Metalloprotease type III effectors that specifically cleave JNK and NF-κB
Enteropathogenic E. coli secretes multiple effector proteins that enable host-cell colonization. Two effectors, NleC and NleD, are here shown to act as endopeptidases, cleaving p65 and JNK to inhibit the inflammatory response.
Keywords: enteropathogenic E. coli , JNK, NF-κB, NleC, NleD
Abstract
Two major arms of the inflammatory response are the NF-κB and c-Jun N-terminal kinase (JNK) pathways. Here, we show that enteropathogenic Escherichia coli (EPEC) employs the type III secretion system to target these two signalling arms by injecting host cells with two effector proteins, NleC and NleD. We provide evidence that NleC and NleD are Zn-dependent endopeptidases that specifically clip and inactivate RelA (p65) and JNK, respectively, thus blocking NF-κB and AP-1 activation. We show that NleC and NleD co-operate and complement other EPEC effectors in accomplishing maximal inhibition of IL-8 secretion. This is a remarkable example of a pathogen using multiple effectors to manipulate systematically the host inflammatory response signalling network.
Introduction
Enteropathogenic Escherichia coli (EPEC) is a typical ‘attaching and effacing' (AE) pathogen. These pathogens make use of a type III protein secretion system (TTSS) to translocate a set of effector proteins into the infected host cell. The injected effectors target different host-cell processes to allow efficient host colonization (summarized in Dean and Kenny, 2009). EPEC carries 21 effector-encoding genes located in the locus of enterocyte effacement, several prophages (PP), and insertion elements (IE) (Iguchi et al, 2009).
Cells have the capacity to detect intruding bacteria by sensing pathogen-associated molecular patterns (PAMPs) such as LPS, CpG DNA, and flagellin. These molecules trigger Toll-like receptors (TLRs) signalling pathways, leading to an inflammatory response via activation of NF-κB and AP-1 transcription factors (Doyle and O'Neill, 2006; Kawai and Akira, 2006). The NF-κB family comprises of five related proteins RelA (p65), RelB, c-Rel, p50 (NF-κB1), and p52 (NF-κB2), which function as homo- or heterodimers. Normally, NF-κBs are retained in the cytoplasm by association with inhibitory proteins termed IκBs. Upon different stimulations, including those mediated by TLRs and TNFα-receptor, IκBs are phosphorylated by the IκB kinase (IKK). This triggers IκB ubiquitination, leading to its proteasome-dependent degradation and consequently allowing translocation of the free NF-κBs to the nucleus. The nuclear NF-κBs regulate the expression of inflammation-associated, and other genes (Karin and Ben-Neriah, 2000; Chen, 2005).
The c-Jun N-terminal kinases (JNKs) are serine/threonine kinases belonging to the MAP kinase family. JNKs are activated by a plethora of extracellular signals and consequently represent essential mediators of signal transduction (Davis, 2000; Shaulian and Karin, 2001; Eferl and Wagner, 2003). The JNK family consists of three genes JNK1, JNK2, and JNK3. JNK1/2 are expressed in most tissues, whereas JNK3 is expressed mainly in the brain. Notably, each of these genes produces several isoforms. JNK activation involves its phosphorylation on threonine-183 and tyrosine-185, located within a region termed the ‘activation loop' (Kallunki et al, 1994). JNK activation can be mediated by several MAP3Ks after stimulation of TLRs, IL-1R, or TNFR (Takeuchi and Akira, 2001). Upon activation, JNKs phosphorylate the proto-oncoprotein c-Jun, a key member of the AP-1 group of transcription factors that regulate cellular proliferation, apoptosis, inflammation, and tumorigenesis (Shaulian and Karin, 2001; Eferl and Wagner, 2003).
EPEC infection triggers an inflammatory response, mainly via a flagellin-dependent pathway (Ruchaud-Sparagano et al, 2007). The pathogen inhibits this response by injecting host cells with several effectors that block the NF-κB pathway. These effectors include NleE that blocks IKKβ activation; NleB that blocks the TNFα-mediated NF-κB activation, upstream to IKKβ activation; and NleH1 that inhibits the translocation of the NF-κB co-factor, RPS3, to the nucleus (Gao et al, 2009; Nadler et al, 2010; Newton et al, 2010). It has been speculated that additional effectors must be involved in blocking the inflammatory response (Gao et al, 2009; Nadler et al, 2010). Indeed, in this report we show that EPEC inject into host cells two additional effectors, NleD and NleC, which are Zn metalloproteases that specifically cleave and inactivate JNK and the p65 subunit of NF-κB, respectively.
Results
EPEC induce JNK cleavage
TNFα induces activation of the MAP3K TAK1 that in turn activates both IKKβ and JNK phosphorylations. We showed previously that NleE and NleB block IKKβ activation and therefore we tested whether these effectors inhibit also JNK activation. To this end we infected HeLa cells with wild-type or various EPEC mutants and tested for JNK activation. We observed that wild-type EPEC and a ΔnleBE mutant, but not a TTSS-deficient mutant (ΔescV), induced cleavage of JNK (Figure 1A; Supplementary Figure S1A and data not shown). We concluded that EPEC inject into the infected cells effector protein(s) other than NleE and NleB that induce JNK cleavage. These findings are similar to previous observations (Ruchaud-Sparagano et al, 2007).
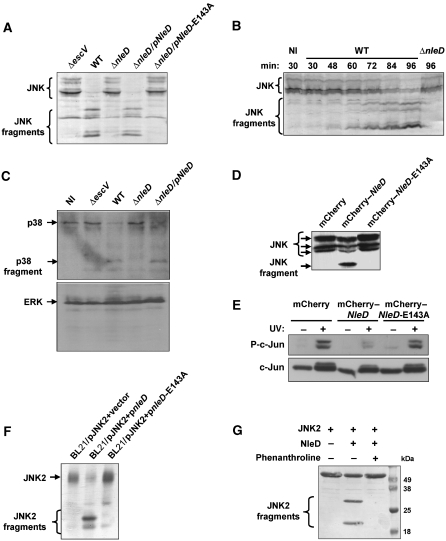
Figure 1.
