Abstract
The MSL5 gene, which codes for the splicing factor BBP/ScSF1, is essential in Saccharomyces cerevisiae, yet previous analyses failed to reveal a defect in assembly of (pre)-spliceosomes or in vitro splicing associated with its depletion. We generated 11 temperature-sensitive (ts) mutants and one cold-sensitive (cs) mutant in the corresponding gene and analyzed their phenotypes. While all mutants were blocked in the formation of commitment complex 2 (CC2) at non-permissive and permissive temperature, the ts mutants showed no defect in spliceosome formation and splicing in vitro. The cs mutant was defective in (pre)-spliceosome formation, but residual splicing activity could be detected. In vivo splicing of reporters carrying introns weakened by mutations in the 5′ splice site and/or in the branchpoint region was affected in all mutants. Pre-mRNA leakage to the cytoplasm was strongly increased (up to 40-fold) in the mutants. A combination of ts mutants with a disruption of upf1, a gene involved in nonsense-mediated decay, resulted in a specific synthetic growth phenotype, suggesting that the essential function of SF1 in yeast could be related to the retention of pre-mRNA in the nucleus.
Keywords: KH domain/nonsense-mediated decay/Saccharomyces cerevisiae/snRNP/spliceosome
Introduction
Gene expression is a highly ordered process that has to be controlled temporally and spatially. In eukaryotic cells, the RNA transcript undergoes several processing steps in the nucleus before it is exported to the cytoplasm. Pre-mRNA splicing is required to remove introns from the transcript in the nucleus. Introns often contain stop codons in-frame with the upstream protein coding sequence. Thus, escape of the unspliced pre-mRNA from the nucleus would result in the accumulation of aberrant RNAs in the cytoplasm, possibly leading to the production of truncated and potentially deleterious proteins. Therefore, tight control to prevent undesired pre-mRNA export is an essential requirement for gene expression.
How can the cell distinguish between intron-containing pre-mRNAs, spliced mRNAs and intronless mRNAs to ensure that the first are retained in the nucleus while the latter two are exported to the cytoplasm? In yeast, a pioneering study using a reporter system that allows for the detection of cytoplasmic pre-mRNAs has shown that an intact 5′ splice site and branchpoint are required for the nuclear retention of pre-mRNAs (Legrain and Rosbash, 1989). Moreover, several factors already known for their involvement in the splicing process were found to affect pre-mRNA retention, namely the proteins Prp6 and Prp9 and the U1 snRNA. This led to a model where assembly of splicing complexes onto splicing signals serves as a retention signal for pre-mRNA. However, in some circumstances cells have to allow export of partly spliced or unspliced pre-mRNAs to the cytoplasm. A striking example is provided by the HIV-1 Rev protein, which overcomes the retention of unspliced viral messages by binding to a Rev response element (RRE), thereby promoting export of target RNA independent of its splicing status. Other viruses, like the type D retroviruses, use the cellular protein TAP/Mex67p, which binds to an element in the viral RNA to export the unspliced RNA (reviewed in Nakielny and Dreyfuss, 1999). In yeast, other splicing factors involved in early steps of spliceosome assembly were subsequently also found to be involved in pre-mRNA retention, like MUD2, the homolog of U2AF65, and more recently the cap binding complex CBC (Rain and Legrain, 1997; P.J.Lopez and B.Séraphin, unpublished data). All these factors (except for Prp6; Abovich et al., 1990) are involved in early steps of intron recognition that precede complete spliceosome formation (reviewed in Krämer, 1996). Intron recognition is initiated by the binding of the U1 snRNP to the 5′ splice site (reviewed in Rosbash and Séraphin, 1991). It involves a U1 snRNA–pre-mRNA base-pairing interaction that is strengthened by interaction of some snRNP proteins with neighboring pre-mRNA regions (Puig et al., 1999; Zhang and Rosbash, 1999) and a bridging interaction of the cap binding complex with the methyl-7-guanosine cap of the pre-mRNA (Colot et al., 1996; Lewis et al., 1996). This complex can be detected in native gels following assembly in yeast extracts, and is called commitment complex 1 (CC1) because it commits the pre-mRNA to the splicing pathway (Séraphin and Rosbash, 1989a). In the next step of spliceosome assembly, the branchpoint region in the intron is recognized by the branchpoint binding protein (BBP), originally called splicing factor 1 (SF1; Krämer and Utans, 1991). This binding occurs in a sequence-specific manner (Abovich and Rosbash, 1997; Berglund et al., 1997). In humans, U2AF65 binds to the polypyrimidine tract (Zamore and Green, 1989) and has been shown to interact with mBBP/SF1 and enhance its association to the branchpoint (Abovich and Rosbash, 1997; Berglund et al., 1998a; Rain et al., 1998). In addition, U2AF65 later facilitates the base pairing of U2 snRNA with the branch site (Valcárcel et al., 1996). In Saccharomyces cerevisiae, the closest homolog of U2AF65, Mud2p, requires an intact branchpoint region for binding (Abovich et al., 1994; Rain and Legrain, 1997) and also appears to interact with BBP/ScSF1 (Abovich and Rosbash, 1997; Fromont-Racine et al., 1997; Rain et al., 1998; our unpublished results). A bridging interaction between this complex and the U1 snRNP bound to the 5′ splice site has been established (Séraphin and Rosbash, 1989b; Michaud and Reed, 1993; Abovich and Rosbash, 1997). The exact nature of this bridging is not clear, but proteins of the U1 snRNP have been shown to interact with ScSF1 (Prp40p and Prp39p; Abovich and Rosbash, 1997; Fromont-Racine et al., 1997). This complex, which requires an intact 5′ splice site and branchpoint region for its formation, is called commitment complex 2 (CC2) in yeast and complex E in mammals, and can also be detected in native gels (Séraphin and Rosbash, 1989a; Das and Reed, 1999). Although CC2 is a precursor of mature spliceosomes (Séraphin and Rosbash, 1989a), depletion of BBP/ScSF1, which blocks CC2 formation, has no significant effect on (pre)-spliceosome formation and splicing in vitro (Rutz and Séraphin, 1999). In yeast, Mud2p and BBP/ScSF1 leave CC2, being replaced at the branchpoint by the U2 snRNP. We therefore proposed a recycling model that allows for the efficient recycling of BBP/ScSF1 after the binding of U2 snRNP (Rutz and Séraphin, 1999). This would allow the assembly pathway to continue in the presence of catalytic amounts of BBP/ScSF1. In humans, a protein that may correspond to mBBP/SF1 has also been shown to be replaced at the branchpoint by the binding of U2 snRNP (MacMillan et al., 1994; Chiara et al., 1996). This substitution, which is the first ATP-dependent step in the pathway, most likely requires two accessory complexes of the U2 snRNP, SF3a and SF3b (Krämer, 1996 and references therein; Caspary et al., 1999; Das et al., 1999; Krämer et al., 1999). Prp11/ySF3a66 has been shown to interact genetically with MUD2 (Abovich et al., 1994). hSAP155/SF3b155 was found to contact the pre-mRNA at both sides of the branchpoint and to interact with U2AF65 (Gozani et al., 1998). The binding of U2 snRNP to the branchpoint in the so-called pre-spliceosome is followed by the addition of the U4/5/6 tri-snRNP (reviewed in Moore et al., 1993). Before catalysis can occur, the spliceosome is rearranged and the U1 and U4 snRNPs are released. Two transesterification reactions cut out the intron in a lariat configuration and connect the two exons to yield the mature mRNA (reviewed in Madhani and Guthrie, 1994).
Previously we have shown that although BBP/ScSF1 is essential for vegetative growth in yeast (Abovich and Rosbash, 1997), depletion of this factor to >99% does not result in a block of (pre)-spliceosome formation or splicing in vitro (Rutz and Séraphin, 1999). Still, the requirement of BBP/ScSF1 for stable CC2 formation, its interaction with other splicing factors and its recognition of the branchpoint region clearly indicate a role for this factor in splicing. To analyze the function of BBP/ScSF1, we generated conditional mutants of the MSL5 gene. These allowed for functional analyses in vivo and in vitro. Consistent with our previous results we found no significant defects in formation of mature spliceosomes and splicing in vitro for the 11 temperature-sensitive (ts) mutants. Only the cold-sensitive (cs) mutant showed reduced levels of (pre)-spliceosomes, although in vitro splicing was not affected. The in vivo analysis with sensitive splicing reporters revealed splicing defects in all mutants for introns with non-consensus splice sites, but not for an intron with consensus splice sites. More importantly, analysis of pre-mRNA leakage showed that all msl5 mutants were defective in retention of unspliced RNA. This result was further strengthened by our finding of synthetic lethality between three independent msl5 mutants and a disruption of the nonsense-mediated decay (NMD) pathway.
