Abstract
Cytochrome c oxidase is a respiratory enzyme catalysing the energy-conserving reduction of molecular oxygen to water. The crystal structure of the ba3-cytochrome c oxidase from Thermus thermophilus has been determined to 2.4 Å resolution using multiple anomalous dispersion (MAD) phasing and led to the discovery of a novel subunit IIa. A structure-based sequence alignment of this phylogenetically very distant oxidase with the other structurally known cytochrome oxidases leads to the identification of sequence motifs and residues that seem to be indispensable for the function of the haem copper oxidases, e.g. a new electron transfer pathway leading directly from CuA to CuB. Specific features of the ba3-oxidase include an extended oxygen input channel, which leads directly to the active site, the presence of only one oxygen atom (O2–, OH– or H2O) as bridging ligand at the active site and the mainly hydrophobic character of the interactions that stabilize the electron transfer complex between this oxidase and its substrate cytochrome c. New aspects of the proton pumping mechanism could be identified.
Keywords: ba3-cytochrome c oxidase/MAD phasing/membrane protein/Thermus thermophilus/X-ray structure
Introduction
Cytochrome c oxidase (EC 1.9.3.1) is the terminal respiratory membrane protein complex in eukaryotic and many prokaryotic aerobic organisms. The enzyme catalyses the reduction of molecular oxygen to water concomitant with the oxidation of reduced cytochrome c. The free energy of this reaction is conserved as a transmembrane proton gradient that drives ATP synthesis. Some bacterial terminal oxidases use quinol as substrate (quinol oxidases). Due to the homology of their primary structures, both types of enzyme belong to the superfamily of haem copper oxidases (for reviews, see Ferguson-Miller and Babcock, 1996; Michel et al., 1998).
Recently, the three-dimensional structures of two members of this superfamily have been determined. (i) The crystal structure of the cytochrome c oxidase from Paracoccus denitrificans was solved at 2.8 and 2.7 Å resolution for the four and two subunit enzyme, respectively (Iwata et al., 1995; Ostermeier et al., 1997). (ii) The bovine cytochrome c oxidase was solved initially at 2.8 Å and refined later to 2.3 Å (Tsukihara et al., 1995, 1996; Yoshikawa et al., 1998a). In 1995, we described the crystallization of the ba3-type oxidase from the thermophilic eubacterium Thermus thermophilus (Soulimane et al., 1995). We recognized cytochrome c552 as substrate of the ba3-oxidase and determined its crystal structure at 1.28 Å resolution (Soulimane et al., 1997; Than et al., 1997). Furthermore, the three-dimensional structures of bioengineered soluble copper (CuA)-binding domains derived from subunit II of the Escherichia coli quinol oxidase (Wilmanns et al., 1995) and the T.thermophilus ba3-oxidase (Williams et al., 1999) have been reported.
Two important observations within the current oxidase structures are: (i) a covalent bond between His233 and Tyr237 (ba3-oxidase numbering) at the active site of the cytochrome c oxidase (Ostermeier et al., 1997; Yoshikawa et al., 1998a), which has been confirmed by protein chemical investigations (Buse et al., 1999); and (ii) the structural characterization of proton pathways (Iwata et al., 1995; Tsukihara et al., 1996; Ostermeier et al., 1997; Yoshikawa et al., 1998a) called ‘K-’, ‘D-’ and ‘H-’ (bovine) or ‘E-’ pathways (P.denitrificans). These pathways are named according to the specific residues Lys354P, Asp124P and His413 (bovine) or Glu442P (numbers alone correspond to the actual ba3-oxidase structure and numbers followed by a P correspond to the numbering in the P.denitrificans crystal structure, unless noted otherwise). The ‘K-’ and ‘D-’ pathways were identified initially and their functional importance was supported further by extensive mutagenesis experiments (for a review, see Gennis, 1998a). The understanding of the catalytic function of cytochrome c oxidases, regarding oxygen reduction, proton pumping and electron transfer, has advanced significantly during the last decades. In spite of the known structural data as well as many spectroscopic and kinetic characterizations, many details are, however, still not known and are controversially discussed (Gennis, 1998b; Michel, 1998, 1999; Verkhowsky et al., 1999).
The ba3-type cytochrome c oxidase from the extremely thermophilic eubacterium T.thermophilus HB8 (ATCC 27634) is expressed under limited O2 supply (Keightley et al., 1995). It is known as a two subunit oxidase (Keightley et al., 1995; Soulimane et al., 1995). The amino acid sequence of subunit I shows a clear, but distant homology to other eubacterial members of the superfamily. This has been suggested to be the result of an early gene duplication preceding the separation of bacteria and archaea (Castresana and Saraste, 1995). The archaeal oxidases and the ba3-oxidase described herein belong to the SoxB type, while most other eubacterial oxidases are of the SoxM type. Alignments of subunit II of the T.thermophilus ba3-oxidase with sequences from different species show another typical characteristic of SoxB-type terminal oxidases (Mattar and Engelhard, 1997). It lacks ∼60 N-terminal residues including the first transmembrane-spanning helix. The A-type haem of the ba3-oxidase corresponds to the haem As present in the SoxB-type terminal oxidase from Sulfolobus acidocaldarius: the farnesyl chain is replaced by a geranylgeranyl group (Lübben and Morand, 1994). The site of O2 reduction of the ba3-enzyme, the binuclear haem a3 CuB centre, reveals a number of unusual features (Surerus et al., 1992; Kim et al., 1998; Giuffré et al., 1999), including an unusual pattern of reactivity towards exogenous haem as3 ligands such as CN–, CO, NO or H2O2. Cyanide binds to the ferrous but not to ferric haem as3 of the ba3-oxidase, while ligation to the ferric state is much stronger in other oxidases. The CO affinity of CuB in the reduced ba3-oxidase may be 50–100 times higher than in the bovine heart enzyme. Silietzkyi et al. (1999) showed that the ferric ba3-enzyme does not react with H2O2 and they postulated that the binuclear centre of the oxidized enzyme may be closed and opens after one electron reduction.
The two structurally known cytochrome c oxidases are very similar in sequence (>50% identity) and structure with respect to their main subunits I and II. Interestingly, the ba3-oxidase shows very little sequence homology (<20% identity; Keightley et al., 1995) and it lacks most of the highly conserved amino acid residues that form the proton pathways in the other oxidases. Nevertheless, the enzyme generates an electric transmembrane potential under steady-state conditions and pumps protons. While 1.0 H+/e– is consumed, as is usual, for water formation, the ba3-oxidase shows a reduced H+-pumping efficiency of 0.4–0.5 H+/e– instead of ∼1 H+/e– found for the P.denitrificans oxidase (Kannt et al., 1998).
Considering the low sequence homology as well as the different kinetic characteristics of the ba3-oxidase, the structural analysis of this oxidase and its comparison with other cytochrome oxidases will help us to understand further the general function and mechanistic details of the terminal haem copper oxidases. Specific features that are present only in this oxidase may elucidate the structural requirements for its operation at high temperatures and at low oxygen concentration.