Involvement of NleD in JNK cleavage. (A) NleD is required for JNK degradation. HeLa cells were infected with one of the following EPEC: wild-type (WT), nleD deletion mutant (ΔnleD), nleD deletion mutant complemented with a plasmid expressing wild-type nleD (pKB4345, indicated as pNleD) or nleD deletion mutant complemented with a plasmid expressing mutated nleD (pLG4457, indicated as pNleD-E143A). After 3 h, proteins were extracted and subjected to western blot analysis using anti-JNK antibody. JNK and its degradation fragments are indicated. Cells infected with a TTSS-deficient mutant (ΔescV) were used as negative control. (B) The kinetics of JNK cleavage upon EPEC infection. HeLa cells were infected with EPEC primed to express the TTSS for the indicated periods before proteins were extracted and subjected to western blot analysis using anti-JNK antibody. JNK and its degradation fragments are indicated. Non-infected cells (NI) and cells infected with nleD deletion mutant (ΔnleD) served as controls. (C) NleD induces cleavage of p38, but not that of ERK. HeLa cells were infected as indicated in (A). After 3 h, proteins were extracted and subjected to western blot analysis using anti-p38 or anti-ERK antibodies. ERK, p38 and its degradation fragments are indicated. (D) Ectopically expressed NleD correlates with JNK degradation. HEK293 cells were transfected with one of the following plasmids expressing mCherry–NleD, mCherry–NleD-E143A, or mCherry (pLG4419, pLG4477, and pSC4141, respectively). After 24 h, proteins were extracted and subjected to western blot analysis using anti-JNK antibody. JNK and its degradation fragments are indicated. (E) Ectopically expressed NleD is associated with inhibition JNK activity. HEK293 cells transfected with plasmids expressing mCherry, mCherry–NleD, or mCherry–NleD-E143A, were irradiated with 30 J/m2 of UV and harvested 3 h later. The levels of phospho-c-Jun and total c-Jun were determined by western analysis using anti-c-Jun and anti-phospho-c-Jun antibodies. (F) NleD induces cleavage of JNK in E. coli cytoplasm. E. coli BL21 was co-transformed with plasmid expressing JNK2 and either vector only (pCX341) or plasmid expressing nleD (pEM3654). Co-expression of JNK2 and NleD was induced for 2 h by IPTG, before proteins were extracted and subjected to western blot analysis using anti-JNK antibody. JNK and its degradation fragments are indicated. (G) NleD clips JNK in vitro. Purified JNK2 and NleD were incubated in a reaction mixture at a molar ratio of 40:1, in the presence or absence of the Zn protease inhibitor phenanthroline. The reaction was stopped by addition of SDS loading buffer and proteins separated by SDS–PAGE. Finally, proteins were visualized by Coomassie blue staining. JNK2 and its degradation fragments are indicated. Of note, NleD does not appear in this gel as its concentration is below detection levels.
NleD is required for JNK clipping
To identify the putative effector that induces JNK cleavage, we screened a collection of mutant EPEC with large chromosomal deletions (Nadler et al, 2010; Supplementary Table S1). We found that deletion of the PP4 prophage (Iguchi et al, 2009) renders EPEC deficient in inducing JNK cleavage (Supplementary Figure S1A). In contrast, EPEC mutants with other chromosomal deletions induced JNK cleavage as effectively as wild type (Supplementary Figure S1A). Further deletion analysis identified the nleD gene as required for JNK clipping (Figure 1A). NleD contains a conserved motif, HEXXH, typical of Zn metalloproteases (Supplementary Figure S2; Marches et al, 2005). To test whether this motif is required for JNK clipping, we complemented the nleD deletion mutant with plasmids expressing either a wild-type NleD or a mutated NleD, where the glutamic acid of the HEXXH motif was replaced by alanine (NleD-E143A). We found that wild-type NleD, but not NleD-E143A, restored the capacity of nleD mutant (ΔnleD) to induce JNK clipping (Figure 1A; Supplementary Figure S6). Kinetics analysis showed that JNK cleavage initiated 30 min after the infection and intact JNK could no longer be detected after 150 min (Figure 1B; Supplementary Figure S1B). Similar results were obtained using Caco2 cells that better mimic enterocytes; the primary target host cells of EPEC (Supplementary Figure S3). Taken together, these results indicate that NleD and its putative Zn metalloprotease activity are required for the EPEC-induced cleavage of JNK.
We next tested whether NleD is involved in cleavage of other members of the MAP kinase family; p38 and ERK. We infected HeLa cells with wild-type or various EPEC mutants and tested for cleavage of p38 and ERK. EPEC induced the cleavage of p38 in NleD-dependent manner, but not that of ERK (Figure 1C). We concluded that under infection conditions p38 and JNK, but not ERK, are targeted by NleD. As cleavage of JNK was more robust we further focussed our attention on NleD–JNK interaction.
Ectopic expression of NleD results in JNK cleavage
To examine whether NleD expression is sufficient to induce JNK clipping, we constructed mammalian expression vectors expressing mCherry fused to NleD (mCherry–NleD) or to NleD-E143A (mCherry–NleD-E143A). These plasmids were used to transfect HeLa cells, exhibiting similar transfection efficiency, expression and distribution in the cells, as determined by microscopy. mCherry–NleD expression, but not that of mCherry–NleD-E143A, correlated with JNK cleavage (Figure 1D). These results indicate that NleD is sufficient for induction of JNK cleavage.
To determine whether the cleavage inactivates JNK, we examined if NleD expression affects the phosphorylation state of JNK targets. We focussed on a key JNK target, c-Jun, which is phosphorylated by JNK at serine 63/73 and threonine 91/93 following exposure to UV radiation (Hibi et al, 1993; Derijard et al, 1994; Yogev et al, 2008). HEK293 cells transfected with plasmids expressing either NleD or NleD-E143A, or a control vector, were UV irradiated and then c-Jun phosphorylation levels evaluated. Expression of NleD correlated with marked reduction in the levels of phosphorylated c-Jun, whereas expression of the NleD-E143A mutant had little effect (Figure 1E). These results indicate that NleD expression correlates with both cleavage and inactivation of JNK.
NleD directly cleaves JNK
To investigate whether NleD requires a host co-factor, we co-transformed an E. coli laboratory strain (BL21) with two plasmids: one expressing 6His-JNK2 and the other expressing NleD. Upon co-expression, we found that JNK2 was efficiently clipped in the E. coli cytoplasm in an NleD-dependent manner (Figure 1F), suggesting that NleD does not require a host-specific cell factor. We further tested whether purified 6His-tagged NleD could cut purified JNK2 in vitro (Figure 1G). When wild-type NleD and JNK2 were mixed at a molar ratio of 1:40, JNK was cleaved readily (Figure 1G). Notably, this cleavage was inhibited by the Zn metalloprotease-specific inhibitor phenanthroline (Figure 1G). In summary, our results suggest that NleD is a Zn metalloprotease that cleaves JNK directly.
NleD cuts within the activation loop of JNK2
To define the NleD digestion site we took advantage of a JNK2 construct with an N-terminal HA tag. The activation loop of this protein is flanked by two epitopes, the N-terminal HA epitope and an undefined epitope localized somewhere in the C-terminal domain that is recognized by the anti-JNK monoclonal antibody (Figure 2A). HEK293 cells were transfected with HA-JNK2 expressing plasmid and then infected with wild-type EPEC or nleD mutant. After 2.5 h, immunoprecipitation was performed using anti-HA antibodies. Western blot analysis using either anti-HA (Figure 2B) or anti-JNK (Figure 2C) antibodies was employed to examine the precipitated proteins, which comprised full-length HA-tagged JNK2 and N-terminal JNK2 fragments. An NleD-cleavage derived, N-terminal JNK2 fragment of ∼23 kDa was recognized by anti-HA antibody (Figure 2B). In parallel, distinct C-terminal JNK2 fragments of ∼33 kDa were recognized by anti-JNK antibody (Figure 2C). The co-precipitation of the C- and N-terminal JNK2 fragments indicates they remained associated even upon the cleavage of the HA-JNK2. This association was further confirmed by an in vitro analysis and similar results were obtained when HA-JNK1 was used instead of HA-JNK2 (Supplementary Figures S4 and S5). Based on the predicted sizes of the C- and N-terminal fragments generated by NleD, it appears that NleD cuts JNK2 and JNK1 within the activation loop (Figure 2A; Supplementary Figure S5).