Results
Generation of conditional mutants of MSL5
We first generated a collection of mutated msl5 open reading frames (ORFs) coding for BBP/ScSF1 by amplifying this DNA fragment by mutagenic PCR (Cadwell and Joyce, 1992). A library of ∼13 000 independent clones was created by cloning the mutagenized PCR product together with the MSL5 wild-type promoter region (259 bp) into a yeast centromeric vector (see Materials and methods). This should maintain expression of mutant proteins as close as possible to the wild-type SF1 levels. Sequencing of eight randomly selected clones revealed a mutation rate of ∼1.4% at the nucleotide level (3600 bp analyzed; data not shown). Highly mutagenic conditions were favored because previous analyses failed to reveal conditional mutants in this gene. Consistently, only ∼20% of these plasmids provided sufficient SF1 function to complement an msl5 disruption. The viable clones (>400) were subcloned on yeast extract/peptone/dextrose (YPD) plates and tested for their growth at various temperatures (16, 23, 30 and 37°C). Potential mutants were selected and a second round of plasmid shuffling was performed to ensure a plasmid-linked phenotype (see Materials and methods). Following this selection, 11 clones showed slow or no growth at 37°C (msl5-1 to -4, msl5-7 to -11, msl5-13 and msl5-14) and one clone showed very slow growth at 16°C (msl5-5) compared with the controls containing MSL5 on a plasmid or on the chromosome (data not shown). Some of the ts mutants (e.g. msl5-2, -3 and -4) already showed growth inhibition at 30°C while others (e.g. msl5-11 and -14) were still able to grow with a reduced rate at 37°C (data not shown). Quantitative analysis of growth rate in liquid medium confirmed the results obtained on solid medium (data not shown). Growth was inhibited at the latest 6 h after shift to 37°C for ts mutants and 8 h after shift to 16°C for the cs mutant (data not shown).
Identification of mutations in msl5-2 (ts) and msl5-5 (cs)
Because sequencing of several clones (data not shown) indicated that the mutagenesis procedure had introduced an average of 15 amino acid mutations per ORF, we sought to determine the number and the nature of the mutations required for the generation of an msl5 conditional phenotype. We selected the strongest ts mutant (msl5-2) and the cs mutant (msl5-5) for this analysis. Both mutant ORFs were sequenced entirely to determine the position of mutations. This revealed the presence of 17 mutations in msl5-2 and 13 mutations in msl5-5 (Figure 1). Then, using standard cloning procedures and three conveniently located restriction sites (BsmAI, Mva1269I and BglII), four domains corresponding approximately to the N-terminus, the KH domain, the Zn-knuckle region and the C-terminus (see Figure 1) of the MSL5 ORF were exchanged one by one or in combinations between the mutants and the wild type. The resulting chimeric constructs were re-introduced into yeast and tested for their effect on growth at different temperatures. The N- and the KH domain of msl5-2 were both required to confer the ts phenotype to the otherwise wild-type ORF. The KH- and the Zn domain of msl5-5 were both necessary to retain the cs phenotype. Since the KH domain in both cases contained a single mutation, this mutation was clearly required for the phenotype (Figure 1). To reduce the number of mutations further, additional constructs that contained different combinations of mutations in addition to the mutation in the KH domain (see Materials and methods) were generated. The final construct contained the minimal number of mutations required for generating a onditional phenotype. For msl5-2, two mutations were required in the N domain (R60>G, I72>N) and one mutation in the KH domain (P155>S). For the msl5-5 mutant, the mutation E258>V was found to be required together with the mutation V195>D in the KH domain to confer the cs phenotype.
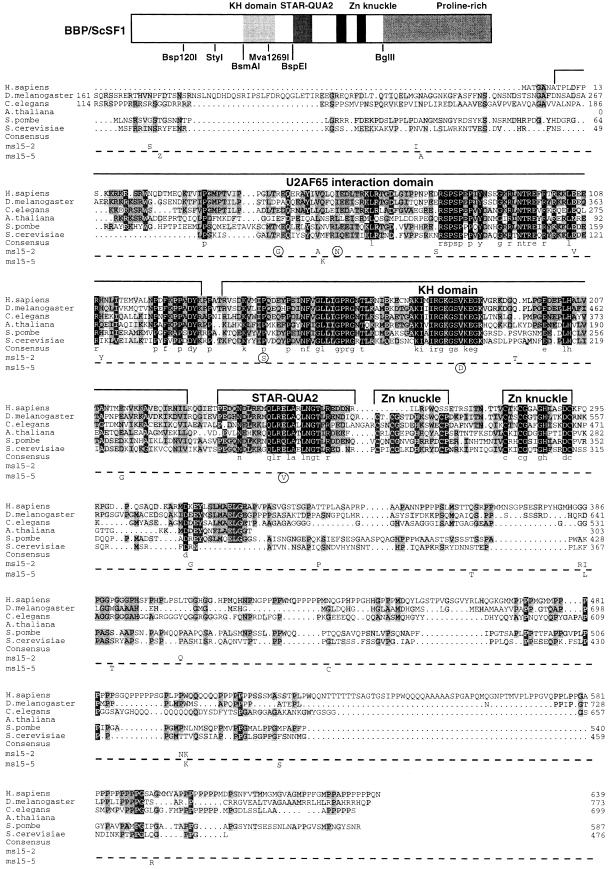
Fig. 1. Amino acid sequence alignment of SF1 orthologs and position of mutations in msl5-2 and msl5-5. Putative homologs of SF1 from Homo sapiens, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana (partial sequence), Schizosaccharomyces pombe and Saccharomyces cerevisae were aligned using the program ClustalX (Thompson et al., 1997) and manual refinement. Amino acids are shaded according to the degree of conservation. Functional domains and structural motifs are indicated above the alignment. The U2AF65 interaction domain is drawn according to the results published for HsSF1 (Rain et al., 1998). The positions of point mutations in the cs mutant msl5-5 (below the dashed line) and in the ts mutant msl5-2 (above the dashed line) are indicated below the S.cerevisiae SF1 sequence. Mutations that were sufficient and necessary to retain a growth defect are encircled (compare with Figure 2). Sequences have the following DDBJ/EMBL/GenBank accession Nos: AtSF1, AB023044; CeSF1, AJ243905; DmSF1, AJ243904; HsSF1, Y08765; ScSF1, U53877; SpSF1, SPTREMBL O74555. The partial protein sequence of A.thaliana SF1 was generated by comparison of the genomic sequence with a profile (derived from an alignment of the known SF1 orthologs with the program PROFILEWEIGHT; Thompson et al., 1994) using the program PairWise (Birney et al., 1996). Restriction enzymes used for domain swapping between mutants and wild-type ORF are shown below the cartoon, which depicts the structural organization of BBP/ScSF1.
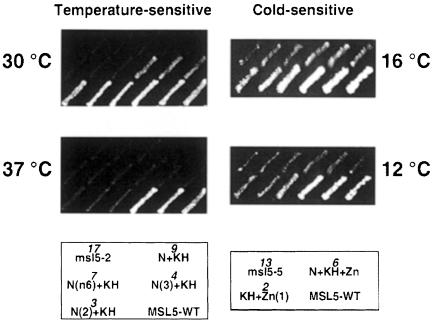
For both mutants msl5-2 and msl5-5, the reduction in the number of mutations was found to reduce the severity of the growth defect, e.g. while the original msl5-2 mutant was barely growing at 30°C the final mutant containing only three point mutations was nearly wild type at 30°C and showed residual growth at 37°C (Figure 2). Similarly, the original cs mutant was cold sensitive at 16°C while the mutant containing only two point mutations showed still very slow growth at 12°C (Figure 2). This indicates that the combination of several mutations accounts for the strong phenotype observed. This may explain why MSL5 has so far escaped genetic screens for conditional mutants affecting splicing (Hutchison et al., 1969; Vijayraghavan et al., 1989; Noble and Guthrie, 1996). Indeed, accumulation of such a large number of mutations is very unlikely to be achieved with classical mutagenesis techniques.
Fig. 2. Growth analysis of minimal mutants derived from msl5-2 and msl5-5. Partial mutants of msl5-2 (ts) and msl5-5 (cs) were generated by domain swapping between the original mutants and the wild type or by site-directed generation of mutations by PCR. Mutant derivatives were compared with the original mutants (msl5-2 or msl5-5) and the wild type (MSL5-WT) for their growth at different temperatures (as indicated on side of the plates). The names of the different mutant derivatives and the number of mutations (in italics) are indicated in the cartoon below in the same position as on the replica plates. Three subclones of each strain are shown side by side.
In summary, we could show that several amino acid mutations are required for the strong growth phenotype of two msl5 mutants. The most relevant mutations were located in evolutionarily conserved domains of the protein.
ts mutants of MSL5 show no effect on (pre)-spliceosome formation and in vitro splicing
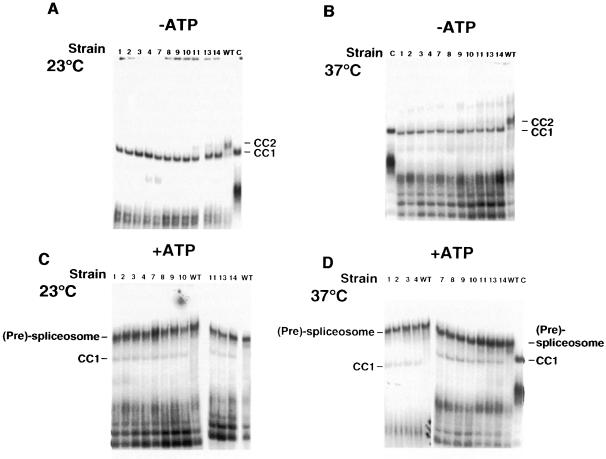
The ts msl5 mutant strains and an isogenic wild-type control strain (BSY809) containing MSL5 on a plasmid were grown at permissive temperature (23°C) and equal numbers of cells were removed before and 6 h after a temperature shift to 37°C. Splicing extracts were prepared and analyzed for the formation of splicing complexes and for in vitro splicing. To test for commitment complex formation, extracts were incubated with labeled pre-mRNA in the absence of ATP. The resulting complexes were analyzed by native gel electrophoresis (Séraphin and Rosbash, 1989a). Extracts from the mutant cells grown at the permissive temperature were blocked in CC2 formation and accumulated CC1 (Figure 3A, lanes 1–4 and 7–14), while the wild-type control showed normal levels of CC2 (Figure 3A, lane WT). The same result was obtained for extracts prepared from cells grown at non-permissive temperature (Figure 3B). This result confirmed that BBP/ScSF1 has a crucial role for the formation of CC2. Furthermore, this demonstrated that even very weak mutants that showed only little growth defect at 37°C and no growth defect at 30°C (like msl5-11 and -14; data not shown) were already blocked in CC2 formation at the permissive temperature (Figure 3A).
