U2 snRNA is inducibly pseudouridylated at novel sites by Pus7p and snR81 RNP
Modification of stable RNAs has always been considered constitutive, but this identification of certain snRNA sites being de novo pseudouridylated in response to environmental conditions offers precedent for an intriguing new concept of dynamic RNA modification as regulatory means.
Keywords: inducible RNA modification, pseudouridylation, Pus7, SnR81 RNP, U2 snRNA
Abstract
All pseudouridines identified in RNA are considered constitutive modifications. Here, we demonstrate that pseudouridylation of Saccharomyces cerevisiae U2 snRNA can be conditionally induced. While only Ψ35, Ψ42 and Ψ44 are detected in U2 under normal conditions, nutrient deprivation leads to additional pseudouridylation at positions 56 and 93. Pseudouridylation at position 56 can also be induced by heat shock. Detailed analyses have shown that Pus7p, a single polypeptide pseudouridylase known to modify U2 at position 35 and tRNA at position 13, catalyses Ψ56 formation, and that snR81 RNP, a box H/ACA RNP known to modify U2 snRNA at position 42 and 25S rRNA at position 1051, catalyses Ψ93 formation. Using mutagenesis, we have demonstrated that the inducibility can be attributed to the imperfect substrate sequences. By introducing Ψ93 into log-phase cells, we further show that Ψ93 has a role in pre-mRNA splicing. Our results thus demonstrate for the first time that pseudouridylation of RNA can be induced at sites of imperfect sequences, and that Pus7p and snR81 RNP can catalyse both constitutive and inducible pseudouridylation.
Introduction
Pseudouridylation (Ψ) is one of the major post-transcriptional modifications found in stable RNAs, including tRNAs, rRNAs and spliceosomal snRNAs (Reddy and Busch, 1988; Bjork, 1995; Massenet et al, 1998; Ofengand and Fournier, 1998; Yu et al, 2005; Grosjean, 2009). In fact, there are ∼100 pseudouridines in mammalian rRNAs (Ofengand and Fournier, 1998), and a total of 24 in vertebrate spliceosomal snRNAs (Reddy and Busch, 1988; Massenet et al, 1998; Karijolich et al, 2009). Even in lower eukaryotic organisms, such as Saccharomyces cerevisiae, ∼50 pseudouridines are found in rRNAs (Ofengand and Fournier, 1998), and six are identified in spliceosomal snRNAs, including three in U2 (see Figure 1) (Massenet et al, 1998). Over the years, many of these pseudouridines have been assigned specific functions. For instance, at least some pseudouridines in rRNAs have an important function in ribosome biogenesis and protein synthesis (King et al, 2003; Liang et al, 2007; Piekna-Przybylska et al, 2008). Many pseudouridines in U2 snRNA contribute significantly to pre-mRNA splicing (Yu et al, 1998; Donmez et al, 2004; Zhao and Yu, 2004b, 2007).
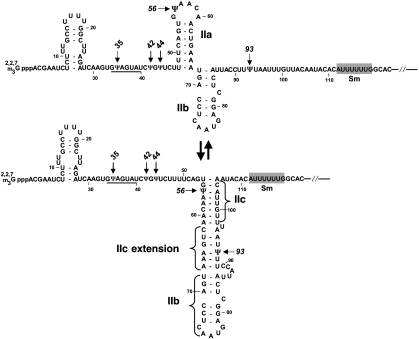
Figure 1.
S. cerevisiae U2 snRNA. A partial sequence and two alternate secondary structures of yeast U2 are shown. The three constitutive pseudouridines (Ψ35, Ψ42 and Ψ44) as well as the two inducible pseudouridines (Ψ56 and Ψ93) are also shown. The Sm-binding site (shaded boxes) and the branch site recognition sequence (underlined) are indicated. According to Hilliker et al (2007) and Perriman and Ares (2007), toggling between two alternate structures, stem-loop IIa and stem IIc, is required for splicing.
There exist two distinct molecular mechanisms (RNA dependent and RNA independent) that are capable of site-specifically pseudouridylating RNAs (Ganot et al, 1997; Ni et al, 1997; Massenet et al, 1999; Ma et al, 2003, 2005). While RNA-independent pseudouridylation is catalysed by single polypeptide enzymes, which both specify the substrate and catalyse the isomerization reaction (Massenet et al, 1999; Ma et al, 2003), RNA-dependent mechanism requires protein–RNA complexes, known as box H/ACA sno (small nucleolar) or sca (small Cajal body specific) RNPs (Ganot et al, 1997; Ni et al, 1997; Ma et al, 2005). Box H/ACA RNP consists of a unique box H/ACA RNA and a set of four core proteins: Cbf5p/Nap57/Dyskerin, Nhp2p, Gar1p and Nop10p (Yu et al, 2005). The box H/ACA RNA forms a unique ‘hairpin-hinge-hairpin-tail' structure (Figure 3A), in which each of the two hairpins contains a pseudouridylation pocket that directs the conversion of a specific uridine to pseudouridine in the substrate RNA (Figure 3A). Both RNA-dependent and RNA-independent mechanisms are used in yeast U2 pseudouridylation. While Pus7p and Pus1p modify, in an RNA-independent manner, positions 35 and 44, respectively (Massenet et al, 1999; Ma et al, 2003), snR81 box H/ACA RNP, introduces pseudouridylation at position 42 (Ma et al, 2005).
In contrast to protein and DNA modifications, which are mostly inducible, all pseudouridines identified thus far, including the three (Ψ35, Ψ42 and Ψ44) in yeast U2 snRNA, are considered constitutive and permanent modifications (Reddy and Busch, 1988; Massenet et al, 1998). Given that the RNA modification enzymes are constantly available in the cell, it is widely believed that modifications are introduced into an RNA once (or shortly after) it is synthesized (Yu et al, 1997, 2001; Jady et al, 2003), and perhaps remain in the RNA molecule thereafter. Thus, unlike protein and DNA modifications, RNA modifications are not expected to be subjected to regulation.
In the current work, we have demonstrated for the first time that, in addition to the three constitutive pseudouridines (Ψ35, Ψ42 and Ψ44), there are at least two pseudouridines (Ψ56 and Ψ93) that can be conditionally induced in S. cerevisiae U2 snRNA. We have shown that snR81 RNP and Pus7p are responsible for inducible U2 pseudouridylation at positions 93 and 56, respectively. We have also shown that a near-perfect (rather than perfect) consensus substrate sequence is necessary for induction.
Results
Pseudouridylation of yeast U2 RNA occurs at novel sites in response to environmental stress
To explore the possibility that spliceosomal snRNA modification can be induced, we exposed yeast cells to two widely used stresses—heat-shock and nutrient deprivation (growing cells to saturation/stationary phase or using nutrient-depleted media). Cells were collected before and after stress, and RNAs were isolated for pseudouridylation assays (Bakin and Ofengand, 1993; Zhao and Yu, 2004a).
Remarkably, using the standard semi-quantitative primer-extension-based assay (CMC modification followed by primer extension) (Bakin and Ofengand, 1993), we detected two clear pseudouridine signals at positions 56 and 93 in U2 snRNA isolated from confluent (or nutrient-deprived) cells (Figure 2A). When the cells were heat shocked, we also detected Ψ56 (but not Ψ93) (Figure 2B). These two sites (positions 56 and 93) were previously identified as ‘unmodified' uridines in yeast U2 snRNA (Massenet et al, 1998).
