Abstract
The plastid genome is known to be transcribed by a plastid-encoded prokaryotic-type RNA polymerase (PEP) and by a nucleus-encoded phage-type RNA polymerase (NEP). The spinach plastid rrn operon promoter region harbours three different, overlapping promoters. Two of them are of the prokaryotic type. The third promoter is a non-consensus-type NEP promoter. We separated three different transcriptional activities from spinach chloroplasts: PEP, the phage-type RNA polymerase NEP-1, and a third, hitherto undescribed transcriptional activity (NEP-2). NEP-2 specifically transcribes the rrn operon in the presence of the transcription factor CDF2. CDF2 was previously shown to recruit PEP to the rrn promoter to repress transcription. Together, our results suggest the existence of a third RNA polymerase in plastids and a mechanism of rDNA transcriptional regulation that is based on the interaction of the transcription factor CDF2 with two different transcriptional systems.
Keywords: CDF2/rDNA transcription/plastids/regulation/RNA polymerases
Introduction
In all organisms, the transcription of rDNA genes plays a central role in the overall regulation of ribosome biosynthesis. In eukaryotes as in prokaryotes, rDNA transcription is tightly coordinated with cell growth rates and environmental conditions, and multiple control mechanisms are engaged in rRNA expression. Escherichia coli rRNA, for instance, is synthesized from seven non-contiguous operons that all have two tandem promoters that are regulated differently (Condon et al., 1995).
Plastid genomes of most higher plants contain two rDNA operons that are localized in the inverted repeat regions of the circular DNA molecules (Kössel, 1991). It has been suggested that two different types of RNA polymerase are implicated in the transcription of these plastid rDNA genes (Iratni et al., 1994; Allison et al., 1996), but very little is known about the regulatory mechanisms that determine rDNA gene expression. Sequencing data have shown that one of the plastid enzymes is of the prokaryotic type and is plastid encoded (Hu and Bogorad, 1990; Hu et al., 1991). This enzyme is called PEP (plastid-encoded plastid RNA polymerase). The second enzyme is of the phage type and is nucleus encoded (Hedtke et al., 1997). This enzyme is called NEP (nucleus-encoded plastid RNA polymerase). However, the diversity of existing biochemical results (Lerbs et al., 1985; Zaitlin et al., 1989; Lakhani et al., 1992; Lerbs-Mache, 1993; Pfannschmidt and Link, 1994, 1997) can only be explained partially by the existence of two enzymes and rather suggests the existence of a third RNA polymerase or multiple subunit recombinations to form different types of enzymes.
Actually mapped (Sexton et al., 1990; Vera and Sugiura, 1995; Hajdukiewicz et al., 1997; Iratni et al., 1997; Kapoor et al., 1997; Silhavy and Maliga, 1998) and/or analysed (Gruissem and Zurawski, 1985; Kim and Mullet, 1995; Wu et al., 1997; Sriraman et al., 1998a,b; Liere and Maliga, 1999) plastid promoter regions can be classified into three different categories: PEP, NEP and exceptional promoters (Hess and Börner, 1999). PEP promoters are prokaryotic-type promoters of the σ70 type that are recognized by PEP. The majority of actually characterized NEP promoters are of the ATAGAAT consensus type and are probably transcribed by the phage-like NEP. Exceptional promoters are promoters that do not harbour the σ70 type or the ATAGAAT consensus sequences. This raises the question of how and by which enzyme exceptional promoters are recognized.
We used the spinach plastid rDNA PC promoter to analyse this question. This promoter belongs to the group of exceptional promoters. It overlaps with two different PEP promoters (Iratni et al., 1997), but its sequence has nothing in common with the hitherto established NEP consensus sequences. Recently, we have demonstrated the formation of two different DNA–protein complexes at the rrn promoter region (Baeza et al., 1991; Iratni et al., 1994). The so-called small complex (S) is the association of a transcription factor, named CDF2, to a well defined region upstream of the PC transcription start site. The large complex (L) corresponds to CDF2 in tight association with PEP. We have proposed a mechanism of transcriptional regulation in which CDF2 serves to recruit inactive PEP to the PEP-P2 promoter, thus repressing rDNA transcription. However, we could not show by which enzyme the rrn operon is transcribed at PC. Also, the question of why the two prokaryotic-type promoters, P1 and P2, have been conserved during evolution, although they are not used for transcription, has not been answered.
In the present paper, we show that the rrn operon is transcribed at PC by a NEP enzyme and that CDF2 acts as initiation factor for rDNA transcription. As is known for nucleolar and prokaryotic rDNA transcription, plastid rDNA transcription is also tightly coordinated with cell growth rates and environmental conditions. Immunological analyses indicate that the rDNA-transcribing NEP enzyme does not correspond to the previously characterized phage-type enzyme of 110 kDa, suggesting the existence of a third enzyme in chloroplasts. We propose a model for rDNA transcription regulation that is based on the interaction of CDF2 with two different transcription systems.
Results
PC is recognized by NEP
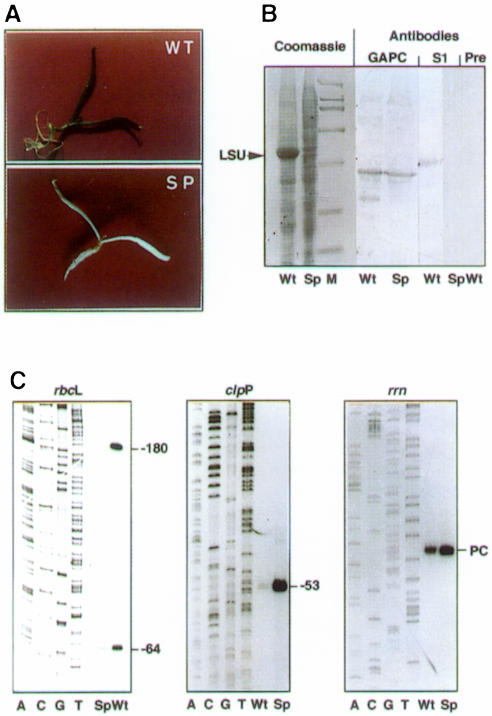
In order to analyse whether PC is recognized by a nucleus-encoded RNA polymerase or by the plastid-encoded enzyme, we used the recently described approach of ribosome depletion of plastids by antibiotic treatment (Kapoor et al., 1997; Hübschmann and Börner, 1998; Zubko and Day, 1998). In plastids that lack ribosomes, PEP cannot be made and any transcription is due to NEP (Hess et al., 1993). Spinach plantlets were grown for 1 week under sterile conditions on an agarose medium in the absence or presence of 500 µg/ml spectinomycin (Figure 1A). Ribosome depletion in the spectinomycin-treated white plantlets was confirmed by the disappearance of the large subunit (LSU) of Rubisco and the disappearance of the plastid ribosomal protein S1 (Figure 1B). The analysis of rbcL gene expression, a gene that is under the control of a PEP promoter, confirmed the lack of PEP. The analysis of an mRNA (clpP) that is under the control of an NEP promoter confirmed the presence of NEP in spectinomycin-treated plantlets (Figure 1C). RNA that corresponds to initiation of transcription at the rrn PC promoter was enhanced in ribosome-depleted plants, i.e. transcription is carried out by a nucleus-encoded enzyme (Figure 1C, compare clpP and rrn).
