Abstract
During early stages of cotranslational protein translocation across the endoplasmic reticulum (ER) membrane the ribosome is targeted to the heterotrimeric Sec61p complex, the major component of the protein-conducting channel. We demonstrate that this interaction is mediated by the 28S rRNA of the eukaryotic large ribosomal subunit. Bacterial ribosomes also bind via their 23S rRNA to the bacterial homolog of the Sec61p complex, the SecYEG complex. Eukaryotic ribosomes bind to the SecYEG complex, and prokaryotic ribosomes to the Sec61p complex. These data indicate that rRNA-mediated interaction of ribosomes with the translocation channel occurred early in evolution and has been conserved.
Keywords: protein translocation/ribosome/rRNA/SecY/Sec61
Introduction
Protein translocation across the ER membrane can occur either co- or post-translationally. In the cotranslational pathway, a polypeptide chain is synthesized on a membrane-bound ribosome; the elongating chain is transferred from a tunnel in the ribosome through a channel in the membrane into the lumen of the ER (Crowley et al., 1994; Mothes et al., 1994; Beckmann et al., 1997). In the posttranslational pathway, the chain is first completed in the cytosol before being transported across the membrane. In both cases the same membrane channel is used, formed by three or four copies of the evolutionarily conserved heterotrimeric Sec61p complex. In the cotranslational pathway, the Sec61p channel associates with the ribosome (Görlich and Rapoport, 1993; Kalies et al., 1994; Hanein et al., 1996; Beckmann et al., 1997), and in the posttranslational pathway, at least in the yeast Saccharomyces cerevisiae, it associates with the tetrameric Sec62/63p membrane protein complex to form a heptameric Sec complex (Deshaies et al., 1991; Panzner et al., 1995; Hanein et al., 1996).
How the ribosome binds to the Sec61p complex during cotranslational translocation is unknown, and it is unclear whether the interaction involves ribosomal RNA (rRNA), protein or both. Non-translating mammalian ribosomes can bind with high affinity to the Sec61p complex in mammalian ER membranes (Rolleston, 1972; Borgese et al., 1974; Kalies et al., 1994). This interaction is salt sensitive, and the binding site is likely to be located in the large ribosomal subunit (Rolleston, 1972; Borgese et al., 1974; Unwin, 1977). The association of non-translating ribosomes with the Sec61p complex mimics early stages of cotranslational protein translocation (Jungnickel and Rapoport, 1995; Beckmann et al., 1997; Neuhof et al., 1998). When the signal sequence of a growing nascent chain has just emerged from the ribosome, the ribosome–nascent chain complex is targeted by the signal recognition particle (SRP) to the membrane (Walter and Blobel, 1981) and initially binds to the Sec61p complex in a salt-sensitive manner (Jungnickel and Rapoport, 1995). After further chain elongation, the signal sequence is recognized by the channel and inserts into it, and the ribosome–membrane interaction becomes salt resistant (Jungnickel and Rapoport, 1995). Such tightly bound ribosomes can only be removed by both releasing the nascent chain from the ribosome (e.g. by puromycin) and high salt treatment (Adelman et al., 1973).
Bacteria have a close homolog of the Sec61p complex in their cytoplasmic membrane, the heterotrimeric SecYEG complex. The largest subunits of both complexes span the membrane 10 times. They share sequence similarity in particular cytosolic loops and some trans-membrane domains, but not in regions on the other side of the membrane (Görlich et al., 1992). The smallest subunits of the trimeric complexes also show sequence similarity in a cytosolic region (Hartmann et al., 1994). These similarities suggest that the eukaryotic and prokaryotic complexes interact with a conserved cytosolic component, perhaps the ribosome. Eukaryotic and bacterial ribosomes have similar structures, in particular their RNAs share sequence similarities (Gutell et al., 1993; Schnare et al., 1996; Frank, 1997), but whether the SecYEG complex actually binds ribosomes has not yet been demonstrated. In fact, the existence of cotranslational translocation in bacteria is controversial. A tight ribosome–membrane interaction as in mammals has not been observed (Smith et al., 1978), and many secretory proteins cross the membrane in a manner that is uncoupled from chain elongation (Randall, 1983). On the other hand, nascent inner membrane proteins are targeted to the membrane by bacterial SRP, suggesting a pathway similar to the cotranslational process in eukaryotes (Ulbrandt et al., 1997).
Here, we have found that ribosome binding is indeed a general feature of the Sec61p and SecYEG complexes, and that the 28/23S RNA of the large ribosomal subunit is responsible for the interaction.