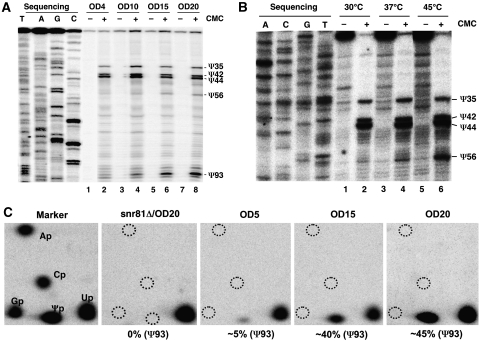
Figure 2.
Conditionally inducible formation of Ψ56 and Ψ93. (A) S. cerevisiae cells were grown in YPD to various ODs (indicated at the top), total RNA was recovered and pseudouridylation was assayed (by CMC modification followed by primer extension). Samples of even-numbered lanes were treated with CMC and samples of odd-numbered lanes were not treated with CMC. Signals corresponding to Ψ35, Ψ42, Ψ44, Ψ56 and Ψ93 are indicated. U2 sequencing is also shown on the left. (B) S. cerevisiae cells were first grown at 30°C, then were kept at 30°C (lanes 1 and 2) or shifted to 37°C (lanes 3 and 4) or 45°C (lanes 5 and 6) for 30 min. Total RNA was recovered and pseudouridylation assay (CMC modification followed by primer extension) was performed (see (A)). (C) RNA was recovered from cells at various OD600 (indicated). U2 snRNA was site-specifically radiolabelled at position 93 (see Materials and methods). After digestion with nuclease T2, cleaved RNA samples were analysed by thin layer chromatography (TLC). Relative percentages of pseudouridylation at position 93 were calculated (under each panel).
To better measure this inducible pseudouridylation, we carried out a quantitative pseudouridylation assay, namely site-specific cleavage/labelling followed by nuclease digestion and TLC (Zhao and Yu, 2004a). We checked position 93 of U2 before and after cells entered stationary phase (nutrient deprivation) (Figure 2C). Whereas U2 RNA isolated from late log-phase cells showed very little pseudouridylation at position 93 (OD5, OD600 of 5), U2 RNA from cells in the post-diauxic growth period (OD15) and stationary phase (OD20) exhibited strong pseudouridylation (∼40 and ∼45%, respectively) at position 93. Undoubtedly, U2 pseudouridylation at position 93 is induced in response to changes in growth conditions.
Pus7p and snR81 RNP are required for inducible pseudouridylation at positions 56 and 93, respectively
Inspection of the sequences surrounding positions 56 and 93 revealed some elements that are similar but not identical to those of position 35 within U2 and position 1051 within 25S rRNA, respectively (Figure 3A and B). Thus, we suspected that Pus7p, which naturally modifies U2 at position 35, might be responsible for Ψ56 formation. Likewise, snR81 RNP (a box H/ACA RNP), whose 3′ pseudouridylation pocket naturally guides the formation of Ψ1051 within 25S rRNA, might catalyse Ψ93 formation. To test this hypothesis, we used wild-type and mutant strains (including the pus7Δ, snr81Δ and cbf5D95A that is defective in box H/ACA RNP-catalysed pseudouridylation) to measure U2 pseudouridylation at positions 56 and 93 under the induced growth conditions (i.e. nutrient-deprivation conditions).
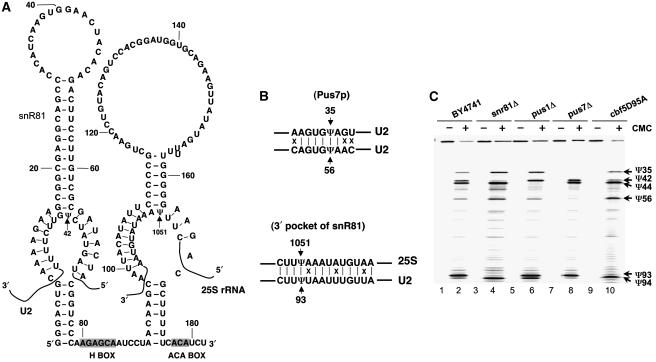
Figure 3.
Pus7p and snR81 RNP are required for the formation of Ψ56 and Ψ93, respectively. (A) The sequence and secondary structure of snR81 box H/ACA RNA are shown. Boxes H and ACA are indicated (shaded boxes). The two internal loops, including the 5′ one that guides the formation of Ψ42 in U2 and the 3′ one that directs the formation of Ψ1051 in 25S rRNA, are also shown. (B) Sequences surrounding Ψ35 (target of Pus7p) and Ψ56 of U2 are aligned and sequences flanking Ψ1051 of 25S rRNA (target of 3′ pocket of snR81) and Ψ93 of U2 are also aligned. Identical (a vertical line) and different (X) nucleotides are indicated. (C) Wild-type (BY4741) (lanes 1 and 2), snr81Δ (lanes 3 and 4), pus1Δ (lanes 5 and 6), pus7Δ (lanes 7 and 8) and cbf5D95A (lanes 9 and 10) strains were all grown to saturation. CMC-primer-extension-based pseudouridylation assay was then carried out (see legend to Figure 2A). Signals corresponding to Ψ35, Ψ42, Ψ44, Ψ56, Ψ93 and Ψ94 are indicated.
As shown in Figure 3C, while pseudouridylation at positions 93 and 56 was detected in the wild-type strain (BY4741), no such inducible pseudouridylation was detected in the respective mutant strains. Specifically, no Ψ93 was in the snr81Δ strain or cbf5D95A strain, but Ψ56 remained unchanged (lanes 4 and 10). As expected, snr81Δ also affected the formation of Ψ42, a constitutive substrate of snR81 RNP (lanes 4 and 10). In the pus7Δ strain, Ψ93 was detected, whereas Ψ56 and Ψ35 (a constitutive target of Pus7p) were not (lane 8). Deletion of PUS1, a protein pseudouridylase catalysing U2 pseudouridylation at position 44, abolished Ψ44 formation but did not affect the formation of Ψ56 and Ψ93 (lane 6). Interestingly, in the absence of snR81 or when CBF5 was mutated, Ψ94 appeared (lanes 4 and 10). Thus, at least one of them (either Ψ93, Ψ94) is modified during nutrient deprivation, implying the importance of the modification in this region. Our data suggested that Pus7p and snR81 RNP were required for induced modification at positions 56 and 93, respectively.
Pus7p and snR81 RNP directly catalyse inducible pseudouridylation at positions 56 and 93, respectively
Given that Pus7p and snR81 RNP (its 5′ pseudouridylation pocket; see Figure 3A) also target, respectively, positions 35 and 42 of U2 for pseudouridylation (Ma et al, 2003, 2005), it is possible that induced pseudouridylation at positions 56 and 93 is dependent on the prior pseudouridylation at positions 35 and 42, and that Pus7p and snR81 RNP might not directly act on nucleotides 56 and 93. To test this possibility, we used mutant snR81 RNAs, where either the 5′ pocket or the 3′ pocket was randomly mutated.
