Abstract
Aims
Human thoracic aortic aneurysms (TAAs) are characterized by extracellular matrix breakdown associated with progressive smooth muscle cell (SMC) rarefaction. These features are present in all types of TAA: monogenic forms [mainly Marfan syndrome (MFS)], forms associated with bicuspid aortic valve (BAV), and degenerative forms. Initially described in a mouse model of MFS, the transforming growth factor-β1 (TGF-β1)/Smad2 signalling pathway is now assumed to play a role in TAA of various aetiologies. However, the relation between the aetiological diversity and the common cell phenotype with respect to TGF-β signalling remains unexplained.
Methods and results
This study was performed on human aortic samples, including TAA [MFS, n = 14; BAV, n = 15; and degenerative, n = 19] and normal aortas (n = 10) from which tissue extracts and human SMCs and fibroblasts were obtained. We show that all types of TAA share a complex dysregulation of Smad2 signalling, independent of TGF-β1 in TAA-derived SMCs (pharmacological study, qPCR). The Smad2 dysregulation is characterized by an SMC-specific, heritable activation and overexpression of Smad2, compared with normal aortas. The cell specificity and heritability of this overexpression strongly suggest the implication of epigenetic control of Smad2 expression. By chromatin immunoprecipitation, we demonstrate that the increases in H3K9/14 acetylation and H3K4 methylation are involved in Smad2 overexpression in TAA, in a cell-specific and transcription start site-specific manner.
Conclusion
Our results demonstrate the heritability, the cell specificity, and the independence with regard to TGF-β1 and genetic backgrounds of the Smad2 dysregulation in human thoracic aneurysms and the involvement of epigenetic mechanisms regulating histone marks in this process.
Keywords: Marfan syndrome, Transforming growth factor-β1, Smooth muscle cells, Smad, Histone acetylation
1. Introduction
The transforming growth factor-β1 (TGF-β) signalling pathway is a ubiquitous transduction system expressed in many cell types and involved in numerous biological activities, including extracellular matrix synthesis, cell survival, and differentiation.1 TGF-β1 initiates several intracellular pathways, including the Smad pathway.2 Smads are transcriptional factors critical for translating immediate signals from TGF-β receptors to the nucleus. Combinatorial interactions of Smads and variability of association with transcriptional factors induce a context-dependent transcriptional regulation and a substantial diversity in the Smad response.3–5 Recent studies have demonstrated altered regulation of TGF-β1 signalling in both syndromic (FBN1,6 TGFBR2,7 GLUT108) and non-syndromic [aneurysms associated with bicuspid aortic valve (BAV) and degenerative9] TAA, and have suggested that the TGF-β pathway could play an important role in TAA pathology, due to its involvement in the control of SMC survival and extracellular matrix integrity10,11 via a postulated increase in TGF-β1 bioavailability.12,13 TAA is characterized by an important aetiological diversity in humans including (i) monogenic forms associated with mutations in FBN1,14 TGFBR1/27,15 [Marfan syndrome (MFS) and Loeys–Dietz syndrome (LDS)], MYH11,16,17 ACTA2,18 and GLUT10,8 presenting a large functional diversity, i.e. structure of the extracellular matrix, contractile and transporter functions,19 (ii) aneurysms associated with BAV,20 or (iii) degenerative forms linked to ageing.21,22 Increases in Smad2 activation and its nuclear translocation have been observed in TAA, regardless of aetiology.6,7,9,23–25 However, several contradictory results suggest a complex dysregulation of the TGF-β/Smad signalling pathway.26 Smad2 activation and the increase in stored TGF-β1 occur concomittently in the aneurysmal media but the location of phosphorylated Smad2 and the sites of TGF-β1 storage and bioavailability do not coincide.9 Indeed, the activation of Smad2 is homogeneously observed throughout the media, whereas the latent form of TGF-β1 is present mainly in the outer media, near the adventitia. The increase in phosphorylated Smad2 is also present in patients bearing mutations in TGFBR2,9,24 despite the fact that these mutations induce a loss of function of the TGF-β receptors.15
These preliminary observational data suggested an autonomization of Smad expression as a specific arterial SMC phenotype associated with TAA. The purpose of the present study was to elucidate the dysregulation of Smad2 signalling and to associate the modifications of Smad2 expression with an epigenetic regulatory process in aortic SMCs from syndromic and non-syndromic human TAAs.
2. Methods
2.1. Patients and aortic specimens
The clinical research protocol was approved by the local Ethics Committee (CPP 05 04 32, Ambroise Paré, Boulogne, France, April 2005; updated in March 2008) and was in accordance with the principle of the Declaration of Helsinki.27 All patients gave informed consent.
Aneurysmal ascending aortic specimens were collected during aortic surgery (Hôpital Bichat). Forty-six specimens were divided into three groups according to their clinical features and genetic background: MFS related to mutations in FBN1 (n = 12, mean age 39 ± 15 years) or TGFBR2 (n = 2, mean age 44 ± 11 years), BAV (n = 15, mean age 59 ± 13 years), and degenerative ascending aortic aneurysm (n = 19, mean age 68 ± 20 years). The clinical data associated with this series have been reported elsewhere.28 All specimens were from aneurysms of >5 cm in diameter. Normal ascending aortas were obtained from organ transplant donors (n = 10) with the authorization of the French Biomedicine Agency (PFS09-007) and in accordance with the Declaration of Helsinki. Aneurysmal tissues were sampled in the outer curvature, the most dilated part of the ascending aorta. Aortic tissue preparation consisted of an immediate dissection to separate medial and adventitial layers followed by either freezing or enzymatic digestion to obtain SMC and fibroblast cultures.
2.2. Primary cultures and in vitro stimulation
Medial and adventitial samples were incubated in either collagenase and elastase 0.1% or collagenase 0.3% solutions, respectively (3 h, 37°C), to obtain SMCs and fibroblasts to be cultured.29 All cells were routinely cultured in smooth muscle cell (SMC) medium (Promocell) containing 5% foetal calf serum (FCS), gentamicin-sulfate (50 mg/mL), amphotericin B (50 µg/mL), insulin (5 mg/mL), and growth factors (human epidermal and fibroblast growth factors, respectively 0.5 and 2 µg/mL). Passages 3–6 were used for experiments. All stimulations were performed after 24 h incubation in serum-free medium. SMCs and fibroblasts were then incubated with TGF-β1 and/or anti-TGF-β1 (Sigma-Aldrich, St Louis, MO, USA) for 24 h.