Results
Ribosome binding is evolutionarily conserved
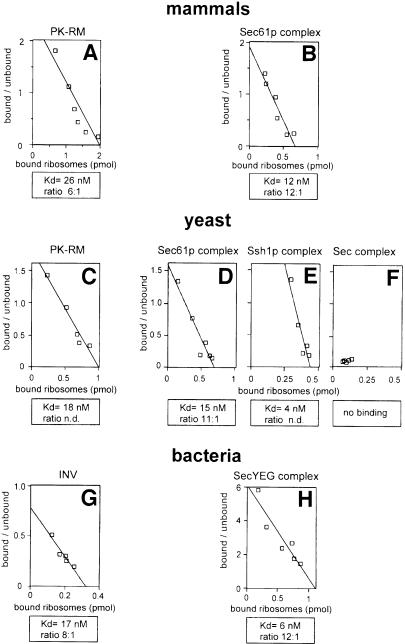
To determine ribosome binding to membranes, we used a flotation assay (Borgese et al., 1974; Kalies et al., 1994). Radiolabeled ribosomes were mixed with increasing concentrations of unlabeled ribosomes and incubated with membranes. After flotation of the membranes in a sucrose gradient, the amounts of radioactive ribosomes in the floated and non-floated fractions were determined. Scatchard plot analysis allows the estimation of both the apparent dissociation constant and the number of binding sites. With this assay, we have shown previously that the mammalian Sec61p complex binds mammalian ribosomes and is in fact responsible for essentially all binding sites in native ER membranes (Kalies et al., 1994). In agreement with these results, we observed binding of mammalian ribosomes to both intact canine pancreas ER membranes, previously stripped of ribosomes by treatment with puromycin and high salt (PK-RM) (Figure 1A), and to the purified mammalian Sec61p complex reconstituted into proteoliposomes (Figure 1B). The dissociation constants were similar to those determined before (Figure 1A and B; Table I; see also Kalies et al., 1994). The number of binding sites in intact membranes, relative to the amount of Sec61p complex contained in them (determined by quantitative immunoblotting), suggests that several copies of Sec61p complex molecules form one ribosome binding site (Figure 1A). In the case of reconstituted Sec61p complex, the number is higher (Figure 1B), as expected from the existence of two orientations of the protein complex in the lipid bilayer.
Fig. 1. Ribosome binding in mammals, yeast and bacteria. Radiolabeled canine (A and B), S.cerevisiae (C–F) or E.coli (G and H) ribosomes were incubated with ribosome-stripped microsomal membranes (PK-RM) (A and C), inverted vesicles from E.coli (INV) (G), or proteoliposomes containing the purified membrane protein complexes indicated (B, D–F and H). Bound and unbound ribosomes were separated and Scatchard plot analysis performed to estimate the apparent dissociation constant (Kd) and the ratio of moles of membrane protein complex, determined by immunoblotting, per mole of bound ribosomes under saturation conditions.
Table I. Ribosome binding is evolutionarily highly conserved.
| Protein complexa | Ribosomes (C.familiaris) | (S.cerevisiae) | (E.coli) |
|---|---|---|---|
| Sec61 (C.familiaris) | 12 nMb | 14 nM | 36 nM |
| 12:1c | 8:1 | 10:1 | |
| Sec61 (S.cerevisiae) | 14 nM | 15 nM | 90 nM |
| 8:1 | 11:1 | 9:1 | |
| Ssh1 (S.cerevisiae) | 6 nM | 4 nM | 44 nM |
| n.d. | n.d. | n.d. | |
| SecYEG (E.coli) | 2 nM | n.d. | 6 nM |
| 14:1 | n.d. | 12:1 |
aThe binding of canine (C.familiaris), yeast (S.cerevisiae) or bacterial (E.coli) ribosomes was determined with proteoliposomes containing the purified membrane protein complexes indicated.
b,cThe apparent dissociation constants and ratios of moles of membrane protein complex per mole of bound ribosomes under saturation conditions were obtained by Scatchard plot analysis.
n.d., not determined.
We next examined the interaction of yeast ribosomes with yeast ER membranes and purified yeast Sec61p complex (Figure 1C and D). The dissociation constants were almost the same as with mammalian components (Figure 1C and D; Table I). The number of binding sites estimated for the purified Sec61p complex indicated again that several correctly oriented molecules are required to bind one ribosome (Figure 1D). In agreement with the characteristics of the mammalian system, liposomes lacking protein did not bind ribosomes, and the interaction with yeast ER membranes was salt sensitive, essentially vanishing at 400–500 mM salt (data not shown). Ribosome binding to a purified Sec61p complex lacking the β-subunit was indistinguishable from that to the intact Sec61p complex (data not shown). This is in agreement with previous data that showed that the β-subunit of the mammalian Sec61p complex is dispensable for ribosome interaction (Kalies et al., 1998), and is consistent with the fact that deletion of the subunit in yeast is not lethal (Finke et al., 1996).
The heterotrimeric Ssh1p complex, which is structurally related to the Sec61p complex, has also been proposed to be involved in cotranslational translocation in S.cerevisiae (Finke et al., 1996). In agreement with this, we found that the reconstituted, purified Ssh1p complex binds yeast ribosomes with characteristics very similar to those of the Sec61p complex (Figure 1E). On the other hand, the heptameric Sec complex, involved in posttranslational translocation (Panzner et al., 1995), did not bind ribosomes (Figure 1F). Thus, by its association with the Sec62/63p complex, the Sec61p complex may not only be activated for posttranslational translocation, but may also be inactivated for cotranslational translocation.
Next we tested whether bacterial ribosomes can bind to the bacterial SecYEG complex. We found that Escherichia coli ribosomes bind tightly to both inverted vesicles of E.coli cytoplasmic membranes (Figure 1G), and to reconstituted SecYEG complex purified from E.coli (Figure 1H). The binding was as strong as with the eukaryotic systems (Figure 1; Table I), and the number of binding sites again indicated that several correctly oriented SecYEG molecules are required for the binding of one ribosome (Figure 1H). As in eukaryotes, very little binding was seen at salt concentrations >500 mM (data not shown). We conclude that ribosome binding is an evolutionarily conserved, general property of the Sec61p/SecYEG complex, consistent with the sequence similarity observed in several cytosolic domains of the subunits. These data suggest that bacterial membrane-bound ribosomes are not solely linked via the nascent chain (Smith et al., 1978).
