Abstract
Background
Ideally, disease modifying therapies for Alzheimer disease (AD) will be applied during the ‘preclinical’ stage (pathology present with cognition intact) before severe neuronal damage occurs, or upon recognizing very mild cognitive impairment. Developing and judiciously administering such therapies will require biomarker panels to identify early AD pathology, classify disease stage, monitor pathological progression, and predict cognitive decline. To discover such biomarkers, we measured AD-associated changes in the cerebrospinal fluid (CSF) proteome.
Methods and Findings
CSF samples from individuals with mild AD (Clinical Dementia Rating [CDR] 1) (n = 24) and cognitively normal controls (CDR 0) (n = 24) were subjected to two-dimensional difference-in-gel electrophoresis. Within 119 differentially-abundant gel features, mass spectrometry (LC-MS/MS) identified 47 proteins. For validation, eleven proteins were re-evaluated by enzyme-linked immunosorbent assays (ELISA). Six of these assays (NrCAM, YKL-40, chromogranin A, carnosinase I, transthyretin, cystatin C) distinguished CDR 1 and CDR 0 groups and were subsequently applied (with tau, p-tau181 and Aβ42 ELISAs) to a larger independent cohort (n = 292) that included individuals with very mild dementia (CDR 0.5). Receiver-operating characteristic curve analyses using stepwise logistic regression yielded optimal biomarker combinations to distinguish CDR 0 from CDR>0 (tau, YKL-40, NrCAM) and CDR 1 from CDR<1 (tau, chromogranin A, carnosinase I) with areas under the curve of 0.90 (0.85–0.94 95% confidence interval [CI]) and 0.88 (0.81–0.94 CI), respectively.
Conclusions
Four novel CSF biomarkers for AD (NrCAM, YKL-40, chromogranin A, carnosinase I) can improve the diagnostic accuracy of Aβ42 and tau. Together, these six markers describe six clinicopathological stages from cognitive normalcy to mild dementia, including stages defined by increased risk of cognitive decline. Such a panel might improve clinical trial efficiency by guiding subject enrollment and monitoring disease progression. Further studies will be required to validate this panel and evaluate its potential for distinguishing AD from other dementing conditions.
Introduction
Clinicopathological studies suggest that Alzheimer's disease (AD) pathology (amyloid plaque formation, followed by gliosis and neurofibrillary tangle formation) begins 10–15 years before the onset of very mild dementia [1], [2]. This period of ‘preclinical AD’ could provide an opportunity for disease modifying therapies to prevent or forestall the synaptic and neuronal losses associated with cognitive impairment [3]–[5]. However, before such interventions can be developed and judiciously administered, accurate tools must be in place to diagnose and monitor the pathophysiological condition of individuals with preclinical AD and very early stage AD dementia. Clinical examination cannot detect preclinical disease or measure cellular and molecular changes within the brain, and, in general, has limited accuracy when diagnosing the very earliest symptomatic stages of AD. Therefore, there is an urgent need to identify biomarkers that can do so. Because its composition is rapidly and directly influenced by the brain, the cerebrospinal fluid (CSF) proteome represents an appealing source for such biomarkers.
Indeed, a few CSF proteins have already shown promise as diagnostic biomarkers for clinical AD (dementia of the Alzheimer type [DAT]) and even preclinical AD. Lower mean levels of CSF Aβ42 and higher mean levels of tau and phosphorylated tau can distinguish groups with DAT from cognitively normal controls [6], [7]. Unfortunately, value ranges for each biomarker show substantial overlap between groups.
Recently, using positron-emission tomography PET imaging with Pittsburgh Compound B (PIB) to measure brain amyloid in vivo, we and others have demonstrated that low CSF Aβ42 can serve as an indicator of amyloid deposition [8]–[13], and that CSF tau levels correlate positively with in vivo brain amyloid load [11], [14]. Importantly, both of these associations are independent of clinical diagnosis [8]–[11], though CSF tau does correlate with more sensitive measures of cognition [14]. These findings suggest that the overlap of biomarker values between clinical groups may, in part, reflect “contamination” of control groups by cognitively normal individuals exhibiting amyloid plaques and early neurodegeneration (preclinical AD), low CSF Aβ42 and elevated CSF tau. Supporting this notion, elevated ratios of tau/Aβ42 and p-tau181/Aβ42 (consistent with the presence of amyloid plaques and neurodegeneration) have been associated with increased risk of converting from cognitive normalcy to mild cognitive impairment or dementia [9], [15], and with increased rate of cognitive decline among those with very mild dementia [16]. Together, these findings suggest that CSF biomarkers can describe neuropathological state and trajectory. They also suggest that a pathological staging system based on biomarkers might be a favorable alternative or adjunct to clinical staging for guiding treatment decisions or designing clinical trials.
Beyond amyloid plaque formation, other features of AD pathophysiology might also be exploited as therapeutic targets, sources of diagnostic biomarkers, or measures of disease progression. In addition to Aβ42 and tau, many other candidate AD biomarkers have been identified by either targeted or unbiased proteomics screens [17]–[27]. Only a few of these studies have tested large, well-characterized cohorts, however. Even fewer have evaluated biomarkers for their ability to distinguish the very early stages of AD pathophysiology. Thus, there remains a critical need for validated AD biomarkers that can properly categorize individuals by early pathological stage; such markers may have potential for monitoring neuropathological decline and, thereby, for evaluating response to disease-modifying therapies.
The goal of this study, therefore, is to identify such CSF protein biomarkers for AD using the unbiased proteomic technique of two-dimensional difference-in-gel electrophoresis (2D-DIGE) coupled with liquid chromatography and tandem mass spectrometry (LC-MS/MS), and to evaluate them further in a larger independent cohort using quantitative enzyme-linked immunosorbent assays (ELISA). Our findings suggest that a small ensemble of novel biomarkers may be able to distinguish several stages of cognitive decline in early AD, and improve the ability of current leading biomarkers tau and Aβ42 to discriminate early symptomatic AD from cognitive normalcy.
Methods
Ethics Statement
The study protocols were approved by the institutional review boards of the University of Washington, the Oregon Health and Science University, the University of Pennsylvania, the University of California San Diego, and Washington University. Written informed consent was obtained from all participants at enrollment. All aspects of this study were conducted according to the principles expressed in the Declaration of Helsinki.
Participant Selection for Discovery Cohort
Participants (n = 48), community-dwelling volunteers from University of Washington [n = 18], Oregon Health and Science University [n = 11], University of Pennsylvania [n = 11], and University of California San Diego [n = 8], were 51–87 years of age and in good general health, having no other neurological, psychiatric, or major medical diagnoses that could contribute to dementia, nor use of exclusionary medications (e.g. anticoagulants) within 1–3 months of lumbar puncture (LP). Cognitive status was evaluated based on criteria from the National Institute of Neurological and Communicative Diseases and Stroke-Alzheimer's Disease and Related Disorders Association [28]. In the morning after overnight fasting, CSF was obtained by LP, collected and aliquoted in polypropylene tubes, and immediately frozen at −80°C. Participants who were cognitively normal (Clinical Dementia Rating [CDR] of 0 [n = 24]) [29], or had mild “probable AD” (CDR 1) (n = 24), were selected from a larger group of 120 individuals on the basis of CSF Aβ42 (relatively high and low values, respectively), and, when possible, CSF tau (relatively low and high values, respectively) to increase the likelihood of CDR 1 participants having and CDR 0 participants not having AD pathology. CSF Aβ42 and tau levels for the discovery cohort were all measured in a single laboratory using well-established ELISA assays ([30] and Innotest, Innogenetics, Ghent, Belgium). Although quantitative thresholds were not defined prior to sample selection, the lowest CDR 0 value and the highest CDR 1 value for CSF Aβ42 in this ‘discovery cohort’ were 609 and 361 pg/mL, respectively; ranges for CSF tau were 141–461 pg/mL for CDR 0 and 215–1965 pg/mL for CDR 1.
Participant Selection for Validation Cohort
Participants (n = 292), community-dwelling volunteers enrolled at the Knight Alzheimer Disease Research Center at Washington University (WU-ADRC), were ≥60 years of age and met the same exclusion criteria as the discovery cohort. The study protocol was approved by the Human Studies Committee at Washington University, and written and verbal informed consent was obtained from participants at enrollment. Cognitive status was determined as with the discovery cohort. Participants who were cognitively normal (CDR 0, n = 198), very mildly demented (CDR 0.5, n = 65) or mildly demented (CDR 1, n = 29) at the time of LP were selected without regard to previously measured biomarkers. Some CDR 0.5 participants met criteria for mild cognitive impairment (MCI) and some showed even milder impairment, and could be considered “pre-MCI” [31]. All CDR 1 individuals had received a diagnosis of DAT (See Table 1 for demographic characteristics). Apolipoprotein E (APOE) genotypes were determined by the WU-ADRC Genetics Core. Fasted CSF (20–30 mL) was collected, gently mixed, centrifuged, aliquoted and frozen at −80°C in polypropylene tubes [9].
Table 1. Demographic, clinical, genotype characteristics of validation cohort.
| Characteristic | CDR 0 | CDR 0.5 | CDR 1 |
| Number of Participants | 198 | 65 | 29 |
| Gender (% Female) | 63% | 54% | 52% |
| APOE genotype, % ε4 positive | 35% | 51% | 59% |
| Mean MMSE score (SD) | 28.9 (1.3) | 26.3 (2.8) | 22.3 (3.9) |
| Mean age at LP (SD), years | 71.0 (7.3) | 73.8 (6.8) | 76.5 (6.2) |
| Mean CSF Aβ42 (SD), pg/mL | 605 (240) | 446 (230) | 351 (118) |
| Mean CSF tau (SD), pg/mL | 304 (161) | 539 (276) | 552 (263) |
| Mean CSF p-tau181 (SD), pg/mL | 55 (25) | 85 (44) | 77 (38) |
Abbreviations: CDR, Clinical Dementia Rating; CDR 0, cognitively normal; CDR 0.5, very mild dementia; CDR 1 mild dementia; APOE, apolipoprotein E; MMSE, Mini-Mental State Examination; LP, lumbar puncture; SD, standard deviation; CSF, cerebrospinal fluid; Aβ42, amyloid beta 42 peptide; p-tau181, tau phosphorylated at threonine 181.
Multi-Affinity Immunodepletion of CSF
A pooled CSF sample, containing an equivalent volume from every ‘discovery’ cohort sample, was prepared as an internal standard for 2D-DIGE to facilitate the matching of gel features, and to allow normalization of the intensity of each gel feature among different gels. To enrich for proteins of low-abundance prior to 2D-DIGE, each CSF sample was depleted of six highly-abundant proteins (albumin, IgG, IgA, haptoglobin, transferrin, and α-1-antitrypsin) by immunoaffinity chromatography (Agilent Technologies, Palo Alto, CA) according to the manufacturer's instructions and as described previously [32]. Depleted samples were then concentrated using 10 kDa exclusion filters to retain larger molecules. As a ‘benchmark’ of immunodepletion column performance, an aliquot of reference CSF was depleted after every group of seven experimental chromatographic depletions. Non-depleted reference CSF, depleted CSF and the proteins that were retained by the column were analyzed by 2D-DIGE as previously described [32], [33]; gel images obtained from all reference CSF depletion analyses were similar (data not shown), indicating consistent column performance over time.