2.3. siRNA transfection
To verify TGF-β1/Smad2 target gene specificity, we inhibited Smad2 expression by using an siRNA sequence targeting human Smad2. Calcium phosphate-mediated transfection was performed. siRNA duplexes were incubated with CaCl2 (2.5 M) and then added to HBSP solution (190 mM NaCl, 0.75 mM Na2HPO4, 6 mM glucose, 5 mM KCl, 25 mM HEPES). Human SMCs were transfected with a final concentration of 200 nM siRNA. TGF-β1 (5 ng/mL, 24 h) was added 48 h later. Finally, Smad2 and target gene [connective tissue growth factor (CTGF) and α-actin] expressions were quantified by qPCR 72 h after transfection.
2.4. Aortic tissue extraction
Extractions (protein, mRNA, and chromatin) were performed directly from frozen aortic media and adventitia stored at −80°C. Aortic samples were first cryogenically pulverized in liquid nitrogen, using a freezer mill (model 6750 SPEX SamplePrep). For protein, mRNA, and chromatin isolation, homogenization of 100–300 mg of crushed aortic powder was performed in the corresponding extraction buffer.
2.5. Reverse transcription-PCR
Total RNA was extracted from SMCs or fibroblasts, using the EZNA kit (Omega Biotek), according to the manufacturer's directions. Reverse transcription was performed using kits from Invitrogen (Carlsbad, CA, USA). Real-time PCR was performed in the LightCycler system with SYBR Green detection (Roche Applied Science) using specific primers (see Supplementary material online, Table S1). The mRNA levels were normalized to GAPDH mRNA.
2.6. Immunoblotting
Proteins were extracted from medial or adventitial tissues or SMCs. Tissues or cells were homogenized in a lysis buffer [50 mM Tris (pH 8), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 5 mM EDTA], containing protease inhibitors and a serine/threonine and tyrosine phosphatase inhibitors (Sigma-Aldrich). The protein concentration from each sample was determined (Thermo Fisher Scientific, Rockford, IL, USA). Extracts were separated by 10% sodium dodecyl sulfate–polyacrilamide gel electrophoresis (SDS–PAGE). Proteins were transferred to polyvinylidene difluoride membranes, blocked with 5% BSA-TBS-T [Tris-buffered saline (pH 7.4)-0.1% Tween 20] for 1 h. Membranes were then incubated overnight (4°C) with primary antibodies: p-smad2/3 (0.5 µg/mL), Smad2 (0.5 µg/mL), Smad7 (1 µg/mL; all from Santa Cruz Biotechnology), Smad4 (1 µg/mL, Cell Signaling Technology), α-actin (28 µg/mL, Dako), SM-myosin (1 µg/mL, Abcam), or GAPDH (1.6 µg/mL, Biovalley) and then washed with TBS-T and incubated with peroxidase-conjugated anti-rabbit, anti-mouse, or anti-goat IgG (Jackson Laboratories) for 1 h. The signal was detected using a chemiluminescence kit (ECL+ kit; Amersham). Positive immunoreactive bands were quantified by densitometry. Protein levels were normalized to GAPDH protein levels. Immunoprecipitation (IP) of phosphorylated TGFβR1 (pTGFβR1) was performed. An amount of 300 µg of total protein extracts were incubated with TGFβR1 antibody (2 µg, Santa Cruz Biotechnology) for 2 h at room temperature, and then incubated with Bio-Adembeads Protein G 0433 (30 µL/2 µg TGFβR1 antibody, Ademtech) overnight at 4°C. Immunoprecipitates were washed five times in cold lysis buffer-TBS-T (1:1) and then resuspended in sample buffer for analysis by SDS–PAGE. Phospho-serine polyclonal antibody (2 µg/mL, Abcam) was used for immunoblotting.
2.7. Chromatin immunoprecipitations
Chromatin immunoprecipitations (ChIPs) were performed on frozen aneurysmal (Marfan: n = 3; degenerative: n = 3; BAV: n = 3) and control (n = 5) media and adventitia. Tissue homogenates were cross-linked with 1% formaldehyde for 15 min, and glycine (final concentration of 125 mM) was added. Chromatin shearing and IPs were performed using histone ChIP kit (Diagenode), following the manufacturer's instructions. Chromatin shearing was verified by agarose gel migration, and chromatin fragment lengths between 200 and 800 base pairs were obtained. IPs were performed with H3K9/14ac, H3K4me, H3K9me2, H3K9me3, H4K5/8/12/16ac, and H4K20me (1 µg/IP, Diagenode). After IP, DNA was isolated from chromatin and purified. DNA from non-immunoprecipitated sheared chromatin (INPUT) was also purified. PCR was then performed using specific primers (see Supplementary material online, Table S1).
2.8. Statistical analysis
Values are expressed as means ± SEM. For results expressed as box plots, the median is shown. Upper and lower limits of boxes represent inter-quartiles (25th and 75th), whereas upper and lower bars show percentiles. The significance of differences between groups was tested using the Mann–Whitney non-parametric test. A value of P ≤ 0.05 was considered significant.
See also Supplementary material online, Methods, for TGFβR2 transfection and DNA methylation methods.