We present here the structure determination of the ba3-type cytochrome c oxidase from T.thermophilus at 2.4 Å resolution, the first intrinsic membrane protein to be solved by multiple anomolous dispersion (MAD) phasing. The structure reveals the presence of a novel subunit IIa spanning the membrane. The model of this oxidase is analysed and compared with the P.denitrificans and bovine aa3-cytochrome oxidase structures, showing: (i) modified proton pumping pathways; (ii) one oxygen atom (µ-oxo species, hydroxide ion or water molecule) as the bridging ligand between CuB and haem as3; (iii) the exchange of the highly conserved Glu278P to Ile235 close to the haem as3 CuB centre, which optimizes the O2 input channel; (iv) an alternative electron transfer path leading directly from CuA to CuB; and (v) that the interaction of the ba3-oxidase with its substrate cytochrome c552 is based mainly on hydrophobic interactions.
Results and discussion
Overall structure
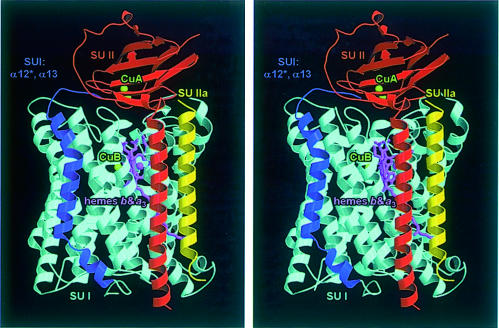
An overall view of the T.thermophilus ba3-cytochrome oxidase complex (84 884 Da, 764 residues) parallel to the membrane is presented in Figure 1. The final model is composed of three protein subunits, I, II and IIa, and forms a bundle of 15 transmembrane helices (54 × 50 Å) as well as a small periplasmic domain (35 × 29 × 20 Å). The main part of the complex is formed by subunit I with 13 transmembrane helices, which binds the haems b and as3 as well as CuB. It is associated with one transmembrane helix each of subunit II and subunit IIa. Except for the N-terminal 13 residues and residues 495–501 of subunit I, the two N-terminal residues of subunit II, one N-terminal residue of subunit IIa and very few solvent-exposed side chains, all amino acids have well-defined electron density. All N-termini of the three subunits are on the cytoplasmic (negative) side of the membrane and all C-termini are on the periplasmic side. The periplasmic domain of subunit II contains the primary electron acceptor, CuA. Haem b and haem as3 are located in the hydrophobic core of subunit I ∼15 Å from the periplasmic surface. The distances from the CuA atom CU1 (located closer to the membrane) to the haem b Fe, haem as3 Fe and CuB are 19.0, 21.8 and 21.6 Å, respectively. The distance between the haem b Fe and haem as3 Fe is 13.9 Å. The CuB is located 4.4 Å away from haem as3 Fe. The structure also shows the presence of three nonyl-β-d-glucoside detergent molecules. No phospholipid molecules and no additional metal ions have been seen in the structure as expected from biochemical analyses. No P, Mg, Ca or Zn could be detected using IPC-AES (data not shown).
Fig. 1. Stereo ribbon plot of the ba3-cytochrome c oxidase from T.thermophilus viewed parallel to the membrane. The 12 transmembrane helices of subunit I that are also present in the other oxidases are shown in cyan, and the additional transmembrane segment (helices α12* and α13) in dark blue. Subunit II is represented in red, and the new subunit IIa, which corresponds to the first transmembrane helix of subunit II of other oxidases, in yellow. The haem prosthetic groups are depicted as stick models in purple, and the copper atoms as green balls. This figure was created with MOLSCRIPT (Kraulis, 1991) and rendered with RASTER3D (Merritt and Murphy, 1994).
Subunit I
The 13th transmembrane helix of subunit I is an unusual property in the oxidase superfamily; subunit I consists of only 12 transmembrane helices in most other haem copper oxidases. This 13th helix does not superimpose with any of the 22 transmembrane helices of P.denitrificans or the 28 α-helices of the bovine heart oxidase. The loops connecting the 13 helices of subunit I are shorter than in the other cytochrome oxidases. This is a typical property of thermostable proteins (Adams and Kelly, 1998). Edman degradation of the crystals shows that the gene-deduced N-terminal sequence (MAVRASEIS) is missing from the crystals. The following four N-terminal residues (RVYE) are disordered and not visible in the electron density map. The previously described pores A, B and C (Iwata et al., 1995) can be identified if the molecule is viewed from the periplasmic side. Pore A is blocked by several aromatic residues, pore B by the binuclear centre haem as3 CuB and pore C by haem b. The C-terminal part (551–560) of subunit I wraps around the top of this subunit and is located at the edge of the interface between subunits I and II on the top of pore A.
Haem b and haem as3 CuB centre
Haem b is the simplest protohaem containing a low-spin iron with the two histidine residues His72/α2 and His386/α10 as axial ligands at distances of 2.2 Å each. The coordination sphere of the high-spin haem as3 iron consists of five protein constituents and is completed by the bridging ligand at the active site. The iron is in the plane of the haem and the distance to the axial histidine ligand (His384/α10) is 3.3 Å, a considerably larger distance than described in the current oxidase structures (2.1 Å for the P.denitrificans and 1.9 Å for the bovine heart oxidases). This finding is in accordance with recent resonance Raman data (Kim et al., 1998; Gerscher et al., 1999). Haem as3 is characterized by the presence of a formyl group at C8 and a hydrophobic hydroxyethylgeranylgeranyl moiety instead of a hydroxyethylfarnesyl side chain on C2. For the aa3-oxidase from P.denitrificans, the hydroxyethylfarnesyl group leaves pore B and penetrates into the lipid bilayer, while in the ba3-oxidase structure the geranylgeranyl side chain is straight and reaches the cytoplasmic side, but it does not interfere with the proton pathways.
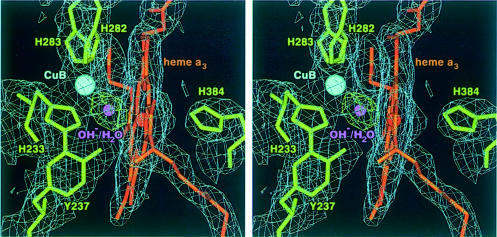
Figure 2 shows the final model and the 2Fobs – Fcalc and Fobs – Fcalc electron density maps in the vicinity of the binuclear haem as3 CuB centre. The electron density of the three histidine ligands to CuB, His283/α7, His282/α7 and His233/α6, as well as the covalent bridge between the imidazole Nε2 (His233/α6) and the Cε2 of the phenol ring (Tyr237/α6) is well defined. The distance between the haem as3 Fe and the Tyr237 OH is 5.6 Å. The observed Fobs – Fcalc difference electron density between haem as3 and CuB is located symmetrically between the two metals and is of almost spherical shape. It is best interpreted as one oxygen atom (µ-oxo species, hydroxide ion or water molecule). This oxygen atom is at a distance of 2.3 Å from each of the two metals in the final refined structure.
