Abstract
Wnt growth factors regulate a variety of developmental processes by altering specific gene expression patterns. In vertebrates β-catenin acts as transcriptional activator, which is needed to overcome target gene repression by Groucho/TLE proteins, and to permit promoter activation as the final consequence of Wnt signaling. However, the molecular mechanisms of transcriptional activation by β-catenin are only poorly understood. Here we demonstrate that the closely related acetyltransferases p300 and CBP potentiate β-catenin-mediated activation of the siamois promoter, a known Wnt target. β-catenin and p300 also synergize to stimulate a synthetic reporter gene construct, whereas activation of the cyclin D1 promoter by β-catenin is refractory to p300 stimulation. Axis formation and activation of the β-catenin target genes siamois and Xnr-3 in Xenopus embryos are sensitive to the E1A oncoprotein, a known inhibitor of p300/CBP. The C-terminus of β-catenin interacts directly with a region overlapping the CH-3 domain of p300. p300 could participate in alleviating promoter repression imposed by chromatin structure and in recruiting the basal transcription machinery to promoters of particular Wnt target genes.
Keywords: β-catenin/p300/CBP acetylase/T-cell factor/transcriptional activation/Wnt signaling
Introduction
Members of the Wnt family of growth factors modulate numerous developmental processes in a variety of organisms (Cadigan and Nusse, 1997). Deregulation of Wnt signaling events is also implicated in the development of cancer (Miller et al., 1999). On the basis of whether they are able to transform cultured cells in vitro or to induce dorso-anterior body axis duplication in Xenopus embryos, Wnt genes have been grouped into distinct classes. The functional distinction is reflected by the activation of separate intracellular signaling pathways by different Wnts and their receptors, relatives of the Drosophila Frizzled proteins (Cadigan and Nusse, 1997; Miller et al., 1999). A subset of Frizzled receptors engage β-catenin as effector protein (Cadigan and Nusse, 1997; Miller et al., 1999). β-catenin and its Drosophila ortholog Armadillo belong to a family of proteins that are characterized by the presence of multiple copies of a 42 amino acid motif, the so-called Armadillo repeat (Peifer et al., 1994). In addition to their role in transmitting Wnt signals, β-catenin and Armadillo carry out essential functions in cell–cell adhesion as components of cadherin–catenin complexes (Aberle et al., 1996).
Wnt factors control the signaling function of β-catenin by modulating the activity of a multiprotein complex containing the adenomatous polyposis coli (APC) tumor suppressor protein, Axin, and glycogen synthase kinase-3β (GSK-3β) (reviewed by Cadigan and Nusse, 1997; Miller et al., 1999). The APC–Axin–GSK-3β complex serves as a molecular relay station that either directs β-catenin towards degradation (Aberle et al., 1997) or allows its accumulation in the nucleus, where it associates with members of the TCF family of HMG-box factors to alter the expression of certain target genes (reviewed by Eastman and Grosschedl, 1999; Miller et al., 1999). Which route β-catenin takes depends on differential phosphorylation by GSK-3β at its N-terminus, which is reduced in response to Wnt signals (Cadigan and Nusse, 1997; Miller et al., 1999). Aside from Wnts, cell–substrate interactions and the integrin-linked kinase are also capable of inhibiting GSK-3β activity and inducing β-catenin–TCF-mediated transcriptional activation (Novak et al., 1998). Additional levels of control are provided by the LIT-1/Nemo-like kinase, which regulates DNA-binding and the subcellular distribution of TCF factors (Ishitani et al., 1999; Meneghini et al., 1999; Rocheleau et al., 1999), and by the dCBP acetylase, which regulates the interaction of dTCF with Armadillo in Drosophila (Waltzer and Bienz, 1998).
Although capable of binding to DNA in a sequence-specific manner, TCF proteins possess no intrinsic ability to modulate transcription (Eastman and Grosschedl, 1999). Rather, TCFs can be viewed as chromosomal docking sites for auxiliary proteins, which upon interacting with TCFs alter the transcriptional activity of a nearby promoter. β-catenin is an example for an activating interaction partner of TCFs (van de Wetering et al., 1997). In contrast, association of TCFs with the Groucho/TLE or CtBP proteins leads to promoter repression (Cavallo et al., 1998; Levanon et al., 1998; Roose et al., 1998; Brannon et al., 1999). Groucho/TLE proteins are tightly associated with chromatin and are known to interact with histones and the histone deacetylase Rpd3 (Palaparti et al., 1997; Chen et al., 1999a). This suggests that transcriptional repression by Groucho/TLE involves the formation of a specialized chromatin structure. How β-catenin overcomes repression by Groucho/TLE and activates target genes is largely unknown. Previous attempts to identify cofactors that assist β-catenin in its function as transcriptional activator have led to the identification of the TATA-binding protein (TBP), which can directly and indirectly interact with several of the transactivating elements in β-catenin (Bauer et al., 1998; Hecht et al., 1999). Thus, β-catenin may function in part by recruiting general transcription factors to target gene promoters. However, the interactions with TBP are unlikely to represent the sole basis for transactivation by β-catenin. Although TBP is ubiquitously present, nuclear localization of β-catenin and its interaction with TCF factors are not always sufficient for reporter gene activation (Prieve and Waterman, 1999). Moreover, some of the β-catenin transactivating elements stimulate gene expression but apparently do not interact with TBP (Hecht et al., 1999). In addition, different signals lead to the activation of β-catenin–TCF-dependent transcription in distinct cellular settings and the target genes of β-catenin are likely to play different roles in developmental or cell cycle regulation. β-catenin may not regulate all these genes in the same way, but affect their expression through diverse mechanisms and possibly employ various cofactors.
Nuclear histone acetyltransferases (HATs) play important roles in gene regulation (Workman and Kingston, 1998). They catalyze the reversible acetylation of histones to alleviate chromatin-mediated restrictions imposed on promoter activation. In addition, a growing number of non-histone substrate proteins for HATs has been identified (Grant and Berger, 1999). Among the known HATs are GCN5 and the related P/CAF, the nuclear hormone receptor coactivators SRC-1 and ACTR, the TBP-associated TAFII250, Adenovirus E1A-associated p300 and the closely related CREB-binding protein (CBP) (Grant and Berger, 1999). CBP and p300 can form larger protein complexes that include other acetylases and serve as coactivators of numerous transcription factors. They can accomplish their task by utilizing their intrinsic HAT activity but also as adapter proteins that link activators to the basal transcription machinery (Shikama et al., 1997; Grant and Berger, 1999). A common feature of many p300/CBP-dependent activators is that their activities are regulated by developmental or extracellular cues (Shikama et al., 1997).
Here we report that β-catenin can synergize with the p300 and CBP transcriptional coactivator proteins to stimulate the activity of particular promoters in vertebrates. In vitro, β-catenin and p300 physically interact through specific regions located at the C-terminus of β-catenin and the CH-3 domain of p300. Although the intrinsic acetylase activity of p300 is not required for its function as coactivator of β-catenin, the synergism between β-catenin and p300 may play a role in alleviating repression of Wnt-target genes induced by Groucho/TLE proteins and chromatin structure.
Results
p300 is a promoter-specific coactivator of β-catenin
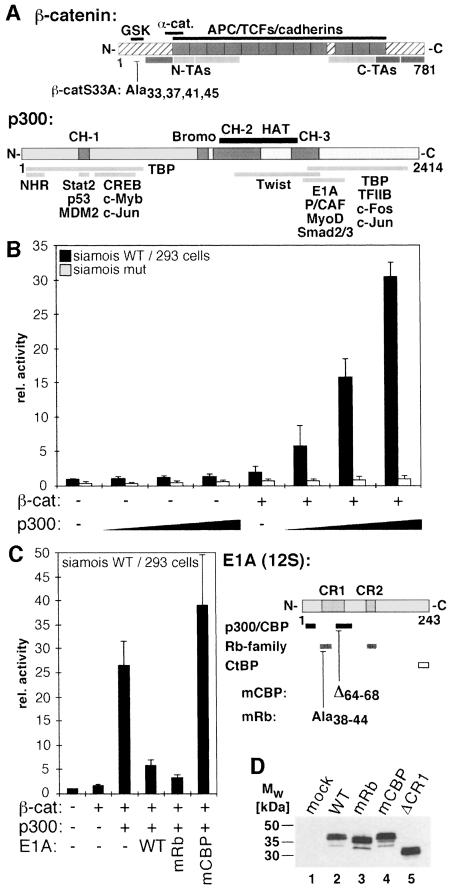
To test whether β-catenin cooperates with p300 we transfected human 293 kidney cells with increasing amounts of a plasmid coding for human p300 in the presence of a mutant form of β-catenin (β-catS33A), to see if this would stimulate the activity of a luciferase reporter gene driven by the siamois promoter. β-catS33A carries four alanine substitutions in the GSK-3β recognition site (Aberle et al., 1997) (Figure 1A). This leads to constitutive activation of β-catenin–TCF target genes (Morin et al., 1997). 293 cells were chosen because they express TCF factors as interaction partners for β-catenin (Porfiri et al., 1997), and their endogenous p300 is largely neutralized due to the presence of the Adenovirus E1A oncoprotein (Eckner et al., 1994). When β-catS33A and p300 were expressed simultaneously, reporter gene activity increased in a dosage-dependent manner up to an additional 15-fold over levels of β-catenin alone (Figure 1B). Expression of p300 had no effect in the absence of β-catenin or on a mutant promoter lacking TCF binding sites (Figure 1B). The synergistic activation of the siamois promoter by p300 and β-catenin could be abrogated by expression of a dominant negative form of the TCF family member LEF-1 lacking the catenin binding site (not shown). These observations demonstrate that p300 requires the β-catenin–TCF complex for its stimulatory effect.
