Bypassing of stems versus linear base-by-base inspection of mammalian mRNAs during ribosomal scanning
During ribosomal scanning intact mRNA stem loops close to the start codon can pass the mRNA entry, but not the Exit channel, resulting in incorrect mRNA positioning and defective translation. The DExH-box helicase DHX29 and the initiation factor eIF1 resolve these aberrant translation initiation complexes.
Keywords: Ded1, DHX29, helicase, scanning, translation initiation
Abstract
Initiation codon selection in eukaryotes involves base-by-base inspection of the 5′-untranslated region of mRNA by scanning ribosomal 43S preinitiation complexes. We employed in vitro reconstitution to investigate factor requirements for this process and report that in the absence of eIF1 and DHX29, eIFs 4A, 4B and 4G promote efficient bypassing of stable stems by scanning 43S complexes and formation of 48S initiation complexes on AUG codons immediately upstream and downstream of such stems, without their unwinding. However, intact stems are not threaded through the entire mRNA Exit channel of the 40S subunit, resulting in incorrect positioning of mRNA upstream of the ribosomal P site in 48S complexes formed on AUG codons following intact stems, which renders them susceptible to dissociation by eIF1. In 48S complexes formed on AUG codons preceding intact stems, the stems are accommodated in the A site. Such aberrant complexes are destabilized by DHX29, which also ensures that mRNA enters the mRNA-binding cleft in a single-stranded form and therefore undergoes base-by-base inspection during scanning.
Introduction
On most eukaryotic mRNAs, translation initiation occurs by the scanning mechanism (Jackson et al, 2010). The first step is assembly of a 43S preinitiation complex comprising a 40S ribosomal subunit, eIF2/GTP/Met-tRNAiMet, eIF3, eIF1 and eIF1A. 43S complexes attach to the 5′-proximal region of mRNA and scan to the initiation codon where they form 48S initiation complexes with established codon–anticodon base pairing. Although 43S complexes alone can bind to the 5′-end of an unstructured 5′-UTR and scan to the initiation codon, revealing their intrinsic ability to move along mRNA, ribosomal attachment and scanning on structured 5′-UTRs requires eIFs 4A, 4B and 4F, factors associated with RNA unwinding (Pestova and Kolupaeva, 2002). eIF4F comprises eIF4E (a cap-binding protein), eIF4A (a DEAD-box RNA helicase) and eIF4G (a scaffold for eIF4E and eIF4A, which also binds eIF3). The weak helicase activity of eIF4A is enhanced by eIF4G and eIF4B: eIF4G acts by aligning the DEAD-box motifs of eIF4A in a productive conformation (Schütz et al, 2008), whereas eIF4B might prevent mRNA reannealing and promote unidirectional eIF4A movement (Marintchev et al, 2009).
The mRNA path on bacterial 30S subunits (Yusupova et al, 2001) comprises three regions: the Entry channel (∼12 nts), the exposed interface surface (the A, P and E sites), and the Exit channel (∼12 nts). These regions are conserved between bacterial and eukaryotic small ribosomal subunits, and the mRNA path on the 40S subunit is very similar to that in bacteria (Pisarev et al, 2008). mRNA enters the 40S subunit between the head and the shoulder and passes through a layer of ribosomal proteins (rp) including rpS2 and rpS3 and then through a layer of rRNA including helices (h) 18 in the body and 34 in the neck. h18 and h34 form a latch that is closed in free 40S subunits but opens upon binding of eIF1 and eIF1A, which occupy the areas of P and A sites, respectively (Lomakin et al, 2003; Passmore et al, 2007; Yu et al, 2009). Before exiting between the head and the platform, mRNA passes through another tunnel formed by elements that include rpS5 and h23, whereas further upstream, it is positioned close to rps S14/S26/S28 and to the 3′-end of 18S rRNA. It is not known if eIFs 4A/4G/4B act at the 40S subunit's leading edge by unwinding mRNA before it enters the mRNA-binding cleft, or if they bind near its trailing edge and assist scanning by ‘pulling' mRNA through the 40S subunit. Although eIFs 4A/4B/4G can mediate scanning through stems of ΔG⩽−13.1 kcal/mol, movement of mammalian 40S subunits through stems of ΔG>−19 kcal/mol requires an additional DExH-box protein, DHX29 (Pisareva et al, 2008). Silencing of DHX29 impairs translation, resulting in polysome disassembly and accumulation of mRNA-free 80S ribosomes (Parsyan et al, 2009). DHX29 acts synergistically with eIFs 4A/4G/4B, and binds directly to 40S subunits, likely at the mRNA entrance (Pisareva et al, 2008). The mechanism by which DHX29 assists scanning is also unknown.
Efficient scanning also depends on adoption by 40S subunits of a scanning-competent conformation induced by eIF1 and eIF1A: omission of eIF1A reduces the intrinsic scanning ability of 43S complexes, whereas omission of eIF1 almost abrogates it (Pestova and Kolupaeva, 2002). eIF1 also has a key role in maintaining the fidelity of initiation codon selection, enabling 43S complexes to discriminate against non-AUG triplets, and AUG triplets in suboptimal context or located within 8 nts of the 5′-end (Pestova and Kolupaeva, 2002; Lomakin et al, 2006; Pisarev et al, 2006). In a current model, eIF1 acts by antagonizing conformational changes that occur upon codon–anticodon base pairing and switch ribosomal complexes from ‘open' (scanning competent) to ‘closed' conformations (Lorsch and Dever, 2010).
The yeast DEAD-box helicase Ded1 and its mammalian homologue DDX3 have also been implicated in initiation (Tarn and Chang, 2009). Mutational inactivation of Ded1 severely reduces polysomes and leads to accumulation of 80S monosomes (Chuang et al, 1997; de la Cruz et al, 1997). Ded1 is likely a more potent helicase than eIF4A (Marsden et al, 2006), and their functions are not redundant (Chuang et al, 1997; de la Cruz et al, 1997). This has led to a suggestion that eIF4A functions in 43S complex attachment to mRNA, whereas Ded1 promotes scanning (Marsden et al, 2006). In contrast, DDX3's involvement in initiation is controversial. Although some reports indicated that depletion of DDX3 inhibited general translation (Lee et al, 2008) or specifically affected translation of mRNAs with long or structured 5′-UTRs (Lai et al, 2008), others show that protein synthesis in a cell line expressing a ts DDX3 mutant was normal at non-permissive temperatures (Fukumura et al, 2003), and that silencing DDX3 even enhanced general translation (Shih et al, 2008).
Although eIF4A, DHX29 and Ded1 have convincingly been implicated in initiation, their mechanisms of action are largely unknown. Moreover, many mechanistic aspects of scanning are also poorly understood. Thus, it is not clear what happens when a scanning 43S complex encounters a stable stem: can an intact stem at least to some extent penetrate into the mRNA-binding cleft or is it stopped at its entry, and do changes to the structure of the mRNA-binding channel induced by eIF1, eIF1A and presumably DHX29 influence mechanistic aspects of movement of structured mRNAs through the mRNA-binding cleft of the 40S subunit?
Using an in vitro reconstitution approach, we investigated 48S complex formation on mRNAs containing stable stems and AUG triplets at different positions relative to them. We report that in the absence of DHX29 and eIF1, eIFs 4A/4B/4G promote efficient bypassing of stable stems by scanning 43S complexes, and formation of 48S complexes on AUGs immediately upstream and downstream of such stems without their unwinding. However, the intact stems likely cannot be threaded through the entire Exit channel, resulting in incorrect positioning of mRNA upstream of the P site that renders 48S complexes formed on AUGs downstream of intact stems susceptible to dissociation by eIF1. In 48S complexes formed on AUGs preceding stems, the stem and an adjacent mRNA region between the stem and the AUG are accommodated in the A site. Such complexes are destabilized by DHX29, which also ensures that structured mRNAs enter the mRNA-binding cleft in a single-stranded form and undergo linear base-by-base inspection during scanning. We also found that Ded1 promoted scanning on 5′-UTRs containing moderately stable stems and could act cooperatively with eIFs 4A/4B/4G. However, the activity of eIFs 4A/4B/4G in promoting scanning through more stable stems was higher when they were combined with DHX29 than with Ded1. No translational activity of DDX3 was observed in any circumstances.