Fig. 2. Stereo representation of the binuclear centre haem as3 CuB including the final 2Fobs – Fcalc electron density map contoured at 1.0σ (blue). Haem as3, the histidine ligands and the covalently linked Tyr237 are shown as stick models in orange and green, respectively. The covalent bond between Tyr237 and His233 is well defined in the electron density. The Fobs – Fcalc difference electron density (contoured at 5.0σ, green) between the haem as3 iron and CuB (cyan) is of almost spherical shape and is best interpreted as one oxygen atom (O2–, OH– or H2O; purple), located equidistant between the two metal atoms. This figure was prepared with MAIN (Turk, 1992).
In the bovine structure (Yoshikawa et al., 1998a), the bridging ligand was interpreted as an O–O for the resting state of the enzyme, while in the case of the P.denitrificans structure two ligands, one water molecule at the haem a3 Fe and one hydroxide ion at the CuB within hydrogen bonding distance, were favoured (Ostermeier et al., 1997). The identification of the correct ligation at the oxygen-binding site in the oxidized state of the enzyme is important for the elucidation of the proton pumping mechanism (Michel, 1998). We have tested these models in the ba3-oxidase and found that the introduction of either an O–O moiety or two ligands at this site leads to a significantly poorer fit to the observed electron density, with very high B-factors for the second oxygen atom (33.4 Å2 for the one oxygen species; 34.6 and 51.3 Å2 for the O–O model; and 31.7 and 58.1 Å2 for the H2O/OH– model). Recently published electrochemically induced UV/VIS difference spectra (Hellwig et al., 1999) suggest that no peroxy ligand is bound in the oxidized (as prepared) state of the ba3-oxidase.
Subunit II, cytochrome c-binding site, CuA centre and the novel subunit IIa
Subunit II has only one transmembrane helix (residues 4–38), which is firmly bound to subunit I. It is the only subunit with a polar domain, which consists of a 10-stranded β-barrel (β1 71–75, β2 78–86, β3 89–91, β4 95–97, β5 102–108, β6 114–118, β7 124–127, β8 133–138, β9 143–148 and β10 161–166). This domain is located at the periplasmic side of the membrane and shows similarities to the class I copper proteins, but with a binuclear CuA site. The membrane-spanning helix α1 and the first β-sheet are connected by two short helices (α2 and α3, Figure 3) and a long loop. In the structure of the engineered soluble domain (Williams et al., 1999), this so-called ‘linker region’ is located far from the transmembrane part as deduced from a comparison with other oxidase structures. The authors suggest that the structure of this linker region is influenced by crystal lattice forces (Williams et al., 1999). The current structure provides the native conformation of both the transmembrane helix of subunit II and this linker region.
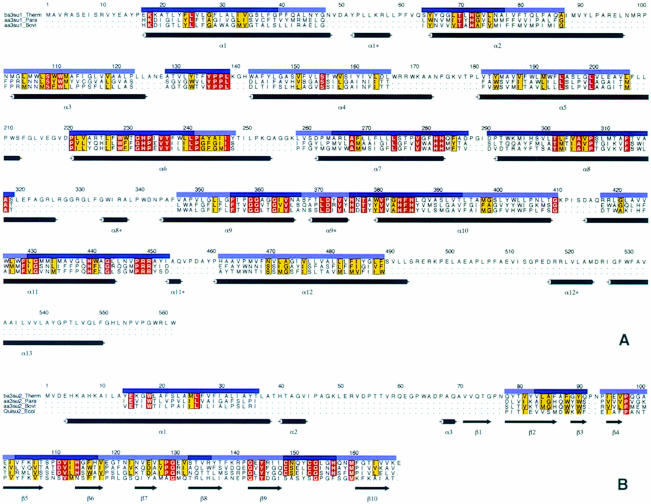
Fig. 3. Structure-based sequence alignment of subunits I (A) and II (B) of the T.thermophilus ba3-oxidase with the aa3-oxidases from P.denitrificans and bovine heart as well as the engineered soluble CuA-binding domain of the quinol oxidase from E.coli. The transmembrane helices are numbered α1–13 for subunit I and α1 for subunit II. Short connecting helices of subunit I are marked with an asterisk (*) and the β-sheets of subunit II are numbered β1–10. The alignment is shown only for those residues that occupy comparable positions in space (∼60%), and the quality of this structural alignment is indicated with dark and light blue bars for residues at almost identical positions (r.m.s.d. of these Cα atoms 1.1 Å) and those that can be identified as being at similar positions within the structure (r.m.s.d. of these Cα atoms 2.0 Å), respectively. All other residues show no structural similarity between the T.thermophilus ba3-oxidase and other cytochrome oxidases. Strictly conserved amino acids are shown in red, and those with similar chemical characteristics in yellow. This figure was created with ALSCRIPT (Barton, 1993).
The CuA site is symmetrical with respect to the two histidine (His114 and His157) and the two cysteine (Cys149 and Cys153) ligands. The distance between the two copper atoms is 2.4 Å. The imidazole rings are not co-planar as described for the engineered soluble CuA domain (Williams et al., 1999), but the angle between these two planes is smaller than in other cytochrome c oxidases (∼25°). The two thiolate groups of Cys149 and Cys153 act as bridging ligands between the copper atoms. CU2 of the CuA centre is coordinated by Cys149, Cys153, His114 and Met160, with distances of 2.4, 2.4, 2.1 and 2.5 Å, respectively. The distances are in good agreement with those found for the structure of the engineered soluble domain (Williams et al., 1999). The second copper of CuA (CU1) is coordinated by Cys149, Cys153, His157 and Gln151. The distances are 2.5 (2.3), 2.3 (2.3), 2.1 (1.9) and 2.8 (2.6) Å on average longer than those observed for the 1.6 Å structure of the engineered soluble domain (values given in parentheses).
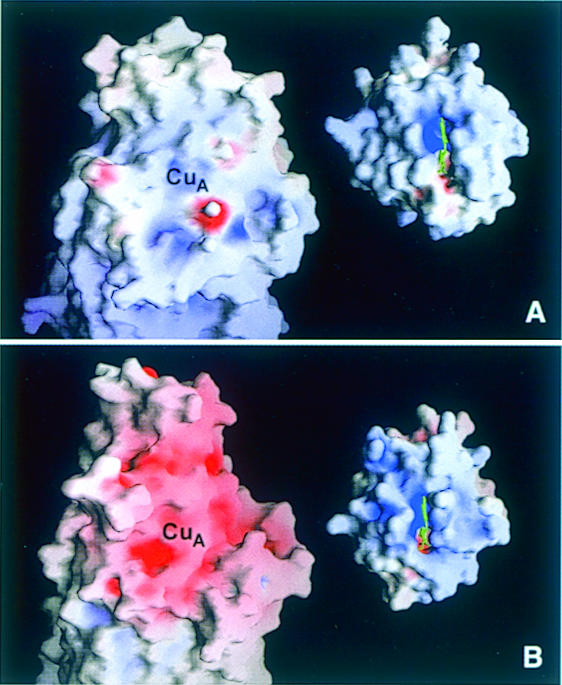
Ludwig and co-workers (Witt et al., 1998a,b) proposed that the highly conserved residues Trp121P and Tyr122P might be involved in the electron transfer from cytochrome c to CuA and that Glu126P, Asp135P, Asp159P and Asp178P are part of the potential binding site for cytochrome c. As shown in Figure 3, Trp121P and Tyr122P are replaced by Phe88 and Gly89, respectively. The aromatic ring of Phe86 occupies almost the same space as Tyr122P and might, together with Phe88, substitute for the otherwise conserved residues. The remaining acidic amino acid residues, except for Asp111, are not conserved in the ba3-oxidase, and the proposed cytochrome c-binding surface of the CuA domain contains, in contrast to the P.denitrificans and bovine enzymes, almost only hydrophobic residues. This corresponds to the hydrophobic character of the surface around the exposed haem edge of cytochrome c552 (Than et al., 1997; Figure 4). We conclude that the interactions between cytochrome c552 and the ba3-oxidase are mainly hydrophobic. This is in good agreement with the generally lowered stability of electrostatic interactions at higher temperatures and it is supported further by the experimental data of Giuffré et al. (1999) which show that, in contrast to the bovine and P.denitrificans oxidases, no tight electrostatic complex is formed between the ba3-oxidase and its substrate cytochrome c552.
