Tracking and visualizing the circadian ticking of the cyanobacterial clock protein KaiC in solution
The authors add a dynamic perspective to earlier reported structures of the cyanobacterial oscillator protein KaiC. Mutant analyses combined with molecular modelling provide novel mechanistic insights into the biochemical ‘ticking' of the circadian clock.
Keywords: ATPase, circadian clock, phosphorylation, small-angle X-ray scattering, temperature compensation
Abstract
The circadian clock in cyanobacteria persists even without the transcription/translation feedbacks proposed for eukaryotic systems. The period of the cyanobacterial clock is tuned to the circadian range by the ATPase activity of a clock protein known as KaiC. Here, we provide structural evidence on how KaiC ticks away 24 h while coupling the ATPase activity in its N-terminal ring to the phosphorylation state in its C-terminal ring. During the phosphorylation cycle, the C-terminal domains of KaiC are repositioned in a stepwise manner to affect global expansion and contraction motions of the C-terminal ring. Arg393 of KaiC has a critical function in expanding the C-terminal ring and its replacement with Cys affects the temperature compensation of the period—a fundamental property of circadian clocks. The conformational ticking of KaiC observed here in solution serves as a timing cue for assembly/disassembly of other clock proteins (KaiA and KaiB), and is interlocked with its auto-inhibitory ATPase underlying circadian periodicity of cyanobacteria.
Introduction
Circadian clocks are endogenous timing systems that have evolved ubiquitously to enable organisms to adapt to daily alterations in the environment. Cyanobacteria, which exhibit daily regulation of nitrogen fixation, photosynthesis, and amino-acid uptake, are among the simplest organisms known to possess a circadian oscillator (Golden et al, 1997). The central oscillator of the cyanobacterium Synechococcus elongatus PCC 7942 comprises three clock proteins termed KaiA, KaiB, and KaiC (Ishiura et al, 1998). KaiC is rhythmically phosphorylated and then dephosphorylated in vivo even in the absence of transcription–translation feedback regulation (Tomita et al, 2005), a mechanism proposed to underlie various eukaryotic circadian systems.
In vitro reconstruction of the KaiC phosphorylation cycle, achieved simply by incubating KaiA, KaiB, and KaiC in the presence of ATP (Nakajima et al, 2005), has provided a means of studying the detailed mechanisms of the Kai-protein oscillator. KaiA enhances the auto-phosphorylation of KaiC (Iwasaki et al, 2002), whereas KaiB attenuates the effects of KaiA (Kitayama et al, 2003; Xu et al, 2003). During the KaiC phosphorylation cycle, the three Kai proteins are repeatedly assembled into hetero-multimeric complexes and then disassembled (Kageyama et al, 2006; Akiyama et al, 2008). The frequency of the phosphorylation cycle is closely correlated with the ATP hydrolysis rate exhibited by KaiC (Terauchi et al, 2007). The ATPase activity of KaiC is kept constant across a wide range of temperatures by an as yet unknown mechanism, whereas ordinary biochemical reactions are accelerated several fold upon raising the temperature by 10°C. A recent study pointed out a physiological linkage between KaiC ATPase activity and the gating of cell division (Dong et al, 2010). The temperature-compensated ATPase activity of KaiC, which is far lower than that reported for typical molecular motors, has a central function in maintaining slow and regular oscillations.
KaiC, a 58-kDa protein of 519 amino acids, is a dumbbell-shaped molecule composed of tandemly duplicated N-terminal (C1) and C-terminal (C2) domains, each of which includes ATPase motifs (Walker motifs A and B) (Ishiura et al, 1998). Six protomers of KaiC are assembled into a hexamer to attain a double-doughnut shape (Hayashi et al, 2003; Pattanayek et al, 2004). A recombinant KaiC having only the C1 domain forms a C1 ring (Mori et al, 2002), and retains approximately 70% of the basal ATPase activity of full-length KaiC (Terauchi et al, 2007). During the KaiC phosphorylation cycle, KaiC is auto-phosphorylated and auto-dephosphorylated both on Ser431 and Thr432 in the C2 domain in a programmed sequence (Nishiwaki et al, 2004, 2007; Xu et al, 2004; Rust et al, 2007) as follows: KaiCS/pT → KaiCpS/pT → KaiCpS/T → KaiCS/T (where ‘S' represents Ser431, ‘pS' represents phosphorylated-Ser431, ‘T' represents Thr432, and ‘pT' represents phosphorylated-Thr432). These features apparently imply a strict division of labour within the KaiC hexamer. However, the ATPase activity of KaiCS/T-mimicking mutant KaiCA/A is higher than KaiCpS/pT-mimicking mutant KaiCD/E (Terauchi et al, 2007). This indicates the presence of functional coupling between the C1 and C2 rings. Therefore, uncovering the structural basis for this functional coupling is of great importance.
Although an X-ray crystallographic study has provided visualization of the detailed structure of phospho-mimicking KaiC mutants (Pattanayek et al, 2009), only subtle structural changes are localized near the dual phosphorylation site in the C2 ring. Thus, the molecular mechanisms by which ATPase and phosphorylation regulate one another remain unknown. Here, we use time-resolved fluorescence spectroscopy, small-angle X-ray scattering (SAXS), and mutational analyses to track and visualize the ATPase/phosphorylation-coupled ticking of KaiC in solution.