Results
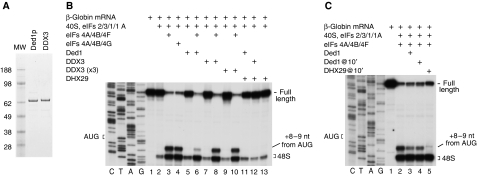
To investigate the individual and synergistic activities of eIFs 4A/4B/4G, Ded1, DDX3 and DHX29 in promoting scanning and base-by-base inspection of structured 5′-UTRs by 43S complexes, we determined their ability to mediate 48S complex formation on mRNAs containing defined stems of various stabilities and AUG codons at different positions relative to them, using a mammalian in vitro reconstituted translation system. To assess the influence of the conformation of the mRNA-binding cleft, experiments were done with and without eIF1 and eIF1A. In these studies we used native DHX29, recombinant Ded1 and DDX3 (Figure 1A) that were biochemically active and possessed RNA-dependent ATPase activity (data not shown), native eIF4F and recombinant eIF4A, eIF4B and N-terminally truncated ΔeIF4G682–1599, which we refer to as eIF4G below. Assembly of 48S complexes was monitored by the appearance of characteristic toe-prints +15–17 nts downstream of the initiation codon.
Figure 1.
Comparison of the activities of eIFs 4A/4B/4F, DHX29, Ded1 and DDX3 in 48S complex formation on β-globin mRNA. (A) Purified recombinant Ded1 and DDX3 resolved by SDS–PAGE. (B, C) Toe-printing analysis of 48S complex formation on native β-globin mRNA in the presence of indicated combinations of factors. Positions of the initiation codon, of the full-length cDNA and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict the corresponding DNA sequences.
Activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in 48S complex formation on β-globin mRNA
We first investigated 48S complex formation on native capped β-globin mRNA, which does not have an unstructured 5′-terminal region (Lockard et al, 1986) that would allow 43S complexes to bind without prior unwinding. Ded1 and DDX3 individually or with DHX29 did not promote 48S complex formation (Figure 1B, lanes 5, 7, 9, 11 and 12) and did not influence the efficiency of 48S complex formation mediated by eIFs 4A/4B/4F (Figure 1B, lanes 3, 6, 8 and 10). The inability of Ded1 and DDX3 to promote 48S complex formation, which contrasted with the activity of eIFs 4A/4B/4F, could not be attributed eIF4F's specific affinity to the cap because the activities of eIFs 4A/4B/4G and 4A/4B/4F were similar (Figure 1B, lanes 3 and 4). Interestingly, like DHX29 (Pisareva et al, 2008), Ded1 reduced the intensity of aberrant toe-prints +8–9 nts downstream of the AUG codon, albeit less efficiently, particularly when it was added to preassembled 48S complexes (Figure 1C). DDX3 did not influence +8–9 toe-prints even at elevated concentrations (Figure 1B, lanes 8 and 10).
Importantly, as β-globin mRNA does not contain an unstructured 5′-terminal region that would allow 43S complexes to attach directly, the inability of Ded1 and DDX3 to promote 48S complex formation on it might, as in the case of DHX29 (Pisareva et al, 2008), reflect their inability to promote ribosomal attachment rather than a lack of translational activity.
Individual and synergistic activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in ribosomal scanning
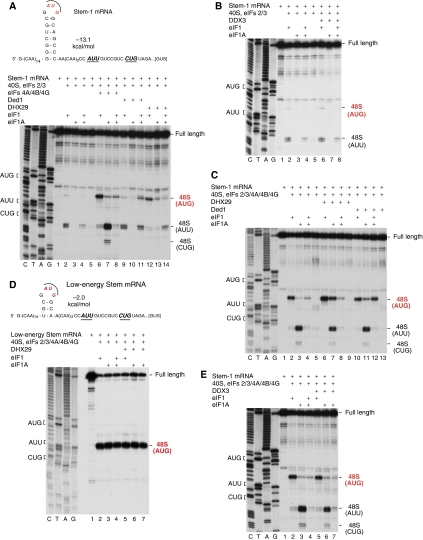
To investigate the activity of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in promoting scanning independently of a role in ribosomal attachment, we used model (CAA)n-Stem-GUS mRNAs comprising a GUS reporter and 5′-UTRs with 43 unstructured 5′-terminal nucleotides (CAA repeats) that allow helicase-independent attachment of 43S complexes (Pestova and Kolupaeva, 2002), followed by stems of different stabilities (Figures 2A and 3A). To verify that it is base-by-base inspection of mRNAs that is being monitored, AUG codons in good context (with purines at −3 and +4 positions) were inserted into the loop of the hairpins rather than downstream of them, to exclude the potential for formation of 48S complexes through bypassing of the stem by scanning 43S complexes. The absence of near-cognate codons preceding the AUG triplet also allowed the dependence of 48S complex formation on eIF1 and eIF1A to be assayed.
Figure 2.
Individual and synergistic activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in promoting scanning through 5′-UTRs with an internal stem of moderate stability. (A–E) Toe-printing analysis of 48S complex formation on (A–D) Stem-1 and (E) low-energy Stem mRNAs in the presence of indicated combinations of factors. Initiation codons and the positions of full-length cDNAs and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict corresponding DNA sequences.
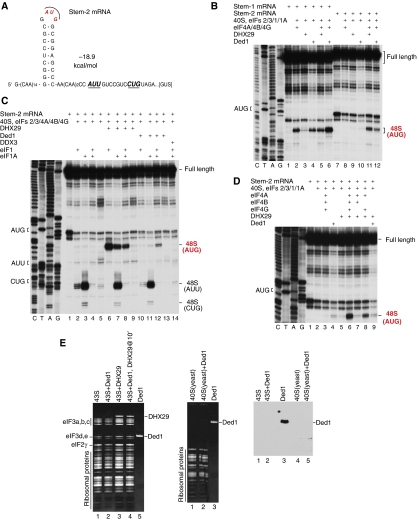
Figure 3.
Individual and synergistic activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in promoting scanning through 5′-UTRs with a highly stable internal stem. (A) Sequence of the 5′-UTR of Stem-2 mRNA. (B–D) Toe-printing analysis of 48S complex formation on (B) Stem-1 and (B–D) Stem-2 mRNAs in the presence of indicated combinations of factors. Initiation codons and the positions of full-length cDNAs and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict corresponding DNA sequences. (E) Association of Ded1 (all panels) and DHX29 (left panel) with mammalian 43S complexes (left and right panels) and yeast 40S subunits (central and right panels) was assayed by sucrose density gradient (SDG) centrifugation. Gradient fractions that corresponded to ribosomal peaks were analysed by SDS–PAGE and fluorescent SYPRO staining (left and central panels) or western blotting using anti-Ded1 antibodies (right panel).
First, the individual activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 were compared in 48S complex formation on Stem-1 mRNA containing a GC-rich stem of moderate stability (ΔG=−13.1 kcal/mol; Figure 2A). In the absence of helicases, low-level 48S complex formation occurred on the AUG and on the AUU 10 nts downstream of Stem-1 in the presence of eIF1 alone, which was reduced further when eIF1 was combined with eIF1A (Figure 2A, lanes 2 and 4). No 48S complexes formed without eIF1 (Figure 2A, lanes 3 and 5). eIFs 4A/4B/4G promoted 48S complex formation efficiently on the AUG in the presence of eIF1 and moderately in the presence of eIF1A or eIF1/1A (Figure 2A, lanes 6–8). Surprisingly, in the presence of eIF1A alone, eIFs 4A/4B/4G also mediated high-level 48S complex formation on the AUU and even moderately on the CUG further downstream (Figure 2A, lane 7). Thus, in the presence of eIF1A, eIFs 4A/4B/4G did not ensure efficient inspection of Stem-1 by scanning 43S complexes, but instead promoted 48S complex formation downstream of the stem. In the presence of eIF1 and eIF1A together (but not individually) Ded1-mediated 48S complex formation on the AUG at a level similar to that observed in the presence of eIFs 4A/4B/4G (Figure 2A, lanes 9–11), whereas DHX29 stimulated 48S complex formation on the AUG most strongly in the presence of eIF1 alone, moderately in the presence of eIF1A alone, and weakly in the presence of both factors (Figure 2A, lanes 12–14). Interestingly, Ded1 also allowed more efficient 48S complex formation on the AUU in the presence of both eIF1 and eIF1A than eIFs 4A/4B/4G (Figure 2A, lanes 8 and 11). Moreover, initiation on the first AUG codon of mRNAs containing unstructured 5′-UTRs with two AUG triplets was considerably more efficient in the presence of Ded1 than of eIFs 4A/4B/4G, irrespective of the nucleotide context of the first AUG triplet (Supplementary Figure S1). Thus, Ded1-mediated scanning was less discriminating with respect to the context/sequence of the initiation codon than scanning mediated by eIFs 4A/4B/4G, and thus less leaky. Importantly, neither Ded1 nor DHX29 promoted efficient 48S complex formation downstream of Stem-1 in the presence of eIF1A alone (Figure 2A, lanes 10 and 13). eIFs 4A/4B/4G, Ded1 or DHX29 did not stimulate 48S complex formation when both eIF1 and eIF1A were absent (data not shown). DDX3 was not active in any circumstances (Figure 2B).
