Abstract
Stimulation of GCN4 mRNA translation due to phosphorylation of the α-subunit of initiation factor 2 (eIF2) by its specific kinase, GCN2, requires binding of uncharged tRNA to a histidyl-tRNA synthetase (HisRS)-like domain in GCN2. GCN2 function in vivo also requires GCN1 and GCN20, but it was unknown whether these latter proteins act directly to promote the stimulation of GCN2 by uncharged tRNA. We found that the GCN1–GCN20 complex physically interacts with GCN2, binding to the N-terminus of the protein. Overexpression of N-terminal GCN2 segments had a dominant-negative phenotype that correlated with their ability to interact with GCN1–GCN20 and impede association between GCN1 and native GCN2. Consistently, this Gcn– phenotype was suppressed by overexpressing GCN2, GCN1–GCN20 or tRNAHis. The requirement for GCN1 was also reduced by overexpressing tRNAHis in a gcn1Δ strain. We conclude that binding of GCN1–GCN20 to GCN2 is required for its activation by uncharged tRNA. The homologous N-terminus of Drosophila GCN2 interacted with yeast GCN1–GCN20 and had a dominant Gcn– phenotype, suggesting evolutionary conservation of this interaction.
Keywords: eIF2α kinase/GCN2/regulation/translation/yeast
Introduction
Protein synthesis in mammalian cells is down-regulated by phosphorylation of translation initiation factor 2 (eIF2) in response to starvation, stress or viral infection (Clemens, 1996). Phosphorylation of the α-subunit of eIF2 converts it from a substrate to an inhibitor of its guanine nucleotide exchange factor, eIF2B, decreasing formation of the ternary complex comprised of eIF2, GTP and methionyl-tRNAMet (Met-tRNAiMet). Because the ternary complex is responsible for transferring Met-tRNAiMet to the ribosome, this elicits a general reduction in translation initiation. Four different mammalian eIF2α kinases have been identified that are regulated by different stimuli: HRI is activated by hemin deprivation, PKR by double-stranded RNA in virus-infected cells (Clemens, 1996), PEK or PERK by unfolded proteins in the endoplasmic reticulum (Shi et al., 1998; Harding et al., 1999) and mouse GCN2 by serum starvation (Berlanga et al., 1998; Sood et al., 2000). There are also GCN2 homologs in Neurospora crassa (Sattlegger et al., 1998), Drosophila melanogaster (Santoyo et al., 1997; Olsen et al., 1998) and Saccharomyces cerevisiae. Thus, GCN2 may be the most widespread enzyme in this kinase subfamily, and possibly its founding member (Dever, 1999). GCN2 was first identified in yeast by its role in translational induction of GCN4, a transcriptional activator of amino acid biosynthetic enzymes. Four short open reading frames (uORFs) in the GCN4 mRNA leader underlie a specialized reinitiation mechanism that elicits increased GCN4 translation in response to reductions in ternary complex levels too small to block general translation. Accordingly, eIF2α phosphorylation in amino acid-starved yeast cells leads specifically to induction of a transcriptional activator capable of ameliorating amino acid limitation (Hinnebusch, 1996).
GCN2 exists as a latent kinase during growth on nutrient-rich medium and is thought to be activated by uncharged tRNAs that accumulate in amino acid-starved cells. This model is based partly on the fact that GCN2 contains a region homologous to the type II aminoacyl-tRNA synthetase for histidine (HisRS) located immediately C-terminal to the kinase domain. Amino acids in the GCN2 HisRS domain corresponding to key residues required for tRNA binding by class II synthetases (the m2 motif) are required for GCN2 kinase activity in vivo and in cell extracts, and for binding of tRNA to the isolated HisRS domain in vitro (Wek et al., 1995; Zhu et al., 1996). In addition, mutations in aminoacyl-tRNA synthetases lead to GCN2-dependent derepression of GCN4 and its target genes without a starvation for amino acids (Hinnebusch, 1996). The HisRS-like domain is conserved in all known GCN2 homologs (Santoyo et al., 1997; Olsen et al., 1998; Sattlegger et al., 1998; Berlanga et al., 1999; Sood et al., 2000), suggesting that uncharged tRNA is an activating ligand for these enzymes in Drosophila and mammals, as well as in fungi. GCN2 contains several additional domains that are conserved in all known GCN2 homologs (Figure 2, top). The extreme C-terminal segment is required for ribosome binding (Ramirez et al., 1991; Zhu and Wek, 1998) and contains the principal dimerization determinants in yeast GCN2 (Qiu et al., 1998). Immediately N-terminal to the kinase domain is a region of unknown function that resembles a truncated kinase domain with no specific homology to the eIF2α kinases. N-terminal to this pseudokinase domain (ΨK) is a charged region (+/–) and highly conserved N-terminal (CNT) domain, both of unknown function.
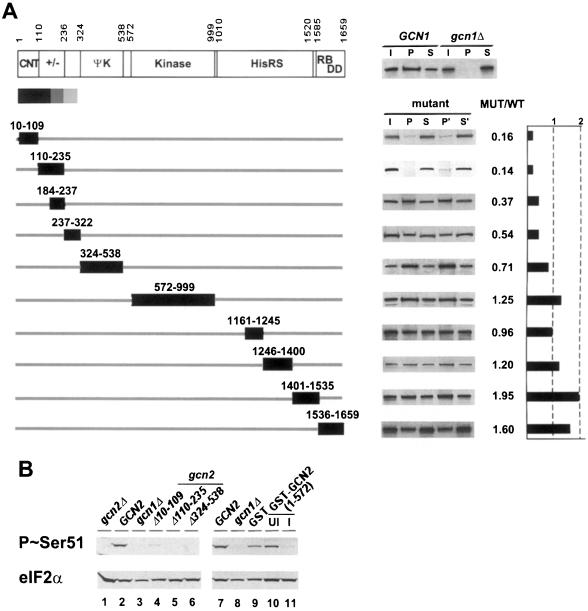
Fig. 2. N-terminal deletions in GCN2 impair its interaction with the GCN1/20 complex. (A) GCN1 antibodies were used to co-immunoprecipitate GCN2 from 500 µg of whole-cell extracts prepared from gcn2Δ strain H1149 transformed with high-copy plasmids bearing wild-type GCN2 (WT) or the indicated gcn2 deletion alleles. The amino acids removed by each deletion are shown by the numbered boxes beneath the schematic depiction of the GCN2 sequence on the top. The latter shows the amino acid positions of GCN2 functional domains designated as in the text, except for RB/DD which signifies the ribosome-binding/dimerization domain at the C-terminus. For each construct, the results of Western analysis of GCN2 for duplicate immunoprecipitations are shown on the right, with input (I), pellet (P) and supernatant (S) fractions as described in Figure 1. The amount of GCN1 detected in the pellet fractions did not vary significantly from one mutant extract to the next (data not shown). For each gcn2 mutant, a control co-immunoprecipitation was performed in parallel using an extract from a transformant expressing wild-type GCN2 from the high copy number plasmid p630, allowing quantitative comparisons of the recovery of mutant and wild-type GCN2 proteins in immune complexes with GCN1. The amount of each mutant GCN2 protein co-immunoprecipitating with GCN1 is expressed in the graph on the right as a fraction of the amount of wild-type GCN2 that co-immunoprecipitated with GCN1. To the right of the GCN2 schematic at the top is a typical result showing co-immunoprecipitation of wild-type GCN2 with GCN1 from a GCN1 but not a gcn1Δ extract. (B) Lanes 1–6: exponentially growing cultures of transformants of gcn2Δ strain H1149 bearing vector alone (lane 1) or high-copy plasmids containing either wild-type GCN2 (lane 2) or the indicated gcn2 deletion alleles (lanes 4–6), or a transformant of gcn1Δ strain H2081 bearing the high-copy GCN2 plasmid (lane 3), were grown for 1 h in minimal (SD) medium containing 40 mM 3AT. Whole-cell extracts were prepared and subjected to SDS–PAGE and immunoblot analysis using phosphospecific antibodies raised against an eIF2α peptide bearing phosphoserine 51 (top panel) or polyclonal antibodies against eIF2α (bottom panel). Lanes 7–11: extracts were prepared from transformants of GCN2 strain H1511 expressing GST alone or GST–GCN2(1–572), described below in Figure 5B, grown in Sgal medium (lanes 9 and 11) to induce (I) expression of the GST fusions or in uninducing conditions (SD medium; UI; lane 10). Lanes 7 and 8 contain the same extracts as in lanes 2 and 3, re-examined as controls.
Genetic studies have identified two trans-acting positive effectors of GCN2, known as GCN1 and GCN20, which interact with one another in vivo and are required for phosphorylation of eIF2α by GCN2 in starved cells (Marton et al., 1993; Vazquez de Aldana et al., 1995; Hinnebusch, 1996). Because they are dispensable for eIF2α phosphorylation in yeast cells expressing the human eIF2α kinase PKR in place of GCN2, it appears that GCN1 and GCN20 stimulate GCN2 kinase activity rather than inhibiting an eIF2α phosphatase. Neither protein is required for the autokinase or eIF2α kinase activities of GCN2 in immune complex assays in vitro, indicating that they are dispensable for GCN2 catalytic function per se (Marton et al., 1993; Vazquez de Aldana et al., 1995).