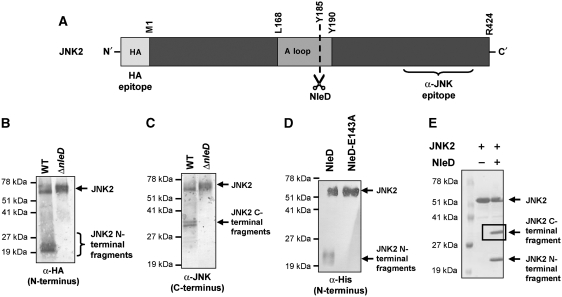
Figure 2.
NleD clips JNK within the activation loop. (A) Schematic diagram of JNK2. The location of the HA and anti-JNK antibody epitopes, activation loop (A loop), T and Y phosphorylation sites and NleD-cleavage site are indicated. (B, C) NleD cleaves JNK2 in vivo. HeLa cells were transfected with plasmids expressing HA-tagged JNK2. At 25 h post-transfection, the cells were infected with wild-type (WT) or nleD deletion mutant (ΔnleD) EPEC. After 2.5 h, proteins were extracted from the infected HeLa cells, immunoprecipitated using anti-HA antibody and subjected to western blot analysis using anti-HA (B) or anti-JNK (C) antibodies. JNK2 and its degradation fragments are indicated. Negative control using non-transfected cells confirmed the antibody specificity (data not shown). (D, E) NleD cleaves JNK2 in vitro. Purified, N-terminally tagged, 6His-JNK2 was incubated with purified NleD or NleD-E143A. After 60 min, the reaction was stopped using SDS loading buffer. To estimate the size of the N-terminal fragment of the clipped JNK2, the reaction mixture was subjected to western blot analysis using anti-6His antibody (D). Intact JNK2 and its N-terminal fragments are indicated. To determine the size of the two JNK2 fragments, the reaction mixture was analysed also by SDS–PAGE followed by Commassie blue staining (E). JNK2 and its degradation fragments are indicated. The framed C-terminal band was subjected to N-terminal sequencing analysis.
To better define the NleD-cleavage site, purified N-terminally tagged 6His-JNK2 was incubated with purified NleD and the exact cleavage location was determined by N-terminus sequencing. Initially, the reaction products were characterized by immunoblotting using anti-6His antibody (Figure 2D) and in parallel, by SDS–PAGE followed by Coomassie blue staining (Figure 2E); the former to detect specifically N-terminal products and the latter all generated products. Notably, similarly sized C- and N-terminal JNK fragments were observed in this in vitro study as in the in vivo analysis described above (compare Figure 2B and C with 2D and E). Furthermore, similar results were obtained when p38 was used as substrate instead of JNK (data not shown). To determine the precise cleavage point within JNK, a band corresponding to the C-terminal product of JNK (Figure 2E) was subjected to N-terminus sequencing using the Edman degradation method. The obtained sequence was YVVTR, indicating that NleD cleaved JNK after residue P184 within the TPY motif (Supplementary Figures S7 and S8). Notably, JNK activation requires phosphorylation of both T183 and Y185 (Supplementary Figure S8). These results further indicate that NleD directly cleaves JNK and define the cleavage site.
NleD inhibits the pro-apoptotic activity of JNK
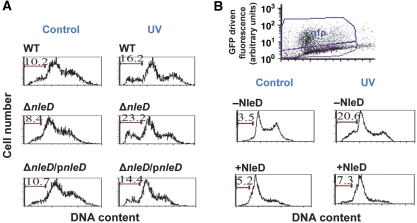
Activated JNK exhibit marked pro-apoptotic activity and UV-induced apoptosis is strictly JNK dependent (Tournier et al, 2000). We thus tested whether NleD inhibits UV-induced JNK-dependent apoptosis. HeLa cells were infected for 3 h with different EPEC strains to allow effectors injection, washed, UV irradiated, or not, and incubated for additional 4 h in medium supplemented with gentamicin. Then the apoptosis levels were measured using flow cytometry analysis of DNA content. Judging by the sub-G1 DNA content, 8.4–10.7% of the non-irradiated cells become apoptotic upon infection with the different EPEC strains (Figure 3A). In contrast, non-infected cells exhibited only 3% apoptosis (data not shown). These findings confirm early reports that EPEC induce apoptosis (Hemrajani et al, 2010 and references within). Nevertheless, exposure to UV, of cells infected with wild-type EPEC, nleD mutant, and complemented mutant, resulted in differential increase in apoptosis, 1.6-, 2.76-, and 1.35-fold, respectively (Figure 3A). These results suggest that upon infection, NleD clearly, but modestly, inhibited the JNK-dependent apoptosis.
Figure 3.
NleD inhibits JNK-dependent apoptosis. (A) HeLa cells were infected with wild-type EPEC, nleD mutant and complemented mutant. After infection the cells were washed, UV irradiated (30 J/m2), and harvested 4 h later. The DNA was stained with propidium iodide and cells were analysed by flow cytometry. Cells containing sub-G1 DNA are indicated by bars and their percentage in the population is indicated above the bars. Irradiated cells are indicated by ‘UV' and non-irradiated control cells by ‘Control'. (B) RKO cells were transfected with plasmid expressing NleD–GFP fusion protein and UV irradiated 24 h later (30 J/m2), or not (marked as Control). After additional 12 h, cells were harvested and analysed by flow cytometry. The non-transfected population (marked as −NleD), and the GFP expressing, transfected cells (+NleD), were gated, as exhibited in the upper panel, and the DNA content in each gated population was determined. Cells containing sub-G1 DNA are indicated by bars and their percentage in the population is indicated above the bars.
In addition to NleD, EPEC inject the host cells with an array of pro-and anti-apoptotic proteins including Map, EspF, NleH1, NleH2, and EspZ (Dean and Kenny, 2009; Hemrajani et al, 2010; Shames et al, 2010). The combined activity of these effectors may determine the apoptosis level upon infection. Thus, to isolate the effect of NleD from that of the other effectors and simplify the interpretation, we transiently expressed NleD fused to GFP (NleD–GFP) in RKO cells, expressing wild-type p53. The transfected cells were UV irradiated and the sub-G1 DNA content of cells was analysed. We compared the fraction of apoptotic cells among the NleD–GFP expressing cells to that of other cells, from the same transfection plates, that did not express NleD–GFP (Figure 3B, upper panel). As expected from cells expressing p53, transfection per se resulted in some G1 cell cycle arrest (Renzing and Lane, 1995). Nevertheless, 12 h after irradiation, the increase in apoptosis in NleD expressing cells was very small (1.4-fold) compared to cells that did not express NleD (5.88-fold). These results further support the notion that NleD inhibits JNK-dependent apoptosis, presumably via JNK inactivation.