2D-DIGE
2D-DIGE was performed as described previously [32], [33]. Briefly, CDR 0 and CDR 1 samples were randomly paired. 50 micrograms of protein from each paired sample and from an aliquot of the pooled CSF sample were labeled with one of three N-hydroxysuccinimide cyanine dyes. The labeled proteins and 100 micrograms of unlabeled protein from each sample were mixed and equilibrated with an immobilized pH gradient strip for isoelectric focusing (first dimension), after which the strip was treated with reducing and alkylating solutions prior to SDS-PAGE (second dimension). Cy2, Cy3 and Cy5-labeled images were acquired on a Typhoon 9400 scanner (GE Healthcare, United Kingdom) at excitation/emission wavelengths of 488/520, 532/580, and 633/670 nm, respectively.
Gel Image and Statistical Analysis
The comparative two-dimensional gel analysis was performed using an established experimental design [34] in which the high variation between gels is minimized by including a common, labeled pooled sample in all gels. Intra-gel feature detection, quantification and inter-gel matching and quantification were performed using the Differential In-Gel Analysis (DIA) and Biological Variation Analysis (BVA) modules of DeCyder software v 6.5 (GE Healthcare), respectively, as described previously [32]. This process (DIA analysis) resulted in approximately 5,000 gel features per gel image. In five gels, one sample contained significant amounts of hemoglobin indicating possible blood contamination. Therefore, all images from gels with these hemoglobin-containing samples were removed from further analysis. Remaining gel images were separated into three sets: standard (pool of all samples), CDR 0 and CDR 1. The pooled sample image with the largest number of well-resolved gel features was chosen as a master image. Gel features in each remaining pooled sample image were hand matched to gel features in the master image. For each gel feature that was matched across >50% of the gels (n = 764), a Student's t-test (α = 0.05) was performed to determine the statistical significance of CDR 0/CDR 1 ratios, using the DeCyder EDA (Extended Data Analysis) module. To maximize discovery rate and minimize type II error, no multiple test correction was applied. The image intensity data for the statistically significant gel features (n = 119) were then subjected to unsupervised hierarchical clustering (DeCyder EDA module).
Protein/Peptide Identification by LC-MS/MS
Gel features with significant intensity differences were targeted by a robotic gel sampling system (ProPic; Genomics Solutions, Ann Arbor, MI) and transferred into 96 well plates for in-gel digestion with trypsin using a modification of a method [35] described previously [33]. Aliquots of these digests were processed for and analyzed by LC-MS/MS using a capillary LC (Eksigent, Livermore CA) interfaced to a nano-LC-linear quadrupole ion trap Fourier transform ion cyclotron resonance mass spectrometer (nano-LC-FTMS) [36] QStar [37] or LTQ [36]. The tandem spectra were searched against the National Center for Biotechnology Information non-redundant protein database NR (downloaded on 02-18-2007) using MASCOT, version 2.2.04 (Matrix Sciences, London). The database searches were constrained by allowing for trypsin cleavage (with up to two missed cleavage sites), fixed modifications (carbamidomethylation of Cys residues) and variable modifications (oxidation of Met residues and N-terminal pyroglutamate formation). Protein identifications were considered genuine if at least two peptides were matched with individual MASCOT ion scores ≥40.
Using nano-LC-MS/MS, multiple proteins were identified in the majority of individual gel features. The frequent observation of multiple proteins in single gel features was attributed to the sensitivity and greater peptide coverage that can be achieved with nano-LC-MS methods as compared to, for example, MALDI-MS analysis of peptides from gel features. Assignment of the major protein(s) from each gel feature was achieved using quantitative proteomics from spectra counting [38]. The detection of multiple proteins within single gel features could also be attributed to artifacts and technical issues associated with 2D gel electrophoresis: 1) incomplete resolution of proteins by gel electrophoresis (due to similar charge and size characteristics, excessive abundance of neighboring proteins, or artifactual streaking); 2) changes in molecular weight associated with cyanine dye labeling, particularly for lower molecular weight proteins; and 3) sample ‘carryover’ during robotic gel sampling or during nano-LC-MS/MS.
All relevant proteomics data are detailed in Table S1.
Enzyme Linked Immunosorbent Assays (ELISAs) and Statistical Analyses
CSF samples were analyzed by ELISA in duplicate for Aβ42, total tau, and phospho-tau181 (Innotest, Innogenetics, Ghent, Belgium) after one freeze-thaw cycle, and in triplicate for all other ELISAs after two freeze-thaw cycles. Samples were evaluated using commercially available ELISAs for NrCAM (R&D Systems Inc., Minneapolis, MN), YKL-40 (Quidel Corporation, San Diego, CA), apolipoprotein E (Medical and Biological Laboratories Company, Ltd., Nagoya, Japan), clusterin/apolipoprotein J (ALPCO Diagnostics, Salem, NH), pigment epithelium-derived factor (PEDF)/serpin-F1 (Chemicon International Inc./ Millipore Corporation, Billerica, MA), beta-2 microglobulin (ALPCO Diagnostics), ceruloplasmin (Assaypro, St. Charles, MO), chromogranin A (ALPCO Diagnostics, low binding capacity manufacturing protocol), transthyretin (Assaypro), and cystatin C (US Biological, Swampscott, MA), according to manufacturer's instructions, with adjustments for the analysis of CSF. A sandwich ELISA was developed for carnosinase I using goat anti-human carnosinase I antibody (2 µg/mL, R&D Systems Inc.) for capture, rabbit anti-human carnosinase I antibody (1 µg/mL, Sigma-Aldrich Corporation, St. Louis, MO) for detection, goat anti-rabbit:horseradish peroxidase (1∶5000, Upstate Biologicals Inc./Millipore Corporation) for reporting, and TMB (3,3′,5,5′-tetramethylbenzidine) Super Slow (Sigma-Aldrich Corporation) for color development; recombinant carnosinase I (R&D Systems Inc.) was used as standard.
Statistical analyses were performed using commercially available software: SAS 9.2 (SAS Institute Inc., Cary, NC) for Receiver Operating Characteristic (ROC)/area under curve (AUC) calculations and logistic regression analyses, and SPSS 18 (SPSS Inc., Chicago, IL) for all other analyses.
Comparisons between CDR 0 and CDR 1 groups of the ‘discovery’ cohort (one sample was unavailable for re-evaluation, n = 47) were performed using unpaired t-test. For the ‘validation’ cohort (n = 292), correlations with age and gender were evaluated using the Spearman rho correlation coefficient (α = 0.05). Chi-square analyses were performed to evaluate need for adjustment for observed correlations. Comparisons between the three CDR groups were performed using one-way analysis of variance (ANOVA), with Bonferroni and LSD post-hoc tests for pair-wise group comparisons, with the following exceptions: one-way ANOVA with Welch's correction was applied for markers (transthyretin) demonstrating unequal variances (Levene <.05); markers correlating with age (tau, p-tau181, Aβ42, YKL-40) were evaluated by analysis of covariance (ANCOVA) adjusting for age, followed by Bonferroni and LSD post-hoc tests. Multiple post-hoc tests were applied in recognition of their different levels of stringency (Bonferroni > LSD), and their non-uniform popularity among statisticians. For CDR 0 vs >0 comparisons and CDR 1 vs <1 comparisons, unpaired t-test was used; Welch's correction for unequal variances was applied for YKL-40, p-tau181, tau, and Aβ42. For each biomarker measured in the larger ‘validation’ cohort, the ROC curve and the AUC were calculated for predicting CDR 0 versus CDR>0. A stepwise logistic regression analysis was used to identify an optimal combination of these biomarkers for this data set. These analyses were repeated for CDR 1 vs CDR<1.
Results
Sample Processing and 2D-DIGE Analysis
To identify new candidate biomarkers for AD, we utilized an unbiased proteomics approach, 2D-DIGE LC-MS/MS [32], [33], to compare the relative concentrations of CSF proteins in individuals with mild “probable AD” (CDR 1, n = 24) to those in individuals with normal cognition (CDR 0, n = 24). The two clinical groups were selected on the basis of relative biomarker values for CSF Aβ42 and tau (see Methods), and differed somewhat with respect to age at LP and gender (CDR 0: 64.8±8.8 yrs, 38% female; CDR 1: 72.8 yrs ±7.9 yrs, 54% female). Five samples showed evidence of blood contamination by 2D-DIGE; the five gels containing these samples were excluded from subsequent image analyses. The remaining individual sample images (n = 38, from 19 gels) were aligned using the BVA module (described under Methods).
Among the 764 gel features that were present in >50% of the gels, 119 were found to have significant intensity differences between CDR 0 and CDR 1 groups (Student's t-test [α = 0.05]) (Figure 1). The image intensity data for these 119 gel features were subjected to unsupervised hierarchical clustering (EDA module, DeCyder software) and the gel features themselves were analyzed for protein composition.
Figure 1. Two-dimensional difference in gel electrophoresis (2D-DIGE) of cerebrospinal fluid immunodepleted of six high abundance proteins.
Representative 2D-DIGE (grayscale) image with labeled locations of 119 gel features that differed in intensity between CDR 0 and CDR 1 groups. Gel features are numbered 1 through 119, and relevant information about each is listed in Table 2 and in Table S1. Approximate molecular weight (in kilodaltons [kDa]) is indicated along the right border; isoelectric point ranges from 3 (left) to 11(right) and is non-linear (not shown). The large, intense, protein spots commonly attributed to transthyretin are boxed; a subset of the differentially abundant gel features in which transthyretin was identified by mass spectrometry is circled.
Protein Identification by LC-MS/MS
LC-MS/MS identified single dominant proteins in 78 of the 119 gel features (Table 2). In 29 gel features, our analyses identified two or more co-dominant proteins. The 12 remaining gel features were not annotated from the nano-LC-MS/MS data. Among the characterized gel features, there was considerable redundancy in protein identifications, with some proteins appearing in multiple gel features. Such ‘redundant’ gel features, likely representing a modified form or variant of the same ‘parent’ protein, generally migrated with some proximity on 2D-gel electrophoresis (Figure 1). Forty-seven unique proteins were identified (Table 2). Thirteen of these unique proteins had been identified in our previous studies [32], [33] (including chromogranin B, cystatin C, prostaglandin H2 D-isomerase/beta trace, neuronal pentraxin receptor, gelsolin, beta-2 microglobulin, carnosinase I, angiotensinogen, apolipoprotein H, secretogranin III, alpha-1-antichymotrypsin, chitinase 3-like 1/YKL-40, and kininogen I) and others have been reported by other groups [17], [19], [20], [23], [25], [27]. These previous reports provide supporting evidence that this list of proteins may contain viable candidate biomarkers for AD that are worthy of pursuit in validation experiments.