When the snr81Δ strain was transformed with snR81 carrying a mutated 3′ pocket, no pseudouridylation was detected at position 93 in nutrient-deprived cells (Figure 4A, lane 2); however, Ψ42 was detected (lane 2). In contrast, when the snr81Δ strain was transformed with snR81 containing a mutated 5′ pocket, Ψ93 formation was efficiently induced upon entry into stationary phase (lane 4); however, as expected, Ψ42 formation was blocked (lane 4).
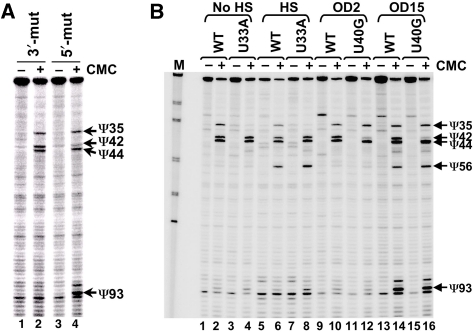
Figure 4.
Induced pseudouridylation at positions 56 and 93 is independent of prior constitutive pseudouridylation at other sites. (A) snR81 RNA, containing a mutated 3′ pseudouridylation pocket (lanes 1 and 2) or a mutated 5′ pseudouridylation pocket (lanes 3 and 4), was expressed in an snr81-deleted strain. Upon entry into stationary phase, cells were collected, RNA isolated and pseudouridylation assay (CMC modification followed by primer extension) was carried out. In lanes 1 and 3, CMC was omitted; in lanes 2 and 4, samples were treated with CMC. Signals corresponding to Ψ35, Ψ42, Ψ44 and Ψ93 are indicated. (B) Pseudouridylation assay as in (A). Instead of expressing a snR81 mutant, wild-type U2 (lanes 1, 2, 5, 6, 9, 10, 13 and 14) and two mutant U2 substrates were expressed in the cell. In lanes 3, 4, 7 and 8, U2 contained a U33A mutation; in lanes 11, 12, 15 and 16, U2 contained a U40G mutation. Before RNA isolation, cells were allowed to grow to log phase (lanes 1–12, OD2) or stationary phase (lanes 13–16, OD15). In lanes 5–8, cells were further heat shocked before being lysed for RNA isolation. Samples of even-numbered lanes were treated with CMC and samples of odd-numbered lanes were not treated with CMC. Signals corresponding to Ψ35, Ψ42, Ψ44, Ψ56 and Ψ93 are indicated.
We also took a reciprocal approach by changing the U2 sequence (rather than the guide RNA sequence). Specifically, a viable U2 mutant, in which the uridine at position 40 was changed to guanosine (U40G), was created. This mutant could barely be pseudouridylated by snR81 RNP at position 42 due to the alteration in complementarity between the guide RNA and the substrate (Yang et al, 2005). As shown in Figure 4B (42 was present in the wild-type strain (lanes 2, 6, 10 and 14), but was barely detected in the U40G strain (lanes 12 and 16). Upon nutrient deprivation, Ψ93 (Ψ56 as well) was efficiently induced, regardless of whether Ψ42 was present (lane 14) or absent (lane 16).
To check whether there is an interdependent relationship between Ψ35 and Ψ56, we created a U2 that was mutated at position 33 (U33A) such that position 35 was no longer a Pus7p substrate (Behm-Ansmant et al, 2003; Urban et al, 2009) (direct mutations cannot be initiated at position 35 because such mutations are lethal). As shown in Figure 4B, while Ψ35 was detected in the wild-type strain (lanes 2, 6, 10 and 14), no Ψ35 formation was observed in the U33A mutant strain (lanes 4 and 8). Upon heat shock (HS), Ψ56 was induced in both wild-type (lane 6) and U33A (lane 8) strains regardless of whether Ψ35 was present or absent.
Taken together, our results demonstrate that Pus7p, which catalyses U2 pseudouridylation at position 35, also catalyses pseudouridylation at position 56 during nutrient deprivation or heat shock. Likewise, wild-type snR81 RNP, which catalyses U2 pseudouridylation at position 42 (using the 5′ pocket) and 25S rRNA pseudouridylation at position 1051 (using the 3′ pocket), also catalyses pseudouridylation at position 93 (using the 3′ pocket) under nutrient deprivation conditions. Thus, induced pseudouridylation is independent of prior pseudouridylation at the other position (Ψ35 or Ψ42), suggesting strongly that Pus7p and snR81 RNP directly modify Ψ56 and Ψ93, respectively, under induced conditions.
An imperfect consensus sequence is necessary for induction
Our sequence inspection (Figure 3B) suggested that the imperfect consensus sequence at positions 56 and 93 may contribute to induced modification. To directly test this hypothesis, we carried out a series of mutagenesis analyses, focusing primarily on Ψ93 of U2 and its enzyme snR81 RNP.
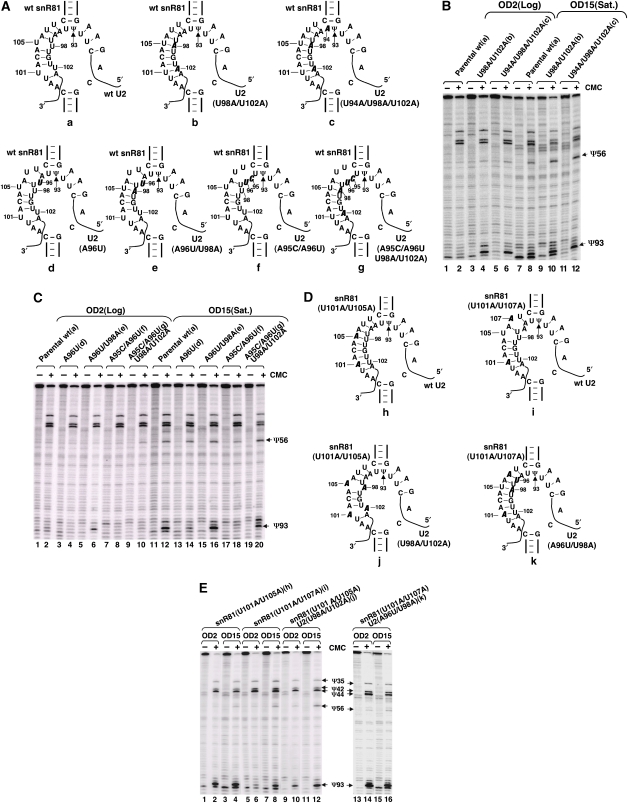
We first mutated U2 snRNA in the Ψ93 region that participates in base pairing with the guide sequence of the 3′ pocket of snR81. We made changes at two positions (U98A/U102A) such that the new U2 sequence matched perfectly with the snR81 guide sequence (Figure 5Ab). Consequently, Ψ93 formation became constitutive as the Ψ93 signal was detected both in the log phase and in the stationary phase (Figure 5B, lanes 4 and 10). Likewise, mutations at three positions (U94A/U98A/U102A), which similarly led to perfect base pairing with the guide sequence (Figure 5Ac), resulted in constitutive formation of Ψ93 (Figure 5B, lanes 6 and 12). In contrast, when we introduced mutations into U2 (A96U or A95C/A96U) to weaken the base pairing between U2 and the snR81 guide sequence (Figure 5Ad and Af), no Ψ93 was detected, even when cells were grown to saturation (stationary phase) (Figure 5C, lanes 4 and 14, and lanes 8 and 18). Importantly, when the mutations (U98A/U102A) that improved base pairing were combined with the mutations (either A96U or A95C/A96U) that weakened the pairing (Figure 5Ae and Ag), induced Ψ93 formation was restored––no (or very mild) Ψ93 signal was detected in the log phase (Figure 5C, lanes 6 and 10), but the signal was dramatically enhanced when the cells were grown to saturation (Figure 5C, lanes 16 and 20). It should be noted that although the combined mutations fixed one or two original mismatches, they created new mismatches. As a result, a total of two mismatches were left in the base-pairing region (imperfect consensus sequence), a situation similar to the pairing between the wild-type U2 and wild-type snR81 RNA (see Figure 5Aa and C, lanes 2 and 12).