3. Results
3.1. Smad2 activation and overexpression in TAA
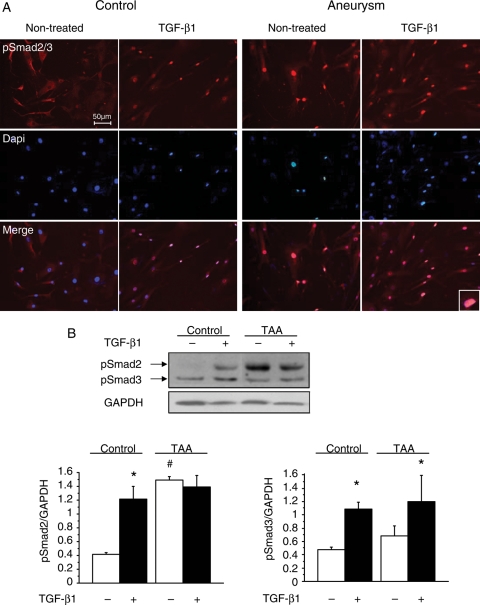
Smad activation was measured in extracts of the medial layer from TAA and ascending aorta. In agreement with previous studies,6,7,9 pSmad2 levels were increased in the TAA medial layer (Figure 1A and B) regardless of aetiology: FBN1, TGFBR2, BAV, and degenerative, compared with normal media (control) (P < 0.0001). Total Smad2 protein levels were also increased in TAA medial extracts (P < 0.001). A positive correlation was observed between pSmad2 and Smad2 protein expression (R2 = 0.68; P < 0.0001) (Figure 1C). Smad2 activation and overexpression were specific downstream to the TGF-β receptors since the activity and expression levels of the other Smad-family proteins remained unchanged (pSmad3, Smad4, and Smad7) (Figure 1A; Supplementary material online, Figure S1A). Similarly, non-Smad pathways, RhoA and MAPkinase pathways,2 were not affected in TAA samples. RhoA target expression (smooth muscle α-actin and smooth muscle-myosin) and MAPkinase activation were similar in TAA and control medial extracts (Figure 1A; Supplementary material online, Figure S1B). The response to exogenous TGF-β1 and the constitutive nature of the Smad2 dysregulation were next analysed in control and TAA-derived SMCs. In control SMCs, basal levels of pSmad2 were low and treatment with TGF-β1 induced the phosphorylation and the nuclear translocation of Smad2. In contrast, a constitutive nuclear staining of pSmad2 was observed independently of the addition of TGF-β1 in TAA-derived SMCs (Figure 2A). As for medial extracts, this constitutive activation was specific to Smad2 since a normal Smad3 phosphorylation induced by TGF-β1 was observed in TAA-derived SMC cultures (Figure 2B).
Figure 1.
Smad2 activation and overexpression in aortic extracts and SMC cultures. (A) Immunoblots performed with aortic medial extracts showing increased phosphorylated Smad2 and total Smad2 levels in TAA extracts compared with controls. No differences in pSmad3, α-actin, and SM-myosin protein levels are observed. (B) Quantification of pSmad2 (relative pSmad2 protein level: TGFBR2: 2.14 ± 0.94; FBN1: 3.03 ± 1.47; degenerative: 2.92 ± 1.64; BAV: 2.35 ± 1.38 vs. control: 0.64 ± 0.26; *P < 0.0001) and Smad2 (TGFBR2: 1.60 ± 0.19; FBN1: 1.92 ± 1.4; degenerative: 1.86 ± 1.36; BAV: 1.85 ± 1.28 vs. control: 0.44 ± 0.15; *P < 0.001). Results are expressed as relative protein levels: pSmad2/GAPDH or Smad2/GAPDH. (C) Positive correlation between pSmad2 and Smad2 protein levels in TAA (all aetiologies included). R2 = 0.68. No positive correlation was observed in control extracts (R2 = 0.003).
Figure 2.
Constitutive activation and phosphorylation of Smad2 in TAA. (A) Constitutive activation of Smad2 in cultured aneurysmal SMCs. Immunostaining of pSmad2/3, performed after stimulation by TGF-β1 (5 ng/mL; 24 h), shows a nuclear accumulation of pSmad2/3 induced by TGF-β1 in control SMCs. Increased nuclear accumulation of pSmad2/3 is observed in aneurysmal SMCs (magnified in inset) independently of the addition of exogenous TGF-β1. Nuclear counterstaining: DAPI. Scale bar: 50 µm. (B) pSmad2/3 immunoblotting performed with protein extracts from control and aneurysmal SMCs. Results are expressed as means ± SEM. TGF-β1 induces a significant increase in pSmad2 levels in control SMCs (*P < 0.01) and in pSmad3 levels in both control and aneurysmal SMCs (*P < 0.01). A constitutive increase in pSmad2 is observed in aneurysmal SMCs (#P < 0.001) compared with non-treated SMCs. Results expressed as pSmad2/GAPDH or pSmad3/GAPDH.
3.2. Constitutive nature and specificity of the Smad2 activation
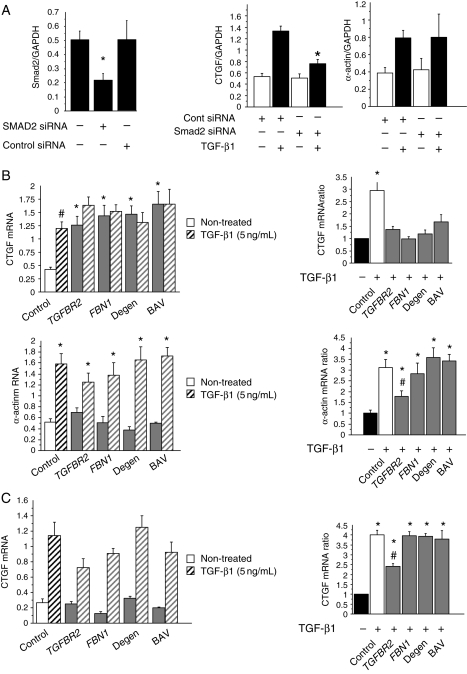
To further explore the specificity of the Smad2 activation, Smad2 (CTGF) and non-Smad/RhoA (α-actin) targets were quantified.2,30 Transfection of control SMCs with Smad2-siRNA specifically decreases Smad2 expression (60%) (Figure 3A). Smad2 silencing was associated with a significant decrease in CTGF expression in response to TGF-β1, whereas α-actin levels were not modified (Figure 3A).
Figure 3.
