Abstract
Regulation of ornithine decarboxylase in vertebrates involves a negative feedback mechanism requiring the protein antizyme. Here we show that a similar mechanism exists in the fission yeast Schizosaccharomyces pombe. The expression of mammalian antizyme genes requires a specific +1 translational frameshift. The efficiency of the frameshift event reflects cellular polyamine levels creating the autoregulatory feedback loop. As shown here, the yeast antizyme gene and several newly identified antizyme genes from different nematodes also require a ribosomal frameshift event for their expression. Twelve nucleotides around the frameshift site are identical between S.pombe and the mammalian counterparts. The core element for this frameshifting is likely to have been present in the last common ancestor of yeast, nematodes and mammals.
Keywords: antizyme/frameshifting/polyamines/yeast
Introduction
The efficiency of +1 ribosomal frameshifting at a specific codon is used as a sensor to regulate polyamine levels in mammalian cells. The frameshifting occurs in decoding the gene antizyme 1, which has two partially overlapping open reading frames (ORFs). Protein sequencing showed that the reading-frame shift occurs at the last codon of ORF1, causing a proportion of ribosomes to enter ORF2 to synthesize a transframe protein (Matsufuji et al., 1995). ORF2 encodes the main functional domains (Matsufuji et al., 1990; Miyazaki et al., 1992) of antizyme but has no ribosome initiation site of its own. The antizyme 1 protein binds to ornithine decarboxylase (ODC) (Murakami et al., 1992a; Li and Coffino, 1993, 1994), inhibits it (Heller et al., 1976) and targets it for degradation by the 26S proteosome without ubiquitylation (Murakami et al., 1992b, 1999). ODC catalyzes the first and usually rate-limiting step in the synthesis of polyamines, conversion of ornithine to putrescine. Putrescine is a substrate for the synthesis of spermidine and spermine. Because of its inhibition of ODC, antizyme 1 is a negative regulator of the synthesis of polyamines. In addition, antizyme 1 is a negative regulator of the polyamine transporter (Mitchell et al., 1994; Suzuki et al., 1994; Sakata et al., 1997). As discovered by Matsufuji and colleagues (Gesteland et al., 1992) and Rom and Kahana (1994), increasing polyamine levels elevate frameshifting in decoding antizyme 1 mRNA and so increase the level of antizyme 1. Since antizyme 1 negatively regulates the synthesis and uptake of polyamines, the frameshifting is the sensor for an autoregulatory circuit. A second mammalian paralog of antizyme, antizyme 2, has very similar properties to antizyme 1, including the regulatory frameshifting, but does not stimulate degradation of ODC under certain conditions where antizyme 1 is active (Ivanov et al., 1998a; Zhu et al., 1999; Y.Murakami, S.Matsufuji, I.P.Ivanov, R.F.Gesteland and J.F.Atkins, in preparation). Just like antizyme 1, antizyme 2 mRNA is ubiquitously expressed in the body but is 16 times less abundant than mRNA of antizyme 1 (Ivanov et al., 1998a). In addition to antizyme 1 and 2, mammals have a third paralog of the gene, antizyme 3 (also encoded by two ORFs), which is expressed only during spermatogenesis (Ivanov et al., 2000). Zebrafish also have multiple antizyme genes, which differ in their expression patterns and activities (Saito et al., 2000).
Numerous studies have addressed the regulation of fungal ODC in response to exogenously added polyamines. In the cases examined, Physarum polycephalum (Mitchell and Wilson, 1983), Saccharomyces cerevisiae (Fonzi, 1989; Toth and Coffino, 1999) and Neurospora crassa (Barnett et al., 1988; Williams et al., 1992), added polyamines, especially spermidine, result in significant repression of ODC activity. The mechanisms of repression seem to vary from fungus to fungus and are apparently different from the mechanism of polyamine-dependent regulation of ODC in higher eukaryotes. In some cases, the existence of an antizyme-like protein has been suggested but has either been disproved, as in the case of N.crassa (Barnett et al., 1988), or has never been substantiated, as is the case with S.cerevisiae.
As expected from their small cationic nature and ability to neutralize negative charges locally, polyamines play key roles in processes ranging from the functioning of certain ion channels (Williams, 1997), nucleic acid packaging, DNA replication, apoptosis, transcription and translation. The role of polyamines can be complex as illustrated by the transfer of the butylamine moiety of spermidine to a lysine residue to form hypusine in mammalian translation initiation factor eIF-5A, the only known substrate for this reaction (Tome et al., 1997; Lee et al., 1999). Spermine negatively regulates the growth of prostatic carcinoma cells at their primary site (Smith et al., 1995), but at later stages of tumor progression it fails to induce antizyme, which correlates with cells becoming refractory to spermine (Koike et al., 1999). Lack of antizyme function is also important in the early de-regulation of cellular proliferation in oral tumors (Tsuji et al., 1998) and probably others. The levels of polyamines are altered in many tumors, and inhibitors of polyamine synthesis are being tested for antiproliferative and cell death effects. The synthesis of ODC varies during the cell cycle in normal cells (Linden et al., 1985; Fredlund et al., 1995). It is induced by many growth stimuli and is constitutively elevated in transformed cells (Pegg, 1988; Auvinen et al., 1992) with some phosphorylated ODC being translocated to the surface membrane where it is important for mitotic cytoskeleton rearrangement events (Heiskala et al., 1999).
Antizyme is one example of certain mRNA-contained signals that can elevate specific frameshifting >1000-fold above the background level of normal translational errors. In addition to antizyme, frameshifting is also involved in the decoding of some bacterial and yeast genes and especially in many mammalian Retroviruses and Coronaviruses, plant viruses and bacterial insertion sequences (Atkins et al., 1999). The site of frameshifting in both mammalian antizyme 1 and 2 mRNAs is UCC UGA, where quadruplet translocation occurs at UCCU (underlined) to shift reading to the +1 frame, immediately before the UGA stop codon of the initiating frame (Matsufuji et al., 1995; Ivanov et al., 1998a). For the frameshifting to occur with an efficiency of 20% or more, it is important that the 3′ base of the quadruplet is the first base of a stop codon. Other important features are a pseudoknot just 3′ of the shift site and a specific sequence 5′ of the shift site (Matsufuji et al., 1995; Ivanov et al., 1998a). A pseudoknot 3′ of the shift site is a common stimulator for eukaryotic –1 frameshifting, but the synthesis of antizyme is the only known case utilizing +1 frameshifting.
Comparative analysis of RNA sequences from different organisms is informative about important features and the different options selected by evolution. Since most of the known examples of programmed frameshifting are in viruses or chromosomal mobile elements, the opportunity for comparison of frameshift cassettes in divergent organisms where the time of divergence can be approximated is limited. A start has been made with the frameshifting required for bacterial release factor 2 expression (Persson and Atkins, 1998), but antizyme provides the first opportunity for such a comparison in eukaryotes. Antizyme genes in genetically tractable lower eukaryotes would be helpful for understanding the functionally important interactions responsible for autoregulatory programmed frameshifting.