Figure 5.
Imperfect interactions between enzyme and substrate are necessary for induced pseudouridylation. (A) Shown are interactions between the wild-type 3′ pocket of snR81 and the wild-type and various mutant U2 substrates. (a) Wild-type U2; (b) U2 with U98A/U102A mutations; (c) U2 with U94A/U98A/U102A mutations; (d) U2 with an A96U mutation; (e) U2 with A96U/U98A mutations; (f) U2 with A95C/A96U mutations; (g) U2 with A95C/A96U/U98A/U102A mutations. Bold and italicized letters represent mutations. (B) Pseudouridylation assay (CMC modification followed by primer extension) upon expression of wild-type U2 (a) (lanes 1, 2, 7 and 8), U2 with U98A/U102A mutations (b) (lanes 3, 4, 9 and 10) and U2 with U94A/U98A/U102A mutations (c) (lanes 5, 6, 11 and 12). Lanes 1–6, total RNA was isolated from log-phase cells; lanes 7–12, total RNA was isolated from stationary-phase cells. (C) Pseudouridylation assay as in (B) except that different mutant U2 RNAs (as indicated at the top of each lane) were used. In lanes 1–10, log-phase cells were used; in lanes 11–20, stationary-phase cells were used. (D) Interactions of mutant 3′ pseudouridylation pocket of snR81 with various U2 are shown. (h) snR81 with U101A/U105A mutations interacting with wild-type U2; (i) snR81 with U101A/U107A mutations interacting with wild-type U2; (j) snR81 with U101A/U105A mutations interacting with U2 containing U98A/U102A mutations; (k) snR81 with U101A/U107A mutations interacting with U2 containing A96U/U98A mutations. Mutations are indicated by bold and italicized letters. (E) Pseudouridylation assay as in (B) except that various snR81 RNAs and U2 RNAs, as indicated at the top of each lane, were used. In lanes 1, 2, 5, 6, 9, 10, 13 and 14, log-phase cells were used; in lanes 3, 4, 7, 8, 11, 12, 15 and 16, stationary-phase cells were used.
We also mutated the 3′ pocket of snR81 to test the hypothesis that the imperfect base pairing is necessary for induced pseudouridylation. First, we made mutations at two positions (U101A/U105A) such that the new guide sequence matched perfectly with the substrate at position 93 (Figure 5Dh), with the attempt to make Ψ93 formation uninducible (or constitutive). Indeed, when this guide RNA was expressed in the snR81Δ strain, Ψ93 formation was detected in both log-phase cells and stationary-phase cells (Figure 5E, lanes 2 and 4), regardless of growth conditions. We then made another mutant (U101A/U107A) in which mutation at one site was retained (U101A) and mutation at the other site (U105A) was switched to a different site (U107A). This mutant was expected to fix one original mismatch while creating a new mismatch, thus resulting in a total of two mismatches (Figure 5Di). Consequently, expression of this mutant guide RNA led to induced Ψ93 formation (Figure 5E, compare lane 6 with lane 8).
We then further tested the importance of imperfect base pairing for induced Ψ93 formation by combining mutant snR81 with mutant U2. When snR81 (U101A/U105A) and U2 (U98A/U102A) were co-expressed, two mismatches were expected (Figure 5Dj), and these two mismatches should be similar to the mismatches observed when wild-type snR81 and wild-type U2 were used. Indeed, as expected, a mild Ψ93 signal was detected when cells were in log phase; the signal was significantly enhanced when the cells were grown to saturation (stationary phase) (Figure 5E, compare lane 10 with lane 12). We also coupled snR81 (U101A/U107A) with U2 (A96U/U98A), with an attempt to form a perfect match between the two (Figure 5Dk). As expected, we observed the constitutive formation of Ψ93 (Figure 5E, lanes 14 and 16). Taken together, we conclude that the imperfect base pairing (with two mismatches) between snR81 RNA and U2 is necessary for induced Ψ93 formation.
We also tested U1051, of 25S rRNA, using mutant snR81 (U101A/U105A). According to our conclusion (imperfect pairing is necessary for induced modification), it was expected that pseudouridylation of U1051 could be induced when snR81 (U101A/U105A) was used. Surprisingly, however, no pseudouridine was detected at U1051, even when cells were grown to saturation (data not shown). This result suggests that while imperfect pairing is necessary, it is not sufficient for induced pseudouridylation (see Discussion).
Induced pseudouridines are functionally relevant
The induced pseudouridylation at positions 56 and 93 suggested that they might be functionally relevant under stressed conditions. To test this hypothesis, we decided to develop a functional phenotypic assay. However, it is difficult to monitor cell growth under the heat-shock and nutrient-deprived (stationary phase) conditions. To facilitate our analysis, we took a reverse approach. Instead of blocking the induced pseudouridylation (via deletion of enzymes) and analysing the phenotype under induced conditions, we decided to force pseudouridylation to occur constitutively at position 93, thus allowing a phenotypic analysis under the log-phase growth conditions.
We took advantage of mutant snR81 (U101A/U105A), which matched perfectly with the sequence surrounding position 93 (Figure 5D) and was capable of directing Ψ93 formation even when cells were grown to log phase in fresh rich medium (see Figure 5D and E). We co-transformed the cup1Δ strain with snR81 (U101A/U105A) and a plasmid containing an ACT1–CUP1 reporter. Splicing of ACT1–CUP1 pre-mRNA would lead to the production of Act1–Cup1 fusion protein, which in turn would confer copper resistance (Lesser and Guthrie, 1993). The transformed cells were grown in the fresh medium to log phase and were subsequently plated on selective medium with or without copper. In the meantime, RNA was isolated from log-phase cells and subjected to primer-extension assay (to check splicing of ACT1–CUP1 pre-mRNA) (Lesser and Guthrie, 1993; Siatecka et al, 1999).