Specificity of Smad2 activation. (A) Smad2 silencing by siRNA targeting Smad2 in control SMCs. Smad2 siRNA transfection decreases Smad2 expression (P < 0.0001). Smad2 mRNA relative level was quantified by qPCR (Smad2 siRNA: 0.0217 ± 0.05; cont siRNA 0.5 ± 0.133; non-transfected SMCs: 0.5 ± 0.06). Transfected SMCs were treated with TGF-β1 (5 ng/24 h). Smad2 silencing induces a significant lack of CTGF expression in response to TGF-β1, whereas it has no effect on α-actin expression (normalization by GAPDH mRNA expression). (B) Effect of TGF-β1 treatment on CTGF (Smad2 target) and α-actin (non-Smad2 target) expression. All data are means ± SEM. CTGF mRNA levels are significantly increased with TGF-β1 treatment of control SMCs (#P < 0.001). Basal CTGF levels are significantly increased in aneurysmal SMCs compared with controls (*P < 0.0001). No difference is observed between aetiologies. Increased α-actin levels are induced by TGF-β1 treatment in both control and aneurysmal SMCs (*P < 0.001) (left panel). Results are represented as the ratio between treated and non-treated levels and expressed as a fold-increase compared with the non-treated condition (right panel). *Significant increase in CTGF and α-actin ratio compared with non-treated level (P < 0.0001), #significant decrease in α-actin ratio in TGFBR2 compared with control and aneurysmal SMCs (P < 0.05). (C) Quantification of CTGF mRNA levels in control and aneurysmal fibroblasts (control: n = 5; TGFBR2: n = 2; FBN1: n = 4; degenerative: n = 4; BAV: n = 3). *Significant increase in CTGF ratio compared with non-treated level (P < 0.01), #significant decrease in CTGF ratio in TGFBR2 compared with control and aneurysmal fibroblasts (P < 0.05).
Increased basal levels of CTGF were observed in TAA-derived SMCs compared with controls (P < 0.001) (Figure 3A; Supplementary material online, Figure S2), regardless of aetiology. In contrast, no differences in basal levels of α-actin were observed. CTGF expression was not modulated by TGF-β1 in TAA-derived SMCs, contrasting with the α-actin expression (Figure 3B). The effect of TGF-β1 on α-actin expression was significantly reduced in SMCs bearing TGFBR2 mutations compared with controls (P < 0.01). Nevertheless, the persistence of a high-basal CTGF expression was observed in association with these genetic defects. Besides its constitutive nature, Smad2 activation is characterized by its cell specificity and heritability. TGF-β1 treatment induced a similar increase in Smad2 activation in control and aneurysmal fibroblasts, except for samples belonging to the TGFBR2 subgroup. In this case, the regulation of fibroblast CTGF expression by TGF-β1 was significantly decreased (∼50%) compared with fibroblasts from controls and from the other types of TAA (Figure 3C).
3.3. Autonomization of the Smad2 activation
To understand the relationship between the constitutive activation of Smad2 and the activity of TGFβR2, we performed transient transfections with wild-type or mutated [Q508Q (stop codon) and R537C (missense)] TGFβR2 in 293 cells (see Supplementary material online, Methods; Figure S3). A lack of phosphorylation and nuclear translocation of Smad2/3 was observed in Q508Q-TGFβR2- and R537C-TGFβR2-transfected 293 cells, incubated with TGF-β1, indicating a loss of function, further supporting the functional dissociation of Smad2 activation from TGF-β receptors.15 Quantification of TGFβR2 and TGFβR1 mRNA levels revealed no significant differences in TAA-derived SMCs compared with controls. Next, we estimated WT and mutated TGFβR2 mRNA expression in SMCs bearing Q508Q and R537C mutations. In these cases, expression of WT and mutated TGFβR2 mRNA was equivalent (see Supplementary material online,Figure S3).
TGF-β receptor complex functionality was estimated by measuring pTGFβR1 protein levels in medial extracts. pTGFβR1 levels were similar in control and TAA medial extracts, except for those carrying TGFBR2 mutations (Q508Q and R537C) in which the pTGFβR1 level was significantly lower compared with both controls and other forms of TAA (P < 0.05) (Figure 4A). In control SMC cultures, incubation with TGF-β1 induced a significant increase in pTGFβR1, CTGF, and α-actin levels (P < 0.01), an effect abrogated by the anti-TGF-β1 antibody (Figure 4B). In TAA-derived SMCs (except TGFBR2 subgroup), incubation with TGF-β1 induced a similar increase in pTGFβR1 and α-actin levels (but not in CTGF mRNA) compared with controls. In the case of TGFBR2 mutations, a defect in TGFβRI phosphorylation and α-actin expression induced by TGF-β1 was observed. Nevertheless, a constitutive increase in CTGF levels was detected. These results provide evidence of the autonomy of Smad2 activation with respect to TGFβR1/2 activity in human TAA-derived SMCs.
Figure 4.
TGF-β receptor activity. (A) Estimation of TGFβR1/2 activity by measurement of TGFβR1 phosphorylation. Results are expressed as the ratio of pTGFβR1/TGFβR1. *Significant decrease in TGFβR1 phosphorylation in aneurysmal media from patients with TGFBR2 mutations (Q508Q and R357C) compared with controls and other types of TAA (P < 0.05). (B) Dissociation between Smad2 activation and TGF-β receptor activity in aneurysmal SMCs. *P < 0.01, treated vs. non-treated conditions. #P < 0.05 vs. control.
3.4. Heritability and cell specificity of Smad2 perturbation
Basal Smad2 mRNA expression was constitutively increased in TAA-derived SMCs compared with control SMCs (P < 0.0001) (Figure 5A), regardless of aetiology. In TAA-derived SMCs, mRNA quantification performed over three passages (passages 3–5) showed an increase in both Smad2 and CTGF expression which was maintained over several cycles of cell division. Next, control SMCs were incubated with conditioned medium from TAA-derived SMCs. The incubation did not induce increases in either activation (CTGF mRNA level) or expression of Smad2 (Figure 5B). Similar results were obtained with TAA-derived fibroblasts incubated with conditioned medium from TAA-derived SMCs (see Supplementary material online,Figure S4A). These results eliminate any autocrine TGF-β1 signalling as the cause of the constitutive Smad2 activation and demonstrate that the Smad2 perturbation, observed in all types of TAA, consists of a constitutive activation and overexpression of Smad2 that are independent of extracellular TGF-β1 and of TGF-β receptor activity.
Figure 5.
Heritability and cell specificity of the Smad2 overexpression and activation. (A) Smad2 and CTGF mRNA levels are quantified over three successive passages between passages 3 and 5. SMCs were cultured in free-serum SMC medium 24 h before RNA extraction. Increased Smad2 and CTGF expression is observed in aneurysmal SMCs compared with controls, whatever the passage (*P < 0.001). (B) Incubation of control SMCs with conditioned medium from aneurysmal SMCs (24 h), followed by the quantification of CTGF and Smad2 mRNA expression. No effect of the incubation is visualized on Smad2 activation (CTGF mRNA level) and Smad2 expression. (C) Quantification of Smad2 mRNA levels in aneurysmal and control fibroblasts shows no difference in Smad2 expression. (D) Representation of SMAD2. SMAD2 has two TSSs. Three mRNA variants are synthesized: 1a, 1b, and 1a + 1b*. Quantification of each variant is performed using specific primers (arrows). Representative migration showing a specific overexpression of Smad2 mRNA variants 1a and 1a + 1b in aneurysmal SMCs compared with controls, whatever the aetiology.