Results
Identification of an antizyme gene in Schizosaccharomyces pombe
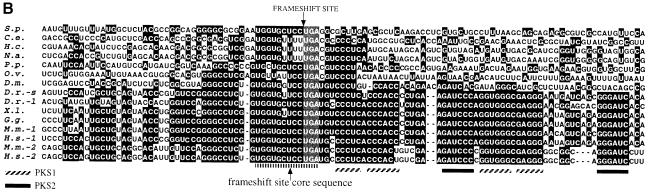
A search for DNA sequences encoding protein sequences homologous to Drosophila melanogaster antizyme (Ivanov et al., 1998b) and Homo sapiens antizyme 1 identified the same S.pombe anonymous cDNA clone (DDBJ/EMBL/GenBank accession No. D89228). The similarity is limited (∼10% identity, 24% similarity to both human antizyme 1 and D.melanogaster antizyme); however, it is highest in regions that are most highly conserved among the previously identified antizymes (Figure 1A). Closer examination of the cDNA nucleotide sequence provided further evidence that it encodes an S.pombe homolog of antizyme. The initiating AUG codon for the ORF that is similar to higher eukaryotic antizymes (ORF2 of those genes) is not the 5′-most AUG in this cDNA. In fact, there are eight AUGs closer to the 5′ end. The first or the second AUGs would initiate translation of an ORF (ORF1) that overlaps the longer downstream ORF (ORF2) such that a +1 translational frameshifting event in the overlap would generate a protein product analogous to the products of antizyme genes from higher eukaryotes. Furthermore, the last 12 nucleotides of ORF1 (UGG-UGC-UCC-UGA) are identical to the last 12 nucleotides of mammalian antizyme 1 ORF1s, including the frameshift site. Eleven of these 12 nucleotides are identical to the corresponding regions of all previously identified antizyme genes (Figure 1B). Previous experiments with the mammalian frameshift sequence tested in S.pombe have shown that this short 12 nucleotide sequence, by itself, is sufficient to stimulate measurable levels (up to 0.5%) of +1 frameshifting (Ivanov et al., 1998c). To confirm the ORF configuration of the putative S.pombe antizyme gene, a region corresponding to the two overlapping ORFs plus ∼80 nucleotides of the 5′ UTR and 370 nucleotides of the 3′ UTR, was amplified from both S.pombe genomic DNA and a cDNA library. The sequence of the amplified DNA confirmed that there are indeed two overlapping ORFs with the deduced configuration. This sequence (DDBJ/EMBL/GenBank accession No. AF217277) differs from the previously sequenced cDNA clone by three nucleotides (two in the coding region and one in the 3′ UTR); one changes an alanine codon to proline, another is a silent mutation within a proline codon. Since the sequences from the cDNA library and genomic DNA are identical, we conclude that the differences with clone No. D89228 are most likely due to strain variation. This gene contains no introns within the amplified region.
Fig. 1. Sequence comparison of different antizyme genes. (A) Amino acid comparison. Black background indicates amino acid identity among at least eight proteins. Shaded background indicates amino acid similarity among at least eight proteins. An arrow indicates the position of the frameshift site and the two most highly conserved regions are underlined. (B) Nucleotide sequence comparison of the frameshift sites. Black background indicates nucleotide identity among at least eight antizyme genes. The frameshift site is indicated. The stems of the vertebrate antizyme 3′ RNA pseudoknot and the ‘core’ sequence are underlined (PKS1 = vertebrate antizyme RNA pseudoknot stem 1; PKS2 = stem 2 of the same pseudoknot). Species abbreviations are as follows: S.p., Schizosaccharomyces pombe; C.e., Caenorhabditis elegans; H.c., Haemonchus contortus; N.a., Necator americanus; P.p., Pristioncus pacificus; O.v., Onchocerca volvulus; D.m., Drosophila melanogaster; D.r., Danio rerio; X.l., Xenopus laevis; G.g., Gallus gallus; M.m., Mus musculus; H.s., Homo sapiens.
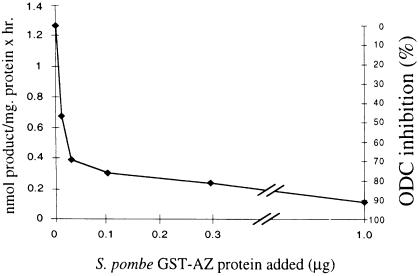
The S.pombe protein was tested for antizyme activity using a gene fusion with glutathione S-transferase (GST). In this construct, ORF1 and ORF2 of antizyme are fused in-frame by deleting the T nucleotide that encodes U of the stop codon of ORF1. This GST–antizyme fusion gene was expressed in Escherichia coli and the protein was purified by affinity chromatography. ODC inhibitory activity was tested by incubating the recombinant antizyme protein with an S.pombe crude extract and then assaying the mixture for ODC activity. The results (Figure 2) show that the recombinant protein can inhibit S.pombe ODC. GST alone (1 µg) does not inhibit S.pombe ODC (data not shown). In light of these results, the S.pombe gene will be called S.pombe ODC antizyme (SPA). Interestingly, the S.pombe ODC was also inhibited by mouse antizyme 1 and antizyme 2 (both expressed as GST fusions); however, the yeast fusion protein did not inhibit mouse ODC (data not shown).
Fig. 2. Inhibition of S.pombe ODC by SPA. Various amounts of affinity-purified GST–SPA were mixed with S.pombe ODC-active extract and assayed for ODC activity.
Deletion and overexpression of SPA
Although the effects of overexpression of antizyme on cellular physiology have been tested previously in mammalian cells, the physiological changes associated with complete absence of antizyme activity have not yet been investigated because of the complication of multiple antizymes. The single S.pombe antizyme provides the chance to explore a knockout. SPA deletion strains were generated by replacing the two ORFs of the gene with the ORFs of either URA4 or LEU2 (see Materials and methods). Complete deletion of SPA (both ORFs) did not affect the viability of S.pombe cells in rich (YE) or minimal (MM) media. Temperature had no differential effect on mutant and wild-type cell growth. Similarly, the growth rates, mating efficiencies and overall morphology of the knockout strains are apparently indistinguishable from those of wild-type cells (results not shown).
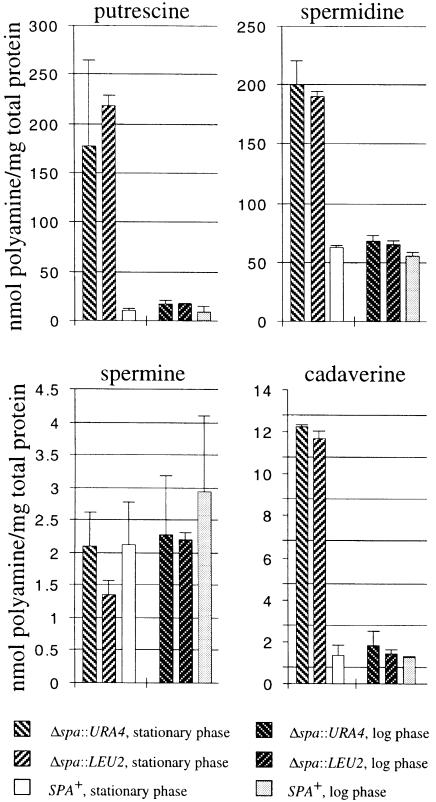
In wild-type S.pombe cells the most abundant polyamine is spermidine followed by putrescine (Figure 3). Spermine and cadaverine are found in much smaller amounts. This distribution of polyamine content is very similar to that in other fungi for which polyamine concentrations have been measured (for references, see review by Tabor and Tabor, 1985). The effect of SPA deletion on cellular polyamine contents was examined in both exponentially growing and stationary phase cells (Figure 3). The cellular concentrations of putrescine, spermidine and cadaverine (but not spermine) were higher in the knockout strains than in wild-type cells. The greatest effect was seen on putrescine and cadaverine content, with smaller effects on spermidine, presumably because eukaryotic ODC activity directly catalyzes decarboxylation of both ornithine and lysine to produce putrescine and cadaverine, respectively (Pegg and McGill, 1979), but subsequent regulatory events affect homeostasis of spermidine and spermine. The effect of inactivating antizyme on the polyamine contents in exponentially growing cells is modest (<2-fold in all cases). The effect becomes very pronounced in cells in stationary phase with up to 40- and 10-fold increases of putrescine and cadaverine contents, respectively, in the knockout strains.