Fig. 1. Characterization of normal and spectinomycin-grown spinach plantlets. (A) Wild type (WT) and spectinomycin-grown (SP) spinach plantlets. (B) Total proteins prepared from Wt and Sp spinach plantlets were analysed by SDS–PAGE and Coomassie staining of the gel and by immunoblotting using antibodies raised against spinach cytoplasmic glyceraldehyde-3-phosphate dehydrogenase (GAPC) as marker protein for constitutive protein synthesis, or against plastid ribosomal protein S1 (S1) as marker protein for ribosomes. Pre corresponds to the control made with preimmune serum. (C) RNA analysis by primer extension (Baeza et al., 1991) of Wt and Sp spinach plantlets (the rbcL Wt and Sp cDNAs have been inversely loaded on the gel compared with the corresponding clpP and rrn cDNAs). The accompanying sequence ladders were made using the same primers. Primers were 5′-AGATATCAGTATCTAGGG-3′ (rbcL), 5′-GCATCTTCCTCTCCAGG-3′ (clpP) and 5′-TTCATAGTTGCATTACT-3′ (rrn). Transcription start sites are –180 (rbcL), –53 (clpP) and PC (rrn). The –64 transcript (rbcL) is a processing intermediate.
Plastid rrn expression depends on growth conditions
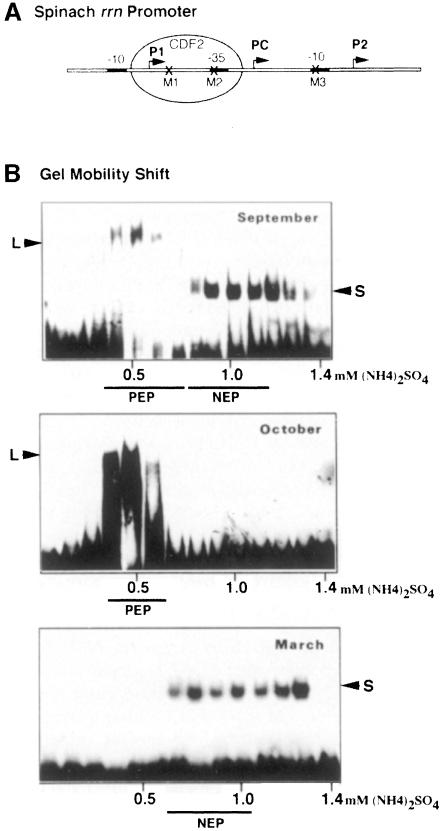
Having shown that PC is recognized by NEP, we wanted to purify the corresponding enzyme. Figure 2 shows gel mobility shift (GMS) assays using plastid extracts of spinach leaves that were grown under different climatic conditions.
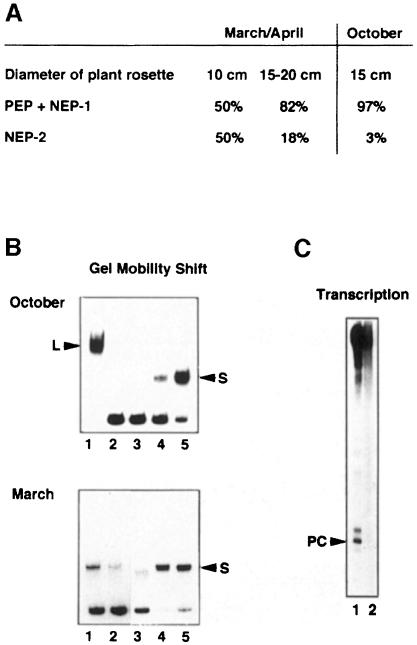
Fig. 2. Changes of plastid transcriptional activities and DNA–protein interactions in relation to spinach growth conditions. (A) Schematic representation of the spinach rrn promoter region. The three transcription start sites, P1, PC and P2, are indicated by arrows. The localizations of the three point mutations, M1, M2 and M3, are marked by crosses and the CDF2 factor is represented as an oval. (B) Analysis of promoter–protein interactions. Proteins of 75 000 g chloroplast supernatants were fractionated by heparin–Sepharose chromatography (Baeza et al., 1991). Eight microlitres of each eluted fraction were analysed for the presence of DNA binding proteins using the radioactively labelled rrn promoter region by GMS. Transcriptionally active fractions are underlined and the predominant enzymes are indicated. The same type of elution pattern was reproduced over a period of 3 years. L, CDF2 + PEP; S, CDF2.
L and S complexes were formed on the plastid rDNA promoter region when extracts were prepared from young, rapidly growing summer spinach plants, which were sown in August and harvested at the beginning of September (Figure 2B, top). It has previously been shown that the L complex fractions contain mainly PEP and recognize specifically the rbcL promoter but not the rrn promoter (Iratni et al., 1994). When analysed by GMS, the second peak of transcriptional activity (NEP) contains CDF2 (S complex) as well as the phage-like monomeric enzyme (Lerbs-Mache, 1993; Hedtke et al., 1997). Several attempts to purify the phage-like enzyme from these fractions have been without success. The transcriptional activity was completely lost during further purification.
When plants from the same field were harvested in October, i.e. at maturity, only the L complex had formed (Figure 2B, middle), i.e. CDF2 was not present in a free form in plastids, instead being complexed with PEP and thus repressing rrn transcription (Iratni et al., 1994). Transcriptional extracts prepared from this type of plant transcribe the rbcL promoter but do not transcribe the rrn operon (not shown).
Spinach sown in October develops into the rosette stage and grows through the end of November. During winter, growth and plastid transcriptional activity are both virtually arrested (not shown). At the beginning of March in the following year the plants began to grow again. This reprise of growth following hibernation is correlated with a re-induction of transcriptional activity and a reappearance of the small complex (Figure 2B, bottom). According to its elution profile from the heparin–Sepharose column this transcriptional activity should correspond preferentially to the phage-type enzyme, i.e. to NEP (Lerbs-Mache, 1993; Hedtke et al., 1997). Formation of the L complex on the rDNA promoter is not observed using protein extracts obtained from these plants.
The changes of the elution profiles of the transcriptionally active fractions together with the changes observed in rrn promoter–protein complex formation indicate large differences in transcription activities between spinach harvested in October or in March. They also indicate that spinach harvested in March should contain preferentially NEP, i.e. the enzyme that transcribes the rrn operon at PC.