We next assayed the ability of DHX29, Ded1 and DDX3 to act synergistically with eIFs 4A/4B/4G. eIFs 4A/4B/4G and DHX29 synergistically stimulated 48S complex formation on the AUG when eIF1 or eIF1A was present individually, whereas synergy between eIFs 4A/4B/4G and Ded1 required both factors (Figure 2C, lanes 6–8, 10–12). As no synergy between eIFs 4A/4B/4G and DHX29 occurred in the presence of eIF1 and eIF1A, the activity of eIFs 4A/4B/4G was higher in combination with Ded1 than with DHX29 when both eIF1 and eIF1A were present (Figure 2C, lanes 8 and 12). As in the case of individual helicases, combinations of them also did not promote 48S complex formation when both eIF1 and eIF1A were absent (Figure 2C, lanes 9 and 13). With both combination of helicases, efficient 48S complex formation on AUU and CUG triplets downstream of Stem-1 again occurred in the presence of eIF1A alone (Figure 2C, lanes 3, 7 and 11), although enhanced 48S complex assembly on the AUG in the presence of DHX29 was accompanied by a moderate reduction in 48S complex formation downstream of the stem (Figure 2C, lane 7). Importantly, elimination of GC pairs at the base of Stem-1 resulted in efficient recognition of the AUG in the loop of the stem irrespective of the presence of DHX29 and consequently, in abrogation of 48S complex formation on downstream AUU and CUG codons (Figure 2D). This confirmed that efficient recognition of the downstream AUU on Stem-1 mRNA in the presence of eIF1A alone was due to a blockage of linear scanning and skipping of the AUG in the hairpin by 43S complexes. DDX3 did not influence the activity of eIFs 4A/4B/4G in any circumstances (Figure 2E).
To compare the relative strength of helicases individually and in combination, 48S complex formation was assayed on Stem-2 mRNA containing a more stable stem (ΔG=−18.9 kcal/mol) (Figure 3A). If both eIF1 and eIF1A were present, eIFs 4A/4B/4G, Ded1 and DHX29 could individually promote 48S complex formation on the AUG of Stem-1 mRNA, albeit with different efficiencies (Figure 3B, lanes 2–4), but not on Stem-2 mRNA: this required eIFs 4A/4B/4G to be combined with DHX29 or Ded1 (Figure 3B, lanes 8–12). As expected, this process strictly required eIF4G and was stimulated by eIF4B (Figure 3D, lanes 6–8). eIFs 4A/4B/4G and DHX29 functioned synergistically whether eIF1 and eIF1A were present individually or together (Figure 3C, lanes 6–8), but synergy between eIFs 4A/4B/4G and Ded1 again required both factors (Figure 3C, lanes 10–12). Interestingly, the activity of eIFs 4A/4B/4G alone and with DHX29 in promoting 48S complex formation on both Stem-1 and Stem-2 mRNAs was higher when eIF1 was present alone than with eIF1A (Figures 2B and D and 3C, lanes 6 and 8). The most likely explanation for this is that together, eIF1 and eIF1A sense deviations in the context of this AUG at positions other than the −3 and +4 consensus purines, and thus do not permit high-level 48S complex formation on it. Notably, in the presence of eIF1 and eIF1A, eIFs 4A/4B/4G were more active in 48S complex formation on Stem-2 mRNA with DHX29 than with Ded1 (Figure 3C, lanes 8 and 12), whereas the situation on Stem-1 mRNA was reversed (Figure 2C). Initiation on the AUG codon in the loop of a stem would require unwinding of the stem as well as successful recognition of the AUG codon by the scanning complex. A possible explanation for the difference in the activities of Ded1 and DHX29 in promoting initiation on the AUGs of Stem-1 and Stem-2 mRNAs could be that Stem-1 can be efficiently unwound by both sets of factors but the AUG is better recognized in the presence of Ded1 (which would be consistent with the reduced leaky scanning during Ded1-mediated 48S complex formation described above), whereas more efficient unwinding of Stem-2 can be achieved in the presence of DHX29. The step that limits initiation on the internal AUG of Stem-1 mRNA would therefore be initiation codon recognition, whereas initiation on the more stable Stem-2 mRNA would be limited by unwinding of the hairpin. Importantly, efficient eIF4A/4B/4G-mediated 48S complex formation on AUU and CUG triplets downstream of Stem-2 again occurred in the presence of eIF1A alone (Figure 3C, lanes 3, 7 and 11). There was no synergy between Ded1 and DHX29 (Figure 3D, lanes 4, 5 and 9), and DDX3 did not influence the activity of eIFs 4A/4B/4G in any circumstances (Figure 3C, lane 14; data not shown).
Although Ded1 and DHX29 showed some functional similarities, unlike DHX29, Ded1 did not bind stably to mammalian 43S complexes or to yeast 40S subunits, and preincubation of 43S complexes with Ded1 did not influence their association with DHX29 (Figure 3E). This might indicate a difference in the mechanisms of coupling of the helicase activities of Ded1 and DHX29 with ribosomal complexes.
In conclusion, in the presence of eIF1 and eIF1A, Ded1 (but not DDX3) was able to promote scanning on mRNAs containing moderately stable GC-rich stems and could act synergistically with eIFs 4A/4B/4G. However, eIFs 4A/4B/4G promoted scanning through more stable stems more efficiently with DHX29 than with Ded1. Importantly, in the presence of eIF1A alone, eIFs 4A/4B/4G did not mediate efficient inspection of stems by scanning 43S complexes, but instead promoted high-level 48S complex formation on downstream near-cognate codons.
Bypassing of stable stems by scanning 43S complexes
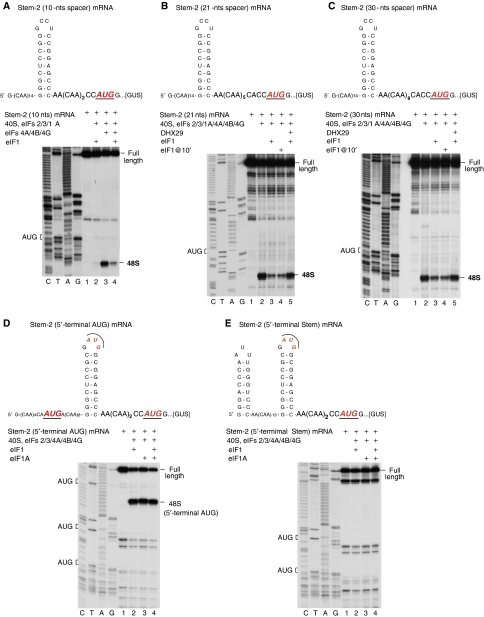
The efficient 48S complex formation mediated by eIFs 4A/4B/4G in the presence of eIF1A on near-cognate codons downstream of stable stems without their base-by-base inspection poses two important questions: (i) can formation of such complexes tolerate eIF1 if a near-cognate codon is replaced by an AUG triplet and (ii) do they form by 5′-end-dependent scanning or by internal ribosomal entry? To address the first question, 48S complex formation was assayed on mRNA in which the AUG in Stem-2 was eliminated, and the AUU downstream of the stem was replaced by AUG (Figure 4A, upper panel). Efficient 48S complex formation on the new AUG again occurred only in the absence of eIF1 (Figure 4A, lanes 3 and 4). As eIFs 4A/4B/4G did not promote inspection of this stem (Figure 3C), it likely remained intact in 48S complexes assembled on the downstream codon. The proximity (10 nts) of the stem to the AUG could affect correct positioning of mRNA in the Exit portion of the mRNA-binding cleft, and thus result in sensitivity of 48S complexes to dissociation by eIF1 (Pestova and Kolupaeva, 2002). However, increasing the separation between the stem and the AUG to 21 or 30 nts did not eliminate the sensitivity of 48S complex formation to eIF1 (Figure 4B and C). Moreover, even delayed addition of eIF1 to preassembled 48S complexes led to their dissociation (Figure 4B and C, lane 4). On the other hand, inclusion of DHX29 substantially enhanced 48S complex formation in eIF1's presence (Figure 4B and C, compare lanes 3, 4 with 5).