Results
Fluorescence detection of KaiC conformational changes
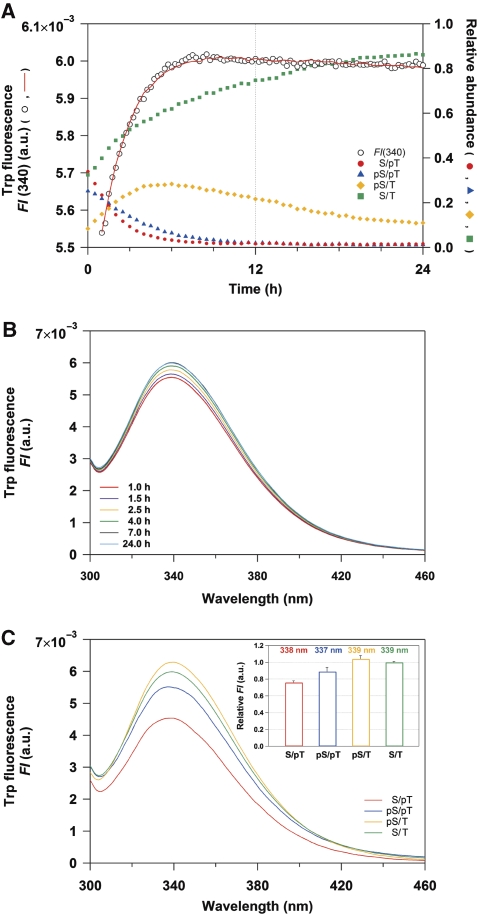
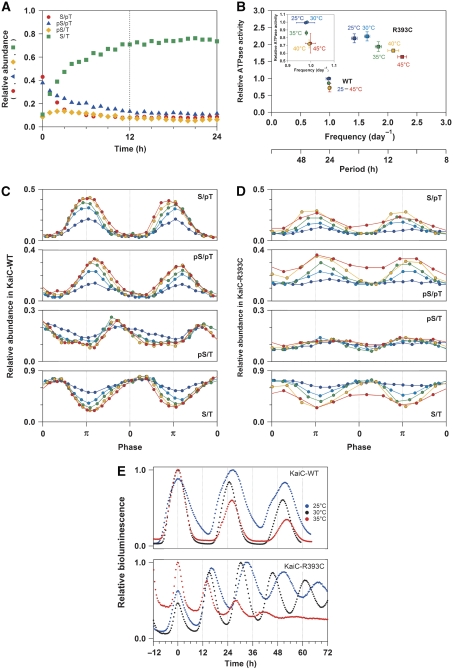
We investigated the structure of KaiC in solution along the reaction sequence of the phosphorylation cycle to elucidate the structural basis for the C1/C2 coupling. The auto-dephosphorylation reaction of wild-type KaiC (KaiC-WT) was initiated by a temperature jump from ‘on ice (∼1°C)' to 30°C, and its potential structural transition was monitored with time-resolved fluorescence spectroscopy in real time. KaiC-WT intrinsically possesses three tryptophan (Trp) residues, which provide sensitive fluorescent probes for monitoring local structural changes. One Trp residue is located in the C1 domain (W92), whereas the other two Trp residues are located in the C2 domain (W331 and W462). As reported previously (Nishiwaki et al, 2007; Rust et al, 2007), a decrease in the KaiC-WTS/pT population within 4 h was followed by the auto-dephosphorylation of KaiC-WTpS/pT into KaiC-WTS/T via KaiC-WTpS/T (Figure 1A). Concomitantly with this shift in the phosphorylation states, the fluorescence intensity at 340 nm (FI(340)) became gradually elevated with a subsequent slight decrease to be nearly time independent after 24 h (Figure 1A). Similar biphasic relaxation was also observed at other emission wavelengths in the time-resolved fluorescence spectra (Figure 1B), which were deconvoluted successfully into the four KaiC phosphorylation states (Figure 1C) as confirmed by a reasonable fit to FI(340) (Figure 1A). Significant differences in FI of approximately 17% were observed during transitions from KaiC-WTS/pT to KaiC-WTpS/pT as well as from KaiC-WTpS/pT to KaiC-WTpS/T (Figure 1C, inset). This demonstrates that clear structural changes of KaiC occur during the auto-dephosphorylation reaction.
Figure 1.
Tracking the KaiC auto-dephosphorylation reaction of KaiC-WT using time-resolved fluorescence spectroscopy. (A) Time course of Trp fluorescence (open circles) at 340 nm compared with the relative abundance of KaiC-WTS/pT (red circles), KaiC-WTpS/pT (blue triangles), KaiC-WTpS/T (orange diamonds), and KaiC-WTS/T (green squares). The red line represents a global fit of Equation (1) to FI(340) using pre-determined concentration profiles of the four phosphorylation states (see Materials and methods). (B) Time-resolved fluorescence spectra of Trp excited at 295 nm for KaiC-WT. For clarity, only 6 of the 93 time points are displayed. (C) Trp fluorescence spectra deconvoluted into four phosphorylation states of KaiC-WT. The inset shows emission maxima and relative FI of the deconvoluted spectra. See also Supplementary Figure S3.
Tracking global shape changes in KaiC hexamer
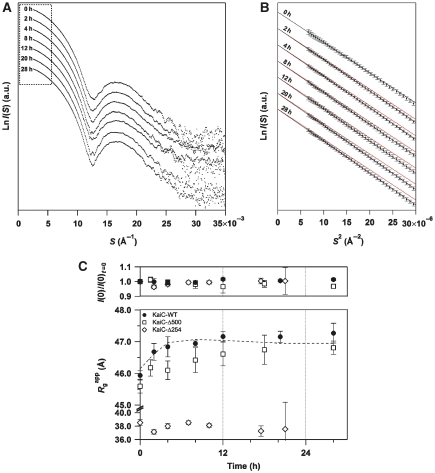
Structural ticking of KaiC in solution was further characterized by tracking its auto-dephosphorylation reaction using time-resolved SAXS. Concomitantly with the shift in the phosphorylation states (Supplementary Figure S1), X-ray scattering from KaiC-WT revealed a clear sign of structural transition (Figure 2A), which was best verified by the time evolution of the slope in the Guinier plot (Figure 2B). The slope related to the particle shape of KaiC became steeper in a time-dependent manner (from top to bottom in Figure 2B), relative to the reference red lines generated by longitudinal shifting of the initial slope. The apparent radius of gyration (Rgapp) and the forward scattering intensity, I(0), were determined from the slope and intercept of the linear fits, respectively (Figure 2B), and are each plotted in Figure 2C as a function of time. Whereas the relative molecular mass estimated from I(0) was nearly constant for 30 h (upper panel in Figure 2C), KaiC-WT became less compact as confirmed by a gradual increase in Rgapp from 45.9 to 47.2 Å (lower panel in Figure 2C). The gradual increase in Rgapp while I(0) remains constant is indicative of global changes in the shape of the KaiC hexamer during the auto-dephosphorylation reaction.
Figure 2.
Tracking the KaiC auto-dephosphorylation reaction using time-resolved SAXS. (A) Temporal evolution of the entire SAXS curve. Natural logarithm of scattering intensity (LnI(S)) is plotted against scattering angular momentum (S). The plots from top to bottom represent the measurements at 0, 2, 4, 8, 12, 20, and 28 h, respectively (see also Supplementary Figure S1). Each plot is longitudinally shifted for clarity of presentation. The region surrounded by the dotted box is expanded as a Guinier plot in Figure 2B. (B) Temporal evolution of Guinier plots. Natural logarithm of scattering intensity (LnI(S)) is plotted against squared scattering angular momentum (S2). Each black line shows the linear fit to LnI(S) using the S range from 0.0026 Å to Smax <1.3/2πRg. The slope, which relates to the particle shape of KaiC, becomes steeper relative to the reference line in red (generated by shifting the linear fit at 0 h longitudinally to facilitate detection of changes) in a time-dependent manner (from top to bottom). Rgapp and I(0) were determined from the slope and intercept, respectively, of the linear fit using Equation (2), and were plotted in Figure 2C as a function of time. At each time point, the SAXS curve was independent of the KaiC concentration, indicating little inter-particle interference in the measured range (0.9–1.4 mg ml−1). (C) Kinetic traces of apparent radius of gyration (Rgapp) and forward scattering intensity (I(0)) for KaiC-WT (circles), KaiC-Δ500 (squares), and KaiC-Δ254 (diamonds). The broken line denotes reproduced results using Equation (3), Ai(t) in Supplementary Figure S1, and the Rg values of a mutant set (KaiCA/E, KaiCD/E, KaiCD/T*, and KaiCS/A*) (Figure 3C). The results are presented as mean±s.d. from three or more independent experiments.