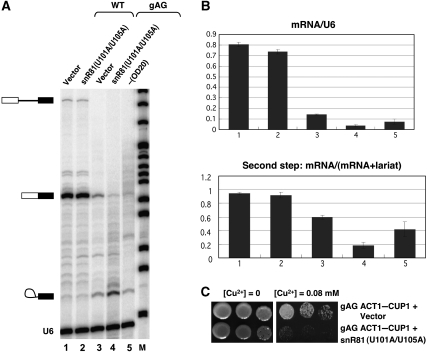
Although no significant changes were detected in the splicing of the wild-type ACT1–CUP1 pre-mRNA (Figures 6A and B, compare lane 1 with lane 2), the splicing of a second-step mutant ACT1–CUP1 pre-mRNA, in which the 3′ end of the intron was changed from UAG to gAG (Hilliker et al, 2007), was markedly affected (Figure 6A and B, compare lane 3 with lane 4). Specifically, the second-step splicing defect was exacerbated when U93 was changed to Ψ93, as assessed by increased accumulation of the lariat intermediate and decreased production of mature mRNA (Figure 6A and B, compare lanes 4 and 3). Consistently, cells lacking or containing the snR81 (U101A/U105A) grew equally well in medium without copper; however, cells containing the snR81 mutant developed an exacerbated growth defect phenotype in copper-containing medium (Figure 6C).
Figure 6.
Ψ93 formation contributes to splicing regulation. (A) Yeast cup1Δ strain was transformed with a plasmid carrying an ACT1–CUP1 reporter pre-mRNA gene (either wild-type, lanes 1 and 2, or gAG mutant, lanes 3–5) (Hilliker et al, 2007), along with either a vector only (lanes 1 and 3) or a plasmid containing snR81 (U101A/U105A) gene specifically targeting position 93 of U2 (lanes 2 and 4). Cells (for lanes 1–4) were harvested during early/mid-log phase. Total RNA was recovered and primer extension was carried out (using an ACT1–CUP1-specific primer and a U6-specific primer), allowing for checking pre-mRNA splicing efficiency. In lane 5, cells were transformed with only the gAG/A mutant pre-mRNA (no guide RNA co-transformation), but the cells were harvested in stationary phase (OD600=20) (rather than in log phase). Primer-extension products corresponding to pre-mRNA, mRNA, lariat and U6 are indicated. (B) Splicing efficiency was quantified, based on three independent experiments. mRNA level relative to U6 level (upper panel), and the ratio of mRNA to (mRNA+lariat), reflecting the efficiency of second step of splicing (lower panel), are shown. (C) cup1Δ cells, transformed with the gAG/A mutant pre-mRNA along with an empty vector or a plasmid containing an artificial box H/ACA guide RNA gene specifically targeting position 93 of U2 (corresponding to lanes 3 and 4 in (A, B)), were plated on a synthetic medium containing no copper ([Cu2+]=0) or 0.08 mM copper ([Cu2+]=0.08).
For comparison, we also transformed the cells with the second-step mutant ACT1–CUP1 alone (without guide RNA), grew cells to the stationary phase (OD20), and isolated RNA for primer extension. Our experiment detected a slightly more accumulation of the lariat intermediate and less production of the mature ACT1–CUP1 mRNA (Figure 6A and B, compare the ratio of mRNA/lariat intermediate in lanes 3 and 5), consistent with the above results where the log-phase cells containing both snR81 (U101A/U105A) and the second-step mutant ACT1–CUP1 were analysed (lane 4). While the mechanism by which induced pseudouridylation affects splicing is still unclear (see Discussion), we nonetheless conclude that Ψ93 has a role in pre-mRNA splicing regulation in yeast cells.
Discussion
In the current work, we have identified at least two novel pseudouridines (Ψ56 and Ψ93) in yeast U2 snRNA. These pseudouridines are conditionally inducible (induced upon nutrient starvation or heat shock). We have also identified the pseudouridylases responsible for these modifications: Pus7p catalyses Ψ56 formation, and snR81 RNP modifies position 93. Furthermore, we have shown that the imperfect consensus sequences are critical for initiating induced modification. Our discovery of inducible pseudouridylation raises a number of interesting questions that will certainly prompt a reassessment of the mechanisms and functions of U2 pseudouridylation and RNA modification in general.
Imperfect consensus sequence is necessary but not sufficient for induction
As discussed earlier, the sequence surrounding position 56 is similar, but not identical to that surrounding position 35, a constitutive pseudouridylation site catalysed by Pus7p. Likewise, the sequence flanking position 93 is not exactly the same as that of position 1051 of 25S rRNA, a natural pseudouridylation site directed by the 3′ pocket of snR81 (Figure 3A and B). In fact, the 3′ pseudouridylation pocket of snR81 snRNA, while matches perfectly with the sequence surrounding position 1051 of 25S rRNA, matches imperfectly with the sequence surrounding position 93. In the latter case, there are three mismatches, two of which are in the middle of the base-pairing duplex. Under normal conditions, such mismatched pairing would not lead to modification (Xiao et al, 2009). However, they appear to be critical for induced pseudouridylation. In fact, our experimental data have indicated that an imperfect base-pairing interaction, which occurs naturally (see Figure 5Aa) or artificially (introduced by mutations, see Figure 5Ae and Ag and 5Di and Dj), is necessary for the induction of Ψ93 formation.
Although necessary, however, the imperfect consensus sequence (or imperfect base pairing) appears to be insufficient for the induction. This assertion is derived from our U40G mutant results, in which we showed that U-to-G mutation at position 40 abolished Ψ42 formation in both log-phase cells and stationary-phase cells. In other words, the imperfect interaction (introduced by mutation) between the mutant sequence surrounding position 42 and the guide sequence of snR81 (5′ pocket) failed to induce Ψ42 formation, suggesting that the imperfect consensus sequence is not sufficient for induction. Similarly, imperfect pairing between the 3′-pocket-mutant snR81 (U101A/U105A) and the sequence surrounding U1051 of 25S rRNA did not lead to the induced formation of Ψ1051. Thus, there must be more components that contribute to induction, in addition to imperfect sequence or base pairing. With regards to Ψ42, we note that the 5′ pocket of snR81 catalyses Ψ42 formation and that the 3′ pocket of snR81 guides Ψ93 formation. It is therefore possible that some factor(s), specifically associated with the 3′ pocket, is involved in induced Ψ formation. With regards to failed pseudouridylation of U1051 by mutant snR81 (U101A/U105A), although the 3′ pocket of snR81 is involved, there are at least two major differences between 25S rRNA and U2. First, while U2 is transcribed by Pol II, 25S rRNA is transcribed by Pol I. Second, unlike U2, which is localized to the Cajal bodies and nucleoplasm, 25S rRNA resides in the nucleoli. It is possible that a specific factor(s) (e.g. a chaperone), which is only expressed and present in Cajal bodies (or the nucleoplasm) under induced conditions, participates in induced modification. As a consequence, 25S rRNA, which resides exclusively in the nucleoli, is not exposed to the factor(s), hence no induced formation of Ψ1051. To clarify this problem, further experiments are necessary.
Mechanisms of induced pseudouridylation by Pus7p and snR81 RNP
It is extremely interesting that Pus7p and snR81 RNP, which, under normal conditions, do not recognize positions 93 and 56, respectively, loosen their specificities to include these inducible sites upon induction. What is the molecular mechanism behind the induced pseudouridylation?