We next examined whether ribosome binding is sufficiently conserved to allow cross-species interactions. Indeed, mammalian ribosomes bound just as well to the yeast Sec61p or Ssh1p complexes, or even the bacterial SecYEG complex, as they do to the mammalian Sec61p complex (Table I). Yeast ribosomes also interacted equally well with the yeast and mammalian membrane protein complexes. Escherichia coli ribosomes bound more strongly to the bacterial SecYEG complex than to the eukaryotic Sec61p and Ssh1p complexes, indicating some specificity, but even the binding in the heterologous systems was significant. In all cases, the number of binding sites was independent of the type of ribosomes used in the assay. Heterologous interactions could also be detected when E.coli ribosomes were bound to intact canine PK-RMs, or when canine ribosomes were bound to E.coli inverted vesicles. The number of binding sites was again independent of the source of the ribosomes. In addition, they were the same if the binding reaction was performed at 150 mM, rather than 100 mM salt (data not shown), indicating that the salt concentration was high enough to suppress non-specific interactions (Kalies et al., 1994). Taken together, these data show that the interacting components in the ribosomes and the channels must be highly conserved.
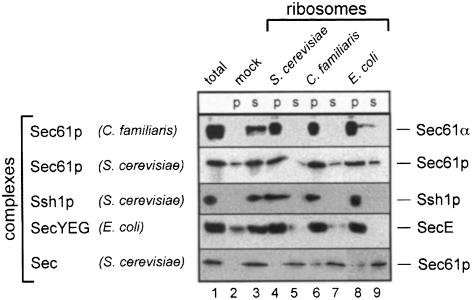
To test these conclusions further, we employed a different ribosome binding assay (Figure 2). Ribosomes were mixed with purified membrane protein complexes in detergent solution, the ribosomes were sedimented through a sucrose cushion, and the amount of complex in the pellet and supernatant fractions was determined by immunoblotting. Regardless of the source of ribosomes, most of the mammalian Sec61p complex, yeast Sec61p complex, yeast Ssh1p complex and E.coli SecYEG complex was found in the pellet (p) fraction. If ribosomes were omitted (mock), the percentage of sedimented membrane protein complexes was small (Figure 2, lane 2 versus 3). The validity of the assay is supported by the observation that the amount of membrane complex in the pellet was dependent on the amount of ribosomes added, and that 1 µΜ aurintricarboxylic acid, an inhibitor of ribosome binding to PK-RM (Borgese et al., 1974), also inhibited the binding in the soluble assay (data not shown). In addition, as in the flotation assay, the yeast Sec complex involved in posttranslational translocation did not interact with ribosomes from any species (Figure 2). Thus, these data confirm that the interaction between ribosomes and the Sec61p/SecYEG complex is highly conserved in evolution.
Fig. 2. Ribosome binding in detergent solution. Canine (C.familiaris), yeast (S.cerevisiae) or bacterial (E.coli) ribosomes were incubated in detergent solution with the purified membrane protein complexes indicated. Ribosomes were added in a 3-fold molar excess over membrane protein complex. As a control, a mock incubation in the absence of ribosomes was performed. After sedimentation of the ribosomes, membrane proteins in the pellets (p) and supernatants (s) were detected by SDS–PAGE and immunoblotting, using antibodies directed against the proteins indicated on the right. Lane 1 shows the input material.
Ribosome binding through rRNA
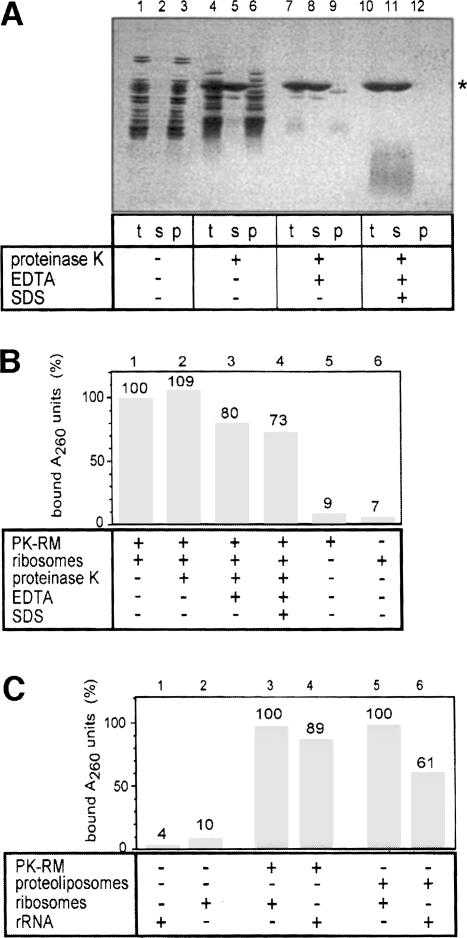
To test whether ribosomal proteins are required for the ribosome–membrane interaction, we initially used an approach similar to that employed to test the involvement of protein in the peptidyltransferase activity of the ribosome (Noller et al., 1992). Mammalian ribosomes were treated with high concentrations of proteinase K under different conditions (Figure 3A) and tested for binding to ER membranes (Figure 3B). When native ribosomes were treated with proteinase K, many ribosomal proteins were degraded (Figure 3A, lane 1 versus 4) but, upon sedimentation of the ribosome remnants, all polypeptide fragments were still seen (Figure 3A, lane 6). When the protease treatment was carried out in the presence of EDTA, a few bands were found in the pellet (Figure 3A, lane 9), but when performed in EDTA plus SDS, no protein bands were detectable (lane 12). In contrast, ∼95% of the rRNA was recovered in the pellet fractions (data not shown). The ribosome remnants were tested in a flotation assay for their ability to bind to mammalian ER membranes (PK-RM), using UV absorption to follow the rRNA (Figure 3B). Even the harshest protease treatment reduced ribosome binding by <30%. In the absence of membranes, neither ribosomes (Figure 3B, lane 7) nor extracted rRNA (not shown) were found in the floated fraction. These data thus suggest that rRNA is responsible for the interaction of ribosomes with the Sec61p complex.