In contrast with SMCs, no difference in basal CTGF mRNA expression was found between aneurysmal and control fibroblasts (Figure 5C).
3.5. Smad2 mRNA variant expression in TAA
The transmission to daughter cells31 and the cell-specificity of the dysregulation of Smad2 expression in TAA strongly suggest the implication of epigenetic mechanisms. Two putative transcription start sites (TSSs) have been identified in the Smad2 promoter: 1a TSS and 1b TSS.32 Depending on the TSS implicated and possible alternative splicing, three transcriptional variants containing the same coding region but different 5′ untranslated regions were generated: 1a, 1b, and 1a + 1b (Figure 5D). The expression of each updated transcriptional variant was estimated by PCR. Transcriptional variants 1a and 1a + 1b were overexpressed in TAA-derived SMCs compared with controls. In contrast, no difference in 1b variant expression was found in TAA-derived SMCs vs. control. These data indicate that the constitutive Smad2 overexpression was associated with an increase in the expression of Smad2 variants, depending only on 1a TSS. This increase was not dependent on variations in Smad2 mRNA stability (see Supplementary material online, Figure S4B), suggesting an activation of the Smad2 promoter.
3.6. Epigenetic modifications on histone tails control Smad2 overexpression in TAA
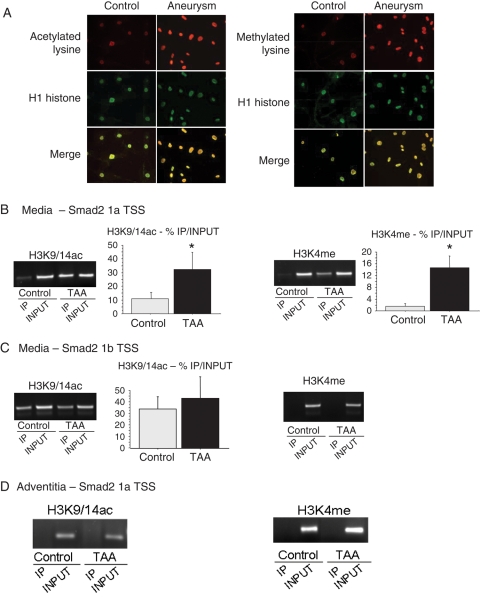
Modifications of epigenetic markers carried by DNA or histones have already been reported to be associated with an increase in promoter activity.33,34 Consequently, we investigated epigenetic markers on the Smad2 promoter in TAA and control aortic media and adventitia. We first hypothesized that the CpG-rich regions of the Smad2 promoter could be hypomethylated. However, for both TAA and control DNA, the Smad2 promoter appeared unmethylated by bisulfite sequencing (see Supplementary material online, Methods; Figure S5), suggesting that differential DNA methylation is not implicated in Smad2 promoter activation in TAA. In contrast, increased acetylated and methylated lysine staining was observed in TAA-derived SMC cultures, and colocalized with histone H1 staining in the nucleus (Figure 6A). These results suggest the involvement of covalent modifications of histone tails, triggering chromatin remodelling in TAA. Our next step was to determine whether epigenetic markers were differentially present on the alternate Smad2 promoters. Precipitation of H3K9/14ac (ChIP) revealed an increase in H3 acetylation on the Smad2 promoter, in the vicinity of the 1a TSS in TAA extracts compared with controls (P < 0.01) (Figure 6B). Similar results were obtained performing ChIP assays with an H3K4me-specific antibody (P < 0.05). No changes in H3K9/14 acetylation and H3K4 methylation levels have been observed associated with the 1b TSS Smad2 promoter (Figure 6C). H3 acetylation and methylation were specifically associated with the 1a TSS Smad2 promoter, corresponding to the specific increase in 1a and 1a + ab transcriptional variants. Moreover, these epigenetic markers on histone H3 were specific of the TAA media and were not observed in TAA adventitial extracts (Figure 6D). These observations correlated with the cell specificity of Smad2 expression. Contrasting with H3 histone, H4 acetylation and methylation (H4K5/8/12/16ac and H4K20me) levels were similar in TAA extracts and controls. Furthermore, no differences in repressive epigenetic histone H3 modifications (H3K9me2, H3K9me3) were found between TAA medial extracts and controls (see Supplementary material online, Figure S6). Thus, we have observed a specific increase in histone H3 acetylation (H3K9/14ac) and methylation (H3K4me) in the medial layer of TAAs of all aetiologies.
Figure 6.
Increase in histone acetylation (H3K9/14) and methylation (H3K4) on Smad2 promoter. (A) Increased acetylated and methylated lysine staining (red) is observed in aneurysmal SMCs and colocalizes with H1 histone staining (green) in the nucleus. Scale bar: 50 µm. (B–D) ChIP performed with aneurysmal and control media (B and C) or adventitia (D). After ChIP, PCR is performed with primers annealing specific sequences upstream to 1a TSS (B and D) or 1b TSS (C). Representative migrations of immunoprecipitated (IP) and non-immunoprecipitated (INPUT) samples are shown for each precipitation. An increase in H3K9/14ac and H3K4me is observed upstream to the 1a TSS, only in aneurysmal SMCs (all aetiologies included, no difference between aetiologies). Results are expressed as %IP/INPUT. *Significant increase in H3K9/14ac (TAA: 36.4 ± 13.3 vs. control: 13.6 ± 1.9; P < 0.01) and H3K4me (TAA: 14.7 ± 10.5 vs. control: 1.8 ± 1.6; P < 0.05) is observed in TAA (all aetiologies included) compared with control.