NleC, but not NleD, is required for inhibition of TNFα-induced IL-8 transcription
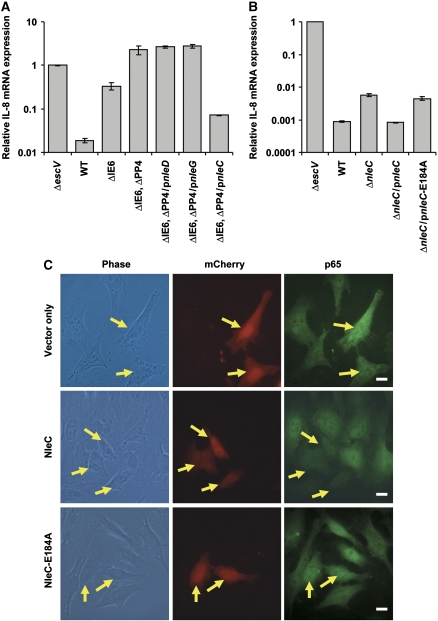
Maximal induction of IL-8 expression by TNFα involves NF-κB and JNK/AP-1 (Roger et al, 1998; Kang et al, 2007). EPEC infection has been shown to interfere with this induction of IL-8 (Nadler et al, 2010). To delineate whether NleD has a role in repressing IL-8 transcription during EPEC infection, we compared the capacity of two strains to repress TNFα-induced IL-8 transcription: a mutant with the IE6 region deleted (IE6 contains the nleBE genes, previously reported to inhibit IL-8 expression); and a mutant with both IE6 and PP4 regions deleted, the latter region containing nleD. HeLa cells were infected with the relevant strains, exposed to TNFα and then harvested to assay IL-8 mRNA levels by qPCR. The ΔIE6 mutant was partially deficient in repressing IL-8 expression, whereas the double mutant (ΔIE6 and ΔPP4) was completely deficient (Figure 4A). These results could suggest that nleD, a gene within the PP4 region that we had shown to inactivate JNK, contributes to IL-8 repression. However, complementation analysis using expression plasmids revealed that nleD was not the gene within the PP4 region mediating repression of IL-8 transcription (Figure 4A). We therefore tested other PP4 genes, including nleC and nleG, for PP4 complementation. Importantly, nleC, and not nleD or nleG, was found to influence IL-8 repression (Figure 4A). Similarly, deleting the nleD gene did not reduce the capacity of EPEC to inhibit IL-8 transcription (Supplementary Figure S9), but deletion of nleC rendered EPEC strongly attenuated in repression of IL-8 transcription (Figure 4B). Like NleD, also NleC contains a conserved Zn metalloprotease signature motif, HEXXH (Supplementary Figure S10). Importantly, only a plasmid expressing NleC but not one expressing NleC-E184A (a mutant in the HEXXH motif) restored the ability of the nleC mutant EPEC to block TNFα-induced IL-8 transcription (Figure 4B; Supplementary Figure S6). These results show that NleC is a mediator of EPEC-dependent repression of IL-8 transcription, which possibly exerts this effect via proteolysis of a key component in the NF-κB pathway.
Figure 4.
NleC expression correlates with inhibition of the NF-κB pathway. (A) Deletion analysis to identify the EPEC gene that represses IL-8 induction. HeLa cells were infected with one of the following EPEC: EPEC with a deleted IE6 region (ΔIE6), EPEC with deleted IE6 and PP4 regions (ΔIE6 and ΔPP4), or the latter complemented with plasmids expressing either wild-type NleD, NleG, or NleC (pnleD, pnleG, and pnleC, respectively). Cells infected with TTSS mutant (ΔescV) and wild-type EPEC (WT) served as negative and positive controls, respectively. HeLa cells were infected with the relevant EPEC for 2 h to allow injection of effectors before stimulation with TNFα for 3 h. Then RNA was extracted from the HeLa cells and real-time PCR performed to quantify IL-8 mRNA levels. Experiments were performed in duplicates and a typical experiment out of three is shown. Error bars indicate s.d. (B) NleC is required for inhibition of TNFα-induced IL-8 expression. HeLa cells were infected with nleC mutant, or this mutant complemented with plasmids expressing wild-type NleC (pnleC) or NleC-E184A mutant (pnleC-E184A). TTSS mutant (ΔescV) or wild-type EPEC (WT) served as negative and positive controls, respectively. HeLa cells were infected with these strains as described in (A), RNA was extracted from the HeLa cells and real-time PCR performed to quantify IL-8 mRNA levels. The indicated values are relative to IL-8 RNA levels in cells infected with ΔescV mutant. Experiments were performed in duplicates and a typical experiment out of four is shown. Error bars indicate the s.d. (C) NleC reduces p65 levels in vivo. HeLa cells transfected with plasmid expressing mCherry, mCherry–NleC or mCherry–NleC-E184A (red) were treated with TNFα for 30 min, after which they were fixed and visualized using anti-p65 antibody (green). Yellow arrows indicate cells expressing mCherry proteins. Bar represents 20 μm.
Ectopically expressed NleC is localized predominantly to the nucleus and associated with reduced levels of p65
We next tested whether NleC represses NF-κB. To this end we examined if NleC inhibits translocation of NF-κB to the nucleus. HeLa cells were transfected with vectors expressing mCherry, mCherry fused to NleC (mCherry–NleC), or mCherry fused to NleC-E184A (mCherry–NleC-E184A). In the following day, we exposed the cells to TNFα and evaluated p65 localization and levels by fluorescent microscopy using anti-p65 antibody. The three vectors exhibited similar transfection efficiency (∼20%) and expression levels of mCherry (data not shown). The mCherry–NleC and mCherry–NleC-E184A exhibited different localization from that of mCherry and were localized predominantly to the nucleus (Supplementary Figure S11). Interestingly, the p65 staining was considerably diminished in cells expressing mCherry–NleC, whereas strong p65 staining was evident in cells expressing mCherry or mCherry–NleC-E184A (Figure 4C). Quantification revealed that only 6% (n=64) of the mCherry–NleC expressing cells exhibited clear p65 staining. In contrast, in cells expressing mCherry or mCherry–NleC-E184A, the fraction of cells clearly stained with anti-p65 (96%, n=53 and 97%, n=67, respectively) was significantly (P<0.001) higher. These results suggest that NleC expression is associated with a significant reduction in p65 levels and that NleC is localized mainly to the nucleus.
NleC induces p65 cleavage
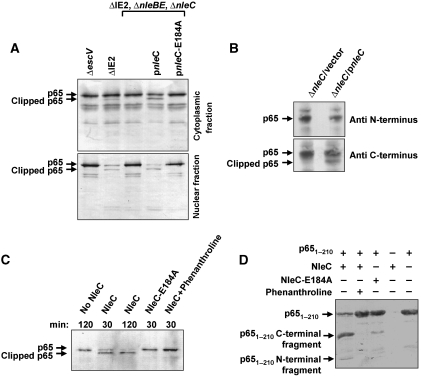
To test the hypothesis that NleC induces p65 proteolysis, we infected HeLa cells with various EPEC strains, then separated cytoplasmic proteins from nuclear proteins and subjected these two fractions to western analysis using anti-p65 antibodies. The ΔIE2 strain was used as wild type in these experiments, as this strain exhibits wild-type phenotype with respect to NF-κB inhibition (Nadler et al, 2010). We found that ΔIE2 EPEC, but not a TTSS-deficient mutant (ΔescV), induced marked reduction in nuclear p65 levels and partial clipping of both nuclear and cytoplasmic p65 (Figure 5A). To isolate the effect of NleC from that of the co-injected NleB and NleE, we took advantage of a triple mutant ΔnleBE, ΔnleC. As expected, this mutant was deficient in causing reduction of nuclear p65 and in p65 clipping (Figure 5A). Importantly, the capacity of this triple mutant to reduce nuclear levels of p65 and stimulate p65 clipping was restored by a plasmid expressing NleC, but not by a plasmid expressing NleC-E184A (Figure 5A). Similar results were obtained using Caco2 cells, which mimic enterocytes better than HeLa cells (Supplementary Figure S12). These data indicate that injected NleC reduces nuclear p65 levels in conjunction with p65 cleavage.
