Membrane curvature during peroxisome fission requires Pex11
Pex11p is required for peroxisome proliferation. This study demonstrates that the N-terminus of Pex11p forms an amphipathic helix that generates membrane curvature required for peroxisome fission.
Keywords: amphipathic helix, lipid composition, membrane curvature, peroxisome proliferation, Pex11
Abstract
Pex11 is a key player in peroxisome proliferation, but the molecular mechanisms of its function are still unknown. Here, we show that Pex11 contains a conserved sequence at the N-terminus that can adopt the structure of an amphipathic helix. Using Penicillium chrysogenum Pex11, we show that this amphipathic helix, termed Pex11-Amph, associates with liposomes in vitro. This interaction is especially evident when negatively charged liposomes are used with a phospholipid content resembling that of peroxisomal membranes. Binding of Pex11-Amph to negatively charged membrane vesicles resulted in strong tubulation. This tubulation of vesicles was also observed when the entire soluble N-terminal domain of Pex11 was used. Using mutant peptides, we demonstrate that maintaining the amphipathic properties of Pex11-Amph in conjunction with retaining its α-helical structure are crucial for its function. We show that the membrane remodelling capacity of the amphipathic helix in Pex11 is conserved from yeast to man. Finally, we demonstrate that mutations abolishing the membrane remodelling activity of the Pex11-Amph domain also hamper the function of full-length Pex11 in peroxisome fission in vivo.
Introduction
Peroxisomes are ubiquitous single membrane-bound organelles that have an important function in many metabolic pathways. The main functions of peroxisomes include β-oxidation of fatty acids and detoxification of hydrogen peroxide (van den Bosch et al, 1992). Moreover, peroxisomes are crucial for efficient biosynthesis of β-lactam antibiotics in filamentous fungi (Kiel et al, 2005, 2009; van den Berg et al, 2008; Sprote et al, 2009). In Penicillium chrysogenum, two enzymes of the penicillin biosynthetic pathway are localized to peroxisomes where they perform the final steps in the formation of this secondary metabolite (Muller et al, 1991; Lamas-Maceiras et al, 2006). The importance of peroxisomes is highlighted by the fact that peroxisomes are indispensable in mammals (Gould and Valle, 2000), plants (Schumann et al, 2003) and Trypanosomes (Guerra-Giraldez et al, 2002).
The maintenance of the peroxisome population during vegetative cell reproduction requires continuous multiplication of these organelles. Peroxisomes may form de novo from the endoplasmic reticulum (ER) or multiply by division of pre-existing organelles (Thoms and Erdmann, 2005; Hettema and Motley, 2009). In this latter mechanism, peroxisomes undergo extensive changes in the shape of their surrounding membrane. The current model predicts that the initial organelle elongation event is followed by the actual fission step, mediated by GTPases from the dynamin-related protein family (e.g. Dnm1, Vps1, Drp1) (Thoms and Erdmann, 2005).
Pex11 is a peroxisomal membrane protein and the first protein identified to be involved in peroxisome proliferation (Erdmann and Blobel, 1995; Marshall et al, 1995). In most organisms, the number and size of peroxisomes can be prescribed by modulation of the Pex11 protein levels (Fagarasanu et al, 2007). The molecular details of the function of Pex11 in peroxisome proliferation are, however, still largely unknown.
In general, maintenance of the unique shape of organelles is obtained by regulation of their membrane properties. One of the current models predicts that Pex11 is involved in peroxisome elongation/tubulation. Several mechanisms have been proposed for the induction and regulation of membrane curvature (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006). One of these is insertion of amphipathic α-helices into one leaflet of the lipid bilayer, thus generating membrane asymmetry, resulting in bending of the membrane (Drin and Antonny, 2009).
Here, we show that the N-terminus of Pex11 contains a conserved amphipathic helix, tentatively termed Pex11-Amph. We show that this α-helical motif can bind to membranes and alter the shape of liposomes with a lipid composition resembling that of the peroxisomal membrane, thereby causing extensive tubulation. Using directed mutagenesis, we demonstrate that the amphipathic properties of Pex11-Amph are crucial for the function of Pex11 in peroxisome fission in vivo.
Results
Pex11 contains a conserved N-terminal amphipathic helix
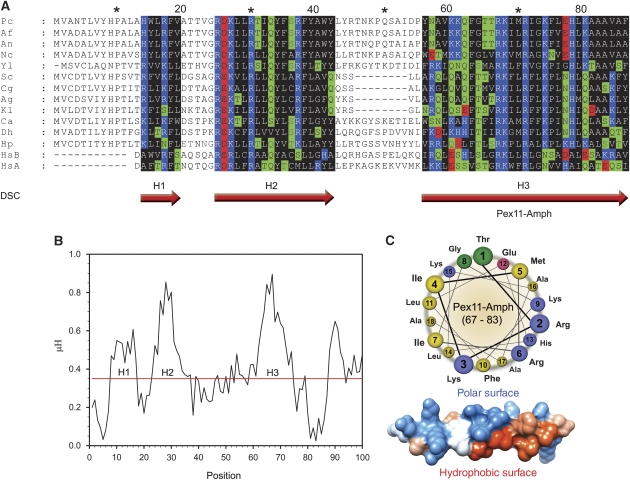
The analysis of the function of Pex11 in peroxisome proliferation was initiated by a sequence comparison of known Pex11 proteins for the presence of membrane deforming motifs. Using multiple sequence alignments of Pex11 proteins from different species, we identified an N-terminally conserved motif of approximately 25 residues, designated Pex11-Amph (H3 in Figure 1A). This motif is predicted to form an α-helix comprising of hydrophobic and polar (mainly positively charged) residues (Figure 1A). Analysis of the hydrophobic moment of the P. chrysogenum Pex11 N-terminus revealed three regions with amphipathic properties, of which Pex11-Amph is the most conspicuous one (Figure 1B). Helical wheel and 3D surface representations of Pex11-Amph confirm that this region indeed is predicted to contain hydrophobic and hydrophilic surfaces of approximately equal size (Figure 1C).
Figure 1.
Pex11 contains a conserved N-terminal amphipathic helix. (A) Sequence alignment of N-terminal regions of Pex11 proteins from various species. Putative α-helices were predicted using the DSC programme and are marked with red arrows (H1–H3). Residues in predicted α-helices are coloured based on the physico-chemical properties of amino acids as follows: hydrophilic, charged: D, E (red), K, R, H (blue); hydrophilic, neutral: S, T, Q, N (green); hydrophobic: A, V, L, I, M, W, F, Y, G, P (black). The conserved helix H3 consists of hydrophobic and polar, positively charged residues arranged in a recurrent manner. Abbreviations and accessions numbers used in sequence alignments: Pc—Penicillium chrysogenum, AAQ08763; Af—Aspergillus fumigatus, EAL88627; An—Aspergillus nidulans, EAA65086; Nc—Neurospora crassa, XP_960428; Yl—Yarrowia lipolytica, CAG81724; Sc—Saccharomyces cerevisae, CAA99168; Cg—Candida glabrata, CAG58440; Ag—Ashbya gossypii, AAS54890; Kl—Kluyveromyces lactis, CAG99119; Ca—Candida albicans, EAK92906; Db—Debaryomyces hansenii, CAG84534; Hp—Hansenula polymorpha, DQ645582; HsA—Homo sapiens Pex11α, AAH09697; HsB—Homo sapiens Pex11β, AAH11963. Asterisk and numbers mark amino acids positions in the alignment. (B) Plot of hydrophobic moments (μH) for the N-terminus of PcPex11. The positions of predicted α-helices are marked with H1–H3. The highest value of the μH was obtained for the long putative helix H3, suggesting strong amphipathic properties for this motif. (C) Helical wheel representation and 3D model of the portion of the putative helix H3 with the strongest amphipathic properties, tentatively termed Pex11-Amph (PcPex11 amino acids 67–83). In the helical wheel representation, the amino acids are coloured according to the physico-chemical properties of the side chains (hydrophobic—yellow; polar, positively charged—blue; polar, negatively charged—pink; polar, uncharged—green). In the 3D model of an ideal α-helix built on the basis of the Pex11-Amph sequence, the computed surfaces are marked as follows: hydrophobic (red), positively charged (blue), other amino acids (white).