Table 2. Proteins identified by 2D-DIGE LC-MS/MS with differential abundance in CDR 1 vs. CDR 0 CSF.
| Spot | BVA | GI number(s) | Protein | Change | p value | Protein |
| 1 | 4709 | 31543193 | hypothetical protein LOC146556 | −1.36 | 7.02E-04 | 1 |
| 2 | 5659 | 4502807 | chromogranin B | −1.31 | 1.18E-03 | 2 |
| 3 | 4683 | 4502101 | annexin I | −1.31 | 9.54E-04 | 3 |
| 4 | 4608 | 62089004 | chromogranin B | −1.24 | 6.49E-03 | |
| 181387 | cystatin C | 4 | ||||
| 134464 | secretogranin-2 | 5 | ||||
| 5 | 4297 | 4502807 | chromogranin B | −1.26 | 0.0157 | |
| 6 | 4545 | −1.34 | 3.86E-03 | |||
| 7 | 4695 | 4502807 | chromogranin B | −1.27 | 0.0115 | |
| 8 | 4044 | 4502807 | chromogranin B | −1.32 | 2.15E-03 | |
| 9 | 1314 | 1621283 | neuronal cell adhesion molecule (NrCAM) | −1.22 | 0.0119 | 6 |
| 10 | 1320 | 1621283 | neuronal cell adhesion molecule (NrCAM) | −1.33 | 6.31E-04 | |
| 11 | 1382 | 6651381 | neuronal cell adhesion molecule (NrCAM) | −1.28 | 9.53E-04 | |
| 12 | 1383 | 6651381 | neuronal cell adhesion molecule (NrCAM) | −1.25 | 6.64E-03 | |
| 13 | 4033 | 4502807 | chromogranin B | −1.21 | 0.0419 | |
| 14 | 4191 | 4502807 | chromogranin B | −1.23 | 0.0107 | |
| 15 | 4293 | 4502807 | chromogranin B | −1.33 | 4.64E-03 | |
| 825635 | calmodulin | 7 | ||||
| 16 | 4266 | 62089004 | chromogranin B | −1.22 | 0.0315 | |
| 17 | 4615 | −1.22 | 0.0188 | |||
| 18 | 4677 | −1.3 | 9.63E-03 | |||
| 19 | 4906 | 5454032 | S100 calcium binding protein A1 | −1.3 | 1.36E-04 | 8 |
| 62898141 | prosaposin | 9 | ||||
| 627391 | brain-associated small cell lung cancer antigen/NCAM-140/CD56 | 10 | ||||
| 17136078 | VGF nerve growth factor inducible precursor | 11 | ||||
| 20 | 5014 | 443295 | transthyretin | −1.3 | 2.10E-03 | 12 |
| 21 | 4884 | 224917 | apolipoprotein CIII | −1.34 | 9.78E-04 | 13 |
| 337760 | prosaposin/cerebroside sulfate activator protein | |||||
| 22 | 3423 | 39654998 | chain A, Hr1b Domain From Prk1 | −1.27 | 0.0133 | 14 |
| 32171249 | prostaglandin H2 D-isomerase/beta trace | 15 | ||||
| 23 | 3470 | 17402888 | neuronal pentraxin receptor | −1.25 | 7.23E-03 | 16 |
| 114593356 | extracellular superoxide dismutase (SOD3) | 17 | ||||
| 24 | 4954 | 34616 | beta-2 microglobulin | −1.3 | 4.15E-03 | 18 |
| 25 | 3436 | 32171249 | prostaglandin H2 D-isomerase | −1.22 | 0.0266 | |
| 178775 | proapolipoprotein | 19 | ||||
| 39654998 | chain A, Hr1b Domain From Prk1 | |||||
| 26 | 3714 | −1.27 | 0.03 | |||
| 27 | 4922 | 39654998 | chain A, Hr1b Domain From Prk1 | −1.27 | 0.0194 | |
| 28 | 3786 | 2072129 | chromogranin A | −1.38 | 8.96E-03 | 20 |
| 29 | 4076 | 7341255 | brain acetylcholinesterase putative membrane anchor | −1.25 | 0.0375 | 21 |
| 30 | 4111 | 62089004 | chromogranin B | −1.28 | 0.0206 | |
| 31 | 4167 | 4502807 | chromogranin B | −1.29 | 0.0207 | |
| 32 | 2652 | 28373309 | gelsolin | −1.23 | 0.0346 | 22 |
| 33 | 1313 | 6651381 | neuronal cell adhesion molecule (NrCAM) | −1.19 | 8.08E-03 | |
| 34 | 1372 | 1620909 | ceruloplasmin | −1.19 | 9.00E-03 | 23 |
| 1483187 | inter-alpha-trypsin inhibitor family heavy chain-related protein | 24 | ||||
| 31874098 | hypothetical protein (NrCAM) | |||||
| 6651381 | neuronal cell adhesion molecule (NrCAM) | |||||
| 35 | 1387 | 68534652 | neuronal cell adhesion molecule (NrCAM) | −1.29 | 8.16E-05 | |
| 1620909 | ceruloplasmin | |||||
| 36 | 4808 | 337760 | prosaposin/cerebroside sulfate activator protein | −1.22 | 0.0114 | |
| 37 | 1319 | 68534652 | neuronal cell adhesion molecule (NrCAM) | −1.19 | 0.0198 | |
| 1942284 | ceruloplasmin | |||||
| 38 | 1386 | 6651381 | neuronal cell adhesion molecule (NrCAM) | −1.29 | 1.24E-03 | |
| 39 | 1353 | 21706696 | calsyntenin 1 | −1.22 | 0.0417 | 25 |
| 40 | 1329 | 1621283 | neuronal cell adhesion molecule (NrCAM) | −1.22 | 4.61E-03 | |
| 41 | 2456 | 5802984 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 | −1.13 | 0.0449 | 26 |
| 42 | 2550 | 20178323 | pigment epithelium-derived factor precursor (PEDF)/Serpin-F1/EPC-1 | −1.15 | 0.022 | 27 |
| 43 | 2125 | 21071039 | carnosinase 1 | −1.21 | 0.0245 | 28 |
| 44 | 2131 | 21071039 | carnosinase 1 | −1.19 | 0.049 | |
| 45 | 2152 | 21071039 | carnosinase 1 | −1.15 | 0.0366 | |
| 46 | 5614 | 21071039 | carnosinase 1 | −1.18 | 0.0109 | |
| 47 | 2166 | 21071039 | carnosinase 1 | −1.21 | 0.0122 | |
| 48 | 2328 | 416180 | man9-mannosidase/α1,2-mannosidase IA | −1.16 | 0.0464 | 29 |
| 49 | 3360 | −1.15 | 0.045 | |||
| 50 | 3447 | 32171249 | prostaglandin H2 D-isomerase/beta trace | −1.19 | 0.0334 | |
| 51 | 3546 | 1621283 | neuronal cell adhesion molecule (NrCAM) | −1.17 | 0.0368 | |
| 32171249 | prostaglandin H2 D-isomerase/beta trace | |||||
| 52 | 4745 | 443295 | transthyretin | −1.26 | 0.0181 | |
| 53 | 3032 | 11056046 | nectin-like molecule-1/SynCAM3/TSLL1 | −1.13 | 0.0472 | 30 |
| 54 | 3718 | 39654998 | chain A, Hr1b Domain From Prk1 | −1.14 | 0.0455 | |
| 32171249 | prostaglandin H2 D-isomerase/beta trace | |||||
| 55 | 4902 | 14277770 | apolipoprotein C-Ii | −1.19 | 0.0495 | 31 |
| 337760 | prosaposin/cerebroside sulfate activator protein | |||||
| 2072129 | chromogranin A | |||||
| 56 | 3290 | 409725 | carbonic anhydrase IV | −1.14 | 0.0141 | 32 |
| 57 | 4379 | 17942890 | transthyretin | −1.15 | 0.0219 | |
| 39654998 | chain A, Hr1b Domain From Prk1 | |||||
| 34999 | cadherin 2 precursor | 33 | ||||
| 58 | 4388 | 32171249 | prostaglandin H2 D-isomerase/beta trace | −1.14 | 0.0218 | |
| 39654998 | chain A, Hr1b Domain From Prk1 | |||||
| 443295 | transthyretin | |||||
| 59 | 2192 | 21071039 | carnosinase 1 | −1.34 | 6.56E-03 | |
| 532198 | angiotensinogen | 34 | ||||
| 5531817 | secretogranin III | 35 | ||||
| 9665262 | EGF-containing fibulin-like extracellular matrix protein 1/Fibulin-3 | 36 | ||||
| 177933 | alpha-1-antichymotrypsin | 37 | ||||
| 4504893 | kininogen 1 | 38 | ||||
| 36573 | vitronectin | 39 | ||||
| 60 | 5336 | 443295 | transthyretin | −1.17 | 0.0301 | |
| 61 | 3009 | 178855 | apolipoprotein J/clusterin | −1.26 | 0.0288 | 40 |
| 4557325 | apolipoprotein E | 41 | ||||
| 62 | 3042 | 4557325/178853 | apolipoprotein E | −1.21 | 0.047 | |
| 338305 | apolipoprotein J/clusterin | |||||
| 63 | 3016 | 338305 | apolipoprotein J/clusterin | −1.32 | 6.69E-05 | |
| 64 | 3050 | 4557325/178853 | apolipoprotein E | −1.24 | 5.19E-04 | |
| 178855 | apolipoprotein J/clusterin | |||||
| 65 | 3075 | 4557325/178853 | apolipoprotein E | −1.42 | 5.59E-06 | |
| 178855 | apolipoprotein J/clusterin | |||||
| 66 | 3038 | 4557325/178853 | apolipoprotein E | −1.41 | 2.84E-05 | |
| 178855 | apolipoprotein J/clusterin | |||||
| 67 | 3301 | 178849 | apolipoprotein E | −1.4 | 1.29E-05 | |
| 68 | 3182 | 4557325/178853 | apolipoprotein E | −1.41 | 3.43E-04 | |
| 178855 | apolipoprotein J/clusterin | |||||
| 69 | 2443 | 532198 | angiotensinogen | −1.2 | 6.85E-03 | |
| 70 | 2493 | 4503009 | carboxypeptidase E precursor | −1.23 | 6.09E-03 | 42 |
| 71 | 5621 | 532198 | angiotensinogen | −1.17 | 0.0434 | |
| 72 | 5624 | 532198 | angiotensinogen | −1.22 | 0.0147 | |
| 73 | 5622 | 553181 | angiotensinogen | −1.17 | 0.04 | |
| 74 | 5625 | 532198 | angiotensinogen | −1.16 | 0.0423 | |
| 75 | 5627 | −1.22 | 0.0113 | |||
| 76 | 2849 | 4557325 | apolipoprotein E | −1.28 | 6.26E-03 | |
| 77 | 5009 | 443295 | transthyretin | −1.24 | 0.0268 | |
| 78 | 5033 | 443295 | transthyretin | −1.27 | 4.59E-03 | |
| 79 | 5078 | 443295 | transthyretin | −1.2 | 0.0144 | |
| 80 | 2958 | 4504067 | aspartate aminotransferase 1 | −1.22 | 8.60E-03 | 43 |
| 81 | 3657 | 32171249 | prostaglandin H2 D-isomerase/beta trace | −1.22 | 3.07E-03 | |
| 82 | 3867 | −1.28 | 0.0437 | |||
| 83 | 3176 | 4557325 | apolipoprotein E | −1.63 | 3.03E-04 | |
| 84 | 3228 | 4557325 | apolipoprotein E | −1.4 | 1.39E-03 | |
| 443295 | transthyretin | |||||
| 85 | 3074 | 4557325/178853 | apolipoprotein E | −2.36 | 4.41E-09 | |
| 86 | 5647 | 4557325 | apolipoprotein E | −2.35 | 2.92E-07 | |
| 87 | 3224 | 4557325/178853 | apolipoprotein E | −2.13 | 6.36E-07 | |
| 443295 | transthyretin | |||||
| 88 | 3126 | 4557325/178853 | apolipoprotein E | −1.93 | 7.55E-06 | |
| 89 | 5297 | −1.44 | 0.0473 | |||
| 90 | 3083 | 4557325 | apolipoprotein E | −1.7 | 2.82E-05 | |
| 91 | 2218 | 112911 | alpha-2-macroglobulin | 1.22 | 0.0282 | 44 |
| 92 | 2226 | 6573461 | apolipoprotein H | 1.27 | 0.0305 | 45 |
| 93 | 2252 | 112911 | alpha-2-macroglobulin | 1.26 | 0.0267 | |
| 4557327 | apolipoprotein H | |||||
| 94 | 3255 | 1.24 | 0.0315 | |||
| 95 | 3630 | 178775 | proapolipoprotein | 1.24 | 0.0287 | |
| 32171249 | prostaglandin H2 D-isomerase/beta trace | |||||
| 39654998 | chain A, Hr1b Domain From Prk1 | |||||
| 96 | 2229 | 177933 | alpha-1-antichymotrypsin | 1.42 | 3.09E-03 | |
| 97 | 2235 | 177933 | alpha-1-antichymotrypsin | 1.35 | 0.0388 | |
| 98 | 2261 | 177933 | alpha-1-antichymotrypsin | 1.3 | 6.04E-03 | |
| 99 | 2262 | 177933 | alpha-1-antichymotrypsin | 1.25 | 0.0294 | |
| 100 | 2220 | 1.29 | 0.0158 | |||
| 101 | 3084 | 1.16 | 0.0211 | |||
| 102 | 3508 | 32171249 | prostaglandin H2 D-isomerase/beta trace | 1.22 | 9.21E-03 | |
| 103 | 2825 | 23512215 | chitinase 3-like 1/YKL-40/HC-gp39 | 1.41 | 0.0167 | 46 |
| 104 | 2863 | 4557018 | chitinase 3-like 1/YKL-40/HC-gp39 | 1.5 | 0.0144 | |
| 105 | 2846 | 29726259 | chitinase 3-like 1/YKL-40/HC-gp39 | 1.46 | 7.88E-03 | |
| 106 | 2843 | 23512215 | chitinase 3-like 1/YKL-40/HC-gp39 | 1.32 | 0.0241 | |
| 107 | 3030 | 4557325 | apolipoprotein E | 2.46 | 3.70E-05 | |
| 108 | 3152 | 4557325/178853 | apolipoprotein E | 2.39 | 8.73E-07 | |
| 109 | 3203 | 178853 | apolipoprotein E | 3.23 | 3.13E-07 | |
| 110 | 3185 | 4557325/178853 | apolipoprotein E | 1.9 | 9.72E-04 | |
| 443295 | transthyretin | |||||
| 111 | 3069 | 338305 | apolipoprotein J/clusterin | 1.5 | 6.40E-04 | |
| 112 | 3079 | 1.64 | 4.47E-04 | |||
| 113 | 3133 | 178853 | apolipoprotein E | 1.49 | 8.66E-04 | |
| 338057 | apolipoprotein J/clusterin | |||||
| 114 | 3151 | 178853 | apolipoprotein E | 1.28 | 9.25E-03 | |
| 338057 | apolipoprotein J/clusterin | |||||
| 115 | 3249 | 4557325 | apolipoprotein E | 1.37 | 2.46E-03 | |
| 178855 | apolipoprotein J/clusterin | |||||
| 443295 | transthyretin | |||||
| 116 | 3118 | 4557325/178853 | apolipoprotein E | 1.64 | 9.96E-04 | |
| 117 | 5698 | 178855 | apolipoprotein J/clusterin | 1.73 | 5.82E-04 | |
| 118 | 2819 | 40737343 | C4B3 | 2 | 0.038 | 47 |
| 119 | 3137 | 4557325 | apolipoprotein E | −2.5 | 8.52E-07 |
Column 1, coded protein spot ID (as in Figure 1).