GCN1 is a 297 kDa cytoplasmic protein with an internal segment related in sequence to translation elongation factor 3 (EF3). EF3 is a member of the ATP-binding cassette (ABC) family of proteins, and has ribosome-stimulated ATPase activity. It is thought to function at each round of elongation to stimulate release of uncharged tRNA from the ribosomal exit (E) site and binding of charged tRNA complexed with EF1α⋅GTP to the acceptor (A) site (Triana-Alonso et al., 1995). GCN1 is similar to EF3 in a region of unknown function N-terminal to the ABC domains in EF3, but it lacks the signature sequences of ABC proteins for nucleotide binding (Marton et al., 1997). GCN20 also belongs to the ABC family of proteins and is highly related to EF3 in the ABC domains; however, the non-homologous N-terminal segment of GCN20 is sufficient for its stable binding to GCN1 and regulatory function in vivo. The similarity of GCN1 and GCN20 to ABC proteins originally prompted the idea that the GCN1–GCN20 complex (GCN1/20) could function as an amino acid transporter and regulate GCN2 indirectly by controlling amino acid availability and, hence, the efficiency of tRNA charging. The particularly strong homology between GCN1 and EF3, combined with our finding that GCN1 and GCN20 are associated with polysomes in cell extracts, led us to propose instead that GCN1/20 functions on the ribosome in regulating GCN2 (Marton et al., 1997). In Escherichia coli, the relA protein detects uncharged tRNA paired with a cognate codon in the ribosomal A-site to mediate stringent control of ribosome and amino acid biosynthesis in response to amino acid starvation (Cashel and Rudd, 1987). By analogy with this system, we suggested that GCN2 could be activated by uncharged tRNA in the ribosomal A-site, and that GCN1 might perform an EF3-like function to promote A-site binding of tRNAs or transfer of uncharged tRNAs from the ribosome to the HisRS-like domain of GCN2 (Marton et al., 1997).
These models imply that GCN1/20 comes into proximity with GCN2 on the ribosome. However, biochemical evidence for physical association between GCN1/20 and GCN2 has been lacking, leaving open the possibility of an indirect mechanism. The results presented here make the latter possibility very unlikely by demonstrating direct binding of GCN1/20 to GCN2 in vivo. Furthermore, we have localized GCN1/20 binding to a GCN2 segment comprised of the evolutionarily conserved N-terminal domain and adjacent +/– region. We present strong genetic evidence that the CNT and +/– domains mediate complex formation with GCN1/20 in vivo, and that this interaction is important for activation of GCN2 by uncharged tRNA. The CNT and +/– domains of Drosophila GCN2 also interacted with GCN1/20 in yeast, implying that GCN1 and GCN20 homologs identified in higher eukaryotes (Vazquez de Aldana et al., 1995; Marton et al., 1997) are most likely to regulate their cognate GCN2 proteins.
Results
GCN1 and GCN20 can be co-immunoprecipitated with GCN2 independently of the protein kinase or ribosome-binding activities of GCN2
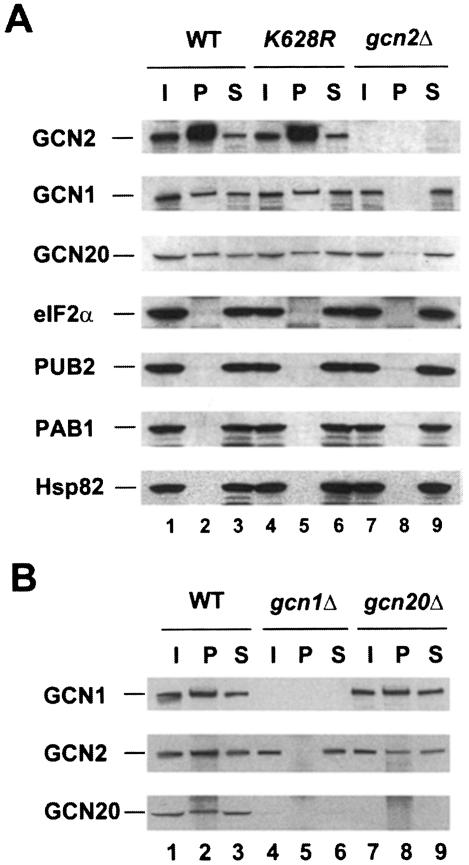
In order to demonstrate that GCN2 physically interacts with the GCN1/20 complex, we overexpressed GCN2 from a high-copy plasmid in an attempt to drive its interaction with GCN1/20 by mass action. Whole-cell extracts from gcn2Δ strains bearing a high-copy plasmid encoding GCN2 were prepared in a low-salt buffer lacking detergents and immunoprecipitated with GCN2 antibodies. Western analysis of the immune complexes showed that 5–10% of the GCN1 and GCN20 was co-immunoprecipitated with GCN2 (Figure 1A, lane 2). Importantly, when GCN2 was absent in the extract (lane 8), or if pre-immune serum was employed (data not shown), little or no GCN1 and GCN20 were immunoprecipitated. The immune complexes containing GCN2 harbored no detectable eIF2α, the substrate for GCN2, or the unrelated poly(A)-RNA binding protein PAB1 (Figure 1A). These findings indicate that GCN1 and GCN20 were co-immunoprecipitated specifically with GCN2 from cell extracts. It was suggested recently that GCN2 forms a complex with the yeast Hsp90 homolog known as HSP82 (Donze and Picard, 1999); however, we did not detect any HSP82 co-immunoprecipitating with GCN2 in these experiments (Figure 1A). Presumably, the GCN2–HSP82 complex is unstable in cell extracts.
Fig. 1. GCN2 can be co-immunoprecipitated in a complex with GCN1 and GCN20 from cell extracts. (A) GCN2 was immunoprecipitated with antibodies against GCN2 from 500 µg of whole-cell extracts prepared from gcn2Δ strain H1894 transformed with either the vector YEp24 (gcn2Δ) or high-copy plasmids containing wild-type GCN2 (WT) or a catalytically inactive mutant (K628R). Following SDS–PAGE separation of the proteins and transfer to nitrocellulose, the membranes were probed for the different proteins indicated on the left. I and S indicate one-tenth of the input extract and supernatant, respectively, and P represents the total pellet. (B) GCN1 antibodies were used to co-immunoprecipitate GCN2 and GCN20 with GCN1 from 500 µg of whole-cell extracts prepared from strain H1402 (wild-type) and the isogenic strains H2081 (gcn1Δ) and H2563 (gcn20Δ). Western blotting was performed with antibodies against the proteins indicated on the left.
We observed no significant differences in the amounts of GCN1 or GCN20 that co-immunoprecipitated with GCN2 from cells grown under histidine starvation versus non-starvation conditions, nor in the presence or absence of uncharged total yeast tRNA added to the extracts at 10–100 µg/ml (data not shown). Thus, complex formation by these proteins may be constitutive in vivo (see below). Interactions of GCN1 and GCN20 with GCN2 were unaffected by the gcn2-K628R mutation in GCN2, which destroys kinase activity (Wek et al., 1990), indicating that a functional kinase domain in GCN2 is not required for complex formation (Figure 1A, lanes 2 and 5). Because GCN2 and the GCN1/20 complex were shown previously to be associated with ribosomes (Ramirez et al., 1991; Zhu and Wek, 1998), their co-immunoprecipitation from extracts might have resulted from independent binding of these proteins to the same ribosomes without any direct contact between them. One argument against this possibility is that substantial binding of GCN1 and GCN20 to ribosomes in vitro required addition of exogenous ATP (Marton et al., 1997), whereas no ATP was added prior to the immunoprecipitations shown in Figure 1. Secondly, we observed no detectable PUB2, an integral 60S subunit ribosomal protein (Anderson et al., 1993), co-immunoprecipitating with GCN2 (Figure 1A, lane 2). Finally, we show below that the ribosome-binding domain of GCN2 is dispensable for its co-immunoprecipitation with GCN1 and GCN20 from cell extracts.
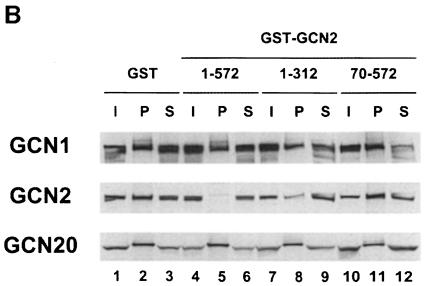
To determine whether the interaction between GCN2 and GCN1/20 could be observed without overexpressing GCN2, the co-immunoprecipitation experiments were repeated with extracts from strains containing wild-type chromosomal GCN2 using polyclonal serum against GCN1 for the immunoprecipitations. As shown in Figure 1B, native GCN2 and GCN20 were co-immunoprecipitated with GCN1 in a manner dependent on the presence of GCN1 in the extract (compare lanes 2 and 5). Based on the efficiencies of immunoprecipitation, we estimated that a minimum of 40–60% of the GCN2 and 40–50% of the GCN20 was associated with GCN1 in the wild-type extracts. GCN2 was co-immunoprecipitated with GCN1 from the gcn20Δ extract, but at a lower yield than from the GCN20 extract (Figure 1B, lanes 2 and 8). Thus, the presence of GCN20 in the complex may stabilize the interaction between GCN1 and GCN2. Because the steady-state level of GCN20 is greatly reduced in a gcn1Δ strain (Vazquez de Aldana et al., 1995) (Figure 1B, lanes 1 and 4), we could not determine whether GCN2 can interact with GCN20 in the absence of GCN1 in this type of experiment.