Figure 5.
NleC induces p65 cleavage, which is associated with reduced nuclear p65 levels. (A) NleC induces p65 cleavage in vivo. HeLa cells were infected for 3 h with the ΔIE2 ΔnleBE, ΔnleC mutant EPEC that was complemented, or not, with plasmids expressing NleC or mutated NleC (NleC-E184A), as indicated. Proteins were extracted from the infected HeLa cells, separated into cytosolic and nuclear fractions and subjected to western blot analysis using anti-p65 antibody. An EPEC mutant deficient in TTSS activity (ΔescV) and EPEC deleted of the IE2 region (ΔIE2) served as negative and positive controls, respectively; the latter was used here as wild type. Intact and clipped p65 are indicated. (B) NleC induces clipping the N-terminal end of p65. HeLa cells were infected for 3 h with the ΔnleC mutant EPEC that was complemented with plasmids expressing NleC or vector only, as indicated. Proteins were extracted from the infected HeLa cells and subjected to western blot analysis using anti-N-terminus of p65 or anti-C-terminus of p65 antibodies. Intact and clipped p65 are indicated. (C) NleC induced p65 cleavage in vitro. The cytosolic fraction of HeLa extracts was combined with NleC, NleC-E184A, or buffer alone in the presence or absence of phenanthroline, a Zn metalloprotease inhibitor, as indicated. The incubation time is indicated above each lane. Reaction products were visualized by western blot analysis using anti-p65 antibody. Intact and clipped p65 are indicated. (D) NleC directly cuts p651–210. Purified p651–210 and NleC were incubated in a reaction mixture at a molar ratio of 40:1, in the presence or absence of phenanthroline. The reaction was stopped by addition of SDS loading buffer and proteins separated by SDS–PAGE and visualized by Coomassie blue staining. p651–210 and its degradation fragments are indicated. NleC does not appear in this gel.
NleC directly cleaves p65
To determine where p65 is cleaved by NleC, HeLa cells were infected with nleC mutant (ΔnleC) complemented with either plasmid only (vector) or vector expressing wild-type NleC (pnleC). After 3 h, cells were extracted and lysates were subjected to a western blot analysis using two different anti-p65 antibodies; one raised against the p65 N-terminus region and the other against the p65 C-terminus region. Importantly, both antibodies reacted with the full-length p65, but only the anti-p65 C-terminus antibody reacted with the cleaved form of p65 (Figure 5B). These findings indicate that NleC mediates the clipping of the p65 N-terminal domain.
To confirm our premise that NleC directly cleaved p65 and determine the precise cleavage site, we tested whether NleC cleaves p65 in vitro. As we were unable to generate full-length purified p65 due to its poor stability and solubility in E. coli (data not shown), we employed extracts of HeLa cells, where we mixed with purified NleC, or NleC-E184A, and performed western blot analysis using anti-p65 antibodies to detect the reaction products (Figure 5C). In this assay NleC, but not NleC-E184A, effectively induced p65 cleavage and phenanthroline inhibited the NleC-induced p65 clipping (Figure 5C). To test whether NleC directly cuts p65 and map the cleavage site, we constructed a plasmid expressing the N-terminal Rel homology domain of p65 (residues 1–210 of p65). The p651–210 fragment was soluble and stable in E. coli and the purified p651–210 fragment was efficiently cleaved by purified NleC, but not in the presence of the metalloprotease inhibitor phenanthroline (Figure 5D). Moreover, the cleavage activity of the purified NleC-E184A mutant was strongly attenuated. To determine the exact cleavage point of NleC within p65, the cleaved p651–210 was subjected to N-terminal sequencing. The obtained sequence, EGRSA, indicates that NleC cleaved p65 within its conserved DNA-binding domain after residue C38 (Supplementary Figure S13). Further analysis suggested that NleC cleaves other NF-κB family members containing this conserved sequence (Supplementary Figure S14). The nleD mutant fails to cleave JNK, p38, or Erk although it carries intact nleC gene (Figure 1A and C; Supplementary Figure S3), suggesting that MAPKs are not good NleC substrates. We further verified the NleC and NleD specificity, in vitro, using purified NleD and NleC (Supplementary Figure S15). Taken together, these results indicate that NleC directly cuts and inactivates p65 and probably other Rel members.
Interplay between NleC and NleBE in blocking p65-dependent transcription
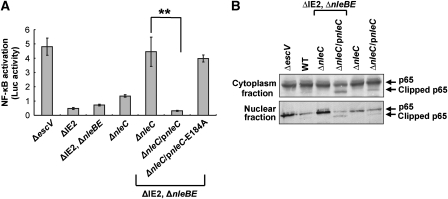
Residues 1–37 of p65 are required for DNA binding (Supplementary Figures S13 and S16; Toledano et al, 1993; Chen et al, 1998a, 1998b). To substantiate the notion that NleC abolishes the capacity of NF-κB to promote specific transcription, we transfected cells with a plasmid containing NF-κB-dependent promoter fused to the luciferase reporter gene (luc) (Yaron et al, 1998). These cells were infected with different EPEC mutants and their ability to activate luc expression, via the EPEC PAMPs, was determined. The TTSS mutant (escV) clearly induced luc expression, while the wild-type strain (ΔIE2) strongly inhibits this activation (Figure 6A). Furthermore, the presence of native, or plasmid expressed, NleC, or NleBE, was sufficient for strong repression of NF-κB. Yet, under native expression levels both NleC and NleBE were required for complete NF-κB repression (Figure 6A). These results further confirmed that both, NleC and NleBE, can repress NF-κB-dependent transcription.
Figure 6.
Interplay between NleC and NleBE, in mediating NF-κB repression. (A) NleC and NleBE are required for complete repression of NF-κB. HeLa cells were transfected with a mix of plasmid expressing luc via an NF-κB promoter and a plasmid expressing renilla-luc expressed from constitutive promoter. The cells were next infected with different EPEC strains, and after 3 h, infection was stopped by replacing the media with DMEM supplemented with gentamicin. At 7 h post-infection, the levels of luc and renilla-luc expression were determined. Shown is a reprehensive experiment out of two that gave similar results. The assay was performed in triplicates. Error bars indicate the s.d. (**P-value<0.05). (B) NleBE expression is associated with attenuation the NleC-dependent p65 cleavage. HeLa cells were infected with various EPEC strains as indicated. After 3 h, proteins were extracted, separated to cytoplasmic and nuclear fractions and subjected to western analysis using anti-p65 C-terminal antibody. The intact and clipped p65 are indicated.
We further tested whether NleBE would interfere with the NleC-dependent p65 cleavage. To this end we infected HeLa cells with various EPEC strains, separated cytoplasmic proteins from nuclear proteins and subjected these two fractions to western analysis using anti-p65 C-terminal antibodies. The results show that in the presence of NleBE, the NleC-dependent p65 cleavage was attenuated; both in the cytoplasm and in the nucleus (Figure 6B). These results further highlight the interplay between these effectors, but the involved mechanism is yet to be elucidated. NleBE is known to stabilize IκB (Nadler et al, 2010; Newton et al, 2010), and it is possible that IκB protects p65 from cleavage by preventing its nuclear localization or by limiting the access of NleC to the bound p65.