Fig. 3. Polyamine contents of SPA+ and Δspa strains in logarithmic growth or stationary phase in MM media.
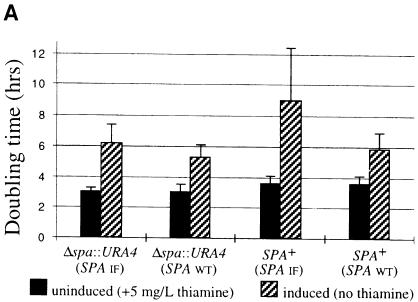
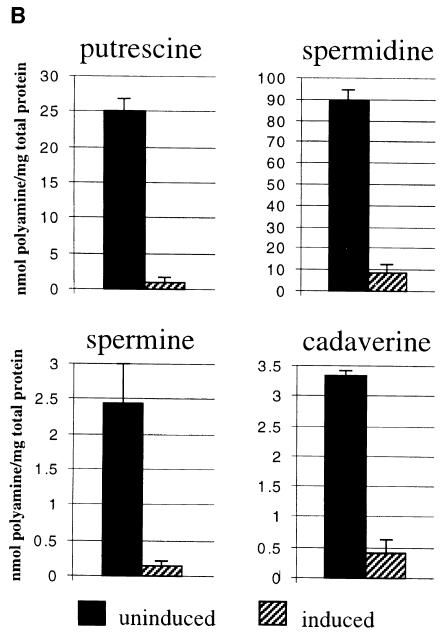
To test overexpression of SPA, two versions of the gene were cloned into pREP3 expression vector behind a strong, thiamine-repressible promoter (nmt1). One had the wild-type SPA sequence while in the second, ORF1 and ORF2 are fused in-frame. SPA wild type and an SPA deletion strain were transformed with each of the overexpression constructs. Derepression of the nmt1 promoter is a gradual process since it requires dilution of the intracellular pool of thiamine (the repressor) through cell division. After 2.5 days of exponential growth under derepressed conditions, yeast strains transformed with either SPA overexpression construct show significant increases in doubling time (Figure 4A). The growth inhibition is greater with the construct expressing the in-frame version of SPA and after prolonged incubation (5–7 days); these cells cease growth and accumulate in G1 as determined by flow cytometry (data not shown). The fact that the in-frame overexpression construct, which differs by a single nucleotide from the wild-type construct, confers a more severe phenotype is consistent with the hypothesis that translational frameshifting is required for expression of SPA. The growth phenotype associated with SPA overexpression is only partially relieved by adding 100 µM putrescine to the media (1 mM had no further effect) (data not shown). To see whether the slower growth is correlated with aberrant polyamine levels the polyamine contents of the deletion strain carrying in-frame SPA overexpression vector were measured under derepressed and repressed conditions, in both cases after 2 days of exponential growth (Figure 4B). As expected, overexpression of SPA results in significant reduction in the intracellular levels of all four polyamines. After longer (4–5 days) incubation under derepressed conditions, no putrescine and cadaverine can be detected (data not shown).
Fig. 4. Effects of SPA overexpression [with wild type (SPA WT) orin-frame (SPA IF) constructs] on cell growth of Δspa::URA4 and SPA+ strains (A) and cellular polyamine contents of Δspa::URA4 strain overexpressing SPA IF construct (B) (all in MM media).
Translational frameshifting during expression of SPA
Previously, we developed an assay for measuring antizyme translational frameshifting in both S.cerevisiae (Matsufuji et al., 1996) and S.pombe (Ivanov et al., 1998c). Briefly, the nucleotide sequence to be assayed is inserted between GST and lacZ, such that ORF1 of the assayed sequence is fused in-frame to GST, while ORF2 is fused in-frame to lacZ. β-galactosidase activity provides a measure of frameshifting efficiency. To determine whether translational frameshifting occurs in the overlap of ORF1 and ORF2 of SPA, a region of SPA including all but the first codon of ORF1 plus 180 nucleotides downstream of the ORF1 stop codon was tested. +1 frameshifting occurred at 2.2% compared with a construct in which ORF1 and ORF2 are fused in-frame. This result is consistent with +1 frameshifting being crucial for expression of SPA.
Previous experiments have shown that the frameshift cassette of mammalian antizyme 1 can direct efficient +1 frameshifting when tested in S.pombe. The reverse experiment was conducted here. The SPA gene was translated in vitro in rabbit reticulocyte lysate and its resulting frameshift efficiency measured. With no addition of polyamines, frameshifting efficiency is ∼1.5%. Addition of spermidine to the translation mixture to a final concentration of 1 mM results in a 3.7-fold increase in frameshifting to ∼5.5%, a level even higher than that observed in the endogenous system in vivo (autoradiogram not shown).
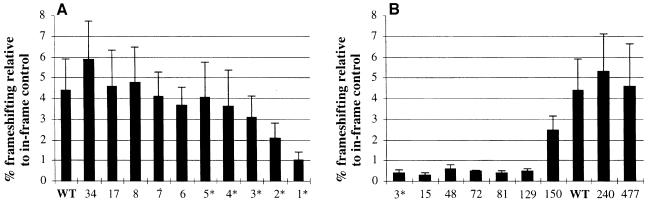
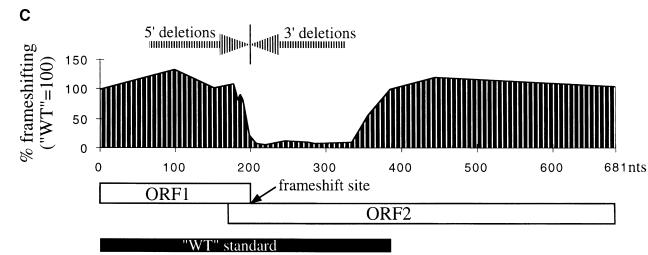
The observed efficiency of frameshifting with the SPA frameshifting cassette in vivo in S.pombe is significantly more than that expected from its limited nucleotide similarity to the antizyme frameshift sites of higher eukaryotes. This prompted a search for additional stimulatory elements within the SPA frameshift cassette. The following experiments were done in a strain carrying deletion of SPA (high polyamines) because it gives higher frameshifting and higher β-galactosidase activity in general; however, we obtained similar ratios for mutant to wild-type frameshifting efficiency in a strain with the intact SPA gene. Deleting 5′ sequences up to the third to last sense codon of ORF1 has little or no effect on frameshifting efficiency. Deleting all but the last sense codon (UCC) of ORF1 leads to a 4- to 5-fold reduction in frameshifting efficiency (Figure 5A). This implies that the conservation of the six nucleotides 5′ of the UCC-UGA frameshift site is due to their importance for stimulating +1 frameshifting. It also suggests that no additional ORF1 sequences of SPA stimulate the +1 recoding event. The 180 nucleotide 3′ region was searched for possible structure by computer RNA folding algorithms plus visual inspection. The algorithms predicted several minimal structures in that region. 3′ deletion constructs (constructs del.3,3′–81,3′) tested the importance of any putative structure on the frameshifting efficiency. The results (Figure 5B and C) show that all of these deletions lead to a significant (∼10-fold) reduction in +1 frameshifting, indicating the presence of a major 3′ stimulatory element in the 180 nucleotide region immediately following the frameshift site of SPA. However, the results indicate that none of the putative RNA structures in this region are sufficient for the activity of this element. Several additional 3′ deletions delineated the boundaries of this stimulatory element from the frameshift site to 150 and 180 nucleotides downstream (since construct del.150,3′ stimulates 5.5-fold more +1 frameshifting than del.129,3′, 150 nucleotides downstream probably contain most of the 3′ stimulator).