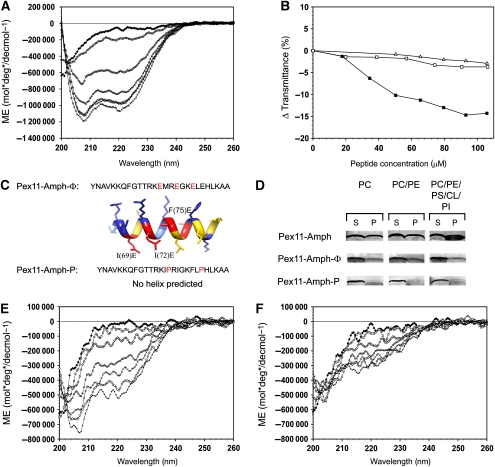
To analyse whether Pex11-Amph can fold into an α-helix, a peptide corresponding to residues 56–83 of P. chrysogenum Pex11 (termed the Pex11-Amph peptide) was synthesized and analysed by circular dichroism (CD) spectroscopy. As has been observed before for other amphipathic helices (Low et al, 2008), the Pex11-Amph peptide did not form any secondary structures in an aqueous solution. However, the addition of increasing amounts of the secondary structure inducer 2,2,2-trifluoroethanol (TFE) caused folding of the Pex11-Amph peptide into an α-helix. Maximal α-helix formation was obtained at 40% of TFE and further increase of the TFE concentration did not significantly change the CD spectrum (Figure 2A). These data show that the Pex11-Amph peptide indeed has the potential to fold into an α–α helix.
Figure 2.
Interaction of the Pex11-Amph with model membranes. (A) The secondary structure of the Pex11-Amph peptide was analysed using CD spectroscopy. The spectrum shows that this peptide is unstructured in phosphate buffer (♦). Addition of increasing amounts of 2,2,2-trifluoroethanol (TFE) induces changes in the spectrum, resulting in minima at 208 and 220 nm, typical for α-helical structure; 10% TFE (□), 20% TFE (◊), 30% TFE (Δ), 40% TFE (x), 50% TFE (○), 60% TFE (+). (B) Turbidimetric changes induced by the addition of the Pex11-Amph peptide to SUV suspensions. Addition of increasing amounts of the Pex11-Amph peptide to neutral vesicles did not significantly alter the turbidity of the solution (Δ—PC liposomes; ϕ—PC/PE liposomes). However, titration of PC/PE/PS/PI/CL SUVs (▪) with the Pex11-Amph peptide resulted in decreased transmittance of the solution. (C) Sequences of mutant peptides Pex11-Amph-Φ and Pex11-Amph-P. Amino-acid substitutions are marked in red. The Pex11-Amph-Φ peptide is predicted to still form an α-helix with a strongly reduced hydrophobic area as depicted by the 3D model of an ideal α-helix. (D) Binding assays of the Pex11-Amph peptide with SUVs of varying phospholipid content (see Supplementary Table III). After incubation with the peptide, the SUVs were collected by centrifugation. The pellet was suspended in buffer (1/10 volume of the initial mixture). Equal volumes of the supernatant (S) and resuspended pellet (P) fractions were loaded per lane. The figure shows a silver stained SDS–polyacrylamide gel. The Pex11-Amph peptide co-sediments with both neutral and negatively charged vesicles, but significantly more peptide binds to the SUVs when negatively charged liposomes resembling the peroxisomal membrane are used (30% peptide bound). Mutant variants of the Pex11-Amph are affected in membrane binding. The Pex11-Amph-Φ peptide co-sediments with liposomes at a reduced level relative to the wild-type peptide. The Pex11-Amph-P mutant peptide does not associate with neutral liposomes and only very small amounts of the mutant peptide co-sedimented with negatively charged vesicles. (E) CD spectrum of the Pex11-Amph-Φ peptide, showing that this mutant peptide is unstructured in phosphate buffer (⧫), similarly to Pex11-Amph. In the presence of increasing amounts of TFE, the CD spectrum of the Pex11-Amph-Φ peptide displays minima at 208 and 220 nm, indicating that some α-helical structure was formed at these conditions; 10% TFE (□), 20% TFE (⋄), 30% TFE (Δ), 40% TFE (x), 50% TFE (○), 60% TFE (+). (F) The CD spectra of the Pex11-Amph-P peptide show that this mutant peptide does not form an α-helix in either phosphate buffer (⧫) or in TFE; 10% TFE (□), 20% TFE (◊), 30% TFE (Δ), 40% TFE (x), 50% TFE (○), 60% TFE (+).
The amphipathic helix of P. chrysogenum Pex11 interacts in vitro with membranes and alters their shape
The presence of positive charges on the polar surface of the Pex11-Amph suggests a capacity of the peptide to bind to negatively charged membranes. Incubation of Pex11-Amph with neutral small unilamellar vesicles (SUVs) resulted in co-sedimentation of a minor portion of the peptide with the liposomes. In contrast, enhanced amounts of the Pex11-Amph peptide co-sedimented with negatively charged SUVs, composed such that they strongly resemble the phospholipid composition of peroxisomal membranes (Wriessnegger et al, 2007) (Figure 2D).
The influence of Pex11-Amph binding on the shape/size of SUVs of different phospholipid content was analysed using turbidimetry measurements. Addition of the Pex11-Amph peptide to neutral vesicles did not influence the turbidity of the solution. In contrast, incubation of vesicles containing a phospholipid composition mimicking that of peroxisomal membranes with increasing amounts of the Pex11-Amph peptide did influence the turbidity of the solution, leading to a significant decrease in transmittance (Figure 2B). These data suggest that the Pex11-Amph peptide interacts with SUVs resembling the phospholipid composition of the peroxisomal membrane and upon binding may influence liposome shape and size.
Interaction of the Pex11-Amph peptide with negatively charged membranes results in tubulation of SUVs
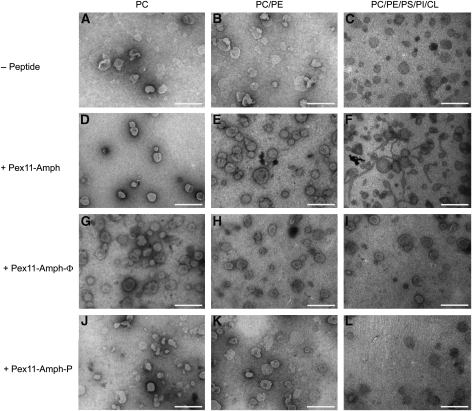
The nature of the changes in turbidity induced by Pex11-Amph binding to membranes was subsequently analysed by transmission electron microscopy using SUVs of different phospholipid content incubated in the presence of the Pex11-Amph peptide. The data revealed that addition of the peptide to neutral vesicles had no influence on their shape and size (Figure 3D and E), which is in agreement with the turbidity measurements. However, in experiments using SUVs with a phospholipid content resembling that of peroxisomal membranes, hypertubulation was evident (Figure 3F). The tubules reached lengths over 10 μm long and 40–50 nm in diameter. These data show that the Pex11-Amph peptide may efficiently remodel membranes in vitro.
Figure 3.
The Pex11-Amph peptide tubulates SUVs of lipid composition similar to peroxisomal membranes. Electron micrographs of SUVs following incubation with the Pex11-Amph peptide. The PC, PC/PE and PC/PE/PS/PI/CL liposomes do not form any elongated structures in the absence of Pex11-Amph (A–C). Also, addition of the Pex11-Amph peptide to SUVs consisting of PC (D) or PC/PE (E) did not alter the shape of these vesicles. Addition of the Pex11-Amph peptide resulted in extensive tubulation of vesicles that had a phospholipid content resembling that of the peroxisomal membrane (F). Mutant peptides Pex11-Amph-Φ (G–I) and Pex11-Amph-P (J–L) were unable to tubulate both neutral and negatively charged vesicles. The bars represent 500 nm.