NleB, E, C, D co-operate in repression of IL-8 secretion
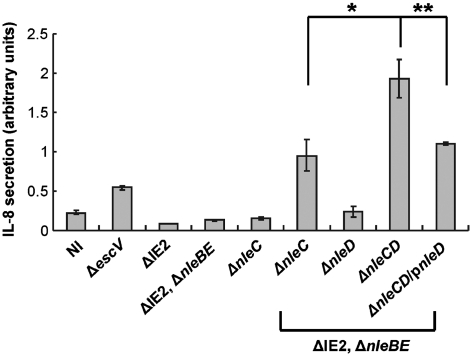
JNK/p38 may enhance IL-8 production by a post-transcriptional mechanisms (Jijon et al, 2002; Yu et al, 2003; Jandhyala et al, 2010). We therefore tested the effect of the effectors on IL-8 production. HeLa cells were infected with different EPEC strains and their influence on the levels of secreted IL-8, in the medium, was tested by ELISA. Increase in IL-8 secretion was detected in cells infected with the TTSS (escV) mutant, likely due to host-cell activation by the bacterial PAMPs. In contrast, upon infection with wild-type EPEC (ΔIE2), IL-8 secretion was strongly repressed (Figure 7). Furthermore, both nleBE and nleC mutants still exhibit strong repression of IL-8 secretion. Only upon deletion of nleC and nleBE, secretion of IL-8 was restored (Figure 7) and repression of secretion was resumed upon complementation of this mutant with either NleBE or NleC (Supplementary Figure S17). Importantly, deletion of all four genes (nleBE, nleC, and nleD) resulted in further increase in IL-8 secretion and complementation of this quadruple mutant with nleD restored partial inhibition of IL-8 secretion (Figure 7). Interestingly, cells infected with the triple (nleBEC) or quadruple (nleBECD) mutants secret more IL-8 than cells infected with the TTSS mutant (escV). This might reflect the notion that structural components of the TTSS needle complex are PAMPs (Auerbuch et al, 2009; Miao et al, 2010), which might further activate IL-8 secretion by the infected cells. Taken together, these results demonstrate that NleBE, NleC, and NleD co-operate to achieve maximal inhibition of IL-8 secretion. NleBE and NleC probably mediate this inhibition by blocking NF-κB-dependent IL-8 transcription, and NleD might inhibit JNK/p38-dependent IL-8 production.
Figure 7.
NleE, NleB, NleC, and NleD are required for maximal EPEC-induced repression of IL-8 secretion. HeLa cells were infected with different EPEC strains as indicated. After 3 h, cells were washed and the media was replaced by DMEM supplemented with gentamicin. At 19 h post-infection, the media was harvested from the respective wells, cleared and the amount of secreted IL-8 determined by ELISA. The relative amounts of secreted IL-8 are shown. Experiments were performed in triplicates and a typical experiment out of two is shown. Standard deviation (bars) (*P-value<0.1, **P-value<0.05).
Discussion
Upon infection, bacterial PAMPs stimulate host-cell TLRs, which in turn trigger an inflammatory response. TLR activation is associated with complex signalling that culminates in the activation of two transcription factors families, the NF-κB and AP-1 (Takeuchi and Akira, 2001) (Figure 8). Activated AP-1 and NF-κB modify the transcription pattern of hundreds of genes. The ultimate outcome of NF-κB and AP-1 activation is multifaceted and depends on other coincident signalling inputs and specific cell type. In general, both NF-κB and AP-1 are considered to function synergistically to induce inflammation. However, whereas the AP-1 c-Jun stimulates apoptosis, NF-κB induces expression of anti-apoptotic factors (Barkett and Gilmore, 1999; Kucharczak et al, 2003) (Figure 8).
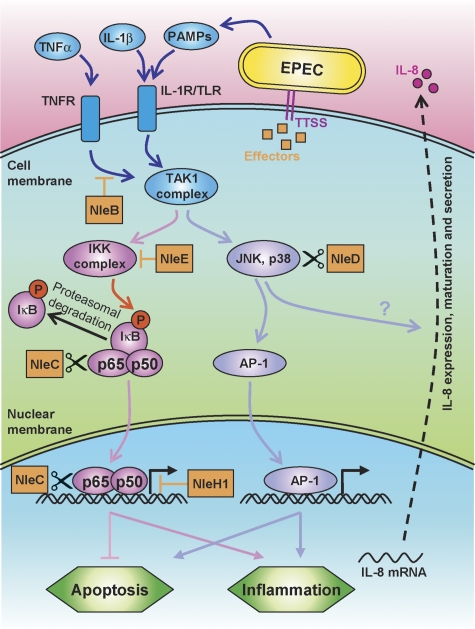
Figure 8.
A model of the anti-inflammatory activity of NleBCDE and NleH1. Schematic diagram of the signalling cascades activated by PAMPs, IL-1, and TNF is shown. The different EPEC effectors (orange boxes) interact with this signalling network at multiple points. NleB inhibits the TNFR signalling upstream to the TAK1 complex, NleE prevents IKK activation, NleC cleaves and inactivates cytoplasmic and nuclear p65, and NleH1 inhibits the interaction of NF-κB with some promoters. Finally, NleD cuts and inactivates JNK and p38. These effectors function in concert to repress IL-8 secretion and to modulate the inflammation and apoptosis processes.
Using its type III secretion system, EPEC injects host cells with multiple effectors that manipulate the complex TLR signalling network. Recent studies have demonstrated that two EPEC effectors, NleB and NleE, are required for NF-κB inhibition (Nadler et al, 2010; Newton et al, 2010) (Figure 8). An additional effector that interferes with the NF-κB pathway is NleH1 (Gao et al, 2009). Here, we show that NleC and NleD are additional anti-inflammatory effectors of EPEC. NleC is a Zn endopeptidase that specifically cleaves p65, inactivate it and perhaps also destabilize it. Under native conditions, the amount of injected NleC, NleB, and NleE is very low (Nadler et al, 2010 and unpublished data) and each of these effectors only partially inhibits TNFα-induced IL-8 expression (Nadler et al, 2010; Newton et al, 2010; Figure 4A), but together they achieve full inhibition of NF-κB. Notably, upon their overexpression, resulting in increased injection, either NleC or NleBE, is sufficient to mediate complete inhibition of IL-8 expression (Nadler et al, 2010; Newton et al, 2010; Figure 4A). These observations led us to suspect that the relatively low-native doses of injected NleC, NleB, and NleE represent a selected level, operational but not toxic.
The second Zn endopeptidase effector, NleD, specifically cuts and inactivates JNK and p38, but not ERK. Notably, NleD cleaves JNK at a sequence in the activation loops, which is highly conserved in JNK and p38, but missing in ERK. AvrPphB, a cysteine protease effector of plant pathogens also inactivates a host kinase (PBS1) by cleavage of its activation segment (Shao et al, 2003). Interestingly, NleD co-operate with NleE, NleB, NleC to inhibit IL-8 production, likely by inactivation of JNK and/or p38. In addition, NleD counteracts JNK-dependent apoptosis, most likely via direct JNK inactivation. Experimental infection using calves suggested that NleD is required for colonization by EHEC EDL933 (Dziva et al, 2004), but further attempts to confirm these findings and to identify other phenotypes associated with nleC or nleD mutants failed (Marches et al, 2005). Thus, additional analysis is required to elucidate the NleD and NleC function in vivo.