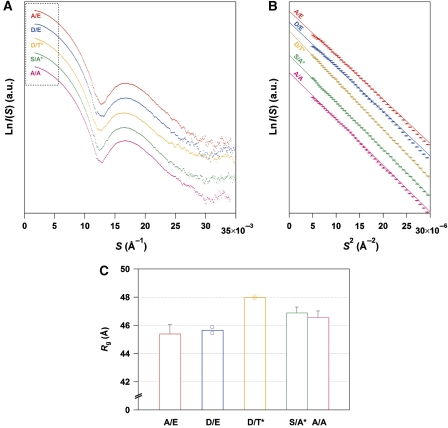
To determine the key step by which the KaiC hexamer undergoes structural changes, we measured the SAXS curves of KaiC mutants designed to mimic its phosphorylation state. Although the entire SAXS curve of each mutant was virtually indistinguishable from the others (Figure 3A), the difference in Rg among the mutants could be clearly ascertained by the slopes in the Guinier plot (Figure 3B and C). Close structural mimicking was validated by an agreement between the experimental time course of Rgapp for KaiC-WT (filled circles in lower panel of Figure 2C) and a simulation (broken line) using the Rg values of the phospho-mimicking mutants (Figure 3C). This agreement also implies that subunit exchange between two KaiC hexamers (Kageyama et al, 2006) will make only a limited contribution, if any, to the observed increase in the Rg values. A substantial difference of 2.3±0.4 Å was observed between the KaiCpS/pT-mimicking mutant KaiCD/E and the KaiCpS/T-mimicking mutant KaiCD/T* (the asterisk indicates dephosphorylation induced by incubation for 4 h at 30°C) (Nishiwaki et al, 2007). The 5% increase in Rg indicates the occurrence of a large conformational change in the KaiC hexamer because of its intrinsically quasi-spherical shape (Supplementary Figure S2; Supplementary Table S1; Supplementary Experimental Procedures). While the value of Rg increased by 2 Å in nearly one step from KaiCpS/pT to KaiCpS/T (Figure 3C), the value of FI increased by approximately 17% at each step from KaiCS/pT to KaiCpS/T (Figure 1C, inset). These fluorescence and SAXS observations indicate local changes in the environments of the Trp residues for each transition from KaiCS/pT to KaiCpS/T, and a global change in the shape of the KaiC hexamer during the transition from KaiCpS/pT to KaiCpS/T.
Figure 3.
SAXS curves of phospho-mimicking KaiC mutants. (A) Comparisons among entire SAXS curves at infinite dilution. Natural logarithm of scattering intensity (LnI(S)) is plotted against scattering angular momentum (S). The plots from top to bottom represent the SAXS curves of KaiCA/E (red), KaiCD/E (blue), KaiCD/T* (orange), KaiCS/A* (green), and KaiCA/A (magenta), respectively. Each plot is longitudinally shifted for clarity of presentation. The region surrounded by the dotted box is expanded as a Guinier plot in Figure 3B. (B) Comparisons among Guinier plots at infinite dilution. Natural logarithm of scattering intensity (LnI(S)) is plotted against squared scattering angular momentum (S2). Each thin line shows the linear fit to LnI(S) using the S range from 0.0022 Å to Smax <1.3/2πRg. The slope for KaiCD/T* is much steeper than those for other KaiC mutants, demonstrating a less compact conformation with a larger Rg. (C) Radius of gyration (Rg) of phospho-mimicking KaiC mutants at infinite dilution. KaiCD/T* and KaiCS/A* mutants were incubated for 4 h at 30°C to mimic KaiC-WTpS/T and KaiC-WTS/T, respectively (Nishiwaki et al, 2007). For KaiCD/E and KaiCD/T* mutants, results from two independent measurements are plotted as open circles together with their mean as bars. For other mutants, results are presented as mean±s.d. from three or more independent experiments. See also Supplementary Figure S2 and Supplementary Table S1.
To identify the structural unit responsible for the transition of the KaiC hexamer, we recorded the time course of Rgapp for the truncated KaiC mutants. While KaiC-Δ500 lacking the C-terminal tail displayed a gradual increase in Rgapp similar to that observed for KaiC-WT, KaiC-Δ254 lacking both the C2 domain and C-terminal tail showed time-independent Rgapp values (lower panel of Figure 2C). The gradual increase in Rgapp for KaiC-Δ500 relative to that for KaiC-WT is attributable to a slower and limited accumulation of KaiC-Δ500pS/T during the auto-dephosphorylation reaction (Supplementary Figure S1). The slightly smaller Rg value for KaiC-Δ500 than that for KaiC-WT is qualitatively consistent with compaction upon truncation of the C-terminal tails. Furthermore, KaiC-W92F retained the same relaxation of FI as observed for KaiC-WT (Supplementary Figure S3), suggesting that W331 and W462 in the C2 domain are responsible for the higher FIs in KaiCpS/T and KaiCS/T. These SAXS and fluorescence observations suggest that the C2 ring undergoes a drastic structural change in a stepwise manner compared with the C1 ring.
Visualizing expansion and contraction motions of C2 ring
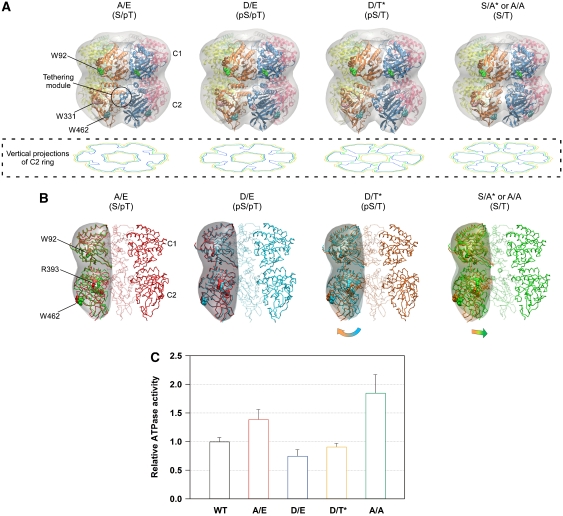
On the basis of the observations above, we constructed low-resolution SAXS models of the KaiC hexamer by rigid-body refinement of a known X-ray crystal structure (Pattanayek et al, 2006) against the SAXS curves of the phospho-mimicking KaiC and truncated KaiC-Δ254 mutants under a constraint of P6 symmetry (Figure 4; Supplementary Figure S4; Supplementary Experimental Procedures). As shown in vertical projections of the C2 ring (Figure 4A), the C2 domains that interact with each other in both KaiCS/pT and KaiCpS/pT are rearranged outward in both KaiCpS/T and KaiCS/T so as to be more loosely packed. The consequences of this structural transition are the expansion of the C2 ring and the disruption of some of the interactions between the two adjacent protomers (Figure 4B). The enlarged radius of the C2 ring is consistent with the increase of Rg in KaiCpS/T (Figure 3C). The loose protomer–protomer contact is expected to cause partial hydration of W331 in KaiCpS/T, as suggested by the subtle red shift of the fluorescence emission (Figure 1C, inset) and the lower fluorescence quenching by the surrounding amino acids (Figure 1C). The size of the expanded C2 ring is partly reduced in the transition from KaiCpS/T to KaiCS/T by a slight inward rearrangement of the C2 domains (Figure 4B), and is subsequently minimized in the transition from KaiCS/T to least fluorescent KaiCS/pT by a close repacking of the C2 domains (Figure 4A). These models demonstrate a structural change in the KaiC hexamer, which occurs through expansion and contraction motions of the C2 ring.