PEP, NEP-1 and NEP-2
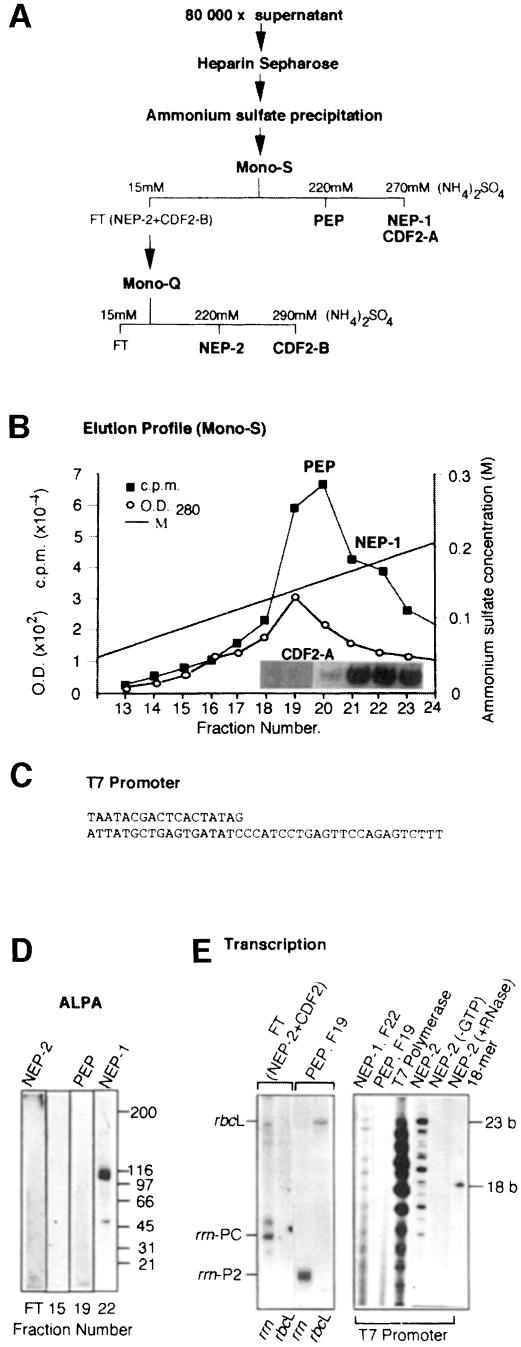
To obtain more information on the enzyme that transcribes the rrn operon in plastids, transcriptional activities from such winter spinach (March) have been further purified by Mono-S (Figure 3) and Mono-Q (Figure 3) FPLC column chromatography (see Materials and methods). The purification procedure is schematically represented in Figure 3A. The measurement of (non-specific) transcriptional activity showed two partially overlapping peaks on Mono-S (Figure 3B), designated PEP and NEP-1, as well as some activity in the flow-through fraction, designated FT (not shown). These three transcriptional activities were further characterized by in vitro transcription and/or GMS studies. GMS assays were performed using the plastid rDNA promoter. For in vitro transcription studies we used three different templates: the cloned spinach rbcL and rrn promoter regions (Iratni et al., 1994; Figure 2A) and an oligonucleotide-assembled T7 RNA polymerase promoter (Figure 3C).
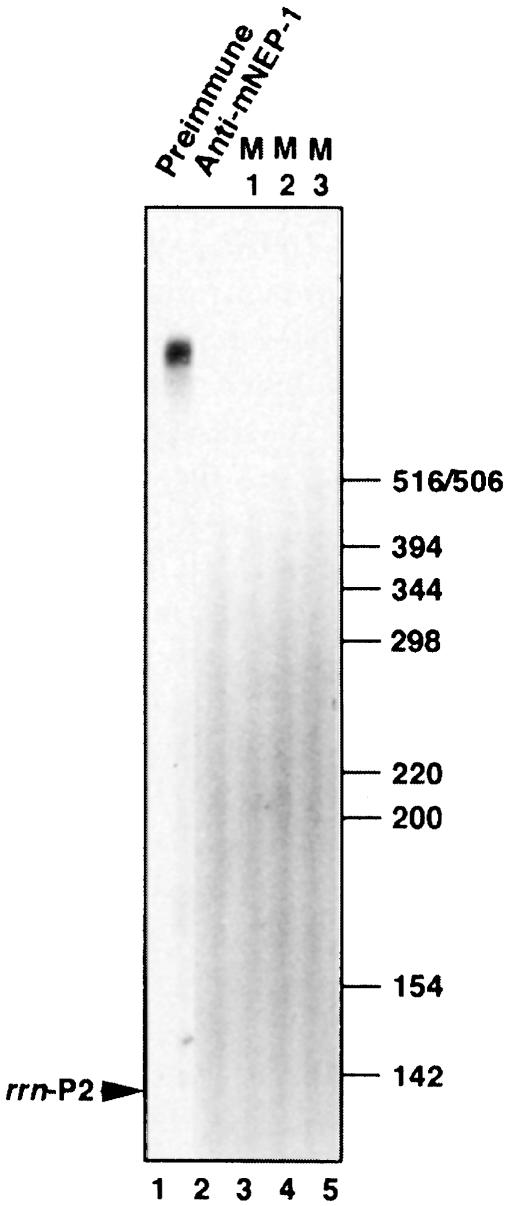
Fig. 3. Separation of three different RNA polymerases. (A) Schematic representation of the separation of three different plastid RNA polymerase activities and two different CDF2 factors. (B) Elution profile of PEP and NEP-1 on a Mono-S FPLC column. The inset shows the localization of CDF2-A in the Mono-S-separated fractions by GMS using the rrn promoter region. (C) Oligonucleotide-assembled T7 RNA polymerase-specific template. (D) Identification of the 110 kDa, phage-like NEP-1 by ALPA. Western blots of the flow-through (FT), F15, F19 and F22 of the Mono-S column were analysed. Each blot was treated with mNEP-1 antibodies (see Materials and methods) in the first binding reaction and with the same fraction that was blotted on the filter (FT, F15, F19 or F22) in the second binding reaction, followed by RNA polymerase assays. The NEP-1 filter was exposed for 1 h, the other filters have been exposed for 16 h during autoradiography. (E) Characterization of promoter specificity of the FT (NEP-2), PEP (F19), NEP-1 (F22) and T7 RNA polymerase by in vitro transcription using the cloned rrn or rbcL promoter region, or the oligo-assembled T7 RNA polymerase promoter. Transcription products of the rrn and rbcL corresponding transcripts were separated in 6% polyacrylamide gels (left-hand side), and transcription products of the T7 promoter in 19% polyacrylamide gels (right-hand side). The 18mer corresponds to a radioactively labelled primer of 18 bases. Control experiments omitting one nucleotide in the transcription reaction (–GTP) or followed by RNase treatment (+RNase) have been systematically made and are shown for NEP-2 as an example.
The 110 kDa phage-like RNA polymerase (NEP-1), which was first characterized by a biochemical approach (Lerbs-Mache, 1993) and more recently by cDNA sequencing (Hedtke et al., 1997), elutes from the Mono-S column in the same fraction as CDF2. This can be concluded from Western blot analysis of all Mono-S fractions (not shown) and GMS assays using the rrn promoter region (Figure 3B, inset of CDF2-A). As expected, CDF2-A is sensitive to M1 and insensitive to M2 and M3 (not shown). That NEP-1 indeed corresponds to the 110 kDa phage-like RNA polymerase was further verified by antibody-linked polymerase assays (ALPA; Lerbs et al., 1985; Lerbs-Mache, 1988; Figure 3D) and transcription of the oligonucleotide-assembled T7 promoter (Figure 3E, right-hand side).
On the other hand, as expected and already shown (Iratni et al., 1994), the rrn P2 promoter and the rbcL promoter are transcribed by PEP (fraction 19, Figure 3E). PEP is inhibited by Tagetin (Figure 4A) and does not recognize the T7 promoter (Figure 3E). The FT fraction of the Mono-S column (NEP-2) also transcribes the T7 promoter; however, the transcription pattern differs somewhat from that of T7 RNA polymerase and NEP-1 in that longer RNAs are preferentially made (Figure 3E).