Fig. 3. Ribosome binding occurs through rRNA. (A) Mammalian ribosomes were proteolyzed under different conditions and centrifuged through a sucrose cushion. Aliquots of the total (t), supernatant (s) and pellet (p) fractions were analyzed by SDS–PAGE and staining with Coomassie Blue. The band marked with a star is proteinase K. (B) Ribosome remnants in the pellet fraction (p) of (A) were tested for their binding to ribosome-stripped microsomes (PK-RM). Binding of the rRNA was followed by absorption at 260 nm (A260 units). The binding of untreated ribosomes was set to 100%. (C) The binding of total rRNA, isolated by extraction with phenol–chloroform, to PK-RM and proteoliposomes containing purified Sec61p complex was tested. The molar ratio of ribosomes (B and C), ribosomal remnants (B) or rRNA (C) to trimeric Sec61 complexes was ∼1:2.
To test this directly, we extracted the rRNA from mammalian ribosomes with phenol–chloroform and tested its binding to mammalian PK-RM and reconstituted Sec61p complex (Figure 3C). In both cases, the binding of isolated rRNA was significant. As with ribosomes, the binding was salt sensitive, essentially disappearing above 500 mM (data not shown). Binding of rRNA could be detected both by UV absorption (Figure 3C) or by using end-labeled radioactive RNA (not shown). In the latter case, it could be demonstrated that the binding was prevented by an excess of unlabeled rRNA, but not of mRNA coding for preprolactin (not shown).
The binding site is contained in the large rRNA
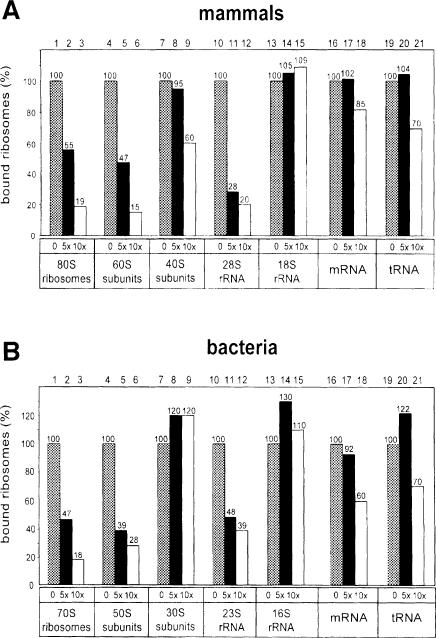
In agreement with previous data (Rolleston, 1972; Borgese et al., 1974; Unwin, 1977), we found that the large subunit of mammalian ribosomes is responsible for membrane binding. In competition experiments, unlabeled mammalian 60S ribosomal subunits were more potent than 40S subunits in preventing the binding of radiolabeled 80S ribosomes to reconstituted mammalian Sec61p complex (Figure 4A, columns 5 and 6 versus 8 and 9). The weak competition seen with the 40S subunits (Figure 4A, column 9) is likely to be due to contamination with large subunits, as some 28S RNA could be seen in RNA gel electrophoresis (data not shown). When similar competition experiments were performed with isolated RNAs, the 28S rRNA of the large subunit was a competitor for ribosome binding, in contrast to the 18S RNA of the small subunit, which did not inhibit at all (Figure 4A, columns 11 and 12 versus 14 and 15). mRNA coding for preprolactin and tRNA showed only little inhibition (Figure 4A, columns 16–21). These data confirm that RNA is responsible for the interaction of ribosomes with the Sec61p complex, and indicate that binding occurs through specific sites in the 28S RNA of the large subunit.
Fig. 4. The large ribosomal subunit and its RNA inhibit ribosome binding. (A) Radiolabeled mammalian ribosomes were mixed with a 5- or 10-fold molar excess of the unlabeled competitor indicated. Binding to proteoliposomes containing the purified mammalian Sec61p complex was determined in a flotation assay. The molar ratio between labeled ribosomes and Sec61 complexes was 1:10. Percentage binding relative to that in the absence of competitor is shown. (B) As in (A) except that the binding of radiolabeled E.coli ribosomes to proteoliposomes containing the SecYEG complex was determined in the presence of various competitors.