Figure 4.
48S complex formation on mRNAs with AUG triplets downstream of stable stems depending on the presence of eIF1 and DHX29. Toe-printing analysis of 48S complex formation on (A–C) mRNAs-containing AUG codons 10, 21 and 30 nucleotides downstream of Stem-2, respectively, (D) mRNA-containing AUG triplets 15 nts downstream of the 5′-end of mRNA, in the loop of Stem-2, and 10 nts downstream of the stem, and (E) mRNA containing an additional GC-rich stem at the extreme 5′-end and two AUG triplets, in the loop of Stem-2 and 10 nts downstream from the stem, respectively, in the presence of indicated combinations of factors. Initiation codons and the positions of full-length cDNAs and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict corresponding DNA sequences.
Apparently, mRNA is not properly fixed in the mRNA-binding cleft of 48S complexes formed downstream of the stem if it is not unwound, rendering them susceptible to dissociation by eIF1. To investigate if these complexes nonetheless form by scanning, two further mRNAs were used. Both contained AUGs in Stem-2 and 10 nts downstream of the stem, but the first also contained an AUG 15 nts from the 5′-end (Figure 4D, upper panel), whereas the second had a stable 5′-terminal stem (Figure 4E, upper panel). If 48S complex formation on the AUG downstream of the stem occurs by 5′-end-dependent scanning, introduction of an upstream AUG or a 5′-terminal stem should abrogate initiation on it. No 48S complexes formed on the AUG downstream of the stem on either mRNA (Figure 4D and E), confirming that 48S complex assembly on the downstream AUG occurs by 5′-end-dependent scanning.
These data suggest that eIFs 4A/4B/4G enable scanning 43S complexes to bypass stable stems and to form 48S complexes downstream of them, but that intact stems likely cannot be threaded through the entire Exit portion of the mRNA-binding cleft, resulting in incorrect ribosomal positioning of mRNA upstream of the P site and consequent susceptibility of 48S complexes to dissociation by eIF1. DHX29, on the other hand, enables 43S complexes to penetrate into the stem (Figure 3C), so that in its presence, 48S complexes form on initiation codons downstream of the stem following its unwinding, and the correct positioning of unwound mRNA renders them resistant to dissociation by eIF1.
Linear base-by-base inspection of mRNA by scanning 43S complexes
The data described above suggest that in the absence eIF1, eIFs 4A/4B/4G enable scanning 43S complexes to bypass stable stems and to form 48S complexes downstream of them. This, in turn, raises the following questions (i) does eIF1 prevent intact stems from entering the mRNA-binding cleft or does it permit entry, but acts only later, while codon–anticodon base pairing is being established and (ii) if eIF1 does not prevent intact stems from entering the mRNA-binding cleft, then what is the mechanism that ensures that structured mRNAs enter the mRNA-binding cleft in a single-stranded form and undergo linear base-by-base inspection?
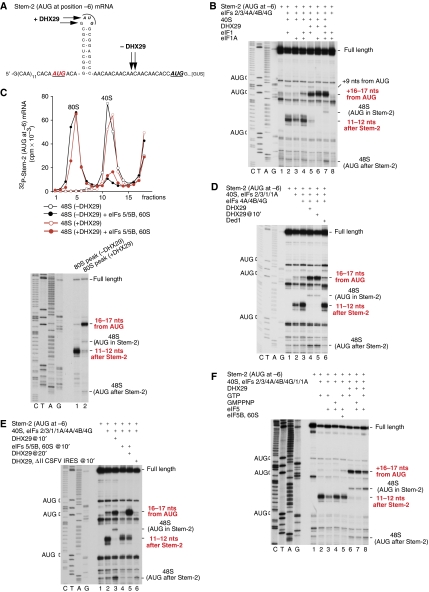
To address these questions, we used an mRNA in which an AUG at position −6 relative to Stem-2 was introduced in addition to AUGs in the stem and 21 nts downstream of it (Figure 5A). We hypothesized that if an intact stem can be stably accommodated in the Entrance portion of the mRNA-binding cleft downstream of the P site, then 48S complexes formed on the AUG preceding the stem should yield toe-prints not +15–17 nts downstream of this codon, but instead a few nts downstream of the stem. Consistent with our hypothesis, eIFs 4A/4B/4G promoted efficient 48S complex formation on the AUG preceding the stem yielding toe-prints +11–12 nts downstream of the stem (with additional minor stops at +13–15 nts) (Figure 5B, lanes 2–4), and only a very small proportion of complexes with conventional +16–17 nts toe-prints assembled in the presence of both eIF1 and eIF1A (Figure 5B, lane 4). The toe-prints +11–12 nts downstream of the stem were caused by bound 40S subunits and did not appear in their absence (Figure 5B, lane 8). A small amount of 48S complexes with toe-prints +11–12 nts downstream of the stem formed even in the absence of eIFs 4A/4B/4G (Figure 5D, lanes 2 and 3). Importantly, eIF1 even stimulated formation of 48S complexes with toe-prints +11–12 nts downstream of the stem (Figure 5B, compare lanes 2, 4 with lane 3) showing that it does not prevent intact stems from entering the mRNA-binding cleft.
Figure 5.
48S complex formation on mRNAs with AUG triplets upstream of stable stems depending on the presence of DHX29. (A) Sequence of the 5′-UTR of Stem-2 (AUG at –6) mRNA, containing an AUG triplet 6 nts before Stem-2 in addition to two other AUG triplets, in the loop of the stem and 21 nts downstream from it, respectively. (B, D–F) Toe-printing analysis of 48S complex formation on Stem-2 (AUG at –6) mRNA in the presence of indicated combinations of translation components. Initiation codons and the positions of full-length cDNAs and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict corresponding DNA sequences. Positions of toe-prints that were observed with/without DHX29 are indicated with arrows on the sequence of mRNAs in panels (A, F). (C) Ribosomal subunit joining on 48S complexes assembled on 32P-labelled Stem-2 (AUG at –6) mRNA with/without DHX29, assayed by SDG centrifugation followed by Cerenkov counting (upper panel), and toe-printing analysis of the 80S-containing fractions (lower panel).
Strikingly, addition of DHX29 resulted in almost exclusive assembly of 48S complexes with +16–17 nts toe-prints, indicative of correct ribosomal positioning of unwound mRNA downstream of the P site (Figure 5B, lanes 5–7). Consistent with the ability of eIF1 and eIF1A to increase scanning processivity, a small amount of 48S complexes also formed on the two other downstream AUGs in the presence of DHX29 and both eIF1 and eIF1A (Figure 5B, lane 7). Upon incubation with eIF5, eIF5B and 60S subunits, 48S complexes assembled with and without DHX29 (and thus characterized by different toe-prints) efficiently formed 80S complexes that yielded toe-prints at the same positions (Figure 5C). Both types of 80S complexes were puromycin reactive (data not shown). Importantly, delayed addition of DHX29 to preassembled 48S complexes also led to exclusive formation of complexes with +16–17 nts toe-prints (Figure 5D, lanes 4 and 5). Conversion by DHX29 of aberrant 48S complexes with +11–12 nts. toe-prints into correctly assembled complexes could result either from changes induced by DHX29 in the former while they remained bound to the mRNA, or from their destabilization, mRNA release and its subsequent entry into a new round of initiation. To distinguish between these scenarios, DHX29 was added to preassembled 48S complexes simultaneously with an mRNA competitor, the ΔII CSFV IRES lacking Domain II, which in contrast to the wt IRES can efficiently bind to DHX29-associated 43S complexes (Pisareva et al, 2008). This led to disappearance of 48S complexes with toe-prints +11–12 nts downstream of the stem without assembly of complexes with +16–17 nts toe-prints (Figure 5E, compare lanes 2, 3 and 6), suggesting that DHX29 first destabilizes 48S complexes with intact stems in the mRNA-binding channel downstream of the P site, after which properly assembled 48S complexes form in a new round of initiation. 48S complexes containing intact stems were protected from DHX29-induced dissociation by joining with 60S subunits (Figure 5E, lanes 4 and 5). In contrast, Ded1 did not suppress formation of 48S complexes containing an intact stem and characterized by toe-prints +11–12 nts downstream of it (Figure 5D, lane 6; Figure 6A and B, lane 8).