Fig. 1. β-catenin utilizes p300 as coactivator at the siamois promoter. (A) Schematic representation of β-catenin and p300. In β-catenin (hatched bar) the Armadillo repeats (gray boxes) and interaction domains for various binding partners are marked. The GSK-3β target sequence (GSK) and the transactivating elements (N-TAs, C-TAs) are also shown. Autonomous TAs: dark gray; auxiliary TAs: light gray. In β-catS33A serine/threonine residues at positions 33, 37, 41 and 45 in the GSK-3β target sequence were substituted with alanines. In p300 the cysteine/histidine-rich elements CH-1, CH-2, and CH-3, the Bromo domain (gray boxes), the HAT domain (black bar) and regions interacting with nuclear hormone receptors (NHR), other transcription factors and E1A are denoted. (B) 293 cells were transfected with luciferase reporter plasmids harboring either the WT siamois promoter or a deletion mutant lacking the TCF binding sites (1.0 µg), the lacZ expression plasmid pCMVβ (Promega) (0.5 µg), a construct for expression of β-catS33A (0.5 µg) and increasing amounts of expression vector for p300 (0.5, 1.0, 2.0 µg) as indicated. Luciferase activities were determined 40 h post transfection, normalized against β-galactosidase values and expressed as relative activity compared with cells transfected with the WT siamois reporter alone (rel. activity: 1). (C) 293 cells were transfected with the WT siamois reporter, pCMVβ and expression vectors for β-catS33A (0.5 µg), p300 (2.0 µg) and WT E1A or mutant E1A (0.2 µg) as indicated. Reporter activity without β-catenin and p300: 1. A diagram of 12S-E1A is depicted. Conserved regions 1 and 2 (CR1, CR2), domains involved in binding Rb-family members, p300/CBP and the CtBP transcriptional repressor, and the location of the mRb and mCBP mutations are indicated. (B, C) Average values from at least three independent experiments are shown with standard deviations. (D) SW480 cells were transfected with expression vectors (2.5 µg each) for the E1A proteins as indicated. Whole cell lysates were prepared 40 h after transfection and E1A expression levels were analyzed by Western blotting with a monoclonal antibody. Mw: molecular weight markers.
Without cotransfection of p300, β-catS33A only weakly stimulated the siamois promoter, most likely because of the inhibition of endogenous p300 by E1A in 293 cells (Eckner et al., 1994). To analyze further the functional cooperation of β-catenin and p300 we challenged β-catenin–p300-mediated activation of the siamois promoter by overexpression of wild type (WT) or mutant E1A (Figure 1C). E1AmRb contains alanine substitutions at positions 38–44, which selectively disturb interactions with the retinoblastoma protein and E1AmCBP lacks residues 64–68, which compromises the interaction with p300/CBP (Bannister and Kouzarides, 1995). Both WT E1A and E1AmRb efficiently abrogated siamois promoter activation by β-catenin and p300 (Figure 1C). In contrast, E1AmCBP had no inhibitory effect even though its expression levels are above or similar to those of WT E1A or E1AmRb (Figure 1D). The apparent E1A-sensitivity of β-catenin-dependent transactivation and the differential effects of E1AmRb and E1AmCBP further support the assumption that β-catenin can utilize p300 as cofactor for the activation of the siamois promoter.
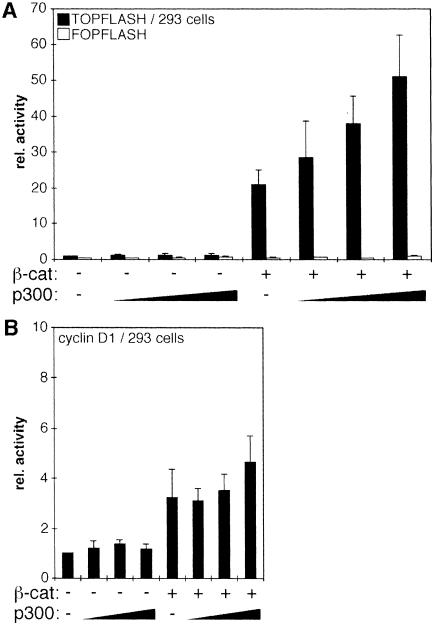
To test whether the p300 requirement of β-catenin was unique to the siamois promoter we performed transfection experiments with the TOPFLASH/FOPFLASH plasmids (Korinek et al., 1997) and another known Wnt target, the cyclin D1 promoter (Shtutman et al., 1999; Tetsu and McCormick, 1999). In TOPFLASH, luciferase expression is driven by four TCF binding motifs in front of the c-fos minimal promoter. In the control plasmid FOPFLASH, the TCF binding motifs are mutated. The cyclin D1 promoter fragment harbors sequences from –973 to +139 relative to the transcriptional start site and includes the β-catenin–TCF response element around position –75 (Shtutman et al., 1999; Tetsu and McCormick, 1999). As in previous studies, expression of mutant β-catenin was sufficient to increase TOPFLASH activity by 15- to 20-fold (Figure 2A) (Morin et al., 1997; Porfiri et al., 1997; Aoki et al., 1999; Vleminckx et al., 1999). Activity of the cyclin D1 luciferase reporter was raised 3- to 4-fold by β-catS33A. Little if any further increase in cyclin D1 promoter activity was conferred by coexpression of p300 (Figure 2B). Activity of the TOPFLASH reporter, however, could be stimulated up to an additional 3-fold in the presence of p300 (Figure 2A). These findings indicate that β-catenin has the capacity to activate transcription both in a p300-dependent and in a p300-independent manner as dictated by a specific promoter context.
Fig. 2. Promoter-specific differences in transcriptional activation mediated by β-catenin and p300. (A) 293 cells were transfected with the TOPFLASH or FOPFLASH reporter plasmids (1.0 µg), pCMVβ (0.5 µg), a construct for expression of β-catS33A (0.5 µg) and increasing amounts of expression vector for p300 (0.5, 1.0, 2.0 µg) as indicated. Luciferase activities were determined as before. (B) Same experiment as in (A) but with a reporter construct harboring the cyclin D1 promoter. Reporter activity in (A) and (B) without β-catenin and p300: 1.
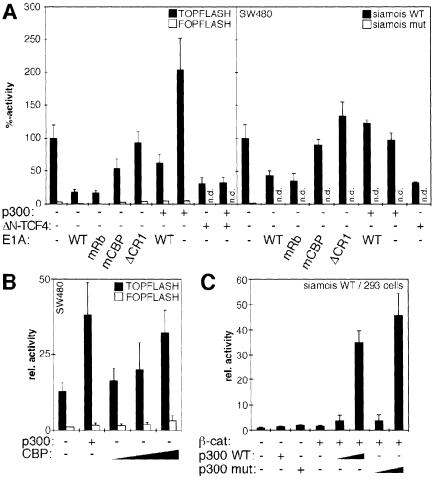
To see whether p300 and β-catenin also cooperated in other cells we performed transfection experiments with the human colon carcinoma cell line SW480. A mutant APC tumor suppressor protein in these cells abolishes the β-catenin control mechanism (Korinek et al., 1997). This leads to constitutive transcriptional activation by endogenous WT β-catenin as revealed by high levels of reporter gene activity in cells transfected with TOPFLASH or the WT siamois promoter construct compared with cells transfected with the corresponding luciferase plasmids without functional β-catenin–TCF response elements (Figure 3A) (Korinek et al., 1997). If transcriptional activation by β-catenin in SW480 involves p300, then TOPFLASH or siamois promoter activity should be sensitive to E1A. Indeed, expression of E1A or E1AmRb causes a 4- to 5-fold reduction in TOPFLASH activity and a 2.5-fold decrease in siamois promoter activity (Figure 3A). To evaluate the efficiency of inhibition we also analyzed a dominant negative form of TCF4 (ΔN-TCF4), which is unable to bind β-catenin. ΔN-TCF4 diminished promoter activity to an extent similar to E1A or E1AmRb (Figure 3A). E1AmCBP with a small deletion in the p300/CBP binding site had no effect on the siamois promoter but resulted in a 2-fold decrease in TOPFLASH activity. E1AΔCR with a complete deletion of conserved region 1 behaved neutrally at both promoters although it is expressed at levels similar to the other E1A mutants (Figure 1D). Inhibition of TOPFLASH or siamois promoter activity by E1A could be rescued by coexpression of p300 (Figure 3A). In contrast, promoter inhibition by ΔN-TCF4 was unaffected by excess p300. With the TOPFLASH construct, expression of p300 actually increased reporter gene activity. Thus, unlike the situation in 293 cells, the p300 requirement of β-catenin for activation of a reporter gene construct in SW480 cells is more pronounced with TOPFLASH than with the siamois promoter. This suggests that there are not only promoter-specific but also cell-type-specific aspects to the functional cooperation between β-catenin and p300. Nonetheless, our results so far show that β-catenin can utilize p300 as a promoter-specific transcriptional coactivator in different cell lines.