Similar conclusions could be derived for the bacterial system. In competition experiments with radiolabeled 70S ribosomes from E.coli, unlabeled 50S subunits were almost as potent competitors as unlabeled 70S subunits, while unlabeled 30S subunits were inactive (Figure 4B, columns 2, 3, 5 and 6 versus 8 and 9). The isolated 23S RNA of the large ribosomal subunit was a reasonable competitor, in contrast to the 16S RNA of the small subunit (Figure 4B, columns 11 and 12 versus 14 and 15). At 5-fold molar excess, mRNA or tRNA also did not act as competitors, although at higher excess a moderate inhibition was seen (Figure 4B, columns 16–21). We conclude that the RNA of the large ribosomal subunit must contain the conserved binding site for interaction with the Sec61p/SecYEG complex.
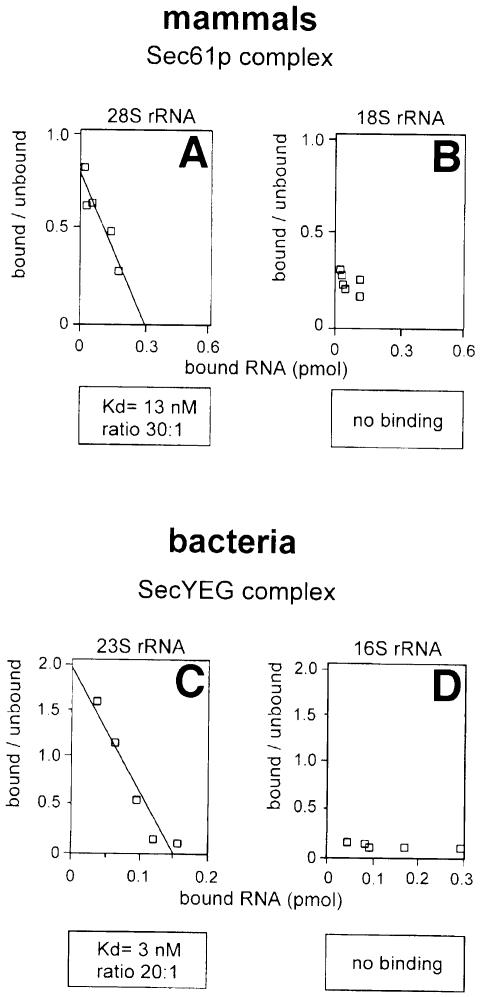
To test the interaction between rRNAs and the Sec61p/SecYEG complex directly, we purified mammalian 28S and 18S RNAs and bacterial 23S and 16S RNAs, end-labeled them with radioactive nucleotides, and used them in a flotation binding assay with reconstituted, purified membrane protein complexes (Figure 5A and B). Scatchard plot analysis indicates that the apparent dissociation constants for the large rRNAs were similar to those obtained for intact ribosomes (13 and 3 nM for the 28 and 23S RNAs, respectively). In contrast, the binding of the 18S RNA was only slightly above background and that of the 16S RNA was negligible. These data therefore confirm that the large rRNA from both eukaryotic and bacterial ribosomes is largely, if not solely responsible for the interaction with the Sec61p/SecYEG complex.
Fig. 5. Large rRNAs bind with the same affinity as ribosomes. (A) Mammalian 28S rRNA or (B) 18S rRNA was radiolabeled and its binding to proteoliposomes containing the purified mammalian Sec61p complex determined in a flotation assay. (C) The binding of bacterial 23S rRNA or (D) 16S rRNA to proteoliposomes containing the purified SecYEG complex was determined in a similar way. Scatchard plot analysis was carried out to estimate the apparent dissociation constants (Kd) and the ratio of moles of membrane protein complex per mole of bound ribosomes under saturating conditions.
Discussion
Here we have made the surprising observation that the interaction between the ribosome and the protein- conducting channel in the membrane is largely mediated by the RNA of the large ribosomal subunit. The interaction has been highly conserved during evolution; eukaryotic ribosomes bound to the prokaryotic channel and, vice versa, prokaryotic ribosomes to the eukaryotic channel. This is consistent with the sequence similarities between eukaryotic and prokaryotic channel components and ribosomes.