Figure 4.
Expanding and contracting motions of the C2-ring coupled to the ATPase activity of KaiC. (A) Low-resolution models of the KaiC hexamer. The protomer–protomer arrangement of the known X-ray crystal structure of KaiC was refined against the SAXS data under the assumption of P6 symmetry (see also Supplementary Figure S4). Smooth envelopes were calculated at 25 Å resolution to visualize molecular shapes using the SITUS software package (Wriggers and Chacon, 2001). Residues displayed as space-filling models indicate the approximate positions of three Trp residues, W92 (green), W331 (pink), and W462 (cyan). The potential tethering module is highlighted by a circle. The contour image below each model is the vertical projection (voxel spacing of 5 Å) of the C2 ring (low: orange, medium: green, high: blue). (B) Cross-sectional view of the KaiC models along the P6 symmetric axis. Panels from left to right correspond to KaiCS/pT (red), KaiCpS/pT (cyan), KaiCpS/T (orange), and KaiCS/T (green). To visualize the changes in the domain orientation, one protomer of the former phosphorylation state is superimposed upon one protomer of each model using the C1 ring. Smooth envelopes at a 25 Å resolution were drawn only for the compared protomers. Residues displayed as space-filling models point to approximate positions of W92, W462, and R393 in KaiC. (C) ATPase activity of phospho-mimicking KaiC mutants. The rates of ATP hydrolysis are plotted as relative to that of KaiC-WT at 30°C. As KaiC-WT includes four different phosphorylation states during the assay (e.g. Figure 1A), its ATPase activity set to unity is intermediate between the lowest KaiCD/E activity (0.75±0.11) and the highest KaiCA/A activity (1.85±0.32). Data for KaiCA/A and KaiCD/E are taken from Terauchi et al (2007). The results are presented as mean±s.d. from five or more independent experiments.
ATPase interlocked with dynamic motions of C2 ring
A correlation between the dynamic motion of the C2 ring with the ATPase activity was inspected by measuring steady-state ATPase activities for phospho-mimicking KaiC mutants (Figure 4C). The ATPase activity is elevated in the transition from KaiCpS/T to KaiCS/T, as supported by the higher activity of the KaiCS/T-mimicking mutant KaiCA/A than that of the KaiCpS/T-mimicking mutant KaiCD/T*. The elevation of the ATPase activity is accompanied by the partial C2-ring contraction resulting from the inward rearrangement of the C2 domain (Figure 4B). The ATPase activity is then restrained in each phosphorylation step from KaiCS/T, and is eventually minimized in the KaiCpS/pT-mimicking mutant KaiCD/E including the compact C2 ring. Interestingly, the ATPase activity of the KaiCpS/T-mimicking mutant KaiCD/T* was found to be similar to that of the KaiCpS/pT-mimicking mutant KaiCD/E, suggesting that the ATPase activity is less sensitive to the expansion of the C2 ring.
KaiCpS/T with an expanded C2 ring as a key state of in vitro oscillation
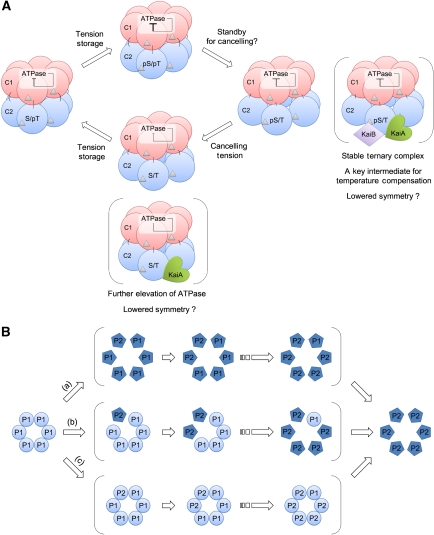
Another notable finding from the present structural models is the presence of a structural unit tethering the C2 domains in an appropriate configuration. At least in solution, the C2 domains in KaiC are marginally arranged in the hexameric configuration on the scaffolding C1 ring, as demonstrated by their inability to form a hexameric C2 ring in the absence of the C1 domains (Hayashi et al, 2006). In the SAXS-based models (Figure 4A), a structural region consisting of an N-terminus of α8 helix and of α9 helix remains in close proximity to a region consisting of β4-α3 and β5-α5 loops in the adjacent C2 protomer throughout the reaction cycle. These regions likely function as a potential module that tethers the C2 domains to a hexameric configuration by adjusting the relative distance and orientation of the neighbouring C2 protomer (highlighted by the circle in Figure 4A). In fact, Arg393 is one of the amino-acid residues involved in the potential tethering module, and its substitution by Cys (KaiC-R393C) resulted in a selective destabilization of KaiC-R393CpS/T during its auto-dephosphorylation (Figure 5A) as compared with KaiC-WT (Figure 1A). Considering the marginal stability and dynamic nature of the C2 ring, the protomer–protomer interaction through the potential tethering module should be important not only in keeping the C2 domains swung out in KaiCpS/T, but also in maintaining a stable population of KaiCpS/T.
Figure 5.
Modulation of the temperature-compensation mechanism in KaiC-R393C. (A) Relative abundance of KaiC-R393CS/pT (red circles), KaiC-R393CpS/pT (blue triangles), KaiC-R393CpS/T (orange diamonds), and KaiC-R393CS/T (green squares) during the auto-dephosphorylation reaction. (B) Temperature dependence of the relationship between the ATPase activity of KaiC alone (in the absence of KaiA and KaiB) and period/frequency of in vitro phosphorylation cycle. The ATPase activities of KaiC-WT (circles) and KaiC-R393C (squares) at 25°C (blue), 30°C (cyan), 35°C (green), 40°C (orange), and 45°C (red) were plotted relative to that of KaiC-WT at 30°C. The inset depicts the expanded view for KaiC-WT. (C, D) Relative abundance of KaiC-WT and KaiC-R393C, respectively, during the phosphorylation cycle at 25°C (blue), 30°C (cyan), 35°C (green), 40°C (orange), and 45°C (red). The curves were plotted as a function of phase angle. The phase angle of π corresponds to peaks of KaiC-WTS/pT or KaiC-R393CS/pT (see also Supplementary Figure S5). (E) Bioluminescence rhythms of KaiC-WT and KaiC-R393C at 25°C (blue), 30°C (black), and 35°C (red). For clarity of comparison of the temperature dependency of the period, each bioluminescence rhythm is shifted horizontally so that the first peak matches 0 h.