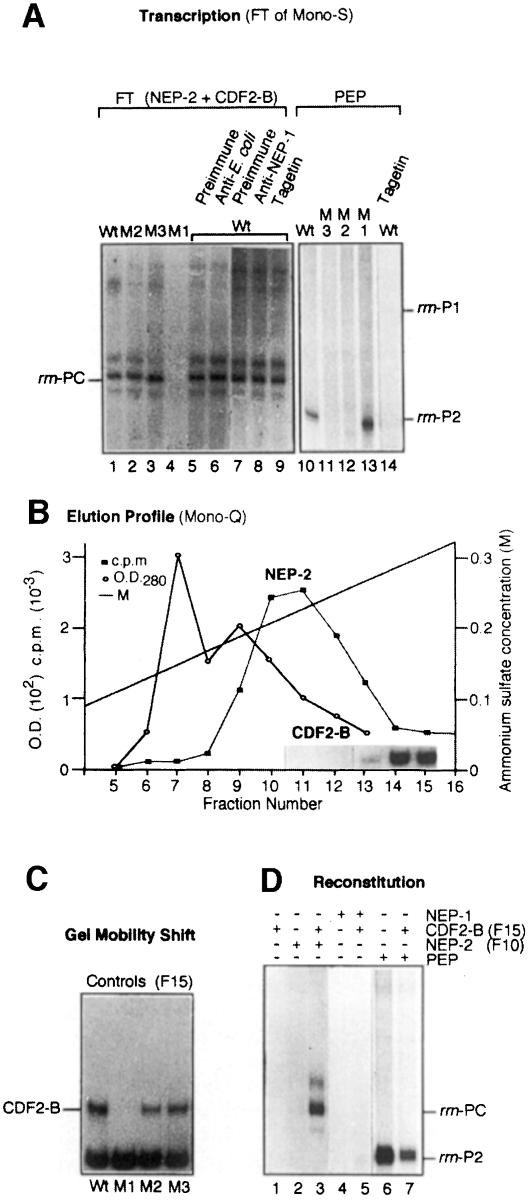
Fig. 4. Characterization of CDF2-B as a transcription initiation factor for NEP-2. (A) Comparison of in vitro transcription products of the cloned rrn promoter region and its mutations by NEP-2 and PEP. The wild-type promoter (Wt, lanes 1, 5–10 and 14) and the three mutations M1–M3 (lanes 2–4 and 11–13) were transcribed either by the FT fraction (lanes 1–9) or by PEP (lanes 10–14) in the presence of preimmune serum (lanes 5 and 7), E.coli RNA polymerase antibodies (lane 6), mNEP-1 antibodies (lane 8) or Tagetin (2U, lanes 9 and 14). (B) Elution profile of NEP-2 and CDF2-B on a Mono-Q FPLC column. The localization of CDF2-B in the Mono-Q-separated fractions by GMS using the rrn promoter region is indicated as an inset. (C) Characterization of CDF2-B by GMS using the three rrn mutations M1–M3. (D) Reconstitution of rrn transcription at PC. The cloned rrn promoter region was transcribed by CDF2-B (fraction 15 of Mono-Q, lanes 1, 3, 5 and 7), NEP-2 (fraction 10 of Mono-Q, lanes 2 and 3), NEP-1 (fraction 22 of Mono-S, lanes 4 and 5) and PEP (fraction 19 of Mono-S, lanes 6 and 7). Autoradiography of plastid RNA polymerase transcription products was for 16 h.
In contrast to the PEP- and NEP-1-containing fractions, the FT fraction of the Mono-S column transcribes the rrn PC promoter but not the rbcL promoter (Figure 3E). Some CDF2 activity, as measured by GMS, was also observed in this fraction (not shown). To distinguish this activity from the Mono-S-bound activity we named it CDF2-B. Although the FT fraction recognizes the T7 promoter, we could not detect the 110 kDa phage-like RNA polymerase in this fraction either by Western blot analysis of SDS–PAGE-separated proteins (not shown) or by ALPA assays (Figure 3D). Transcription initiation at the rrn PC promoter (schematically represented in Figure 2A) is not inhibited by Tagetin, a specific inhibitor of PEP, or by antibodies raised against E.coli RNA polymerase or the maize plastid phage-like enzyme (Figure 4A, lanes 5–9). Noticeably, transcription at the rrn PC promoter by NEP-2 is completely abolished if CDF2-B cannot bind to the template, as obtained by the mutation M1 (Figure 4A, compare lanes 1 and 4; Figure 4C, compare Wt and M1). The two mutations in the E.coli-like P2 promoter elements (M2 and M3) do not influence CDF2-B binding (Figure 4C, Wt, M2, M3) or transcriptional activity (Figure 4A, lanes 1–3). In contrast, PEP initiates transcription at P2 faithfully on M1, but not on M2 and M3 (Figure 4A, lanes 10–13). PEP activity is completely inhibited by Tagetin (Figure 4A, lanes 14–16). In all these respects, NEP-2 behaves oppositely to PEP, confirming that NEP-2 and PEP are principally different enzymes. Furthermore, our results suggest that CDF2-B is a transcription initiation factor for NEP-2.
CDF2-B acts as an initiation factor for NEP-2
To verify the hypothesis that CDF2-B acts as an initiation factor for NEP-2, we separated CDF2-B from NEP-2 by chromatography on a Mono-Q FPLC column (Figure 4B). The enzyme, as measured by non-specific transcription, elutes at a lower salt concentration (Figure 4B, fractions 10–12) than CDF2-B (inset showing GMS analysis in Figure 4B, fractions 14 and 15). Reconstitu<\\jy>tion assays were performed using fractions 10 (for NEP-2) and 15 (for CDF2-B). This experiment shows that while neither of the two fractions alone were able to support transcription of the rrn operon (Figure 4D, lanes 1 and 2), transcription could be recovered when CDF2-B was added to NEP-2 (Figure 4D, lane 3). CDF2-B was also tested in reconstitution assays with PEP (fraction 19 of Mono-S) and NEP-1 (fraction 22 of Mono-S). Addition of CDF2-B to PEP inhibits transcription initiation at the P2 promoter as expected (Figure 4D, lanes 6 and 7), again confirming that NEP-2 and PEP are different enzymes. Addition of CDF2-B to NEP-1 does not lead to initiation at PC (Figure 4D, lanes 4 and 5), also indicating that NEP-1 and NEP-2 are different enzymes.
NEP-1 is different to NEP-2
Mono-S-purified NEP-1 (fraction 22) produces only short RNAs, as shown by using the oligonucleotide-assembled T7 promoter (Figure 3E). Using supercoiled templates harbouring a cloned promoter region, no transcription is observed (Figure 4D and not shown). In this respect, NEP-1 differs from NEP-2. To analyse the activity of NEP-1 in more detail, transcriptionally active Mono-S fractions were further purified on a 16S-agarose affinity column using the conditions previously described (Trifa et al., 1998). Figure 5 shows the transcription pattern of the 16S-agarose-purified NEP-1 activity (lane 1). The enzyme produces exclusively long transcripts. The synthesis of these transcripts is inhibited in the presence of mNEP-1 antibodies (Figure 5, lane 2), indicating that these transcripts are produced by NEP-1. Furthermore, the production of these transcripts is abolished on all three promoter mutations (Figure 5, lanes 3–5, M1–M3), showing differences between NEP-1 and NEP-2.