We confirm that the large ribosomal subunit is responsible for the interaction of the ribosome with the ER membrane (Rolleston, 1972; Borgese et al., 1974; Unwin, 1977). This is in agreement with the recent structure of complexes of the yeast Sec61p complex and non-translating ribosomes determined by electron microscopy (Beckmann et al., 1997). The channel was found to bind to the large ribosomal subunit, and only one thin connection was seen. More recent electron microscopy data confirm that there are only a few (probably four) connections between the ribosome and the channel (J.F.Ménétret, A.Neuhof, D.G.Morgan, K.Plath, I.Akey, M.Radermacher, T.A.Rapoport and C.W.Akey, unpublished results). Together with the present findings, this suggests that a small number of RNA domains are involved in the interaction. The salt sensitivity indicates that the interaction is electrostatic, but since mRNA, tRNA, DNA (Kalies et al., 1994) and the small rRNAs do not bind, specific nucleotide sequences in the 28/23S RNA must be important. There are in fact several domains outside the peptidyltransferase center that are highly conserved in sequence between bacterial and eukaryotic ribosomes (Gutell et al., 1993; Schnare et al., 1996). Since the binding activity of the 28 or 23S RNAs was little perturbed by phenol–chloroform extraction, RNA domains that are either extremely stable or reform rapidly in aqueous solution may be prime candidates for the interaction with the Sec61p/SecYEG complex. Indeed, the large rRNA contains several evolutionarily conserved and stable base-paired stem loops (Brimacombe, 1995). Good candidates for domains within the large rRNA that could be responsible for the interaction with the membrane channel are the helices H5, H6, H7, H46, H50, H52, H53 and H101. On the basis of computer fitting of the RNA into the three-dimensional structure of the large ribosomal subunit of E.coli, these domains are predicted to be on the ribosomal surface close to the exit site of the nascent chain on the ribosome (R.Brimacombe, personal communication). We speculate that one or several of these regions form a specific structure that allows certain phosphate groups to interact with positively charged residues in conserved cytosolic loops of the Sec61p/SecYEG complex. As demonstrated by freeze-fracture electron microscopy, ribosomes can induce the formation of oligomeric channel structures from Sec61p complex molecules reconstituted into proteoliposomes (Hanein et al., 1996). It remains to be seen whether single Sec61p complex molecules interact differently with ribosomes than the oligomers, and whether the large rRNA can bind to both forms. In fact, the lower number of binding sites per Sec61 molecule in experiments with purified rRNA (Figure 5) compared with experiments using intact ribosomes (Figure 1 and Table I) could be due to an decreased ability of the rRNA to induce the formation of oligomers. It should also be noted that we have only analyzed a reaction that corresponds to the initial membrane binding of a ribosome–nascent chain complex; it is possible that at later stages of the translocation process ribosomal components other than the 28/23S rRNA may participate in the ribosome–channel interaction.
The strong interaction between E.coli ribosomes and the SecYEG complex suggests that some cotranslational translocation in prokaryotes occurs in a manner similar to that in eukaryotes. The previous conclusion that tightly bound ribosomes do not exist in bacteria was based on the fact that puromycin released a large percentage of ribosomes from membranes (Smith et al., 1978), but it remained unclear whether these ribosomes were actually synthesizing exported proteins. It is also possible that only the transport of certain membrane proteins needs a tight ribosome–membrane interaction (Seluanov and Bibi, 1997; Ulbrandt et al., 1997). How the ribosome–channel interaction might be regulated in bacteria is unclear. Our data in yeast demonstrate that the Sec61p complex can be converted from a cotranslational into a posttranslational translocation machine by its association with the Sec62/63p complex. In bacteria, the cytoplasmic SecA protein might play an analogous role for the SecYEG complex.
Finally, our results have evolutionary implications. rRNA has been shown to be responsible for the basic function of the ribosome, the peptidyltransferase activity (Noller et al., 1992), suggesting that it is extremely ancient. That it is the rRNA that contacts the translocation channel thus suggests that this interaction is also very old. RNAs can perform many enzymatic reactions and may have been the original biological catalytic components (Cech, 1993; Orgel, 1998). On the other hand, it is clear that compartmentation by a surrounding membrane must also be ancient because it would make all reactions much more efficient. Transport across membranes to gain essential water-soluble nutrients might not have been possible by the hydrophilic RNA and may have necessitated the existence of membrane proteins, which must therefore be an early achievement, perhaps an even earlier one than soluble proteins. A direct coupling between the RNA that performed peptide synthesis and a peptide-conducting membrane channel may have allowed the cotranslational integration of membrane proteins that would otherwise aggregate in an aqueous environment. Ribosomal proteins may have co-evolved to increase the efficiency of basic reactions, to help in assembly or to prevent undesired interactions of the RNA. Interestingly, in E.coli the secY gene is found on an operon with genes coding for ribosomal proteins. This arrangement of genes has been conserved in most eubacteria and archaea. Once established, the interaction between the ribosome and the protein-conducting channel appears to have been highly conserved during evolution.
Materials and methods
Preparation of microsomes, proteoliposomes, ribosomes and rRNA
Microsomal membranes from canine pancreas, stripped of ribosomes by treatment with puromycin and high salt (PK-RM), and yeast PK-RM were prepared as described (Görlich et al., 1992). Inverted vesicles from E.coli were isolated as reported (Douville et al., 1995). The reconstitution of proteoliposomes containing purified membrane protein complexes, and the isolation of mammalian ribosomes were performed as described (Görlich et al., 1992; Görlich and Rapoport, 1993; Panzner et al., 1995). Yeast ribosomes were isolated similarly from cytosol and treated with puromycin/high salt. Mammalian ribosomal subunits were separated on a 10–25% sucrose gradient in HKM800 [50 mM HEPES–KOH pH 7.6, 800 mM potassium acetate, 10 mM magnesium acetate, 1 mM dithiothreitol (DTT)] by centrifugation at 20°C at 35 000 r.p.m. in a Beckman SW41 rotor. Escherichia coli ribosomes were isolated by centrifugation at 100 000 r.p.m. for 20 min at 2°C in a TLA 100.3 rotor. Ribosomes were treated with 1 mM puromycin and high salt, centrifuged and resuspended in HKM150 (50 mM HEPES–KOH pH 7.6, 150 mM potassium acetate, 5 mM magnesium acetate, 1 mM DTT). Ribosomal subunits of E.coli were a kind gift of Dr R.Brimacombe. Ribosomes and subunits were radiolabeled with 35S-labeling reagent (SLR; Amersham) as described (Kalies et al., 1994).