It should be noted that a lower population of KaiCpS/T results in a modulation of the temperature-compensation mechanism embedded in KaiC. As shown in Figure 5B, the in vitro phosphorylation rhythm of KaiC-WT is temperature compensated (Supplementary Figure S5A and C). Whereas the populations of KaiC-WTS/pT and KaiC-WTpS/pT at the phase angle of 3π/2 were found to increase as the temperature was increased, the KaiC-WTS/T population decreased so as to maintain the KaiC-WTpS/T population essentially unaltered (Figure 5C). On the other hand, the phosphorylation cycle of KaiC-R393C, which repeats at a higher frequency than that of KaiC-WT, was found to be accelerated in a temperature-dependent manner (Figure 5B; Supplementary Figure S5B and D). This was also confirmed in vivo (Figure 5E). A time-averaged population of the KaiC-R393CpS/T decreased to the trough level of KaiC-WTpS/T (Figure 5C and D). The ATPase activity of KaiC-R393C is highly unlikely to be a source of the observed temperature dependency of the phosphorylation cycle, because it is compensated in a wide temperature range from 25 to 45°C, as is KaiC-WT (Figure 5B). Rather, the enhanced temperature dependency is attributed to the reduced population of KaiC-R393CpS/T resulting from structural frustration near the potential tethering module.
Direct observation of ticking KaiC
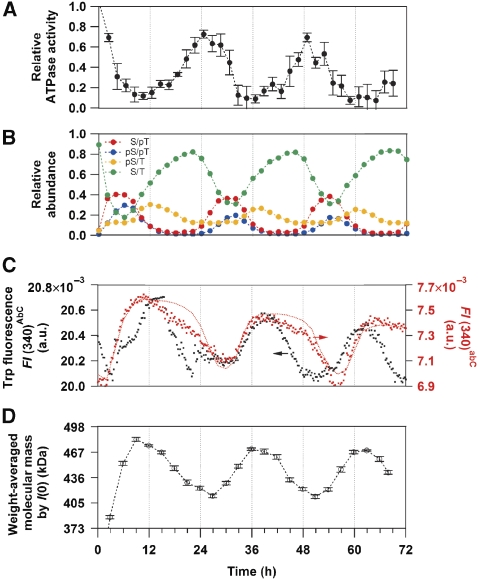
We discovered that the Trp fluorescence of a solution containing KaiA, KaiB, and KaiC exhibited a circadian oscillation (FI(340)AbC, black circles in Figure 6C). One possibility is that the oscillation comes from KaiA, as it possesses one Trp residue intrinsically in its N-terminal domain (W10), whereas KaiB has no Trp residues. Another possibility is simply that the oscillation originates from KaiC through the interconversion among the four phosphorylation states with the different FI(340) (Figure 1C). The latter possibility is likely, because the peaks and troughs of FI(340)AbC shown in Figure 6C are matched to the accumulation of the more fluorescent KaiCpS/T and the less fluorescent KaiCS/pT, respectively (Figures 1C and 6B). This interpretation is further validated by measuring the Trp fluorescence of a solution containing KaiA-W10F, KaiB, and KaiC (FI(340)abC). A removal of the fluorescent contribution from KaiA greatly diminished the FI(340) (see legend and right axis in Figure 6C). However, both the oscillatory period and amplitude of the fluorescence signal were not affected significantly by W10F substitution in KaiA (red circles in Figure 6C), clearly demonstrating that the oscillatory component of FI(340)AbC mainly comes from the expansion and contraction motions of the C2 ring in KaiC.
Figure 6.
Circadian ticking of KaiC in relation to the temporal patterns of ATPase activity, phosphorylation, and assembly/disassembly in the presence of KaiA, KaiB, and KaiC at 30°C. (A) Relative ATPase activity of KaiC. (B) Relative abundance of the phosphorylated/dephosphorylated states of KaiC. (C) Trp fluorescence intensities at 340 nm using KaiA-WT (FI(340)AbC, black circles) or KaiA-W10F mutant (FI(340)abC, red circles). Given that the Trp fluorescence from all molecules of KaiA-WT remained unchanged during the cycle, their contribution to FI(340)AbC in Figure 6C is calculated to be about 14.7 × 10−3. The red dotted line represents a result simulated by using Equation (1), Ai(t) upon coincubation of KaiA-W10F with KaiB and KaiC-WT, and the deconvoluted FI(340) shown in Figure 1C (plotted with a slight baseline offset; FI(t,340)−3.59 × 10−6 t+1.64 × 10−3). (D) Weight-averaged molecular mass estimated from forward scattering intensity, I(0), taken from Akiyama et al (2008).
Interestingly, the peak of FI(340)AbC is related to the timing of KaiC-WTpS/T accumulation (Figure 6B and C). At the same time, the peak time of FI(340)AbC closely coincides with that of I(0) (Akiyama et al, 2008) and of immunoprecipitation analysis (Nishiwaki et al, 2007), each of which is indicative of the accumulation of ternary Kai complexes (Figure 6D). These correlations suggest that KaiA and/or KaiB are recruited to bind to the expanded C2 ring of fluorescent KaiCpS/T, consistent with low-resolution models of Kai complexes (Akiyama et al, 2008). The rhythmic ATPase activity of KaiC was approximately antiphase to FI(340)AbC (Figure 6A), suggesting that the lower ATPase activity is a fundamental property of KaiCpS/T irrespective of the presence (Figure 6A) or absence of KaiA and KaiB (Figure 4C).
Discussion
To our knowledge, this is the first direct evidence for the dynamic conformational transition of the KaiC hexamer in solution. The C2 ring of KaiC is robustly expanded and then contracted along the programmed sequence of the KaiC phosphorylation cycle. These structural changes have not been observed in the recent X-ray crystal structures of phospho-mimicking KaiC mutants (Pattanayek et al, 2009). We believe, however, that this apparent discrepancy between in solution and in crystalline phases should be an inevitable consequence of confining the KaiC hexamer carrying the dynamic C2 domains into the crystal lattice. In fact, the previous biochemical study suggests that the C2 ring of the KaiC hexamer is marginally stabilized in solution (Hayashi et al, 2006). Furthermore, the dynamic shape transition of the KaiC hexamer can be traced in real time by measuring oscillatory Trp fluorescence of a solution containing KaiA, KaiB, and KaiC (Figure 6C).
The observation of rearrangement of the ring architecture is not unique to KaiC, but is often confirmed in other AAA/AAA+ proteins with SAXS. Valosin-containing protein p97, a homohexameric ATPase belonging to the AAA family, undergoes massive rearrangements of the protomers to alter overall ring shape during the ATP hydrolysis cycle (Davies et al, 2005). The AAA+ ATPase domain of NtrC1 forms a heptameric ring, and both the pore size and ring thickness vary in a manner that is dependent upon the status of bound nucleotides (Chen et al, 2007). Repeated reorganizations of the ring structure are thus a fundamental property of AAA/AAA+ ATPases including KaiC. However, the magnitude of the structural change in KaiC (up to 4% change in Rg) is not as large as those of these specific examples (up to 9% difference in Rg for p97) (Davies et al, 2005). The relatively modest transitions of the C2 ring may be related to the functional uniqueness of KaiC. Most of the ATPases have likely become adapted to diverse biological requirements by converting the chemical energy of ATP hydrolysis into mechanical work carried out on substrates. On the other hand, the substrate for KaiC is likely to be KaiC itself. It has been proposed that the ATP hydrolysis energy is stored on the KaiC hexamer to restrain its own ATPase activity and to maintain slow and regular oscillatory reactions (∼15 ATP KaiC−1 day−1) (Terauchi et al, 2007). The relatively modest but notable conformational transition of the C2 ring is seemingly compatible with the restrained ATPase activity of KaiC.