Fig. 5. Characterization of NEP-1 and mNEP-1 antibodies. The transcription plasmid pTZ19-Ta was transcribed by 16S-agarose-purified NEP-1 in the presence of preimmune serum (lane 1) and in the presence of mNEP-1 antibodies (lane 2). Lanes 3–5 have been loaded with transcription assays using the three 16S promoter mutations (M1–M3). The migration of molecular size markers is indicated on the right-hand side.
Developmental and environmental changes of rDNA transcription
In order to confirm that plastid rDNA transcription is regulated by growth and environmental conditions we compared Mono-S-bound (PEP + NEP-1) and Mono-S-FT transcriptional activities (NEP-2) from actively growing and slowly growing spinach plants. For this reason, spinach was harvested in March at the beginning of the growth period (10 cm diameter plant rosette) and 2 weeks later (15–20 cm diameter rosette) from the same field. The transcriptional activities from the 15–20 cm plants were compared with spinach of about the same size that had stopped growth and was harvested in October. As shown in Figure 6A, young, actively growing spinach (10 cm) contains much more NEP-2 activity than older spinach (15–20 cm). In addition, plants of the same size but ‘growth-reactivated’ in March/April have more NEP-2 activity than plants harvested in October when growth is already very slow. The presence of CDF2-A and CDF2-B in these plants, which are of the same size but have been grown under different environmental conditions, was analysed by GMS assays (Figure 6B). While CDF2-A is present in both spinach plants (compare lanes 3–5, top and bottom), CDF2-B is only detectable in March-grown spinach (compare lane 2, top and bottom). Accordingly the rDNA promoter is only recognized by the NEP-2 enzyme of the March spinach (Figure 6C).
Fig. 6. Quantitative changes of PEP + NEP-1/NEP-2 and CDF2-B during spinach growth. Transcriptional extracts were prepared from 2 kg of spinach harvested in March/April or October using the same purification procedure. (A) Transcriptional activities corresponding to the total volumes of Mono-S-bound (PEP and NEP-1) and Mono-S flow-through fractions (NEP-2) were determined using filter binding assays of newly synthesized RNA (see Materials and methods). The activity of NEP-2 is indicated as a percentage of all three activities. (B) The presence of CDF2-A and CDF2-B was determined by GMS using spinach harvested in October (top) and in March/April (bottom). Heparin–Sepharose (lane 1), Mono-S FT (CDF2-B, lane 2) and Mono-S fractions 20–22 (CDF2-A, lanes 3–5) were analysed by GMS using the spinach rDNA promoter region. (C) In vitro transcription of the spinach rDNA promoter region using the Mono-S flow-through fraction from spinach harvested in March/April (lane 1) and October (lane 2).
Discussion
Plastids are green-life-specific organelles having evolved from eubacteria-like endosymbionts related to cyanobacteria. With a genome size of ∼150 kbp the circular plastid DNA molecule is >30 times smaller than the genome of one of its ancestors living today (Kaneko et al., 1996). Intriguingly, for transcription of this small genome, higher plant plastids need different types of RNA polymerase. In addition, many plastid transcription units are preceded by different types of promoters, suggesting competition or co-transcription of the same operon by the different enzyme types. At present, only little is known about the regulation of gene expression of such multi-type promoter regions. We are using the spinach plastid rrn promoter region, which harbours three different promoters, as a model system to analyse the role(s) of the different plastid RNA polymerases in plastid gene expression.
Spinach growth conditions
Spinach is a very attractive model to analyse the role(s) of NEP and PEP enzymes on multi-type promoter regions during plant and plastid development and in response to environmental changes, because it is one of the plants that can grow under quite different climatic conditions. The most interesting is the winter spinach, which is harvested in March. It is planted from September up to the end of October, develops into the rosette stage and keeps on growing until the end of November. Then it stops growth and largely ceases plastid transcriptional activity during wintertime. At the beginning of March in the following year it starts growing again. This re-growth is connected with an increase in plastid transcriptional activity. Thus, we have a situation of large changes and rearrangements of plastid transcriptional complexes that occur in mature chloroplasts depending on growth conditions (see Figure 2). In addition, it had been shown for monocotyledons that at the leaf base, i.e. during early phases of plastid development, transcription is very high for the rrn operon (Baumgartner et al., 1993). Re-growth of spinach after hibernation might be somehow comparable to such an ‘early stage of plastid development’ with respect to transcription, because in both cases the transcription machinery has to be ‘turned on’. Therefore, we found it especially interesting to analyse plastid rDNA transcription during such a ‘turn on’ period and to compare it with rDNA expression in plants that stop growth (‘turn-off’ period, October spinach).
Model of transcriptional regulation of the plastid spinach rrn operon by interaction of CDF2 with two types of RNA polymerase
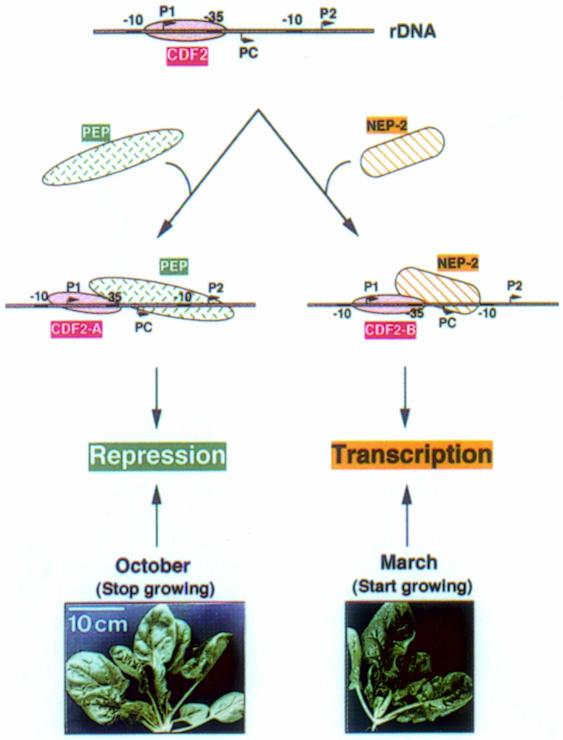
The results presented above, together with our previously obtained data (Iratni et al., 1994) and unpublished results, suggest the following model for plastid rDNA transcription (Figure 7). Initiation at PC requires a nucleus-encoded enzyme, which we have termed NEP-2, and the transcription factor CDF2. Repression of transcription at P2 is brought about by PEP and CDF2 (Iratni et al., 1994; Figure 4D). The abundance of NEP-2 seems to be related to plant growth conditions. The activity of NEP-2 is relatively high in young spinach plants, corresponding to a higher need for ribosomes in actively growing plants. During maturation, the levels of NEP-2 decrease (Figure 6).
Fig. 7. Schematic representation of activation and repression of the plastid rrn operon.