The amphipathic properties of Pex11-Amph are crucial for its membrane remodelling activity
The significance of the amphipathic properties of Pex11-Amph for membrane remodelling was studied using mutant peptides. In one of these, the Pex11-Amph-Φ mutant peptide, mutations I69E, I72E and F75E were introduced. Although the overall charge of this peptide changed, these residues were chosen as they still allow the peptide to form a helix. Moreover, these mutations do not affect the positive charge at one side of the amphipathic helix, but only reduce the hydrophobic surface. Furthermore, we used a second mutant peptide (Pex11-Amph-P with mutations M70P and E77P) that contains two proline residues predicted to hamper the formation of the helical structure (Figure 2C). The secondary structure of both mutant peptides was analysed by CD spectroscopy. The data indicate that in phosphate buffer, both mutant peptides are unstructured, similar as observed for the Pex11-Amph peptide (Figure 2E and F). As expected, the addition of increasing amounts of TFE to the Pex11-Amph-Φ peptide promoted α-helix formation, albeit to a lower extent than the WT peptide, and reached a maximum in 60% TFE (Figure 2E). In contrast, the presence of two proline residues in the Pex11-Amph-P peptide completely abolished the ability of the peptide to form an α-helix, even in 60% TFE (Figure 2F).
Subsequently, the mutant peptides were analysed for their membrane binding and remodelling activity. When incubated in the presence of neutral vesicles, only a minor portion of the Pex11-Amph-Φ peptide co-sedimented with these vesicles. Similar data were obtained for SUVs mimicking the phospholipid composition of peroxisomal membranes. The Pex11-Amph-P peptide was unable to interact with all types of membranes tested (Figure 2D). Addition of increasing amounts of either the Pex11-Amph-Φ (Supplementary Figure S1A) or the Pex11-Amph-P peptide (Supplementary Figure S1B) did not alter the turbidity of the liposome suspension.
Both mutant peptides were subsequently analysed by electron microscopy for their tubule-forming activity. Incubation of SUVs with the mutant peptides Pex11-Amph-Φ (Figure 3G–I) and Pex11-Amph-P (Figure 3J–L) had no effect on the shape of the vesicles, implicating that these mutations abolished the tubulating activity of the Pex11-Amph.
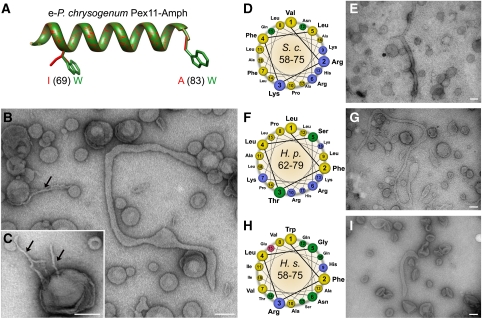
To further confirm the membrane remodelling activity of Pex11-Amph, a gain-of-function mutant peptide was designed (e-Pex11-Amph). In this mutant, peptide 2 residues that form the hydrophobic side of the helix were changed to tryptophanes (I69W, A83W) (Figure 4A). The obtained peptide was predicted to form an amphipathic α-helix with the membrane-facing interface occupied by bulkier hydrophobic residues, which could lead to increased tubulating activity (Masuda et al, 2006). The secondary structure of the e-Pex11-Amph peptide was analysed using CD spectroscopy. Similarly to the Pex11-Amph, the e-Pex11-Amph mutant peptide was unfolded in phosphate buffer and formed an α-helix after addition of TFE (Supplementary Figure S2A), suggesting that introduction of the mutation has not altered the secondary structure of the peptide. Next, the membrane remodelling activity of the e-Pex11-Amph peptide was analysed with electron microscopy. Addition of the e-Pex11-Amph peptide to liposomes with a phospholipid composition resembling the peroxisomal membrane leads to extensive tubulation of vesicles (Figure 4B). Apart from tubules similar to the ones observed for the Pex11-Amph peptide (40–50 nm in diameter), tubules with much smaller diameter (10–15 nm) were frequently observed after incubation of the e-Pex11-Amph peptide with SUV's (Figure 4B and C). The mutant peptide e-Pex11-Amph was also able to induce the formation of multiple tubules from a single liposome (Figure 4C). Moreover, the e-Pex11-Amph-induced tubules of smaller diameter were often found in network-like clusters (Supplementary Figure S3).
Figure 4.
The membrane remodelling activity of different Pex11-Amph peptides. (A) Superimposition of a 3D model of ideal α-helices of Pex11-Amph (red) and the e-Pex11-Amph mutant (green) with marked mutations. The mutant peptide e-Pex11-Amph contains bulkier tryptophane residues on the hydrophobic interface of the amphipathic helix. (B) Electron micrographs of SUVs of a phospholipid content resembling peroxisomal membranes following incubation with the e-Pex11-Amph peptide. Besides typical tubules observed for Pex11-Amph, low diameter tubules could often be observed (black arrow). (C) The e-Pex11-Amph peptide also induces formation of multiple low diameter tubules developing from single liposomes (black arrows). (D) Helical wheel representation of the region comprising the Pex11-Amph from S. cerevisiae (residues 58–75 of S.c.Pex11) showing the hydrophobic (yellow) and polar, positively charged (blue) interfaces of the helix. (E) Electron micrographs of PC/PE/PS/PI/CL SUVs following incubation with Sc-Pex11-Amph peptide showing extensive tubulation of liposomes. (F) Helical wheel representation of the Pex11-Amph from H. polymorpha (residues 62–79 of Hp-Pex11) revealing the amphipathic properties of the α-helix with a charged polar surface similar to the Pex11-Amph motifs from P. chrysogenum and S. cerevisiae. (G) Incubation of Hp-Pex11-Amph with liposomes with phospholipid content resembling that of the peroxisomal membrane causes efficient tubulation of vesicles. (H) Representation of Pex11-Amph from H. sapiens (residues 58–75 of Hs-Pex11α) in the helical wheel mode shows the amphipathic properties of this motif. In contrast to fungal Pex11-Amph, the corresponding motif in human Pex11α contains a polar interface that is less enriched in basic amino acids. (I) Electron micrograph of PC/PE/PS/PI/CL liposomes after addition of the Hs-Pex11-Amph peptide showing that the amphipathic helix of human Pex11α is active in vesicle tubulation. The observed tubules were usually shorter than for other Pex11-Amph peptides. Scale bars represent 100 nm.
These data imply that the presence of the α-helix as well as conservation of its amphipathic nature are crucial for Pex11-Amph function and suggest that insertion of the hydrophobic surface of Pex11-Amph into the membrane may be responsible for the observed tubulating activity.
The function of Pex11-Amph is conserved among species
To study whether the membrane remodelling activity of Pex11-Amph is evolutionary conserved, peptides comprising the identified N-terminal amphipathic helices of Pex11 from Saccharomyces cerevisiae, Hansenula polymorpha and Homo sapiens (see Figure 1A; Supplementary Figure S4) were synthesized. Similarly to P. chrysogenum Pex11-Amph, amphipathic α-helices from both yeast species contain polar interfaces enriched in basic residues (Figure 4D and F). In contrast, the hydrophilic side of the N-terminal amphipathic helix of human Pex11α is mainly composed of polar, uncharged residues (Figure 4H). The secondary structure of the peptides containing Pex11-Amph motifs from different species was analysed by CD spectroscopy. All three peptides were unfolded in phosphate buffer and efficiently formed α-helical structures after addition of TFE, reaching maxima at around 30–40% TFE (Supplementary Figure S2B–D). Next, the membrane remodelling activity of the Pex11-Amph peptides was tested with electron microscopy using liposomes with a phospholipid content resembling peroxisomal membranes. All three peptides efficiently tubulated vesicles. However, in comparison with the long tubules formed by the Pex11-Amph peptides from P. chrysogenum, S. cerevisiae and H. polymorpha (Figures 3F, 4E and G), tubules induced by Pex11-Amph from H. sapiens were generally shorter (Figure 4I).