Fig. 5. Effects of 5′ and 3′ deletions (relative to the frameshift site) on the +1 frameshifting efficiency of SPA. The constructs were transformed in Δspa::LEU2 ura4-D18 leu1-32 ade6-M216 h– strain (high polyamine levels). (A) 5′ deletion data. Numbers on the x-axis indicate SPA ORF1 sense codons left [wild type (WT) = 66 codons left]. (B) 3′ deletion data. x-axis numbers indicate nucleotides left 3′ of the frameshift site (WT = 180 nucleotides left). * marks constructs for which in-frames were not made. In these cases frameshifting was calculated relative to the closest available in-frame construct. (C) Graphical representation of the deletion data in which the deletions are shown to scale relative to the coding region of SPA (ORF1 + ORF2). The position and size of the WT standard is also depicted. The frameshift site is indicated by an arrow.
In the experiments described above, two of the characteristics of the autoregulatory circuit of mammalian antizyme 1 were confirmed: SPA inhibition of ODC and the +1 translational frameshifting. The key question left is whether the recoding event is responsive to polyamine levels in cells. As shown above, overexpression of SPA leads to significant reduction of polyamine levels in S.pombe. An SPA+ strain was co-transformed with an SPA wild-type overexpressing plasmid (cells overexpressing wild-type SPA grow slowly but continuously) and a construct that monitors the +1 frameshifting from an SPA frameshift sequence. The +1 frameshifting was compared with that in SPA non-overexpressing cells (in both cases frameshifting was measured relative to in-frame control). The results (Figure 6) show a significant reduction (6.5-fold) in frameshifting efficiency in SPA-overproducing cells that correlates with a decrease of polyamine content (4.5-fold for putrescine and 3.9-fold for spermidine). This indicates that polyamines modulate the frameshifting efficiency of SPA. An alternative but less likely possibility is that SPA overexpression reduces frameshifting because high levels of SPA transcript titrate some factor limiting for frameshifting.
Fig. 6. Effect of polyamine depletion on SPA +1 frameshifting. Polyamine depletion is achieved by overexpression of the wild-type version of SPA. The same cultures were assayed both for frameshifting and polyamine content. Numbers above columns indicate fold reduction of frameshifting and polyamine content compared with cells that do not overexpress SPA.
The SPA frameshift signals direct 2-fold more frameshifting in Δspa::LEU2 cells (4.4%) than in SPA+ cells (in both cases the measurement is done during stationary phase); however, the relatively high standard deviations for both measurements make it difficult to draw firm conclusions from this particular result.
Identification of antizyme genes in nematodes
A search of Caenorhabditis elegans expressed sequence tag (EST) sequences with mammalian antizyme 1 sequence identified 20 clones. These sequences could be deconvoluted into a contiguous cDNA sequence. Primers designed on the basis of this sequence were used to PCR amplify and subclone this cDNA from a C.elegans cDNA library. The sequence of the subcloned cDNA was confirmed (DDBJ/EMBL/GenBank accession No. AF217278); the subsequently released genomic sequence of this C.elegans gene (DDBJ/EMBL/GenBank accession No. AF040659) confirms our cDNA data. The amino acid sequence deduced from the cDNA sequence revealed that the longer ORF has similarity to previously reported antizyme sequences (overall 27% identity, 39% similarity to human antizyme 1; 19% identity, 34% similarity to Drosophila antizyme). These similarities are higher than that of SPA to these two antizyme genes and again are concentrated in the regions most highly conserved among previously identified antizymes (Figure 1A). Just like mammalian antizymes, the longer ORF (ORF2) lacks an appropriate in-frame initiation codon, and expression could be provided by initiation in a short upstream overlapping ORF (ORF1) leading to +1 ribosomal frameshifting in the overlap. The putative C.elegans antizyme frameshift site (the nucleotides proximal to the end of ORF1) has 18 of 26 nucleotides identical to the consensus sequence for antizyme frameshift sites (Figure 1B).
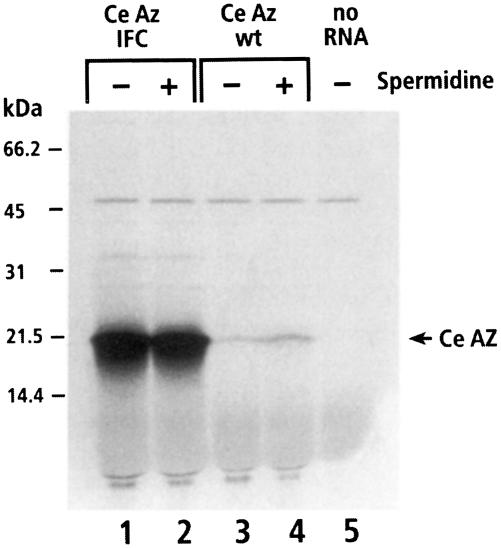
Frameshifting for expression of C.elegans antizyme was investigated in heterologous systems. Two constructs containing the entire antizyme cDNA, one with the wild-type sequence and one with a single nucleotide deletion that fuses ORF1 to ORF2 in-frame (in-frame control), were transcribed in vitro and the RNA was translated in rabbit reticulocyte lysate. The products were examined by SDS–PAGE (Figure 7). The main product from both constructs has an apparent Mr of 21 kDa, slightly greater than the predicted Mr of 17.7 kDa [aberrant, slower than expected, mobility is observed with antizyme proteins from other species (Ivanov et al., 1998a)]. From the ratio of wild-type to in-frame product, we estimate that the efficiency of frameshifting of C.elegans antizyme in reticulocyte lysate is ∼0.8%, which is somewhat lower than SPA frameshifting in the same system. Addition of spermidine to the translation reactions almost doubles the efficiency of frameshifting to ∼1.5% (the exact numbers are not easy to determine because of difficulty in defining background values). The frameshifting properties of C.elegans antizyme mRNA were also tested in vivo in S.pombe cells. A sequence including all but the first codon of ORF1 plus 180 nucleotides downstream was inserted between GST and lacZ of the PIU-LAC plasmid. Comparison of the β-galactosidase activity of cells (Δspa::LEU2 strain) transformed with the wild-type construct and the in-frame control constructs indicated 3.5% +1 frameshifting. From the frameshifting observed in the heterologous systems, as well as the sequence considerations discussed above, we conclude that expression of this C.elegans gene requires ribosomal frameshifting.