The transcription factor CDF2 exists in two different forms, CDF2-A and CDF2-B. Depending on the physiological stage of spinach, CDF2-A co-purifies with NEP-1 on Mono-S (Trifa et al., 1998; Figure 3B) and forms a stable repressive complex with PEP (Iratni et al., 1994; R.Iratni, unpublished results). CDF2-B co-purifies with NEP-2 on Mono-S and acts as initiation factor for NEP-2 (Figure 4D). It represses transcription of the rrn operon at P2 by PEP in vitro (Figure 4D). The efficiency of plastid rDNA transcription could be regulated by competition of either PEP/CDF2-A or NEP-2/CDF2-B for DNA binding. The difference found in NEP-2 activity between March/April and October spinach (which are of about the same size) is not yet completely clear. It might be explained by differences in temperature and/or light intensities or other factors. A more detailed analysis would need large space to grow spinach in sophisticated growth chambers.
The regulation of transcriptional activity of NEP-1 is not clear at the moment. As purified from Mono-S the enzyme seems to be unable to escape into the elongation cycle and produces only short transcripts (Figures 3E, 4D and data not shown). Therefore, we used the oligonucleotide-11-assembled T7 promoter and ALPA assays to demonstrate transcription by this enzyme. This incapacity to produce longer transcripts might be due either to the presence of inhibitory substances analogous to the inhibition of T7 RNA polymerase by T7 lysozyme (Zhang and Studier, 1997), or to the absence of substances that help to change conformation and to enter into the elongation cycle. Larger transcripts can be made if we further purify NEP-1 on 16S-agarose (Figure 5).
Also, it is not yet clear what causes the difference between CDF2-A and CDF2-B. According to their different affinities to either Mono-S or Mono-Q, both factors should be different. This difference could be due to different polypeptide compositions or to post-translational protein modifications. Both factors seem to be the same size in GMS assays and they are both sensitive to the same base changes in the CDF2 binding site (M1), indicating similarities in DNA recognition. A detailed analysis to compare the DNA–protein interactions between the two factors is under way. However, our efforts to purify the proteins that form the CDF2 complexes have failed so far because of the instability of the complexes during purification.
Regulatory proteins that activate as well as repress transcription have already been described (Choy et al., 1995; Monsalve et al., 1996). These factors exert their functions on different, but closely spaced promoters by interacting with the same RNA polymerase—that of E.coli. The difference that distinguishes CDF2 from these other factors is that CDF2 interacts on the same promoter with two different RNA polymerases, thus activating or repressing transcription. So far, we have not found in vivo initiation of transcription at one of the PEP promoters, i.e. PEP never transcribes the rrn operon in spinach. Therefore, we suggest that the P2 promoter has been conserved during evolution not for initiation but to ensure regulation of rrn expression, i.e. it functions in down-regulation of rrn expression under specific conditions. The spinach plastid rrn operon is the first hitherto described example of transcriptional regulation of this type.
Another peculiarity common to plastid and eukaryotic rDNA transcription is species specificity, which might be linked to rapid evolution of rDNA gene promoters in eukaryotic systems (Bell et al., 1989; Doelling and Pikaard, 1996). In plants, we distinguish between three types of rDNA transcription: (i) in tobacco, rice and barley, transcription starts at the P2 promoter (Allison et al., 1996; Hübschmann and Börner, 1998; Silhavy and Maliga, 1998); (ii) in spinach and mustard, transcription starts at the PC promoter (Iratni et al., 1994, 1997; Pfannschmidt and Link, 1997); and (iii) in Arabidopsis, transcription starts at both of these promoters (Sriraman et al., 1998b). The reason for these start site variations is not yet understood, but it seems to be more likely that these differences in plastid rDNA transcription are related to species-specific differences of transcription factors (Iratni et al., 1997; Sriraman et al., 1998b) than to rapid evolution of promoter structures, since the plastid genome sequences are highly conserved in different plant species.
Characterization of NEP-2
The results of our in vitro transcription studies and immunological analysis are summarized in Table I. They show that there are differences but also some similarities between the three enzyme activities. This raises the question of whether these three activities correspond to three different core enzymes or whether at least two of these activities use the same core enzyme composed of different transcription factors. This question is best addressed by using specific transcription inhibitors that act independently from the initiation process. Rifampicin, for instance, reacts during the initiation process, and different σ factors might influence the sensitivity of the RNA polymerase to the drug (Wegrzyn et al., 1998). Therefore, different sensitivities to rifampicin cannot be taken as an indication of the existence of different core enzymes. Tagetin is more specific for distinguishing between different RNA polymerases because it blocks RNA polymerase during elongation (Mathews and Durbin, 1994), i.e. its action is independent of different initiation factors. On the basis of different Tagetin sensitivities we can conclude that PEP has another core enzyme than NEP-1 and NEP-2. The question of whether NEP-1 and NEP-2 are made of the same core enzyme (the phage-like RNA polymerase) is more difficult to answer. Both enzymes, NEP-1 and NEP-2, recognize the oligonucleotide-assembled T7 promoter in vitro (Figure 3E; Table I). However, strong evidence that NEP-1 and NEP-2 are two different enzymes comes from transcription of the 16S promoter mutations and from the antibody results. (i) Transcription by NEP-1 is abolished on all three mutations (M1–M3, Figure 5). In contrast, transcription by NEP-2 is only abolished on mutation M1 (Figure 4A). (ii) NEP-1 and NEP-2 respond differently to the mNEP-1 antibodies in in vitro transcription assays. The activity of NEP-1 is inhibited, the activity of NEP-2 is not inhibited (Figures 5 and 4A). (iii) By using ALPA assays and Western analysis the phage-like enzyme is only detected in the Mono-S-recovered fractions. It is not detectable either in the FT fraction or in the Mono-Q-eluted fractions (Figure 3E and our unpublished results).
Table I. Characterization of the three transcriptional activities.
| PEP | NEP-1 | NEP-2 | |
|---|---|---|---|
| Recognition of rbcL | + | (+)a | – |
| Recognition of rrn P2 | + | – | – |
| Recognition of PC | – | (?) | + |
| Recognition of T7 promoter | – | + | + |
| Inhibition by Tagetin | + | – | – |
| Presence of 110 kDa phage-like RNA polymerase (ALPA and Western) | – | + | – |
| Inhibition of transcription by mNEP-1 antibodies | – | + | – |
| Sensitivity of transcription to M2 and M3 | + | + | – |
| Sensitivity of transcription to M1 | – | + | + |
aLow transcription of rbcL probably results from overlapping PEP activity.
The existence of a third RNA polymerase in plastids, besides the prokaryotic type and the phage type, is not unlikely. The characterization of internal promoter regions (Gruissem et al., 1986; Wu et al., 1997; Sriraman et al., 1998a) as well as the importance of TATA-like sequence elements in plastid promoter regions (Link, 1984; Eisermann et al., 1990) support this assumption and raise the question of the existence of a third, eukaryotic-type RNA polymerase in plastids. Whether NEP-2 corresponds to a eukaryotic-type RNA polymerase or to a modified phage-type enzyme that has different properties to NEP-1 cannot be decided at present. However, whatever NEP-2 represents as enzyme, it is different from PEP, and the mode of rDNA transcription regulation in plastids described here is unique in so far as a transcription factor interacts with two different enzymes to adapt rRNA synthesis to growth conditions.