These data suggest that the membrane remodelling activity of the N-terminal amphipathic helix of Pex11 proteins is conserved from yeast to human.
The soluble N-terminal domain of Pex11 is active in membrane remodelling
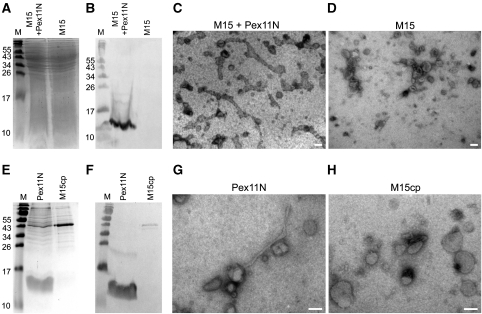
We next tested whether the N-terminal amphipathic helix of P. chrysogenum Pex11 also tubulates vesicles in the context of the entire soluble N-terminal domain of the protein (Pex11N). All Pex11 proteins are predicted to contain an N-terminal domain facing the cytosol and a C-terminal region containing two transmembrane spans (Anton et al, 2000) (cf. Supplementary Figure S5A). To this purpose, we produced the first 97 amino acids of P. chrysogenum Pex11 with a C-terminal His-tag in Escherichia coli M15. Western blotting using α-His-tag antibodies revealed that the protein was properly synthesized (indicated as a band of approximately 12 kDa) and absent in the lysate of the empty host strain (Figure 5A and B).
Figure 5.
Membrane remodelling activity of the N-terminal domain of P. chrysogenum Pex11. Coomassie brilliant blue stained SDS–PAA gel (A) and western blot decorated with α-His-tag antibodies (B) of bacterial lysates prepared from E. coli M15 cells producing Pex11N (M15+Pex11N) or the empty host (M15). The western blot shows the presence of a protein band of approximately 12 kDa, which represents Pex11N. Equal amounts of protein were loaded per lane. Electron micrograph of SUVs with a phospholipid content resembling peroxisomal membranes incubated with the bacterial lysate containing Pex11N (M15 + Pex11N; C) showing extensive tubulation of liposomes. Tubulation was not observed when the control lysate prepared from the empty M15 host was used (D). Coomassie brilliant blue stained gel (E) and the corresponding western blot decorated with α-His antibodies (F) showing the protein fraction enriched in Pex11N (Pex11N) and the corresponding control fraction obtained from the empty M15 host (M15cp). Pex11N is detected as a band of approximately 12 kDa both on the Coomassie stained gel and on the western blot. Equal amounts of proteins were loaded per lane. Electron micrographs of peroxisome-like SUVs transformed into tubules after addition of Pex11N protein (G). In the control sample (H; M15cp), liposome tubulation was never observed. The scale bars represent 100 nm.
Addition of the cell lysate of the E. coli strain producing Pex11N to liposomes with a phospholipid content resembling peroxisomal membranes resulted in vesicle tubulation (Figure 5C). This was not observed when a lysate of the empty E. coli strain (M15) was used (Figure 5D).
To further confirm that the tubulation activity was due to Pex11N, we prepared protein fractions highly enriched in Pex11N by affinity and ion exchange chromatography (Figure 5E and F). As a control, the corresponding fractions obtained from the empty E. coli M15 lysate were used. Western blotting using α-His-tag antibodies confirmed the presence of the 12-kDa band in the purified fraction, which was absent in the control (Figure 5E and F). As shown in Figure 5G and H, the Pex11N fraction, but not the control, caused extensive tubulation of liposomes. These data imply that Pex11-Amph also functions in vesicle tubulation in the context of the entire soluble domain of Pex11.
The amphipathic helix of Pex11 is responsible for organelle tubulation during peroxisome fission in vivo
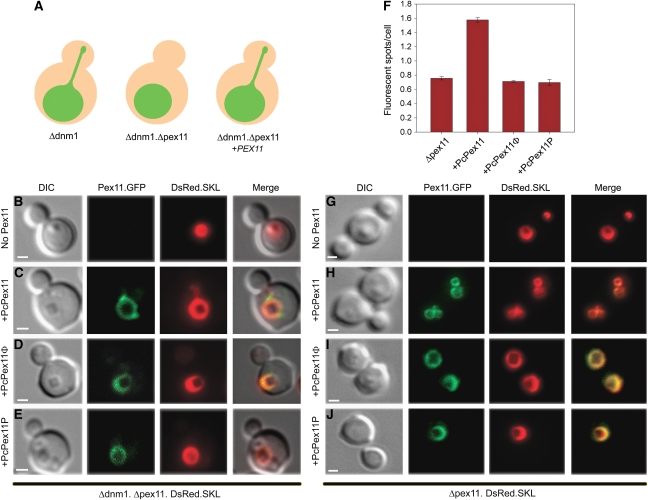
The role of the Pex11-Amph region in Pex11-induced membrane deformation was analysed in vivo using the methylotrophic yeast H. polymorpha as a model organism. In this analysis, we took advantage of the fact that in H. polymorpha dnm1 cells, long peroxisomal extensions are formed that may protrude into the bud, but are not separated from the mother organelle due to the absence of Dnm1 (Nagotu et al, 2008). The formation of these extensions is Pex11 dependent as they are not formed in H. polymorpha dnm1 pex11 double mutant cells that usually contain a single, enlarged peroxisome per cell (Figure 6A and B). The observation that production of P. chrysogenum Pex11 stimulates proliferation of peroxisomes in H. polymorpha (Kiel et al, 2005) fully validates the use of H. polymorpha as a model system to study the significance of Pex11-Amph in Pex11-induced peroxisome membrane remodelling. To this end, we expressed P. chrysogenum PEX11•GFP under control of the inducible H. polymorpha amine oxidase promoter in H. polymorpha dnm1 pex11 cells, which also produced DsRed•SKL to mark peroxisomes. Fluorescence microscopy analysis demonstrated that production of the fusion protein resulted in the formation of peroxisome extensions akin to those observed in H. polymorpha dnm1 cells (Figure 6C). Furthermore, P. chrysogenum Pex11•GFP accumulated at the site of extension formation, which is in agreement with the localization data of H. polymorpha Pex11 (Nagotu et al, 2008) (Figure 6C). Subsequently, we introduced point mutations in the amphipathic helix of PcPex11•GFP at the same positions as in the mutant peptides (see above), resulting in the mutant proteins PcPex11-Φ•GFP (with mutations I69E, I72E and F75E) and PcPex11-P•GFP (with mutations M70P and E77P). The mutant genes were introduced in H. polymorpha dnm1 pex11 cells and the resulting transformants analysed by fluorescence microscopy. The data (Figure 6D and E) indicate that both mutant proteins normally localized to the peroxisomal membrane. However, the formation of peroxisome extensions was completely impaired.
Figure 6.