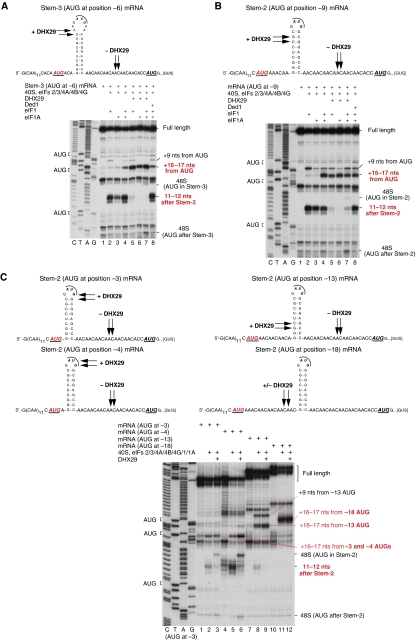
Figure 6.
Influence of the position of the AUG triplet relative to the downstream stable stem on 48S complex formation in the presence and in the absence of DHX29. Toe-printing analysis on mRNAs containing (A) the extended Stem-3 and three AUG triplets, 6 nts before Stem-3, in the loop of Stem-3 and 21 nts downstream of Stem-3, and (B, C) Stem-2 and three AUG triplets, (B) 9 nts, (C) 3, 4, 13 or 18 nts before Stem-2, in the loop of Stem-2 and 21 nts downstream of Stem-2, respectively, in the presence of indicated combinations of factors. Initiation codons and the positions of full-length cDNAs and of assembled ribosomal complexes are shown on the sides of each panel. Lanes C/T/A/G depict corresponding DNA sequences. Positions of toe-prints that were observed with/without DHX29 are indicated with arrows on the sequence of all mRNAs.
We next assayed the influence of eIF5 on initiation codon selection in conditions that do not permit ribosomal subunit joining (i.e. in the absence of eIF5B and 60S subunits). In the presence of GMPPNP, inclusion of eIF5 in reaction mixtures did not influence 48S complex formation whether or not DHX29 was present (Figure 5F, compare lanes 2 and 6 with lanes 4 and 7). However, inclusion of eIF5 in the presence of GTP reduced assembly of 48S complexes on the AUG preceding the stem in the absence of DHX29 (Figure 5F, lane 3), and resulted in a decrease in 48S complex formation on the AUG preceding the stem and a concomitant increase in 48S complex formation on downstream AUGs in the presence of DHX29 (Figure 5F, lane 8). Thus in the first instance, ribosomal complexes that fail to recognize the first AUG are unable to access downstream AUGs because of the absence of DHX29, whereas in its presence, inclusion of eIF5 leads to increased leaky scanning past the first AUG triplet. These results were consistent with the leaky scanning phenotype observed on deletion of eIF5B in yeast (Choi et al, 1998). When subunit joining is permitted, there was no strong decrease in the formation of 48S complexes on the first AUG in the absence of DHX29 (Figure 5F, lane 5) and no increase in their assembly on downstream AUGs in its presence (Figure 5C, lane 2; Figure 5E, lane 5).
As expected, lengthening the stem did not alter the position of toe-prints downstream of it when 48S complexes were assembled without DHX29 (Figure 6A, lanes 2–4). Again, addition of DHX29 resulted in formation of 48S complexes with +16–17 nts toe-prints (Figure 6A, lanes 5–7). However, changing the position of the AUG from −6 to −3, −4, −9 or −13 relative to the stem also did not change the position of toe-prints downstream of the stem observed in DHX29's absence (Figure 6B, lanes 2–4; Figure 6C, lanes 2, 5 and 8), suggesting that the intact stem has a ‘preferred' position in the mRNA-binding channel that determines the position of the toe-print. The length of the mRNA Entry channel, which is about 10–12 nts (Yusupova et al, 2001), and the fact that 48S complexes assembled on the AUG immediately preceding the stem (−3 position) also yield the +11–12 nts toe-prints indicate that the stem and a region between it and the AUG are accommodated in the A site. In contrast, 48S complexes that formed on mRNA with the AUG at position −18 (in which case the stem would be outside of the mRNA-binding channel) yielded only canonical +16–17 nts toe-prints irrespective of DHX29's presence (Figure 6C, lanes 11 and 12).
Interestingly, placement of the AUG at positions −9 or −13 also yielded relatively strong toe-prints +9 nts downstream of the codon (Figure 6B and C). Only a very weak +9 nts toe-print was seen when the AUG was closer to the stem or was separated from it by 18 nts (Figures 5B and 6A and C). Ribosomal complexes with +8–9 nts toe-prints have been observed on various mRNAs (Figure 1B; Pisareva et al, 2008). They most likely represent stable initiation complexes, in which mRNA is not correctly positioned along the entire length of the Entrance area of the mRNA-binding cleft, which allows reverse transcriptase to penetrate further. Separation of the AUG and the stem by ⩾9, but <17 nts, likely encouraged such complexes to form.
In conclusion, these data indicate that eIF1 can assess correct positioning of mRNA only upstream of the P site, whereas DHX29 has the function of ensuring that structured mRNAs enter the mRNA-binding cleft in a single-stranded form and undergo linear base-by-base inspection by scanning 43S complexes.
Discussion
Individual and synergistic activities of eIFs 4A/4B/4G, DHX29, Ded1 and DDX3 in 48S complex formation on mRNAs with structured 5′-UTRs
Consistent with Ded1's reported function in initiation (Chuang et al, 1997; de la Cruz et al, 1997) and in particular, with its suggested role in scanning (Marsden et al, 2006), we found that individually, Ded1 could promote scanning on a 5′-UTR containing a 43-nt long unstructured 5′-terminal region and a moderate GC-rich stem (ΔG=−13.1 kcal/mol). Like DHX29, it could also act cooperatively with eIFs 4A/4B/4G. However, the activity of eIFs 4A/4B/4G in promoting scanning through more stable stems (ΔG=−18.9 kcal/mol) was higher with DHX29 than with Ded1. Interestingly, the activity of Ded1 required both eIF1 and eIF1A, whereas eIFs 4A/4B/4G and DHX29 were able to promote efficient scanning when they were present individually. Moreover, unlike DHX29, Ded1 did not bind stably to mammalian or yeast 40S subunits and did not prevent assembly of 48S complexes containing intact stems in the mRNA-binding cleft downstream of the P site. Taken together, these data suggest that the mechanisms of action of DHX29 and Ded1 might not be identical. This poses the question whether yeast has a DHX29 orthologue. BLAST searches identified YLR419w as the protein most closely related to DHX29 in Saccharomyces cerevisiae, but reverse searches showed that YLR419w is closer to mammalian DHX57 and DHX36 than to DHX29. In any case, homology between these proteins is limited to their C-terminal two thirds. YLR419w is not essential (Shiratori et al, 1999) and a YLR419w deletion strain has no apparent translational defect, YLR419w does not associate with 40S subunits in vivo, and purified YLR419w does not bind yeast or mammalian 40S subunits in vitro (VP, TP, A Komar, unpublished data). These data indicate that YLR419w is likely not a functional DHX29 orthologue. Importantly, most yeast mRNAs have short (<100 nt long) 5′-UTRs without strong secondary structure (Lawless et al, 2009), and the level of scanning processivity mediated by Ded1 that was observed in the mammalian reconstituted system might be sufficient to ensure initiation in yeast. Moreover, Ded1's helicase activity might be better coupled with yeast than with mammalian ribosomal complexes, in which case Ded1 might promote more processive scanning in yeast than in the mammalian reconstituted system.
The observed lack of activity of DDX3 contradicts some reports (Lai et al, 2008; Lee et al, 2008), but is consistent with others (Fukumura et al, 2003; Shih et al, 2008). Although the recombinant DDX3 used in our studies was biochemically active, we cannot exclude that its lack of activity in 48S complex formation might be due to partial misfolding, or to the absence of potentially important post-translational modifications or of a cofactor that could couple its helicase activity with ribosomal movement. DDX3's lack of translational activity seems to be difficult to reconcile with complementation by DDX3 and related mammalian proteins of the lethality of a chromosomal ded1 deletion or the cold-sensitive lethality of a ded1 mutation (Chuang et al, 1997; Mamiya and Worman, 1999; Fukumura et al, 2003), but given the numerous overlapping functions of DDX3 and Ded1 in processes other than translation (e.g. splicing, mRNA export and formation of translationally repressed messenger ribonucleoprotein particles; Tarn and Chang, 2009), these complementation studies do not provide unambiguous proof for a common role for Ded1 and DDX3 in translation.