Fig. 4. Solid surface representation of the electrostatic potential of the proposed cytochrome c-binding surface near the mixed valence CuA site of cytochrome c oxidases (left) and the corresponding front face of cytochrome c (right) for (A) the T.thermophilus ba3-oxidase and its substrate cytochrome c552 (Than et al., 1997) and (B) the bovine heart aa3-oxidase (Tsukihara et al., 1996) and horse heart cytochrome c (Bushnell et al., 1990). Colouring is according to the calculated electrostatic potential of the oxidized oxidases and the reduced cytochromes, and contoured from –40 kT/e (intense red) to 40 kT/e (intense blue). This figure was prepared with GRASP (Nicholls et al., 1993).
Surprisingly, after placing all of the visible residues of subunits I and II into the experimental electron density, one additional transmembrane-spanning helix was found. The corresponding protein subunit was isolated and its amino acid sequence was determined by automated Edman degradation (data not shown). It consists of 34 amino acid residues, forming one transmembrane helix. This helix superimposes with the first transmembrane helix of subunit II of the two structurally known cytochrome c oxidases, but with opposite polarity. The presence of this transmembrane helix seems to be important for the function of cytochrome c oxidases. As was deduced from their sequences, the SoxB-type quinol and cytochrome c oxidases from S.acidocaldarius and Natronobacterium pharaonis both lack the first transmembrane helix in subunit II. They also contain an additional small polypeptide, which was termed SoxD and subunit IV for these enzymes, respectively (Lübben et al., 1992; Mattar and Engelhard, 1997). The sequence of this short subunit from the N.pharaonis enzyme shows 38% identity to subunit IIa of the T.thermophilus oxidase (data not shown). There is no indication for the presence of additional subunits in the T.thermophilus enzyme which may correspond to either apocytochrome b (SoxC) of the S.acidocaldarius oxidase or subunit III of the N.pharaonis oxidase.
Electron transfer
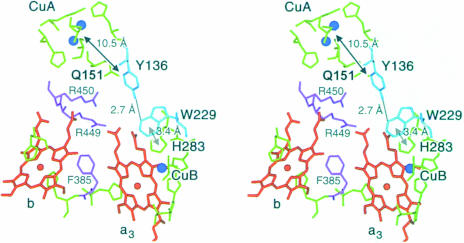
The structure-based sequence alignment of subunit I (Figure 3) between the phylogenetically distant ba3-oxidase and the P.denitrificans and bovine heart oxidases shows that functionally vital residues, such as haem and Cu ligands or the residues proposed for the electron transfer from CuA to the haems, are conserved. In addition to these residues, a highly conserved motif (YPPL, located between α3 and α4) can be discerned from the alignment. Tyr136 forms a tight hydrogen bond to Trp229, which is also conserved. The π-system of this residue shows, in turn, a parallel stacking interaction with the π-system of His283, one ligand for CuB. These residues might form an electron transfer pathway from CuA directly to CuB, which, except for the relatively long distance between CuA and Tyr136 (10.5 Å), involves several π-systems and very few strong hydrogen and coordinative bonds as well as the above-mentioned π-stacking interaction (Figure 5). The electrons may be passed from CuA to the Tyr136 π-system by tunnelling effects (Moser et al., 1992; Page et al., 1999), which can be as fast as 109/s for distances of ∼10 Å, or via the CuA ligand Gln151. The two prolines and the leucine are probably conserved for structural reasons in order to provide the correct orientation of Tyr136. In accordance with the above-mentioned conservation of these residues, the same structural arrangement is found in the P.denitrificans and bovine heart oxidases, and a possible involvement in electron transfer has been suggested (Iwata et al., 1995; Tsukihara et al., 1996).
Fig. 5. Stereo representation of the electron transfer pathways in the ba3-cytochrome c oxidase from T.thermophilus. The haems b and as3 are shown in orange, the copper atoms in blue and the metal-ligating amino acid residues in green. The two arginine residues (Arg450 and Arg449) and Phe385, which are involved in the electron transfer from CuA via haem b to the active site haem as3 CuB are depicted in purple. The residues that form the newly postulated electron transfer pathway leading from CuA to the aromatic ring system of Tyr136 and from there via a hydrogen bond and Trp229 to the CuB ligand His238 are represented in cyan, including the corresponding distances. This figure was prepared with MAIN (Turk, 1992).
The current understanding of the oxygen reduction mechanism at the binuclear centre requires the input of at least one of the electrons via CuB (Hill, 1994; Michel, 1998). The electron transfer from CuA via haem a/b to haem as3 at the binuclear centre is well established (Hill, 1994), and the corresponding residues (Arg450, Arg449 and Phe385) are conserved in the ba3-oxidase. We propose the above-described pathway via Tyr136, Trp229 and His283 as an additional electron transfer pathway which should be used for electrons that are provided from CuB to the catalytic oxygen intermediates. To our knowledge, such a direct electron transfer from CuA to CuB has never been discussed, and experimental data will be needed to prove its mechanistic significance. Kinetic experiments of internal electron transfer (Hill, 1994) show that electrons that are passed through haem a3 are transferred via haem a/b, not supporting a model of direct electron transfer from CuA to haem a3, as proposed by Yoshikawa et al. (1998b), but not excluding the possibility of a direct electron transfer from CuA to CuB. Since the redox state of CuB is difficult to assess spectrophotometrically (Hill, 1994), the direct observation of internal electron transfer steps is usually limited to the other three redox centres.