4. Discussion
Our results demonstrate, for the first time, the heritability, the cell specificity, and the independence with regard to extracellular ligands (TGF-β1) and genetic background of the Smad2 dysregulation in a human chronic disease. This signalling misfunction of Smad2 results in its activation and its overexpression in human TAAs, regardless of aetiology. The specific Smad2 overexpression is associated with modifications of the histone H3 marker pattern in the vicinity of 1a TSS on the Smad2 promoter, in pathological SMCs. The involvement of such an epigenetic mechanism provides evidence of the possible establishment of a sustained modified phenotype of SMCs in chronic cardiovascular diseases.
The perturbations of Smad2 signalling and expression are present in the media of patients with MFS or LDS, regardless of the mutations involved (FBN1 and TGFBR2), as described previously.7 Our article demonstrates clearly a functional dissociation between the Smad2 activation and the activity of the TGF-β1 receptors in the human aneurysmal media. The loss of function induced by the TGFBR2 mutation is effective in adventitial fibroblasts or in vascular SMCs for the non-Smad2/TGF-β1 pathway, but is overwhelmed in TAA-derived SMCs. SMCs from all types of TAA acquire autonomy in Smad2 activation with regard to the canonical TGF-β1 pathway involving the interaction of TGF-β1/TGF-β receptors. These findings also explain the lack of colocalization within the aneurysmal tissue between the retained TGF-β1 and the activation of Smad2.9 An important observation of this study is the signalling specificity of the dysregulation exclusively relating to Smad2 and its downstream pathway. Our results, associating Smad2 and CTGF perturbations, are in accordance with previous studies.30 Moreover, Smad2 silencing proves clearly the independence of α-actin expression with respect to Smad2. Smooth muscle α-actin (ACTA2) is known to be dependent on TGF-β1 via the RhoA signalling pathway.2 The fact that α-actin expression increases in human TAA SMCs proves that these cells can ‘respond’ to TGF-β1 and that the dysregulation is limited to the Smad2 pathway. CTGF, known to be a TGF-β1/Smad target gene, can itself activate the TGF-β1 signalling pathway by its direct binding to TGF-β1. This interaction is associated with an activation of TGF-β1 signalling.35 Thus, although CTGF overexpression can lead to TGF-β1 signalling activation, the present data do not support the idea of an autocrine loop (Figure 5). In contrast, they strengthen the concept of autonomous intracellular Smad2 activation.
The autonomous and constitutive overexpression of Smad2 is associated with basal hyperacetylation and hypermethylation of lysine residues. More precisely, this modulation of histone markers concerns specifically H3K9/14ac and H3K4me on the Smad2 promoter. These epigenetic markers are associated with an increase in transcription activation,36,37 strongly suggesting a direct association between histone H3 acetylation and methylation and Smad2 overexpression. Moreover, a spatial specificity is observed on the Smad2 promoter since H3K9/14ac and H3K4me are only increased in the vicinity of the 1a TSS on which the transcription variants overexpressed in TAA depend (1a and 1a + 1b). The present study focuses on the transcriptional regulation inducing Smad2 overexpression. However, this process can be associated with modifications in Smad2 protein stability. Indeed, modulation of Smad stability (degradation via the ubiquitin–proteasome pathway) plays a major role in the negative feedback. Smad ubiquitination is catalysed by enzymatic systems, including the ubiquitin ligase E3 and associated proteins, and Smurf (Smad ubiquitin regulatory factor) 1 and 2.38,39 Smurf targets C-terminally phosphorylated Smad2 which involves ubiquitination and degradation by proteasomes. A deficit in this process appears not to be the main mechanisms responsible for the global increase in Smad2 and pSmad2. Nevertheless, the investigation of protein stability could permit the evaluation of the potential cooperation between these two mechanisms.
The activation and the overexpression of Smad2 are specific of TAA-derived SMCs from the media. Such dysregulation was not observed in the adjacent adventitial fibroblasts. Furthermore, the increase in H3 acetylation and methylation is specifically observed in the media but not in the adventitia. This cell specificity raises the question of the trigger of the epigenetic activation of Smad2, which is a crucial point in the understanding of the global process of aneurysm progression, suggesting that the genetic background, i.e. the FBN1 and TGFBR2 mutations, is not the direct cause of the Smad2 perturbation but could initiate long-term environmental modifications leading to epigenetic reprogramming. Vascular SMCs possess plasticity which allows them to modulate their phenotype in response to changes in environmental cues, such as mechanical stress, matrix integrity, and cell–matrix interaction.40,41 In TAA, the aetiological diversity suggests several potential triggers: haemodynamic/rheological modifications, protease activation, extracellular matrix degradation, mutations, or any combination of these events. Phenotypic modulation is associated with specific gene expression which could be regulated by epigenetic marks, chromatin remodelling, and specific SMC transcription factor binding.42
We clearly observed an increase in basal Smad2 levels in both medial extracts and cultured SMCs. This overexpression presents a strong heritable character since it is sustained over several passages without the addition of any cytokines or growth factors. The maintenance of this Smad2 overexpression over several cycles of cell division and the participation of epigenetic regulation on the Smad2 promoter are consistent with the notion of epigenetic memory.31,43 Our study demonstrates the implication of epigenetic mechanisms (sustained modulation of histone marks) in the development of a pathological phenotype of the vascular SMCs in human TAA. Such phenotypic modulation can be associated with the overexpression of the embryonic form of the smooth muscle myosin which was previously observed in the media of TAA, regardless of aetiology.9 These observations indicate the possible involvement of a de-differentiation process in TAA-derived SMCs, induced by environmental or genetic perturbations. De-differentiation and phenotypic modulation are processes that are consistent with the notions of autonomy, heritability, cell specificity, and epigenetic regulation44 observed in chronic pathologies such as TAA.31
Together, these results demonstrate that all types of TAA present the same modes of dysregulation of Smad2 expression and activation, which are constitutive, heritable, and cell specific. This constitutive overexpression is associated with modifications of the histone H3 marker patterns in the vicinity of 1a TSS on the Smad2 promoter. The importance of epigenetic reprogramming and how chromatin modulation regulates SMC reprogramming might provide new insights into the onset and progression of TAA and other chronic vascular diseases.41
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Funding
D.G. was supported by a grant of the Medical Research Foundation (FRM). This study was supported by the French National Research Agency and the EU FP-7-integrated project ‘Fighting Aneurysmal Disease’ (FAD, www.fighting-aneurysm.org/). Funding to pay the Open Access publication charges for this article was provided by INSERM.