The Pex11-Amph region is responsible for Pex11-mediated peroxisome tubulation during organelle fission. (A) Schematic drawing of the yeast fission model used in this study. In dividing H. polymorpha cells lacking the dynamin-related protein Dnm1, actual fission of peroxisomes is blocked. In those cells, an intermediate state in peroxisome proliferation, namely a peroxisomal elongation extending from the mother cell into the bud, is observed. Subsequent deletion of PEX11 in H. polymorpha dnm1 cells disturbs the formation of peroxisomal extensions, but this can be functionally complemented by (P. chrysogenum) PEX11. (B) H. polymorpha dnm1 pex11 cells grown on methanol-containing media usually contain a single, large peroxisome (labelled with DsRed•SKL). Extensions to the bud are not formed. (C) Expression of WT P. chrysogenum PEX11•GFP in H. polymorpha dnm1 pex11 cells restores the formation of peroxisomal extensions. Pex11•GFP is localized on the peroxisomal membrane and accumulates at the basis of the peroxisomal extension marked by DsRed•SKL. Also, PcPex11-Φ•GFP (D) and PcPex11-P•GFP (E) localize to the peroxisome membrane in H. polymorpha dnm1 pex11 cells. However, in these cells, the formation of peroxisomal extensions is not restored. (F–J) The Pex11-Amph is crucial for proliferation of peroxisomes. P. chrysogenum PEX11•GFP and mutants P. chrysogenum PEX11-Φ•GFP or PEX11-P•GFP were introduced into H. polymorpha pex11 cells producing DsRed•SKL to mark peroxisomes. For quantitative analysis of peroxisome numbers, two independent cultures were grown on methanol-containing media and the number of fluorescent spots per cell was counted in 150 cells each. H. polymorpha pex11 cells contain usually a single enlarged peroxisome per cell, but a number of peroxisome-deficient cells can be also observed (average number per cell 0.76; F, G). Expression of P. chrysogenum PEX11•GFP in H. polymorpha pex11 cells causes a significant increase in peroxisome numbers, as multiple organelles can be observed (Student's t-test: P=0.0023; average 1.58) (F, H). Upon expression of either of the mutant genes P. chrysogenum PEX11-Φ•GFP (I) or PEX11-P•GFP (J) in H. polymorpha pex11 cells, the number of organelles was similar to that in the PEX11 deletion strain (averages 0.71 and 0.70, respectively) and significantly different from H. polymorpha pex11 cells producing WT P. chrysogenum PEX11•GFP (Student's t-test: P=0.0015 and 0.0037, respectively). Bars in (F) represent SEM. The bars in B–E and G–J represent 1 μm.
The role of the Pex11-Amph motif was also tested in H. polymorpha pex11 cells, which still contained Dnm1. In these cells, the number of peroxisomes is strongly reduced as compared with wild type (Figure 6F and G) (Krikken et al, 2009). First, we introduced P. chrysogenum PEX11•GFP under control of the inducible H. polymorpha amine oxidase promoter in H. polymorpha pex11 cells, producing DsRed•SKL. Fluorescence microscopy revealed that expression of P. chrysogenum PEX11•GFP in this strain resulted in an increase in peroxisome numbers compared with pex11 control cells (Figure 6F and H). Next, we introduced the mutant proteins PcPex11-Φ•GFP and PcPex11-P•GFP in H. polymorpha pex11. Production of both mutant proteins did not result in increasing peroxisome numbers (Figure 6F, I and J).
These data imply that the Pex11-Amph region is important for peroxisome proliferation in vivo.
Discussion
In this paper, we demonstrate that Pex11 proteins contain a conserved amphipathic helix at the N-terminus (tentatively termed Pex11-Amph) that interacts with membranes and is capable of mediating membrane curvature/elongation, in particular when using vesicles of a phospholipid composition resembling that of peroxisomal membranes. A characteristic feature of these membranes is their negative charge. Effective binding of the Pex11-Amph region to these negatively charged membranes is likely related to the fact that the Pex11-Amph contains positive charges on the polar surface of the helix. The finding that the interaction of the Pex11-Amph with these vesicles resulted in changes in vesicle shape is consistent with the observed increased turbidity of the samples after peptide supplementation. The Pex11-Amph peptide was unable to remodel liposomes consisting solely of PC and PC/PE probably because of the reduced affinity of the positive polar face of the peptide for neutral membranes. Formation of long tubules was shown previously for other membrane binding and bending amphipathic helices, such as those from clathrin, Bin-1, the viral protein Tip and artificial peptides using liposomes with a lipid composition resembling Golgi membranes (Lee et al, 2001; Ford et al, 2002; Low et al, 2008; Min et al, 2008).
Pex11 proteins are major factors involved in peroxisome proliferation in all species studied thus far. Current models indicate that Pex11 proteins function at the initial stages of peroxisome fission, causing changes in the organelle shape (Nagotu et al, 2010). We show that the Pex11-Amph regions from the yeast species S. cerevisiae and H. polymorpha, from the filamentous fungus P. chrysogenum as well as from H. sapiens, can efficiently induce membrane curvature. This implies that the membrane remodelling activity of Pex11-Amph is evolutionary conserved.
In vivo membranes undergo continuous remodelling events, often accompanied by temporary stabilization of specific shapes. Many cellular processes rely on changes in the shape of the membrane. Moreover, the unique shape of organelles like the ER is maintained by a complex interplay between membrane interacting proteins and organelle membranes (Shibata et al, 2009). One of the mechanisms to induce membrane curvature is insertion of an amphipathic helix into one leaflet of the lipid bilayer, causing membrane asymmetry and resulting in membrane bending (Campelo et al, 2008). Among the proteins that utilize this mechanism are N-BAR domain-containing proteins. These proteins use an N-terminal amphipathic α-helix to induce membrane curvature during endocytosis (Dawson et al, 2006; Gallop et al, 2006). Membrane curvature is further stabilized by interaction with the banana-shaped BAR domains (Low et al, 2008; Yin et al, 2009). Recently, Bif-1, an N-BAR domain protein, was proposed to be responsible for shape regulation of the autophagosomal membrane during autophagy (Takahashi et al, 2009). Insertion of an amphipathic helix is also used by the ENTH domain-containing protein epsin to bend membranes during the initial steps of clathrin-coated vesicle formation during endocytosis (Ford et al, 2002; Kweon et al, 2006). Moreover, the small GTPases Arf and Sar1 utilize the same mechanism to induce membrane curvature during COPI and COPII vesicular transport (Lee et al, 2005; Krauss et al, 2008; Lundmark et al, 2008).
The mechanism by which Pex11-Amph remodels the membranes was analysed using mutant peptides. A Pex11-Amph-Φ peptide with a reduced hydrophobic surface was unable to efficiently bind and tubulate liposomes, which is in line with previous data reported for mutant epsin proteins (Ford et al, 2002). Moreover, the α-helical structure appears to be a prerequisite for the binding and bending activity of the Pex11-Amph peptide, as insertion of prolines at key positions in the peptide prevented membrane deformation. Introduction of two bulkier tryptophane residues into the hydrophobic interface of Pex11-Amph resulted in increased tubulating activity and the formation of tubules with a smaller diameter. Together, these data suggest that the conserved amphipathic α-helical domain at the N-terminus of P. chrysogenum Pex11 remodels membranes using a mechanism resembling that of amphipathic helices in other proteins. In such proteins, often additional domains are present that stabilize membrane curvature or interact with scaffolding proteins. Taking this into account, it is tempting to speculate that other regions of Pex11 might not only be required to determine the subcellular location of the protein, but also have a function in membrane remodelling or, alternatively, other proteins may be involved.
In line with our in vitro data, we observed that the same mutations in the Pex11-Amph peptides that affect the tubulation activity of this region affected peroxisome proliferation in vivo when introduced in full-length Pex11 (Figure 5F and J). Although we cannot fully exclude that our in vivo data are due to an effect of these mutations on protein–protein interactions or protein folding, this is very unlikely given the conserved tubulation properties of this region from yeast to humans.
Pex11 has a crucial function during proliferation of peroxisomes. The tubulation event is considered to represent the initial event in peroxisome multiplication (Thoms and Erdmann, 2005), a step that is followed by fission (Nagotu et al, 2008). Our data suggest that the organelle elongation step is crucially dependent on Pex11 and that its membrane remodelling activity is related to the N-terminal amphipathic helix. Deletion of the N-terminal 84 residues of human Pex11β, which comprises the conserved Pex11-Amph motif, abolished its peroxisome proliferating activity (Kobayashi et al, 2007), which is in line with our findings, showing that the N-terminal domain of P. chrysogenum Pex11 is active in membrane remodelling. Moreover, the N-terminus of Pex11β was shown to be involved in oligomerization of Pex11 proteins (Kobayashi et al, 2007). It has been suggested that oligomeric forms of Pex11 may not be active in peroxisome proliferation (Marshall et al, 1996). Thus, it is tempting to speculate that in oligomeric Pex11 protein complexes, the Pex11-Amph region cannot interact with the membrane (Supplementary Figure S5A). In this scenario, a Pex11 activation step is required that includes dissociation of Pex11 oligomers. This would be followed by insertion of the amphipathic helices of the released monomers into the peroxisome membrane, resulting in initial membrane deformation (Supplementary Figure S5B). In addition to this, it was recently shown that curved membranes contain defects in phospholipid packaging and that the exposed hydrophobic parts of the membrane are efficiently recruiting amphipathic helices (Hatzakis et al, 2009). Additionally, membrane curvature was also suggested to be the driving force for segregation of lipids and proteins and lead to formation of specific membrane domains (Huang et al, 2006; Vogel and Sheetz, 2006; Reynwar et al, 2007; Mukhopadhyay et al, 2008). Thus, possible initiation of the membrane curvature by Pex11-Amph will be followed by a massive concentration of Pex11 proteins in this region, a process that is dependent on the insertion of the amphipathic helix (Supplementary Figure S5C). Such a scenario might explain our previous observation that Pex11 assembles at the site of the peroxisomal extension in H. polymorpha dnm1 cells (Nagotu et al, 2008; cf. Figure 5C) and suggests that the formation of Pex11-enriched domains is required to trigger tubulation of the organelle.