Fig. 7. Frameshifting of C.elegans antizyme (AZ) mRNA in reticulocyte lysates. Translation products were separated on a 17.5% SDS–PAGE gel. Normally in vitro translation reactions contain 0.5 mM spermidine (‘–’, 0.5 mM and ‘+’, 1.0 mM spermidine). IFC, in-frame control.
Searching the EST database with the newly discovered C.elegans antizyme identified antizyme orthologs in four other nematode species. In two cases (Necator americanus and Haemonchus contortus), the cDNA sequences in the database were sufficient to make contigs of the complete coding regions. In the other two cases [Onchocerca volvulus (DDBJ/EMBL/GenBank accession No. AF217279) and Pristioncus pacificus (DDBJ/EMBL/GenBank accession No. AF217280)] the complete cDNA sequences were obtained by PCR amplifying and sequencing the full genes from cDNA libraries. As with the previously identified eukaryotic antizyme genes, the ORF configuration of the newly found nematode orthologs implies the necessity for +1 frameshifting for synthesis of full-length protein.
The C.elegans antizyme mRNA frameshift site UUU-UGA is unique, differing from the UCC-UGA of previously known antizyme mRNAs. The C.elegans antizyme gene shares this feature with N.americanus and H.contortus but not with P.pacificus and O.volvulus antizymes. The phylogenetic tree of nematode antizyme protein sequences matches exactly the phylogenetic relationship (Blaxter, 1998) of the nematodes expressing them, indicating that these gene sequences are the result of divergent evolution within the nematode lineage (data not shown). These results also show that the UUU-UGA frameshift site evolved after the last common ancestor of P.pacificus and C.elegans but before the divergence of C.elegans, N.americanus and H.contortus (probably 450–500 million years ago).
The ability of UUU-UGA sequence to direct +1 frameshifting was further tested in a mammalian system in the context of the mammalian antizyme mRNA (i.e. in the presence of the 3′ RNA pseudoknot and 5′ stimulator). A BMV-coat-protein–antizyme 1 gene fusion construct, which has a TCC-TGA to TTT-TGA substitution, was transcribed and then translated in a rabbit reticulocyte lysate. Eleven percent frameshift efficiency was seen in the absence of exogenously added polyamines, 2.2 times the efficiency seen with the UCC-UGA transcript. The frameshift efficiency becomes 18% when 0.6 mM spermidine is added, which is 1.3 times that with the wild type (Matsufuji et al., 1995). Similar results were obtained in cultured mammalian (Cos7) cells transfected with TTT-TGA mutant construct, the frameshift being higher than that of wild-type construct in both high- and low-polyamine conditions (our unpublished results). These results demonstrate that the putative C.elegans frameshift site (UUU-UGA) is, if anything, shiftier than UCC-UGA in the antizyme 1 context and is subject to polyamine stimulation.
Discussion
The results presented show that the yeast S.pombe has a homolog of mammalian antizyme. This is the first documented example of antizyme-type regulation of ODC in a lower eukaryote.
Deleting SPA from the yeast genome has no detectable effect on viability or any other overt phenotypic effect but, as expected, it results in altered accumulation of polyamines in the cell. Interestingly, the effect is most pronounced in cells in stationary phase, where the knockout cells accumulate up to 40 times more putrescine than wild-type counterparts. This compares with a <2-fold increase of putrescine in exponentially growing cells. A likely explanation for this observation is that the usual rate of ornithine decarboxylation in exponentially growing cells is close to capacity given ‘normal’ concentrations of substrate, enzyme and product. At the same time, all newly synthesized polyamines are continuously diluted through cell growth and division at a rate that is almost identical to the rate of maximum capacity synthesis. Cells in stationary phase can no longer dilute newly synthesized polyamines, and more importantly lack an effective antizyme- independent mechanism of shutting off ODC. This suggests that SPA is the primary regulator of ODC activity in S.pombe, not only during cell growth (short term regulation) but also in non-dividing cells (longer term regulation).
Overexpression of SPA (5–7 days derepression) leads to complete depletion of intracellular putrescine. This result implies that in S.pombe ornithine decarboxylation is the only source of putrescine synthesis (the pathway from arginine via agmatine is not utilized). The complete depletion of cadaverine in SPA overexpressing cells suggests that ODC is the only enzyme in S.pombe that can decarboxylate lysine, which is also the case in rat tissues (Pegg and McGill, 1979).
It is somewhat perplexing that addition of putrescine to the media leads to only partial relief of the growth phenotype associated with SPA overexpression. There are two likely explanations. (i) Perhaps S.pombe imports putrescine poorly. (ii) Alternatively, like the mammalian system, maybe SPA inhibits not only ODC but also the polyamine transporter. Further experiments will help to distinguish between these two models.
It is unclear how widespread the antizyme gene is within the fungal kingdom. We have identified and cloned antizyme homologs from two other fission yeasts (Schizosaccharomyces octosporus and Schizosaccharomyces japonicus) and from two distantly related fungi (Botryotinia fuckeliana and Emericella nidulans) (our unpublished results). The antizyme frameshift site of the latter two fungi has evolved in a unique way different from all other known antizymes, but nevertheless even these two distantly related fungi have conserved the autoregulatory +1 frameshifting. The fact that the yeast S.pombe has an antizyme gene suggests the possibility that the higher eukaryotic metazoans may all have an antizyme gene.
The only previously reported antizyme activity in unicellular organisms is from E.coli, but recent analyses suggest that E.coli does not have a true antizyme (Ivanov et al., 1998d). This makes SPA the first bona fide antizyme in a unicellular organism.
The remarkable similarity of the core sequence important for antizyme frameshifting from S.pombe to humans could be due to convergent or divergent evolution. The near identity of this sequence in worms, Drosophila, Xenopus, zebrafish and humans argues against convergent evolution, as if antizyme frameshifting arose in a common ancestor perhaps more than one billion years ago.
Three cis-acting RNA elements are known to stimulate mammalian antizyme 1 frameshifting. One is a 50 nucleotide sequence immediately 5′ of the shift site (Matsufuji et al., 1995; our unpublished results). A second stimulator is the UGA stop codon of ORF1 and the third is an RNA pseudoknot starting 3 nucleotides 3′ of the UGA stop codon. Among frameshift sites of the previously identified antizymes from mammals all the way to Drosophila, there is substantial similarity in the sequences immediately 5′ of the shift site. Sixteen of the last 18 nucleotides of ORF1 are completely conserved in these genes. Schizosaccharomyces pombe and C.elegans antizymes have 9 of 9 and 6 of 9 (14 out of 19 in O.volvulus) nucleotides identical to the consensus, respectively. For the 5′ sequences, generally, the more distantly related two antizymes are, the more the similarity is confined to the 3′ end of that region. Our SPA ORF1 deletion data show that mutation of nucleotides that are part of the 5′ consensus sequence leads to reduced frameshifting efficiency. This is another indication that conservation of nucleotide sequence in this region is because of its importance for stimulating efficient +1 frameshifting. It is quite striking that in all antizyme gene sequences identified so far, including a number of unpublished ones, ORF1 ends with a UGA stop codon. This is particularly surprising since any of the other two stop codons can substitute for UGA to stimulate antizyme 1 frameshifting, although slightly less efficiently, in vitro (Matsufuji et al., 1995) and in vivo (our unpublished results).