Proton pathways, O2 channel and water pool
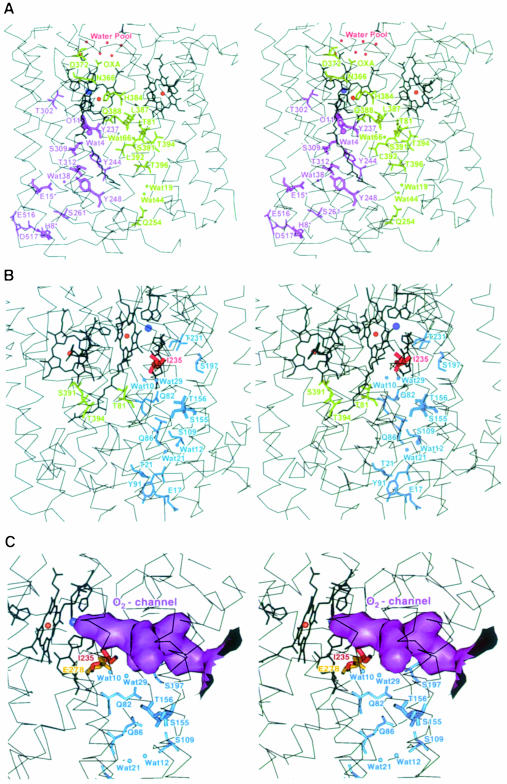
The crystal structure of the ba3-oxidase reveals three possible proton pathways, which originate at the cytoplasmic side of the membrane and consist of several polar amino acid residues and crystallographically well-defined water molecules connected by hydrogen bonding networks. In principle, they could provide for the transfer of protons from the cytoplasmic side to either the periplasmic side of the membrane or the active site. Two of these pathways have similarities to the previously identified ‘K- and D-pathways’ (Iwata et al., 1995); however, most of the key residues are not conserved.
The first pathway is formed by residues that belong mainly to the transmembrane helices α6 and α8 (Figure 6A) and leads to the conserved active site tyrosine Tyr237 (α6), which is covalently linked to the CuB ligand His233. Its spatial location within the molecule is comparable with the previously described ‘K-pathway’. The otherwise highly conserved residues Thr351P and Lys354P, which in the other oxidases, upon mutation to non-polar residues, result in either a less active or a completely inactive form (Gennis, 1998a), are replaced by Ser309 and Thr312, respectively. Other residues that are part of this hydrogen bonding network include the haem as3 OH, Tyr244, Tyr248 and two internal water molecules, Wat38 and Wat4. The lower part of this pathway is formed by Asp517, His8 (SUII), Glu516, Glu15 (SUII) and Ser261. In spite of the generally low homology of this pathway to its counterparts in the bovine and P.denitrificans structures, Glu15 of subunit II is part of this hydrogen bonding network in all three structures and may be more important for the function of the oxidases than previously assumed. This finding is also in agreement with recent data of Ma et al. (1999).
Fig. 6. Stereo representation of the proton pathways, the O2 channel and the water pool in the ba3-cytochrome c oxidase from T.thermophilus including the Cα traces of the transmembrane helices (thin black lines), the haems and metal ligands (thick black lines) as well as the metal atoms as red and blue balls for the haem iron and copper atoms, respectively. Residues that belong to subunit II are marked by an asterisk. (A) The new ‘Q-pathway’ leading from the cytoplasmic side of the membrane via the haem as3 coordinating His384 to the water pool on top of the haem propionates is represented in green, and the pathway that corresponds to the classical ‘K-pathway’ is shown in purple. (B) Residues of the proton pathway, which corresponds to the classical ‘D-pathway’, are shown in blue, Thr81 and Thr394 that may connect it to Ser391 of the ‘Q-pathway’ are in green and Ile235 in red. (C) Solid surface representation of the O2 input channel (purple), which extends towards the active site including the upper part of the ‘D-pathway’(blue) and Ile235 (red). The highly conserved amino acid Glu278P (yellow), which is indispensable for the proton pumping activity of other oxidases, occupies there the same space as Ile235. This figure was prepared with MAIN (Turk, 1992).
The second pathway (Figure 6B) leads from Glu17 (α1), via Tyr91 (α2), Thr21 (α1), two water molecules, Wat21 and Wat12, and a number of polar residues such as Ser109 (α3), Gln86 (α2), Ser155 (α4), Thr156 (α4) and Gln82 (α2), to an internal cavity that is relatively close to the active site (12.6 Å) and filled with the water molecules Wat10 and Wat29. The further path of the protons is less clear. They could approach the binuclear centre either directly from this internal cavity or via Ser197 and Thr231. Alternatively, the protons may be transferred via Thr81 and Thr394 to Ser391 of the third pathway, which will be discussed below. The distance between the side chain Oγ1 atoms of the two threonines as resolved in the crystal structure is ∼8 Å, which is relatively large. However, if one considers rotation of their side chains, this distance can be reduced to ∼5 Å, which is significantly shorter than the distance between either Wat10/29 or Thr231 and the active site. The spatial location of this pathway within the molecule is comparable with the previously described ‘D-pathway’. However, even fewer residues are conserved within the primary structure than for the ‘K-pathway’. In particular, one of the key residues, Glu278P, is replaced in sequence and space by the hydrophobic Ile235. Even though this glutamate is indispensable for the functionality of the ‘D-pathway’ and the proton pumping activity in other oxidases (Gennis, 1998a), the ba3-oxidase does pump protons, but with reduced efficiency (Kannt et al., 1998).
The presence of Ile235 in the ba3-oxidase optimizes the formation of a hydrophobic pore leading from the middle of the membrane to the binuclear centre (Figure 6C). A considerably smaller hydrophobic pore could be identified in the P.denitrificans structure and was discussed as an oxygen input channel (Riistama et al., 1996). Since the ba3-oxidase is expressed only under limited oxygen supply, in addition to the reduced solubility of gaseous oxygen in water at elevated temperatures, the evolutionary development of an optimized oxygen input channel seems appropriate. This finding is in good agreement with the primary structures of the archaeal S.acidocaldarius aa3-quinol oxidase and N.pharaonis ba3-cytochrome oxidase, where Glu278P is replaced by valine. These organisms grow also under low oxygen tension and at high temperatures. The replacement of Glu278P by Ile235 or valine eliminates one of the key residues discussed as being essential for proton pumping in other oxidases and may be responsible for the reduced proton pumping efficiency of the ba3-oxidase. The formation of an optimized, hydrophobically coated oxygen input channel and hence the supply of the oxidase with sufficient amounts of substrate is probably more important for the function of the enzyme under limited oxygen concentration than proton pumping.
The third proton pathway, termed the ‘Q-pathway’, starts at the cytoplasmic side of the membrane with Gln254 and leads via two internal water molecules, Wat44 and Wat19, Thr396, the carbonyl oxygen of Leu392, Wat66 and Ser391 to the above-mentioned junction to the ‘D-pathway’ close to Thr394 and Thr81 (Figure 6A). From there, it continues via Gln388, the carbonyl oxygen of Leu387, the haem as3 ligand His384, Asn366, Asp372 and the propionate of the haem as3 pyrole ring A to a water pool, which is located above the haem propionates. It is equivalent neither to the so-called ‘H-pathway’, which was described in detail for the bovine oxidase (Yoshikawa et al., 1998a), nor to the similar ‘E-pathway’ (Pfitzner et al., 1998) reported for the P.denitrificans oxidase (Ostermeier et al., 1997). The residues involved in the hydrogen bond network of the ‘H- and E-pathways’ are not conserved in the ba3-oxidase structure. This ‘Q-pathway’ could either transfer protons independently or be involved in the pumping of protons that originate from the ‘D-pathway’. In the latter case, such protons would be translocated to Ser391 of the ‘Q-pathway’ via Thr81 and Thr394. Interestingly, this pathway includes the axial haem as3 histidine ligand His384, which shows a weaker bond to the haem as3 iron, as compared with the other oxidase structures. One might postulate that His384 undergoes structural changes during the catalytic cycle resulting in an active participation in proton pumping. The lowered H+-pumping activity of the ba3-oxidase may then be correlated with either a completely different H+-pumping mechanism including this His384 and not Glu278P or with the differences in the ligation of haem as3 by His384.