The potential tethering module has a critical function during the dynamic structural transition of KaiC, especially in holding the C2 domains swung out away from the KaiCpS/T hexamer. Our results clearly suggest that the expanded C2 ring in KaiCpS/T serves as a timing cue for recruiting KaiA and KaiB to the KaiCpS/T hexamer (Figure 6). As confirmed by the R393C mutation, undesirable structural frustrations forced to the potential tethering module of KaiCpS/T results in a considerable reduction of the KaiCpS/T pool (Figure 5D), thereby modulating the temperature-compensation mechanism (Figure 5B and E). We previously reported that the ATPase activity was fully temperature compensated in KaiC alone, but was slightly affected by temperature when both KaiA and KaiB were also present (Terauchi et al, 2007). These observations are presumably a consequence of attenuating the temperature sensitivity of assembly/disassembly processes by coupling them to the robustly temperature-compensated ATPase activity through the expanded C2 ring of KaiCpS/T. Although the crosstalk between the ATPase activity in the C1 ring and the phospho-dependent assembly/disassembly in the C2 ring is seemingly bidirectional in KaiCpS/T, a determinant of the temperature-compensated period is encapsulated in the ATPase activity of KaiC (Terauchi et al, 2007).
The current observations provide clues to the puzzling relationship between the ATPase activity and phosphorylation of KaiC. The ATPase activity of KaiC is elevated by a slight inward rearrangement of the C2 domains during the transition from KaiCpS/T to KaiCS/T (Figure 4). The C2-ring contraction may perturb the ATP hydrolysis in the C2 ring, but the contribution of the perturbation to the overall ATPase activity should be relatively small and limited, if any, because the ATPase activity of the C2 domain is intrinsically lower than that of the C1 domain (Terauchi et al, 2007; Murakami et al, 2008). According to the X-ray crystallographic structure (Pattanayek et al, 2004), one end of a polypeptide loop connecting the C1 and C2 domains is anchored to an ATP-binding region of the C1 ring. Therefore, rearrangements of catalytic side-chains associated with the ATP hydrolysis in the C1 ring can be synchronized with the contraction of the C2 ring, and vice versa, through a mechanical pull-up/down of the loop (Figure 7A). A pair of catalytic Glu residues (E78 and E79) is one of the structural units regulating the ATPase activity in the C1 domains (Murakami et al, 2008), and R226 might function as an Arg finger bringing a water molecule into the active site (Kagawa et al, 2004). These critical residues would be repositioned between more and less active configurations during the hydrolysis cycle, and the rate of repositioning would be adjusted dependent on the strength of the tension imposed on the KaiC hexamer. It is worth mentioning that this kind of intramolecular regulation of the ATPase activity is linked to physiological phenomena such as the closure of the cell division gate in cyanobacteria (Dong et al, 2010).
Figure 7.
Schematic drawing of interconversions of the KaiC hexamer in solution. (A) Proposed model for circadian ticking of the KaiC hexamer. The C1 and C2 domains in each protomer of KaiC are schematically drawn as red and blue spheres, respectively. Silver triangles represent bound nucleotides. Determinant of the temperature-compensated period is encapsulated in the auto-inhibitory ATPase of KaiC (Terauchi et al, 2007), which is interlocked with expansion and contraction motions of the C2 ring. The models in the parenthesis represent the KaiC hexamers interacted with KaiA and/or KaiB. KaiC-WTpS/T complexed with KaiA and KaiB is likely a key state of temperature compensation of assembly and disassembly processes. The ATPase activity could be further elevated by lowering the symmetry of the KaiC-WTS/T hexamer upon binding of KaiA to its C2 domains and C-terminal tails. (B) Schematic drawing of potential asymmetric transitions of KaiC hexamers. A hypothetical hexamer composed of circular protomers in a hypothetical phosphorylation state ‘P1' is converted to a hypothetical hexamer composed of pentagon protomers in a hypothetical phosphorylation state ‘P2' via three different pathways. The circular protomers are different in shape/configuration from the pentagon protomers. (a) Cooperative conformational switching triggered by the first transition of the phosphorylation state in a single protomer. (b) Continuous transition through multiple states with asymmetric shapes. (c) Cooperative conformational switching triggered by the last conversion of the phosphorylation state in a single protomer.
In contrast, the ATPase activity of KaiC is less sensitive to the C2-ring expansion in the transition from KaiCpS/pT to KaiCpS/T (Figure 4C). These contrasting observations suggest that a mechanism regulating the ATPase activity is not as simple as an on/off (contraction/expansion) switching process. Once elevated, the ATPase activity of KaiC is gradually but monotonously restrained as the phosphorylation proceeds. This observation seems to be compatible with the hypothesis that the energy released upon ATP hydrolysis is stored in KaiC as structural tension, which downregulates its own ATPase activity (Terauchi et al, 2007). In this context, KaiCpS/pT can be interpreted as a maximally tensed state. The temporal expansion of the C2 ring in KaiCpS/T, just before the ATPase elevation, may represent a standby process for cancelling structural tensions. Our results indicate that the auto-inhibitory mechanism proposed for KaiC better describes the structural ticking observed here in solution (Figure 7A).
To present the simplest model reconciled with the current observation, potential asymmetries of phosphorylation and protomer configuration within the KaiC hexamer are not included in Figure 7A. Some of the reaction steps in Figure 7A might be subdivided into elementary reactions as schematically shown in Figure 7B. However, both the fluorescence and SAXS observations of the auto-dephosphorylation reaction of KaiC alone could be interpreted as the interconversion among only the four different symmetric states (Figures 1A and 2C). We can speculate that the asymmetric species is rather short lived at least in the absence of KaiA and KaiB ((b) in Figure 7B), otherwise, that the KaiC hexamer is maintained roughly symmetric by an as yet unknown cooperative switching mechanisms ((a) and (c) in Figure 7B).
The asymmetry in the KaiC hexamer could be more frequent in the presence of KaiA and KaiB, given non-symmetrical interactions among the Kai proteins. A sign of the asymmetry was detected in the late stage of the dephosphorylation process (24 and 48 h in Figure 6C), in which FI(340) simulated by using only four symmetric species (red dotted line, see legend of Figure 6) slightly deviated from the experimental FI(340)abC (red circles). A plausible explanation is the presence of high- and low-fluorescent states of KaiC-WTS/T. Even the identity of the phosphorylation state, an asymmetric tension imposed on the KaiC hexamer by binding of KaiA and/or KaiB, could result in the different fluorescence intensity. In fact, our previous studies suggest the progressive formation of KaiA:KaiC complexes at incubation time of approximately 24 and 48 h (Kageyama et al, 2006; Nishiwaki et al, 2007; Akiyama et al, 2008). In addition, the previous study indicated that the ATPase activity of KaiCA/A is further elevated in the presence of KaiA (Murakami et al, 2008). The discrepancy is thus an indication of the asymmetry in the KaiC-WTS/T hexamer with the elevated ATPase by binding of KaiA to its C2 domains and C-terminal tails. Further visualization of the ATPase activity in relation to inter-ring and inter-domain interactions at higher time and spatial resolution is in progress with the objective of understanding the coupled ATPase and phosphorylation activities in KaiC.