Column 2, biological variation analysis (BVA) number for spot generated by Decyder software.
Column 3, GI accession number(s) assigned to proteins identified by MASCOT.
Column 4, name of protein identified by MASCOT.
Column 5, fold-change in protein abundance; negative values indicate decreases in CDR 1 vs. CDR 0.
Column 6, p value of the CDR 1 versus CDR 0 comparison (Student's t test).
Column 7, consecutive numbering identifying proteins as unique.
Unsupervised Clustering Analysis
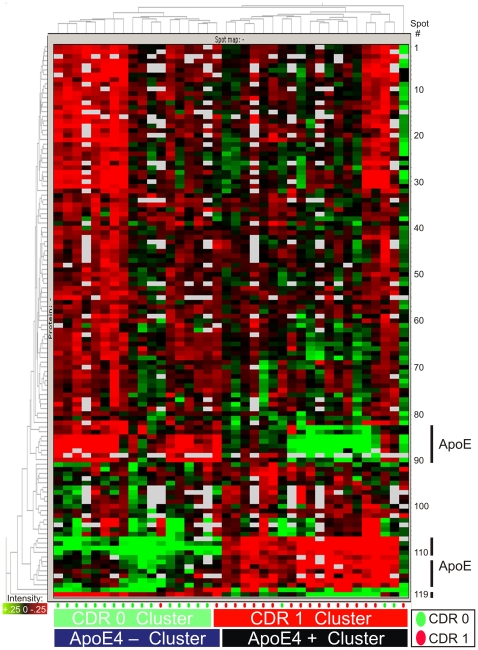
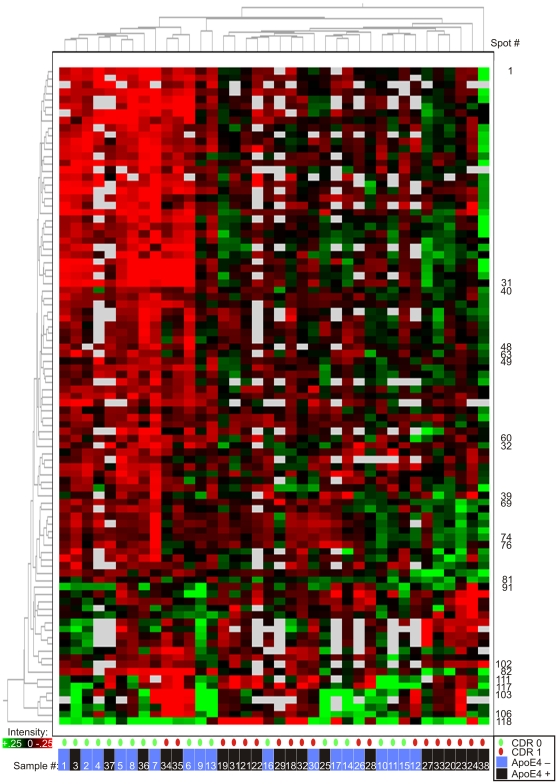
The intensity data from the 119 gel features of interest were subjected to an unsupervised clustering analysis to evaluate their ability to segregate the CDR 0 and CDR 1 samples, and to assess their collective potential as a diagnostic biomarker panel (Figure 2). The ‘heatmap’ generated from this analysis appeared to segregate CDR 0 and CDR 1 individuals (indicated by green and red ovals, respectively) almost completely, with only four participants ‘misclassified.’ However, closer examination revealed an additional layer of segregation on the basis of APOE genotype (indicated by ‘ApoE 4+ Cluster’ and ‘ApoE 4 – Cluster’) which showed perfect sample segregation. Given that the APOE-ε4 allele is a dominant genetic risk factor for AD, some clustering of individuals by APOE genotype might be expected simply from successful segregation of CDR 0 and CDR 1 individuals. However, we hypothesize that the apoE protein exerts a dominant clustering influence through the markedly different electrophoretic profiles of its different isoforms derived from APOE-ε2, APOE-ε3 and APOE- ε4 alleles (illustrated in Figure S1). ApoE was present in 24 of the 119 gel features found to differ in intensity between the CDR groups, and was found to be the primary protein in 12 of these gel features. This heterogeneous electrophoretic mobility of apoE results from the inherent charge differences of the three major apoE isoforms (-E2, -E3, -E4) and the appearance of each isoform as an array of multiple distinct gel features caused by post-translational modifications. These isoform-specific differences are reflected in the prominent red and green clusters, located within the lower third of Figure 2 (corresponding to gel features 83–90, 107–117, and 119), that correlate very closely with participant APOE genotypes. Recognizing this correlation, we hypothesized that APOE genotypes were in large part driving the clustering of participant samples in Figure 2. To test this hypothesis, we performed a second unsupervised clustering analysis, including only those gel features from the initial analysis that did not contain apoE protein (Figure 3). Although this ‘apoE-free’ analysis segregated CDR 1 and CDR 0 groups less completely, it appropriately re-clustered (by CDR status) several samples (#12, 36, 37) that were aberrantly segregated in Figure 2, potentially due to their APOE genotypes. Moreover, clustering of participant samples into APOE genotype subgroups in Figure 3 appears negligible. The underlying benefit of this ‘apoE-free’ analysis is that it reveals the sample-clustering potential of other gel features, which was previously obscured by the inclusion of apoE-containing gel features. As can now be better visualized in Figure 3, gel features appearing within the upper three-fourths of the heatmap appear to show greater intensity in CDR 1 samples; the converse is true of gel features within the lower fourth. It is important to note that measurements of Aβ42 and tau (two proteins measured by ELISA and not detected by 2D-DIGE) were not included in these clustering analyses; because these ‘discovery’ samples were selected for this study on the basis of CSF Aβ42 and tau levels, such inclusion would presumably yield perfect or near-perfect segregation by CDR status in this ‘discovery’ cohort. Therefore, this analysis reflects the potential of these candidate biomarkers to segregate CDR 0 and CDR 1 individuals independent of any contribution from current leading CSF biomarkers Aβ42 and tau. It does not address whether these biomarker candidates might improve upon the utility of Aβ42 and tau, however.
Figure 2. Unsupervised clustering of CSF samples by 2D-DIGE data from the 119 statistically significant gel features.
(Student's t-test, α = 0.05, present in >50% of images). Five gels containing hemoglobin (n = 10 samples) were excluded. Columns represent samples; rows, numbered 1 through 119 from top to bottom, represent gel features depicted in Figure 1. Gel feature intensity is encoded colorimetrically from red (low intensity) to green (high intensity); white indicates absent data. CDR status of individuals at time of CSF collection is encoded below by small green (CDR 0) and red (CDR 1) ovals; CDR 0 and CDR 1 clusters are indicated below by green and red bars, respectively. APOE-ε4 allele status of individuals and groups, alike, is indicated by black (possessing ApoE4 protein, or one or two APOE-ε4 alleles) or blue (possessing no ApoE4 protein, or no APOE-ε4 alleles) bars. Rows representing gel features containing ApoE protein are indicated along the lower right border.
Figure 3. Unsupervised clustering of CSF samples by 2D-DIGE data, excluding gel features containing apoE protein.
All other statistically significant gel features (Student's t-test α = 0.05, present in >50% of images) are retained. As in Figure 2, five gels containing hemoglobin (n = 10 samples) were excluded. Columns represent samples, numbered according to their original positions in Figure 2. Rows represent gel features, numbered as in Figure 2; unlabeled rows are in consecutive order from upper number to lower number, with interruptions in sequence indicated by labels. ApoE-containing features are removed. Gel feature intensity is encoded colorimetrically from red (low intensity) to green (high intensity); white indicates absent data. CDR status of participants at time of CSF collection is encoded below, by small green (CDR 0) and red (CDR 1) ovals. APOE-ε4 status (as described for Figure 2) is indicated by blue (ApoE4 negative) or black (ApoE4 positive) bars, below. Clustering pattern of samples (numbered consecutively in order of appearance in Figure 2, from left to right) relative to Figure 2 is indicated by white numerals, below.
Validation of Candidate Biomarkers by ELISA
Before evaluating a subset of these candidate biomarkers in a larger independent sample set, we first assessed the capacity of protein-specific quantitative ELISAs to detect significant differences between the CDR 0 and CDR 1 groups of the original ‘discovery’ cohort. When possible, to facilitate future reproduction of our findings by other groups and potential translation to clinical use, we applied commercially available ELISA kits.