The described modifications at the proton pathways, i.e. the probably lowered proton uptake activity of the ‘D-pathway’, are in good agreement with the lowered proton pumping efficiency of the ba3-oxidase (Kannt et al., 1998) and a proton pumping mechanism that is based on electrostatic repulsion and charge compensation, as discussed for instance by Mitchell and Rich (1994), Rich (1995) or Michel (1998). Less efficient proton uptake from the cytoplasmic side would result in a lowered electrostatic repulsion of the protons at or near the active site and hence less efficient proton pumping. In contrast, mechanisms where the primary proton pumping step is linked directly to the redox changes at one of the redox centres and must always result in the pumping of one proton per electron transferred through this site seem less favourable. Such a mechanism should always lead to the pumping of four protons per dioxygen molecule consumed by the enzyme, which does not agree with the experimental data for the ba3-oxidase. However, it cannot be excluded that multiple electron transfer pathways exist. If they are used to various extents in different oxidases, then the lowered proton pumping efficiency could be explained alternatively by these effects.
The above-mentioned water pool on top of the haem propionates is conserved among all structurally known oxidases, and many surrounding residues are either invariant or replaced by amino acids with very similar chemical properties. It is connected to the bulk solvent on the periplasmic side of the membrane via several well-developed pathways consisting of polar amino acid side chains and additional water molecules that are hydrogen bonded. This accumulation of water molecules was also identified for the P.denitrificans oxidase, and its involvement in the proton exit channel(s) was supported further by mutagenesis experiments (Ostermeier et al., 1997; Puustinen and Wikström, 1999). Based on the general existence of this water pool in all oxidases, its localization on top of the haem propionates and the obviously fast equilibrium with the bulk solvent, we propose it as the primary acceptor not only for pumped protons but also for the water molecules formed during the catalytic turnover of the enzyme.
Materials and methods
Purification and crystallization
The purification of ba3-oxidase was described briefly by Giuffré et al. (1999). Further details and a protein chemical description of the terminal oxidase will be given elsewhere. For the batch crystallization, 0.5 ml of the protein solution (8 mg/ml) in 0.4% nonyl-β-d-glucoside-containing 10 mM Tris–HCl buffer pH 7.0 was transferred into a 1.5 ml Eppendorf tube, adjusted to 6% polyethylene glycol 2000 as precipitant, and sealed. Crystals appear after 5 days. Crystals were also grown by vapour diffusion in sitting drops containing 6 µl of protein solution and 2.5 µl from the 1 ml reservoir solution containing 14% polyethylene glycol 2000 as precipitant in 20 mM Bis–Tris buffer pH 7.0 at 20°C. Both crystallization methods led to crystals of about equal quality, but of different size and form (data not shown). They were harvested after 1 week, mounted in 1 mm glass capillaries and analysed using X-ray rotation photography. The crystals were 0.3–2 mm in size and belong to the space group P41/3212. The cell constants changed during data collection at room temperature (a = b = 112–116 Å, c = 174–190 Å). Hence, the finding of a cryo condition was indispensable.
Cryogenic data collection and structure determination
X-ray diffraction data for MAD phasing were collected at the wiggler beamline BW6 at DORIS (DESY Hamburg, Germany). In a first step, the crystal was mounted in a standard cryo-loop using paraffin oil as cryoprotectant without freezing. The crystal lattice undergoes a transformation due to evaporation of solvent through the oil layer. This transformation was followed via diffraction images. The crystal was frozen after the quality of the images had reached its optimum, thereby preserving the system in this optimal state. This procedure led to an increase in diffraction power from initially ∼3 Å to 2.1 Å, and a decrease in cell constants to a = b = 112.11 Å, c = 161.41 Å. The crystals contain one molecule of ba3-oxidase per asymmetric unit. This results in a Matthews coefficient (Matthews, 1968) of 2.98 Å3/Da, which corresponds to a solvent content of 59% in their transformed state (native crystals: 3.50 Å3/Da, 65% solvent). With a complete data set, it was not possible to solve the structure applying Patterson search techniques employing the two known cytochrome c oxidase structures (Tsukihara et al., 1996; Ostermeier et al., 1997).
MAD data were measured from one frozen crystal at 100 K at five different wavelengths comprising the Cu and Fe K-absorption edges as well as one remote point (Table I). Diffraction data were collected up to 2.4 Å in frames of 0.2° through a continuous range of 70° followed by another continuous range of 70° for measuring Friedel opposites in inverse beam geometry using a 130 mm MAR CCD detector (Mar Research, Hamburg, Germany). These data were processed with DENZO/SCALEPACK (Otwinowski and Minor, 1993).
Table I. Summary of data collection.
| Data seta | Wavelength (Å) | Rsym (%)b | Completeness |
Unique reflections | |
|---|---|---|---|---|---|
| Overall | Anomalous | ||||
| Remote | 1.07 | 4.3 (14.7) | 99.6 | 99.5 | 39 379 |
| Fe-f′ | 1.7399 | 4.7 (18.6) | 99.8 | 99.8 | 40 384 |
| Fe-f′′ | 1.7371 | 4.7 (29.5) | 99.9 | 99.9 | 40 293 |
| Cu-f′ | 1.3805 | 4.7 (30.5) | 99.7 | 99.6 | 40 219 |
| Cu-f′′ | 1.3782 | 4.8 (29.3) | 99.8 | 99.8 | 40 351 |
Anomalous and dispersive difference Patterson syntheses were calculated from each of the MAD data sets resulting in a clear solution for one of the iron sites. Subsequent phasing and analysis of the residual maps using SHARP (de La Fortelle and Bricogne, 1997) gave rise to the second Fe and the three Cu sites as well as the correct enantiomorph (space group P43212). The final phases were calculated with SHARP using all five wavelengths, followed by 130 cycles of density modification with SOLOMON (CCP4, 1995) as implemented in the SHARP interface. The resulting electron density map was of exceptional quality (Figure of merit after SHARP: acentrics, 0.713; centrics, 0.609; after SOLOMON: 0.825) and most of the polypeptide chain could be built, including the side chains in the initial cycle of model building. The atomic model was built using MAIN (Turk, 1992) and refined in CNS v0.3 (Brünger et al., 1998). Phase combination of MAD and model phases using SFALL (CCP4, 1995) and SIGMAA (CCP4, 1995) followed by density modification using either DM (CCP4, 1995) or SOLOMON proved useful in the determination of some less well ordered residues during the initial cycles of model building. The target parameters of Engh and Huber (1991) were used throughout the refinement. During the final refinement cycles, bulk solvent correction and overall anisotropic B-factor scaling were applied. Water molecules were added automatically with ARP (Lamzin and Wilson, 1997) and optimized further during the final cycles of model inspection. The model quality is summarized in Table II and was checked with PROCHECK (Laskowski et al., 1993) and SFCHECK (CCP4, 1995). The coodinates were deposited with the Protein Data Bank (PDB accession No. 1EHK).