The 3′ pseudoknot that stimulates frameshifting in antizyme 1 is highly conserved in all known vertebrate antizymes, including mammalian antizyme 2 (Figure 1B). None of the invertebrate antizyme mRNAs identified so far, including those presented here, has a sequence in the equivalent region that can be simply folded to a comparable RNA structure. However, sequences immediately 3′ of the frameshift site are conserved between invertebrates and vertebrates. The conservation of this region between Drosophila and the vertebrate counterparts has already been noted (Ivanov et al., 1998b). The C.elegans antizyme gene contains the sequence YGYCCCYCA (Y = pyrimidine) in this region, which is identical to the consensus. The antizyme genes from the other four nematodes also have a similar sequence (Figure 1B). The significance of this similarity is not clear [in fact, sequences in this region appear to play no role in antizyme 1 in vitro frameshifting outside of the RNA pseudoknot context (Matsufuji et al., 1995)].
Only two examples are known where RNA elements 3′ of the frameshift site stimulate +1 frameshifting. One is the RNA pseudoknot of mammalian antizyme 1 and the second is a short RNA sequence immediately following the frameshift site of Ty3 (Farabaugh et al., 1993). Additional examples would be very helpful in deciphering the role such elements play in the mechanism of +1 frameshifting. It is currently not known how many and which of the invertebrate antizyme genes contain 3′ frameshift stimulators. The results presented here show that an S.pombe 3′ stimulator enhances frameshifting ∼10-fold. This stimulator appears completely different from the 3′ RNA pseudoknot in vertebrates. Our deletion experiments indicate that none of the predicted RNA structures contained within the minimally required 3′ region [up to 150–180 nucleotides downstream of the frameshift site (Figure 5C)] are sufficient to confer the stimulatory effect. The SPA 3′ stimulator may act directly through sequence or may have an unusual RNA structure involving non-Watson–Crick base pairing. More detailed mutagenesis combined with phylogenetic analysis would be required to discern the nature of the 3′ stimulator of SPA.
The nematode antizymes were analyzed for the presence of possible 5′ or 3′ stimulators flanking the core frameshift site. Computer RNA folding programs did not identify any potentially interesting structure. More importantly, phylogenetic analysis with the five identified nematode antizymes failed to identify any conservation of primary RNA sequence (or for that matter potential secondary structure) outside of the core region that is shared between two or more members. This could indicate that no such extra cis-acting stimulators exist in nematode antizymes or that they are located in a very different place within the mRNA, for example the 3′ untranslated region (the latter suggestion is not supported by our sequence analysis).
A common mechanism for frameshifting is re-pairing of the peptidyl tRNA in the new reading frame. However, an alternative mechanism whereby the peptidyl tRNA merely occludes the first base of the next codon, has been documented for yeast Ty3 frameshifting (Farabaugh et al., 1993). Results of experiments with some mutants of the mammalian antizyme 1 shift site pointed to an occlusion mechanism (Matsufuji et al., 1995). However, the mechanism with the wild-type, UCC-UGA, shift site is not clear. For C.elegans antizyme the UUU-UGA sequence would be an obvious candidate for a re-pairing since Phe-tRNA could pair perfectly with UUU in both frames. But with UCC-UGA the Ser-tRNA first reading UCC could at best pair two out of three with CCU. This important problem warrants further investigation.
The frameshift efficiency of SPA frameshift site is lower than that observed with mammalian antizyme 1 even when both are tested in the same organism (S.pombe) [for the frameshift efficiency of antizyme 1 cassette in S.pombe, see Ivanov et al. (1998c)]. It is possible that the observed efficiencies for S.pombe antizyme are artificially low because the constructs do not include all the cis-acting stimulatory elements. On the other hand there is no reason why a lower level of frameshifting does not correctly reflect the evolved balance with the other characteristics of the complex system such as relative protein stabilities.
Like other core cellular processes, the antizyme polyamine regulatory scheme is conserved from yeast S.pombe to human. It is not obvious why this very special mechanism is so exquisitely preserved over vast evolutionary time. Perhaps there is another whole aspect to the system that our experiments do not yet detect. From this viewpoint it would seem very important to exploit the genetics systems of S.pombe and C.elegans to understand more thoroughly the physiological effects of perturbing the antizyme system.
Materials and methods
DNA manipulation and sequencing
The SPA gene was amplified using the following primers: 5′-CAAAACAAGTTTTCATTATTGGTTTTTTTTAAATCAATCCCC (sense) and 5′-CGTAAATCCAATCTAAATTTAATCTTCAACTAAATCATGAAAAGCCTC (antisense). The S.pombe cDNA library used as a template in the amplification was kindly provided by R.Rowley (University of Utah). The C.elegans antizyme gene was amplified using the following primers: 5′-CCCAGGAATTCCTCGAGTATTTTGA GTATAATTTTAC (sense) and 5′-CGGCCGCTCGAGTTAGACCTT GTAGCTCATGATG (antisense). This same amplified DNA was used to make the constructs for in vitro transcription and translation of C.elegans antizyme by cloning it into pTZ18U plasmid using the SacI and HindIII sites incorporated in the two primers. The in-frame construct was made using a two-step PCR. The cDNA sequences of O.volvulus and P.pacificus antizyme genes were obtained by performing 5′ and 3′ RACE PCR with cDNA libraries, which were kindly provided by Ralf Sommer (P.pacificus) and Susan Haynes (O.volvulus). The SPA overexpression constructs were made by amplifying the gene with the primers 5′-GCATCCGAATTCCCAAATCCAAGCATCATACGCC (sense) and 5′-GCATCCGGATCCGCCAGTGTTCTTACTTTGAGA TGC (antisense), and then inserting BamHI-digested product between the MscI and BamHI sites of pREP3 plasmid. The in-frame construct was made by two-step PCR and subsequently all in-frame SPA constructs described below were made by one-step PCR using this plasmid’s DNA as a template. To make the constructs for frameshift assays in S.pombe, DNA fragments with a given nucleotide length (as described in the main text), were amplified from both the SPA and C.elegans antizyme constructs described above. These fragments were then cloned between the KpnI and BstEII sites of PIU-LAC plasmid (Ivanov et al., 1998c). The PCR primers included an ‘AC’ spacer between the 5′ cloning site (BstEII) and the antizyme sequences in order to correct the reading frame. The in vivo frameshifting assays in S.pombe (strains ura4-D18 leu1-32 ade6-M216 h– and Δspa::LEU2 ura4-D18 leu1-32 ade6-M216 h–) were done as described (Ivanov et al., 1998c). The plasmid for GST–SPA expression was made by PCR amplifying SPA (all but the first codon of ORF1 through the downstream ORF2) from an in-frame template and cloning the product into the EcoRI and XhoI restriction sites of pGEX-5X-3 plasmid. The antizyme frameshift site in the BMV-coat-antizyme fusion construct (C3NE) (Matsufuji et al., 1995) was mutated with a two-step PCR. To generate the two knockout strains, Δspa::URA4 and Δspa::LEU2, both ORFs of SPA were replaced exactly with the ORF of either URA4 or LEU2. To accomplish this, two pairs of primers amplified URA4 and LEU2 such that 50–60 nucleotides, which normally flank the two ORFs of SPA, flank the ORFs of the two genes. The amplified DNA products were gel purified and 2 µg of each were used to electroporate into ura4-D18 leu1-32 ade6-M216 h– cells. URA+ and LEU+ transformants were selected by growth on URA– and LEU– media, respectively. PCR screen and partial sequencing, with primers flanking the regions used for the homologous recombination, confirmed the SPA disruptions. All DNA clones were sequenced with automated sequencing machines (ABI 100).