Fig. 3. Splicing complex analysis of ts mutants. Extracts from cells grown at permissive (23°C) (A and C) or non-permissive (37°C) temperature (B and D) were incubated with labeled pre-mRNA for 30 min at 30°C in the absence (–) (A and B) or presence (+) of ATP (C and D). The resulting complexes were analyzed in native gels. The allele number of the corresponding mutant strain or the wild-type control (WT) is indicated above each lane. A control reaction was performed with a wild-type extract and a labeled pre-mRNA lacking the branchpoint sequence (C) to obtain a marker for the migration of CC1. Note that the wild-type extract used in lane C was prepared with a different method (Newman, 1994), which yields more concentrated extracts. This results in a slight retardation of the migration of CC1 and of the unspecific complex in the lower part of the gel.
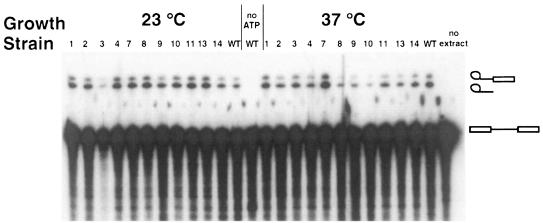
Spliceosome assembly was assayed by gel electrophoresis after incubation at 30°C in the presence of ATP. At the non-permissive temperature, it was not possible to perform the complex assembly assays because aggregates formed that were not resolved in the native gel (data not shown). Since the native gel system used to detect commitment complexes does not resolve pre-spliceosomes from spliceosomes (Séraphin and Rosbash, 1989a), the band corresponding to these species was dubbed (pre)-spliceosome. The levels of (pre)-spliceosome were not reduced in the mutant strains compared with the wild type for both growth temperatures (Figure 3C and D). However, for both temperatures, reactions with the mutant extracts contained an additional complex, which was absent in reactions containing the wild-type extract. This complex comigrated with CC1 (Figure 3D, compare with the marker in lane C). It is noteworthy that a similar phenotype has been observed following biochemical and/or genetic depletions of BBP/ScSF1 (Rutz and Séraphin, 1999). To test whether (pre)-spliceosomes assembled in the mutant extracts were functional, we analyzed intermediates and products of in vitro splicing reactions containing an RP51A intron substrate by denaturing gel electrophoresis. These reactions were performed at 25°C because in vitro splicing was already inhibited at higher temperatures for the wild-type extract (data not shown). Comparison with a wild-type extract showed variability in the efficiency of in vitro splicing for some mutants at the non-permissive temperature (Figure 4, e.g. msl5-10), while at 23°C no significant differences could be observed. Because the quality of the individual extracts has a strong influence on the resulting splicing activity (Cheng et al., 1990) it is not possible to draw quantitative conclusions from this experiment (e.g. note that several mutants with a strong growth defect, like msl5-2, were not reduced in splicing efficiency). However, qualitatively, it is striking that all mutant extracts were able to splice an exogenous reporter pre-mRNA.
Fig. 4. In vitro splicing analysis of ts mutants. Extracts from cells grown at permissive (23°C) or non-permissive (37°C) temperature were incubated with labeled pre-mRNA for 30 min at 25°C in the presence of ATP. RNA was extracted and analyzed in a denaturing gel. The allele number of the corresponding mutant strain or the wild-type control (WT) is indicated above each lane. Control reactions in the absence of ATP (no ATP) or extract (no extract) were analyzed in parallel. The observed RNA species (from top to bottom: lariat intermediate, intron lariat, pre-mRNA) are indicated by cartoons on the side. Exon 1 and mRNA were obscured by degradation products from the pre-mRNA.
Overall, these analyses demonstrated a role of BBP/ScSF1 in CC2 formation, but failed to indicate an effect of the ts mutations on (pre)-spliceosome assembly or activity. These results are consistent with, and confirm, the results obtained following depletion of this factor (Rutz and Séraphin, 1999). Moreover, the analysis of 11 independent mutants showed that this phenotype is related to the function of BBP/ScSF1 and is not allele specific or due to limitations of the depletion methods.
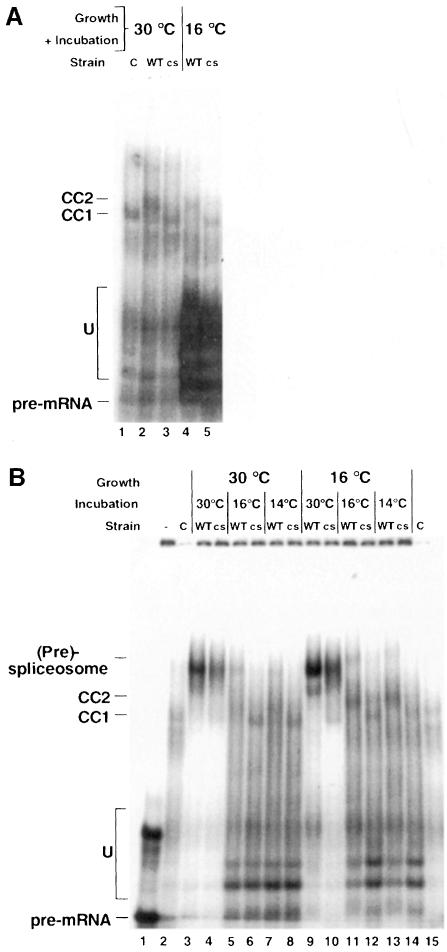
The cs mutant of MSL5 blocks (pre)-spliceosome formation at non-permissive temperature
The cs mutant (msl5-5) was grown together with the isogenic wild-type strain (BSY809) at permissive temperature (30°C) and cells were collected before and 27 h after a temperature shift to 16°C. Extracts were prepared from the different samples. For the analysis of commitment complex formation, extracts were incubated with labeled pre-mRNA substrate in the absence of ATP. The incubation temperature was identical to the temperature of previous growth. At both temperatures a block in CC2 formation and accumulation of CC1 were observed for the mutant, while the wild type showed normal levels of CC2 (Figure 5A, compare lanes 2 and 4 with lanes 3 and 5). This was similar to the phenotype observed for the ts mutants.
Fig. 5. (A) Commitment complex analysis of the cs mutant. Extracts from cells grown at permissive (30°C) or non-permissive (16°C) temperature were incubated with labeled pre-mRNA for 30 min at 30 or 16°C in the absence of ATP. Reactions containing extracts from the mutant strain (cs) or the wild-type control (WT) are indicated above each lane. A control reaction was performed with the wild-type extract and a labeled pre-mRNA lacking the branchpoint sequence (C) to obtain a marker for the migration of CC1. (B) (Pre)-spliceosome analysis of cs mutant. Extracts from cells grown at permissive (30°C) or non-permissive (16°C) temperature were incubated with labeled pre-mRNA for 30 min at 30, 16 or 14°C in the presence of ATP. Labeling of the gel is as in (A).
To analyze (pre)-spliceosome formation, we performed assembly reactions at three different incubation temperatures (30, 16 and 14°C) for extracts from cells grown at permissive and non-permissive temperatures. We observed nearly identical patterns for both growth temperatures (Figure 5B, compare lanes 3–8 and 9–14). In both cases slightly reduced levels of (pre)-spliceosome were observed for the cs mutant when the extracts were incubated at 30°C (Figure 5B, compare lane 3 with 4 and 9 with 10). However, when the incubation was performed at 16 or 14°C (pre)-spliceosome formation was abolished in the mutant extracts while the wild-type extracts showed significant, but strongly reduced levels of (pre)-spliceosomes (Figure 5B, lanes 5–8 and 11–14). As already observed in the analysis of commitment complexes and also for the ts mutants, CC1 accumulated at the non-permissive temperature in the mutant strains. In contrast, the wild-type strain showed some accumulation of CC2 in addition to (pre)-spliceosomes at the low temperatures (Figure 5B, lanes 5, 7, 11 and 13).
This experiment revealed that the cs mutant of msl5, in contrast to the ts mutants, shows a block of (pre)-spliceosome formation that can be induced in vitro and does not depend on the prior growth temperature of the cells.
We tested the activity of the extracts for splicing in vitro at two different temperatures (25 and 16°C). Surprisingly, we could not observe any difference in splicing comparing mutant with wild-type extracts (data not shown). This suggests that a low level of mature spliceosomes can still assemble and that the complexes seen in vitro are not rate limiting for the formation of spliced products.
This extends our observation of a block in CC2 formation and no significant defects in in vitro splicing to the cs mutant. However, the cs mutant, in contrast to the ts mutants, showed a defect in (pre)-spliceosome formation or stability at the non-permissive temperature.