Fig. 3. (A) Endogenous β-catenin in SW480 cells utilizes p300 as coactivator. SW480 cells were transfected with pCMVβ (0.5 µg), the TOPFLASH/FOPFLASH (1.5 µg) or siamois promoter constructs (2.5 µg) and expression vectors for p300 (2.0 µg) or dominant negative ΔN-TCF4 (2.0 µg) as indicated. To interfere with p300-mediated transcriptional activation WT E1A, E1AmRb, E1AmCBP or E1AΔCR1 were cotransfected (0.2 µg each). Luciferase activities were determined as before. Activity of the TOPFLASH or WT siamois reporter without exogenous p300, ΔN-TCF4 or E1A was set as 100% (n.d.: not done). (B) Functional similarity of p300 and CBP. SW480 cells were transfected with pCMVβ (0.5 µg), the TOPFLASH/FOPFLASH reporter constructs (1.5 µg) and expression vectors for p300 (4.0 µg) or CBP (1.0, 2.0 and 4.0 µg) as indicated. Luciferase activities, determined as before, are expressed as relative activities compared with cells transfected with FOPFLASH alone. (C) HAT activity of p300 is not required for its function as coactivator of β-catenin. 293 cells were transfected with the siamois reporter (0.5 µg), pCMVβ (0.1 µg), and expression vectors for β-catS33A (0.5 µg), WT p300, or HAT-defective p300mut (0.5 and 2.5 µg) as indicated. Control transfectants received 2.5 µg of empty expression vector or p300 plasmids. Luciferase activities were determined as before. Reporter activity without β-catenin and p300: 1.
Functional similarity between p300 and CBP
CBP and p300 are structurally and functionally highly related (Shikama et al., 1997), but differ in certain aspects (Kawasaki et al., 1998), and dCBP negatively regulates Wnt signaling in Drosophila (Waltzer and Bienz, 1998). To determine whether also in mammalian cells CBP was functionally different from p300 and could repress β-catenin-mediated transcriptional activation, we transfected SW480 cells with the TOPFLASH and FOPFLASH reporter constructs and increasing amounts of an expression vector for mouse CBP. If CBP in contrast to p300 played an inhibitory role, we would have expected to see a decrease in TOPFLASH activity. Instead, CBP caused further stimulation of the reporter gene, although it was somewhat less effective than p300 (Figure 3B). The stimulatory effect of CBP as well as the small quantitative difference between p300 and CBP can also be observed in 293 cells (not shown). Apparently, both p300 and CBP can serve as coactivators of β-catenin in human cells.
Intrinsic HAT activity of p300 is not required for stimulation of β-catenin transactivation
To potentiate β-catenin transactivation, p300 may link β-catenin to the basal transcription machinery or alter chromatin structure by utilizing its intrinsic or associated acetylase activities. As in other cases (Munshi et al., 1998; Chen et al., 1999b), p300 could function both during the activation and the downregulation of inducible genes by selectively acetylating different target proteins. To distinguish between these possibilities we analyzed how a DI to AL exchange of p300 residues 1485 and 1486, which abolishes HAT activity (Martinez-Balbas et al., 1998; data not shown), would affect the synergistic activation of the siamois promoter by p300 and β-catenin. WT and mutant p300 were expressed at similar levels (not shown) and were both capable of cooperating with β-catenin after transfection into 293 cells (Figure 3C). Eliminating intrinsic p300 HAT activity led to slightly higher promoter activation. However, this effect was not observed upon transfection of WT and HAT-defective p300 into SW480 cells (not shown). Thus, the absence of p300 HAT activity does not appear to influence β-catenin–TCF transactivation in mammalian cells, and acetylation of histone or non-histone proteins by p300 itself is not required for its stimulatory effect on β-catenin.
Interactions between β-catenin and p300 in vitro
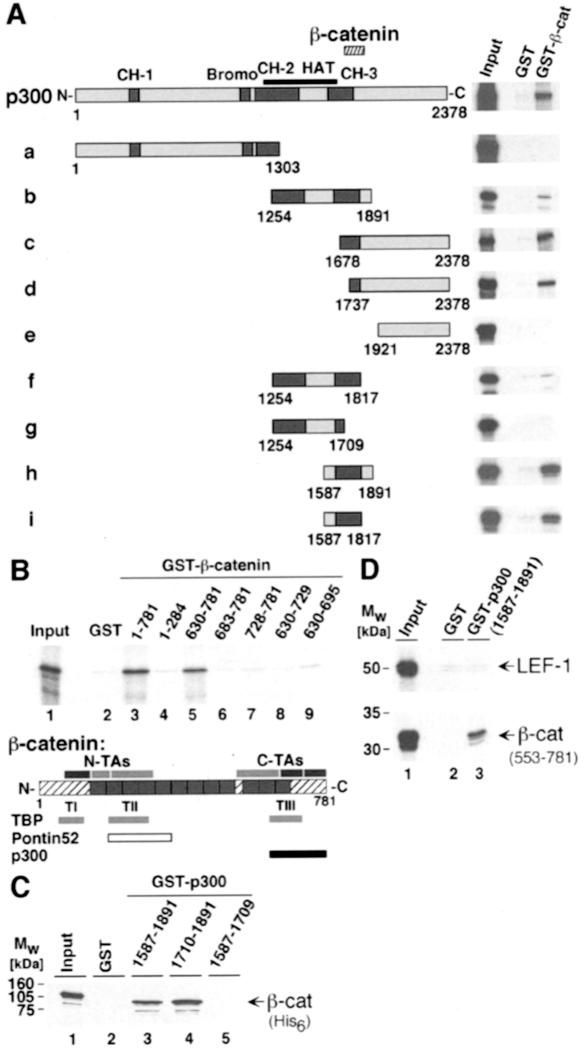
Physical interactions between β-catenin and p300 were analyzed in vitro by pull-down experiments with glutathione S-transferase (GST) fusion proteins. Glutathione–Sepharose beads were coated with recombinant GST or a GST fusion protein with full-length β-catenin and used as affinity matrices for radiolabeled p300 fragments that had been expressed in a reticulocyte lysate system in vitro. The p300 fragment spanning residues 1–2378, which had also been used for the functional assays, specifically bound to the GST–β-catenin fusion and not to the GST control (Figure 4A). Mapping experiments with various deletion mutants show that p300 fragments are capable of interacting with β-catenin as long as they contain at least residues 1737–1817 (Figure 4A, fragments b, c, d, f, h, i). This β-catenin interaction domain partially overlaps with the CH-3 domain of p300.
Fig. 4. Interactions between p300 and β-catenin in vitro. (A) Mapping of the β-catenin interaction domain in p300. GST or GST–β-catenin fusion proteins were bound to GSH–Sepharose beads and incubated with [35S]methionine-labeled p300 fragments. After extensive washing, proteins retained by GST or GST–β-catenin were analyzed together with 10% of the input material by SDS–PAGE and fluorography. The structure of p300 and its derivatives is shown schematically. End points of the p300 fragments and the location of the β-catenin interaction domain (hatched bar) are given. (B) Mapping of the p300 interaction domain in β-catenin. Bacterially expressed GST fusion proteins with β-catenin sequences as indicated were bound to GSH–Sepharose and incubated with a radiolabeled p300 fragment (residues 1737–2378). Material bound to the various GST fusions was analyzed by SDS–PAGE and fluorography. Lane 1: a sample corresponding to 10% of the p300 input material. A diagram of β-catenin giving the location of its transactivating elements as well as the interaction sites TI–TIII for TBP (light gray bar), Pontin52 (open bar) and p300 (black bar) is also shown. (C) Direct interactions between β-catenin and p300. GST or GST fusion proteins with p300 sequences as indicated were bound to GSH–Sepharose and incubated with histidine-tagged β-catenin (β-catHis6) purified from E.coli. Material bound to the various GST–p300 fusions was analyzed by SDS–PAGE and Western blotting. (D) The C-terminus of β-catenin but not LEF-1 interacts with the CH-3 domain of p300 in vitro. Bacterially expressed GST or GST fusion proteins with p300 sequences as indicated were bound to GSH–Sepharose and incubated with radiolabeled LEF-1 or a β-catenin fragment comprising residues 553–781. Material retained on the GST or GST–p300-coated matrices was eluted and analyzed by SDS–PAGE and fluorography. Lane 1, Input: a sample corresponding to 10% of the LEF-1 or β-catenin material used for the assay. Mw: molecular weight markers.