Interaction between GCN2 and the GCN1/20 complex is mediated by the N-terminal region of GCN2
We next investigated which portion of GCN2 is required for its physical interaction with GCN1 and GCN20 using the internally deleted gcn2 alleles shown in Figure 2A. The N-terminus of GCN2 depicted there contains 69 amino acids that were not recognized previously as part of the protein (Roussou et al., 1988; Wek et al., 1989). The AUG triplet located 69 codons 5′ of the reported start codon (henceforth referred to as AUG-1) and the next 68 codons are present in the same reading frame as GCN2 and show strong sequence similarity to the N-terminal sequences of all known GCN2 homologs (data not shown). Furthermore, a frameshift mutation introduced at a SacI site insertion made previously between codons 21 and 22 (Wek et al., 1989) has a 3-aminotriazole-sensitive (3ATs) phenotype (data not shown). Resistance to 3AT, an inhibitor of the histidine biosynthetic enzyme encoded by HIS3, requires GCN2-mediated derepression of GCN4 translation with attendant transcriptional induction of HIS3 by GCN4. To prove that AUG-1 is the GCN2 start codon, we inserted the coding sequences for the c-Myc epitope flanked by unique SacI and MluI sites between codons 1 and 2 of plasmid-borne GCN2, and also introduced a frameshift mutation at the newly created SacI site. The c-Myc-tagged allele without a frameshift mutation complemented the 3ATs phenotype of a gcn2Δ strain, and GCN2 could be detected by Western analysis of extracts from the transformants with antibodies against GCN2 or the c-Myc epitope. In contrast, transformants bearing the frameshift allele were 3ATs and contained no detectable GCN2 protein (data not shown). Reading upstream from AUG-1, multiple stop codons are encountered before reaching the next in-frame AUG. Therefore, we concluded that AUG-1 is most probably the GCN2 initiation codon. Accordingly, our deletion analysis included a mutation that removes the conserved N-terminal region of GCN2 from amino acids 10 to 109.
The mutant GCN2 proteins shown in Figure 2A were expressed from high-copy plasmids and tested for co-immunoprecipitation with GCN1 from cell extracts. Because the fraction of wild-type GCN2 co-immunoprecipitating with GCN1 was somewhat variable, each extract containing a mutant GCN2 protein was examined in triplicate in parallel with the wild-type GCN2 extract, and the fraction of mutant protein immunoprecipitated with GCN1 was compared with that seen for wild-type GCN2. We found that only the deletions of the extreme N-terminal segments of GCN2 [residues 10–109 (the CNT) and 110–235 (+/– domain)] greatly reduced complex formation with GCN1. The next two consecutive deletions shown in Figure 2A, which remove residues 184–237 or 237–322, reproducibly reduced complex formation by ∼50% relative to wild-type GCN2. None of the other internal deletions in GCN2 significantly impaired complex formation with GCN1 (Figure 2A). The gcn2-Δ110–235 product was expressed at ∼50% of wild-type GCN2; however, this small reduction in expression probably does not account for its defective interaction with GCN1 for two reasons. First, the gcn2-Δ1246–1400 product was expressed at only 20% of wild-type levels and showed no binding defect. Secondly, GCN2 was expressed from the chromosomal locus at a level only ∼2% of that produced from the high-copy plasmid, yet native GCN2 interacted strongly with GCN1 (Figure 1B). We conclude that the N-terminal 235 residues in GCN2 are the most critically required for complex formation with the GCN1/20 complex in vivo.
It is noteworthy that the C-terminal 123 residues of GCN2 are completely dispensable for interaction with GCN1 (Figure 2A), as this region is responsible for ribosome association by GCN2 (Ramirez et al., 1991; Zhu and Wek, 1998) and also contains the most important dimerization determinant in the protein (Qiu et al., 1998). Thus, it appears that neither ribosome association nor dimerization by GCN2 is required for its interaction with GCN1. The fact that two internal deletions in the HisRS-like region also did not impair the interaction with GCN1 (Figure 2A) suggests that tRNA binding by GCN2 is not required for complex formation, in accordance with the presence of the GCN2–GCN1 complex in extracts prepared from non-starved cells. Moreover, the Y1119L and R1120L substitutions in the conserved m2 motif of this domain, shown previously to abolish tRNA binding in vitro (Wek et al., 1995), did not reduce co-immunoprecipitation of GCN2 with GCN1 (data not shown). As expected, none of the deletions in GCN2 had any effect on the amount of GCN20 that co-immunoprecipitated with GCN1 (data not shown).
All of the gcn2Δ alleles analyzed in Figure 2A appear to be non-functional, as judged by their failure to complement the 3ATs phenotype of a gcn2Δ mutant (Wek et al., 1989, 1990; Qiu et al., 1998) (data not shown). We verified this conclusion for deletions of the CNT and +/– domains by measuring levels of phosphorylated eIF2α in vivo using antibodies specific for phosphoserine 51 of eIF2α. As shown in Figure 2B (lanes 1–6), deletions of the CNT alone, the CNT together with the +/– domain, or the ΨK domain of GCN2 essentially abolished eIF2α phosphorylation under histidine starvation conditions, just as occurred in an isogenic gcn1Δ strain expressing wild-type GCN2.
Amino acids 1–272 of GCN2 comprise the minimal segment sufficient for interaction with the GCN1/20 complex
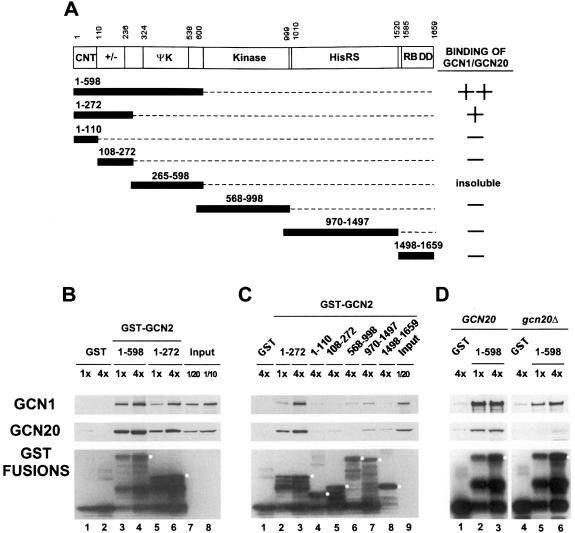
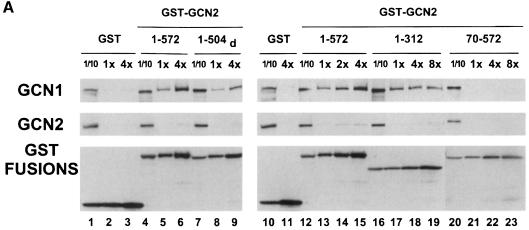
To extend the results of our deletion analysis, we performed in vitro binding experiments with the GST–GCN2 fusions shown in Figure 3A that were expressed in E.coli, immobilized on glutathione–Sepharose beads and used to precipitate native GCN1 and GCN20 from yeast cell extracts (GST pull-down assays). We found that the N-terminal one-third of GCN2 (amino acids 1–598) and a smaller region covering the CNT and +/– domains (amino acids 1–272) bound substantial amounts of GCN1 and GCN20 in the extract (Figure 3B, lanes 3–6). At the highest concentration of GST–GCN2(1–598), we precipitated ∼20% of the GCN1 and GCN20, whereas ∼4-fold higher concentrations of GST–GCN2(1–272) were required to precipitate the same proportions of GCN1 and GCN20 (Figure 3B, compare lanes 3 and 6). Thus, GST–GCN2(1–272) seems to have a lower affinity than GST–GCN2(1–598) for the GCN1/20 complex.
Fig. 3. Precipitation of GCN1 and GCN20 in cell extracts with bacterially expressed GST–GCN2 fusions. (A) The segments of GCN2 contained in different GST fusions are shown as numbered black bars below the schematic depiction of GCN2, along with a summary of their GCN1–GCN20 binding abilities based on the data shown in (B) and (C). (B and C) Similar amounts of the indicated GST–GCN2 fusion proteins, or GST alone, expressed in E.coli were used to precipitate native GCN1 and GCN20 from yeast cell extracts prepared from gcn2Δ strain H2557. Selected fusions were examined at two different relative concentrations (1× or 4×) to verify that binding of GCN1–GCN20 was dose dependent at the selected concentrations of GST fusions. Lanes labeled Input contain one-tenth or one-twentieth of the amount of cell extract used in each binding assay, as indicated. The precipitated proteins were subjected to Western analysis using antibodies against GCN1 (top panel), GCN20 (middle panel) or GST (bottom panel). White dots in the bottom panel mark the predicted sizes of the full-length GST–GCN2 fusions. (D) Binding experiments were performed using bacterially expressed GST–GCN2(1–598) (lanes 2 and 3, and 5 and 6) or GST alone (lanes 1 and 4) and 500 µg of yeast cell extracts prepared from wild-type strain H1511 (GCN20, lanes 1–3) or isogenic gcn20Δ strain H2563 (gcn20Δ, lanes 4–6).
The two GST–GCN2 fusions containing only the CNT (amino acids 1–110) or +/– domain (amino acids 108–272) failed to precipitate GCN1/20 from the extract above the background levels obtained with GST alone (Figure 3C, lanes 4 and 5). The fusion containing the HisRS segment (970–1497) interacted with GCN1 and GCN20 above background levels, but this interaction was very weak compared with that seen for the 1–272 and 1–598 segments of GCN2 (lane 7 versus lanes 2 and 3). Excluding the fusion containing amino acids 265–598, which was insoluble and could not be tested, none of the other fusions showed significant binding to both GCN1 and GCN20. Combining these data with the results of co-immunoprecipitation experiments in Figure 2, we conclude that the CNT and +/– segments together comprise a domain in GCN2 that is necessary and sufficient for specific binding to native GCN1 and GCN20 in cell extracts.