One possible mechanism for induced pseudouridylation is that the enzyme structures (or the substrate structures) may be altered when the cellular environment is appropriate to induce pseudouridylation; consequently, the similar yet non-identical sites, positions 93 and 56, become the substrates. Mechanistically, induced conditions (heat-shock or nutrient deprivation) may activate (or block) a signalling pathway(s) that in turn triggers the expression (or re-distribution) of a protein(s) (or chaperone) that helps the enzymes (or the substrates) re-structure and modify the imperfect sites.
An alternate mechanism for induced pseudouridylation is that changes in conditions might escalate the expression of the enzymes, which would push modification at imperfect sites. However, our preliminary results indicated that there is no escalation in snR81 level upon condition changes (data not shown), suggesting that the induced overexpression of the enzymes is unlikely. However, we realize that no change in steady-state levels of enzymes does not exclude the possibility of a massive re-distribution of the enzymes to the site of modification, and the resultant high concentrations of enzymes might be able to push pseudouridylation at imperfect sites. It is true that Pus7p and snR81 RNP are available to U2 RNA when cells are grown under both uninduced and induced conditions because these enzymes bind specifically to and modify the constitutive pseudouridylation sites in U2, that is, positions 35 and 42 (guided by the 5′ pocket of snR81), respectively. However, the concentrations of the enzymes might be low under uninduced conditions. As a consequence, the low level of enzymes might be able to modify only the perfect sites. Upon induction (changes in conditions), more enzymes might be re-located to the site of modification, forcing modification at the imperfect sites. It should also be noted that synthesis of rRNA and tRNA is downregulated under nutrient deprivation or heat-shock conditions. It is thus possible that under these conditions, Pus7p and snR81 RNP, which also modify rRNA and tRNA, are freed up, making them more readily available for U2 snRNA. To clarify this point, localization analyses under induced and uninduced conditions may be necessary.
Why would starved cells or heat-shocked cells induce pseudouridylation in U2 and how might induced pseudouridylation affect function?
It is well established that in order to survive harsh conditions, such as starvation or heat shock, cells downregulate gene expression in general (although the production of many specific gene products is enhanced), including downregulation of pre-mRNA splicing and protein synthesis (Herman, 2002; Gray et al, 2004). This response keeps most gene products at a minimum level, thereby conserving energy. It is conceivable that induced pseudouridylation in U2, an important component of spliceosome, may contribute to this downregulation pathway, perhaps by negatively impacting pre-mRNA splicing.
In regard to splicing, the Staley lab and the Ares lab have recently proposed an attractive model for the splicing mechanism involving toggling between two alternate U2 structures (stem-loop IIa and stem IIc) during splicing (Hilliker et al, 2007; Perriman and Ares, 2007) (Figure 1). Stem-loop IIa is favoured for U2 to bind to the branch site, whereas the alternate stem IIc is necessary for catalysis. To convert stem-loop IIa to stem IIc, an eight-nucleotide sequence (nt 98–105) downstream of stem II forms a duplex with the loop sequence (nt 54–61) of stem-loop IIa, thus resulting in the disruption of stem IIa and the creation of stem IIc. Conversely, the reverse process leads to the conversion of stem IIc to stem-loop IIa.
Interestingly, Ψ93 and Ψ56 both fall into the regions of stem-loop IIa and stem IIc, suggesting that these two inducible pseudouridines have a role in splicing, perhaps by modulating the changes between stem-loop IIa and stem IIc. In this regard, Ψ56 is part of stem IIc. In the case of Ψ93, it lies just upstream of nt 98–105, and may participate, along with its surrounding sequence (nt 91–96), in additional base pairing with the sequence (nt 62–67) that is part of stem IIa, thus disrupting stem IIa and forming an extension of stem IIc (see Figure 1). From a thermostability perspective, such additional base-pairing interactions (stem IIc extension) would help compete against the stem IIa structure (Figure 1), thus facilitating the conversion from stem IIa to stem IIc. In this regard, it has been reported that the Ψ-A pair is more stable than the U–A pair, and that the Ψ-A pair could also bring rigidity to the structure (Charette and Gray, 2000; Newby and Greenbaum, 2002a, 2002b; Kolev and Steitz, 2006; Dai et al, 2007). Together, Ψ56 and Ψ93 may thus tip the balance towards stem IIc formation, and perhaps stabilize the structure, thereby inhibiting splicing. This hypothesis is consistent with our experimental results demonstrating that Ψ93 indeed exacerbated the splicing defect of a second-step mutant pre-mRNA (Figure 6). However, it should be noted that toggling between stem IIa and stem IIc is necessary not only for the second step, but also for the first step of splicing. Although we observed the negative effect of Ψ93 on the splicing of a second-step mutant, we failed to detect a similar effect of Ψ93 on the splicing of several first-step mutants tested. It appears that the mechanism by which induced pseudouridylation affects pre-mRNA splicing is far more complex than we expected. A detailed functional analysis is underway.
We also note that the effect of Ψ93 on splicing is rather mild. However, we suspect that there are a lot more induced modifications (including pseudouridines) that are yet to be identified in spliceosomal snRNAs under stressed conditions (see below). Based on previous reports that pseudouridines often function synergistically or cooperatively (King et al, 2003; Zhao and Yu, 2004b, 2007), induced pseudouridines, if tested in combination, will likely have a much larger functional impact.
How abundant are induced modifications in spliceosomal snRNAs and other RNAs?
Our discovery of Ψ56 and Ψ93 in U2 raises the issue of the existence and abundance of inducible modifications in spliceosomal snRNAs (and RNAs in general). Given that pre-mRNA splicing can be regulated in response to environmental stress and that spliceosomal snRNA modifications (including pseudouridylation and 2′-O-methylation) can influence pre-mRNA splicing (Yu et al, 1998; Donmez et al, 2004; Zhao and Yu, 2004b, 2007; Yang et al, 2005), we suspect that inducible modifications occur frequently in spliceosomal snRNAs in response to environmental stress. This hypothesis is consistent with our experimental data showing that limited search (we have checked only for pseudouridylation in the first 100 nucleotides of U2 in response to only two growth condition changes) identified at least two inducible pseudouridines (Ψ56 and Ψ93, and possibly Ψ91 and Ψ94 as well, see Figures 3 and 4), suggesting a potentially high frequency of inducible modifications. Also, according to our results, inducible pseudouridylation does not require the substrate to have a perfect match with the consensus sequence (Figures 3A and 5). For instance, the sequence surrounding position 93 matches imperfectly with its guide, the 3′ pseudouridylation pocket of snR81 snRNA (Figures 3A and 5). Using this criterion, we have aligned the guide sequences of known yeast snoRNAs with yeast spliceosomal snRNAs, and have identified a number of imperfect (close-to perfect) matches, suggesting a potential inducible enzyme–substrate relationship. Expanding the hypothesis to other RNAs, one can identify an even larger number of imperfect matches, thus suggesting that inducible modifications are abundant and widespread.
In short, our findings have opened the door to a number of experiments towards identifying more inducible RNA modifications, and understanding their molecular mechanisms and regulatory functions.