To determine which part of β-catenin interacts with p300 we performed a set of pull downs with GST fusion proteins harboring various regions of β-catenin. The p300 fragment used here consisted of residues 1737–2378 (fragment d in Figure 4A). Only the GST fusions with full-length β-catenin or with residues 630–781 were able to bind the p300 fragment (Figure 4B, lanes 3 and 5). No interactions were detectable with N-terminal residues 1–284 of β-catenin or C-terminal portions shorter than residues 630–781. Thus, the sequence requirements for the binding of p300 to β-catenin differ from those of TBP or Pontin52 (Bauer et al., 1998; Hecht et al., 1999), suggesting that the p300–β-catenin interaction occurs independently of TBP or Pontin52. Yet, p300 interacts with a domain of β-catenin that contains several transactivating elements, and the integrity of which is crucial for the function of β-catenin as signaling molecule (van de Wetering et al., 1997; Hsu et al., 1998; Vleminckx et al., 1999).
The finding that the CH-3 domain of dCBP could bind to murine LEF-1 (Waltzer and Bienz, 1998) raised the possibility that the interaction between p300 and β-catenin might be mediated indirectly through LEF-1 or other TCF factors present in the reticulocyte lysate used for the previous pull-down experiments. Although the location of the p300 binding region in β-catenin argues against this, we analyzed this issue further. As shown in Figure 4C, GST fusions with residues 1587–1891 or only 1710–1891 of p300 bound recombinant histidine-tagged β-catenin-His6 (Figure 4C, lanes 3 and 4). No interaction was seen with GST or a control GST fusion with residues 1587–1709 of p300 (Figure 4C, lanes 2 and 5). We also found that the GST fusion containing the CH-3 domain of p300 did not bind murine LEF-1, whereas under the same conditions the C-terminal residues 553–781 of β-catenin did (Figure 4D). These results confirm our conclusions from the mapping experiments shown in Figure 4A and indicate that β-catenin and p300 can directly interact in vitro.
Interactions between β-catenin and p300 in vivo
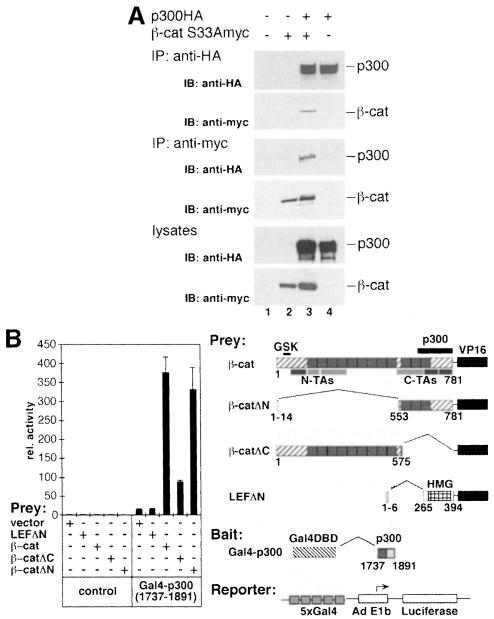
The functional synergism of β-catenin and p300 and their ability to interact in vitro imply that they also interact in vivo. Evidence for this is provided by co-immunoprecipitation experiments (Figure 5A). HA-tagged p300 and myc-tagged β-catS33A were expressed either individually or in combination in 293 cells. Upon immunoprecipitation of p300 from cellular lysates of transfected cells, β-catS33Amyc could be detected by Western blotting in the immunoprecipitate and vice versa (Figure 5A, lane 3). This argues that multiprotein complexes that contain both p300 and β-catenin exist in vivo.
Fig. 5. Interactions between p300 and β-catenin in vivo. (A) Co-immunoprecipitation of p300 and β-catenin. 293 cells were transfected with expression plasmids for HA-tagged p300 and myc-tagged β-catS33A as indicated. Cell extracts were prepared 40 h after transfection and used for immunoprecipitations with anti-HA or anti-myc antibodies. Immunoprecipitated material and a fraction of each lysate was resolved by SDS–PAGE and analyzed by Western blotting with antibodies as shown. IP: immunoprecipitation; IB: immunoblot. (B) Mammalian two-hybrid analyses of interactions between β-catenin and p300. NIH 3T3 cells were transfected with pCMVβ (0.05 µg), the Gal4-dependent pG5E1bLuc reporter construct (0.1 µg) and empty control vectors or expression vectors for Gal4–p300 (1737–1891) (0.5 µg), LEFΔN–VP16 (0.5 µg) and β-catenin–VP16 fusions (0.5 µg) as indicated. Luciferase activities, determined as before, are expressed as relative activity compared with cells transfected with the pG5E1bLuc reporter and empty expression vector (rel. activity: 1). The diagram on the right shows the structure of the β-catenin–VP16 and the LEFΔN–VP16 prey fusions, the Gal4–p300 (1737–1891) bait and the pG5E1bLuc reporter. In β-catenin, the GSK recognition sequence, N-terminal and C-terminal transactivating elements (N-TAs, C-TAs) and the p300 interaction domain (black bar) are marked. LEFΔN–VP16 contains the HMG-box (hatched box) but lacks the β-catenin binding site. The Gal4–p300 fusion harbors the C-terminal portion of the CH-3 domain (dark gray box). VP16: VP16 transactivation domain.
To confirm this independently, we analyzed the interaction of p300 and β-catenin in a mammalian two-hybrid system in NIH 3T3 fibroblasts, which express only low levels of endogenous β-catenin. A p300 fragment coding for residues 1737–1891 was fused to the DNA-binding domain (DBD) of the Gal4 protein and served as bait (Figure 5B). Prey constructs contained different regions of β-catS33A fused to the VP16 transactivation domain (Figure 5B). Western blot experiments were used to verify that the β-catenin fusion proteins were all expressed at similar levels (not shown). In the absence of the Gal4–p300 fusion none of the prey proteins activated the pG5E1bLuc reporter, which contains five Gal4-binding sites to drive luciferase expression from the Adenovirus E1b promoter (Figure 5B). Although the Gal4–p300 bait activated the reporter construct to some extent by itself, the full-length β-catenin–VP16 fusion enhanced reporter gene expression an additional 27-fold when combined with Gal4–p300 (1737–1891). Thus, the p300 domain that interacted with β-catenin in vitro is also capable of mediating an interaction with β-catenin in vivo.
Regions in β-catenin involved in this interaction with p300 were delineated by two-hybrid studies with β-catenin–VP16 fusions lacking either N-terminal residues 15–552 (β-catΔN) or C-terminal residues 576–781 (β-catΔC). This two-hybrid system relies on the activity of the VP16 domain as indicator of a bait–prey interaction and is largely independent of transactivating elements in β-catenin since even full-length β-catenin constructs lacking the VP16 transactivation domain hardly activated the reporter gene (not shown). As expected, the β-catΔN fusion, which contains the p300 in vitro interaction site, stimulated reporter gene expression in the presence of the Gal4–p300 fusion (Figure 5B). This indicates that the C-terminus of β-catenin is sufficient to mediate an interaction with p300 in vivo. Interestingly, we observed a 5-fold activation of the reporter gene when the β-catΔC and the Gal4–p300 fusions were cotransfected. Thus, N-terminal portions of β-catenin appear also to mediate interactions with p300. It is unlikely that these contacts are mediated by LEF-1 because LEFΔN–VP16 did not interact with the Gal4–p300 fusion, as indicated by the lack of reporter stimulation (Figure 5B). Although expression levels of the LEFΔN–VP16 fusion are lower than those of the β-catenin–VP16 fusions (not shown), these findings are consistent with the results of our GST-binding experiments (Figure 4D) and indicate that neither LEF-1, nor probably any other TCF, is required for the interaction between β-catenin and p300. More likely, some of its N-terminal transactivating elements can indirectly link β-catenin to p300 in vivo, because compared with the C-terminus, the N-terminus of β-catenin appears to interact less efficiently with p300 in the two-hybrid assay, and we did not detect a direct interaction between p300 and the N-terminus of β-catenin in vitro.