Of the eleven ELISAs applied to the ‘discovery’ cohort (n = 47, one sample was unavailable for validation), six (NrCAM, YKL-40, chromogranin A, carnosinase I, transthyretin, cystatin C) showed statistically significant or near-significant differences between CDR 0 and CDR 1 groups (Figure 4); five others (PEDF, beta-2 microglobulin, clusterin/apoJ, ceruloplasmin, apoE) did not.
Figure 4. Quantitative ELISAs for 11 biomarker candidates applied to ‘discovery’ cohort CSF samples (n = 47).
Each assay performed in triplicate; mean value reported for each sample. The six assays represented in the upper two rows (A. YKL-40, B. Transthyretin, C. NrCAM, D. Chromogranin A, E. Carnosinase I, and F. Cystatin C) measured differences between CDR 0 and CDR 1 groups (unpaired t-test); the five assays represented in the lower two rows (G. ApoE, H. PEDF, I. Clusterin, J. Ceruloplasmin, K. β-2 microglobulin) did not.
The six ELISAs that measured differences between the CDR 0 and CDR 1 CSF samples of the ‘discovery’ cohort were subsequently applied to a larger, independent set of CSF samples (n = 292) collected from volunteer participants studied by the WU-ADRC. This ‘validation’ cohort included a CDR 0.5 group in addition to CDR 0 and CDR 1 groups, allowing for biomarker assessment in the very early clinical stage of AD. Demographic, clinical, and genetic characteristics of these individuals at time of sample collection are presented in Table 1. Unlike the ‘discovery’ cohort, this ‘validation’ cohort was not preselected on the basis of prior biomarker values (CSF Aβ42 and tau), although assays for CSF Aβ42, tau and p-tau181 were performed.
Because the age and gender compositions differed among the clinical groups of the ‘validation cohort,’ we evaluated each of these 9 biomarkers (six novel candidates, Aβ42, tau, and p-tau181) for age and gender correlations in order to apply covariate analyses appropriately. Correlating with age were tau (r = 0.318, p<0.0001), p-tau181 (r = 0.2216, p<0.001), Aβ42 (r = −0.2334, p<0.0001) and YKL-40 (r = 0.4001, p<0.001); no biomarkers correlated with gender (p>0.05).
As shown in Figure 5, statistically significant differences between clinically defined groups were measured for Aβ42, tau, p-tau181, NrCAM, YKL-40, chromogranin A, and carnosinase I; for transthyretin and cystatin C, non-significant trends were measured. These differences appeared in three patterns: Aβ42 showed a pronounced decrease from CDR 0 to CDR 0.5 and a lesser reduction from CDR 0.5 to CDR 1; tau, p-tau181, and YKL-40 showed increases that were equivalent in CDR 0.5 and CDR 1 relative to CDR 0; NrCAM, chromogranin A, and carnosinase I showed decreases relative to CDR 0 only in CDR 1, and not in CDR 0.5.
Figure 5. Six biomarker candidates and established biomarkers tau, p-tau181 and Aβ42 in ‘validation’ cohort CSF (n = 292).
Each candidate biomarker assay was performed in triplicate, with one mean value reported for each sample; assays for tau, p-tau181 and Aβ42 were performed in duplicate. In addition to A. tau, B. p-tau181 and C. Aβ42 (top row), four assays (D. YKL-40, E. carnosinase I, F. chromogranin A, G. NrCAM) measured statistical differences between clinically defined groups, as indicated; H. transthyretin and I. cystatin C did not reach criterion (α = 0.05) for any comparisons. * p<0.05; * * p<0.01; * * * p< 0.001; * * * * p<0.0001; solid circle p<0.05 by LSD only; double solid circle p<0.05 by unpaired t-test and Mann-Whitney, not by unpaired t-test with Welch's correction.
Diagnostic Utility of Validated Candidate Biomarkers
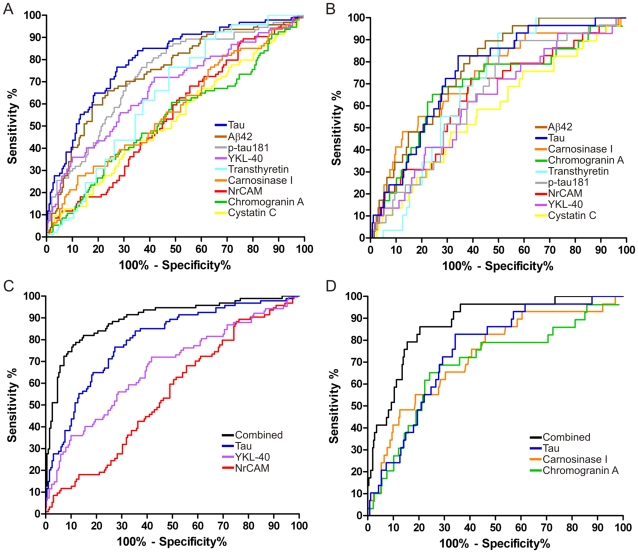
To evaluate and compare the potential of the validated candidate biomarkers and Aβ42, tau, and p-tau181 for identifying either very mild to mild dementia (combined CDR 0.5 and CDR 1) or mild dementia (CDR 1), ROC curves and AUCs were calculated for each biomarker using data from the ‘validation’ cohort (Figure 6A, B, Tables 3, 4). Stepwise logistic regression analyses indicated that, among the nine biomarkers under consideration, YKL-40, NrCAM and tau yielded the highest AUC (0.896) in discriminating cognitive normalcy (CDR 0) from very mild to mild dementia (CDR>0) (Figure 6C, Table 3); for discriminating mild dementia (CDR 1) from CDR<1, carnosinase I, chromogranin A and tau yielded the highest AUC (0.876) (Figure 6D, Table 4).
Figure 6. Receiver Operating Characteristic (ROC) curves of ELISA data from ‘validation’ cohort.
Simple ROC analyses were performed for each biomarker to distinguish A. CDR>0 from CDR 0 (“earlier diagnosis”) and B. CDR 1 from CDR<1 (“early diagnosis”). Stepwise logistic regression models were used to identify combinations of these biomarkers that would distinguish C. CDR>0 from CDR 0 (“earlier diagnosis”), AUC = 0.90 and D. CDR 1 from CDR<1 (“early diagnosis”), AUC = 0.88.
Table 3. Receiver Operating Characteristic Curve Areas for CDR 0 vs <0 Comparison.
| Biomarker | Area Under Curve | Standard Error | 95% Confidence Interval |
| Tau | 0.8004 | 0.0279 | 0.7457–0.8551 |
| Aβ42 | 0.7429 | 0.0315 | 0.6812–0.8046 |
| p-tau181 | 0.7339 | 0.0315 | 0.6721–0.7956 |
| YKL-40 | 0.6717 | 0.0349 | 0.6033–0.7401 |
| Transthyretin | 0.6190 | 0.0331 | 0.5541–0.6838 |
| Carnosinase I | 0.5735 | 0.0365 | 0.5020–0.6450 |
| NrCAM | 0.5422 | 0.0355 | 0.4726–0.6118 |
| Chromogranin A | 0.5303 | 0.0373 | 0.4572–0.6034 |
| Cystatin C | 0.5297 | 0.0366 | 0.4579–0.6014 |
| Logistic Regression | 0.8955 | 0.0212 | 0.8539–0.9372 |
ROC analyses of ‘validation’ cohort ELISA data were performed for each biomarker to distinguish CDR>0 from CDR 0 (“earlier diagnosis”). A stepwise logistic regression model, applied to identify a complementary combination of these biomarkers that would optimize accuracy (maximize area under the curve [AUC]) without including additional non-contributory biomarkers, accepted tau, YKL-40 and NrCAM and yielded an AUC of 0.8955 (“Logistic Regression,” lowest row).
Table 4. Receiver Operating Characteristic Curve Areas for CDR 1 vs <1 Comparison.
| Biomarker | Area Under Curve | Standard Error | 95% Confidence Interval |
| Aβ42 | 0.7690 | 0.0376 | 0.6953–0.8427 |
| Tau | 0.7502 | 0.0420 | 0.6679–0.8325 |
| Carnosinase I | 0.7277 | 0.0512 | 0.6273–0.8281 |
| Chromogranin A | 0.6879 | 0.0566 | 0.5771–0.7988 |
| Transthyretin | 0.6605 | 0.0380 | 0.5860–0.7350 |
| p-tau181 | 0.6512 | 0.0483 | 0.5566–0.7458 |
| NrCAM | 0.6411 | 0.0553 | 0.5326–0.7495 |
| YKL-40 | 0.6271 | 0.0532 | 0.5228–0.7313 |
| Cystatin C | 0.5752 | 0.0565 | 0.4645–0.6858 |
| Logistic Regression | 0.8762 | 0.0314 | 0.8147–0.9377 |
ROC analyses of ‘validation’ cohort ELISA data were performed for each biomarker to distinguish CDR 1 from CDR<1 (“early diagnosis”). A stepwise logistic regression model, applied to identify a complementary combination of these biomarkers that would optimize accuracy (maximize area under the curve [AUC]) without including additional non-contributory biomarkers, accepted tau, carnosinase I and chromogranin A, yielding an AUC of 0.8762 (“Logistic Regression,” lowest row).
Discussion
Using an unbiased proteomics approach (2D-DIGE LC-MS/MS), this study identified 47 novel candidate CSF protein biomarkers for early AD. Subsequently, by evaluating a subset of these candidate biomarkers by ELISA, this study validated the utility of four candidate biomarkers for distinguishing groups with mild, very mild, or no dementia (CDR 1, 0.5, 0, respectively). Further statistical analyses demonstrated that these biomarkers could improve the accuracy of ‘established’ biomarkers Aβ42 and tau for the diagnosis of early AD.
The results from the 2D-DIGE LC-MS/MS portion of this study suggest that many of the recognized neuropathological changes of AD are represented by changes in the CSF proteome. Most of the 47 candidate biomarker proteins identified in this study can be placed into structural and/or functional categories (e.g. synaptic adhesion, synaptic function, dense core synaptic vesicle proteins, inflammation/complement, protease activity/inhibition, apolipoproteins, etc.) associated with accepted neuropathophysiological changes in AD (Table 5). Unsupervised clustering analyses of these 2D-DIGE data, performed without the influence of CSF Aβ42, tau, p-tau181 and APOE genotype, additionally suggest that these biomarker candidates collectively show utility for discriminating groups with and without mild DAT (Figure 3).