Materials and methods
Cell culture, growth conditions and isolation of RNA
S. cerevisiae strains (wild-type and various mutants) were grown at 30°C in YPD or selective media to OD∼2–4 (∼6 × 107 cells/ml). Cells were then heat shocked at either 37 or 45°C for 30 min, or allowed to continue to grow to saturation (OD∼15–20) (∼6 × 108 cells/ml). The heat-shocked cells or the cells at different growth stages were immediately collected, broken open by bead beating. Total RNA was isolated with TRIzol (Invitrogen) according to the manufacture's instruction. The purified RNA was ready for pseudouridylation assays.
Primer-extension-based pseudouridylation assay
Primer-extension-based pseudouridylation assay was carried out exactly as previously described (Bakin and Ofengand, 1993; Ma et al, 2003). Briefly, ∼10 μg of total cellular RNA, purified from corresponding yeast strains, were treated with CMC at 37°C for 20–30 min in a 30-μl mixture, which contained ∼10 μg of yeast total RNA, 50 mM Bicine, pH 8.3, 0.17 M CMC, 4 mM EDTA and 7 M urea. After CMC modification, the RNA was incubated at 37°C for 1.5–2 h in bicarbonate buffer (50 mM, pH 10.4). RNA was recovered and primer extension was performed, as previously described (Yu et al, 1998). The primer used here was a 5′ 32P-radiolabelled oligodeoxynucleotide complementary to nucleotides 104–126 of S. cerevisiae U2. Signals corresponding to pseudouridines were visualized by autoradiography.
Site-specific quantitative pseudouridylation assay
To quantify U2 pseudouridylation at position 93, we purified, using oligonucleotide affinity chromatography (Zhao and Yu, 2007), U2 from total RNA isolated from cells with an OD600 of 5, 15 or 20, and subsequently labelled U2 RNA at position 93 for TLC analysis. Briefly, 40 mg of total RNA were mixed with 2000 pmol of a biotinylated antisense U2 oligonucleotide (complementary to nucleotides 27–49 of U2) in 50 μl NET buffer containing 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.05% (v/v) NP-40. After denaturing, the sample was mixed with 200 μl streptavidin beads (Pierce) with 4 mM of Mg2+. Following a 3-h rotation at 37°C, the beads were precipitated and washed with NET-2 (with 4 mM MgCl2) four times. U2 was then eluted with 2 × 400 μl of 2.4 M of tetraethylammonium chloride at 65°C for 15 min. After PCA extraction, U2 was ethanol precipitated.
Purified U2 was then subjected to site-specific RNase H cleavage directed by 2′-O-methyl RNA–DNA chimera (10 pmol of UmUmAmdAdAdAdGGmTmAmAmTmGmAmGmCmCm for cleavage between position 93 and position 94) (Zhao and Yu, 2004a). The cleaved 5′-half was gel purified and its 3′ end was subsequently ligated, in the presence of a bridging oligodeoxynucleotide, to a 5′ [32P]-radiolabelled RNA oligo (863SR: [32P]UAAUGGAAUAGGACC), and thus, the 3′ side of position 93 became radiolabelled. After gel purification, the ligated RNA was treated with nuclease T2 to completion. The digested sample was dotted on a cellulose TLC PEI membrane (EM science), which was then chromatographed in two dimensions in the standard solvents (first dimension, isobutyric acid/ammonium/water (50:1:29, v/v/v), and second dimension, isopropanol/HCl/water (70:15:15, v/v/v)). The relative intensities of uridylate and pseudouridylate were quantified by PhosphorImager.
Construction of guide RNAs and mutant yeast strains containing point mutations within U2 snRNA
To construct mutant snR81 box H/ACA RNAs (see Figures 4A and 5D), we used PCR to create slightly altered snR81 DNA templates, where either the 5′ pocket was mutated or the 3′ pocket was mutated. The DNA templates were then cloned into vector yEPlac195 (Alexandrov et al, 2006).
Site-directed mutagenesis was used to create mutant U2 DNA templates (see Figures 4B and 5A) containing a point mutation at various positions, according to the manufacturer's instruction (Invitrogen). The mutant templates were then cloned into yEPlac195 (Alexandrov et al, 2006). The resultant U2 plasmids were transformed into the yeast strain DM1001-snrΔ (Mat α ura3Δ his3 lys2 ade2–101 snr20::LYS2 pU2URA-2[pRS316(URA3 CEN SNR20)] snr81::KanMX) (Yang et al, 2005). After selection against Ura under 5FOA medium, the strains containing a point mutation at various positions within U2 RNA was created. These strains were then transformed with various snR81 constructs, and tested for induced pseudouridylation in U2 RNA. In addition to these mutant strains, several previously constructed strains (BY4741, snr81D, pus1D, pus7D, cbf5D95A, DM1001) were also used (see Figure 3) (Madhani and Guthrie, 1992; Zebarjadian et al, 1999; Ma et al, 2003, 2005; Yang et al, 2005).
Splicing assay
We used copper-resistance growth assay coupled with primer extension to assess pre-mRNA splicing in vivo. YCL46 [MATa cup1::ura3 leu2 trp1 lys2 ade2 his3] (Lesser and Guthrie, 1993) was transformed with the plasmid containing the wild-type or mutant (gAG/A) ACT1–CUP1 reporter gene (under control of the GPD promoter, LEU+) as well as the plasmid containing a position 93-specific guide RNA, snR81 (U101A/U105A) (also under control of the GPD promoter, URA+), by incubating at 42°C for 30 min in OST buffer (100 mM lithium acetate, 40% (w/v) PEG3350) as described (Yang et al, 2005). To assay copper sensitivity, strains were grown in selective media to mid-log phase in –Ura –Leu media, then diluted to OD600=0.1 and OD600=0.01. In all, 9 μl of cells were dotted on respective selective plates with or without copper. The plates were incubated at 30°C for 3–4 days and photographed.
Splicing was analysed by primer extension (Lesser and Guthrie, 1993; Siatecka et al, 1999) using total RNA as template. Total RNA was isolated from cells at mid-log phase (see above). The primer, 5′ GGC ACT CAT GAC CTT C 3′, was complementary to the CUP1 exon sequence. Reaction products were loaded on a 8% polyacrylamide, 8 M urea gel (acrylamide:bis=19:1, EM science) and visualized via autoradiography. Relative levels of spliced product (mRNA) and spliced intermediate (lariat) were quantified by Phosphorimager (Molecular Dynamics).
Supplementary Material
Acknowledgments
We thank Eric Phizicky, Beth Grayhack, Christine Guthrie, David McPheeters, Jonathan Staley, for kind gifts of plasmids and strains used in this study. We also thank the members of the Yu laboratory for valuable discussions. This work was supported by grant GM62937 (to Y-TY) from the National Institute of Health.