Conditional mutants of MSL5 are affected in the splicing of weak introns in vivo and show increased pre-mRNA leakage to the cytoplasm
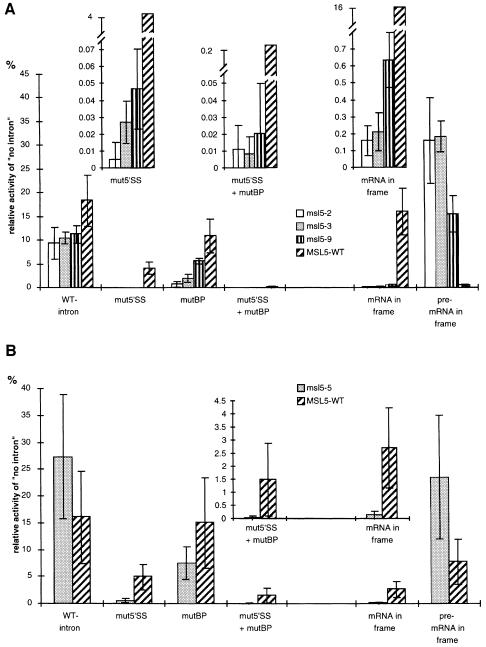
Three ts mutants (msl5-2, -3 and -9) and the cs mutant (msl5-5) were selected to analyze their in vivo effects on splicing and pre-mRNA leakage. Two different reporter systems were used: for the analysis of subtle splicing defects, reporters containing the RP51A intron or mutants thereof inserted within the reading frame of the lacZ gene (Teem and Rosbash, 1983; Jacquier et al., 1985) were utilized. For the analysis of pre-mRNA leakage, a set of synthetic introns designed in a way that either the pre-mRNA or the mRNA codes for β-galactosidase was employed (Legrain and Rosbash, 1989). This last system allows for the detection of pre-mRNAs that have been exported to the cytoplasm where they are translated.
The msl5 mutant strains and an isogenic wild-type strain transformed with these reporters were assayed for β-galactosidase activity after induction of the reporter and incubation at the non-permissive temperature for 2 h (ts) or 4 h (cs). This revealed only a 2-fold decrease in the splicing of the RP51A intron in the ts mutants (Figure 6A, WT-intron). However, splicing of introns containing a mutated 5′ splice site (mut5′SS) or a mutated branchpoint region (mutBP) or the combination of both was much more affected comparing the mutant strains with the wild type (10- to 100-fold for mut5′SS, Figure 6A). The difference in splicing efficiency observed for the three msl5 mutants correlated well with the severity of their respective growth defects (Figure 6A, mut5′SS and mutBP; data not shown). Analysis of the remaining eight mutants confirmed these observations (data not shown).
Fig. 6. Splicing and pre-mRNA leakage analyses. (A) ts mutants msl5-2, -3 and -9. The reporters contained the RP51A intron in the wild-type form (WT-intron) or with a mutation in the 5′ splice site (mut5′SS), in the branchpoint (mutBP) or in both (mut5′SS + mutBP) interrupting the reading frame of the lacZ gene. Another set of reporters contained a synthetic intron, which was designed in a way that either the mRNA (mRNA in frame) or the pre-mRNA (pre-mRNA in frame) would code for β-galactosidase. Activity is expressed as the relative activity of a construct containing no intron in the lacZ gene. The insets show enlarged details of the plots where necessary (note the different scales). (B) cs mutant msl5-5. The analysis was performed as in (A), but cells were grown at 30°C, transferred for 30 min at 16°C before induction for 4 h at 16°C.
For a more sensitive analysis of the splicing phenotype, RNA was extracted from the three mutants and an isogenic wild-type strain after induction of the RP51A-derived reporters as described above. Primer extension was performed with a primer complementary to exon 2 of both the reporter and the endogenous RP51A gene. All BBP/ScSF1 mutants showed a decrease in splicing efficiency for the mut5′SS- and mutBP-containing reporters, as judged by the decrease in mRNA levels and accumulation of pre-mRNA (data not shown). This was verified by quantitative analysis of the gel and calculation of the mRNA/pre-mRNA ratio (Table I), the best indicator of splicing efficiency (Pikielny and Rosbash, 1985). While the wild-type intron was spliced only slightly less efficiently in the mutants than in the wild type (1.5- to 3-fold decrease), the decrease in splicing efficiency was obvious for the reporters containing mutations in the splice sites (∼10-fold in mut5′SS). As already observed for the β-galactosidase activity, the severity of the mutant phenotypes correlated with the defect in splicing efficiency (compare msl5-2 and -9). This demonstrates that BBP/ScSF1 is involved in the splicing of introns in vivo, but indicates that this function is more pronounced for the splicing of introns with weak consensus splice sites. Furthermore, this is consistent with the lack of phenotype of the in vitro splicing analysis performed with the wild-type RP51A intron (Figure 3).
Table I. Splicing efficiency of various reporters in msl5 mutant and wild-type strains.
| Strain | msl5-2 | msl5-3 | msl5-9 | MSL5-WT | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| reporter | WT | 5′ | BP | 5′ + BP | – | WT | 5′ | BP | 5′ + BP | – | WT | 5′ | BP | 5′ + BP | – | WT | 5′ | BP | 5′+ BP | – |
| m/p | 11 | 0.4 | 3.2 | 0.3 | – | 24 | 0.4 | 4.6 | 0.3 | – | 23 | 1.1 | 4.1 | 0.02 | – | 34 | 5.8 | 15 | 1.6 | – |
Reporters contained the RP51A intron in the wild-type form (WT), with a mutation in the 5′ splice site (5′), in the branchpoint region (BP) or in both (5′ + BP). As a control for background from endogenous RNA, cells containing an empty vector (–) were analyzed. The signals corresponding to pre-mRNA and mRNA after primer extension were quantified and the ratio of mRNA/pre-mRNA (m/p) was calculated (Pikielny and Rosbash, 1985).
The second set of reporters was used to analyze the effect of BBP/ScSF1 activity on pre-mRNA leakage to the cytoplasm. Strikingly, a 20- to 40-fold increase in leakage of the pre-mRNA to the cytoplasm was detected in the mutants at the non-permissive temperature (Figure 6A, pre-mRNA in frame). This reveals a role for BBP/ScSF1 in nuclear pre-mRNA retention. The large increase of pre-mRNA export was accompanied by a strong reduction of pre-mRNA splicing of the reporter (Figure 6A, mRNA in frame). The reduction of pre-mRNA splicing observed with this reporter is consistent with our conclusion that BBP/ScSF1 particularly affects splicing of introns that are inefficiently spliced (see above), as reported for the intron in these constructs (Legrain and Rosbash, 1989). Again, the severity of the splicing and pre-mRNA leakage phenotype correlated well with the growth defects of the different mutants. This observation could be extended to the remaining eight ts mutants (Figure 6A; data not shown).
Analysis of the cs mutant showed no significant effect on splicing of the RP51A intron (Figure 6B). However, splicing of reporters containing mutant versions of this intron was significantly reduced (Figure 6B, mut5′SS, mutBP and mut5′SS + mutBP). This confirmed the observations made in the ts mutants. Splicing of the reporter containing the inefficient synthetic intron was also severely inhibited (Figure 6B, mRNA in frame). In the pre-mRNA leakage assay the cs mutant showed an ∼3-fold increase compared with the wild type (Figure 6B, pre-mRNA in frame). However, the wild-type strain also showed an increase in pre-mRNA leakage when compared with the 37°C experiment (compare Figure 6A and B). This could indicate that pre-mRNA retention is less stringent at lower temperatures. Consistent with this hypothesis, a strain disrupted for mud2, which is known to show increased pre-mRNA leakage, has significantly reduced growth rate at 12 and 16°C compared with a wild-type strain (data not shown).
Taken together, the cs mutant had a similar phenotype to the ts mutants, but seemed to be more similar to the intermediate strength ts mutants (like msl5-9). This is in agreement with the growth assay, where the cs mutant still grew very slowly at 16°C while most ts mutants did not grow at 37°C.
In summary, analysis of the conditional mutants of msl5 revealed two major phenotypes: first, an increase in the leakage of pre-mRNA to the cytoplasm, and secondly, a decrease in splicing efficiency of introns with non-consensus splice sites. This phenotype was observed for all 12 mutants, although the strength of the defect in splicing and pre-mRNA leakage varied between individual mutants. The defects in splicing and leakage correlated well with the respective growth phenotypes.