Materials and methods
Strains and growth conditions
The H. polymorpha strains used in this study are listed in Table I. H. polymorpha cells were grown on YPD or mineral media as described before (Nagotu et al, 2008). E. coli DH5α was used for cloning purposes. E. coli M15 cells (Qiagen) were used for production of heterologous protein. Cells were grown at 37°C on LB medium supplemented with the appropriate antibiotics.
Table 1. Hansenula polymorpha strains used in this study.
| Strain | Description | Reference |
|---|---|---|
| dnm1 pex11 DsRed•SKL | Deletion of DNM1 and PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL | Nagotu et al (2008) |
| dnm1 pex11 DsRed•SKL+PcPex11•GFP | Deletion of DNM1 and PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11 containing the P. chrysogenum PEX11•GFP fusion gene | This study |
| dnm1 pex11 DsRed•SKL+PcPex11-Φ•GFP | Deletion of DNM1 and PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11-Φ containing the mutant P. chrysogenum PEX11-Φ•GFP fusion gene | This study |
| dnm1 pex11 DsRed•SKL+PcPex11-P•GFP | Deletion of DNM1 and PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11-P containing the mutant P. chrysogenum PEX11-P•GFP fusion gene | This study |
| pex11 | Deletion of PEX11 | Krikken et al (2009) |
| pex11 DsRed•SKL | Deletion of PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL | This study |
| pex11 DsRed•SKL+PcPex11•GFP | Deletion of PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11 containing the P. chrysogenum PEX11•GFP fusion gene | This study |
| pex11 DsRed•SKL+PcPex11-Φ•GFP | Deletion of PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11-Φ containing the mutant P. chrysogenum PEX11-Φ•GFP fusion gene | This study |
| pex11 DsRed•SKL+PcPex11-P•GFP | Deletion of PEX11 with integration of plasmid pHIPZ4-DsRed-T1-SKL and integration of plasmid LMOPPcPex11-P containing the mutant P. chrysogenum PEX11-P•GFP fusion gene | This study |
Molecular biology techniques
Plasmids and oligonucleotides used in this study are listed in Supplementary Tables I and II. Recombinant DNA manipulations and transformation of H. polymorpha were performed as described before (Sambrook and Maniatis, 1989) (Faber et al, 1992, 1994). All PCR fragments were sequenced (Service XS). In silico analysis of DNA sequences and construction of vector maps was carried out using Clone Manager 5 software (Scientific and Educational Software, Durham).
In silico analysis
Multiple sequence alignments of protein sequences were prepared using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/) and visualized with GeneDoc (http://www.psc.edu/biomed/genedoc). Secondary structure prediction was carried out using the DSC program available on the PAT server (http://abcis.cbs.cnrs.fr/htbin-post/pat/new/wpat.pl). The hydrophobic moment of the sequence of PcPex11 was calculated using the EMBOSS server (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=hmoment). The helical wheel representation of the Pex11-Amph region was prepared using Java Applet (http://cti.itc.virginia.edu/~cmg/Demo/wheel/wheelApp.html). Representations of the Pex11-Amph, Pex11-Amph-Φ and e-Pex11-Amph sequences as 3D models of α-helices were prepared using UCSF Chimera software (Pettersen et al, 2004).
Construction of plasmid LMOPPcPex11
Plasmid LMOPPcPex11 encoding full-length P. chrysogenum Pex11 (PcPex11) fused C-terminally to eGFP was prepared by Multisite Gateway technology (Invitrogen) as follows: first, by PCR a 778-bp DNA fragment comprising the P. chrysogenum PEX11 coding sequence lacking a stop codon was amplified with primers BB-JK001 and BB-JK002 using P. chrysogenum PEX11 cDNA (Kiel et al, 2000) as a template. The fragment was recombined with vector pDONR221, resulting in plasmid pBBK-002. Subsequently, the plasmids pDONR-P4-P1r-PAMO, pBBK-002, pDONR-P2r-P3-eGFP-TAMO and pDEST-R4-R3-NAT were recombined resulting in plasmid LMOPPcPex11. This plasmid and mutant plasmids LMOPPcPex11Φ and LMOPPcPex11P were linearized with AdeI and transformed into H. polymorpha dnm1 pex11 DsRed•SKLcells. Transformants were selected based on their ability to grow on medium containing nourseothricin. Correct integration was analysed by colony PCR. For construction of the H. polymorpha pex11 DsRed•SKL strain and H. polymorpha pex11 DsRed•SKL strains expressing PcPex11•GFP or either of the mutants PcPex11-Φ•GFP and PcPex11-P•GFP, plasmids pHIPZ4-DsRed-T1-SKL (linearized with NotI) and either LMOPPcPex11, LMOPPcPex11Φ or LMOPPcPex11P (all linearized with AdeI) were co-transformed into H. polymorpha pex11 cells. Transformants were selected based on their ability to grow on medium containing nourseothricin and zeocin. Production of DsRed•SKL and PcPex11•GFP was confirmed by fluorescence microscopy.
Site-directed mutagenesis of Pex11
For introduction of targeted mutations in PcPex11, the QuickChange Lightning Site-Directed Mutagenesis kit (Stratagene) was used. To construct plasmids producing mutant forms of PcPex11, PcPex11-Φ (with mutations I69E, I72E and F75E) and PcPex11-P (with mutations M70P and E77P), the LMOPPcPex11 vector was used as a template together with the mutagenic oligonucleotides LMOp062, LMOp063, LMOp064 and LMOp065, generating plasmids LMOPPcPex11Φ and LMOPPcPex11P, respectively. Correctness of introduced mutations was confirmed by sequencing.
Peptide design and synthesis
The peptide Pex11-Amph (YNAVKKQFGTTRKIMRIGKFLEHLKAAA), corresponding to residues 56–83 of PcPex11, the mutant peptides Pex11-Amph-Φ (YNAVKKQFGTTRKEMREGKELEHLKAA), Pex11-Amph-P (YNAVKKQFGTTRKIPRIGKFLPHLKAA), e-Pex11-Amph (YNAVKKQFGTTRKWMRIGKFLEHLKAAW), S. cerevisiae Pex11-Amph (residues 49–78: ARQLQAQFTTVRKFLRFLKPLNHLQAAAKFY), H. polymorpha Pex11-Amph (residues 54–86: YLVRRLQDLFTLSRKPLRALKPLKHLKALSVTV) and H. sapiens Pex11-Amph (residues 44–75 of Pex11α: LKKLESSVSTGRKWFRLGNVVHAIQATEQSI) were synthesized by Pepscan. The purchased peptides were HPLC purified and quality was assessed with LC/MS. To prepare stock solutions, peptides were dissolved in Liposome buffer (20 mM HEPES, 150 mM NaCl, pH 7.4). The concentrations of peptides were determined using absorbance of UV light.
Production and purification of Pex11N
The DNA sequence encoding the first 97 amino acids of P. chrysogenum Pex11 was amplified using the LMOPPcPex11 plasmid as a template and primers LMOp066 and LMOp067 containing NcoI and BglII sites, respectively. The obtained PCR product and expression vector pQE60 (Qiagen) were digested with NcoI and BglII and ligated, resulting in plasmid pQE60-Pex11N, encoding the N-terminal region of P. chrysogenum Pex11 fused to a His-tag at the C-terminus (Pex11N). Correctness of the plasmid was confirmed by sequencing.