Although the exact polypeptide compositions of NEP-1, NEP-2 and PEP are not yet established, most of our experiments indicate that NEP-1, NEP-2 and PEP represent three well distinguishable transcriptional activities playing different roles in plastid genome expression. In this respect, it is very puzzling that a small genome like that of plastids needs three different transcriptional activities to be properly transcribed. The cyanobacterial ancestor of plastids had only one prokaryotic-type RNA polymerase. The diversity of enzymes in higher plant plastids is therefore likely to be intimately linked to the evolutionary integration processes that have produced present-day photosynthetic eukaryotic cells. The purification of the two forms of CDF2 and of NEP-2, and the cloning of the corresponding genes, should provide clues to understanding these processes.
Materials and methods
Preparation of transcriptionally active spinach chloroplast extracts
Heparin–Sepharose-purified chloroplast supernatant was prepared as previously described (Baeza et al., 1991). Transcriptionally active fractions were pooled, precipitated with 80% (NH4)2SO4, re-dissolved in dialysis buffer [20 mM Tris–HCl pH 7.8, 15 mM (NH4)2SO4, 20% (v/v) glycerol, 1 mM dithiothreitol (DTT), 1 mM EDTA] and desalted using a 5 ml Hitrap Desalting column (flow rate: 1 ml/min) connected to a Pharmacia FPLC system. Subsequently, the enzyme was purified on a Mono-S HR 5/5 column (Pharmacia) using a 15 mM to 1 M linear (NH4)2SO4 gradient generated by the GP 250 gradient programmer at a flow rate of 0.25 ml/min. Transcriptionally active fractions were dialysed as indicated above. The flow-through of the Mono-S column was further separated on a Mono-Q HR 5/5 column using the same gradient as for Mono-S, at a flow rate of 0.5 ml/min.
RNA polymerase assays (filter binding)
The enzyme activity (corresponding to 10 µl of each enzyme fraction) was measured in a final volume of 100 µl containing 44 mM Tris pH 7.8, 14 mM MgCl2, 300 µM each of ATP, GTP and CTP (Boehringer), 15 µM UTP including 5 µCi of 5,6-[3H]UTP (30–60 Ci/mmol, Amersham) and 10 µg of denatured calf thymus DNA. After 15 min of incubation at 30°C, newly synthesized RNA was precipitated on DE81 filter discs (Whatman) and the incorporation of UMP into RNA was measured by liquid scintillation counting.
Mobility shift assays
GMS assays were carried out according to Baeza et al. (1991). Labelled rDNA promoter fragment (1 ng) was incubated with 8 µl of chloroplast protein extract in a 20 µl reaction mixture containing 44 mM Tris pH 8.0, 10 mM NaCl, 0.4 mM EDTA, 0.8 mM DTT, 1 µg of poly(dI–dC) at 30°C for 15 min. DNA–protein complexes were subsequently analysed on 4% polyacrylamide gels under non-denaturing conditions.
Antibody preparation
The preparation of an antibody directed against maize plastid phage-like enzyme (mNEP-1) has been described in detail (Chang et al., 1999). Briefly, amino acids 835–951 from the highly conserved C-terminal part of the maize rpoT2 gene, fused to a His6-tag, were overproduced in E.coli. The His-tagged protein was purified on a Ni column and used to produce polyclonal antibodies in rabbits. This antibody recognizes T7 RNA polymerase and inhibits transcription by T7 RNA polymerase. It also recognizes and inhibits the maize mitochondrial phage-like enzyme (Chang et al., 1999).
In vitro transcription
In vitro transcription reactions were performed at 30°C in 25 µl assays. The reaction mixture contained 44 mM Tris–HCl pH 8.0, 20 µM DTT, 14 mM MgCl2, 8 µM EDTA, 50 mM NaCl, 300 µM each of GTP, ATP and CTP, and 5 µM UTP including 20 µCi of [α-32P]UTP, 200 ng of DNA, 8 µl of enzyme extract and 2 µl of factor fractions. Reactions were stopped after 15 min by extraction with phenol–chloroform and precipitated with ethanol. Transcription products were analysed on 8% acrylamide/urea gels.
Antibody-linked polymerase assays
ALPA assays were performed as previously described (Lerbs et al., 1985; Lerbs-Mache, 1988). Briefly, Western blots of RNA polymerase containing plastid protein fractions were incubated with antibodies raised against the maize NEP-1 enzyme or the E.coli RNA polymerase holoenzyme in the first binding reaction. After extensive washing, spinach NEP-1, NEP-2 or E.coli RNA polymerase was coupled in its native form via the free Fab sites to the denatured RNA polymerase subunits on the Immobilon-P membranes (second binding reaction). Polypeptides that had bound transcriptionally active RNA polymerase in the second binding reaction were subsequently revealed by in vitro transcription and autoradiography of the membrane.
Acknowledgments
Acknowledgements
We are grateful to Danny Reinberg for helpful discussions during the preparation of the manuscript. We thank H.Pesey for technical assistance and photographic work. The work was supported by the European Community in the framework of the Biotech program BIO4-CT97-2245.
References
- Allison L.A., Simon,L.D. and Maliga,P. (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J., 15, 2802–2809. [PMC free article] [PubMed] [Google Scholar]
- Baeza L., Bertrand,A., Mache,R. and Lerbs-Mache,S. (1991) Characterization of a protein binding sequence in the promoter region of the 16S rRNA gene of the spinach chloroplast genome. Nucleic Acids Res., 19, 3577–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner B.J., Rapp,J.C. and Mullet,J.E. (1993) Plastid genes encoding the transcription/translation apparatus are differentially transcribed early in barley (Hordeum vulgare) chloroplast development. Plant Physiol., 101, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S.P., Pikaard,C.S., Reeder,R.H. and Tjian,R. (1989) Molecular mechanisms governing species-specific transcription of ribosomal RNA. Cell, 59, 489–497. [DOI] [PubMed] [Google Scholar]
- Chang C.-C., Sheen,J., Bligny,M., Niwa,Y., Lerbs-Mache,S. and Stern,D.B. (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell, 11, 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choy H.E., Park,S.W., Aki,T., Parrack,P., Fujita,N., Ishihama,A. and Adhya,S. (1995) Repression and activation of transcription by Gal and Lac repressors: involvement of α subunit of RNA polymerase. EMBO J., 14, 4523–4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon C., Squires,C. and Squires,C.L. (1995) Control of rRNA transcription in Escherichia coli. Microbiol. Rev., 59, 623–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doelling J.H. and Pikaard,C.S. (1996) Species-specificity of rRNA gene transcription in plants manifested as a switch in RNA polymerase specificity. Nucleic Acids Res., 24, 4725–4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisermann A., Tiller,K. and Link,G. (1990) In vitro transcription and DNA binding characteristics of chloroplast and etioplast extracts from mustard (Sinapis alba) indicate differential usage of the psbA promoter. EMBO J., 9, 3981–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W. and Zurawski,G. (1985) Identification and mutational analysis of the promoter for a spinach chloroplast transfer RNA gene. EMBO J., 4, 1637–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruissem W., Elsner-Menzel,C., Latshaw,S., Narita,J.O., Schaffer,M.A. and Zurawski,G. (1986) A subpopulation of spinach chloroplast tRNA genes does not require upstream promoter elements for transcription. Nucleic Acids Res., 14, 7541–7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P.T.J., Allison,L.A. and Maliga,P. (1997) The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J., 16, 4041–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B., Börner,T. and Weihe,A. (1997) Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis. Science, 277, 809–811. [DOI] [PubMed] [Google Scholar]