Table 5. Candidate CSF biomarkers reflect AD-related pathophysiologic changes.
| Functional/Structural Category | Protein | References |
| Adhesion molecules | N-Cadherin | [39]-[45] |
| NrCAM | [19], [46]-[49] | |
| Calsyntenin | [47], [50]-[53] | |
| Neuronal Pentraxin Receptor | [47], [54] | |
| Brain Associated Small Cell Lung Cancer Antigen (NCAM-140/CD56) | [55] | |
| Nectin-like molecule-1/TSLL1/SynCam3 | [56]-[58] | |
| Dense core vesicles | Chromogranin A | [19], [20], [22], [59]-[62] |
| Chromogranin B | [60], [62] | |
| Secretogranin II | [60]-[63] | |
| Secretogranin III | [59], [64], [65] | |
| VGF NGF Inducible precursor | [20], [22], [23], [66]-[69] | |
| Carboxypeptidase E | [70]-[75] | |
| Synaptic/Neuronal metabolism | Aspartate aminotransferase I | [76]-[82] |
| Synaptic Function | S100A1 | [83] |
| Neuronal Pentraxin Receptor | [27], [47], [54] | |
| Brain Acetylcholinesterase Putative Membrane Anchor (CutA1) | [84], [85] | |
| Calsyntenin | [47], [50]-[53] | |
| Neuroprotection | PEDF (Serpin-F1) | [86]-[96] |
| Annexin I | [97]-[99] | |
| Prosaposin | [20], [100]-[103] | |
| Secretogranin II | [104]-[106] | |
| Carnosinase I | [33], [107]-[111] | |
| Extracellular superoxide dismutase (SOD3) | [112]-[114] | |
| Apoptosis/Actin remodeling | Gelsolin | [115]-[121] |
| Prk-1 (PKN) | [122]-[126] | |
| Synaptic plasticity/Learning and memory | VGF NGF inducible precursor | [20], [22], [23], [66]-[69] |
| NrCAM | [19], [46]-[49] | |
| β3GnT1 | [49], [127], [128] | |
| Carnosinase I | [33], [107]-[111] | |
| Carbonic Anhydrase IV | [129]-[131] | |
| S100A1 | [132] | |
| Carboxypeptidase E | [70]-[75] | |
| Calmodulin | [133]-[136] | |
| Extracellular superoxide dismutase (SOD3) | [114] | |
| Inflammation/Complement | *YKL-40/Chitinase 3-Like 1 | [137]-[148] |
| PEDF (Serpin-F1) | [86]-[96] | |
| Annexin I | [97]-[99] | |
| IHRP/ITIH4 | [149], [150] | |
| Vitronectin | [151]-[155] | |
| *Complement C4B3 | [156]-[161] | |
| Kininogen I | [162], [163] | |
| Chromogranin A | [19], [20], [22], [59]-[62] | |
| Secretogranin III | [59], [64], [65] | |
| Apolipoprotein J | [27], [152], [156], [157], [164]-[167] | |
| Beta 2-microglobulin | [168]-[171] | |
| Extracellular superoxide dismutase (SOD3) | [172] | |
| Prostaglandin metabolism | *Prostaglandin H2 D Isomerase/Beta-trace | [162], [173]-[175] |
| Amyloid beta peptide binding/Amyloidogenesis | *Apolipoprotein A1 (proapolipoprotein) | [176], [177] |
| Apolipoprotein E | [178] | |
| Apolipoprotein J | [27], [152], [156], [157], [164]-[167] | |
| Transthyretin | [87], [173], [175], [179]-[184] | |
| Gelsolin | [115]-[121] | |
| Vitronectin | [151]-[155] | |
| Cystatin C | [22], [173], [185]-[188] | |
| *Prostaglandin H2 D Isomerase/Beta-trace | [162], [173]-[175] | |
| *α-2-macroglobulin | [19], [189]-[194] | |
| *α-1-antichymotrypsin | [33], [195]-[199] | |
| Protease activity | *α-1-antichymotrypsin | [33], [195]-[199] |
| *α-2-macroglobulin | [19], [189]-[194] | |
| Cystatin C | [22], [173], [185]-[188] | |
| Carboxypeptidase E | [70]-[75] | |
| Matrix proteins | Fibulin 3 (EFEMP1) | [200]-[202] |
| Vitronectin | [151]-[155] | |
| Phospholipase activity | Annexin I (Lipocortin) | [97]-[99] |
| Prosaposin | [20], [100]-[103] | |
| Apolipoproteins | *Apolipoprotein A1 (proapolipoprotein) | [24], [165], [166], [176], [177], [203], [204] |
| Apolipoprotein CII | [25], [166], [205] | |
| Apolipoprotein CIII | [25], [206], [207] | |
| Apolipoprotein E | [24], [27], [165], [204] | |
| Apolipoprotein J | [27], [152], [156], [157], [164]-[167] | |
| *Apolipoprotein H | [19], [25], [165], [208], [209] | |
| Calcium binding/homeostasis | Calmodulin | [134]-[136] |
| S100A1 | [83], [210] | |
| Annexin I (Lipocortin) | [97]-[99] | |
| Calsyntenin | [47], [50]-[53] | |
| Gelsolin | [115]-[121] | |
| Metal (Copper and Iron) Binding | Carnosinase I | [33], [107]-[111] |
| Ceruloplasmin | [211]-[217] | |
| Brain Acetylcholinesterase Putative Membrane Anchor (CutA1) | [84], [85] | |
| Chaperone complex/activity | S100A1 | [218] |
| Transthyretin (prealbumin) | [24], [87], [173], [175], [179]-[184] | |
| Endoplasmic Reticulum - Associated Degradation | Man9-mannosidase | [219]-[221] |
| Extracellular and Intraneuronal pH | Carbonic Anhydrase IV | [129]-[131] |
| Carnosinase I | [33], [107]-[111] | |
| Glycobiology (lactosamine synthesis) | β3GnT1 | [49], [127], [128] |
| Hemodynamics | Angiotensinogen | [172], [222] |
| Extracellular superoxide dismutase (SOD3) | [172] | |
| Thyroid hormone transport | Transthyretin (prealbumin) | [24], [87], [173], [175], [179]-[184] |
| Unknown | Hypothetical protein |
CSF biomarkers are grouped according to reported function(s) and, when appropriate, cellular locations. Asterisks (*) indicate those biomarkers found to be increased in AD CSF; the vast majority were decreased.
In the second phase of this study, designed to measure a subset of candidate biomarker proteins in two independent sample sets by ELISA, four of the eleven candidate biomarkers that were tested showed capacity to distinguish clinical groups. However, seven candidate biomarkers did not show statistically significant differences between clinical groups in either the smaller ‘discovery’ cohort or the larger ‘validation’ cohort. Superficially, this ‘failure rate’ might cast doubt on the list of candidate biomarkers identified through 2D-DIGE. However, it is important to note that 2D-DIGE is sensitive to changes in concentrations of minor protein isoforms and post-translational modifications that may not significantly alter the global concentrations of a ‘parent’ protein, which would be measured by ELISA. Therefore, it is not surprising that some of the candidate biomarker ELISAs did not replicate the findings from 2D-DIGE. Transthyretin provides a prime example: all of the significant gel-features ascribed to transthyretin (gel features # 20, 52, 57, 58, 60, 77, 78, 79, 84, 87, 110, 115; Table 2) showed unusual electrophoretic patterns and were dwarfed by the canonical transthyretin gel features that did not individually show statistical differences (Figure 1). In fact, whereas most of the significant transthyretin 2D-DIGE gel features were decreased in AD, the global transthyretin levels measured by ELISA in the ‘discovery’ and ‘validation’ cohorts were actually mildly increased in groups with cognitive impairment (CDR>0) relative to those without (CDR 0) (Figures 4 and 5). To measure the sub-species of transthyretin that were identified by 2D-DIGE as decreasing in AD will require assays that specifically target relevant post-translational modifications and exclude other forms of transthyretin. Similarly, other 2D-DIGE biomarker candidates may also require specifically tailored assays for accurate, high-throughput measurement.
Nevertheless, four candidate biomarkers were successfully validated in both cohorts, and two others showed non-significant trends by ELISA in the larger ‘validation’ cohort (Figure 5). This larger cohort represented three different cognitive stages: normalcy, very mild dementia, and mild dementia (CDR 0, CDR 0.5, CDR 1, respectively), and revealed different patterns of CSF biomarker levels, vis-a-vis cognitive status. The CSF concentration of YKL-40, an astrocytic marker of plaque-associated neuroinflammation [137]–[148], is increased by the very earliest stage of clinical disease (CDR 0.5). Transthyretin [24], [87], [173], [175], [179]–[184] and cystatin C [22], [173], [185]–[188], two proteins with neuroprotective qualities that are implicated in preventing amyloidogenesis of Aβ peptide, show a similar pattern. In contrast, the concentrations of NrCAM, a synaptic adhesion molecule [19], [46]–[49], chromogranin A, a dense core synaptic vesicle protein [19], [20], [22], [59]–[62], and carnosinase I, a neuronal dipeptidase responsible for degradation of the anti-oxidant and metal-chelating dipeptide carnosine [33], [107]–[111] do not decline until mild dementia ensues (CDR 1).
Like the current leading CSF biomarkers for AD (Aβ42, tau and p-tau181), all of these biomarker candidates show ranges with substantial overlap between clinically defined groups. This issue of overlapping values, common among candidate AD CSF biomarkers reported to date, suggests that any one biomarker will be insufficient to accurately identify early AD, and that an ensemble of complementary biomarkers will be required to provide adequate sensitivity and specificity. Therefore, to identify an optimal combination of these biomarkers that can distinguish the early clinical stages of AD from cognitive normalcy, we applied stepwise logistic regression analyses to the ELISA data from our ‘validation’ cohort (Figure 6, Tables 3 and 4). These analyses suggest that four candidate AD biomarkers (YKL-40, NrCAM, chromogranin A, carnosinase I) can improve the ability of tau to classify individuals into CDR 0, CDR 0.5 and CDR 1 groups with appreciable accuracy.
It may appear counter-intuitive that Aβ42 and p-tau181, which individually discriminate very mild AD and mild AD from cognitively normal groups quite well, were not incorporated into either ‘optimal’ biomarker panel by the stepwise logistic regression analyses. Likewise, NrCAM was included in the optimal CDR 0 vs CDR>0 biomarker panel (AUC 0.896) even though its mean levels did not independently show a statistical difference between CDR 0 and CDR>0 groups. In considering this outcome, it may be worth noting that if NrCAM, transthyretin, chromogranin and cystatin C are removed from consideration, the stepwise logistic regression model for the CDR 0 vs CDR>0 comparison yields an ‘optimal’ biomarker panel that includes only tau, Aβ42 and carnosinase I, with an AUC of 0.849 (not shown). In this restricted analysis, the paired contribution of Aβ42 and carnosinase I to tau is apparently greater than that of YKL-40. These analyses illustrate how ‘unpredictable’ and context-dependent optimal biomarker combinations can be, and suggest that biomarker complementarity may be more important to consider than each biomarker's independent performance, when choosing a biomarker panel. Of course, it will be necessary to replicate these findings in additional independent cohorts. It will also be essential to evaluate a greater number of candidate biomarkers in similar fashion, in order to construct a biomarker panel with even greater accuracy.
Another worthwhile feature to consider when evaluating and selecting CSF biomarkers is relative concentration in the blood (plasma, serum), because biomarker measurements in CSF can be artifactually influenced by subtle blood contamination at the time of lumbar puncture; from this perspective, ideal CSF biomarkers show CSF concentrations that are equal to or greater than those in blood. An additional reason to assess plasma/serum concentrations of candidate CSF biomarkers is to determine if venipuncture, which is more easily performed than lumbar puncture, might yield equivalent information. Among the six CSF biomarkers identified by stepwise logistic regression analysis in the current study, Aβ42 and tau [8]–[11], YKL-40 [137], and chromogranin A [223] show higher levels in CSF than in plasma; carnosinase I levels appear similar in CSF and serum [110]; NrCAM levels appear higher in serum than in CSF, although the forms of NrCAM present in these fluids may differ [224]. Concerning independent utility as biomarkers for AD, only plasma YKL-40 and serum NrCAM have shown promise [137], [225], albeit inferior to that of CSF YKL-40 and NrCAM demonstrated here. Plasma tau concentrations in AD and controls are below the level of detection of the most commonly used tau assays, and plasma Aβ42 [8]–[11] and plasma chromogranin A (R.Perrin et al., unpublished data) concentrations show no significant differences among CDR groups. Serum carnosinase activity likewise has not shown significant differences between AD and controls in one small study [111], though a difference between AD and mixed dementia (including vascular dementia) has been reported [111]. To our knowledge, an evaluation of plasma or serum carnosinase I concentrations in the context of AD has not yet been performed or reported. Further assessment of the potential of these and other proteins as candidate AD biomarkers in plasma or serum, complete with evaluation of their performance as ensembles, remains an important task for future studies. Currently, however, this panel of six biomarkers appears likely to show much greater promise in its application to CSF.