To verify our conclusion that GCN20 is not essential for binding of GCN1 to GCN2, we carried out pull-down experiments with the GST–GCN2(1–598) fusion using extracts from wild-type or gcn20Δ strains. As shown in Figure 3D, this fusion bound GCN1 from the gcn20Δ extract, albeit with reduced efficiency compared with that seen using the GCN20 extract (compare lanes 5 and 6 with 2 and 3). These data are consistent with the results in Figure 1B indicating that co-immunoprecipitation of GCN2 with GCN1 occurred at reduced levels from the gcn20Δ versus the GCN20 extract. Similar experiments using a gcn1Δ extract showed only a small amount of GCN20 recovered with the GST–GCN2(1–598) fusion above the background level seen with GST alone. The same result was observed even when the strain contained a high-copy GCN20 plasmid to offset the reduction in GCN20 levels associated with the gcn1Δ allele (data not shown). These results suggest that binding between GCN2 and the GCN1/20 complex is driven primarily by GCN1, but that GCN20 increases the ability of GCN1 to interact with GCN2.
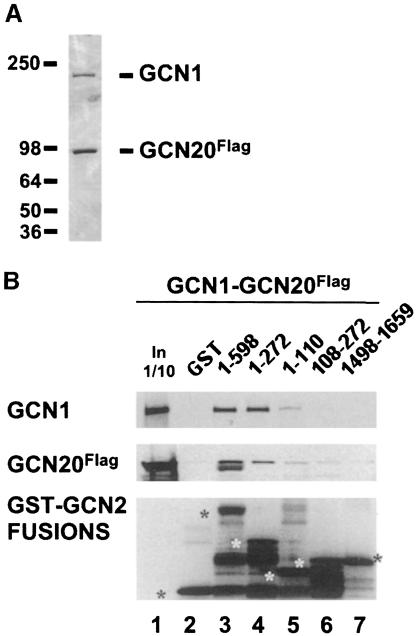
To establish that GCN2 interacts directly with GCN1/20 without the participation of other yeast proteins, we conducted GST pull-down experiments using GST–GCN2 fusions described above and highly purified GCN1/20 complex. The latter was isolated by affinity chromatography from a yeast strain overexpressing GCN1 and FLAG-tagged GCN20 (GCN20Flag), both under the control of a GAL promoter and encoded on high-copy plasmids. SDS–PAGE separation followed by Coomassie staining or Western analysis showed that GCN1 and GCN20Flag were the only major polypeptides present in the purified fraction (Figure 4A). We also verified that the FLAG-tagged GCN20 allele complemented the Gcn– phenotype of a gcn20Δ strain to the same extent as did wild-type GCN20 (data not shown). GCN2 was detected in the purified preparation by Western analysis, but not by Coomassie staining (data not shown). The relatively low yield of GCN2 co-purifying with GCN1/20 was expected because GCN2 was not overexpressed in the strain from which GCN1/GCN20Flag complex was purified. As demonstrated in Figure 4B, GCN1 and GCN20Flag were precipitated by the bacterially expressed GST fusions containing N-terminal GCN2 segments 1–598 or 1–272, but not by the fusions containing segments 108–272 or 1498–1659. A weak interaction was observed for the extreme N-terminal segment (amino acids 1–110) containing only the CNT domain. These results parallel those obtained in pull-down experiments with GCN1/20 in crude extracts, and support the idea that the GCN1/20 complex interacts directly with the N-terminus of GCN2.
Fig. 4. Purified GCN1–GCN20 complex binds specifically to bacterially expressed GST–GCN2 fusions bearing the CNT domain of GCN2. (A) Coomassie staining of 100 ng of affinity-purified GCN1–GCN20 complex. The protein concentration was estimated by comparison with a known amount of purified BSA (fraction V, Boehringer Mannheim) (data not shown). (B) GST pull-down assays were performed using 100 ng of purified GCN1–GCN20 complex shown in (A) and GST–GCN2 fusions bearing the GCN2 residues indicated above each lane. The assays were conducted as described in Figure 3 except that 1 mg of E.coli extract was added to reduce the non-specific binding of GCN1–GCN20 to the GST fusions. The amounts of GCN1 (top panel), GCN20Flag (middle panel) and the GST fusions (bottom panel) recovered in the precipitates were determined by Western analysis. Asterisks in the bottom panel mark the full-length GST fusion proteins. The lane headed ‘In 1/10’ contains one-tenth of the amount of purified GCN1–GCN20 complex used as input in the binding reactions.
We noted that a substantially larger proportion of GCN1 than GCN20Flag in the purified GCN1–GCN20Flag sample was precipitated by GST–GCN2(1–598) (Figure 4B, lanes 3 and 4 versus lane 1). Inspection of the Coomassie staining profile suggests that GCN20Flag is present in molar excess of GCN1 in this preparation (Figure 4A), which can probably be attributed to the fact that GCN20 contained the affinity tag used for purifying the complex. The smaller proportion of GCN20Flag that bound to GST–GCN2(1–598) is consistent with the idea that GCN20 cannot bind directly to GCN2 and that its interaction with GCN2 is mediated by GCN1.
Dominant-negative effect of overexpressing the N-terminal region of GCN2 provides in vivo evidence for complex formation with GCN1/20
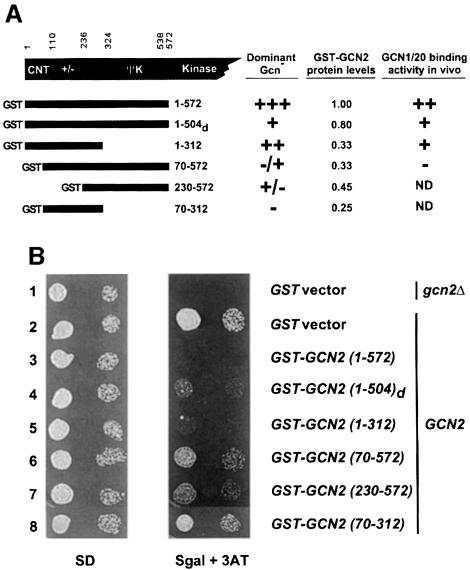
If the N-terminal region of GCN2 is responsible for its interaction with the GCN1/20 complex, then overexpressing this protein segment in yeast cells might compete with native GCN2 for binding to GCN1/20, and thereby interfere with kinase activation. Accordingly, we asked whether overexpression of N-terminal fragments of GCN2 would confer a 3ATS phenotype in a GCN2 strain, indicating a dominant Gcn– phenotype. The GCN2 fragments depicted in Figure 5A were tagged at their C-termini with the c-Myc epitope and expressed as GST fusions from high-copy plasmids under an inducible GAL promoter. Induction of the GST–GCN2(1–572) construct had a dominant-negative Gcn– phenotype, conferring sensitivity to 3AT (Figure 5B, row 3, Sgal + 3AT panel). As expected from this phenotype, galactose induction of GST–GCN2(1–572) reduced the level of eIF2α phosphorylation in vivo (Figure 2B, lanes 10 and 11), confirming the predicted impairment of endogenous GCN2 activity. Eliminating the first 69 GCN2 residues of the GST–GCN2(1–572) construct nearly abolished its dominant Gcn– phenotype [GST–GCN2(70–572)] (Figure 5B, rows 2, 3 and 6), indicating the importance of the CNT domain for the interfering activity of GCN2 residues 1–572. Induction of the GST–GCN2(1–312) construct conferred slightly less 3AT sensitivity than that given by GST–GCN2(1–572) (Figure 5B, rows 3 and 5), in accordance with the relatively weaker binding of GCN1/20 to GCN2 amino acids 1–272 versus amino acids 1–572 shown above (Figure 3B). Induction of the GST–GCN2(230–572) construct, encompassing the entire ΨK domain, had only a modest Gcn– phenotype (Figure 5B, rows 2 and 7). As expected, none of the constructs affected cell growth in galactose-containing medium lacking 3AT (data not shown), or in glucose-containing medium where the proteins are uninduced (Figure 5B, SD panel).
Fig. 5. Dominant-negative effect of overexpressing the N-terminal 572 amino acids of GCN2. (A) Schematic representation of GST–GCN2 fusions containing the indicated GCN2 residues that were expressed in a GCN2 yeast strain under the control of a galactose-inducible promoter. The second construct contains residues 1–504 of D.melanogaster GCN2, which are similar in sequence to yeast GCN2 residues 1–572. The second and third columns contain a summary of the Gcn phenotypes of the fusion constructs measured as described in (B), and the protein levels measured by Western analysis (data not shown), respectively, under galactose-inducing growth conditions. The fourth column contains a qualitative summary of the binding data shown in Figure 6. ND, not determined due to the failure of the fusion to bind the glutathione–Sepharose beads efficiently [GST–GCN2(230–572)] or its low level of expression [GST–GCN2(70–312)]. (B) A transformant of gcn2Δ strain H2557 bearing vector pEGKT (Mitchell et al., 1993) encoding GST alone (top row) and transformants of GCN2 strain H1511 bearing pEGKT or plasmids encoding the indicated GST–GCN2 fusions described in (A) were grown to late logarithmic phase, diluted to an OD600 of 0.15 (first column) and 0.015 (second column) and spotted onto minimal medium containin 2% glucose as carbon source (SD) or minimal medium containing 2% galactose (to induce the GST–GCN2 fusions) and 30 mM 3AT to impose histidine starvation (Sgal + 3AT). The SD and Sgal + 3AT plates were incubated at 37°C for 1 and 3 days, respectively.