Involvement of p300/CBP in Xenopus embryonic development
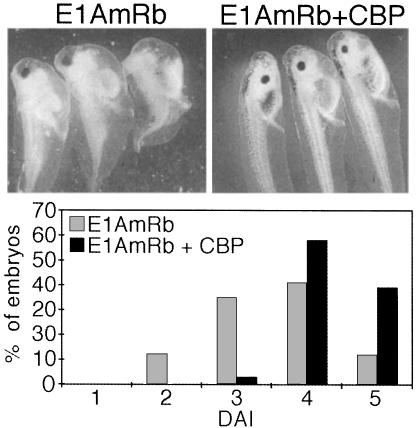
Body axis formation in Xenopus requires activation of siamois and other target genes by β-catenin on the future dorsal side of the embryo (Sokol, 1999). Expression of β-catenin or its activation by Wnt in the ventral marginal zone of developing embryos mimics the endogenous signaling activity and leads to body axis duplication. A cDNA coding for Xenopus p300/CBP has been isolated and the corresponding transcripts are ubiquitously present throughout early embryogenesis (Fujii et al., 1998). Thus, p300/CBP may be involved in early developmental events including the transcriptional activation of natural target genes by β-catenin. To test this we exploited the ability of E1A to interfere with the coactivator function of p300/CBP. In contrast to the previous experiments we now used E1AmRb and E1AmCBP lacking C-terminal residues 178–243, which contain the CtBP binding site and part of the HAT inhibition domain (Brannon et al., 1999; Chakravarti et al., 1999). The inhibitory potential of the E1A mutants towards β-catenin remains largely unaffected by this deletion (not shown) but unwanted effects on developmental processes that in contrast to β-catenin require the HAT activity of p300/CBP or which involve CtBP are minimized. First we analyzed whether E1AmRb or E1AmCBP would affect the ability of XWnt-8 or β-catS33Amyc to induce an ectopic body axis upon ventral expression. Whereas a high proportion of the embryos injected with XWnt-8 or β-catenin RNA alone developed a secondary body axis, only incomplete axis duplication was seen in 21% of the embryos when β-catenin and E1AmRb were coexpressed (Table I). Axis induction by XWnt-8 was also impaired by E1AmRb but higher amounts were required, possibly because Wnts are secreted factors, whereas E1A and β-catenin act intracellularly. As in SW480 cells, also here E1AmCBP had some inhibitory effect, but it was much less efficient than E1AmRb (Table I). Importantly, the impairment of XWnt-8 and β-catenin activity by E1AmRb could be fully reversed by coinjection of CBP RNA (Table I). Injection of CBP RNA alone did not result in axis duplication, and ectopic expression of the E1A mutants or CBP had no effect on the level of β-catS33Amyc protein (not shown). Embryos injected with E1AmRb RNA in the dorsal vegetal zone where endogenous β-catenin signaling activity is first induced during embryonic development, also displayed defects in axial development and showed signs of ventralization as seen after inhibition of embryonic signaling activities, for example by depletion of β-catenin (Sokol, 1999). Again, the inhibitory effect of E1AmRb could be reversed by coexpression of CBP (Figure 6). In conclusion, the differential inhibition of XWnt-8 and β-catS33Amyc by E1AmRb and E1AmCBP and the ability of CBP to rescue E1A-mediated developmental defects indicate that embryonic p300 or CBP are involved both in axis duplication in response to ectopic activity of β-catenin and during formation of the primary body axis.
Table I. Expression of CBP restores the axis inducing activity of β-catenin and Wnt after inhibition by E1A.
| RNA injected (amount) | No. of embryos injected | Total % axis duplication | % Complete axis duplicationa | ||
|---|---|---|---|---|---|
| β-catS33A (0.5 ng) | +β-galNLS | (0.5 ng) | 117 | 81 | 33 |
| +E1AmCBP | (0.5 ng) | 129 | 64 | 14 | |
| +E1AmRb | (0.5 ng) | 126 | 21 | 0 | |
| +E1AmRb | (0.5 ng) | ||||
| +CBP | (2.0 ng) | 106 | 84 | 21 | |
| Wnt-8 (40 pg) | +β-galNLS | (0.5 ng) | 81 | 78 | 42 |
| (2.0 ng) | 86 | 85 | n.a. | ||
| +E1AmCBP | (0.5 ng) | 73 | 78 | 50 | |
| (2.0 ng) | 80 | 45 | n.a. | ||
| +E1AmRb | (0.5 ng) | 69 | 41 | 0 | |
| (2.0 ng) | 91 | 29 | n.a. | ||
| +E1AmRb | (0.5 ng) | ||||
| +CBP | (2.0 ng) | 66 | 82 | 54 | |
| +E1AmRb | (2.0 ng) | ||||
| +CBP | (2.0 ng) | 80 | 61 | n.a. | |
| CBP (2.0 ng) | 51 | 0 |
aDefined as the presence of extra eyes and cement glands.
n.a., not analyzed.
Fig. 6. Formation of the endogenous body axis is sensitive to E1AmRb and can be rescued by coexpression of CBP. Embryos were injected at the four-cell stage in the vegetal zone of the two dorsal blastomeres with RNA for E1AmRb (0.5 ng) alone or together with 2.0 ng of CBP RNA. Ventralization of embryos was scored at the tadpole stage and is expressed as dorso-anterior index (DAI; Kao and Elinson, 1988). Normal embryos have a DAI of 5.
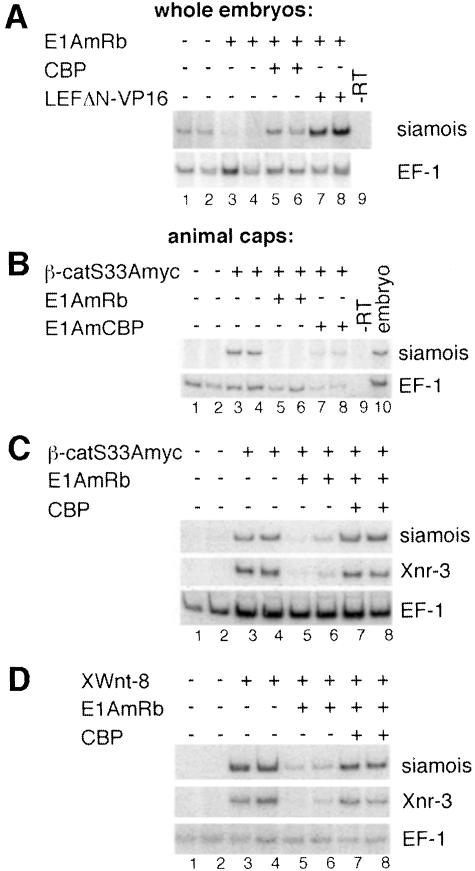
Many developmental regulators synergize with p300/CBP and E1A has pleiotropic effects (Shikama et al., 1997; Kato et al., 1999). Therefore, to establish a molecular link between the requirement for p300/CBP in early embryonic development and transcriptional activation by β-catenin we analyzed how E1A would affect the expression of the siamois gene, which is a direct target of β-catenin. RT–PCR analyses with RNA from whole embryos reveal greatly reduced levels of siamois expression upon injection of E1AmRb RNA in the dorsal vegetal zone (Figure 7A). siamois transcription is rescued by coexpression of CBP and by the chimeric transcriptional activator LEFΔN–VP16. LEFΔN–VP16 is directed to β-catenin–TCF target genes by the LEF-1 HMG box and can functionally replace the β-catenin–TCF complex but it is not or is less E1A sensitive (Vleminckx et al., 1999; not shown). However, unlike CBP, LEFΔN–VP16 does not reverse the morphological defects caused by E1A (not shown). These observations confirm that E1A not only impairs β-catenin function but other signaling processes as well. More importantly, however, they demonstrate that E1A does not generally block siamois inducibility, which points to a defect in transcriptional activation by β-catenin. This is confirmed by results from RT–PCR assays with RNA isolated from animal caps, which allows assessment of a direct effect of E1A on the function of β-catenin. Consistent with the previous results, coexpression of E1AmRb strongly reduced the levels of siamois transcripts induced by β-catenin, whereas E1AmCBP had a weaker effect (Figure 7B). β-catenin-induced transcription of the Xnr-3 gene is also sensitive to E1AmRb (Figure 7C), and, like siamois transcription, can be rescued by coexpression of CBP. Activation of siamois and Xnr-3 in response to XWnt-8 is also E1A-sensitive (Figure 7D). These results strongly suggest that both exogenous and endogenous β-catenin cooperate with murine CBP or the embryonic p300/CBP to activate siamois and other target genes during body axis formation in Xenopus embryonic development.
Fig. 7. Transcriptional activation of β-catenin target genes is sensitive to E1A and can be rescued by CBP. (A) RT–PCR analyses of siamois expression in whole embryos after injection of RNAs for E1AmRb (0.5 ng), LEFΔN–VP16 (0.1 ng) and CBP (2.0 ng) into the dorsal vegetal zone of four-cell stage embryos as indicated. Control reactions were performed with an RNA sample not treated with reverse transcriptase (–RT). (B) RT–PCR analyses of ectopic siamois induction in animal cap explants by injection of 0.5 ng of β-catS33Amyc RNA with or without 2.0 ng of E1AmCBP or E1AmRb RNA. Control reactions were performed with RNA from total embryos or RNA samples that were not treated with reverse transcriptase (–RT). (C and D) Expression of CBP restores β-catenin target gene induction in the presence of E1AmRb. RT–PCR analyses of siamois and Xnr-3 expression induced in animal cap explants by injection of 0.5 ng of β-catS33Amyc RNA (C) or 40 pg of XWnt-8 RNA (D) with or without 0.5 ng of E1AmRb RNA and 2.0 ng of CBP RNA. In (D), the various RNAs were applied by two consecutive injections to ensure an overlapping spatial protein distribution of E1A or CBP and the secreted XWnt-8. (A–D) For each RNA combination two embryos or two pools of animal cap explants were analyzed separately. EF-1 expression was monitored as control for RNA amount and integrity.