ts mutants of MSL5 show a synthetic phenotype with a mutation in the NMD pathway
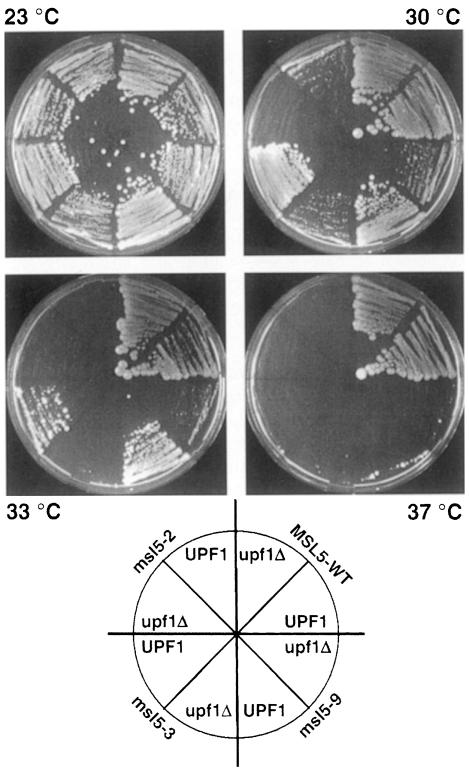
Our observation that conditional mutations in msl5 result in significant pre-mRNA leakage to the cytoplasm suggested that (part of) the essential function of this protein may reside in its ability to prevent escape of unspliced pre-mRNA from the nucleus. Large amounts of cytoplasmic pre-mRNA could lead to the production of truncated polypeptides that could directly or indirectly be toxic to the cell. Since it has been shown that cytoplasmic pre-mRNA that contains in-frame stop codons in the intron is detected and degraded by the NMD pathway (reviewed in Czaplinski et al., 1999; Hilleren and Parker, 1999), we analyzed whether conditional msl5 mutants would have a synthetic phenotype with mutants implicated in the NMD pathway in yeast. While this pathway is not essential in yeast, we reasoned that mutations in this pathway could become deleterious if pre-mRNA leakage was abnormally high and toxic for the cell. Therefore, we generated haploid strains carrying one of the three ts mutants that we had analyzed in vivo (msl5-2, -3 or -9) together with a disruption of upf1, a helicase involved in the NMD pathway (Leeds et al., 1991). These strains were then further analyzed for their growth properties at different temperatures to assay for possible synthetic phenotypes. A difference between strains containing the UPF1 gene and those disrupted for upf1 became obvious at higher temperatures (Figure 7). At 30°C, the msl5-2 mutant did not grow and the msl5-3 and -9 mutants were strongly reduced in growth when upf1 was disrupted, even though the same msl5 mutants were still able to grow in a UPF1 background. At 33°C, msl5-3 did not grow and msl5-9 grew much more slowly when upf1 was disrupted. At 37°C, msl5-2 and -3 did not grow, while msl5-9 showed very slow growth when UPF1 was present and no growth in the context of a upf1 disruption. Importantly, deletion of the UPF1 gene in a wild-type MSL5 background did not affect growth at any temperature (Figure 7). It is noteworthy that the requirement for UPF1 correlates well with the severity of the leakage in the mutants: the msl5-2 mutant that has the strongest growth phenotype is synthetic lethal with upf1-Δ already at 30°C, while msl5-9, which has a more modest growth phenotype and also shows less pre-mRNA leakage, is synthetic lethal only at 37°C. Furthermore, synthetic growth phenotypes were not detected in strains carrying a UPF1 disruption combined with a deletion of either NAM8, LEA1 or MUD2 (data not shown). As NAM8, LEA1 or MUD2 deletions generate splicing phenotypes similar to the one observed in msl5 mutants (Rain and Legrain, 1997; Caspary and Séraphin, 1998; Puig et al., 1999), but no or only limited pre-mRNA leakage to the cytoplasm (Rain and Legrain, 1997; O.Puig, F.Caspary, B.Rutz and B.Séraphin, unpublished data), we conclude that the synthetic phenotype in the upf1-Δ background is specifically related to the strong increase in pre-mRNA leakage rather than to the splicing defect of the msl5 mutants.
Fig. 7. Synthetic lethality of msl5 mutants with upf1-Δ. Strains containing msl5-2, -3 or -9 mutants or a wild-type copy of MSL5 (MSL5-WT) were compared for their growth at different temperatures with isogenic strains carrying in addition a disruption of the upf1 gene (upf1Δ). The orientation of the different strains is indicated in a cartoon below the images.
These results demonstrate that the NMD pathway becomes critical when BBP/ScSF1 function is altered. Our results therefore indicate a tight interplay of splicing, pre-mRNA retention and NMD. Given the strong pre-mRNA leakage phenotype of conditional msl5 mutants, this further suggests that accumulation of unspliced pre-mRNA in the cytoplasm is toxic for the cell. This supports the idea that a function of BBP/ScSF1 is to prevent the negative consequences of pre-mRNA leakage to the cytoplasm.
Discussion
The human splicing factor SF1 was first identified biochemically (Krämer and Utans, 1991; Krämer, 1992). The cloning of the corresponding cDNA (Arning et al., 1996) allowed for more detailed in vitro and in vivo studies of its function and made analyses of homologs in other organisms feasible. In yeast, the gene coding for BBP/ScSF1, MSL5, was found in a synthetic lethal screen with a truncated form of MUD2. BBP/ScSF1 is essential for vegetative growth and contacts the branchpoint region of the intron (Abovich and Rosbash, 1997). More recently, a study in Caenorhabditis elegans showed that the homolog of SF1 in this organism is also required for viability (Mazroui et al., 1999). Despite these accumulating data, genetic and/or biochemical depletion of BBP/ScSF1 in yeast showed no block in the formation of mature spliceosomes or splicing in vitro (Rutz and Séraphin, 1999).
To analyze further the function of BBP/ScSF1 in vivo, we generated conditional mutants of msl5. Using a PCR mutagenesis strategy we could isolate 11 ts mutants and one cs mutant. The mutants showed differences in the severity of their temperature sensitivity and were found not to result from common amino acid changes (Figure 6A; data not shown). These are the first conditional mutants reported for msl5, which so far has escaped all large-scale screens for splicing mutants.
Functionally relevant mutations are located in evolutionarily conserved domains
The definition of ‘minimal’ mutants that still show a growth defect proved the usefulness of our PCR mutagenesis strategy. Only a high mutagenesis rate could create the pattern of mutations required for the strong phenotype observed in some of the mutants. This could indicate that several domains of the protein act synergistically, each of them contributing only partially to the growth phenotype observed.
Clearly the KH domain, which is required for RNA binding (Arning et al., 1996; Berglund et al., 1998b; Rain et al., 1998), is essential for SF1 function. This is reflected by a point mutation being required in this region in both mutants analyzed. The mutation (P155>S) in msl5-2 is located immediately after a predicted β-strand that is present at the same position in five structures of KH domains (Musco et al., 1996, 1997; Lewis et al., 1999, 2000). The proline in this position is one of the most conserved residues in the KH domain (aligned in Lewis et al., 1999) and is absolutely conserved in all members of the STAR family (signal transduction and activation of RNA; Vernet and Artzt, 1997). Two additional mutations are required in the N-terminus of msl5-2. This part of SF1 (amino acids 41–141 in yeast) has been shown to interact with Mud2p/U2AF65 (Rain et al., 1998) and could therefore be required for the cooperative binding to the branchpoint (Berglund et al., 1998a). However, since a complete knock-out of mud2 shows no growth phenotype, this alone can not account for the strong ts phenotype observed in msl5-2. Either the combination with the mutation in the KH domain increases the importance of the interaction with Mud2p or additional interactions are affected by the mutations in this region. One obvious candidate would be the proposed bridging interaction connecting U1 snRNP at the 5′ splice site via Prp40p with the branchpoint region (Abovich and Rosbash, 1997). Alternatively, the mutations could distort structures necessary for the proper folding of the protein. However, previous data suggest that the N-terminal region of BBP/ScSF1 contains a functional domain required for functions other than RNA binding. Indeed, a truncated BBP/ScSF1 protein (amino acids 145–330) shows strong affinity and high specificity for a branchpoint sequence RNA (Berglund et al., 1997, 1998b), while a construct lacking the N-terminus (amino acids 146–476) is not sufficient to complement a disruption of SF1 in yeast (Rain et al., 1998).
The cs mutant contains a mutation (V195>D; Figure 1) in a region that is less conserved among KH domains and also not absolutely conserved between members of the STAR family. However, only conservative substitutions seem to occur in this position in the STAR family (V, I or M). It lies in a flexible loop that could account for the sequence specificity of the KH domain (Lewis et al., 2000). The second mutation (E258>V) lies inside the conserved STAR-QUA2 domain, besides the KH domain another hallmark of the large family of STAR proteins. In a study with both human and yeast BBP/SF1 this region was shown to help RNA binding and specificity when present together with the KH domain (Berglund et al., 1998b), but the exact function of this domain has not yet been determined.
In vitro analysis shows minor defects in(pre)-spliceosome assembly and splicing
Extracts from the msl5 mutants showed very similar in vitro phenotypes compared with the extracts depleted for BBP/ScSF1. They were all defective in CC2 formation, but showed (with the exception of the cs mutant) no effect on (pre)-spliceosome formation and splicing in vitro. The cs mutant extracts were blocked in (pre)-spliceosome assembly at the non-permissive temperature. However, splicing activity was still detectable, suggesting that functional spliceosomes still formed at a very low level. This could indicate a more assembly-related phenotype of the cs mutant, which still allows for splicing but does not allow (pre)-spliceosomes to accumulate to levels detectable in native gels. The fact that BBP/ScSF1 was not found stably associated with pre-spliceosomes (Rutz and Séraphin, 1999) suggests that the mutant form of BBP/ScSF1 affects the assembly of pre-spliceosomes at a very early step. Taken together, these results confirm our previous depletion studies and suggest that the requirement of BBP/ScSF1 in splicing is not universal for all substrates, or that only very small amounts of the functional protein are required for proper splicing.
In vivo analysis reveals a splicing defect of non-consensus introns
We further investigated splicing of the different mutants in vivo using a sensitive reporter assay based on the RP51A intron and mutants thereof. This analysis revealed that splicing of the wild-type intron was only very modestly affected by the mutations in msl5, while introns with mut5′SS and/or mutBP regions showed a strong decrease in splicing efficiency. We conclude that BBP/ScSF1 may be required only for the splicing of introns with weak splice sites or otherwise impaired splicing efficiencies. This observation was not specific for the RP51A intron mutants as it was also observed for a weakly spliced synthetic intron (see above; Legrain and Rosbash, 1989). Because weak introns are not spliced in vitro, we could not test whether this phenotype could be reproduced in vitro.