Western analysis showed that the GST–GCN2(70–572) and GST–GCN2(230–572) constructs were expressed in yeast at about one-third and one-half the level of GST–GCN2(1–572), respectively (data not shown; summarized in Figure 5A); however, the failure of the former constructs to confer 3AT sensitivity is unlikely to result from their reduced protein levels for the following two reasons. First, the GST–GCN2(1–312) fusion produced a dominant Gcn– phenotype even though it too was expressed at only one-third the level of GST–GCN2 (1–572). Secondly, a separate c-Myc-tagged version of GCN2(70–572) lacking the GST moiety was expressed at essentially the same level as GST–GCN2(1–572) but it failed to show any dominant 3AT sensitivity, whereas c-Myc-tagged GCN2(1–572) had a dominant Gcn– phenotype (data not shown). We also verified that the levels of native GCN2, GCN1 and GCN20 were unaffected by overexpression of the various GST–GCN2 fusions depicted in Figure 5 (data not shown).
The five known GCN2 homologs show strong sequence similarity in their CNT domains. Accordingly, we tested whether the N-terminal segment of D.melanogaster had a dominant Gcn– phenotype when overexpressed in GCN2 yeast cells. As shown in Figure 5B, a GST fusion containing residues 1–504 of Drosophila GCN2 conferred 3AT sensitivity, albeit to a lesser degree than that seen for the corresponding fusion bearing yeast GCN2 residues 1–572 (rows 3 and 4, respectively). Considering that GCN1 homologs have been identified in humans (Marton et al., 1997) and Drosophila (E.Sattlegger and A.G.Hinnebusch, unpublished observations), our findings suggest that interaction between the CNT domain of GCN2 and GCN1 is evolutionarily conserved.
We next tested the prediction that GCN1/20 complex should be bound in vivo to the GST–GCN2 fusions that conferred strong dominant Gcn– phenotypes. The results in Figure 6A show that GST–GCN2(1–572) precipitated with a larger fraction of the GCN1 in the cell than did GST–GCN2(1–312) or GST–GCN2(1–504)d (Figure 6A, lanes 4–9 and 12–19). Importantly, the GST–GCN2(70–572) fusion, which conferred a very weak Gcn– phenotype, failed to precipitate with detectable amounts of GCN1 (Figure 6, lanes 21–23). Although this last fusion was expressed at only one-third the level of GST–GCN2(1–572), its expression was comparable with that of GST–GCN2(1–312), which bound substantial amounts of GCN1 even at the lowest amount of extract examined. Thus, the amount of GCN1 found associated with these GST–GCN2 fusions in vivo was correlated with the strengths of their dominant Gcn– phenotypes (see the summary in Figure 5A, last column).
Fig. 6. Dominant-negative GST–GCN2 fusions bind to the GCN1–GCN20 complex in vivo. (A) Extracts were prepared from transformants of the wild-type yeast strain H1511 containing plasmids encoding the GST–GCN2 fusions bearing the GCN2 residues indicated in the column headings, or GST alone, after growth in Sgal medium containing 2% galactose to induce the fusion proteins. Aliquots of extracts containing 50, 200 or 400 µg of protein (1×, 4× or 8×, respectively) were incubated with gluthatione–Sepharose beads to precipitate the GST fusions and the precipitates were probed for GCN1, GCN2 and the GST fusion proteins by Western analysis. Lanes headed ‘1/10’ contain one-tenth of the amount of extract used in the 4× reactions (20 µg). (B) A subset of the extracts employed in (A) were used in co-immunoprecipitation experiments with anti-GCN1 antiserum, exactly as described in Figure 1B.
If overexpression of the GST–GCN2 fusions confers a Gcn– phenotype by sequestering GCN1/20 from native GCN2, then it should decrease the amount of GCN2 associated with GCN1. In agreement with this prediction, overexpression of GST–GCN2(1–572) and GST–GCN2(1–312) led to large and moderate reductions, respectively, in the amount of GCN2 that co-immunoprecipitated with GCN1 (Figure 6B, lanes 2, 5 and 8, GCN2 panel), whereas GST–GCN2(70–572) had no effect on complex formation between endogenous GCN1 and GCN2 (Figure 6B, lanes 2 and 11). The magnitude of the reductions in GCN1–GCN2 association correlated directly with the amounts of GCN1 precipitated with the corresponding GST–GCN2 fusions (Figure 6A). Thus, we conclude that GST–GCN2(1–572) and GST–GCN2(1–312) competed with native GCN2 for interaction with GCN1 in vivo. As expected, overexpressing the GST–GCN2 fusions had no effect on the association of GCN20 with GCN1 (Figure 6B).
In contrast to the relatively large proportion of total GCN1 recovered with GST–GCN2(1–572) (∼20%), only a few percent of the total GCN2 was precipitated with this fusion (Figure 6A, lanes 5 and 6, and 13–15). The GST–GCN2(1–312) fusion also precipitated a trace amount of GCN2, whereas GST–GCN2(1–504)d did not precipitate any detectable yeast GCN2. These findings make it very unlikely that the dominant Gcn– phenotypes of the overexpressed GST–GCN2 fusions result from the formation of heterodimers with native GCN2.
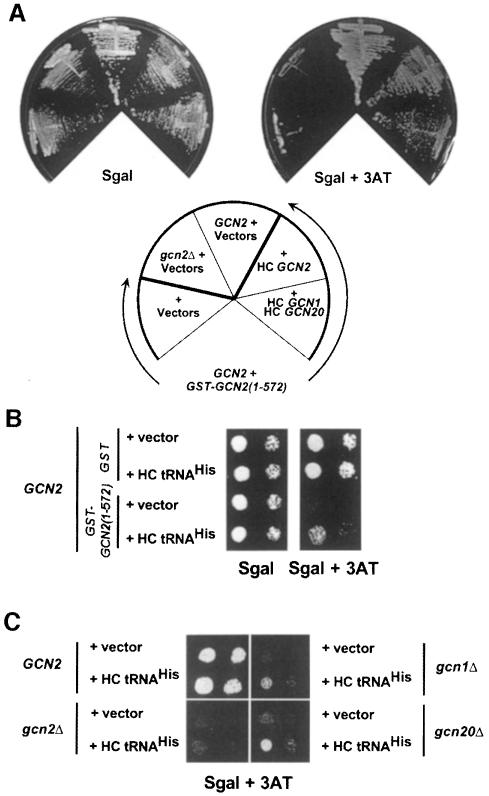
Overexpression of GCN1 and GCN20 can partially suppress the dominant-negative phenotype of GST–GCN2(1–572)
To provide more direct evidence that the dominant Gcn– phenotype of overexpressing GST–GCN2(1–572) arose from its ability to compete with endogenous GCN2 for interaction with GCN1/20, we asked whether its interfering effect could be diminished by overproducing GCN2 or GCN1/20. Towards this end, GST–GCN2(1–572) was induced in GCN2 strains overexpressing GCN1 and GCN20 together, or GCN2 alone, from high-copy plasmids. As expected, overexpression of GCN2 suppressed the 3ATs phenotype of the GST–GCN2(1–572) construct on galactose medium (Figure 7A). Co-overexpression of GCN1 and GCN20 partially suppressed the dominant Gcn– phenotype, consistent with the idea that GST–GCN2(1–572) interferes with endogenous GCN2 by limiting interaction of the latter with GCN1/20 (Figure 7A). The greater ameliorating effect of overproducing GCN2 versus GCN1/20 may be attributable to the fact that GCN2 expression was increased ∼40-fold, whereas GCN1 and GCN20 levels were elevated only ∼3-fold, in the strains expressing these proteins from high-copy plasmids (data not shown). Presumably, the amount of GCN1/20 overexpression was insufficient to titrate all of the GST–GCN2(1–572) present in these strains. These results provide strong genetic evidence that physical interaction between the N-terminal region of GCN2 and the GCN1/20 complex is critical for GCN2 function in amino acid-starved cells.
Fig. 7. Suppression of the dominant-negative phenotype of GST–GCN2(1–572) by overexpression of GCN2, GCN1 or GCN20. (A) Transformants of GCN2 strain H1511 bearing the TRP1 plasmid pB167 encoding the galactose-inducible GST–GCN2(1–572) fusion and high copy number (HC) plasmids encoding GCN2 (URA3 plasmid p630), GCN1 (URA3 plasmid p1834), GCN20 (URA3 plasmid p1748 or LEU2 plasmid p1747) or empty vectors (URA3 vector pRS426 or LEU2 vector pRS425), as indicated, were streaked on Sgal medium or Sgal containing 50 mM 3AT. The two control strains streaked in the upper left-hand sectors were transformants of H1511 (GCN2 + vectors) or gcn2Δ strain H2557 (gcn2Δ + vectors), both bearing empty vectors. The Sgal and Sgal + 3AT plates were incubated at 30°C for 36 h or 4 days, respectively. (B) The GCN2 strain H1511 was transformed with URA3 plasmid pB165 encoding the galactose-inducible GST–GCN2(1–572) fusion and the high copy number LEU2 plasmid p897 bearing the tRNAHis gene, or the corresponding empty vectors (pEGKT and Yep13, respectively). The transformants were cultured and spotted onto Sgal or Sgal containing 50 mM 3AT, as described in Figure 5B, and incubated at 30°C for 2 and 3.5 days, respectively. The first and third columns contain one-tenth the number of cells spotted in the second and fourth columns. (C) Transformants of the isogenic strains H1511 (GCN2, upper left sector), H2557 (gcn2Δ, lower left sector), H2556 (gcn1Δ, upper right sector) and H2558 (gcn20Δ, lower right sector) harboring YEp13 (vector) or p897 (HC tRNAHis) were cultured and spotted onto Sgal containing 25 mM 3AT, as described in Figure 5B, and incubated for 3.5 days at 30°C.