Materials and methods
Expression and purification of Kai proteins
Recombinant Kai proteins were expressed and purified as previously described (Nishiwaki et al, 2004; Akiyama et al, 2008). Kai proteins and molecular mass standards for SAXS were prepared as previously reported (Akiyama, 2010).
KaiC phosphorylation cycle
The KaiC phosphorylation cycle was reconstructed in vitro as previously described (Nakajima et al, 2005; Nishiwaki et al, 2007). In the case of the ATPase, phosphorylation, fluorescence, and SAXS measurements shown in Figure 6, the concentration of each Kai protein was increased three-fold to improve the signal-to-noise ratio in a buffer containing 50 mM Tris, 150 mM NaCl, 0.5 mM EDTA, 5 mM MgCl2, and 3 mM ATP at pH 8.0. The period length of the rhythm was estimated by a non-linear least-squares fitting of a cosine function to each time course of the KaiC phosphorylation ratio (Nakajima et al, 2005).
In vivo bioluminescence assay
The bioluminescence assay and analysis were performed as previously described (Ishiura et al, 1998). Cyanobacterial cells carrying the kaiBC-reporter cassette were cultured on BG-11 solid medium in LL at 36.5 μE m−2 S−1 at 30°C for 3 days. After a dark treatment for 12 h, cells were then transferred to LL at 25, 30, or 35°C. The bioluminescence profiles were monitored using a photomultiplier tube detector.
Fluorescence spectroscopy
Fluorescence emission spectra from Trp residues were collected every 1.0 nm with a 2 s response time and a scan speed of 100 nm min−1 using an excitation wavelength of 295 nm (JASCO, FP-6500). Each sample solution prepared on ice (approximately 1°C) was transferred to an optical cuvette at 30±0.1°C controlled by a Peltier device. The fluorescence signals recorded at emission wavelength λ at time t were normalized by the molar concentration of KaiC to obtain the normalized fluorescence intensity, FI(t, λ).
Under dilute solution conditions, FI(t, λ) is expressed as a linear combination of the contributions from all fluorescent species (i) in the system. Thus, FI(t, λ) during the KaiC auto-dephosphorylation reaction (i=S/pT, pS/pT, pS/T, S/T) is expressed by,
where Ai(t) is the relative abundance of each phosphorylation state. As shown in Figure 1A, Ai(t) can be determined experimentally by sodium dodecyl sulphate–polyacrylamide gel electrophoresis followed by densitometry analysis (Nishiwaki et al, 2007). Thus, FIi(λ) of each phosphorylation state can be determined by globally fitting Equation (1) to a data set of time-resolved fluorescence spectra at each λ.
Small-angle X-ray scattering
SAXS data were measured at RIKEN Structural Biology Beamline I (BL45XU) (Fujisawa et al, 2000; Akiyama et al, 2008) using a charge-coupled device equipped with an X-ray image intensifier (Ito et al, 2005). A series of diluted samples was prepared in the range of 1–3 mg ml−1, and then subjected to SAXS measurements. The recorded SAXS curves were normalized by exposure time, intensity of the incident X-ray (0.9 Å), and protein concentration to obtain the scattering curve, I(S), where S=2sinθ/λ, 2θ is the scattering angle, and λ is the wavelength of the X-ray. The innermost portion of I(S) was fitted under the Guinier approximation (Guinier and Fournet, 1955) to the following equation:
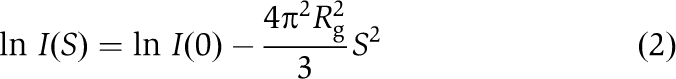 |
where I(0) and Rg are the forward scattering intensity (S=0) and the radius of gyration, respectively. A series of I(0) or Rg values determined at different concentrations was extrapolated to an infinite dilution as previously described (Zimm, 1948; Akiyama et al, 2008).
The forward scattering intensity, I(0), was unchanged during the auto-dephosphorylation reaction of KaiC-WT (upper panel of Figure 2C), ensuring a constant molecular mass for the KaiC hexamer throughout the phosphorylation cycle. In this case, the apparent radius of gyration, Rgapp, under dilute solution conditions is expressed by
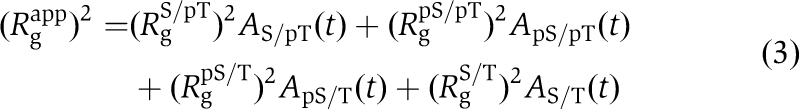 |
where Ai(t) is the same as in Equation (1), and Rgi is the radius of gyration for the respective phosphorylation state (i=S/pT, pS/pT, pS/T, S/T). The broken line in the lower panel of Figure 2C was calculated by substituting the concentration profiles of KaiC-WT (Supplementary Figure S1) for Ai(t) and by substituting the Rg values of the phospho-mimicking mutants (Figure 3C) for Rgi.
Shape reconstructions
Model building of the KaiC hexamer was performed with a rigid-body refinement algorithm implemented in the program, BUNCH (Petoukhov and Svergun, 2005). Each protomer of KaiC is treated as rigid C1 (from 14 to 252) and C2 bodies (from 256 to 497) connected by a flexible hinge. The structures of the two rigid bodies, whose coordinates were taken from a known X-ray crystal structure of KaiC (Pattanayek et al, 2006), were kept unchanged during the refinements. The hinge (from 253 to 255) and terminal regions (from 1 to 13 and from 498 to 519), which are partly missing in the X-ray crystal structure, were presented as dummy residues fulfilling a protein-like geometry. The optimum arrangement of the C1 and C2 domains was refined under an assumption of P6 symmetry against the experimental SAXS curves of the phospho-mimicking mutant (KaiCA/E, KaiCD/E, KaiCD/T*, or KaiCS/A*) and the truncation mutant KaiC-Δ254, so that the resulting model satisfied the two curves simultaneously (Supplementary Experimental Procedures). Multiple reconstructions were performed independently to confirm the reproducibility of the solution. The resulting models, scored with the DAMAVER package (Volkov and Svergun, 2003), were highly similar in shape. The most representative model is shown in Figure 4.
ATPase measurements
ATPase activity of KaiC was measured as described previously (Terauchi et al, 2007) using an ACQUITY UPLC system (Waters). ADP was separated from ATP on a BEH C18 column (2.1 × 50 mm, 1.7 μm) (Waters) at a flow rate of 0.8 ml min−1 with a mobile phase of 20 mM ammonium phosphate, 10 mM tetrabutylammonium hydrogen sulphate (pH 8.5) and 17% (v/v) acetonitrile. The ADP concentrations were calculated from their peak areas.
Supplementary Material
Acknowledgments
SA thanks Drs Kazuki Ito and Takaaki Hikima (RIKEN SPring-8 Center) for kindly supporting the SAXS experiments. The synchrotron radiation experiments were performed at BL45XU in the SPring-8 facility with the approval of RIKEN (Proposal No. 20080026, 20090044, and 20100071). Dr Y Murayama was supported by a fellowship from the Japan Society for the Promotion of Science to Young Scientists. This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan (22687010 to SA, and 19042011, 20570035, and 21023011 to KT), by PRESTO from the Japan Science and Technology Agency (to SA), and by CREST from the Japan Science and Technology Agency (to TK).
