-T, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR et al. (2008) Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 321: 1801–1806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost PJ, Ruland J (2007) Aberrant NF-κB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood 109: 2700–2707 [DOI] [PubMed] [Google Scholar]
- Karin M, Cao Y, Greten FR, Li ZW (2002) NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2: 301–310 [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng W-J, Van Wier S, Tiedemann R, Shi C-X, Sebag M, Braggio E, Henry T, Zhu Y-X, Fogle H, Price-Troska T, Ahmann G, Mancini C, Brents LA, Kumar S, Greipp P et al. (2007) Promiscuous mutations activate the noncanonical NF-κB pathway in multiple myeloma. Cancer Cell 12: 131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern SE, Hruban RH, Hidalgo M, Yeo CJ (2002) An introduction to pancreatic adenocarcinoma genetics, pathology and therapy. Cancer Biol Ther 1: 607–613 [DOI] [PubMed] [Google Scholar]
- Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N (2005) Combinatorial microRNA target predictions. Nat Genet 37: 495. [DOI] [PubMed] [Google Scholar]
- Krikos A, Laherty CD, Dixit VM (1992) Transcriptional activation of the tumor necrosis factor α-inducible zinc finger protein, A20, is mediated by κB elements. J Biol Chem 267: 17971–17976 [PubMed] [Google Scholar]
- Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, Morgan DL, Postier RG, Brackett DJ, Schmittgen TD (2007) Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120: 1046–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, Vose J, Bast M, Fu K, Weisenburger DD, Greiner TC, Armitage JO, Kyle A, May L, Gascoyne RD, Connors JM et al. (2008) Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 359: 2313–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20 [DOI] [PubMed] [Google Scholar]
- Liu M-F, Jiang S, Lu Z, Young KH, Li Y (2010) Physiological and pathological functions of mammalian MicroRNAs. In Comprehensive Toxicology, Vol. Cellular and Molecular Toxicology, McQueen CA (ed.), 2nd edn, 23 London, UK: Elsevier Science [Google Scholar]
- Marinescu V, Kohane I, Riva A (2005) MAPPER: a search engine for the computational identification of putative transcription factor binding sites in multiple genomes. BMC Bioinformatics 6: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A (2006) ABIN-1 binds to NEMO/IKKγ and co-operates with A20 in inhibiting NF-κB. J Biol Chem 281: 18482–18488 [DOI] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I (2006) A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell 126: 1203–1217 [DOI] [PubMed] [Google Scholar]
- Niedick I, Froese N, Oumard A, Mueller PP, Nourbakhsh M, Hauser H, Koster M (2004) Nucleolar localization and mobility analysis of the NF-κB repressing factor NRF. J Cell Sci 117: 3447–3458 [DOI] [PubMed] [Google Scholar]
- Nourbakhsh M, Hauser H (1999) Constitutive silencing of IFN-β promoter is mediated by NRF (NF-κB-repressing factor), a nuclear inhibitor of NF-κB. EMBO J 18: 6415–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourbakhsh M, Kalble S, Dorrie A, Hauser H, Resch K, Kracht M (2001) The NF-κB repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-κB-flanking sequence element. J Biol Chem 276: 4501–4508 [DOI] [PubMed] [Google Scholar]
- Pahl HL (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18: 6853–6866 [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455: 58–63 [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW (1997) Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88: 593–602 [DOI] [PubMed] [Google Scholar]
- Shembade N, Harhaj NS, Parvatiyar K, Copeland NG, Jenkins NA, Matesic LE, Harhaj EW (2008) The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol 9: 254–262 [DOI] [PubMed] [Google Scholar]
- Taganov KD, Boldin MP, Chang K-J, Baltimore D (2006) NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA 103: 12481–12486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang RF, Itakura J, Aikawa T, Matsuda K, Fujii H, Korc M, Matsumoto Y (2001) Overexpression of lymphangiogenic growth factor VEGF-C in human pancreatic cancer. Pancreas 22: 285–292 [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Cimino A, Costinean S, Dumitru CD, Adair B, Fabbri M, Alder H, Liu CG, Calin GA, Croce CM (2007) Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-α stimulation and their possible roles in regulating the response to endotoxin shock. J Immunol 179: 5082–5089 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424: 793–796 [DOI] [PubMed] [Google Scholar]
- Wang XW, Zhan Q, Coursen JD, Khan MA, Kontny HU, Yu L, Hollander MC, O'Connor PM, Fornace AJ Jr, Harris CC (1999) GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci USA 96: 3706–3711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichert W, Boehm M, Gekeler V, Bahra M, Langrehr J, Neuhaus P, Denkert C, Imre G, Weller C, Hofmann HP, Niesporek S, Jacob J, Dietel M, Scheidereit C, Kristiansen G (2007) High expression of RelA/p65 is associated with activation of nuclear factor-κB-dependent signaling in pancreatic cancer and marks a patient population with poor prognosis. Br J Cancer 97: 523–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Arakawa T, Ueda N, Yamamoto S (1995) Transcriptional roles of nuclear factor κB and nuclear factor-interleukin-6 in the tumor necrosis factor-dependent induction of cyclooxygenase-2 in MC3T3-E1 cells. J Biol Chem 270: 31315–31320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.