We reasoned that if association between GCN2 and GCN1/20 is required for activation of GCN2 by uncharged tRNA, then the dominant Gcn– phenotype of the GST–GCN2 fusions might be suppressed by producing higher levels of uncharged tRNA in the cell. This prediction was supported by the results in Figure 7B showing that the 3AT sensitivity conferred by GST–GCN2(1–572) was diminished by a high copy number plasmid encoding tRNAHis, shown previously to overproduce this tRNA and to suppress the Gcn– phenotype of a mutation in the HisRS domain of GCN2 (Vazquez de Aldana et al., 1994). Interestingly, the Gcn– phenotypes of gcn1Δ and gcn20Δ mutants were also partially suppressed by overexpressing tRNAHis in cells starved for histidine by 3AT (Figure 7C, two right panels), whereas the gcn2Δ mutant was not suppressed (lower left panel). These results support the idea that GCN1/20 stimulates GCN2 by promoting its activation by uncharged tRNA.
Discussion
It was established previously that the GCN1/20 complex is required for GCN2 function in yeast under amino acid starvation conditions (Marton et al., 1993, 1997; Vazquez de Aldana et al., 1995). However, prior attempts to demonstrate physical linkage between GCN2 and these positive regulators had failed, and it was unknown whether GCN1/20 acts directly to promote activation of GCN2 in starved cells. Here we presented several lines of biochemical and genetic evidence that this interaction occurs in vivo and is critically required for GCN2 activation by uncharged tRNA. We co-immunoprecipitated GCN2 with GCN1 from cell extracts and showed that this interaction occurred independently of the protein kinase, HisRS and ribosome-binding domains in GCN2. Thus, association of GCN1 with GCN2 did not reflect independent tethering of these proteins to the same ribosomes or tRNA molecules. The CNT and highly charged regions at the N-terminus of GCN2 were necessary and sufficient for interaction with GCN1/20, whether the latter was present in cell extracts or in highly purified form. This is the first biochemical activity assigned to this portion of GCN2, comprising ∼20% of the protein.
The fact that deleting the CNT or +/– domains destroyed GCN2 function in vivo suggests that binding of GCN1/20 to GCN2 is required for kinase activation in starved cells. Additional evidence for this conclusion came from our finding that GST–GCN2(1–572) and GST–GCN2(1–312) expressed in yeast bound to the GCN1/20 complex and conferred a dominant Gcn– phenotype. Analysis of truncated derivatives of these fusions showed that the Gcn– phenotype correlated with binding to GCN1/20. The fact that the same segments of GCN2 were necessary and sufficient for in vivo binding to GCN1/20 and for the dominant Gcn– phenotype strongly suggests that the GST–GCN2 fusions interfered with endogenous GCN2 by sequestering GCN1/20. This interpretation was supported by showing that expressing the GST–GCN2 fusions decreased the amount of endogenous GCN2 associated with GCN1. Furthermore, the Gcn– phenotype of GST–GCN2(1–572) was eliminated by overexpressing GCN2 and diminished by overexpressing GCN1 and GCN20. We conclude that high-level GCN2 function is critically dependent on physical interaction of the CNT and +/– domains with GCN1/20.
The ΨK domain was dispensable for the co-immunoprecipitation of GCN2 with GCN1 and for the GCN1/20-binding activities and dominant Gcn– phenotypes of GST–GCN2 fusions. However, deletion of the ΨK domain led to modest but reproducible decreases in the association between GCN2 and GCN1/20 in each of these assays. Additionally, overexpressing GST–GCN2 fusions containing the ΨK domain but lacking the CNT or +/– domains conferred a slight Gcn– phenotype, although we could not detect any GCN1 co-precipitating with one such fusion [GST–GCN2(70–572)]. Therefore, we suggest that the ΨK domain makes a minor contribution to the binding of GCN1/20 compared with that of the adjacent CNT and charged domains.
What is the functional significance of complex formation between GCN2 and GCN1/20? First, we can state that physical contact between these proteins seems inconsistent with a model in which GCN1/20 would stimulate GCN2 indirectly, e.g. by regulating the size of amino acid pools or amounts of uncharged tRNA in the cytoplasm. Our discovery of a stable interaction between GCN1/20 and GCN2, combined with the fact that all three proteins have ribosome-binding activities, strongly supports the idea that GCN1/20 functions directly to facilitate stimulation of GCN2 by uncharged tRNA on the ribosome. Further support for this hypothesis came from our finding that overexpressing tRNAHis in cells starved for histidine partially restored GCN2 function in gcn1Δ or gcn20Δ mutants, and also in strains where GCN1/20 was sequestered by GST–GCN2(1–572). This epistatic relationship is consistent with the idea that GCN1/20 is required for stimulation of GCN2 by physiological levels of uncharged tRNA. We propose that GCN1/20 binds to the ribosome and, through its interaction with the N-terminus of GCN2, positions the HisRS domain for proper association with uncharged tRNA occupying the A-site. Given their similarities to EF3, and the fact that EF3 facilitates binding and release of tRNAs from the ribosome, GCN1/20 may additionally promote entry of uncharged tRNA to the A-site, or its dissociation from the A-site for interaction with GCN2 (Marton et al., 1997).
Previous studies showed that overexpressing tRNAHis confers a modest derepression of GCN4 independently of GCN2 (Vazquez de Aldana et al., 1994); however, for the experiments in Figure 7, we chose a 3AT concentration where the GCN2-independent derepression of GCN4 was insufficient for growth. Moreover, because there is no detectable eIF2α phosphorylation in gcn1Δ cells (Marton et al., 1993; Vazquez de Aldana et al., 1995) (Figure 2B), the 3AT resistance conferred by overexpressing tRNAHis in the gcn1Δ mutant cannot result from combining low-level GCN2 activity with GCN2-independent derepression. Accordingly, we propose that uncharged tRNAHis, when present at excessively high levels, can bind to the GCN2 HisRS domain and stimulate kinase activity without the intervention of GCN1/20.
It is interesting that deletion of GCN20 appears to weaken the association between GCN1 and GCN2. GCN20 binds to GCN1 in the region that is homologous to EF3. This interaction involves the N-terminal 16% of GCN20 (Marton et al., 1997), which is the only segment of the protein critically required for GCN20 function under amino acid starvation conditions (Vazquez de Aldana et al., 1995). Thus, the N-terminal portion of GCN20 seems to promote the proper conformation or activity of the EF3-like domain in GCN1 in a way that enhances GCN1–GCN2 interaction. The remainder of GCN20 contains two putative ATP-binding domains that are not critically required for GCN2 activation under amino acid starvation conditions (Marton et al., 1997). The results presented here raise the possibility that preventing complex formation between GCN20 and GCN1 could provide a novel means of dampening the GCN1–GCN2 interaction and, hence, activation of GCN2 by uncharged tRNA. Perhaps the GCN20 ATP-binding domains adjust the extent of GCN20–GCN1–GCN2 complex formation according to the levels of other nutrients, integrating this information with amino acid availability to achieve the proper level of GCN2 activation and eIF2α phosphorylation for the conditions at hand.
In all GCN2 homologs analyzed thus far, the CNT domain is highly conserved, as is the position of the +/– domain, although there is limited conservation of specific residues in the latter case. Furthermore, the CNT shows no homology to any other proteins in the databases, defining a domain apparently unique to the GCN2 family of proteins. We found that a GST fusion bearing the CNT and +/– domains from D.melanogaster GCN2 bound to yeast GCN1/20 in vivo and had a dominant Gcn– phenotype. Thus, the GCN1/20-binding function of the CNT is conserved between yeast and Drosophila GCN2. GCN1 (Marton et al., 1997) and GCN20 (Vazquez de Aldana et al., 1995) homologs have been identified in humans. Thus, it seems likely that GCN1/20 homologs in Drosophila and mammals interact with the cognate GCN2 proteins, reflecting a conserved mechanism for activating GCN2 in response to amino acid deprivation. Ostensibly at odds with this idea, mouse GCN2 (mGCN2) expressed in yeast functioned independently of GCN1 (Sood et al., 2000); however, the mGCN2 was being overexpressed and this may have lessened its dependence on GCN1/20. Alternatively, mGCN2 could have a higher intrinsic affinity for uncharged tRNA. It is noteworthy that three different isoforms of mGCN2 were detected in mouse tissues, and the two least abundant and most restricted in their tissue distributions (α and γ) lacked some or all of the predicted binding domain for GCN1/20 (Sood et al., 2000). Perhaps these truncated isoforms are less responsive to uncharged tRNA and regulated in response to other starvation signals.
Materials and methods
Strains and plasmids
The yeast strains and plasmids employed in this study are listed in Tables I and II. Details of their construction are available as supplementary data at The EMBO Journal online.
Table I. Yeast strainsa.
| Yeast strain | Genotype | Reference |
|---|---|---|
| H1402 | a leu2-3 leu2-112 ino1 ura3-52 | Hannig et al. (1990) |
| H1149 | a gcn2Δ leu2-3 leu2-112 ino1 ura3-52 | Wek et al. (1990) |
| H2081 | a gcn1Δ leu2-3 leu2-112 ino1 ura3-52 | Marton et al. (1993) |
| H2563 | a gcn20Δ leu2-3 leu2-112 ino1 ura3-52 | C.Vazquez de Aldana and A.G.Hinnebusch, |
| unpublished observations | ||
| H1894 | a gcn2Δ leu2-3 leu2-112 ino1 ura3-52 | Dever et al. (1993) |
| H2557 | a gcn2Δ leu2-3 leu2-112 trp-Δ63 ura3-52 GAL2 | Marton et al. (1993) |
| H2556 | a gcn1Δ leu2-3 leu2-112 trp-Δ63 ura3-52 GAL2 | Marton et al. (1993) |
| H2558 | a gcn20Δ leu2-3 leu2-112 trp-Δ63 ura3-52 GAL2 | Vazquez de Aldana et al. (1995) |
| H1511 | a leu2-3 leu2-112 trp-Δ63 ura3-52 GAL2 | Foiani et al. (1991) |
aThe first four strains are isogenic and were used in co-immunoprecipitation experiments together with H1894. The last four strains are isogenic and were used in pull-down assays, as well as in the genetic studies.