ODC antizyme assays
Schizosaccharomyces pombe ODC active crude extracts were prepared as follows: S.pombe (strain 1519, leu1-32, h–) provided by R.Rowley was grown to OD600 0.7 in 50 ml of minimal media + LEU. Ten milligrams of lysing enzymes (Sigma) were added, followed by continued incubation for 30 min at 30°C. Cells were harvested and washed once with cold homogenization buffer [25 mM Tris–HCl pH 7, 0.25 M sucrose, 1 mM dithiothreitol (DTT), 20 µM pyridoxal-5-phosphate, 2 mM EDTA] then resuspended in 0.75 ml of homogenization buffer. Cells were broken open and the lysate was clarified by centrifugation at 10 000 r.p.m. for 15 min at 4°C. Extracts were dialyzed overnight in dialysis buffer (25 mM Tris–HCl pH 7.4, 1 mM DTT, 20 µM pyridoxal-5-phosphate, 0.1 mM EDTA). A volume of 25 µl of extract was used for each ODC assay. ODC activity was assayed by measuring the release of 14CO2 from l-[1-14C]ornithine (Amersham) as described (Nishiyama et al., 1988). Each reaction took 1 h. Pre-incubation of S.pombe extract with 0.1 mM difluoromethyl ornithine (DFMO) for 15 min led to >99% inhibition of 14CO2 release.
Polyamine measurements
The cells were collected by centrifugation, washed twice with 1 ml of phosphate-buffered saline (PBS) and then the pellet was frozen at –80°C until use. The pellet was resuspended in 0.1 ml of PBS. An aliquot of the suspension was mixed with an equal volume of 8% perchloric acid, vortexed for 1 min, kept on ice for 5 min and centrifuged at 15 000 r.p.m., 4°C for 5 min. Ten microliters of the supernatant were subjected to polyamine analysis using fluorometry on high-performance liquid chromatography as described previously (Murakami et al., 1989). Protein concentrations were determined with the BCA protein assay kit (Pierce).
In vitro transcription and translation
The experiments with the BMV-coat-antizyme fusion constructs were performed as described previously (Matsufuji et al., 1995). All other plasmid DNA templates were prepared using QIAGEN Miniprep Kit and then digested with HindIII. Transcripts for SPA in vitro translation were made from PCR templates that had a T7 promoter incorporated into the PCR primers. Linearized DNA (1 µg) was used as a template for in vitro transcription with Ambion MEGAshortscriptTM T7 Kit. The DNase-treated RNAs were recovered and resuspended in 40 µl of RNase-free water. One microliter of each specified transcript suspension was used in each in vitro translation reaction [0.5 µl of 1 mM amino acid mix –Met, 7 µl of reticulocyte lysate (Promega), 0.5 µl of [35S]Met (Amersham)] to a total volume of 10 µl. The reactions were stopped by adding 1 µl of RNase (10 mg/ml). The frameshift efficiencies were quantified as described (Ivanov et al., 1998a).
Acknowledgments
Acknowledgements
We thank Roy Rowley for invaluable help with S.pombe. We thank Jasmine Parvaz and Rebecca Sharp for technical help. This work was supported by NIH Genetics Training Grant # 5T32GM07464-23 to I.P.I., Grants-in-Aid from the Japanese Ministry of Education, Science and Culture to Y.M. and S.M., Department of Energy grant DEFG03-99ER62732 to R.F.G. and NIH grant GM48152 to J.F.A.
References
- Atkins J.F., Böck,A., Matsufuji,S. and Gesteland,R.F. (1999) Dynamics of the genetic code. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2nd edn, pp. 637–673. [Google Scholar]
- Auvinen M., Paasinen,A., Andersson,L.C. and Hölttä,E. (1992) Ornithine decarboxylase is critical for cell transformation. Nature, 360, 355–358. [DOI] [PubMed] [Google Scholar]
- Barnett G.R., Seyfzadeh,M. and Davis,R.H. (1988) Putrescine and spermidine control degradation and synthesis of ornithine decarboxylase in Neurospora crassa. J. Biol. Chem., 263, 10005–10008. [PubMed] [Google Scholar]
- Blaxter M. (1998) Caenorhabditis elegans is a nematode. Science, 282, 2041–2046. [DOI] [PubMed] [Google Scholar]
- Farabaugh P.J., Zhao,H. and Vimaladithan,A. (1993) A novel programmed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: frameshifting without tRNA slippage. Cell, 74, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonzi W.A. (1989) Regulation of Saccharomyces cerevisiae ornithine decarboxylase expression in response to polyamine. J. Biol. Chem., 264, 18110–18118. [PubMed] [Google Scholar]
- Fredlund J.O., Johansson,M.C., Dahlberg,E. and Oredsson,T.M. (1995) Ornithine decarboxylase and S-adenosylmethionine decarboxylase expression during the cell cycle of Chinese hamster ovary cells. Exp. Cell Res., 216, 86–92. [DOI] [PubMed] [Google Scholar]
- Gesteland R.F., Weiss,R.B. and Atkins,J.F. (1992) Recoding: reprogrammed genetic decoding. Science, 257, 1640–1641. [DOI] [PubMed] [Google Scholar]
- Heiskala M., Zhang,J., Hayashi,S., Hölttä,E. and Andersson,L.C. (1999) Translocation of ornithine decarboxylase to the surface membrane during cell activation and transformation. EMBO J., 18, 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J.S., Fong,W.F. and Canellakis,E.S. (1976) Induction of a protein inhibitor to ornithine decarboxylase by the end products of its reaction. Proc. Natl Acad. Sci. USA, 73, 1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Gesteland,R.F. and Atkins,J.F. (1998a) A second mammalian antizyme: conservation of programmed ribosomal frameshifting. Genomics, 52, 119–129. [DOI] [PubMed] [Google Scholar]
- Ivanov I.P., Simin,K., Letsou,A., Atkins,J.F. and Gesteland,R.F. (1998b) The Drosophila gene for antizyme requires ribosomal frameshifting for expression and contains an intronic gene for snRNP Sm D3 on the opposite strand. Mol. Cell. Biol., 18, 1553–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Gesteland,R.F., Matsufuji,S. and Atkins,J.F. (1998c) Programmed frameshifting in the synthesis of mammalian antizyme is +1 in mammals, predominantly +1 in fission yeast, but –2 in budding yeast. RNA, 4, 1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.P., Gesteland,R.F. and Atkins,J.F. (1998d) Does antizyme exist in Escherichia coli? Mol. Microbiol., 29, 1521–1522. [DOI] [PubMed] [Google Scholar]
- Ivanov I.P., Rohrwasser,A., Terreros,D.A., Gesteland,R.F. and Atkins,J.F. (2000) Discovery of spermatogenesis, stage specific, ornithine decarboxylase antizyme: antizyme 3. Proc. Natl Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike C., Chao,D.T. and Zetter,B.R. (1999) Sensitivity to polyamine-induced growth arrest correlates with antizyme induction in prostate carcinoma cells. Cancer Res., 59, 6109–6112. [PubMed] [Google Scholar]
- Lee Y.B., Joe,Y.A., Wolff,E.C., Dimitriadis,E.K. and Park,M.H. (1999) Complex formation between deoxyhypusine synthase and its protein substrate, the eukaryotic translation factor 5A (eIF5A) precursor. Biochem. J., 340, 273–281. [PMC free article] [PubMed] [Google Scholar]