Bypassing of stable stems by scanning 43S complexes
The results presented here show that when a scanning 43S complex encounters a stem in the 5′-UTR, it can be either unwound or ‘bypassed'. In the former case, mRNA enters the mRNA-binding channel of the 40S subunit in single-stranded form, and its entire 5′-UTR is inspected base-by-base by the P-site initiator tRNA. The ability of scanning complexes to recognize AUG codons inside stem-loop structures and to assemble 48S complexes on them indicates that after an unwound stem enters the mRNA-binding cleft, it cannot reform while still in contact with the 40S subunit. In the case of stem ‘bypass', the intact stem enters the mRNA-binding region of the 40S subunit and is not subjected to base-by-base inspection. Both processes are stimulated by eIFs 4A/4B/4G, and the outcome (unwinding versus bypassing) is determined by the stability of the stem and depends on the presence of DHX29, which strongly promotes unwinding of mRNA before it enters the mRNA-binding cleft of the 40S subunit.
The mechanisms and factor dependence of 48S complex formation on AUG triplets located upstream and downstream of intact stems, which are determined by the movement through and accommodation in the Entry and Exit channels of unwound stems, differ significantly and will therefore be discussed separately.
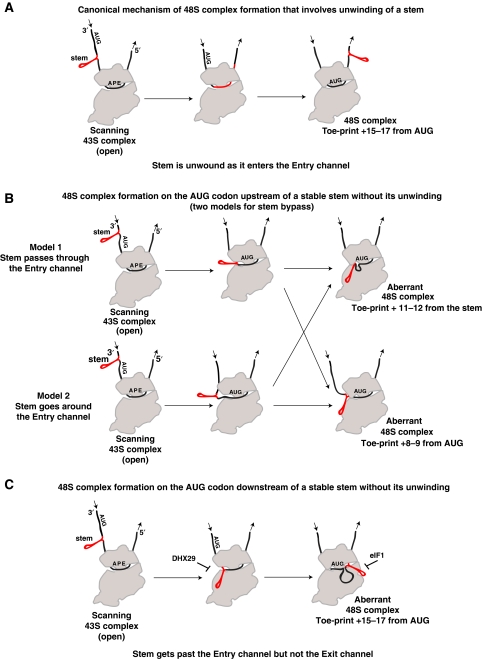
48S complex formation on AUG codons upstream of intact stems. Conventional 48S complexes containing unwound mRNA correctly positioned downstream of the P-site yield toe-prints +15–17 nts downstream of the start codon (Figure 7A). However, initiation in the absence of DHX29 on AUG codons directly upstream of stable stems (from position −3, immediately adjacent to the stem, to position −13) in all cases led to efficient assembly of 48S complexes characterized by toe-prints +11–12 nts downstream of the stem. The mRNA Entry channel in bacterial ribosomes from the entry point at the back/solvent surface of the 30S subunit to the A site is ∼10–12 nts long (Yusupova et al, 2001), and this allows us to conclude (i) that the intact stem cannot be accommodated within the Entry channel, at least in the 48S complex and (ii) that the toe-prints +11–12 nts downstream of the stem corresponds to a 48S complex, in which an intact stem is located between the Entry channel and the P site, whereas the single-stranded mRNA region downstream of the stem is accommodated in the Entry channel (Figure 7B). This places the stem in or around the A site, and in the case of mRNAs in which the AUG is further upstream of the stem (e.g. at positions −9 or −13), the spacer nucleotides must also be looped out in the A-site area. We note that an mRNA stem in the A site has been observed in the crystal structure of the 70S ribosome in complex with a model RNA based on phage T4 gene 32 mRNA (Figure 2 in Yusupova et al, 2001). Importantly, eIF1 does not dissociate 48S complexes with an intact stem in the A site characterized by toe-prints +11–12 nts downstream of the stem and even slightly stimulates their formation. 48S complexes containing intact stems in the A site were able to join 60S subunits, and although it would also be interesting to determine if the A site of such 80S complexes can efficiently accept a cognate aa-tRNA corresponding to the codon immediately following the AUG triplet, such experiments were outside the scope of the present study.
Figure 7.
Models for stem bypass by the scanning 43S complex. (A) Canonical 48S complex formation involving unwinding of a stem. (B, C) Models for 48S complex formation on AUG triplets (B) upstream and (C) downstream of stable stems without stem unwinding. (C) The bypass mechanism should be the same as in (B), except that the stem also moves past the Entry channel and the A and P sites. Stems (either intact or unwound) are shown in red. The positions of the ribosomal A, P and E sites and the 3′/5′ polarity of mRNA are shown in the left panel. The direction of mRNA movement with respect to the 40S subunit is indicated with arrows. The start codon is shown as ‘AUG'. Suppression of stem bypass by DHX29 and of formation of aberrant 48S complexes with an intact stem near the Exit channel by eIF1 are indicated by ‘⊣' in panel (C).
In addition to 48S complexes with toe-prints +11–12 nts downstream of the stem, we observed another type of ‘unconventional' 48S complexes characterized by toe-prints +9 nts downstream of the AUG codon, which formed relatively efficiently on mRNAs with AUGs at positions −9 and −13. The appearance of such complexes was stimulated by eIF1 and to some extent suppressed by eIF1A (this study; Battiste et al, 2000). As suggested previously (Pisareva et al, 2008), in such 48S complexes, mRNA downstream from the AUG is most likely not accommodated along the entire Entry channel, allowing reverse transcriptase to penetrate closer to the start codon (Figure 7B). Our present data indicate that formation of such complexes is enhanced by the presence of a stable stem ⩾9 but <18 nts downstream of the AUG codon. Although in 48S complexes characterized by +9 nts toe-prints, mRNA is not fully accommodated in the entire Entry channel, contacts between segments of the head and body of the 40S subunit (with the ‘latch' between h18 and h34/rpS3 as the most probable candidate) could still form a bridge over its segment 3′ from the P site. The bridging contact between eIF1A and h31 (Yu et al, 2009) cannot be responsible for stopping reverse transcriptase because the +9 nts toe-print was even stronger in eIF1A's absence.
These results raise the question of how the intact stem moves past the Entry channel. Different scenarios compatible with the available data can be envisioned (Figure 7B). One possibility is that the Entry channel in the scanning 43S complex may be more open than in the 48S complex, allowing an intact stem to go through. In this scenario, formation of 48S complexes is accompanied by closure of the Entry channel that ejects the stem, resulting either in 48S complexes with the downstream mRNA not accommodated along the entire Entry channel (toe-prints +9 nts downstream of the AUG codon) or in 48S complexes, in which the Entry channel closes ‘behind' the stem pushing it into the A site, with the single-stranded region downstream of the stem accommodated in the Entry channel (toe-prints +11–12 nts downstream of the stem). Another, somewhat less likely possibility is that when the stem reaches the Entry channel, it cannot go through and is forced to go around it, which would require the Entry channel to open enough for the single-stranded segment of mRNA in front of the stem to be able to slip out of it to allow the stem to go around. In this scenario, the single-stranded region of mRNA behind the stem reenters the Entry channel (toe-prints +11–12 nts downstream of the stem), or does not get back along its entire length (toe-prints +9 nts downstream of the AUG codon).