Table II. Plasmids.
| Plasmid | Allele | Reference |
|---|---|---|
| p630 | Wild type GCN2, 2µ, URA3 | Wek et al. (1990) |
| p2326 | gcn2-K628R in p630 backbone | pB34 in Romano et al. (1997) |
| pB82 | gcn2-Δ572–999 in p630 backbone | Qiu et al. (1998) |
| p733 | gcn2-Δ1161–1245 in p630 backbone | Wek et al. (1989) |
| p732 | gcn2-Δ1401–1535 in p630 backbone | Wek et al. (1989) |
| pB106 | gcn2-Δ1246–1400 in p630 backbone | this study |
| pB47 | gcn2-Δ1536–1659 in p630 backbone | Qiu et al. (1998) |
| p722 | GCN2, CEN6, ARSH4, URA3 | Wek et al. (1989) |
| pB54 | GCN2-myc, unique SacI (5′) and MluI (3′) sites flanking a myc tag inserted betweencodons 1 and 2 of GCN2 in p722 | this study |
| pB121 | GCN2-myc from pB54 in p630 backbone | this study |
| pB123 | gcn2-Δ10–109, unique SacI between codon 1 and 2 of GCN2 and deletion marked withMluI at the deletion junction in pB121 backbone; the myc tag is not present in this construct | this study |
| pB101 | gcn2-Δ237–322, unique SacI between codon 1and 2 of GCN2 and deletion marked withMluI at the deletion junction in pB121 backbone; the myc tag is not present in this construct | this study |
| pB29 | gcn2-Δ110–235 in p630 backbone | this study |
| pB32 | gcn2-Δ324–538 in p630 backbone, with a unique SacI site at the deletion junction | this study |
| pB149 | gcn2-Δ324–538 in p630 backbone, with unique SacI (5′) and MluI (3′) sites at thedeletion junction, separated by two codons encoding Leu and Ala | this study |
| pB152 | gcn2-Δ184–237, codons 184–237 flanked by unique SacI (5′) and MluI (3′) sites introducedinto pB149 | this study |
| pB130 | GST–GCN2(1–598) in pGEX-5X-1 (Pharmacia Biotech.) | this study |
| pB131 | GST–GCN2(1–272) in pGEX-5X-1 | this study |
| pB132 | GST–GCN2(1–110) in pGEX-5X-1 | this study |
| pB133 | GST–GCN2(108–272) in pGEX-5X-1 | this study |
| pHQ551 | GST–GCN2(568–998) in pGEX-5X-1 | Qiu et al. (1998) |
| pHQ531 | GST–GCN2(1498–1659) in pGEX-5X-1 | Qiu et al. (1998) |
| pHQ530 | GST–GCN2(970–1497) in pGEX-5X-1 | gift from Dr H.Qiu |
| pB165 | GST–GCN2(1–598), myc tagged at the 3′ end and under the GAL1 promoter in pEGKT | this study and Mitchell et al. (1993) |
| pB202 | Same as pB165 but containing GST–GCN2(70–598) | this study |
| pB203 | Same as pB165 but containing GST–GCN2(1–272) | this study |
| pB204 | Same as pB165 but containing GST–GCN2(70–272) | this study |
| pB205 | Same as pB165 but containing GST–GCN2(230–572) | this study |
| pB206 | Same as pB165 but containing GST–GCN2(1–504) with codons 1–504 from D.melanogaster GCN2 | this study |
| pB167 | GST–GCN2(1–598), myc tagged in pB201 | this study |
| pB201 | LEU2 version of pEGKT | this study and Mitchell et al. (1993) |
| p897 | tRNAHis in YEp13 | Vazquez de Aldana et al. (1994) |
Co-immunoprecipitation of GCN1/2/20 complexes
Yeast whole-cell extracts (WCEs) were prepared from cultures grown to mid log-phase (OD600 of ∼1.2–1.5) in either YPD or selective SC medium essentially as described previously (Ramirez et al., 1991) except that breaking buffer H-50 (30 mM HEPES pH 7.4, 50 mM KCl), supplemented with protease inhibitors (PIs) 1 µM leupeptin, 1 µM pepstatin, 0.15 µM aprotinin and 100 µM phenylmethylsulfonyl fluoride (PMSF), was used and the final centrifugation was 10 min at 14 000 r.p.m. at 4°C. About 250–500 µg of total protein in the resulting supernatant (WCE) were pre-cleared in the presence of protein A–Sepharose beads [pre-blocked with 5% bovine serum albumin (BSA) and suspended in H-50 + PIs] in a final volume of 200–250 µl of the same buffer. The GCN1/20/2 complexes were immunoprecipitated from the pre-cleared extracts with either 1.5 µl of anti-GCN2 serum (HL2523) or 1.2 µl of anti-GCN1 serum (HL1405) pre-bound to protein A–Sepharose by gently mixing in a Nutator for 2 h at 4°C, and the precipitated proteins were analyzed by 4–12% SDS–PAGE and immunoblot analysis, as described below.
Protein interaction assays using GST–GCN2 fusions expressed in bacteria
Bacterial extracts containing the different GST–GCN2 fusion proteins expressed in E.coli strain BL21 were prepared by sonication according to the protocols suggested by Pharmacia Biotech, except that 50 µl of buffer H-50 + PIs + PIMs per milliliter of original culture were used as the breaking buffer. [PIMs is a cocktail of protease inhibitors described previously (Drysdale et al., 1995) except lacking β-mercaptoethanol.] Triton X-100 to 1% was added to enhance solubility. The yeast extracts used for GST pull-down assays were prepared from the indicated strains grown to an OD600 of 1.5–2 in YPD or SC medium using an adaptation of a method described previously (Drysdale et al., 1998), in which the cells were broken in H-50 + PIs + PIMs, the ammonium sulfate precipitation was omitted and the cell debris was cleared by centrifugation for 1.5 h at 14 000 r.p.m.
Equivalent amounts of the different GST–GCN2 fusions (as estimated by Coomassie blue staining) pre-bound to glutathione–Sepharose beads as suggested by Pharmacia Biotech, in the presence of H-50 + PIs + PIMs, were used in pull-down experiments using 1 mg of pre-cleared yeast WCE in 100–150 µl of H-50 + PIs + PIMs on a Nutator for 2 h at 4°C and the proteins pulled down were analyzed by standard Western blotting. (The WCEs had been pre-cleared as described above for the co-immunoprecipitation experiments, except that glutathione–Sepharose beads were used in this case.)
For GST pull-down assays with purified GCN1/20 complex, the procedure was similar, except that yeast WCE was replaced by ∼100 ng of purified proteins (J.Dong and A.G.Hinnebusch, unpublished observations), supplemented with 1 mg of bacterial extract from E.coli strain DH5 transformed with pUC19 to reduce non-specific binding of the purified GCN1/20 proteins. The protein mixture was pre-adsorbed with glutathione–Sepharose beads in 150 µl of H-50 buffer + PIs + PIMs prior to the pull-down assay. Purification of the GCN1/20 complex will be described elsewhere.
Protein interaction assays using GST–GCN2 fusions expressed in yeast
The different GST–GCN2 fusions were expressed in yeast, by growth in the presence of the required supplements in either SD medium (uninduced conditions) or SGal medium containing 2% galactose and 2% raffinose as the carbon source (induced conditions), respectively, at 30°C to an OD600 of 1.5–2. Extracts were prepared exactly as for the co-immunoprecipitation experiments, except that PIMs protease inhibitors were added to the buffers. Prior to GST pull-down assays, the WCEs, adjusted to a final protein concentration of 400 µg by addition of BSA (fraction V, Boehringer-Mannheim), were pre-cleared using protein A–Sepharose beads (Amersham) in a final volume of 100 µl for 1 h on a Nutator at 4°C. The pull-down was performed with glutathione beads on a Nutator for 30 min at room temperature followed by 1–2 h at 4°C and the proteins bound to the beads were analyzed by SDS–PAGE and immunoblotting.
Protein techniques
For standard Western blot analysis, the membranes were probed with antibodies against GCN1 (H1405, dilution 1:2000) (Vazquez de Aldana et al., 1995), GCN2 (HL2173 and HL2523, 1:2000) (Romano et al., 1997; Qiu et al., 1998), GCN20 (CV1317, 1:2000) (Vazquez de Aldana et al., 1995), eIF2α (1:2000) (Cigan et al., 1989), phosphoserine 51 of eIF2α (1:1000) (DeGracia et al., 1997), PUB2 and PAB1 (1:1000) (a gift from J.Anderson) and Hsp82 (1:5000) (a gift from S.Lindquist). Immune complexes were visualized using donkey anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham) and the enhanced chemiluminescence system (Amersham). To eliminate non-specific signals when probing filters with anti-GCN20 or the HL2173 anti-GCN2 antibodies, yeast WCE prepared from yeast strains lacking GCN20 or GCN2, respectively, was added at 100 µg/ml of diluted antibody in 5% blotto and incubated for 1 h at room temperature prior to use.
Supplementary data
Supplementary data to this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Doug Cavener for the gift of plasmid pYK380, Jim Anderson, Tom Donahue and Susan Lindquist for gifts of antibodies, Henry Levin for advice and suggestions, Tom Dever and Hongfang Qiu for comments on the manuscript, and Bobbie Felix for help in preparation of the manuscript.