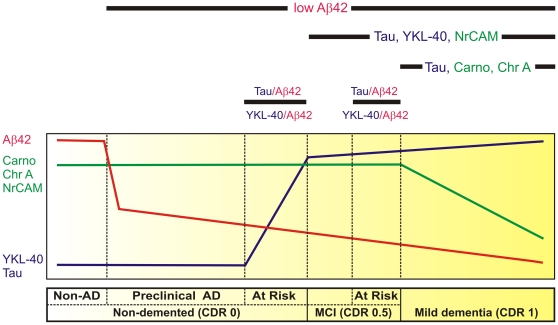
Indeed, by providing proof of concept, this study outlines a scheme to categorize the early stages of AD using CSF protein biomarkers that reflect established features of the pathophysiological evolution of the disease (Figure 7). Building upon previous findings that low CSF Aβ42 can identify cognitively normal individuals with plaques (preclinical AD) [8], [11], and that tau/Aβ42 and YKL-40/Aβ42 ratios can predict risk of developing cognitive impairment [9], [15], [137], this minimal panel of six CSF biomarkers (YKL-40, NrCAM, chromogranin A, carnosinase I, tau and Aβ42) begins to segregate individuals into six clinicopathological categories: normal cognition without amyloid plaques, normal cognition with amyloid plaques (preclinical AD), normal cognition at increased risk to develop dementia (converters), very mild dementia (CDR 0.5), very mild dementia at increased risk for progression, and mild dementia (CDR 1) (Figure 7).
Figure 7. Hypothetical model defines early stages of AD by temporal pattern of CSF protein biomarker levels.
The horizontal bar (below) describes the early clinicopathological progression from cognitive normalcy without AD pathology (‘Non-AD’) to mild dementia in six stages. As depicted by the curves above, Non-AD CSF has high Aβ42 (red line), high chromogranin A (Chr A), carnosinase I (Carno I) and NrCAM (green line), and low YKL-40 and tau (blue line). Reduced CSF Aβ42 correlates with amyloid plaque deposits, the first sign of neuropathologically identifiable AD (‘preclinical AD’) [8]. CSF Aβ42 appears to decrease further as cognition declines from normal (Clinical Dementia Rating [CDR] 0) to very mild cognitive impairment (MCI, CDR 0.5) to mild dementia (CDR 1). When considered as ratios with Aβ42, CSF markers of neuroinflammation (e.g. YKL-40) and neurofibrillary tangle pathology (e.g. tau) appear to increase before and predict the onset of very mild cognitive impairment (MCI, CDR 0.5), defining a CDR 0 group ‘At Risk’ for cognitive decline [9], [15], [137]; YKL-40 and tau also appear to be higher among those who progress rapidly from very mild to mild dementia, defining a CDR 0.5 group ‘At Risk’ for impending cognitive decline [137], [230]. Reductions in synapse-associated (NrCAM, chromogranin A) and neuronal (carnosinase I) proteins, and increases in YKL-40 and tau mirror the progression and anatomical spread of synaptic and neuronal losses, gliosis and tau pathology associated with cognitive decline, and can be used to define CDR 0.5 and CDR 1.
We acknowledge that this minimal panel of biomarkers currently has insufficient sensitivity and specificity for clinical application, particularly because it has not been fully evaluated for its ability to discriminate AD from non-AD causes of dementia (although Aβ42, p-tau181, tau, and specific fragments of chromogranin A and cystatin C have shown some ability to distinguish AD from frontotemporal lobar degeneration [FTLD]) [22], [226], [227]. The incorporation of additional biomarkers that are likely to discriminate early AD from cognitive normalcy, such as those identified in the first phase of this study, or other biomarkers that have already shown promise for distinguishing AD from other leading causes of dementia (e.g. agouti related peptide, eotaxin-3, and hepatocyte growth factor [19], complement C3a des-arg and integral membrane protein 2B CT [22], for FTLDs; and alpha-synuclein [228], apoH and vitamin D binding protein [25] for Lewy body disorders), would likely improve the panel's diagnostic utility. However, even in its current form, this initial panel might show value if applied in the context of clinical trial design, wherein simple enrichment of study populations for characteristics of interest would increase efficiency and power and reduce duration and cost. A biomarker panel like this one might also allow clinical trials to evaluate stage-specific responses to treatment, which may differ. Finally, because most of these biomarkers reflect underlying pathological changes in real time, it is appealing to speculate that these biomarkers may have additional utility for evaluating clinically imperceptible treatment responses (as in [229]) and for monitoring neuropathological – rather than cognitive – decline.
Supporting Information
ApoE protein isoforms appear in different gel features on 2D-DIGE. Overlays of fluorescent 2D-DIGE images from gels representing CSF from two individuals with homozygosity for APOE-ε2 (green) or APOE-ε3 (red) (panel A) and for APOE-ε3 (green) or APOE-ε4 (red) (panel B) illustrate the heterogeneity of signal distribution by isoelectric point and molecular weight among apoE protein isoforms derived from different alleles. In panels C, D, E, F, G, H, signal intensities of individual CSF samples, grouped by genotype (2/2, 3/3 and 4/4 represent homozygotes; 2/3, 3/4 represent heterozygotes) are indicated for six apoE gel features (labeled C, D, E, F, G, H in panels A and B), illustrating that gel features C and D represent apoE2; gel feature E represents multiple forms; gel feature F represents apoE3; and gel features G and H, apoE4.
(TIF)
Mass spectrometry and protein identification data for 2D-DIGE gel features that differ in AD CSF. Results are ordered sequentially by “heat map #” [column A], corresponding to the ‘heat map’ row numbers in Figure 2. “Spot” [column B] refers to BVA number (see Methods). “(Accession) primary protein name” [column C] provides the gi number and protein name from the NCBI database. “Protein molecular weight” [column D] is the gene product molecular weight in Daltons. “Protein score” [column E] is the MASCOT-generated protein score. “Protein ID probability” [column F] indicates Scaffold's percent probability that the protein identification is correct. “Spectral count” [column G] is the number of spectra assigned to the protein by Scaffold. “Unique proteins” [column H] refers to the number of recognized tryptic peptides attributed to the protein by MASCOT. “Peptide sequence” [column I] indicates the amino acid sequence of the tryptic peptide predicted by MASCOT. “MASCOT ion score” [column J] is the MASCOT quality assessment of the peptide sequence assignment. “M/Z (observed)” [column K] is mass/charge ratio. “Mass (observed)” [column L] of peptide is indicated in Daltons. “Mass (theoretical)” [column M] is idealized tryptic peptide mass as predicted by NCBI. “Mass error (ppm)” [column N] is the error in parts per million determined through comparison of theoretical peptide mass to data generated by mass spectrometry. “MS source” [column O] reflects the mass spectrometer that produced the observed data (Q-STAR or LTQ-FT). “Modifications” [column P] lists variable post-translational modifications identified by mass spectrometry peptide sequence analysis.
(XLS)
Acknowledgments
The authors would like to express our appreciation to the Biomarker Core, Clinical Core, Data Management and Statistics Core, Genetics Core, lumbar puncture physicians, and volunteer participants of the Knight ADRC of Washington University in St. Louis, and to the volunteer participants of the University of Washington, the Oregon Health and Science University, the University of Pennsylvania, and the University of California San Diego.
Footnotes
Competing Interests: D.M.H. co-founded C2N Diagnostics, is on the scientific advisory board of C2N Diagnostics, En Vivo, and Satori, consulted for Pfizer, and receives grants that did not support this work from Eli Lilly, Pfizer, and Astra-Zeneca. J.C.M. has consulted for Astra Zeneca, Genentech, and Merck, has received honoraria from ANA Soriano lecture, payment from Journal Watch for preparation of other manuscripts, royalties from Blackwell Publishers and Taylor and Francis, and travel funding for ANA meeting Baltimore. C.M.C. became emeritus at University of Pennsylvania on January 1, 2010, and now works for Avid Radiopharmaceuticals, which did not provide any funding for this study, and did not have any involvement or influence in data production, data analysis, decision to publish, or manuscript preparation. None of the above stated competing interests alter our adherence to all the PLoS ONE policies on sharing data and materials.