Acknowledgements
We would like to thank Mary Osborne-Pellegrin for editing the manuscript and surgeons of the cardiac and vascular surgery departments for providing us with normal and pathological aortic samples. We are also indebted to Sylviane Dennler for providing HA-tagged wild-type and mutated TGFβR2 plasmids. We would like to thank Professor Catherine Boileau for helpful discussion.
Conflict of interest: none declared.
References
- 1.Shi Y, Massague J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. doi:10.1016/S0092-8674(03)00432-X. [DOI] [PubMed] [Google Scholar]
- 2.Derynck R, Zhang YE. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature. 2003;425:577–584. doi: 10.1038/nature02006. doi:10.1038/nature02006. [DOI] [PubMed] [Google Scholar]
- 3.Liu C, Gaca MD, Swenson ES, Vellucci VF, Reiss M, Wells RG. Smads 2 and 3 are differentially activated by transforming growth factor-beta (TGF-beta) in quiescent and activated hepatic stellate cells. Constitutive nuclear localization of Smads in activated cells is TGF-beta-independent. J Biol Chem. 2003;278:11721–11728. doi: 10.1074/jbc.M207728200. doi:10.1074/jbc.M207728200. [DOI] [PubMed] [Google Scholar]
- 4.Massague J, Wotton D. Transcriptional control by the TGF-beta/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. doi:10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmierer B, Tournier AL, Bates PA, Hill CS. Mathematical modeling identifies Smad nucleocytoplasmic shuttling as a dynamic signal-interpreting system. Proc Natl Acad Sci USA. 2008;105:6608–6613. doi: 10.1073/pnas.0710134105. doi:10.1073/pnas.0710134105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, et al. Dysregulation of TGF-beta activation contributes to pathogenesis in Marfan syndrome. Nat Genet. 2003;33:407–411. doi: 10.1038/ng1116. doi:10.1038/ng1116. [DOI] [PubMed] [Google Scholar]
- 7.Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–281. doi: 10.1038/ng1511. doi:10.1038/ng1511. [DOI] [PubMed] [Google Scholar]
- 8.Coucke PJ, Willaert A, Wessels MW, Callewaert B, Zoppi N, De Backer J, et al. Mutations in the facilitative glucose transporter GLUT10 alter angiogenesis and cause arterial tortuosity syndrome. Nat Genet. 2006;38:452–457. doi: 10.1038/ng1764. doi:10.1038/ng1764. [DOI] [PubMed] [Google Scholar]
- 9.Gomez D, Al Haj Zen A, Borges LF, Philippe M, Gutierrez PS, Jondeau G, et al. Syndromic and non-syndromic aneurysms of the human ascending aorta share activation of the Smad2 pathway. J Pathol. 2009;218:131–142. doi: 10.1002/path.2516. doi:10.1002/path.2516. [DOI] [PubMed] [Google Scholar]
- 10.Bonyadi M, Rusholme SA, Cousins FM, Su HC, Biron CA, Farrall M, et al. Mapping of a major genetic modifier of embryonic lethality in TGF beta 1 knockout mice. Nat Genet. 1997;15:207–211. doi: 10.1038/ng0297-207. doi:10.1038/ng0297-207. [DOI] [PubMed] [Google Scholar]
- 11.Owens GK, Wise G. Regulation of differentiation/maturation in vascular smooth muscle cells by hormones and growth factors. Agents Actions Suppl. 1997;48:3–24. doi: 10.1007/978-3-0348-7352-9_1. [DOI] [PubMed] [Google Scholar]
- 12.Matt P, Schoenhoff F, Habashi J, Holm T, Van Erp C, Loch D, et al. Circulating transforming growth factor-beta in Marfan syndrome. Circulation. 2009;120:526–532. doi: 10.1161/CIRCULATIONAHA.108.841981. doi:10.1161/CIRCULATIONAHA.108.841981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.ten Dijke P, Arthur HM. Extracellular control of TGFbeta signalling in vascular development and disease. Nat Rev Mol Cell Biol. 2007;8:857–869. doi: 10.1038/nrm2262. doi:10.1038/nrm2262. [DOI] [PubMed] [Google Scholar]
- 14.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339. doi: 10.1038/352337a0. doi:10.1038/352337a0. [DOI] [PubMed] [Google Scholar]
- 15.Mizuguchi T, Collod-Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, et al. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet. 2004;36:855–860. doi: 10.1038/ng1392. doi:10.1038/ng1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L, Vranckx R, Khau Van Kien P, Lalande A, Boisset N, Mathieu F, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349. doi: 10.1038/ng1721. doi:10.1038/ng1721. [DOI] [PubMed] [Google Scholar]
- 17.Pannu H, Tran-Fadulu V, Papke CL, Scherer S, Liu Y, Presley C, et al. MYH11 mutations result in a distinct vascular pathology driven by insulin-like growth factor 1 and angiotensin II. Hum Mol Genet. 2007;16:2453–2462. doi: 10.1093/hmg/ddm201. doi:10.1093/hmg/ddm201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo DC, Pannu H, Tran-Fadulu V, Papke CL, Yu RK, Avidan N, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493. doi: 10.1038/ng.2007.6. doi:10.1038/ng.2007.6. [DOI] [PubMed] [Google Scholar]
- 19.Milewicz DM, Guo DC, Tran-Fadulu V, Lafont AL, Papke CL, Inamoto S, et al. Genetic basis of thoracic aortic aneurysms and dissections: focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet. 2008;9:283–302. doi: 10.1146/annurev.genom.8.080706.092303. doi:10.1146/annurev.genom.8.080706.092303. [DOI] [PubMed] [Google Scholar]
- 20.Roos-Hesselink JW, Scholzel BE, Heijdra RJ, Spitaels SE, Meijboom FJ, Boersma E, et al. Aortic valve and aortic arch pathology after coarctation repair. Heart. 2003;89:1074–1077. doi: 10.1136/heart.89.9.1074. doi:10.1136/heart.89.9.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isselbacher EM. Thoracic and abdominal aortic aneurysms. Circulation. 2005;111:816–828. doi: 10.1161/01.CIR.0000154569.08857.7A. doi:10.1161/01.CIR.0000154569.08857.7A. [DOI] [PubMed] [Google Scholar]
- 22.Klein DG. Thoracic aortic aneurysms. J Cardiovasc Nurs. 2005;20:245–250. doi: 10.1097/00005082-200507000-00008. [DOI] [PubMed] [Google Scholar]