References
- Anderson J.T., Paddy,M.R. and Swanson,M.S. (1993) PUB1 is a major nuclear and cytoplasmic polyadenylated RNA-binding protein in Saccharomyces cerevisiae. Mol. Cell. Biol., 13, 6102–6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlanga J.J., Herrero,S. and de Haro,C. (1998) Characterization of the hemin-sensitive eukaryotic initiation factor 2α kinase from mouse nonerythroid cells. J. Biol. Chem., 273, 32340–32346. [DOI] [PubMed] [Google Scholar]
- Berlanga J.J., Santoyo,J. and De Haro,C. (1999) Characterization of a mammalian homolog of the GCN2 eukaryotic initiation factor 2α kinase. Eur. J. Biochem., 265, 754–762. [DOI] [PubMed] [Google Scholar]
- Cashel M. and Rudd,K.E. (1987) The stringent response. In Neidhardt,F.C., Ingraham,J.L., Magasanik,B., Low,K.B., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. American Society for Microbiology, Washington, DC, pp. 1410–1438. [Google Scholar]
- Cigan A.M., Pabich,E.K., Feng,L. and Donahue,T.F. (1989) Yeast translation initiation suppressor sui2 encodes the subunit of eukaryotic initiation factor 2 and shares identity with the human subunit. Proc. Natl Acad. Sci. USA, 86, 2784–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens M.J. (1996) Protein kinases that phosphorylate eIF2 and eIF2B and their role in eukaryotic cell translational control. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 139–172. [Google Scholar]
- DeGracia D.J., Sullivan,J.M., Neumar,R.W., Alousi,S.S., Hikade,K.R., Pittman,J.E., White,B.C., Rafols,J.A. and Krause,G.S. (1997) Effect of brain ischemia and reperfusion on the localization of phosphorylated eukaryotic initiation factor 2α. J. Cereb. Blood Flow Metab., 17, 1291–1302. [DOI] [PubMed] [Google Scholar]
- Dever T.E. (1999) Translation initiation: adept at adapting. Trends Biochem. Sci., 24, 398–403. [DOI] [PubMed] [Google Scholar]
- Dever T.E., Chen,J.J., Barber,G.N., Cigan,A.M., Feng,L., Donahue,T.F., London,I.M., Katze,M.G. and Hinnebusch,A.G. (1993) Mammalian eukaryotic initiation factor 2α kinases functionally substitute for GCN2 in the GCN4 translational control mechanism of yeast. Proc. Natl Acad. Sci. USA, 90, 4616–4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O. and Picard,D. (1999) Hsp90 binds and regulates the ligand-inducible α subunit of eukaryotic translation initiation factor kinase Gcn2. Mol. Cell. Biol., 19, 8422–8432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale C.M., Dueñas,E., Jackson,B.M., Reusser,U., Braus,G.H. and Hinnebusch,A.G. (1995) The transcriptional activator GCN4 contains multiple activation domains that are critically dependent on hydrophobic amino acids. Mol. Cell. Biol., 15, 1220–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drysdale C.M. et al. (1998) The Gcn4p activation domain interacts specifically in vitro with RNA polymerase II holoenzyme, TFIID and the Adap–Gcn5p coactivator complex. Mol. Cell. Biol., 18, 1711–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Cigan,A.M., Paddon,C.J., Harashima,S. and Hinnebusch,A.G. (1991) GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 3203–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannig E.H., Williams,N.P., Wek,R.C. and Hinnebusch,A.G. (1990) The translational activator GCN3 functions downstream from GCN1 and GCN2 in the regulatory pathway that couples GCN4 expression to amino acid availability in Saccharomyces cerevisiae. Genetics, 126, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding H.P., Zhang,Y. and Ron,D. (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature, 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Higgins C.F. (1992) ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol., 8, 67–113. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. (1996) Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In Hershey,J.W.B., Mathews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 199–244. [Google Scholar]
- Kamath A. and Chakraburtty,K. (1989) Role of yeast elongation factor 3 in the elongation cycle. J. Biol. Chem., 264, 15423–15428. [PubMed] [Google Scholar]
- Marton M.J., Crouch,D. and Hinnebusch,A.G. (1993) GCN1, a translational activator of GCN4 in S.cerevisiae, is required for phosphorylation of eukaryotic translation initiation factor 2 by protein kinase GCN2. Mol. Cell. Biol., 13, 3541–3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marton M.J., Aldana,C.R.V.d., Qiu,H., Chakraburtty,K. and Hinnebusch,A.G. (1997) Evidence that GCN1 and GCN20, translational regulators of GCN4, function on enlongating ribosomes in activation of the eIF2α kinase GCN2. Mol. Cell. Biol., 17, 4474–4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D.A., Marshall,T.K. and Deschenes,R.J. (1993) Vectors for the inducible overexpression of glutathione S-transferase fusion proteins in yeast. Yeast, 9, 715–722. [DOI] [PubMed] [Google Scholar]
- Olsen D.S., Jordan,B., Chen,D., Wek,R.C. and Cavener,D.R. (1998) Isolation of the gene encoding the Drosophila melanogaster homolog of the Saccharomyces cerevisiae GCN2 eIF-2α kinase. Genetics, 149, 1495–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu H., Garcia-Barrio,M.T. and Hinnebusch,A.G. (1998) Dimerization by translation initiation factor 2 kinase GCN2 is mediated by interactions in the C-terminal ribosome-binding region and the protein kinase domain. Mol. Cell. Biol., 18, 2697–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez M., Wek,R.C. and Hinnebusch,A.G. (1991) Ribosome-association of GCN2 protein kinase, a translational activator of the GCN4 gene of Saccharomyces cerevisae. Mol. Cell. Biol., 11, 3027–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano P.R. et al. (1997) Autophosphorylation in the activation loop is required for full kinase activity in vivo of human and yeast eukaryotic initiation factor 2α kinases PKR and GCN2. Mol. Cell. Biol., 18, 2282–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussou I., Thireos,G. and Hauge,B.M. (1988) Transcriptional–translational regulatory circuit in Saccharomyces cerevisiae which involves the GCN4 transcriptional activator and the GCN2 protein kinase. Mol. Cell. Biol., 8, 2132–2139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo J., Alcalde,J., Mendez,R., Pulido,D. and de Haro,C. (1997) Cloning and characterization of a cDNA encoding a protein synthesis initiation factor-2α (eIF-2α) kinase from Drosophila melanogaster. J. Biol. Chem., 272, 12544–12550. [DOI] [PubMed] [Google Scholar]
- Sattlegger E., Hinnebusch,A.G. and Barthelmess,I.B. (1998) cpc-3, the Neurospora crassa homologue of yeast GCN2, encodes a polypeptide with juxtaposed eIF2α kinase and histidyl-tRNA synthetase-related domains required for general amino acid control. J. Biol. Chem., 273, 20404–20416. [DOI] [PubMed] [Google Scholar]
- Shi Y., Vattem,K.M., Sood,R., An,J., Liang,J., Stramm,L. and Wek,R.C. (1998) Identification and characterization of pancreatic eukaryotic initiation factor 2α-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol., 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sood R., Porter,A.C., Olsen,D., Cavener,D. and Wek,R.C. (2000) A mammalian homologue of GCN2 protein kinase important for translational control by phosphorylation of eukaryotic initiation factor-2α. Genetics, 154, 787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triana-Alonso F.J., Chakraburtty,K. and Nierhaus,K.H. (1995) The elongation factor unique in higher fungi and essential for protein biosynthesis is an E site factor. J. Biol. Chem., 270, 20473–20478. [DOI] [PubMed] [Google Scholar]
- Vazquez de Aldana C.R., Wek,R.C., San Segundo,P., Truesdell,A.G. and Hinnebusch,A.G. (1994) Multicopy tRNA genes functionally suppress mutations in yeast eIF-2α kinase GCN2: evidence for separate pathways coupling GCN4 expression to uncharged tRNA. Mol. Cell. Biol., 14, 7920–7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez de Aldana C.R., Marton,M.J. and Hinnebusch,A.G. (1995) GCN20, a novel ATP binding cassette protein and GCN1 reside in a complex that mediates activation of the eIF-2α kinase GCN2 in amino acid-starved cells. EMBO J., 14, 3184–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Jackson,B.M. and Hinnebusch,A.G. (1989) Juxtaposition of domains homologous to protein kinases and histidyl-tRNA synthetases in GCN2 protein suggests a mechanism for coupling GCN4 expression to amino acid availability. Proc. Natl Acad. Sci. USA, 86, 4579–4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek R.C., Ramirez,M., Jackson,B.M. and Hinnebusch,A.G. (1990) Identification of positive-acting domains in GCN2 protein kinase required for translational activation of GCN4 expression. Mol. Cell. Biol., 10, 2820–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek S.A., Zhu,S. and Wek,R.C. (1995) The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol. Cell. Biol., 15, 4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. and Kaufman,R.J. (1996) Double-stranded (ds) RNA binding and not dimerization correlates with the activation of the dsRNA-dependent protein kinase (PKR). J. Biol. Chem., 271, 1756–1763. [DOI] [PubMed] [Google Scholar]
- Zhu S. and Wek,R.C. (1998) Ribosome-binding domain of eukaryotic initiation factor-2 kinase GCN2 facilitates translation control. J. Biol. Chem., 273, 1808–1814. [DOI] [PubMed] [Google Scholar]
- Zhu S., Sobolev,A.Y. and Wek,R.C. (1996) Histidyl-tRNA synthetase-related sequences in GCN2 protein kinase regulate in vitro phosphorylation of eIF-2. J. Biol. Chem., 271, 24989–24994. [DOI] [PubMed] [Google Scholar]