Pre-mRNA leakage to the cytoplasm
A few splicing mutants have been reported to affect pre-mRNA retention in the nucleus. In contrast to msl5, these mutants (e.g. prp6 and prp9) also showed very pronounced splicing defects detectable in vitro and in vivo (Legrain and Rosbash, 1989; Abovich et al., 1990). Analysis of the msl5 mutants for their ability to retain unspliced pre-mRNA in the nucleus showed that all 12 mutants allowed leakage of unspliced pre-mRNA to the cytoplasm. Furthermore, the strength of this effect correlated well with the severity of the growth defect of the specific mutant. It is important to note that all 12 mutants qualitatively showed the same in vivo and in vitro phenotype regarding splicing and pre-mRNA retention [with the exception of the cs mutant that differed in its block of (pre)-spliceosome formation]. Sequencing of several mutants (data not shown) and the mapping of relevant mutations in two mutants demonstrated that this was not due to identical point mutations present in these mutants (Figure 1). The phenotype for msl5 reported here is therefore not allele specific, but appears to reflect the true function of the gene. Overall, these data suggest that the increased leakage of pre-mRNA to the cytoplasm accounts, at least partially, for the essential phenotype of MSL5.
The MUD2 gene, which functions together with MSL5 in the formation of CC2 (Abovich et al., 1994), also seems to be involved in pre-mRNA retention in the nucleus (Rain and Legrain, 1997). However, since MUD2 is not essential for yeast viability, we were expecting that the splicing and leakage defects of a mud2-Δ mutant would be less pronounced than for the msl5 mutants. Comparison of mud2-Δ and msl5 conditional mutants generated in an isogenic background revealed that mud2-Δ showed only subtle splicing defects (consistent with previous results; Rain and Legrain, 1997) that were always weaker than in the msl5 mutants (data not shown). More importantly, in these strains the leakage of pre-mRNA was increased only ∼5-fold by a mud2 deletion, compared with 20- to 40-fold for the msl5 mutants (data not shown; see also Rain and Legrain, 1997). This strongly suggests that quantitative effects on splicing and pre-mRNA leakage are responsible for the difference in the requirement for the functions of MUD2 and MSL5.
Synthetic lethality in combination with a disruption of the NMD pathway
The importance of BBP/ScSF1 in preventing pre-mRNA leakage is further strengthened by our finding of a synthetic lethal phenotype between three msl5 mutants and a disruption of the NMD pathway. The close correlation between the severity of the pre-mRNA retention defect of the different msl5 mutants and the strength of the synthetic lethal phenotype underlined the importance of pre-mRNA retention for viability. This suggests that the leakage of pre-mRNA in the msl5 mutants causes a lethal phenotype if the cellular control mechanism for these aberrant messages is disrupted.
It is difficult to imagine that an elaborate control mechanism like NMD would have evolved only to prevent the accumulation of aberrant messages from very rare genomic mutation events. Given the importance of splicing in yeast, where about every third message con tains an intron, many of which are located in essential genes (Lopez and Séraphin, 1999), it seems likely that the NMD pathway has evolved to prevent the accumulation of pre-mRNA in the cytoplasm. This idea was first suggested by experiments showing accumulation of the inefficiently spliced CYH2 pre-mRNA in upf1 null mutants (He et al., 1993), and was further confirmed by an analysis of the spatial distribution of pre-mRNAs and their degradation by the NMD pathway in yeast (Long et al., 1995). Our findings strengthen this hypothesis.
In this study we described the generation and analysis of a battery of conditional mutants for the essential yeast gene MSL5, which codes for BBP/ScSF1. We could show in vivo that BBP/ScSF1 is a splicing factor that affects splicing of introns that are spliced inefficiently, but has only a minor effect on the splicing of introns with consensus splice sites. This could indicate a more regulatory function for SF1 in splicing. It will be interesting to find out whether the different isoforms of SF1 found in higher eukaryotes are specific for different introns. More importantly, we found that all mutants analyzed were defective in pre-mRNA retention in the nucleus. We could further strengthen this point by finding a specific synthetic phenotype of three different msl5 mutants with a disruption of the NMD pathway. This indicates an important role for SF1 in the decision as to whether a given pre-mRNA is exported to the cytoplasm or spliced in the nucleus. Also, for this function of SF1 the different isoforms of this protein in higher eukaryotes could confer substrate specificity.
Our results do not allow us to conclude how splicing and pre-mRNA retention are linked. It is possible that these two aspects of pre-mRNA metabolism are two faces of a single process with a kinetic advantage for the splicing process preventing leakage of most pre-mRNA. Alternatively, two separate processes could exist, implicating the presence of a dedicated ‘retention machinery’. Further experiments will be required to understand the integration of pre-mRNA splicing and pre-mRNA retention in eukaryotic nuclei.
Materials and methods
Generation of conditional msl5 mutants
The MSL5 gene (promoter and ORF) was amplified from genomic DNA by PCR using the primers BR34 (5′-TACTGGGCCCTCGAGCCT TCTTGTATACAG-3′) and BR37 (5′-ATACGGATCCAAGCTTTG AATAAAGGTAAG-3′). The PCR product digested with XhoI and BamHI was cloned into the same sites of the URA3 marked yeast shuttle vector pRS416 (Sikorski and Hieter, 1989), creating pBS1703.
To generate a disruption of the MSL5 gene, strategy B described previously (Puig et al., 1998) was followed using primers BR1 (5′-TTTTATTTGTGTTGAAGG-3′), BR2 (5′-TGAAATTACCGA TGAGAA-3′) and BR33 (5′-GCTACAAGTGCTATTCTATCCCAC AATAAAAGGAAATTGTTGATGAAGCTGGAGCTCAAAAC-3′). The product was transformed into the diploid yeast strain BSY320, which is isogenic to MGD453-13D (Séraphin et al., 1988), following the method of Ito et al. (1983). The strategy was designed to remove the complete MSL5 ORF except for the start codon. Transformants were selected on SD-Trp plates and verified by PCR. The resulting strain was called BSY806. This strain was transformed with plasmid pBS1703, which contained the MSL5 gene together with its promoter. Following sporulation and dissection, markers were analyzed by replica plating, and disruption of the MSL5 locus in spores that were ura+ and trp+ was confirmed by PCR. This generated the strains BSY809 (MATa) and BSY810 (MATα).
The ORF of MSL5 was amplified by PCR from genomic DNA with the primers BR36 (5′-TGACTAGTCGACAAGTGCTATTCTATC-3′) and BR37. The amplified fragment was cloned into the SalI and HindIII sites of pBluescriptSK– (Stratagene) and used as a template for the generation of the mutant library. The mutagenic PCR was performed with the T3 and T7 primers according to the method described by Cadwell and Joyce (1992). Briefly, the dNTP concentration was changed to 2 mM dGTP, 2 mM dATP, 10 mM dTTP, 10 mM dCTP; MnCl2 was added to a final concentration of 0.5 mM and the MgCl2 concentration was increased to 4 mM. The concentration of enzyme (Amplitaq; Perkin-Elmer) was increased to 5 U per 25 µl reaction, and 30 amplification cycles were performed with an annealing temperature of 50°C. The MSL5 promoter was amplified with the primers BR34 and BR35 (5′-ACGTAC GTCGACAAATTAGGGAAAAAATTC-3′) from genomic DNA and cloned into the Bsp120I and SalI sites of yeast shuttle vector pRS415, which contains a LEU2 marker (Sikorski and Hieter, 1989), resulting in plasmid pBS1702. The mutagenic PCR was digested with SalI and BamHI, and inserted behind the MSL5 promoter in pBS1702. Eight independent clones were partially sequenced to estimate the mutation frequency. About 13 000 independent clones were collected and used for the preparation of the mutant library named pBS1780. This library was transformed into the yeast strain BSY809. Transformants were selected on SD-Leu plates at 23°C. About 2000 clones were streaked onto SD-Leu plates, grown at 23°C and replica plated onto 5-fluorotic acid-containing plates. The surviving clones (∼400) were streaked on YPD plates, grown again at 23°C and replica plated onto YPD plates that were incubated at 16, 23, 30 and 37°C. Clones that showed a ts growth phenotype were subcloned, checked again for their growth defect and DNA was extracted. The DNA was transformed into E.coli, amplified and retransformed into the yeast strain BSY809 followed by the shuffling strategy described above. The msl5 ORF of four of the mutants was sequenced entirely, three other mutants were sequenced partially.
Mutation mapping
The entire MSL5 ORF present in the plasmids harboring the mutants msl5-2 and -5 was sequenced with four specific primers on a fluorescent sequencer (ALF-system; Pharmacia). The restriction enzymes SalI, BsmAI, Mva1269I, BglII and BamHI were used to split the ORF into four domains. Standard cloning techniques were used to exchange each of these domains or combinations thereof with the corresponding domains of a plasmid containing the MSL5 wild-type ORF. The resulting plasmids were tested by the shuffling strategy described above for their ability to confer the ts or cs phenotype to yeast strain BSY809. The smallest mutant fragment still showing a mutant phenotype was used as the basis for the next mapping step. For msl5-5, the two mutations present in the third domain were separated using the restriction enzyme BspEI, resulting in a final construct that contained one mutation in the KH domain and one mutation in the Zn domain. For msl5-2, the N domain was further dissected by the use of the restriction enzymes Bsp120I and StyI. This resulted in a construct containing four mutations in the N-terminus (between Bsp120I and StyI) and one mutation in the KH domain. The four mutations were further dissected by insertion of products of PCR with mutagenic or wild-type primers on mutant (msl5-2) or wild-type templates between the Bsp120I and StyI sites. This yielded a final construct containing two mutations in the N-terminus and one mutation in the KH domain.