Discussion
Previous studies have shown that β-catenin is an essential nuclear effector of Wnt signals; however, how β-catenin accomplishes target gene activation is largely unclear. By interacting with TCF factors β-catenin may alter promoter architecture and displace Groucho/TLE or CtBP corepressors (Cavallo et al., 1998; Roose et al., 1998; Brannon et al., 1999; Eastman and Grosschedl, 1999; Miller et al., 1999). In addition, there is a strict correlation between the ability of β-catenin to function in Wnt signaling and its ability to transactivate (van de Wetering et al., 1997; Vleminckx et al., 1999) suggesting that β-catenin facilitates further steps during promoter activation. In support of this, β-catenin possesses multiple transactivating elements at its N- and C-termini, which can operate independently from TCFs, and β-catenin can bind to TBP and to RuvBL1/Pontin52, which also interacts with TBP and RNA polymerase II (van de Wetering et al., 1997; Bauer et al., 1998; Hsu et al., 1998; Qiu et al., 1998; Hecht et al., 1999). However, these interactions are not sufficient to explain entirely how β-catenin functions as a transcriptional activator (Hecht et al., 1999; Prieve and Waterman, 1999). Here we have identified p300 as an important coactivator of β-catenin. (i) β-catenin and p300 interact with each other in vitro and in vivo. (ii) Coexpression of p300 stimulates β-catenin-dependent activation of specific promoters in mammalian cells. (iii) The E1A oncoprotein, which is a known inhibitor of p300/CBP, diminishes activation of β-catenin-regulated promoters in transfected mammalian cells and strongly impairs body axis formation and the induction of endogenous β-catenin target genes in Xenopus embryos. E1A-mediated inhibition of β-catenin activity could be reversed by coexpression of p300 or CBP. These observations provide good evidence that β-catenin utilizes p300/CBP as coactivator to perform its function in Wnt signaling.
Other reports also show that inhibition of p300/CBP by dominant negative derivatives or by E1A results in gross alterations of Xenopus embryonic development (Fujii et al., 1998; Kato et al., 1999). Interestingly, while Kato and coworkers, too, reported a lack of axial development, they ascribed this to a defect in mesoderm formation and explicitly stated that Wnt signaling was not affected by E1A (Kato et al., 1999). We find that the contrasting results of our study are due to differences in the amounts of RNA injected and our use of C-terminally deleted E1A with a limited range of inhibitory effects. At the low levels of E1A used by Kato and coworkers we also do not see inhibition of β-catenin activity (not shown). Furthermore, a comparison of full-length and truncated E1A revealed that full-length E1A produces much more complex phenotypes than our shorter versions (not shown). Still, morphological changes in axial development in the presence of E1A cannot be interpreted as exclusive inhibition of β-catenin activity. However, the animal cap assay provides an experimental system where target gene activation solely depends on the activity of β-catenin. The fact that E1A prevents the activation of siamois and Xnr-3 under these conditions, and that CBP expression rescues this defect, strongly supports our idea that β-catenin synergizes with p300/CBP to activate target genes also in their native environment.
A direct interaction between p300 and β-catenin occurs with a C-terminal fragment of β-catenin comprised of residues 630–781. This binding site could not be narrowed down further, indicating that the interaction requires multiple, dispersed contacts. The C-terminus of β-catenin also harbors a previously described TBP-binding site mapping at residues 630–729. In functional tests, however, several short fragments of the β-catenin C-terminus that do not appear to interact with TBP or p300 behave as potent transactivating elements (van de Wetering et al., 1997; Hsu et al., 1998; Hecht et al., 1999). These transactivators also function in yeast cells (Hecht et al., 1999), which possess neither p300 nor CBP. Additional, as yet unidentified, coactivators must act via these regions of β-catenin to enable p300-dependent and -independent transcriptional stimulation of different promoters. The C-terminal part of β-catenin therefore reveals itself as a compact assembly that contains multiple transactivating elements and which provides a multitude of contact sites for transcriptional coactivators including p300/CBP. The ability to interact with and recruit different coactivators may be a necessity to enable β-catenin to function in distinct promoter settings and to facilitate discrete steps during the transcription initiation process.
Multiple functions of p300/CBP in Wnt signaling?
The finding that p300/CBP serves as a coactivator for β-catenin in vertebrates is unexpected given that in Drosophila dCBP has been shown to regulate Wingless-signaling negatively (Waltzer and Bienz, 1998). The apparent discrepancy between the function of vertebrate p300/CBP and dCBP could be explained most easily if vertebrate CBP and p300, or dCBP and its mammalian orthologs were functionally different from each other. Species-specific differences are known for certain aspects of Wnt signaling (Cadigan and Nusse, 1997). Also, CBP and p300 are differentially engaged in retinoic acid and cAMP responses in mammalian cells (Kawasaki et al., 1998). However, our results show that CBP and p300 can both serve as cofactors for β-catenin. On the other hand, we could not detect an interaction between LEF-1 and the CH-3 domain of p300, which corresponds to the dCBP-2 region used by Waltzer and Bienz (1998). Rather, we have demonstrated that β-catenin can interact with p300 in the absence of TCFs. These findings indicate that dCBP differs from p300 and CBP and that in vertebrates p300 or CBP enter the β-catenin–TCF complex through an interaction with β-catenin, whereas dCBP may be brought to a promoter by its interaction with dTCF.
Aside from the difference in the TCF interaction, the genetic studies in Drosophila and our experiments in vertebrates may also reveal a more complicated involvement of p300/CBP in Wnt signaling. For example, upon binding to dTCF, Armadillo may form a ternary complex and reprogram the activity of dCBP that is already present. Alternatively, p300 or CBP may function as coactivators of β-catenin only initially. Over time their activity could change and lead to the downregulation of target genes as reported for the interferon-β enhanceosome, which eventually is destabilized and disassembled by CBP through acetylation of an architectural component, the HMG I(Y) protein (Munshi et al., 1998). Similarly, coactivator complexes associated with the promoter-bound estrogen receptor are dissociated after acetylation by p300/CBP, which leads to the attenuation of the hormone response (Chen et al., 1999b). Thus, one could hypothesize that p300 performs a stimulatory function and also provides a shut-off mechanism in Wnt signaling.
Possible mechanisms for p300/CBP function in the regulation of Wnt target genes
Interactions between p300/CBP and TBP, TFIIB, and RNA helicase A allow for multiple contacts between p300/CBP, general transcription factors and RNA polymerase II (Kwok et al., 1994; Yuan et al., 1996; Nakajima et al., 1997; Felzien et al., 1999). The association of β-catenin and p300/CBP therefore may facilitate the recruitment of the transcription machinery to target gene promoters, and p300/CBP may be a bridging molecule in these processes. However, the involvement of Groucho/TLE repressor proteins in Wnt signaling makes it attractive to explore the potential involvement of acetyltransferases in promoter activation by β-catenin. Groucho/TLE proteins are tightly bound to chromatin and interact with histones in vitro (Palaparti et al., 1997). TUP1, a yeast relative of Groucho/TLE factors, binds specifically to hypoacetylated histones (Edmondson et al., 1996) and a functional interaction between Groucho and the histone deacetylase Rpd3 has been reported (Chen et al., 1999a). Thus, to overcome repression of Wnt target genes, β-catenin may engage a histone acetyltransferase to disrupt Groucho/TLE–chromatin complexes.
CBP and p300 possess HAT activity and can be involved in chromatin modification. In addition, multiple other HATs are associated with p300, including P/CAF, SRC-1 and ACTR (Shikama et al., 1997; Grant and Berger, 1999). Retinoic acid receptors, CREB, STAT-1 and MyoD simultaneously interact with several of these acetylases and selectively utilize their enzymatic activities for promoter stimulation (Puri et al., 1997; Korzus et al., 1998). Thus, in some cases p300 may simply provide a scaffold for the assembly of larger coactivator complexes. E1A can target such complexes in a 2-fold manner. By competitively binding to the CH-3 domain of p300/CBP it may release some of the interacting components and dissociate p300/CBP from activator proteins (Puri et al., 1997; Shikama et al., 1997). In addition, E1A inhibits the enzymatic activities of p300/CBP and P/CAF by binding to their HAT domains (Chakravarti et al., 1999; Hamamori et al., 1999). Because HAT activity of p300 is not required for its coactivator function of β-catenin and the binding domains for E1A and β-catenin in p300 appear to overlap (Eckner et al., 1994) we believe that E1A affects β-catenin activity by simultaneously displacing p300/CBP and associated cofactors from its C-terminus. Thus, even though intrinsic HAT activity of p300 is dispensable for stimulating β-catenin-mediated transactivation, it is still possible that the interaction with p300 is part of a mechanism by which β-catenin alters chromatin structure at target gene promoters. Future experiments will tell whether additional acetylases interact with β-catenin through p300/CBP and whether their activities are needed to overcome promoter repression by Groucho/TLE and chromatin structure.