Table II. Summary of refinement statistics and final stereochemistry.
| Resolution (Å) | 20.0–2.4 |
| No. of non-H protein atoms | 5851 |
| No. of heterogen atoms | 174 |
| No. of water molecules | 119 |
| Rcrysta,b (%) | 22.2 (25.4) |
| Rfreea,c (%) | 26.4 (29.9) |
| Average B (Å)2 | 50.6 |
| R.m.s. deviation bond length (Å) | 0.008 |
| R.m.s. deviation bond angles (°) | 1.6 |
| R.m.s. deviation bonded B-factors (Å)2 | 3.853 |
aValues in parentheses correspond to the last resolution shell from 2.49 to 2.40 Å.
b![]() where Foh and Fch are the observed and the calculated structure factor amplitudes for reflection h.
where Foh and Fch are the observed and the calculated structure factor amplitudes for reflection h.
cRfree was calculated by randomly omitting 5% of the observed reflections from refinement and R-factor calculation.
Structural analysis
The secondary structure of the enzyme was analysed using the program DSSP (Kabsch and Sander, 1983). Single residues inside the transmembrane helices that do not show ideal helical geometry were included in the assignment of the transmembrane helices. The structure of the ba3-oxidase from T.thermophilus was compared with that of the bacterial aa3-oxidase from P.denitrificans (Ostermeier et al., 1997) (PDB accession No. 1AR1), the mitochondrial aa3-oxidase from bovine heart (Tsukihara et al., 1996) (PDB accession No. 2OCC), the periplasmic, soluble fragments of the E.coli quinol oxidase (Wilmanns et al., 1995), which was engineered to contain a CuA centre (PDB accession No. 1CYX) and the ba3-cytochrome c oxidase from T.thermophilus (Williams et al., 1999) (PDB accession No. 2CUA) by r.m.s. alignment of either the five metal atoms (for comparison with the other two cytochrome oxidases), the two CuA atoms and their directly liganding atoms (quinol oxidase) or the Cα atoms (soluble fragment of the ba3-oxidase).
Acknowledgments
Acknowledgement
This work was supported by the Deutsche Forschungsgemeinschaft (Bu 463/3-2).
References
- Adams M.W.W. and Kelly,R.M. (1998) Finding and using hyperthermophilic enzymes. Trends Biotechnol., 16, 329–332. [DOI] [PubMed] [Google Scholar]
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Protein Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buse G., Soulimane,T., Dewor,M., Meyer,H.E. and Blüggel,M. (1999) Evidence for a copper coordinated histidine–tyrosine crosslink in the active site of cytochrome oxidase. Protein Sci., 8, 985–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell G.W., Louie,G.V. and Brayer,G.D. (1990) High-resolution three-dimensional structure of horse heart cytochrome c. J. Mol. Biol., 214, 585–595. [DOI] [PubMed] [Google Scholar]
- Castresana J. and Saraste,M. (1995) Evolution of energetic metabolism: the respiration early hypothesis. Trends Biochem. Sci., 20, 443–448. [DOI] [PubMed] [Google Scholar]
- Collaborative Computing Project No. 4 (1995) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelengh anomalous diffraction methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure and refinement. Acta Crystallogr. A, 7, 392–400. [Google Scholar]
- Ferguson-Miller S. and Babcock,G.T. (1996) Heme/copper terminal oxidases. Chem. Rev., 96, 2889–2907. [DOI] [PubMed] [Google Scholar]
- Gennis R.B. (1998a) Multiple proton-conducting pathways in cytochrome oxidase and a proposed role for the active-site tyrosine. Biochim. Biophys. Acta, 1365, 241–248. [Google Scholar]
- Gennis R.B. (1998b) Cytochrome c oxidase: one enzyme, two mechanisms? Science, 280, 1712–1713. [DOI] [PubMed] [Google Scholar]
- Gerscher S., Hildebrandt,P., Buse,G. and Soulimane,T. (1999) The active site structure of the ba3 oxidase from Thermus thermophilus studied by resonance Raman spectroscopy. Biospectroscopy, 4, 365–377. [DOI] [PubMed] [Google Scholar]
- Giuffré A., Forte,E., Antonini,G., D′Itri,E., Brunori,M., Soulimane,T. and Buse,G. (1999) Kinetic properties of the ba3-oxidase from Thermus thermophilus: effect of temperature. Biochemistry, 38, 1057–1065. [DOI] [PubMed] [Google Scholar]
- Hellwig P., Soulimane,T., Buse,G. and Mäntele,W. (1999) Electrochemical, FTIR, and UV/VIS spectroscopic properties of the ba3-oxidase from Thermus thermophilus. Biochemistry, 38, 9648–9658. [DOI] [PubMed] [Google Scholar]
- Hill B.C. (1994) Modeling the sequence of electron transfer reactions in the single turnover of reduced, mammalian cytochrome c oxidase with oxygen. J. Biol. Chem., 269, 2419–2425. [PubMed] [Google Scholar]
- Iwata S., Ostermeier,C., Ludwig,B. and Michel,H. (1995) Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature, 376, 660–669. [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander,C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- Kannt A., Soulimane,T., Buse,G., Bamberg,E. and Michel,H. (1998) Electrical current generation and proton pumping catalysed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett., 434, 17–22. [DOI] [PubMed] [Google Scholar]
- Keightley J.A., Zimmermann,B.H., Mather,M.W., Springer,P., Pastuszyn,A., Lawrence,D.M. and Fee,J.A. (1995) Molecular genetic and protein chemical characterization of the cytochrome ba3 from Thermus thermophilus HB8. J. Biol. Chem., 270, 20345–20358. [DOI] [PubMed] [Google Scholar]
- Kim Y., Babcock,G.T., Surerus,K.K., Fee,J.A., Dyer,R.B., Woodruff,W.H. and Oertling,W.A. (1998) Cyanide binding and active site structure in heme-copper oxidases: normal coordinate analysis of iron-cyanide vibrations of a3 (2+)CN– complexes of cytochromes ba3 and aa3. Biospectroscopy, 4, 1–15. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lamzin V.S. and Wilson,K.S. (1997) Automated refinement for protein crystallography. Methods Enzymol., 277, 269–305. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Lübben M. and Morand,K. (1994) Novel prenylated hemes as cofactors of cytochrome oxidases. Archaea have modified hemes A and O. J. Biol. Chem., 269, 21473–21479. [PubMed] [Google Scholar]
- Lübben M., Kolmerer,B. and Saraste,M. (1992) An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J., 11, 805–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J. et al. (1999) Glutamate-89 in subunit II of cytochrome bo3 from Escherichia coli is required for the function of the heme-copper oxidase. Biochemistry 38, 15150–15156. [DOI] [PubMed] [Google Scholar]