E. coli M15 cells containing expression plasmid pQE60-Pex11N and empty M15 host cells were grown in LB medium at 37°C, 200 r.p.m. to an optical density of 1 and then induced with 1 mM IPTG followed by 4 h incubation at 30°C, 200 r.p.m. Cells were harvested by centrifugation at 6000 r.p.m., 10 min, 4°C. Cell pellets were suspended in lysis buffer A (50 mM Tris, 8M urea, pH 8.0) or lysis buffer B (100 mM NaH2PO4, 10 mM Tris, 8M urea, pH 8.0) and disrupted by two cycles of stirring with a magnetic stirrer and French press. The soluble fraction was obtained by centrifugation for 1 h at 12°C at 20000 g. Cell extracts in lysis buffer A were used for vesicle tubulation assays. Extracts in buffer B were used for further Pex11N purification.
Lysates in buffer B were incubated with Ni-NTA resin (QIAGEN) for 1 h. The resin was washed with buffer B and proteins were eluted with elution buffer (100 mM NaH2PO4, 10 mM Tris, 8M urea, pH 4.5). The fractions obtained were analysed by SDS–PAGE and western blotting using α-His-tag antibodies (Santa Cruz Biotechnology). Fractions enriched in Pex11N were pooled, concentrated and loaded on Mono-Q resin (Pharmacia Biotech) equilibrated in loading buffer (50 mM Tris, 8M urea, pH 8.0). Bound proteins were eluted with a linear gradient of elution buffer (50 mM Tris, 8M urea, 2M NaCl, pH 8.0). Fractions enriched in Pex11N were concentrated using Amicon Ultra-4 and Microcon YM-3 centrifugation devices (Milipore).
Preparation of SUVs
SUVs were prepared using chloroform solutions of phosphatidylcholine (PC, symmetric, 18:1), phosphatidylethanolamine (PE, symmetric, 18:1), phosphatidylserine (PS, symmetric, 18:1), cardiolipin (CL, symmetric, 18:1) and phospatidylinositol (PI, natural mixture from bovine liver) purchased from Avanti Polar Lipids Inc. To prepare liposomes of desired composition (Supplementary Table III), the required volumes of phospholipid solutions were mixed to a final lipid concentration of 0.8 mg/ml and then the organic solvent was evaporated using a rotary evaporator. The obtained thin lipid film was hydrated in Liposome buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) for 3 h at room temperature. To obtain unilamellar vesicles of the desired diameter, the liposome suspension was extruded 11 times through polycarbonate filters (Avestin) with a 100-nm pore size.
Peptide-binding assay
SUVs were mixed with peptides in Liposome buffer (250 μl) to a final concentration of 0.65 mg/ml and 50 μM, respectively. After incubation for 20 min at room temperature, SUVs were pelleted by ultracentrifugation (20 min, 21°C, 100 000 g) and the pellet was resuspended in 25 μl of liposome buffer. The peptides in the supernatant and pellet (equal volumes) were subsequently subjected to Tricine SDS–PAGE using 15% acrylamide gels and visualized with silver staining using a Silver Stain Plus kit (Bio-rad).
Turbidimetric measurements of the peptide–SUV interaction
SUV solutions (0.4 mg/ml) were titrated with peptides (0 to 100 μM) in Liposome buffer. After each addition of peptide and vigorous mixing the absorbance at 400 nm was recorded for 5 min at room temperature using a Perkin Elmer Lambda 35 spectrophotometer. The obtained absorbance values were converted to transmittance, using the Lambert–Beer equation: A=2–log10%T and changes in transmittance were plotted against the peptide concentration.
Electron microscopy
Peptides, bacterial lysates or purified protein fractions were incubated with SUVs in liposome buffer (20 mM HEPES, 150 mM NaCl, pH 7.4) for 15 min at room temperature before negative staining. The mixtures were subsequently placed on carbon-coated grids and stained with 0.5% uranyl acetate and examined by electron microscopy. Electron microscopy was performed using a CM12 TEM microscope (Philips).
For the analysis of peptides, SUVs were used at a final concentration of 0.4 mg/ml, with a peptide concentration of 100 μM. In experiments using bacterial lysates, the concentration of SUVs was 0.2 mg/ml, the protein concentration 0.02 mg/ml and a final urea concentration of 32 mM. Protein fractions containing Pex11N or the corresponding fractions obtained from cell extracts of the empty M15 host were added to SUVs (0.2 mg/ml) to a final protein concentration of 0.2 mg/ml and a final urea concentration of 0.8 M.
Fluorescence microscopy
Wide-field fluorescence microscopy was performed using a Zeiss Axio Observer Z1 fluorescence microscope (Zeiss). Images were taken using EC-Plan-Neofluar 100 × /1.3 objective and coolsnap HQ2 Camera (Roper scientific Inc). GFP signal was visualized with a 470/40-nm bandpass excitation filter, a 495-nm dichromatic mirror and a 525/50-nm bandpass emission filter. The fluorescence of DsRed was visualized with a 545/25-nm bandpass excitation filter, a 570-nm dichromatic mirror and a 605/70-nm bandpass emission filter. Image analysis was carried out using ImageJ (http://rsb.info.nih.gov/nih-image/) and Adobe Photoshop. For quantification of peroxisome numbers 150 cells from two independent cultures for each strain were analysed. Statistical data analysis was performed using Statistica 8.0 software.
CD spectroscopy
CD measurements were performed on a Jasco J-810 spectropolarimeter. Peptides were dissolved in 10 mM potassium phosphate, 150 mM NaCl, pH 7.3. Spectra were recorded between 200 and 260 nm in a 2-mm cuvette with a peptide concentration of 10 μM and TFE concentrations ranging from 0 to 60%. Machine settings were as follows: 1 nm bandwidth, 1 s response, 0.5 nm data pitch, 100 nm/min scan speed and cell length of 0.1 cm. All CD data presented are the averages from three separate measurements.
Supplementary Material
Additional Information
Acknowledgments
We thank Prof. Dr AJM Driessen and I Kuesters for expert advice in liposome preparation. This project was carried out within the research programme of the Kluyver Centre for Genomics of Industrial Fermentation, which is part of the Netherlands Genomics Initiative/Netherlands Organization for Scientific Research. JAKWK is supported by DSM, Delft, The Netherlands. Part of this work was funded by a Rubicon Fellowship (825.08.023) from the Netherlands Organisation for Scientific Research (NWO) awarded to CW.
Footnotes
The authors declare that they have no conflict of interest.