Total rRNA was isolated from 440 pmol of canine pancreas ribosomes by extraction twice with phenol, once with phenol–chloroform (1:1) and once with chloroform. Ethanol-precipitated RNA was dissolved in HKM25 (50 mM HEPES–KOH pH 7.6, 25 mM potassium acetate, 3 mM magnesium acetate, 1 mM DTT) containing 250 mM sucrose. 28 and 18S rRNA were isolated from 80S ribosomes by electrophoresis in a 1.0% agarose gel using 2% SDS in the sample buffer and purified with an RNA purification kit (RNaid spin kit, Bio 101, Inc., Vista, CA) followed by phenol–chloroform extraction. 23 and 16S rRNA of E.coli were purified directly from ribosomal subunits by phenol–chloroform extraction. rRNA was end-labeled with [γ-32P]ATP.
Proteolysis of ribosomes
Ribosomes (85 pmol) were treated for 1 h at room temperature with 1 mg/ml proteinase K in 0.05 ml of HNM150 (50 mM HEPES–KOH pH 7.6, 150 mM sodium acetate, 5 mM magnesium acetate, 1 mM DTT) containing 0.25 M sucrose. Where indicated, 20 mM EDTA or 0.5% SDS plus 20 mM EDTA was added. The reaction was stopped with 1 mM phenylmethylsulfonyl fluoride (PMSF). The samples were centrifuged through a 0.1 ml cushion of 400 mM sucrose and 0.1 mM PMSF in HNM150 (40 min, 100 000 r.p.m., 18°C, TLA 100) and resuspended in HNM150.
Flotation assay for ribosome and rRNA binding
Radiolabeled ribosomes or rRNA were mixed with increasing amounts of unlabeled ribosomes or rRNA, respectively. Ribosome-stripped membranes (PK-RM, INV) or reconstituted proteoliposomes containing the indicated protein complexes were added in a final volume of 0.03 ml of 0.25 M sucrose in HKM150 or HKM100 (same as HKM150, but 100 mM potassium acetate), respectively. The samples were incubated at 28°C for 30 min. The binding assay for PK-RM and INV was carried out as described (Kalies et al., 1994). The samples containing proteoliposomes made from canine Sec61p complexes were mixed with 0.07 ml of 1.9 M sucrose in HKM150 and overlayered with 0.1 ml of 0.25 M sucrose in HKM150. When proteoliposomes containing purified components from yeast or E.coli were analyzed, HKM100 instead of HKM150 was used. After centrifugation at 100 000 r.p.m. at 4°C for 1 h in a TLA 100 rotor, the upper 0.1 ml of the gradient and the lower 0.1 ml together with the pellet were collected, each mixed with 0.1 ml of water and a scintillation cocktail, and placed in a scintillation counter. The number of binding sites determined in different experiments with similar conditions varied by ∼10%, while the apparent Kd values varied by a factor of ∼2.
Binding of ribosomes in detergent solution
Ribosomes were mixed with the indicated purified membrane protein complexes in 0.03 ml of 0.25 M sucrose and 0.3% DeoxyBigCHAP in HKM100 and incubated for 30 min on ice. The samples were layered on top of 0.1 ml of 0.5 M sucrose cushion in the same buffer and centrifuged for 5 min at 75 000 r.p.m. in a TLA 100 rotor at 4°C. The supernatant and the pellet fractions were analyzed by SDS–PAGE and immunoblotting.
Binding of ribosome remnants
Ribosomes (15 pmol) and ribosome remnants (measured by A260 absorption) were incubated with 10 µl of PK-RM (containing 32 pmol of trimeric Sec61 complex) or an equivalent amount of reconstituted proteoliposomes, respectively, in HNM150 in a final volume of 0.02 ml for 30 min at room temperature. The samples were mixed with 0.07 ml of 2.3 M sucrose in HNM150 and layered under a discontinuous sucrose gradient consisting of 0.08 ml of 1.8 M sucrose in HNM150 and 0.03 ml of HNM150. After centrifugation for 1 h at 100 000 r.p.m. at 18°C (rotor TLA 100) the floated material was collected by removing the upper 0.08 ml. The samples were diluted with 1% SDS to 1 ml and the absorption at 260 nm was determined.
Immunoblotting
Immunoblotting with various antibodies was performed as described (Görlich et al., 1992) with enhanced chemiluminescence (ECL) as a detection system and analysis with a CCD camera (Raytest, Germany). Polyclonal antibodies directed against the following peptides were used: the C-terminus of Sec61α (Görlich et al., 1992), the C-terminus of Sec61p (Panzner et al., 1995), the C-terminus of Ssh1p (Finke et al., 1996) and a peptide comprising residues 64–92 of SecE (Schatz et al., 1991). For quantitative immunoblotting the concentration of purified proteins, determined by sequencing, was used as a standard.
Acknowledgments
Acknowledgements
We thank K.Plath and K.E.S.Matlack for critical reading of the manuscript, R.Brimacombe for providing the ribosomal subunits of E.coli and unpublished data, and S.Kostka and R.Kraft for protein sequencing. The work was supported by a grant to E.H. and K.-U.K. from the DFG (Deutsche Forschungsgemeinschaft).