NleC and NleD are conserved among AE pathogens including EHEC, EPEC, and Citrobacter rodentium. Moreover, using BLAST we have identified closely related proteins, which probably share identical or very similar function, in other pathogens and symbiotic bacteria. The pathogen Salmonella enterica ssp. Arizonae and the insect symbiotic bacteria Hamiltonella defense each encode a putative type III effector homologous to NleD. The HopH1 effector of the plant pathogen Pseudomonas syringae also shows significant but lower similarity to NleD. Homologues of NleC include putative type III effectors of the environmental bacteria Yersinia aldovae, the pathogen S. enterica ssp. enterica serovar Javiana, and the insect symbiotic bacteria Arsenophonus nasoniae. NleC also bears similarity to the putative catalytic domain of the exotoxin of the fish pathogen Photobacterium damselae ssp. piscicida. In conclusion, comparative sequence analysis suggests that NleC and NleD are each representative of novel effector/toxin families with distinct and defined function. The increasing number and diversity of effector/toxin families are a striking reflection of the complexity of host signalling networks and the concomitantly intricate nature of host/pathogen interactions.
Materials and methods
Bacterial strains, plasmids, and primers
Bacterial strains, plasmids, and primers used in this study are listed in Supplementary Tables S1–S3, respectively (Supplementary data). Deletions in the EPEC chromosome were constructed using the primers listed in Supplementary Table S3, as described (Datsenko and Wanner, 2000). For bacterial expression, the genes were cloned in the pSA10, pCX341, pET28a, or pET52 as described (Nadler et al, 2010). For these cloning procedures, genes were amplified by PCR using the primers listed in Supplementary Table S3.
Protein extraction, purification, western blot analysis, and immunoprecipitation
Protein extraction and western blot analysis were performed as described (Nadler et al, 2010). The antibodies used in this study and working conditions as well as the procedures used for purification of NleD, NleC, NleC-E184A, and p651–210 are described in Supplementary data. Agarose-protein-A (Sigma) beads coated with anti-HA or anti-FLAG antibody were used for immunoprecipitation.
In vitro assay for NleD and NleC activity
Purified recombinant JNK2 (15 μg, a gift from D Engeleberg the Hebrew University) or p651–210 (10 μg) was mixed accordingly with purified NleD (0.375 μg) or NleC (0.25 μg) in 40 μl of reaction buffer (50 mM Tris–HCl pH 7.5, 2 mM CaCl2, 50 mM NaCl). In the case of native p65 assay, purified NleC (0.4 μg) was mixed with cleared extracts of HeLa cells in 20 μl of reaction buffer (50 mM Tris pH 7.5, 2 mM CaCl2, 5 mM NaCl). Where specified, 1,10-phenanthroline was added (5 mM, Sigma). Reactions were carried out for the indicated period of time at room temperature and stopped by the addition of SDS loading buffer. Digestion products were visualized using western blot analysis or SDS–PAGE followed by Coomassie staining.
IL-8 expression and secretion
Levels of IL-8 mRNA and secreted IL-8 were determined as described (Nadler et al, 2010).
Flow cytometry
The levels of sub-G1 DNA in cells were determined by flow cytometry as described (Yogev et al, 2006).
Transfection of HeLa cells and p65 staining
HeLa cells were transfected with 1 μg of plasmid DNA using ExGen500 (Fermentas), according to the manufacturer's instructions. After 24 h, the medium was replaced with fresh DMEM with or without 10 ng/ml TNFα. After 1 h, cells were fixed for 10 min (3.7% paraformaldehyde in PBS), washed, perforated (10 min in 0.25% Triton X-100 in PBS), washed again and blocked (2% BSA in TBS) at 4°C overnight. Next, cells were incubated overnight with anti-p65 (SC-372, Santa Cruz) antibodies (1:300 in TBS) before being stained with CY-488 goat anti-rabbit antibodies (Cell Signaling, 1:1000 in TBS) for 1 h. Cells were visualized using fluorescence microscopy.
Luciferase assay
The assay was performed according to the instructions in the Dual-Luciferase Reporter kit (Promega). Briefly, HeLa cells were grown to a 70% confluence in a 24-well plate and than co-transfected with 0.25 μg and 0.025 μg of pNF-κB-luc and pRL-TK Renilla luciferase vector (Promega) DNA, accordingly, using 1 μl of ExGen500 (Fermentas). After 24 h, the medium was replaced with fresh DMEM and the cells were infected for 3 h with a 1:100 dilution of an overnight-cultured EPEC. The medium was then replaced with DMEM supplemented with gentamicin (100 ng/μl) and 2% FCS and cells were incubated for additional 4 h. Luciferase activity was measured with the Dual Reporter Assay kit (Promega) and luciferase activities were normalized by dividing relative light units expressed when the luciferase substrate is present by relative light units expressed when the renilla substrate is present.
Supplementary Material
Acknowledgments
We thank D Engeleberg and A Honigman for plasmids, proteins, and antibodies; and O Schueler-Furman and N London for help with protein modelling. ES is supported by grants from Israel Science Academy of Science and Humanities, Israeli Cancer Research Fund and The Israel Cancer Association and IR and YBN by the German Israeli Foundation (GIF) and IR by the Israel Science Academy of Science and Humanities and the Rosetrees Trust. RZ and CG are supported by the EC 7th Framework Programme (FP7/2007-2013 under grant agreement no. 239182). IR is an Etta Rosensohn Professor of Bacteriology.
Author contributions: KB, LGA, CN, SK, OY, and CG performed the experiments; KB, LGA, OY, CG, ES, RZ, and IR designed the experiments and analysed the results; KB, LGA, GY, and IR wrote the manuscript; YBN, ES, and RZ provided the reagents.
Footnotes
The authors declare that they have no conflict of interest.