48S complex formation on AUG codons downstream of intact stems. Importantly, 48S complexes could form on AUG codons located not only upstream but also downstream of stable stems without their unwinding, and their formation also occurred by 5′-end-dependent scanning. Remarkably, eIF1 inhibited assembly of such complexes both when present during complex formation and when added to preformed 48S complexes, even if the AUG codon was located as far as 30 nts downstream of the stem. eIF1 is the key factor responsible for initiation codon selection in eukaryotes, and it is known to dissociate 48S complexes, in which mRNA does not occupy a sufficient fraction of the Exit channel, such as complexes assembled on AUGs that are ⩽8 nts from the 5′-end of mRNA (Pestova and Kolupaeva, 2002). The mRNA Exit channel is ∼12–14 nts long (Yusupova et al, 2001; Pisarev et al, 2008), and if the intact stem was able to go through its entire length, the stem at position −30 relative to the AUG would no longer be in contact with the scanning complex when the AUG reaches the P site, and a single-stranded mRNA segment behind the stem should be correctly positioned in the mRNA-binding cleft endowing 48S complexes with resistance to eIF1-induced dissociation. However, as such 48S complexes are readily dissociated by eIF1, we propose that when 43S complexes scan past the intact stem, the stem cannot go through the Exit channel and becomes trapped in the vicinity of the E site, and further scanning would result either in looping out of mRNA between the stem and the AUG codon if the region of mRNA in front of the stem remains ribosome bound, or in mRNA dropping off if this region eventually dissociates (Figure 7C). In any case, mRNA upstream of the P site is not correctly fixed in the mRNA-binding channel, rendering 48S complexes susceptible to dissociation by eIF1.
In this respect it is important to note that some exceptions to the linear mode of scanning have been identified, including ribosomal shunting on human hsp70 and adenovirus late mRNAs (Yueh and Schneider, 1996, 2000). Ribosomal complexes bind to the 5′-end of adenovirus mRNAs, scan part of the 5′-UTR but then translocate to the vicinity of the initiation codon, bypassing inserted elements such as hairpins that would impair base-by-base scanning. Although the presence of DHX29 and eIF1 on 43S complexes would appear likely to minimize shunting on these mRNAs, this process is enhanced by heat shock and for adenovirus mRNAs, reportedly by the viral 100 K protein, which binds to eIF4G and the tripartite leader on which shunting occurs (Yueh and Schneider, 2000; Xi et al, 2004), and which could thus function by stabilizing 48S complexes or by counteracting the activities of eIF1 or DHX29. Although only ∼10% of 43S complexes were associated with DHX29 in RRL (Pisareva et al, 2008), DHX29 was most active at substoichiometric concentrations relative to 40S subunits and inhibited initiation at increasing concentrations, suggesting that a low concentration of DHX29 is likely sufficient for all cellular initiation. However, nothing is yet known about potential regulation of levels or the activity of DHX29 in cells. Recent studies suggest that the level of eIF1 is autoregulated by virtue of the poor context of the initiation codon in its own mRNA, so that increased levels of eIF1 discriminate against its own further translation (Ivanov et al, 2010). Although eIF1 mRNA is upregulated in response to stress (Sheikh et al, 1999), the levels of eIF1 protein in stress conditions have not been determined. Thus the existing data do not allow us to speculate with any degree of certainty on whether and under which circumstances shunting on cellular mRNAs could be more widespread than currently appreciated.
Role of DHX29 in ensuring base-by-base inspection of 5′-UTRs by scanning 43S complexes
Whereas in the absence of DHX29, eIFs 4A/4B/4G promote only bypassing of stable stems by scanning 43S complexes, the presence of DHX29 enables scanning 40S subunits to penetrate into stems and inspect them. DHX29 also suppresses formation of aberrant 48S complexes characterized by toe-prints +8–9 nts downstream of the AUG codon, in which the mRNA is not accommodated along the entire Entry channel (Pisareva et al, 2008), as well as 48S complexes with an intact stem in the A site that are characterized by toe-prints +11–12 nts downstream of the stem (this study). Thus, DHX29 promotes unwinding of stems at the Entry channel and ensures that mRNA enters the scanning 43S complex in single-stranded form and is subjected to base-by-base inspection. DHX29 also acts on preassembled aberrant 48S complexes, dissociating some 48S complexes with the +9 nts toe-prints and efficiently eliminating those with the +11–12 toe-prints, containing an intact stem in the A site. In this case, DHX29 appears to act by dissociating the complex first, after which correct complexes form in a new round of initiation, rather than by converting it into a ‘correct' complex directly. However, DHX29 itself does not dissociate aberrant 48S complexes with an intact stem trapped in/near the E site. Such complexes are efficiently dissociated by eIF1, which conversely, does not dissociate 48S complexes with aberrations in mRNA position in/near the A site. A division of labour thus becomes apparent: DHX29 monitors mRNA in the vicinity of the Entry channel, whereas eIF1 monitors it in the vicinity of the Exit channel, in addition to the start codon itself.
What could be the mechanism by which DHX29 performs its function? DHX29 requires NTP hydrolysis for its activity, binds to the 40S subunit and likely contacts h16 (Pisareva et al, 2008). This would place DHX29 at or near the Entry channel, possibly within reach of mRNA as it enters the scanning complex. Although the likely ribosomal position of DHX29 would allow it to take a direct role in unwinding mRNA, DHX29 alone is not a processive helicase and its NTPase activity is stimulated more efficiently by 43S complexes than by RNA (Pisareva et al, 2008). Taking into consideration these properties of DHX29 together with the fact that it can also dissociate preformed 48S complexes with aberrations in mRNA position at the Entry channel and 48S complexes assembled on certain viral IRESs (this study; Pisareva et al, 2008), it seems more likely that DHX29 functions by remodelling ribosomal complexes rather than by direct unwinding of mRNA. In this respect it is relevant that many DExH/D proteins function primarily by remodelling RNA complexes (Pyle, 2008). If DHX29 is bound to h16 next to the Entry channel, it could physically block the access of stems to the channel. However, while blocking the channel could be a part of DHX29's mechanism of action, it alone would not explain why DHX29 requires NTP hydrolysis for its function, or how it can dissociate preformed 48S complexes. A more likely mechanism is that as DHX29 cycles between NTP- and NDP-bound states, it causes the 43S complex itself to cycle between two conformations, which could correspond to opening and closing of the Entry channel. As DHX29 cycles the Entry channel through these alternating conformations, it may indirectly help it to unwind stems entering the channel. It is not yet clear whether DHX29 completely prevents intact stems from getting past the Entry channel or only shifts the equilibrium in favour of unwound stems. Importantly, although it seems more likely that DHX29 works by ribosomal remodelling, we cannot strictly exclude that association with 43S complexes strongly enhances its helicase activity, in which case it may participate directly in mRNA unwinding in addition to remodelling of ribosomal complexes. Determination of the structure of 43S complexes associated with ATP- and ADP-bound forms of DHX29 would help to resolve these questions.
Materials and methods
Purification of initiation factors, Ded1, DDX3, DHX29 and ribosomal subunits, aminoacylation of tRNA and analysis of ribosomal binding of Ded1 are described in Supplementary data.
Plasmids
Vectors for expression of eIF4A, eIF4B, ΔeIF4G682–1599, eIF1, eIF1A, eIF5, Escherichia coli methionyl tRNA synthetase, Ded1 and DDX3, and for transcription of tRNAiMet and ΔII CSFV IRES mRNA have been described (Pestova et al, 1996, 1998, 2000; Iost et al, 1999; Kolupaeva et al, 2000; Pestova and Hellen, 2001; Yedavalli et al, 2004; Lomakin et al, 2006). Vectors for transcription of (CAA)n-Stem-GUS mRNAs comprising 117 nts of GUS coding region and 5′-UTRs containing stems of various stabilities and AUG triplets at different positions were made by inserting appropriate DNA sequences flanked by an upstream T7 promoter and downstream EcoRI restriction site into pJ204 (DNA2.0, Inc.). tRNAiMet and all mRNAs were transcribed using T7 polymerase. 32P-labelled Stem-2 (AUG at −6 position) mRNA (1.5 × 106 c.p.m./μg) was transcribed in the presence of [α32P]ATP (222 Tbq/mmol; MP Biomedicals).
Toe-printing analysis
48S complexes were assembled on native β-globin mRNA (Invitrogen) and in vitro transcribed (CAA)n-Stem-GUS mRNAs containing AUG triplets at different positions. Reaction mixtures (40 μl) containing 4 pmol mRNA, 3 pmol 40S subunits, 7 pmol Met-tRNAiMet, 10 pmol eIF2, 10 pmol eIF3 and different combinations of 10 pmol eIF4A, 7 pmol eIF4B, 4 pmol eIF4F, 10 pmol ΔeIF4G682–1599, 25 pmol eIF1, 25 pmol eIF1A, 10 pmol Ded1, 10 pmol DDX3, 2 pmol DHX29, 5 pmol eIF5, 5 pmol eIF5B, 6 pmol 60S subunits and 10 pmol ΔII CSFV IRES mRNA (as indicated in the figures) were incubated for 10 min at 37°C in buffer A (20 mM Tris pH 7.5, 100 mM KAc, 1 mM DTT, 2.5 mM MgCl2, 0.25 mM spermidine)+1 mM ATP and 0.4 mM GTP. Assembled initiation complexes were analysed by primer extension using AMV reverse transcriptase and appropriate [32P]-labelled primers complementary to coding regions of mRNAs (Pisarev et al, 2007). cDNA products were resolved in 6% polyacrylamide sequencing gels.