Footnotes
The authors declare that they have no conflict of interest.
References
- Akiyama S (2010) Quality control of protein standards for molecular mass determinations by small-angle X-ray scattering. J Appl Cryst 43: 237–243 [Google Scholar]
- Akiyama S, Nohara A, Ito K, Maéda Y (2008) Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell 29: 703–716 [DOI] [PubMed] [Google Scholar]
- Chen B, Doucleff M, Wemmer DE, De Carlo S, Huang HH, Nogales E, Hoover TR, Kondrashkina E, Guo L, Nixon BT (2007) ATP ground- and transition states of bacterial enhancer binding AAA+ ATPases support complex formation with their target protein, sigma54. Structure 15: 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies JM, Tsuruta H, May AP, Weis WI (2005) Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure 13: 183–195 [DOI] [PubMed] [Google Scholar]
- Dong GG, Yang Q, Wang Q, Kim YI, Wood TL, Osteryoung KW, van Oudenaarden A, Golden SS (2010) Elevated ATPase activity of KaiC applies a circadian checkpoint on cell division in Synechococcus elongatus. Cell 140: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Inoue K, Oka T, Iwamoto H, Uruga T, Kumasaka T, Inoko Y, Yagi N, Yamamoto M, Ueki T (2000) Small-angle X-ray scattering station at the SPring-8 RIKEN beamline. J Appl Cryst 33: 797–800 [Google Scholar]
- Golden SS, Ishiura M, Johnson CH, Kondo T (1997) Cyanobacterial circadian rhythms. Annu Rev Plant Physiol Plant Mol Biol 48: 327–354 [DOI] [PubMed] [Google Scholar]
- Guinier A, Fournet G (1955) Small-Angle Scattering of X-Rays. New York: John Wiley & Sons, Inc [Google Scholar]
- Hayashi F, Iwase R, Uzumaki T, Ishiura M (2006) Hexamerization by the N-terminal domain and intersubunit phosphorylation by the C-terminal domain of cyanobacterial circadian clock protein KaiC. Biochem Biophys Res Commun 348: 864–872 [DOI] [PubMed] [Google Scholar]
- Hayashi F, Suzuki H, Iwase R, Uzumaki T, Miyake A, Shen JR, Imada K, Furukawa Y, Yonekura K, Namba K, Ishiura M (2003) ATP-induced hexameric ring structure of the cyanobacterial circadian clock protein KaiC. Genes Cells 8: 287–296 [DOI] [PubMed] [Google Scholar]
- Ishiura M, Kutsuna S, Aoki S, Iwasaki H, Andersson CR, Tanabe A, Golden SS, Johnson CH, Kondo T (1998) Expression of a gene cluster kaiABC as a circadian feedback process in cyanobacteria. Science 281: 1519–1523 [DOI] [PubMed] [Google Scholar]
- Ito K, Kamikubo H, Yagi N, Amemiya Y (2005) Correction method and software for image distortion and nonuniform response in charge-coupled device-based X-ray detectors utilizing X-ray image intensifier. J Appl Phys 44: 8684–8691 [Google Scholar]
- Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T (2002) KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci USA 99: 15788–15793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa R, Montgomery MG, Braig K, Leslie AGW, Walker JE (2004) The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J 23: 2734–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama H, Nishiwaki T, Nakajima M, Iwasaki H, Oyama T, Kondo T (2006) Cyanobacterial circadian pacemaker: Kai protein complex dynamics in the KaiC phosphorylation cycle in vitro. Mol Cell 23: 161–171 [DOI] [PubMed] [Google Scholar]
- Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T (2003) KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J 22: 2127–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Saveliev SV, Xu Y, Stafford WF, Cox MM, Inman RB, Johnson CH (2002) Circadian clock protein KaiC forms ATP-dependent hexameric rings and binds DNA. Proc Natl Acad Sci USA 99: 17203–17208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Miyake A, Iwase R, Hayashi F, Uzumaki T, Ishiura M (2008) ATPase activity and its temperature compensation of the cyanobacterial clock protein KaiC. Genes Cells 13: 387–395 [DOI] [PubMed] [Google Scholar]
- Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyarna T, Kondo T (2005) Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science 308: 414–415 [DOI] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Kitayama Y, Terauchi K, Kiyohara R, Takao T, Kondo T (2007) A sequential program of dual phosphorylation of KaiC as a basis for circadian rhythm in cyanobacteria. EMBO J 26: 4029–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiwaki T, Satomi Y, Nakajima M, Lee C, Kiyohara R, Kageyama H, Kitayama Y, Temamoto M, Yamaguchi A, Hijikata A, Go M, Iwasaki H, Takao T, Kondo T (2004) Role of KaiC phosphorylation in the circadian clock system of Synechococcus elongatus PCC 7942. Proc Natl Acad Sci USA 101: 13927–13932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Mori T, Xu Y, Pattanayek S, Johnson CH, Egli M (2009) Structures of KaiC circadian clock mutant proteins: a new phosphorylation site at T426 and mechanisms of kinase, ATPase and phosphatase. Plos One 4: e7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayek R, Wang JM, Mori T, Xu Y, Johnson CH, Egli M (2004) Visualizing a circadian clock protein: crystal structure of KaiC and functional insights. Mol Cell 15: 375–388 [DOI] [PubMed] [Google Scholar]
- Pattanayek R, Williams DR, Pattanayek S, Xu Y, Mori T, Johnson CH, Stewart PL, Egli M (2006) Analysis of KaiA-KaiC protein interactions in the cyano-bacterial circadian clock using hybrid structural methods. EMBO J 25: 2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI (2005) Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys J 89: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust MJ, Markson JS, Lane WS, Fisher DS, O′Shea EK (2007) Ordered phosphorylation governs oscillation of a three-protein circadian clock. Science 318: 809–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terauchi K, Kitayama Y, Nishiwaki T, Miwa K, Murayama Y, Oyama T, Kondo T (2007) ATPase activity of KaiC determines the basic timing for circadian clock of cyanobacteria. Proc Natl Acad Sci USA 104: 16377–16381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita J, Nakajima M, Kondo T, Iwasaki H (2005) No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science 307: 251–254 [DOI] [PubMed] [Google Scholar]
- Volkov VV, Svergun DI (2003) Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst 36: 860–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wriggers W, Chacon P (2001) Using Situs for the registration of protein structures with low-resolution bead models from X-ray solution scattering. J Appl Cryst 34: 773–776 [Google Scholar]
- Xu Y, Mori T, Johnson CH (2003) Cyanobacterial circadian clockwork: roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J 22: 2117–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Mori T, Pattanayek R, Pattanayek S, Egli M, Johnson CH (2004) Identification of key phosphorylation sites in the circadian clock protein KaiC by crystallographic and mutagenetic analyses. Proc Natl Acad Sci USA 101: 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimm BH (1948) The scattering of light and the radial distribution function of high polymer solutions. J Phys Chem 16: 1093–1099 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.