References
- Auerbuch V, Golenbock DT, Isberg RR (2009) Innate immune recognition of Yersinia pseudotuberculosis type III secretion. PLoS Pathog 5: e1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkett M, Gilmore TD (1999) Control of apoptosis by Rel/NF-kappaB transcription factors. Oncogene 18: 6910–6924 [DOI] [PubMed] [Google Scholar]
- Chen FE, Huang DB, Chen YQ, Ghosh G (1998a) Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature 391: 410–413 [DOI] [PubMed] [Google Scholar]
- Chen YQ, Ghosh S, Ghosh G (1998b) A novel DNA recognition mode by the NF-kappa B p65 homodimer. Nat Struct Biol 5: 67–73 [DOI] [PubMed] [Google Scholar]
- Chen ZJ (2005) Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7: 758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RJ (2000) Signal transduction by the JNK group of MAP kinases. Cell 103: 239–252 [DOI] [PubMed] [Google Scholar]
- Dean P, Kenny B (2009) The effector repertoire of enteropathogenic E. coli: ganging up on the host cell. Curr Opin Microbiol 12: 101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ (1994) JNK1: a protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell 76: 1025–1037 [DOI] [PubMed] [Google Scholar]
- Doyle SL, O'Neill LA (2006) Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol 72: 1102–1113 [DOI] [PubMed] [Google Scholar]
- Dziva F, van Diemen PM, Stevens MP, Smith AJ, Wallis TS (2004) Identification of Escherichia coli O157:H7 genes influencing colonization of the bovine gastrointestinal tract using signature-tagged mutagenesis. Microbiology 150: 3631–3645 [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF (2003) AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3: 859–868 [DOI] [PubMed] [Google Scholar]
- Gao X, Wan F, Mateo K, Callegari E, Wang D, Deng W, Puente J, Li F, Chaussee MS, Finlay BB, Lenardo MJ, Hardwidge PR (2009) Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog 5: e1000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemrajani C, Berger CN, Robinson KS, Marches O, Mousnier A, Frankel G (2010) NleH effectors interact with Bax inhibitor-1 to block apoptosis during enteropathogenic Escherichia coli infection. Proc Natl Acad Sci USA 107: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibi M, Lin A, Smeal T, Minden A, Karin M (1993) Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev 7: 2135–2148 [DOI] [PubMed] [Google Scholar]
- Iguchi A, Thomson NR, Ogura Y, Saunders D, Ooka T, Henderson IR, Harris D, Asadulghani M, Kurokawa K, Dean P, Kenny B, Quail MA, Thurston S, Dougan G, Hayashi T, Parkhill J, Frankel G (2009) Complete genome sequence and comparative genome analysis of enteropathogenic Escherichia coli O127:H6 strain E2348/69. J Bacteriol 191: 347–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala DM, Rogers TJ, Kane A, Paton AW, Paton JC, Thorpe CM (2010) Shiga toxin 2 and flagellin from shiga-toxigenic Escherichia coli superinduce interleukin-8 through synergistic effects on host stress-activated protein kinase activation. Infect Immun 78: 2984–2994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jijon HB, Panenka WJ, Madsen KL, Parsons HG (2002) MAP kinases contribute to IL-8 secretion by intestinal epithelial cells via a posttranscriptional mechanism. Am J Physiol Cell Physiol 283: C31–C41 [DOI] [PubMed] [Google Scholar]
- Kallunki T, Su B, Tsigelny I, Sluss HK, Derijard B, Moore G, Davis R, Karin M (1994) JNK2 contains a specificity-determining region responsible for efficient c-Jun binding and phosphorylation. Genes Dev 8: 2996–3007 [DOI] [PubMed] [Google Scholar]
- Kang SS, Woo SS, Im J, Yang JS, Yun CH, Ju HR, Son CG, Moon EY, Han SH (2007) Human placenta promotes IL-8 expression through activation of JNK/SAPK and transcription factors NF-kappaB and AP-1 in PMA-differentiated THP-1 cells. Int Immunopharmacol 7: 1488–1495 [DOI] [PubMed] [Google Scholar]
- Karin M, Ben-Neriah Y (2000) Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu Rev Immunol 18: 621–663 [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S (2006) TLR signaling. Cell Death Differ 13: 816–825 [DOI] [PubMed] [Google Scholar]
- Kucharczak J, Simmons MJ, Fan Y, Gelinas C (2003) To be, or not to be: NF-kappaB is the answer—role of Rel/NF-kappaB in the regulation of apoptosis. Oncogene 22: 8961–8982 [DOI] [PubMed] [Google Scholar]
- Marches O, Wiles S, Dziva F, La Ragione RM, Schuller S, Best A, Phillips AD, Hartland EL, Woodward MJ, Stevens MP, Frankel G (2005) Characterization of two non-locus of enterocyte effacement-encoded type III-translocated effectors, NleC and NleD, in attaching and effacing pathogens. Infect Immun 73: 8411–8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao EA, Mao DP, Yudkovsky N, Bonneau R, Lorang CG, Warren SE, Leaf IA, Aderem A (2010) Innate immune detection of the type III secretion apparatus through the NLRC4 inflammasome. Proc Natl Acad Sci USA 107: 3076–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler C, Baruch K, Kobi S, Mills E, Haviv G, Farago M, Alkalay I, Bartfeld S, Meyer TF, Ben-Neriah Y, Rosenshine I (2010) The type III secretion effector NleE inhibits NF-kappaB activation. PLoS Pathog 6: e1000743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton HJ, Pearson JS, Badea L, Kelly M, Lucas M, Holloway G, Wagstaff KM, Dunstone MA, Sloan J, Whisstock JC, Kaper JB, Robins-Browne RM, Jans DA, Frankel G, Phillips AD, Coulson BS, Hartland EL (2010) The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS Pathog 6: e1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzing J, Lane DP (1995) p53-dependent growth arrest following calcium phosphate-mediated transfection of murine fibroblasts. Oncogene 10: 1865–1868 [PubMed] [Google Scholar]
- Roger T, Out T, Mukaida N, Matsushima K, Jansen H, Lutter R (1998) Enhanced AP-1 and NF-kappaB activities and stability of interleukin 8 (IL-8) transcripts are implicated in IL-8 mRNA superinduction in lung epithelial H292 cells. Biochem J 330 (Part 1): 429–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud-Sparagano MH, Maresca M, Kenny B (2007) Enteropathogenic Escherichia coli (EPEC) inactivate innate immune responses prior to compromising epithelial barrier function. Cell Microbiol 9: 1909–1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shames SR, Deng W, Guttman JA, de Hoog CL, Li Y, Hardwidge PR, Sham HP, Vallance BA, Foster LJ, Finlay BB (2010) The pathogenic E. coli type III effector EspZ interacts with host CD98 and facilitates host cell prosurvival signalling. Cell Microbiol 12: 1322–1339 [DOI] [PubMed] [Google Scholar]
- Shao F, Golstein C, Ade J, Stoutemyer M, Dixon JE, Innes RW (2003) Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science 301: 1230–1233 [DOI] [PubMed] [Google Scholar]
- Shaulian E, Karin M (2001) AP-1 in cell proliferation and survival. Oncogene 20: 2390–2400 [DOI] [PubMed] [Google Scholar]
- Takeuchi O, Akira S (2001) Toll-like receptors; their physiological role and signal transduction system. Int Immunopharmacol 1: 625–635 [DOI] [PubMed] [Google Scholar]
- Toledano MB, Ghosh D, Trinh F, Leonard WJ (1993) N-terminal DNA-binding domains contribute to differential DNA-binding specificities of NF-kappa B p50 and p65. Mol Cell Biol 13: 852–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier C, Hess P, Yang DD, Xu J, Turner TK, Nimnual A, Bar-Sagi D, Jones SN, Flavell RA, Davis RJ (2000) Requirement of JNK for stress-induced activation of the cytochrome c-mediated death pathway. Science 288: 870–874 [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y (1998) Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature 396: 590–594 [DOI] [PubMed] [Google Scholar]
- Yogev O, Anzi S, Inoue K, Shaulian E (2006) Induction of transcriptionally active Jun proteins regulates drug-induced senescence. J Biol Chem 281: 34475–34483 [DOI] [PubMed] [Google Scholar]
- Yogev O, Saadon K, Anzi S, Inoue K, Shaulian E (2008) DNA damage-dependent translocation of B23 and p19 ARF is regulated by the Jun N-terminal kinase pathway. Cancer Res 68: 1398–1406 [DOI] [PubMed] [Google Scholar]
- Yu Y, Zeng H, Lyons S, Carlson A, Merlin D, Neish AS, Gewirtz AT (2003) TLR5-mediated activation of p38 MAPK regulates epithelial IL-8 expression via posttranscriptional mechanism. Am J Physiol Gastrointest Liver Physiol 285: G282–G290 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.