Spliceosome assembly and in vitro splicing
Yeast splicing extracts were prepared as described previously (Séraphin and Rosbash, 1989a). 32P-labeled pre-mRNA was generated by in vitro transcription of plasmid pBS195 (wild type) or pBS199 (ΔUACUAAC) digested with DdeI (Séraphin and Rosbash, 1991). Native gels and complex assembly reactions were as described (Séraphin and Rosbash, 1989a) at the temperatures indicated. To deplete ATP, the reactions were incubated for 10 min at room temperature with 2 mM glucose prior to addition of labeled pre-mRNA (Liao et al., 1992).
Splicing reactions were as described above, except that the incubation was performed for 30 min at 25°C or the temperature indicated. Pre-mRNA was generated by in vitro transcription of plasmid pBS7 cut with DdeI (Séraphin et al., 1988). The reaction was stopped by addition of 200 µl of PK-buffer (0.1 M Tris–HCl pH 7.5, 12.5 mM EDTA pH 8.0, 150 mM NaCl, 1% SDS) containing 80 µg of proteinase K (Sigma) and 10 µg of Escherichia coli tRNA, and incubation for 20 min at 37°C. RNA was extracted and analyzed in a 5% polyacrylamide–7 M urea gel.
In vivo splicing and pre-mRNA retention analysis
The mutant yeast strains msl5-2, -3, -5 and -9 and an isogenic wild-type strain were transformed with the following reporter plasmids: RP51A wild-type intron (HZ18), mut5′SS (HZ12), mutBP (HZ8) and mut5′SS + mutBP (HZ8 + HZ12) (Jacquier et al., 1985 and references therein). The reporters for pre-mRNA retention analysis were mRNA in-frame (pLG-Acc°), pre-mRNA in-frame (pLG-Nde°Acc°) and no intron (pLG-SD5) (Legrain and Rosbash, 1989). The ts strains (msl5-2, -3 and -9) and the wild-type control were grown at 23°C in synthetic medium without uracil containing 2% lactate pH 5.5, 2% glycerol and 0.05% glucose to an OD600 of 0.5–0.8. Then the cells were transferred to 37°C for 30 min before induction of the reporters by addition of galactose to a final concentration of 2%. Induction was continued for 2 h at 37°C. Cells were harvested, the OD600 was measured and two aliquots were taken for the analysis of β-galactosidase activity as described previously (Séraphin and Kandels-Lewis, 1993). β-galactosidase activity was corrected for the background measured for the plasmid YEp24 (Botstein et al., 1979), which contains no lacZ gene and was expressed as the percentage of the activity of a reporter containing no intron upstream of the lacZ gene (pLG-SD5) in each strain. Three independent clones of each strain were analyzed in duplicate and errors were calculated as the cumulative maximal error from the standard deviations of the measured values. For RNA analysis, RNA was extracted from cells as described (Séraphin and Kandels-Lewis, 1993) and normalized by measuring the OD260. Primer extension was performed using 32P-labeled primer EM38 (Luukkonen and Séraphin, 1997). The extension products were fractionated in a 6% polyacrylamide–7 M urea gel, dried and exposed to a Fluorescent Image Analyzer screen (Fuji). Signals corresponding to pre-mRNA and mRNA were quantified. Analysis of β-galactosidase activity of the cs mutant (msl5-5) was as described above except that cells were grown at 30°C before transfer to 16°C for 30 min followed by a 4 h induction at 16°C.
Synthetic phenotype analysis
To generate a disruption of the UPF1 gene in the desired strain background, strategy B (Puig et al., 1998) was followed using primers BR48 (5′-TTTCTACAGCCATACCTT-3′), BR50 (5′-TGTTAAACT TGGCTTGTG-3′) and BR57 (5′-AAGACCGAATATACTTTTTAT ATTACATCAATCATTGTCATTATCAAATGAAGCTGGAGCTCAA AAC-3′). The product was transformed into the haploid yeast strain MGD453-13D (Séraphin et al., 1988). The strategy was designed to remove the complete UPF1 ORF except for the start codon. Transformants were selected on SD-Trp plates and verified by PCR. The resulting strain was called BSY879. This strain transformed with the LEU2 marked plasmid pRS415 (Sikorski and Hieter, 1989) was mated with strain BSY810 containing the msl5 disruption complemented with a plasmid copy of MSL5. A diploid strain, selected by the simultaneous presence of the LEU2 and URA3 markers, was named BSY883. It was sporulated and dissected. The disruption of both genes, upf1 and msl5, in the resulting spores was checked by the presence of the TRP1 and URA3 markers and by PCR. A strain was selected for further analysis and was called BSY884 (MATα). It was transformed with the LEU2 marker plasmids containing the mutants msl5-2, -3 or -9 or the wild-type MSL5 gene, followed by the shuffling strategy described above to eliminate the wild-type copy of MSL5 on the URA3 marker plasmid. The resulting strains were replica plated on YPD plates that were incubated at 23, 30, 33 and 37°C.
Acknowledgments
Acknowledgements
We thank E.Bouveret, F.Caspary, K.Czaplinski, E.Izaurralde, A.Krämer, S.Kuersten, I.Mattaj, O.Puig, T.Schell, S.Thore and J.Valcárcel for carefully reading the manuscript and helpful discussions. B.S. is on leave from the CNRS.
References
- Abovich N. and Rosbash,M. (1997) Cross-intron bridging and interactions in the yeast commitment complex are conserved in mammals. Cell, 89, 403–412. [DOI] [PubMed] [Google Scholar]
- Abovich N., Legrain,P. and Rosbash,M. (1990) The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol. Cell. Biol., 10, 6417–6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abovich N., Liao,X.C. and Rosbash,M. (1994) The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev., 8, 843–854. [DOI] [PubMed] [Google Scholar]
- Arning S., Grüter,P., Bilbe,G. and Krämer,A. (1996) Mammalian splicing factor SF1 is encoded by variant cDNAs and binds to RNA. RNA, 2, 794–810. [PMC free article] [PubMed] [Google Scholar]
- Berglund J.A., Chua,K., Abovich,N., Reed,R. and Rosbash,M. (1997) The splicing factor BBP interacts specifically with the pre-mRNA branch point sequence UACUAAC. Cell, 89, 781–787. [DOI] [PubMed] [Google Scholar]
- Berglund J.A., Abovich,N. and Rosbash,M. (1998a) A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev., 12, 858–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund J.A., Fleming,M.L. and Rosbash,M. (1998b) The KH domain of the branchpoint sequence binding protein determines specificity for the pre-mRNA branchpoint sequence. RNA, 4, 998–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E., Thompson,J.D. and Gibson,T.J. (1996) PairWise and SearchWise: finding the optimal alignment in a simultaneous comparison of a protein profile against all DNA translation frames. Nucleic Acids Res., 24, 2730–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botstein D., Falco,S.C., Stewart,S.E., Brennan,M., Scherer,S., Stinchcomb,D.T., Struhl,K. and Davis,R.W. (1979) Sterile host yeasts (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene, 8, 17–24. [DOI] [PubMed] [Google Scholar]
- Cadwell R.C. and Joyce,G.F. (1992) Randomization of genes by PCR mutagenesis. PCR Methods Appl., 2, 28–33. [DOI] [PubMed] [Google Scholar]
- Caspary F. and Séraphin,B. (1998) The yeast U2A′/U2B′′ complex is required for pre-spliceosome formation. EMBO J., 17, 6348–6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspary F., Shevchenko,A., Wilm,M. and Séraphin,B. (1999) Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J., 18, 3463–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S.C., Newman,A.N., Lin,R.J., McFarland,G.D. and Abelson,J.N. (1990) Preparation and fractionation of yeast splicing extract. Methods Enzymol., 181, 89–96. [DOI] [PubMed] [Google Scholar]
- Chiara M.D., Gozani,O., Bennett,M., Champion,A.P., Palandjian,L. and Reed,R. (1996) Identification of proteins that interact with exon sequences, splice sites and the branchpoint sequence during each stage of spliceosome assembly. Mol. Cell. Biol., 16, 3317–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot H.V., Stutz,F. and Rosbash,M. (1996) The yeast splicing factor Mud13p is a commitment complex component and corresponds to CBP20, the small subunit of the nuclear cap-binding complex. Genes Dev., 10, 1699–1708. [DOI] [PubMed] [Google Scholar]
- Czaplinski K., Ruiz-Echevarria,M.J., Gonzalez,C.I. and Peltz,S.W. (1999) Should we kill the messenger? The role of the surveillance complex in translation termination and mRNA turnover. BioEssays, 21, 685–696. [DOI] [PubMed] [Google Scholar]
- Das B.K., Xia,L., Palandjian,L., Gozani,O., Chyung,Y. and Reed,R. (1999) Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol. Cell. Biol., 19, 6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das R. and Reed,R. (1999) Resolution of the mammalian E complex and the ATP-dependent spliceosomal complexes on native agarose mini-gels. RNA, 5, 1504–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromont-Racine M., Rain,J.-C. and Legrain,P. (1997) Toward a functional analysis of the yeast genome through exhaustive two-hybrid screens. Nature Genet., 16, 277–282. [DOI] [PubMed] [Google Scholar]