Author contributions:GW designed and carried out the experiments, analysed the data and helped to draft the manuscript. MX designed and performed the experiments and participated in the analysis of the data. CY participated in the initial phase of this project and carried out some of the experiments. YTY formulated research questions, designed experiments, analysed the experimental data and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96 [DOI] [PubMed] [Google Scholar]
- Bakin A, Ofengand J (1993) Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry 32: 9754–9762 [DOI] [PubMed] [Google Scholar]
- Behm-Ansmant I, Urban A, Ma X, Yu YT, Motorin Y, Branlant C (2003) The Saccharomyces cerevisiae U2 snRNA:pseudouridine-synthase Pus7p is a novel multisite-multisubstrate RNA:Psi-synthase also acting on tRNAs. RNA 9: 1371–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjork GR (1995) Biosynthesis and function of modified nucleotides. In tRNA: Structure, Biosynthesis, and Function, Soll D, RajBhandary U (eds) pp 165–205. Washington, DC: ASM Press [Google Scholar]
- Charette M, Gray MW (2000) Pseudouridine in RNA: what, where, how, and why. IUBMB Life 49: 341–351 [DOI] [PubMed] [Google Scholar]
- Dai Q, Fong R, Saikia M, Stephenson D, Yu YT, Pan T, Piccirilli JA (2007) Identification of recognition residues for ligation-based detection and quantitation of pseudouridine and N6-methyladenosine. Nucleic Acids Res 35: 6322–6329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donmez G, Hartmuth K, Luhrmann R (2004) Modified nucleotides in the 5′ end of the human U2 snRNA are required for early spliceosome (E complex) formation in vitro. RNA 10: 1925–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997) Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M (2004) ‘Sleeping beauty': quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 68: 187–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H (2009) Nucleic acids are not boring long polymers of only four types of nucleotides: a guide tour. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution, Grosjean H (ed) pp 1–18. Austin, Texas: Landes Bioscience [Google Scholar]
- Herman PK (2002) Stationary phase in yeast. Curr Opin Microbiol 5: 602–607 [DOI] [PubMed] [Google Scholar]
- Hilliker AK, Mefford MA, Staley JP (2007) U2 toggles iteratively between the stem IIa and stem IIc conformations to promote pre-mRNA splicing. Genes Dev 21: 821–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Darzacq X, Tucker KE, Matera AG, Bertrand E, Kiss T (2003) Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J 22: 1878–1888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J, Huang C, Yu YT (2009) Spliceosomal snRNA pseudouridylation. In DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution, Grosjean H (ed) pp 461–474. Austin, Texas: Landes Bioscience [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ (2003) Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425–435 [DOI] [PubMed] [Google Scholar]
- Kolev NG, Steitz JA (2006) In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat Struct Mol Biol 13: 347–353 [DOI] [PubMed] [Google Scholar]
- Lesser CF, Guthrie C (1993) Mutational analysis of pre-mRNA splicing in Saccharomyces cerevisiae using a sensitive new reporter gene, CUP1. Genetics 133: 851–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XH, Liu Q, Fournier MJ (2007) rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol Cell 28: 965–977 [DOI] [PubMed] [Google Scholar]
- Ma X, Yang C, Alexandrov A, Grayhack EJ, Behm-Ansmant I, Yu YT (2005) Pseudouridylation of yeast U2 snRNA is catalyzed by either an RNA-guided or RNA-independent mechanism. EMBO J 24: 2403–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhao X, Yu YT (2003) Pseudouridylation (Psi) of U2 snRNA in S. cerevisiae is catalyzed by an RNA-independent mechanism. EMBO J 22: 1889–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell 71: 803–817 [DOI] [PubMed] [Google Scholar]
- Massenet S, Motorin Y, Lafontaine DL, Hurt EC, Grosjean H, Branlant C (1999) Pseudouridine mapping in the Saccharomyces cerevisiae spliceosomal U small nuclear RNAs (snRNAs) reveals that pseudouridine synthase pus1p exhibits a dual substrate specificity for U2 snRNA and tRNA. Mol Cell Biol 19: 2142–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massenet S, Mougin A, Branlant C (1998) Posttranscriptional modifications in the U small nuclear RNAs. In Modification and Editing of RNA, Grosjean H (ed) pp 201–228. Washington, DC: ASM Press [Google Scholar]
- Newby MI, Greenbaum NL (2002a) Investigation of Overhauser effects between pseudouridine and water protons in RNA helices. Proc Natl Acad Sci USA 99: 12697–12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newby MI, Greenbaum NL (2002b) Sculpting of the spliceosomal branch site recognition motif by a conserved pseudouridine. Nat Struct Biol 9: 958–965 [DOI] [PubMed] [Google Scholar]
- Ni J, Tien AL, Fournier MJ (1997) Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573 [DOI] [PubMed] [Google Scholar]
- Ofengand J, Fournier M (1998) The pseudouridine residues of rRNA: number, location, biosynthesis, and function. In Modification and Editing of RNA, Grosjean H (ed) pp 229–253. Washington, DC: ASM Press [Google Scholar]
- Perriman RJ, Ares M Jr (2007) Rearrangement of competing U2 RNA helices within the spliceosome promotes multiple steps in splicing. Genes Dev 21: 811–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekna-Przybylska D, Przybylski P, Baudin-Baillieu A, Rousset JP, Fournier MJ (2008) Ribosome performance is enhanced by a rich cluster of pseudouridines in the A-site finger region of the large subunit. J Biol Chem 283: 26026–26036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy R, Busch H (1988) Small nuclear RNAs: RNA sequences, structure, and modifications. In Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles, Birnsteil ML (ed), pp 1–37. Heidelberg: Springer-Verlag Press [Google Scholar]
- Siatecka M, Reyes JL, Konarska MM (1999) Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev 13: 1983–1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban A, Behm-Ansmant I, Branlant C, Motorin Y (2009) RNA sequence and two-dimensional structure features required for efficient substrate modification by the Saccharomyces cerevisiae RNA:{Psi}-synthase Pus7p. J Biol Chem 284: 5845–5858 [DOI] [PubMed] [Google Scholar]
- Xiao M, Yang C, Schattner P, Yu YT (2009) Functionality and substrate specificity of human box H/ACA guide RNAs. RNA 15: 176–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, McPheeters DS, Yu YT (2005) Psi35 in the branch site recognition region of U2 small nuclear RNA is important for pre-mRNA splicing in Saccharomyces cerevisiae. J Biol Chem 280: 6655–6662 [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Narayanan A, Terns RM, Terns MP, Steitz JA (2001) Internal modification of U2 small nuclear (sn)RNA occurs in nucleoli of Xenopus oocytes. J Cell Biol 152: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA (1997) A new method for detecting sites of 2′-O-methylation in RNA molecules. RNA 3: 324–331 [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA (1998) Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Terns RM, Terns MP (2005) Mechanisms and functions of RNA-guided RNA modification. In Topics in Current Genetics, Grosjean H (ed) Vol 12, pp 223–262. New York: Springer-Verlag [Google Scholar]
- Zebarjadian Y, King T, Fournier MJ, Clarke L, Carbon J (1999) Point mutations in yeast CBF5 can abolish in vivo pseudouridylation of rRNA. Mol Cell Biol 19: 7461–7472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT (2004a) Detection and quantitation of RNA base modifications. RNA 10: 996–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT (2004b) Pseudouridines in and near the branch site recognition region of U2 snRNA are required for snRNP biogenesis and pre-mRNA splicing in Xenopus oocytes. RNA 10: 681–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yu YT (2007) Incorporation of 5-fluorouracil into U2 snRNA blocks pseudouridylation and pre-mRNA splicing in vivo. Nucleic Acids Res 35: 550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.