- Li X. and Coffino,P. (1993) Degradation of ornithine decarboxylase: exposure of the C-terminal target by a polyamine-inducible inhibitory protein. Mol. Cell. Biol., 13, 2377–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. and Coffino,P. (1994) Distinct domains of antizyme required for binding and proteolysis of ornithine decarboxylase. Mol. Cell. Biol., 14, 87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linden M., Anehus,S., Langstrom,E., Baldetorp,B. and Heby,O. (1985) Cell cycle phase-dependent induction of ornithine decarboxylase-antizyme. J. Cell Physiol., 125, 273–276. [DOI] [PubMed] [Google Scholar]
- Matsufuji S., Miyazaki,Y., Kanamoto,R., Kameji,T., Murakami,Y., Baby,T.G., Fujita,K., Ohno,T. and Hayashi,S. (1990) Analyses on ornithine decarboxylase antizyme mRNA with a cDNA cloned from rat liver. J. Biochem., 108, 365–371. [DOI] [PubMed] [Google Scholar]
- Matsufuji S., Matsufuji,T., Miyazaki,Y., Murakami,Y., Atkins,J.F., Gesteland,R.F. and Hayashi,S. (1995) Autoregulatory frameshifting in decoding mammalian ornithine decarboxylase antizyme. Cell, 80, 1360–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsufuji S., Matsufuji,T., Wills,N.M., Gesteland,R.F and Atkins,J.F. (1996) Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J., 15, 1360–1370. [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.L. and Wilson,J.M. (1983) Polyamine-stimulated alteration of the ornithine decarboxylase molecule in Physarum polycephalum. Biochem. J., 214, 345–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell J.L.A., Judd,G.G., Bareyal-Leyser,A. and Ling,S.Y. (1994) Feedback repression of polyamine transport is mediated by antizyme in mammalian tissue-culture. Biochem. J., 299, 19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki Y., Matsufuji,S. and Hayashi,S. (1992) Cloning and characterization of a rat gene encoding ornithine decarboxylase antizyme. Gene, 113, 191–197. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Nishiyama,M. and Hayashi,S. (1989) Involvement of antizyme in stabilization of ornithine decarboxylase caused by inhibitors of polyamine synthesis. Eur. J. Biochem., 180, 181–184. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Miyazaki,Y. and Hayashi,S. (1992a) Destabilization of ornithine decarboxylase by transfected antizyme gene expression in hepatoma tissue culture cells. J. Biol. Chem., 267, 13138–13141. [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Kameji,T., Hayashi,S., Igarashi,K., Tamura,T., Tanaka,K. and Ichihara,A. (1992b) Ornithine decarboxylase is degraded by the 26S proteosome without ubiquitination. Nature, 360, 597–599. [DOI] [PubMed] [Google Scholar]
- Murakami Y., Matsufuji,S., Hayashi,S., Tanahashi,N. and Tanaka,K. (1999) ATP-dependent inactivation and sequestration of ornithine decarboxylase by the 26S proteosome are prerequisites for degradation. Mol. Cell. Biol., 19, 7216–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama M., Matsufuji,S., Kanamoto,R., Takano,M., Murakami,Y. and Hayashi,S. (1988) Two-step purification of mouse kidney ornithine decarboxylase. Prep. Biochem., 18, 227–238. [DOI] [PubMed] [Google Scholar]
- Pegg A.E. (1988) Polyamine metabolism and its importance in neoplastic growth and as a target for chemotherapy. Cancer Res., 48, 759–774. [PubMed] [Google Scholar]
- Pegg A.E. and McGill,S. (1979) Decarboxylation of ornithine and lysine in rat tissues. Biochim. Biophys. Acta, 568, 416–427. [DOI] [PubMed] [Google Scholar]
- Persson B.C. and Atkins,J.F. (1998) Does disparate occurrence of autoregulatory programmed frameshifting in decoding the release factor 2 gene reflect an ancient origin with loss in independent lineages? J. Bacteriol., 180, 3462–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rom E. and Kahana,C. (1994) Polyamines regulate the expression or ornithine decarboxylase antizyme in vitro by inducing ribosomal frame-shifting. (Correction published in Proc. Natl Acad. Sci. USA, 91, 9195.) Proc. Natl Acad. Sci. USA, 91, 3959–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T., Hascilowicz,T., Ohkido,I., Kikuchi,Y., Okamoto,H., Hayashi,S., Murakami,Y. and Matsufuji,S. (2000) Two zebrafish antizymes with different expression and activities. Biochem. J., 345, 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakata K., Fukuchi-Shimogori,T., Kashiwagi,K. and Igarashi. K. (1997) Identification of regulatory region of antizyme necessary for the negative regulation of polyamine transport. Biochem. Biophys. Res. Commun., 238, 415–419. [DOI] [PubMed] [Google Scholar]
- Smith R.C., Litwin,M.S. and Zetter,B.R. (1995) Identification of an endogenous inhibitor of prostatic carcinoma cell growth. Nature Med., 1, 1040–1045. [DOI] [PubMed] [Google Scholar]
- Suzuki T., He,Y., Kashiwagi,K., Murakami,Y., Hayashi,S. and Igarashi,K. (1994) Antizyme protects against abnormal accumulation and toxicity of polyamines in ornithine decarboxylase-overproducing cells. Proc. Natl Acad. Sci. USA, 91, 8930–8934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor C.W. and Tabor,H. (1985) Polyamines in microorganisms. Microbiol. Rev., 49, 81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tome M.E., Fiser,S.M., Payne,C.M. and Gerner,E.W. (1997) Excess putrescine accumulation inhibits the formation of modified eukaryotic initiation factor 5A (eIF-5A) and induces apoptosis. Biochem. J., 328, 847–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth C. and Coffino,P. (1999) Regulated degradation of yeast ornithine decarboxylase. J. Biol. Chem., 274, 25921–25926. [DOI] [PubMed] [Google Scholar]
- Tsuji T., Todd,R., Meyer,C., McBride,J., Liao,P.-H., Huang,M.-F., Chou,M.-Y., Donoff,R.B. and Wong,D.T.W. (1998) Reduction of ornithine decarboxylase antizyme (ODC-Az) level in the 7,12-dimethylbenz(a)anthracene-induced hamster buccal pouch carcinogenesis model. Oncogene, 16, 3379–3385. [DOI] [PubMed] [Google Scholar]
- Williams K. (1997) Interactions of polyamines with ion channels. Biochem. J., 325, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L.J., Barnett,G.R., Ristow,J.L., Pitkin,J., Perriere,M. and Davis,R.H. (1992) Ornithine decarboxylase gene of Neurospora crassa: isolation, sequence and polyamine-mediated regulation of its mRNA. Mol. Cell. Biol., 12, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C., Lang,D.W. and Coffino,P. (1999) Antizyme2 is a negative regulator of ornithine decarboxylase and polyamine transport. J. Biol. Chem., 274, 26425–26430. [DOI] [PubMed] [Google Scholar]