- Hess W.R. and Börner,T. (1999) Organellar RNA polymerases of higher plants. Int. Rev. Cytol., 190, 1–59. [DOI] [PubMed] [Google Scholar]
- Hess W.R., Prombona,A., Fieder,B., Subramanian,A.R. and Börner,T. (1993) Chloroplast rps15 and the rpoB/C1/C2 gene cluster are strongly transcribed in ribosome-deficient plastids: evidence for a functioning non-chloroplast-encoded RNA polymerase. EMBO J., 12, 563–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. and Bogorad,L. (1990) Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc. Natl Acad. Sci. USA, 87, 1531–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Troxler,R.F. and Bogorad,L. (1991) Maize chloroplast RNA polymerase: the 78-kilodalton polypeptide is encoded by the plastid rpoC1 gene. Nucleic Acids Res., 19, 3431–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hübschmann T. and Börner,T. (1998) Characterisation of transcription initiation sites in ribosome-deficient barley plastids. Plant Mol. Biol., 36, 493–496. [DOI] [PubMed] [Google Scholar]
- Iratni R., Baeza,L., Andreeva,A., Mache,R. and Lerbs-Mache,S. (1994) Regulation of rDNA transcription in chloroplasts: promoter exclusion by constitutive repression. Genes Dev., 8, 2928–2938. [DOI] [PubMed] [Google Scholar]
- Iratni R., Diederich,L., Harrak,H., Bligny,M. and Lerbs-Mache,S. (1997) Organ-specific transcription of the rrn operon in spinach plastids. J. Biol. Chem., 272, 13676–13682. [DOI] [PubMed] [Google Scholar]
- Kaneko T. et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res., 3, 109–136. [DOI] [PubMed] [Google Scholar]
- Kapoor S., Suzuki,J.Y. and Sugiura,M. (1997) Identification and functional significance of a new class of non-consensus-type plastid promoters. Plant J., 11, 327–337. [DOI] [PubMed] [Google Scholar]
- Kim M. and Mullet,J.E. (1995) Identification of a sequence-specific DNA binding factor required for transcription of the barley chloroplast blue light-responsive psbD-psbC promoter. Plant Cell, 7, 1445–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kössel H. (1991) Structure and expression of rRNA genes. In Mache,R., Stutz,E. and Subramanian,A.R. (eds), The Translational Apparatus of Photosynthetic Organelles. Springer-Verlag, Berlin, Germany, pp. 1–17. [Google Scholar]
- Lakhani S., Khanna,N.C. and Tewari,K.K. (1992) Two distinct transcriptional activities of pea (Pisum sativum) chloroplasts share immunochemically related functional polypeptides. Biochem. J., 286, 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs S., Bräutigam,E. and Parthier,B. (1985) Polypeptides of DNA-dependent RNA polymerase of spinach chloroplasts: characterization by antibody-linked polymerase assays and determination of sites of synthesis. EMBO J., 4, 1661–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1988) Quantification of DNA-dependent RNA polymerase subunits and initiation factor(s) by antibody-linked polymerase assays. FEBS Lett., 234, 392–394. [DOI] [PubMed] [Google Scholar]
- Lerbs-Mache S. (1993) The 110-kDa polypeptide of spinach plastid DNA-dependent RNA polymerase: single-subunit enzyme or catalytic core of multimeric enzyme complexes. Proc. Natl Acad. Sci. USA, 90, 5509–5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liere K. and Maliga,P. (1999) In vitro characterization of the tobacco rpoB promoter reveals a core sequence motif conserved between phage-type plastid and plant mitochondrial promoters. EMBO J., 18, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link G. (1984) DNA sequence requirements for the accurate transcription of a protein-coding plastid gene in a plastid in vitro system from mustard (Sinapis alba L.). EMBO J., 3, 1697–1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.E. and Durbin,R.D. (1994) Mechanistic aspects of tagetitoxin inhibition of RNA polymerase from Escherichia coli. Biochemistry, 33, 11987–11992. [DOI] [PubMed] [Google Scholar]
- Monsalve M., Mencia,M., Rojo,F. and Salas,M. (1996) Activation and repression of transcription at two different phage φ29 promoters are mediated by interaction of the same residues of regulatory protein p4 with RNA polymerase. EMBO J., 15, 383–391. [PMC free article] [PubMed] [Google Scholar]
- Pfannschmidt T. and Link,G. (1994) Separation of two classes of plastid DNA-dependent RNA polymerase that are differentially expressed in mustard. Plant Mol. Biol., 25, 69–81. [DOI] [PubMed] [Google Scholar]
- Pfannschmidt T. and Link,G. (1997) The A and B forms of plastid DNA-dependent RNA polymerase from mustard (Sinapis alba L.) transcribe the same genes in a different developmental context. Mol. Gen. Genet., 257, 35–44. [DOI] [PubMed] [Google Scholar]
- Sexton T.B., Christopher,D.A. and Mullet,J.E. (1990) Light-induced switch in barley psbD-psbC promoter utilization: a novel mechanism regulating chloroplast gene expression. EMBO J., 9, 4485–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silhavy D. and Maliga,P. (1998) Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr. Genet., 33, 340–344. [DOI] [PubMed] [Google Scholar]
- Sriraman P., Silhavy,D. and Maliga,P. (1998a) The phage-type PclpP-53 plastid promoter comprises sequences downstream of the transcription initiation site. Nucleic Acids Res., 26, 4874–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriraman P., Silhavy,D. and Maliga,P. (1998b) Transcription from heterologous rRNA operon promoters in chloroplasts reveals requirement for specific activating factors. Plant Physiol., 117, 1495–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifa Y., Privat,I., Gagnon,J., Baeza,L. and Lerbs-Mache,S. (1998) The nuclear RPL4 gene encodes a chloroplast protein that co-purifies with the T7-like transcription complex as well as plastid ribosomes. J. Biol. Chem., 273, 3980–3985. [DOI] [PubMed] [Google Scholar]
- Vera A. and Sugiura,M. (1995) Chloroplast rRNA transcription from structurally different tandem promoters: an additional novel-type promoter. Curr. Genet., 27, 280–284. [DOI] [PubMed] [Google Scholar]
- Wegrzyn A., Szalewska-Palasz,A., Blaszczak,A., Liberek,K. and Wegrzyn,G. (1998) Differential inhibition of transcription from σ70- and σ32-dependent promoters by rifampicin. FEBS Lett., 440, 172–174. [DOI] [PubMed] [Google Scholar]
- Wu C.-Y., Lin,C.-H. and Chen,L.-J. (1997) Identification of the transcription start site for the spinach chloroplast serine tRNA gene. FEBS Lett., 418, 157–161. [DOI] [PubMed] [Google Scholar]
- Zaitlin D., Hu,J. and Bogorad,L. (1989) Binding and transcription of relaxed DNA templates by fractions of maize chloroplast extracts. Proc. Natl Acad. Sci. USA, 86, 876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. and Studier,F.W. (1997) Mechanism of inhibition of bacteriophage T7 RNA polymerase by T7 lysozyme. J. Mol. Biol., 269, 10–27. [DOI] [PubMed] [Google Scholar]
- Zubko M.K. and Day,A. (1998) Stable albinism induced without mutagenesis: a model for ribosome-free plastid inheritance. Plant J., 15, 265–271. [DOI] [PubMed] [Google Scholar]