References
- Anton M, Passreiter M, Lay D, Thai TP, Gorgas K, Just WW (2000) ARF- and coatomer-mediated peroxisomal vesiculation. Cell Biochem Biophys 32 (Spring): 27–36 [DOI] [PubMed] [Google Scholar]
- Campelo F, McMahon HT, Kozlov MM (2008) The hydrophobic insertion mechanism of membrane curvature generation by proteins. Biophys J 95: 2325–2339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JC, Legg JA, Machesky LM (2006) Bar domain proteins: a role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol 16: 493–498 [DOI] [PubMed] [Google Scholar]
- Drin G, Antonny B (2009) Amphipathic helices and membrane curvature. FEBS Lett 584: 1840–1847 [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G (1995) Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiae lacking the peroxisomal membrane protein Pmp27p. J Cell Biol 128: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber KN, Haima P, Harder W, Veenhuis M, Ab G (1994) Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr Genet 25: 305–310 [DOI] [PubMed] [Google Scholar]
- Faber KN, Swaving GJ, Faber F, Ab G, Harder W, Veenhuis M, Haima P (1992) Chromosomal targeting of replicating plasmids in the yeast Hansenula polymorpha. J Gen Microbiol 138: 2405–2416 [DOI] [PubMed] [Google Scholar]
- Fagarasanu A, Fagarasanu M, Rachubinski RA (2007) Maintaining peroxisome populations: a story of division and inheritance. Annu Rev Cell Dev Biol 23: 321–344 [DOI] [PubMed] [Google Scholar]
- Ford MG, Mills IG, Peter BJ, Vallis Y, Praefcke GJ, Evans PR, McMahon HT (2002) Curvature of clathrin-coated pits driven by epsin. Nature 419: 361–366 [DOI] [PubMed] [Google Scholar]
- Gallop JL, Jao CC, Kent HM, Butler PJ, Evans PR, Langen R, McMahon HT (2006) Mechanism of endophilin N-BAR domain-mediated membrane curvature. EMBO J 25: 2898–2910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ, Valle D (2000) Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet 16: 340–345 [DOI] [PubMed] [Google Scholar]
- Guerra-Giraldez C, Quijada L, Clayton CE (2002) Compartmentation of enzymes in a microbody, the glycosome, is essential in Trypanosoma brucei. J Cell Sci 115: 2651–2658 [DOI] [PubMed] [Google Scholar]
- Hatzakis NS, Bhatia VK, Larsen J, Madsen KL, Bolinger PY, Kunding AH, Castillo J, Gether U, Hedegard P, Stamou D (2009) How curved membranes recruit amphipathic helices and protein anchoring motifs. Nat Chem Biol 5: 835–841 [DOI] [PubMed] [Google Scholar]
- Hettema EH, Motley AM (2009) How peroxisomes multiply. J Cell Sci 122: 2331–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang KC, Mukhopadhyay R, Wingreen NS (2006) A curvature-mediated mechanism for localization of lipids to bacterial poles. PLoS Comput Biol 2: e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel JA, Hilbrands RE, Bovenberg RA, Veenhuis M (2000) Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl Microbiol Biotechnol 54: 238–242 [DOI] [PubMed] [Google Scholar]
- Kiel JA, van den Berg MA, Fusetti F, Poolman B, Bovenberg RA, Veenhuis M, van der Klei IJ (2009) Matching the proteome to the genome: the microbody of penicillin-producing Penicillium chrysogenum cells. Funct Integr Genomics 9: 167–184 [DOI] [PubMed] [Google Scholar]
- Kiel JA, van der Klei IJ, van den Berg MA, Bovenberg RA, Veenhuis M (2005) Overproduction of a single protein, Pc-Pex11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet Biol 42: 154–164 [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Tanaka A, Fujiki Y (2007) Fis1, DLP1, and Pex11p coordinately regulate peroxisome morphogenesis. Exp Cell Res 313: 1675–1686 [DOI] [PubMed] [Google Scholar]
- Krauss M, Jia JY, Roux A, Beck R, Wieland FT, De Camilli P, Haucke V (2008) Arf1-GTP-induced tubule formation suggests a function of Arf family proteins in curvature acquisition at sites of vesicle budding. J Biol Chem 283: 27717–27723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikken AM, Veenhuis M, van der Klei IJ (2009) Hansenula polymorpha pex11 cells are affected in peroxisome retention. FEBS J 276: 1429–1439 [DOI] [PubMed] [Google Scholar]
- Kweon DH, Shin YK, Shin JY, Lee JH, Lee JB, Seo JH, Kim YS (2006) Membrane topology of helix 0 of the Epsin N-terminal homology domain. Mol Cells 21: 428–435 [PubMed] [Google Scholar]
- Lamas-Maceiras M, Vaca I, Rodriguez E, Casqueiro J, Martin JF (2006) Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N-acyltransferase. Biochem J 395: 147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R (2005) Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell 122: 605–617 [DOI] [PubMed] [Google Scholar]
- Lee S, Furuya T, Kiyota T, Takami N, Murata K, Niidome Y, Bredesen DE, Ellerby HM, Sugihara G (2001) De novo-designed peptide transforms Golgi-specific lipids into Golgi-like nanotubules. J Biol Chem 276: 41224–41228 [DOI] [PubMed] [Google Scholar]
- Low C, Weininger U, Lee H, Schweimer K, Neundorf I, Beck-Sickinger AG, Pastor RW, Balbach J (2008) Structure and dynamics of helix-0 of the N-BAR domain in lipid micelles and bilayers. Biophys J 95: 4315–4323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundmark R, Doherty GJ, Vallis Y, Peter BJ, McMahon HT (2008) Arf family GTP loading is activated by, and generates, positive membrane curvature. Biochem J 414: 189–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Dyer JM, Quick ME, Goodman JM (1996) Redox-sensitive homodimerization of Pex11p: a proposed mechanism to regulate peroxisomal division. J Cell Biol 135: 123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM (1995) Pmp27 promotes peroxisomal proliferation. J Cell Biol 129: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda M, Takeda S, Sone M, Ohki T, Mori H, Kamioka Y, Mochizuki N (2006) Endophilin BAR domain drives membrane curvature by two newly identified structure-based mechanisms. EMBO J 25: 2889–2897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438: 590–596 [DOI] [PubMed] [Google Scholar]
- Min CK, Bang SY, Cho BA, Choi YH, Yang JS, Lee SH, Seong SY, Kim KW, Kim S, Jung JU, Choi MS, Kim IS, Cho NH (2008) Role of amphipathic helix of a herpesviral protein in membrane deformation and T cell receptor downregulation. PLoS Pathog 4: e1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay R, Huang KC, Wingreen NS (2008) Lipid localization in bacterial cells through curvature-mediated microphase separation. Biophys J 95: 1034–1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller WH, van der Krift TP, Krouwer AJ, Wosten HA, van der Voort LH, Smaal EB, Verkleij AJ (1991) Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J 10: 489–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagotu S, Saraya R, Otzen M, Veenhuis M, van der Klei IJ (2008) Peroxisome proliferation in Hansenula polymorpha requires Dnm1p which mediates fission but not de novo formation. Biochim Biophys Acta 1783: 760–769 [DOI] [PubMed] [Google Scholar]
- Nagotu S, Veenhuis M, van der Klei IJ (2010) Divide et impera: the dictum of peroxisomes. Traffic 11: 175–184 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Muller MM, Kremer K, Deserno M (2007) Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature 447: 461–464 [DOI] [PubMed] [Google Scholar]
- Sambrook J FE, Maniatis T (1989) Molecular Cloning, a Laboratory Manual, 2nd edn Cold Spring Harbor Laboratory, Cold Spring Harbor [Google Scholar]
- Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci USA 100: 9626–9631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Hu J, Kozlov MM, Rapoport TA (2009) Mechanisms shaping the membranes of cellular organelles. Annu Rev Cell Dev Biol 25: 329–354 [DOI] [PubMed] [Google Scholar]
- Sprote P, Brakhage AA, Hynes MJ (2009) Contribution of peroxisomes to penicillin biosynthesis in Aspergillus nidulans. Eukaryot Cell 8: 421–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y, Meyerkord CL, Wang HG (2009) Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell Death Differ 16: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms S, Erdmann R (2005) Dynamin-related proteins and Pex11 proteins in peroxisome division and proliferation. FEBS J 272: 5169–5181 [DOI] [PubMed] [Google Scholar]
- van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, Garcia-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martin JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, van der Klei IJ, van Peij NN et al. (2008) Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotechnol 26: 1161–1168 [DOI] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RB, Wanders RJ, Tager JM (1992) Biochemistry of peroxisomes. Annu Rev Biochem 61: 157–197 [DOI] [PubMed] [Google Scholar]
- Vogel V, Sheetz M (2006) Local force and geometry sensing regulate cell functions. Nat Rev Mol Cell Biol 7: 265–275 [DOI] [PubMed] [Google Scholar]
- Wriessnegger T, Gubitz G, Leitner E, Ingolic E, Cregg J, de la Cruz BJ, Daum G (2007) Lipid composition of peroxisomes from the yeast Pichia pastoris grown on different carbon sources. Biochim Biophys Acta 1771: 455–461 [DOI] [PubMed] [Google Scholar]
- Yin Y, Arkhipov A, Schulten K (2009) Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure 17: 882–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM (2006) How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol 7: 9–19 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional Information