- 23.Habashi JP, Judge DP, Holm TM, Cohn RD, Loeys BL, Cooper TK, et al. Losartan, an AT1 antagonist, prevents aortic aneurysm in a mouse model of Marfan syndrome. Science. 2006;312:117–121. doi: 10.1126/science.1124287. doi:10.1126/science.1124287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, et al. Aneurysm syndromes caused by mutations in the TGF-beta receptor. N Engl J Med. 2006;355:788–798. doi: 10.1056/NEJMoa055695. doi:10.1056/NEJMoa055695. [DOI] [PubMed] [Google Scholar]
- 25.Robinson PN, Arteaga-Solis E, Baldock C, Collod-Beroud G, Booms P, De Paepe A, et al. The molecular genetics of Marfan syndrome and related disorders. J Med Genet. 2006;43:769–787. doi: 10.1136/jmg.2005.039669. doi:10.1136/jmg.2005.039669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: a paradox in pathogenesis. J Vasc Res. 2009;46:119–137. doi: 10.1159/000151766. doi:10.1159/000151766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.World Medical Association Declaration of Helsinki. Recommendations guiding physicians in biomedical research involving human subjects. Cardiovasc Res. 1997;35:2–3. doi:10.1016/S0008-6363(97)00109-0. [PubMed] [Google Scholar]
- 28.Touat Z, Lepage L, Ollivier V, Nataf P, Hvass U, Labreuche J, et al. Dilation-dependent activation of platelets and prothrombin in human thoracic ascending aortic aneurysm. Arterioscler Thromb Vasc Biol. 2008;28:940–946. doi: 10.1161/ATVBAHA.107.158576. doi:10.1161/ATVBAHA.107.158576. [DOI] [PubMed] [Google Scholar]
- 29.Battle T, Arnal JF, Michel JB. Hyperproliferation of aortic smooth muscle cells and fibroblasts from young SHR rats is not shared by endothelial cells. Clin Exp Pharmacol Physiol. 1994;21:981–989. doi: 10.1111/j.1440-1681.1994.tb02660.x. doi:10.1111/j.1440-1681.1994.tb02660.x. [DOI] [PubMed] [Google Scholar]
- 30.Gressner OA, Lahme B, Siluschek M, Rehbein K, Weiskirchen R, Gressner AM. Connective tissue growth factor is a Smad2 regulated amplifier of transforming growth factor beta actions in hepatocytes—but without modulating bone morphogenetic protein 7 signaling. Hepatology. 2009;49:2021–2030. doi: 10.1002/hep.22850. doi:10.1002/hep.22850. [DOI] [PubMed] [Google Scholar]
- 31.Villeneuve LM, Reddy MA, Lanting LL, Wang M, Meng L, Natarajan R. Epigenetic histone H3 lysine 9 methylation in metabolic memory and inflammatory phenotype of vascular smooth muscle cells in diabetes. Proc Natl Acad Sci USA. 2008;105:9047–9052. doi: 10.1073/pnas.0803623105. doi:10.1073/pnas.0803623105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takenoshita S, Mogi A, Nagashima M, Yang K, Yagi K, Hanyu A, et al. Characterization of the MADH2/Smad2 gene, a human Mad homolog responsible for the transforming growth factor-beta and activin signal transduction pathway. Genomics. 1998;48:1–11. doi: 10.1006/geno.1997.5149. doi:10.1006/geno.1997.5149. [DOI] [PubMed] [Google Scholar]
- 33.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. doi:10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 34.Ogishima T, Shiina H, Breault JE, Terashima M, Honda S, Enokida H, et al. Promoter CpG hypomethylation and transcription factor EGR1 hyperactivate heparanase expression in bladder cancer. Oncogene. 2005;24:6765–6772. doi: 10.1038/sj.onc.1208811. doi:10.1038/sj.onc.1208811. [DOI] [PubMed] [Google Scholar]
- 35.Abreu JG, Ketpura NI, Reversade B, De Robertis EM. Connective-tissue growth factor (CTGF) modulates cell signalling by BMP and TGF-beta. Nat Cell Biol. 2002;4:599–604. doi: 10.1038/ncb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mochizuki K, Takabe S, Goda T. Changes on histone H3 modifications on the GLUT5 gene and its expression in Caco-2 cells co-treated with a p44/42 MAPK inhibitor and glucocorticoid hormone. Biochem Biophys Res Commun. 2008;371:324–327. doi: 10.1016/j.bbrc.2008.04.075. doi:10.1016/j.bbrc.2008.04.075. [DOI] [PubMed] [Google Scholar]
- 37.Nishida H, Suzuki T, Kondo S, Miura H, Fujimura Y, Hayashizaki Y. Histone H3 acetylated at lysine 9 in promoter is associated with low nucleosome density in the vicinity of transcription start site in human cell. Chromosome Res. 2006;14:203–211. doi: 10.1007/s10577-006-1036-7. doi:10.1007/s10577-006-1036-7. [DOI] [PubMed] [Google Scholar]
- 38.Inoue Y, Imamura T. Regulation of TGF-beta family signaling by E3 ubiquitin ligases. Cancer Sci. 2008;99:2107–2112. doi: 10.1111/j.1349-7006.2008.00925.x. doi:10.1111/j.1349-7006.2008.00925.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izzi L, Attisano L. Regulation of the TGFbeta signalling pathway by ubiquitin-mediated degradation. Oncogene. 2004;23:2071–2078. doi: 10.1038/sj.onc.1207412. doi:10.1038/sj.onc.1207412. [DOI] [PubMed] [Google Scholar]
- 40.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. doi:10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 41.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. doi:10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 42.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. doi:10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villeneuve LM, Natarajan R. The role of epigenetics in the pathology of diabetic complications. Am J Physiol Renal Physiol. 299:F14–F25. doi: 10.1152/ajprenal.00200.2010. doi:10.1152/ajprenal.00200.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Park JR, Seo MS, Roh KH, Park SB, Hwang JW, et al. Histone deacetylase inhibitors decrease proliferation potential and multilineage differentiation capability of human mesenchymal stem cells. Cell Prolif. 2009;42:711–720. doi: 10.1111/j.1365-2184.2009.00633.x. doi:10.1111/j.1365-2184.2009.00633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.