References
- Adelman M.R., Sabatini,D. and Blobel,G. (1973) Ribosome membrane interaction. Nondestructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J. Cell Biol., 56, 206–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Borgese N., Mok,W., Kreibich,G. and Sabatini,D.D. (1974) Ribosomal–membrane interaction: in vitro binding of ribosomes to microsomal membranes. J. Mol. Biol., 88, 559–580. [DOI] [PubMed] [Google Scholar]
- Brimacombe R. (1995) The structure of ribosomal RNA: a three-dimensional jigsaw puzzle. Eur. J. Biochem., 230, 365–383. [PubMed] [Google Scholar]
- Cech T.R. (1993) The efficiency and versatility of catalytic RNA: implications for an RNA world. Gene, 135, 33–36. [DOI] [PubMed] [Google Scholar]
- Crowley K.S., Liao,S.R., Worrell,V.E., Reinhart,G.D. and Johnson,A.E. (1994) Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell, 78, 461–471. [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., Sanders,S.L., Feldheim,D.A. and Schekman,R. (1991) Assembly of yeast SEC proteins involved in translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature, 349, 806–808. [DOI] [PubMed] [Google Scholar]
- Douville K., Price,A., Eichler,J., Economou,A. and Wickner,W. (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem., 270, 20106–20111. [DOI] [PubMed] [Google Scholar]
- Finke K., Plath,K., Panzner,S., Prehn,S., Rapoport,T.A., Hartmann,E. and Sommer,T. (1996) A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S.cerevisiae. EMBO J., 15, 1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Frank J. (1997) The ribosome at higher resolution—the donut takes shape. Curr. Opin. Struct. Biol., 7, 266–272. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Rapoport,T.A. (1993) Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell, 75, 615–630. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn,S., Hartmann,E., Kalies,K.-U. and Rapoport,T.A. (1992) A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell, 71, 489–503. [DOI] [PubMed] [Google Scholar]
- Gutell R.R., Gray,M.W. and Schnare,M.N. (1993) A compilation of large subunit (23S and 23S-like) ribosomal RNA structures. Nucleic Acids Res., 21, 3055–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Matlack,K.E., Jungnickel,B., Plath,K., Kalies,K.-U., Miller,K.R., Rapoport,T.A. and Akey,C.W. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Sommer,T., Prehn,S., Görlich,D., Jentsch,S. and Rapoport,T.A. (1994) Evolutionary conservation of components of the protein translocation complex. Nature, 367, 654–657. [DOI] [PubMed] [Google Scholar]
- Jungnickel B. and Rapoport,T.A. (1995) A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell, 82, 261–270. [DOI] [PubMed] [Google Scholar]
- Kalies K.-U., Görlich,D. and Rapoport,T.A. (1994) Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J. Cell Biol., 126, 925–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalies K.-U., Rapoport,T.A. and Hartmann,E. (1998) The β subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol., 141, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W., Prehn,S. and Rapoport,T.A. (1994) Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J., 13, 3973–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof A., Rolls,M.M., Jungnickel,B., Kalies,K.-U. and Rapoport,T.A. (1998) Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell, 9, 103–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller H.F., Hoffarth,V. and Zimniak,L. (1992) Unusual resistance of peptidyl transferase to protein extraction procedures. Science, 256, 1416–1419. [DOI] [PubMed] [Google Scholar]
- Orgel L.E. (1998) The origin of life—a review of facts and speculations. Trends Biochem. Sci. Sci., 23, 491–505. [DOI] [PubMed] [Google Scholar]
- Panzner S., Dreier,L., Hartmann,E., Kostka,S. and Rapoport,T.A. (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell, 81, 561–570. [DOI] [PubMed] [Google Scholar]
- Randall L.L. (1983) Translocation of domains of nascent periplasmic proteins across the cytoplasmic membrane is independent of elongation. Cell, 33, 213–240. [DOI] [PubMed] [Google Scholar]
- Rolleston F.S. (1972) The binding of ribosomal subunits to endoplasmic reticulum membranes. Biochem. J., 129, 721–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz P.J., Bieker,K.L., Ottemann,K.M., Silhavy,T.J. and Beckwith,J. (1991) One of three transmembrane stretches is sufficient for functioning of the SecE protein, a membrane component of the secretory machinery. EMBO J., 10, 1749–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnare M.N., Damberger,S.H., Gray,M.W. and Gutell,R.R. (1996) Comprehensive comparison of structural characteristics in eukaryotic cytoplasmic large subunit (23S-like) ribosomal RNA. J. Mol. Biol., 256, 701–719. [DOI] [PubMed] [Google Scholar]
- Seluanov A. and Bibi,E. (1997) FtsY, the prokaryotic signal recognition particle receptor homologue, is essential for biogenesis of membrane proteins. J. Biol. Chem., 272, 2053–2055. [DOI] [PubMed] [Google Scholar]
- Smith W.P., Tai,P.C. and Davis,B.D. (1978) Nascent peptide as sole attachment of polysomes to membranes in bacteria. Proc. Natl Acad. Sci. USA, 75, 814–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrandt N.D., Newitt,J.A. and Bernstein,H.D. (1997) The E.coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell, 88, 187–196. [DOI] [PubMed] [Google Scholar]
- Unwin P.N. (1977) Three-dimensional model of membrane-bound ribosomes obtained by electron microscopy. Nature, 269, 118–122. [DOI] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1981) Translocation of proteins across the endoplasmic reticulum. III. Signal recognition protein (SRP) causes signal sequence and site specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol., 91, 557–561. [DOI] [PMC free article] [PubMed] [Google Scholar]