- Mattar S. and Engelhard,M. (1997) Cytochrome ba3 from Natronobacterium pharaonis. An archaeal four-subunit cytochrome-c- type oxidase. Eur. J. Biochem., 250, 332–341. [DOI] [PubMed] [Google Scholar]
- Matthews B.W. (1968) Solvent contents of protein crystals. J. Mol. Biol., 33, 491–497. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0: a program for photorealistic molecular graphics. Acta Crysatallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Michel H. (1998) The mechanism of proton pumping by cytochrome c oxidase. Proc. Natl Acad. Sci. USA, 95, 12819–12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel H. (1999) Cytochrome c oxidase: catalytic cycle and mechanisms of proton pumping—a discussion. Biochemistry, 38, 15129–15140. [DOI] [PubMed] [Google Scholar]
- Michel H., Behr,J., Harrenga,A. and Kannt,A. (1998) Cytochrome c oxidase: structure and spectroscopy. Annu. Rev. Biophys. Biomol. Struct., 27, 329–356. [DOI] [PubMed] [Google Scholar]
- Mitchell R. and Rich,P.R. (1994) Proton uptake by cytochrome c oxidase on reduction and on ligand binding. Biochim. Biophys. Acta, 1186, 19–26. [DOI] [PubMed] [Google Scholar]
- Moser C.C., Keske,J.M., Warncke,K., Farid,R.S. and Dutton,P.L. (1992) Nature of biological electron transfer. Nature, 355, 796–802. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) Grasp: graphical representation and analysis of surface properties. Biophys. J., A64, 166. [Google Scholar]
- Ostermeier C., Harrenga,A., Ermler,U. and Michel,H. (1997) Structure at 2.7 Å resolution of the Paracoccus denitrificans two-subunit cytochrome c oxidase complexed with an antibody Fv fragment. Proc. Natl Acad. Sci. USA, 94, 10547–10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1993) DENZO: A Film Processing Program for Macromolecular Crystallography. Yale University Press, New Haven, CT. [Google Scholar]
- Page C.C., Moser,C.C., Chen,X. and Dutton,P.L. (1999) Natural engineering principles of electron tunnelling in biological oxidation–reduction. Nature, 402, 47–52. [DOI] [PubMed] [Google Scholar]
- Pfitzner U., Odenwald,A., Ostermann,T., Weingard,L., Ludwig,B. and Richter,O.-M.H. (1998) Cytochrome c oxidase (heme aa3) from Paracoccus denitrificans: analysis of mutations in putative proton channels of subunit I. J. Bioenerg. Biomembr., 30, 89–97. [DOI] [PubMed] [Google Scholar]
- Puustinen A. and Wikström,M. (1999) Proton exit from the heme-copper oxidase of Escherichia coli. Proc. Natl Acad. Sci. USA, 96, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich P.R. (1995) Towards an understanding of the chemistry of oxygen reduction and proton translocation in the iron–copper respiratory oxidases. Aust. J. Plant Physiol., 22, 479–486. [Google Scholar]
- Riistama S., Puustinen,A., Garcia-Horsman,A., Iwata,S., Michel,H. and Wikström,M. (1996) Channelling of dioxygen into the respiratory enzyme. Biochim. Biophys. Acta, 1275, 1–4. [DOI] [PubMed] [Google Scholar]
- Silietzkyi S., Soulimane,T., Azarkina,N., Vygodina,T.V, Buse,G., Kaulen,A., and Konstantinov,A. (1999) Time-resolved generation of membrane potential by ba3 cytochrome c oxidase from Thermus thermophilus. Evidence for reduction-induced opening of the binuclear centre. FEBS Lett., 457, 98–102. [DOI] [PubMed] [Google Scholar]
- Soulimane T., Gohlke,U., Huber,R. and Buse,G. (1995) Three-dimensional crystals of cytochrome c oxidase from Thermus thermophilus diffracting to 3.8 Å resolution. FEBS Lett., 368, 132–134. [DOI] [PubMed] [Google Scholar]
- Soulimane T., von Walter,M., Hof,P., Than,M.E., Huber,R. and Buse,G. (1997) Cytochrome-c552 from Thermus thermophilus: a functional and crystallographic investigation. Biochem. Biophys. Res. Commun., 237, 572–576. [DOI] [PubMed] [Google Scholar]
- Surerus K.K. et al. (1992) Reaction of cyanide with cytochrome ba3 from Thermus thermophilus: spectroscopic characterization of the Fe(II)a3-CN⋅Cu (II)B-CN complex suggests four 14N atoms are coordinated to CuB. Proc. Natl Acad. Sci. USA, 89, 3195–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Than M.E., Hof,P., Huber,R., Bourenkov,G.P., Bartunik,H.D., Buse,G. and Soulimane,T. (1997) Thermus thermophilus cytochrome-c552: a new highly thermostable cytochrome c structure obtained by MAD phasing. J. Mol. Biol., 271, 629–644. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama,H., Yamashita,E., Tomizaki,T., Yamaguchi,H., Shinzawa-Itoh,K., Nakashima,R., Yaono,R. and Yoshikawa,S. (1995) Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science, 269, 1069–1074. [DOI] [PubMed] [Google Scholar]
- Tsukihara T., Aoyama,H., Yamashita,E., Tomizaki,T., Yamaguchi,H., Shinzawa-Itoh,K., Nakashima,R., Yaono,R. and Yoshikawa,S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science, 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekulgraphik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD thesis, TU München, Germany. [Google Scholar]
- Verkhowsky M.I., Jasaitis,A., Verkhowskaya,M.L., Morgan,J.E. and Wikström,M. (1999) Proton translocation by cytochrome c oxidase. Nature, 400, 480–483. [DOI] [PubMed] [Google Scholar]
- Williams P.A., Blackburn,N.J., Sanders,D., Bellamy,H., Stura,E.A., Fee,J.A. and McRee,D.E. (1999) The CuA domain of Thermus thermophilus ba3-type cytochrome c oxidase at 1.6 Å resolution. Nature Struct. Biol., 6, 509–516. [DOI] [PubMed] [Google Scholar]
- Wilmanns M., Lappalainen,P., Kelly,M., Sauer-Eriksson,E. and Saraste,M. (1995) Crystal structure of the membrane-exposed domain from a respiratory quinol oxidase complex with an engineered dinuclear copper center. Proc. Natl Acad. Sci. USA, 92, 11955–11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt H., Malatesta,F., Nicoletti,F., Brunori,M. and Ludwig,B. (1998a) Tryptophan 121 of subunit II is the electron entry site to cytochrome-c oxidase in Paracoccus denitrificans. J. Biol. Chem., 273, 5132–5136. [DOI] [PubMed] [Google Scholar]
- Witt H., Malatesta,F., Nicoletti,F., Brunori,M. and Ludwig,B. (1998b) Cytochrome-c binding site on cytochrome oxidase in Paracoccus denitrificans. Eur. J. Biochem., 251, 367–373. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S. et al. (1998a) The structure of the 13-subunit oxidized cytochome c oxidase at 2.3 Å. Science, 280, 1723–1730. [DOI] [PubMed] [Google Scholar]
- Yoshikawa S., Shinzawa-Itoh,K. and Tsukihara,T. (1998b) Crystal structure of bovine heart cytochrome c oxidase at 2.8 Å resolution. J. Bioenerg. Biomembr., 30, 7–14. [DOI] [PubMed] [Google Scholar]