Materials and methods
Plasmids
All pro- and eukaryotic expression vectors for β-catenin, TCF4, LEF-1, LEFΔN–VP16, p300, CBP, E1A and the luciferase reporter plasmids have been described (Aberle et al., 1994, 1997; Eckner et al., 1994; Kwok et al., 1994; Bannister and Kouzarides, 1995; Huber et al., 1996; Brannon et al., 1997; Korinek et al., 1997; Aoki et al., 1999; Hecht et al., 1999; Vleminckx et al., 1999), or were derived from published constructs by standard cloning procedures using pGEX4T1 (Amersham Pharmacia), pCS2+ (Turner and Weintraub, 1994), pCS2+LEFΔN-VP16 and pCMVGal4 (Hecht et al., 1999) as recipient vectors. Details of the constructions are available upon request or in the Supplementary data, available in The EMBO Journal Online.
Cell culture, transient transfections and reporter gene assays
Mouse NIH 3T3 cells (ATCC # CRL-1658), human SW480 (ATCC # CCL-228) and 293 cells (ATCC # CRL-1573) were cultured as described (Hecht et al., 1999). For transient transfection 5 × 105 293 or SW480 cells were seeded into 35-mm tissue culture dishes. Four hours after plating, cells received 200 µl of a calcium phosphate–DNA coprecipitate. Total DNA amounts were kept constant by adding empty expression vector as appropriate. NIH 3T3 cells (2 × 105 per 35 mm well) were transfected 2 h after plating with FuGENE6 (Roche Molecular Biochemicals). Luciferase and β-galactosidase activities in cell lysates were determined as before (Hecht et al., 1999).
Immunoprecipitation and Western blotting
For immunoprecipitations 2 × 106 293 cells, seeded into 10-cm dishes, were transfected with 10 µg of expression vector for p300 and 2.5 µg of expression plasmid for β-catS33Amyc. Cells were harvested 40 h later and lysed in 800 µl of 50 mM Tris–HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P-40, 1 mM dithiothreitol, 0.1 mM sodium orthovanadate, 10 mM NaF, 1 mM PMSF, 10 µg/ml leupeptin and 10 µg/ml aprotinin on ice for 30 min. After clearance one half of each lysate was used for immunoprecipitation with 1.0 µg of anti-HA antibody (3F10, Roche Molecular Biochemicals) or anti-myc antibody (9E10) for 3 h at 4°C in the presence of a 12.5 µl bed volume of protein G or protein A–Sepharose (Amersham Pharmacia) preincubated in cell lysates from non-transfected cells supplemented with 1% bovine serum albumin. Immunoprecipitates were washed once with 1 ml of lysis buffer and twice with 1 ml of 20 mM Tris–HCl pH 7.6, 100 mM NaCl, 1 mM EDTA and 0.1% Nonidet P-40 for 10 min at 4°C. Material bound to the beads was eluted in SDS-loading buffer, resolved by SDS–PAGE on 7.5% gels, transferred onto nitrocellulose and analyzed by Western blotting using the ECL detection system (Amersham Pharmacia). Expression of E1A proteins in lysates from transfected cells was analyzed with a mouse monoclonal antibody (DP11; Calbiochem).
GST pull-down assays
GST pull-down assays were carried out as described (Hecht et al., 1999) using ∼2 µg of GST fusion proteins and binding buffer with 1% bovine serum albumin. Binding reactions were supplemented with 1–5 µl (depending on translation efficiency) of SP6 TNT transcription and translation mix (Promega) containing [35S]methionine-labeled p300, β-catenin or LEF-1 derivatives, and binding proceeded for 1 h at 4°C. After extensive washing, proteins retained on the GSH–Sepharose matrix were eluted in SDS–PAGE loading buffer, separated by SDS–PAGE on 8 or 12% gels and visualized by fluorography. In GST pull-downs with recombinant β-catenin, ∼2 µg of β-catenin-His6 (Aberle et al., 1994) were used per reaction. Detection was by Western blotting with a monoclonal anti-β-catenin antibody (Transduction Laboratories).
RT–PCR assay
Total RNA from one embryo or three animal caps was purified with RNAzol B (WAK-Chemie Medical GMBH, Germany) and cDNA was synthesized with oligo(dT) primers (Roche Molecular Biochemicals) as in Vleminckx et al. (1999). One-fifth of the cDNA samples was used for PCR amplification with Taqman Gold polymerase (Perkin Elmer) in the presence of [32P]dCTP (40 nCi/µl) and primers for siamois, Xnr-3 or EF-1 (Vleminckx et al., 1999). Cycling conditions for siamois were: 94°C for 10 min, followed by 28 cycles of 94, 58 and 72°C for 30 s each, and a final incubation at 72°C for 5 min. Conditions for the other primers were similar except for an annealing temperature of 55°C and 25 cycles. Sequences of the Xnr-3 primer pairs were: 5′-GTGAATCCA CTTGTGCAGTT-3′ and 5′-ACAGAGCCAATCTCATGTGC-3′. PCR products were separated on 5% polyacrylamide gels and visualized by autoradiography.
RNA injections
Constructs were linearized and capped RNAs were synthesized using SP6 RNA polymerase (Promega) (Vleminckx et al., 1999). RNAs were injected in a volume of ∼15 nl into the appropriate blastomere(s) of four-cell stage Xenopus embryos except for double injections with XWnt-8 in the animal cap assay. In the latter experiments a first injection of 30 nl containing RNAs for E1A and CBP was followed by a second injection of ∼10 nl XWnt-8 RNA in the same spot. During injection, embryos were kept as described previously (Vleminckx et al., 1999) and scored for axis duplication at stage 16–18 (Nieuwkoop and Faber, 1967).
Acknowledgments
Acknowledgements
We are grateful to H.Clevers, D.Kimelman, R.Moon and R.Müller for gifts of the TCF4 cDNA, the TOPFLASH/FOPFLASH, siamois and cyclin D1 reporter plasmids, and the XWnt-8 expression construct, to R.Eckner, A.Wolffe and R.Janknecht for p300 and CBP expression vectors, to K.-H.Klempnauer for providing us with the E1A expression plasmids, to K.Rose and G.Van Imschoot for excellent technical assistance, and to R.Cassada for critical reading of the manuscript. F.v.R. is a Research Director of the Fund for National Research-Flanders.
References
- Aberle H., Bauer,A., Stappert,J., Kispert,A. and Kemler,R. (1997) β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J., 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aberle H., Butz,S., Stappert,J., Weissig,H., Kemler,R. and Hoschuetzky,H. (1994) Assembly of the cadherin–catenin complex in vitro with recombinant proteins. J. Cell Sci., 107, 3655–3663. [DOI] [PubMed] [Google Scholar]
- Aberle H., Schwartz,H. and Kemler,R. (1996) Cadherin–catenin complex: protein interactions and their implications for cadherin function. J. Cell. Biochem., 61, 514–523. [DOI] [PubMed] [Google Scholar]
- Aoki M., Hecht,A., Kruse,U., Kemler,R. and Vogt,P.K. (1999) Nuclear endpoint of Wnt signaling: Neoplastic transformation induced by transactivating lymphoid-enhancing factor 1. Proc. Natl Acad. Sci. USA, 96, 139–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1995) CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J., 14, 4758–4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A., Huber,O. and Kemler,R. (1998) Pontin52, an interaction partner of β-catenin, binds to the TATA box binding protein. Proc. Natl Acad. Sci. USA, 95, 14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannon M., Brown,J.D., Bates,R., Kimelman,D. and Moon,R.T. (1999) XCtBP is a XTcf-3 co-repressor with roles throughout Xenopus development. Development, 126, 3159–3170. [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts,M., Sumoy,L., Moon,R.T. and Kimelman,D. (1997) A β-catenin–XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev., 11, 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadigan K.M. and Nusse,R. (1997) Wnt signaling: a common theme in animal development. Genes Dev., 11, 3286–3305. [DOI] [PubMed] [Google Scholar]
- Cavallo R.A., Cox,R.T., Moline,M.M., Roose,J., Polevoy,G.A., Clevers,H., Peifer,M. and Bejsovec,A. (1998) Drosophila Tcf and Groucho interact to repress Wingless signalling activity. Nature, 395, 604–608. [DOI] [PubMed] [Google Scholar]
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999a) A functional interaction between the histone deacetylase rpd3 and the corepressor groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999b) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Eastman Q. and Grosschedl,R. (1999) Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr. Opin. Cell Biol., 11, 233–240. [DOI] [PubMed] [Google Scholar]