- Gozani O., Potashkin,J. and Reed,R. (1998) A potential role for U2AF–SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol., 18, 4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F., Peltz,S.W., Donahue,J.L., Rosbash,M. and Jacobson,A. (1993) Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1– mutant. Proc. Natl Acad. Sci. USA, 90, 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P. and Parker,R. (1999) mRNA surveillance in eukaryotes: kinetic proofreading of proper translation termination as assessed by mRNP domain organization? RNA, 5, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison H.T., Hartwell,L.H. and McLaughlin,C.S. (1969) Temperature-sensitive yeast mutant defective in ribonucleic acid production. J. Bacteriol., 99, 807–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukada,Y., Murata,K. and Kimura,A. (1983) Transformation of intact yeast cells treated with alkali cations. J. Bacteriol., 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A., Rodriguez,J.R. and Rosbash,M. (1985) A quantitative analysis of the effects of 5′ junction and TACTAAC box mutants and mutant combinations on yeast mRNA splicing. Cell, 43, 423–430. [DOI] [PubMed] [Google Scholar]
- Krämer A. (1992) Purification of splicing factor SF1, a heat stable protein that functions in the assembly of a presplicing complex. Mol. Cell. Biol. 12, 4545–4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A. (1996) The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu. Rev. Biochem., 65, 367–409. [DOI] [PubMed] [Google Scholar]
- Krämer A. and Utans,U. (1991) Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J., 10, 1503–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krämer A., Grüter,P., Groning,K. and Kastner,B. (1999) Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J. Cell Biol., 145, 1355–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeds P., Peltz,S.W., Jacobson,A. and Culbertson,M.R. (1991) The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev., 5, 2303–2314. [DOI] [PubMed] [Google Scholar]
- Legrain P. and Rosbash,M. (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell, 57, 573–583. [DOI] [PubMed] [Google Scholar]
- Lewis H.A., Chen,H., Edo,C., Buckanovich,R.J., Yang,Y.Y., Musunuru,K., Zhong,R., Darnell,R.B. and Burley,S.K. (1999) Crystal structures of Nova-1 and Nova-2 K-homology RNA-binding domains. Struct. Fold. Des., 7, 191–203. [DOI] [PubMed] [Google Scholar]
- Lewis H.A., Musunuru,K., Jensen,K.B., Edo,C., Chen,H., Darnell,R.B. and Burley,S.K. (2000) Sequence-specific RNA binding by a Nova KH domain: implications for paraneoplastic disease and the fragile X syndrome. Cell, 100, 323–332. [DOI] [PubMed] [Google Scholar]
- Lewis J.D., Görlich,D. and Mattaj,I.W. (1996) A yeast cap binding protein complex (yCBC) acts at an early step in pre-mRNA splicing. Nucleic Acids Res., 24, 3332–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X.C., Colot,H.V., Wang,Y. and Rosbash,M. (1992) Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res., 20, 4237–4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R.M., Elliott,D.J., Stutz,F., Rosbash,M. and Singer,R.H. (1995) Spatial consequences of defective processing of specific yeast mRNAs revealed by fluorescent in situ hybridization. RNA, 1, 1071–1078. [PMC free article] [PubMed] [Google Scholar]
- Lopez P.J. and Séraphin,B. (1999) Genomic-scale quantitative analysis of yeast pre-mRNA splicing: implications for splice-site recognition. RNA, 5, 1135–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luukkonen B. and Séraphin,B. (1997) The role of branchpoint-3′ splice site spacing and interaction between intron terminal nucleotides in 3′ splice site selection in Saccharomyces cerevisiae. EMBO J., 16, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMillan A., Query,C., Allerson,C., Chen,S., Verdine,G. and Sharp,P. (1994) Dynamic association of proteins with the pre-mRNA branch region. Genes Dev., 8, 3008–3020. [DOI] [PubMed] [Google Scholar]
- Madhani H. and Guthrie,C. (1994) Dynamic RNA–RNA interactions in the spliceosome. Annu. Rev. Genet., 28, 1–26. [DOI] [PubMed] [Google Scholar]
- Mazroui R., Puoti,A. and Kramer,A. (1999) Splicing factor SF1 from Drosophila and Caenorhabditis: presence of an N-terminal RS domain and requirement for viability. RNA, 5, 1615–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S. and Reed,R. (1993) A functional association between the 5′ and 3′ splice site is established in the earliest prespliceosome complex (E) in mammals. Genes Dev., 7, 1008–1020. [DOI] [PubMed] [Google Scholar]
- Moore M., Query,C. and Sharp,P. (1993) Splicing of precursors to mRNA by the spliceosome. In Gesteland,R. and Atkins,J. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 303–357. [Google Scholar]
- Musco G., Stier,G., Joseph,C., Castiglione Morelli,M.A., Nilges,M., Gibson,T.J. and Pastore,A. (1996) Three-dimensional structure and stability of the KH domain: molecular insights into the fragile X syndrome. Cell, 85, 237–245. [DOI] [PubMed] [Google Scholar]
- Musco G., Kharrat,A., Stier,G., Fraternali,F., Gibson,T.J., Nilges,M. and Pastore,A. (1997) The solution structure of the first KH domain of FMR1, the protein responsible for the fragile X syndrome [published erratum appears in Nature Struct. Biol., 1997, 4, 840]. Nature Struct. Biol., 4, 712–716. [DOI] [PubMed] [Google Scholar]
- Nakielny S. and Dreyfuss,G. (1999) Transport of proteins and RNAs in and out of the nucleus. Cell, 99, 677–690. [DOI] [PubMed] [Google Scholar]
- Newman A. (1994) Analysis of pre-mRNA splicing in yeast. In Higgins,S. and Hames,B. (eds), RNA Processing: A Practical Approach. Oxford University Press, Oxford, UK, pp. 179–195. [Google Scholar]
- Noble S.M. and Guthrie,C. (1996) Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics, 143, 67–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikielny C.W. and Rosbash,M. (1985) mRNA splicing efficiency in yeast and the contribution of nonconserved sequences. Cell, 41, 119–126. [DOI] [PubMed] [Google Scholar]
- Puig O., Rutz,B., Luukkonen,B.G., Kandels-Lewis,S., Bragado-Nilsson,E. and Séraphin,B. (1998) New constructs and strategies for efficient PCR-based gene manipulations in yeast. Yeast, 14, 1139–1146. [DOI] [PubMed] [Google Scholar]
- Puig O., Gottschalk,A., Fabrizio,P. and Séraphin,B. (1999) Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes Dev., 13, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J.C. and Legrain,P. (1997) In vivo commitment to splicing in yeast involves the nucleotide upstream from the branch site conserved sequence and the Mud2 protein. EMBO J., 16, 1759–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rain J.C., Rafi,Z., Rhani,Z., Legrain,P. and Krämer,A. (1998) Conservation of functional domains involved in RNA binding and protein–protein interactions in human and Saccharomyces cerevisiae pre-mRNA splicing factor SF1. RNA, 4, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosbash M. and Séraphin,B. (1991) Who’s on first? The U1 snRNP–5′ splice site interaction and splicing. Trends Biochem. Sci., 16, 187–190. [DOI] [PubMed] [Google Scholar]
- Rutz B. and Séraphin,B. (1999) Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. RNA, 5, 819–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B. and Kandels-Lewis,S. (1993) 3′ splice site recognition in S.cerevisiae does not require base pairing with U1 snRNA. Cell, 73, 803–812. [DOI] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1989a) Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell, 59, 349–358. [DOI] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1989b) Mutational analysis of the interactions between U1 small nuclear RNA and pre-mRNA of yeast. Gene, 82, 145–151. [DOI] [PubMed] [Google Scholar]
- Séraphin B. and Rosbash,M. (1991) The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA–pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J., 10, 1209–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Séraphin B., Kretzner,L. and Rosbash,M. (1988) A U1 snRNA:pre-mRNA base pairing interaction is required early in yeast spliceosome assembly but does not uniquely define the 5′ cleavage site. EMBO J., 7, 2533–2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teem J.L. and Rosbash,M. (1983) Expression of a β-galactosidase gene containing the ribosomal protein 51 intron is sensitive to the rna2 mutation of yeast. Proc. Natl Acad. Sci. USA, 80, 4403–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Improved sensitivity of profile searches through the use of sequence weights and gap excision. Comput. Appl. Biosci., 10, 19–29. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTALX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcárcel J., Gaur,R., Singh,R. and Green,M. (1996) Interaction of U2AF65 RS region with pre-mRNA of branch point and promotion of base pairing with U2 snRNA. Science, 273, 1706–1709. [DOI] [PubMed] [Google Scholar]
- Vernet C. and Artzt,K. (1997) STAR, a gene family involved in signal transduction and activation of RNA. Trends Genet., 13, 479–484. [DOI] [PubMed] [Google Scholar]
- Vijayraghavan U., Company,M. and Abelson,J. (1989) Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev., 3, 1206–1216. [DOI] [PubMed] [Google Scholar]
- Zamore P.D. and Green,M.R. (1989) Identification, purification and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc. Natl Acad. Sci. USA, 86, 9243–9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D. and Rosbash,M. (1999) Identification of eight proteins that cross-link to pre-mRNA in the yeast commitment complex. Genes Dev., 13, 581–592. [DOI] [PMC free article] [PubMed] [Google Scholar]