Analysis of ribosomal subunit joining
48S complexes were formed by incubating 8 pmol 32P-labelled Stem-2 (AUG at position −6) mRNA with 6 pmol 40S subunits, 14 pmol Met-tRNAiMet, 20 pmol eIF2, 20 pmol eIF3, 30 pmol eIF1, 30 pmol eIF1A, 20 pmol eIF4A, 14 pmol eIF4B and 20 pmol ΔeIF4G682–1599 in the presence/absence of 4 pmol DHX29 for 10 min at 37°C in an 80-μl reaction mixtures containing buffer A+1 mM ATP and 0.4 mM GTP. Assembled 48S complexes were supplemented with 10 pmol 60S subunits, 10 pmol eIF5 and 10 pmol eIF5B and incubated at 37°C for another 10 min. Reaction mixtures were subjected to centrifugation through 10–30% SDGs prepared in buffer A+5 mM MgCl2 in a Beckman SW55 rotor at 53 000 r.p.m. for 75 min. Ribosomal association of [32P] Stem-2 (AUG at position −6) mRNA was measured by Cerenkov counting of an aliquot of each fraction. Fractions that corresponded to 80S ribosomal complexes were also analysed by toe-printing.
Supplementary Material
Acknowledgments
We thank A Komar for yeast 40S subunits; P Linder, G Maga and KT Jeang for the Ded1 and DDX3 expression vectors, and to N Walworth for antibodies against Ded1. This work was supported by NIH Grant GM59660 and HFSP grant RPG0055/2006-C to TVP, NIH Grant AI51340 to CUTH and by Howard Temin K01 CA119107 award from the NCI and a Peter Paul Career Development Professorship from Boston University to AM.
Footnotes
The authors declare that they have no conflict of interest.
References
- Battiste JL, Pestova TV, Hellen CU, Wagner G (2000) The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell 5: 109–119 [DOI] [PubMed] [Google Scholar]
- Choi SK, Lee JH, Zoll WL, Merrick WC, Dever TE (1998) Promotion of met-tRNAiMet binding to ribosomes by yIF2, a bacterial IF2 homolog in yeast. Science 280: 1757–1760 [DOI] [PubMed] [Google Scholar]
- Chuang RY, Weaver PL, Liu Z, Chang TH (1997) Requirement of the DEAD-Box protein Ded1p for messenger RNA translation. Science 275: 1468–1471 [DOI] [PubMed] [Google Scholar]
- de la Cruz J, Iost I, Kressler D, Linder P (1997) The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA 94: 5201–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura J, Noguchi E, Sekiguchi T, Nishimoto T (2003) A temperature-sensitive mutant of the mammalian RNA helicase, DEAD-BOX X isoform, DBX, defective in the transition from G1 to S phase. J Biochem 134: 71–82 [DOI] [PubMed] [Google Scholar]
- Iost I, Dreyfus M, Linder P (1999) Ded1p, a DEAD-box protein required for translation initiation in Saccharomyces cerevisiae, is an RNA helicase. J Biol Chem 274: 17677–17683 [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF (2010) Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci USA 107: 18056–18060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV (2010) The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Pestova TV, Hellen CUT (2000) Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA 6: 1791–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MC, Lee YH, Tarn WY (2008) The DEAD-box RNA helicase DDX3 associates with export messenger ribonucleoproteins as well as tip-associated protein and participates in translational control. Mol Biol Cell 19: 3847–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawless C, Pearson RD, Selley JN, Smirnova JB, Grant CM, Ashe MP, Pavitt GD, Hubbard SJ (2009) Upstream sequence elements direct post-transcriptional regulation of gene expression under stress conditions in yeast. BMC Genomics 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Dias AP, Jedrychowski M, Patel AH, Hsu JL, Reed R (2008) Human DDX3 functions in translation and interacts with the translation initiation factor eIF3. Nucleic Acids Res 36: 4708–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockard RE, Currey K, Browner M, Lawrence C, Maizel J (1986) Secondary structure model for mouse beta Maj globin mRNA derived from enzymatic digestion data, comparative sequence and computer analysis. Nucleic Acids Res 14: 5827–5841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV (2006) The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. EMBO J 25: 196–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Dever TE (2010) Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J Biol Chem 285: 21203–21207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamiya N, Worman HJ (1999) Hepatitis C virus core protein binds to a DEAD box RNA helicase. J Biol Chem 274: 15751–15756 [DOI] [PubMed] [Google Scholar]
- Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G (2009) Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell 136: 447–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsden S, Nardelli M, Linder P, McCarthy JE (2006) Unwinding single RNA molecules using helicases involved in eukaryotic translation initiation. J Mol Biol 361: 327–335 [DOI] [PubMed] [Google Scholar]
- Parsyan A, Shahbazian D, Martineau Y, Petroulakis E, Alain T, Larsson O, Mathonnet G, Tettweiler G, Hellen CU, Pestova TV, Svitkin YV, Sonenberg N (2009) The helicase protein DHX29 promotes translation initiation, cell proliferation, and tumorigenesis. Proc Natl Acad Sci USA 106: 22217–22222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V (2007) The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU (1998) Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859 [DOI] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU (2001) Preparation and activity of synthetic unmodified mammalian tRNAi(Met) in initiation of translation in vitro. RNA 7: 1496–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CU, Shatsky IN (1996) Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol 16: 6859–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV (2006) Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV (2008) Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Unbehaun A, Hellen CU, Pestova TV (2007) Assembly and analysis of eukaryotic translation initiation complexes. Methods Enzymol 430: 147–177 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV (2008) Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyle AM (2008) Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys 37: 317–336 [DOI] [PubMed] [Google Scholar]
- Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U (2008) Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci USA 105: 9564–9569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheikh MS, Fernandez-Salas E, Yu M, Hussain A, Dinman JD, Peltz SW, Huang Y, Fornace AJ Jr (1999) Cloning and characterization of a human genotoxic and endoplasmic reticulum stress-inducible cDNA that encodes translation initiation factor 1(eIF1(A121/SUI1)). J Biol Chem 274: 16487–16493 [DOI] [PubMed] [Google Scholar]
- Shih JW, Tsai TY, Chao CH, Wu Lee YH (2008) Candidate tumor suppressor DDX3 RNA helicase specifically represses cap-dependent translation by acting as an eIF4E inhibitory protein. Oncogene 27: 700–714 [DOI] [PubMed] [Google Scholar]
- Shiratori A, Shibata T, Arisawa M, Hanaoka F, Murakami Y, Eki T (1999) Systematic identification, classification, and characterization of the open reading frames which encode novel helicase-related proteins in Saccharomyces cerevisiae by gene disruption and Northern analysis. Yeast 15: 219–253 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Chang TH (2009) The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol 6: 17–20 [DOI] [PubMed] [Google Scholar]
- Xi Q, Cuesta R, Schneider RJ (2004) Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev 18: 1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT (2004) Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell 119: 381–392 [DOI] [PubMed] [Google Scholar]
- Yu Y, Marintchev A, Kolupaeva VG, Unbehaun A, Veryasova T, Lai SC, Hong P, Wagner G, Hellen CU, Pestova TV (2009) Position of eukaryotic translation initiation factor eIF1A on the 40S ribosomal subunit mapped by directed hydroxyl radical probing. Nucleic Acids Res 37: 5167–5182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ (1996) Selective translation initiation by ribosome jumping in adenovirus-infected and heat-shocked cells. Genes Dev 10: 1557–1567 [DOI] [PubMed] [Google Scholar]
- Yueh A, Schneider RJ (2000) Translation by ribosome shunting on adenovirus and hsp70 mRNAs facilitated by complementarity to 18S rRNA. Genes Dev 14: 414–421 [PMC free article] [PubMed] [Google Scholar]
- Yusupova GZ, Yusupov MM, Cate JH, Noller HF (2001) The path of messenger RNA through the ribosome. Cell 106: 233–241 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.