- Eckner R., Ewen,M.E., Newsome,D., Gerdes,M., DeCaprio,J.A., Lawrence,J.B. and Livingston,D.M. (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev., 8, 869–884. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Felzien L.K., Farrell,S., Betts,J.C., Mosavin,R. and Nabel,G.J. (1999) Specificity of cyclin E-Cdk2, TFIIB and E1A interactions with a common domain of the p300 coactivator. Mol. Cell. Biol., 19, 4241–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii G., Tsuchiya,R., Itoh,Y., Tashiro,K. and Hirohashi,S. (1998) Molecular cloning and expression of Xenopus p300/CBP. Biochim. Biophys. Acta, 1443, 41–54. [DOI] [PubMed] [Google Scholar]
- Grant P.A. and Berger,S.L. (1999) Histone acetyltransferase complexes. Semin. Cell Dev. Biol., 10, 169–177. [DOI] [PubMed] [Google Scholar]
- Hamamori Y., Sartorelli,V., Ogryzko,V., Puri,P.L., Wu,H.Y., Wang,J.Y., Nakatani,Y. and Kedes,L. (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell, 96, 405–413. [DOI] [PubMed] [Google Scholar]
- Hecht A., Litterst,C.M., Huber,O. and Kemler,R. (1999) Functional characterization of multiple transactivating elements in β-catenin, some of which interact with the TATA-binding protein in vitro. J. Biol. Chem., 274, 18017–18025. [DOI] [PubMed] [Google Scholar]
- Hsu S.C., Galceran,J. and Grosschedl,R. (1998) Modulation of transcriptional regulation by LEF-1 in response to wnt-1 signaling and association with β-catenin. Mol. Cell. Biol., 18, 4807–4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber O., Korn,R., McLaughlin,J., Ohsugi,M., Herrmann,B.G. and Kemler,R. (1996) Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech. Dev., 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Ishitani T. et al. (1999) The TAK1-NLK-MAPK-related pathway antagonizes signalling between β-catenin and transcription factor TCF. Nature, 399, 798–802. [DOI] [PubMed] [Google Scholar]
- Kao K.R. and Elinson,R.P. (1988) The entire mesodermal mantle behaves as Spemann’s organizer in dorsoanterior enhanced Xenopus laevis embryos. Dev. Biol., 127, 64–77. [DOI] [PubMed] [Google Scholar]
- Kato Y., Shi,Y. and He,X. (1999) Neuralization of the Xenopus embryo by inhibition of p300/CREB-binding protein function. J. Neurosci., 19, 9364–9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki H., Eckner,R., Yao,T.P., Taira,K., Chiu,R., Livingston,D.M. and Yokoyama,K.K. (1998) Distinct roles of the co-activators p300 and CBP in retinoic-acid-induced F9-cell differentiation. Nature, 393, 284–289. [DOI] [PubMed] [Google Scholar]
- Korinek V., Barker,N., Morin,P.J., van Wichen,D., de Weger,R., Kinzler,K.W., Vogelstein,B. and Clevers,H. (1997) Constitutive transcriptional activation by a β-catenin–Tcf complex in APC–/– colon carcinoma. Science, 275, 1784–1787. [DOI] [PubMed] [Google Scholar]
- Korzus E., Torchia,J., Rose,D.W., Xu,L., Kurokawa,R., McInerney, E.M., Mullen,T.M., Glass,C.K. and Rosenfeld,M.G. (1998) Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science, 279, 703–707. [DOI] [PubMed] [Google Scholar]
- Kwok R.P., Lundblad,J.R., Chrivia,J.C., Richards,J.P., Bachinger,H.P., Brennan,R.G., Roberts,S.G., Green,M.R. and Goodman,R.H. (1994) Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature, 370, 223–226. [DOI] [PubMed] [Google Scholar]
- Levanon D., Goldstein,R.E., Bernstein,Y., Tang,H., Goldenberg,D., Stifani,S., Paroush,Z. and Groner,Y. (1998) Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA, 95, 11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas M.A., Bannister,A.J., Martin,K., Haus-Seuffert,P., Meisterernst,M. and Kouzarides,T. (1998) The acetyltransferase activity of CBP stimulates transcription. EMBO J., 17, 2886–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meneghini M.D., Ishitani,T., Carter,J.C., Hisamoto,N., Ninomiya-Tsuji,J., Thorpe,C.J., Hamill,D.R., Matsumoto,K. and Bowerman,B. (1999) MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature, 399, 793–797. [DOI] [PubMed] [Google Scholar]
- Miller J.R., Hocking,A.M., Brown,J.D. and Moon,R.T. (1999) Mechanism and function of signal transduction by the Wnt/β-catenin and Wnt/Ca2+ pathways. Oncogene, 18, 7860–7872. [DOI] [PubMed] [Google Scholar]
- Morin P.J., Sparks,A.B., Korinek,V., Barker,N., Clevers,H., Vogelstein,B. and Kinzler,K.W. (1997) Activation of β-catenin–Tcf signaling in colon cancer by mutations in β-catenin or APC. Science, 275, 1787–1790. [DOI] [PubMed] [Google Scholar]
- Munshi N., Merika,M., Yie,J., Senger,K., Chen,G. and Thanos,D. (1998) Acetylation of HMG I(Y) by CBP turns off IFN β expression by disrupting the enhanceosome. Mol. Cell., 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop P.D. and Faber,J. (1967) Normal Table of Xenopus laevis (Daudin). Elsevier North Holland Biomedical Press, Amsterdam, The Netherlands. [Google Scholar]
- Novak A., Hsu,S.C., Leung-Hagesteijn,C., Radeva,G., Papkoff,J., Montesano,R., Roskelley,C., Grosschedl,R. and Dedhar,S. (1998) Cell adhesion and the integrin-linked kinase regulate the LEF-1 and β-catenin signaling pathways. Proc. Natl Acad. Sci. USA, 95, 4374–4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palaparti A., Baratz,A. and Stifani,S. (1997) The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J. Biol. Chem., 272, 26604–26610. [DOI] [PubMed] [Google Scholar]
- Peifer M., Berg,S. and Reynolds,A.B. (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell, 76, 789–791. [DOI] [PubMed] [Google Scholar]
- Porfiri E., Rubinfeld,B., Albert,I., Hovanes,K., Waterman,M. and Polakis,P. (1997) Induction of a β-catenin–LEF-1 complex by wnt-1 and transforming mutants of β-catenin. Oncogene, 15, 2833–2839. [DOI] [PubMed] [Google Scholar]
- Prieve M.G. and Waterman,M.L. (1999) Nuclear localization and formation of β-catenin-lymphoid enhancer factor 1 complexes are not sufficient for activation of gene expression. Mol. Cell. Biol., 19, 4503–4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P.L. et al. (1997) Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell, 1, 35–45. [DOI] [PubMed] [Google Scholar]
- Qiu X.B., Lin,Y.L., Thome,K.C., Pian,P., Schlegel,B.P., Weremowicz, S., Parvin,J.D. and Dutta,A. (1998) An eukaryotic RuvB-like protein (RUVBL1) essential for growth. J. Biol. Chem., 273, 27786–27793. [DOI] [PubMed] [Google Scholar]
- Rocheleau C.E., Yasuda,J., Shin,T.H., Lin,R., Sawa,H., Okano,H., Priess,J.R., Davis,R.J. and Mello,C.C. (1999) WRM-1 activates the LIT-1 protein kinase to transduce anterior/posterior polarity signals in C. elegans. Cell, 97, 717–726. [DOI] [PubMed] [Google Scholar]
- Roose J., Molenaar,M., Peterson,J., Hurenkamp,J., Brantjes,H., Moerer,P., van de Wetering,M., Destree,O. and Clevers,H. (1998) The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature, 395, 608–612. [DOI] [PubMed] [Google Scholar]
- Shikama N., Lyon,J. and La Thangue,N.B. (1997) The p300/CBP family: integrating signals with transcription factors and chromatin. Trends Cell Biol., 7, 230–236. [DOI] [PubMed] [Google Scholar]
- Shtutman M., Zhurinsky,J., Simcha,I., Albanese,C., D’Amico,M., Pestell,R. and Ben-Ze’ev,A. (1999) The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA, 96, 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol S.Y. (1999) Wnt signaling and dorso-ventral axis specification in vertebrates. Curr. Opin. Genet. Dev., 9, 405–410. [DOI] [PubMed] [Google Scholar]
- Tetsu O. and McCormick,F. (1999) β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed] [Google Scholar]
- Turner D.L. and Weintraub,H. (1994) Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes Dev., 8, 1434–1447. [DOI] [PubMed] [Google Scholar]
- van de Wetering M. et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- Vleminckx K., Kemler,R. and Hecht,A. (1999) The C-terminal transactivation domain of β-catenin is necessary and sufficient for signaling by the LEF-1/β-catenin complex in Xenopus laevis. Mech. Dev., 81, 49–58. [DOI] [PubMed] [Google Scholar]
- Waltzer L. and Bienz,M. (1998) Drosophila CBP represses the transcription factor TCF to antagonize Wingless signalling. Nature, 395, 521–525. [DOI] [PubMed] [Google Scholar]
- Workman J.L. and Kingston,R.E. (1998) Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem., 67, 545–579. [DOI] [PubMed] [Google Scholar]
- Yuan W., Condorelli,G., Caruso,M., Felsani,A. and Giordano,A. (1996) Human p300 protein is a coactivator for the transcription factor MyoD. J. Biol. Chem., 271, 9009–9013. [DOI] [PubMed] [Google Scholar]