Funding: National Alzheimer's Coordinating Center (NACC) Grant U01 AG16976 (A.M.F.); P50 AG05681 (C.M.R., E.A.G.); P01 AG026276 (C.M.R., E.A.G.); P50 AG05136; P01 AG03991(J.C.M.); P30 NS057105 (D.M.H.); UL1 RR024992 (R.R.T.; C.M.R.); W.M. Keck Foundation (R.R.T.); P41 RR00954 (R.R.T); Dept. of Veterans Affairs (E.R.P.); AG020020 (G.L.); AG23185 (D.R.G.); AG08017 (J.F.Q.); AG08017 (J.A.K.); AG10124 (C.M.C); T32 NS007205 (R.J.P.); and the Charles and Joanne Knight Alzheimer Research Initiative (A.M.F.); Avid Radiopharmaceuticals (C.M.C., employment unrelated to this study). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Braak H, Braak E. Frequency of stages of Alzheimer-related lesions in different age categories. Neurobiol Aging. 1997;18:351–357. doi: 10.1016/s0197-4580(97)00056-0. [DOI] [PubMed] [Google Scholar]
- 2.Morris J, Price J. Pathologic correlates of nondemented aging, mild cognitive impairment, and early stage Alzheimer's disease. J Mol Neurosci. 2001;17:101–118. doi: 10.1385/jmn:17:2:101. [DOI] [PubMed] [Google Scholar]
- 3.Price J, Ko A, Wade M, Tsou S, McKeel D, et al. Neuron number in the entorhinal cortex and CA1 in preclinical Alzheimer's disease. Arch Neurol. 2001;58:1395–1402. doi: 10.1001/archneur.58.9.1395. [DOI] [PubMed] [Google Scholar]
- 4.Barnes LL, Schneider JA, Boyle PA, Bienias JL, Bennett DA. Memory complaints are related to Alzheimer disease pathology in older persons. Neurology. 2006;67:1581–1585. doi: 10.1212/01.wnl.0000242734.16663.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Markesbery W, Schmitt F, Kryscio R, Davis D, Smith C, et al. Neuropathologic substrate of Mild Cognitive Impairment. Arch Neurol. 2006;63:38–46. doi: 10.1001/archneur.63.1.38. [DOI] [PubMed] [Google Scholar]
- 6.Motter R, Vigo-Pelfrey C, Kholodenko D, Barbour R, Johnson-Wood J, et al. Reduction of β-amyloid peptide42 in the cerebrospinal fluid of patients with Alzheimer's disease. Ann Neurol. 1995;38:643–648. doi: 10.1002/ana.410380413. [DOI] [PubMed] [Google Scholar]
- 7.Sunderland T, Linker G, Mirza N, Putnam K, Friedman D, et al. Decreased β-amyloid1-42 and increased tau levels in cerebrospinal fluid of patients with Alzheimer's disease. JAMA. 2003;289:2094–2103. doi: 10.1001/jama.289.16.2094. [DOI] [PubMed] [Google Scholar]
- 8.Fagan A, Mintun M, Mach R, Lee S-Y, Dence C, et al. Inverse relation between in vivo amyloid imaging load and CSF Aβ42 in humans. Ann Neurol. 2006;59:512–519. doi: 10.1002/ana.20730. [DOI] [PubMed] [Google Scholar]
- 9.Fagan A, Roe C, Xiong C, Mintun M, Morris J, et al. Cerebrospinal fluid tau/Aβ42 ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 2007;64:343–349. doi: 10.1001/archneur.64.3.noc60123. [DOI] [PubMed] [Google Scholar]
- 10.Fagan AM, Head D, Shah AR, Marcus D, Mintun M, et al. Decreased cerebrospinal fluid Abeta(42) correlates with brain atrophy in cognitively normal elderly. Ann Neurol. 2009;65:176–183. doi: 10.1002/ana.21559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fagan AM, Mintun MA, Shah AR, Aldea P, Roe CM, et al. Cerebrospinal fluid tau and ptau(181) increase with cortical amyloid deposition in cognitively normal individuals: implications for future clinical trials of Alzheimer's disease. EMBO Mol Med. 2009;1:371–380. doi: 10.1002/emmm.200900048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tolboom N, van der Flier WM, Yaqub M, Boellaard R, Verwey NA, et al. Relationship of cerebrospinal fluid markers to 11C-PiB and 18F-FDDNP binding. J Nucl Med. 2009;50:1464–1470. doi: 10.2967/jnumed.109.064360. [DOI] [PubMed] [Google Scholar]
- 13.Grimmer T, Riemenschneider M, Forstl H, Henriksen G, Klunk WE, et al. Beta amyloid in Alzheimer's disease: increased deposition in brain is reflected in reduced concentration in cerebrospinal fluid. Biol Psychiatry. 2009;65:927–934. doi: 10.1016/j.biopsych.2009.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, et al. Relationships between biomarkers in aging and dementia. Neurology. 2009;73:1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li G, Sokal I, Quinn J, Leverenz J, Brodey M, et al. CSF tau/Aβ42 ratio for increased risk of mild cognitive impairment: A follow-up study. Neurology. 2007;69:631–639. doi: 10.1212/01.wnl.0000267428.62582.aa. [DOI] [PubMed] [Google Scholar]
- 16.Snider BJ, Fagan AM, Roe C, Shah AR, Grant EA, et al. Cerebrospinal fluid biomarkers and rate of cognitive decline in very mild dementia of the Alzheimer type. Arch Neurol. 2009;66:638–645. doi: 10.1001/archneurol.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang J, Goodlett DR, Quinn JF, Peskind E, Kaye JA, et al. Quantitative proteomics of cerebrospinal fluid from patients with Alzheimer disease. J Alzheimers Dis. 2005;7:125–133; discussion 173–180. doi: 10.3233/jad-2005-7205. [DOI] [PubMed] [Google Scholar]
- 18.Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 19.Hu WT, Chen-Plotkin A, Arnold SE, Grossman M, Clark CM, et al. Novel CSF biomarkers for Alzheimer's disease and mild cognitive impairment. Acta Neuropathol. 2010;119:669–678. doi: 10.1007/s00401-010-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonsen AH, McGuire J, Podust VN, Davies H, Minthon L, et al. Identification of a novel panel of cerebrospinal fluid biomarkers for Alzheimer's disease. Neurobiol Aging. 2008;29:961–968. doi: 10.1016/j.neurobiolaging.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Simonsen A, McGuire J, Hansson O, Zetterberg H, Podust V, et al. Novel panel of cerebrospinal fluid biomarkers for the prediction of progression to Alzheimer dementia in patients with mild cognitive impairment. Arch Neurol. 2007;64:366–370. doi: 10.1001/archneur.64.3.366. [DOI] [PubMed] [Google Scholar]
- 22.Simonsen AH, McGuire J, Podust VN, Hagnelius NO, Nilsson TK, et al. A novel panel of cerebrospinal fluid biomarkers for the differential diagnosis of Alzheimer's disease versus normal aging and frontotemporal dementia. Dement Geriatr Cogn Disord. 2007;24:434–440. doi: 10.1159/000110576. [DOI] [PubMed] [Google Scholar]
- 23.Carrette O, Demalte I, Scherl A, Yalkinoglu O, Corthals G, et al. A panel of cerebrospinal fluid potential biomarkers for the diagnosis of Alzheimer's disease. Proteomics. 2003;3:1486–1494. doi: 10.1002/pmic.200300470. [DOI] [PubMed] [Google Scholar]
- 24.Davidsson P, Westman-Brinkmalm A, Nilsson CL, Lindbjer M, Paulson L, et al. Proteome analysis of cerebrospinal fluid proteins in Alzheimer patients. Neuroreport. 2002;13:611–615. doi: 10.1097/00001756-200204160-00015. [DOI] [PubMed] [Google Scholar]
- 25.Abdi F, Quinn J, Jankovic J, McIntosh M, Leverenz J, et al. Detection of biomarkers with a multiplex quantitative proteomic platform in cerebrospinal fluid of patients with neurodegenerative disorders. J Alzheimers Dis. 2006;9:293–348. doi: 10.3233/jad-2006-9309. [DOI] [PubMed] [Google Scholar]
- 26.Choe L DAM, Relkin NR, Pappin D, Ross P, Williamson B, Guertin S, Pribil P, Lee KH. 8-plex quantitation of changes in cerebrospinal fluid protein expression in subjects undergoing intravenous immunoglobulin treatment for Alzheimer's disease. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Finehout EJ, Franck Z, Choe LH, Relkin N, Lee KH. Cerebrospinal fluid proteomic biomarkers for Alzheimer's disease. Ann Neurol. 2007;61:120–129. doi: 10.1002/ana.21038. [DOI] [PubMed] [Google Scholar]
- 28.McKhann G, Drachman D, Folstein M, Katzman R, Price D, et al. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 29.Morris JC. The Clinical Dementia Rating (CDR). Current version and scoring rules. Neurology. 1993;43:2412–2414. doi: 10.1212/wnl.43.11.2412-a. [DOI] [PubMed] [Google Scholar]
- 30.Suzuki N, Cheung TT, Cai XD, Odaka A, Otvos L, Jr, et al. An increased percentage of long amyloid beta protein secreted by familial amyloid beta protein precursor (beta APP717) mutants. Science. 1994;264:1336–1340. doi: 10.1126/science.8191290. [DOI] [PubMed] [Google Scholar]
- 31.Storandt M, Grant E, Miller J, Morris J. Longitudinal course and neuropathologic outcomes in original vs revised MCI and in pre-MCI. Neurology. 2006;67:467–473. doi: 10.1212/01.wnl.0000228231.26111.6e. [DOI] [PubMed] [Google Scholar]
- 32.Hu Y, Malone J, Fagan A, Townsend R, Holtzman D. Comparative proteomic analysis of intra- and interindividual variation in human cerebrospinal fluid. Mol & Cell Proteom. 2005;4:2000–2009. doi: 10.1074/mcp.M500207-MCP200. [DOI] [PubMed] [Google Scholar]
- 33.Hu Y, Hosseini A, Kauwe J, Gross J, Cairns N, et al. Identification and validation of novel CSF biomarkers for early stages of Alzheimer's disease. Proteomics - Clin Appl. 2007;1:1373–1384. doi: 10.1002/prca.200600999. [DOI] [PubMed] [Google Scholar]
- 34.Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, et al. A novel experimental design for comparative two-dimensional gel analysis: two-dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- 35.Havlis J TH, Sebela M, Shevchenko A. Fast-response proteomics by accelerated in-gel digestion of proteins. Anal Chem. 2003;75:1300–1306. doi: 10.1021/ac026136s. [DOI] [PubMed] [Google Scholar]
- 36.King J, Gross J, Lovly C, Rohrs H, Piwnica-Worms H, et al. Accurate mass-driven analysis for the characterization of protein phosphorylation. Study of the human chk2 protein kinase Anal Chem. 2006;78:2171–2181. doi: 10.1021/ac051520l. [DOI] [PubMed] [Google Scholar]
- 37.Bredemeyer A, Lewis R, Malone J, Davis A, Gross J, et al. A proteomic approach for the discovery of protease substrates. Proc Natl Acad Sci USA. 2004;101:11785–11790. doi: 10.1073/pnas.0402353101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 39.Uemura K, Lill CM, Banks M, Asada M, Aoyagi N, et al. N-cadherin-based adhesion enhances Abeta release and decreases Abeta42/40 ratio. J Neurochem. 2009;108:350–360. doi: 10.1111/j.1471-4159.2008.05760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mysore SP, Tai CY, Schuman EM. Effects of N-cadherin disruption on spine morphological dynamics. Front Cell Neurosci. 2007;1:1. doi: 10.3389/neuro.03.001.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bekirov IH NV, Svoronos A, Huntley GW, Benson DL. Cadherin-8 and N-cadherin differentially regulate pre- and postsynaptic development of the hippocampal mossy fiber pathway. Hippocampus. 2008;18:349–363. doi: 10.1002/hipo.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jang YN, Jung YS, Lee SH, Moon CH, Kim CH, et al. Calpain-mediated N-cadherin proteolytic processing in brain injury. J Neurosci. 2009;29:5974–5984. doi: 10.1523/JNEUROSCI.6178-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubota K, Inoue K, Hashimoto R, Kumamoto N, Kosuga A, et al. Tumor necrosis factor receptor-associated protein 1 regulates cell adhesion and synaptic morphology via modulation of N-cadherin expression. J Neurochem. 2009;110:496–508. doi: 10.1111/j.1471-4159.2009.06099.x. [DOI] [PubMed] [Google Scholar]
- 44.Latefi NS, Pedraza L, Schohl A, Li Z, Ruthazer ES. N-cadherin prodomain cleavage regulates synapse formation in vivo. Dev Neurobiol. 2009;69:518–529. doi: 10.1002/dneu.20718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrick C, Fischer A, Srivastava DP, Tronson NC, Penzes P, et al. N-cadherin regulates cytoskeletally associated IQGAP1/ERK signaling and memory formation. Neuron. 2007;55:786–798. doi: 10.1016/j.neuron.2007.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kalus I, Bormann U, Mzoughi M, Schachner M, Kleene R. Proteolytic cleavage of the neural cell adhesion molecule by ADAM17/TACE is involved in neurite outgrowth. J Neurochem. 2006;98:78–88. doi: 10.1111/j.1471-4159.2006.03847.x. [DOI] [PubMed] [Google Scholar]
- 47.Yin GN, Lee HW, Cho JY, Suk K. Neuronal pentraxin receptor in cerebrospinal fluid as a potential biomarker for neurodegenerative diseases. Brain Res. 2009;1265:158–170. doi: 10.1016/j.brainres.2009.01.058. [DOI] [PubMed] [Google Scholar]
- 48.Aisa B, Gil-Bea FJ, Solas M, García-Alloza M, Chen CP, et al. Altered NCAM Expression Associated with the Cholinergic System in Alzheimer's Disease. J Alzheimers Dis. 2010;20:659–668. doi: 10.3233/JAD-2010-1398. [DOI] [PubMed] [Google Scholar]
- 49.Storan MJ, Magnaldo T, Biol-N'Garagba MC, Zick Y, Key B. Expression and putative role of lactoseries carbohydrates present on NCAM in the rat primary olfactory pathway. J Comp Neurol. 2004;475:289–302. doi: 10.1002/cne.20167. [DOI] [PubMed] [Google Scholar]
- 50.Konecna A, Frischknecht R, Kinter J, Ludwig A, Steuble M, et al. Calsyntenin-1 docks vesicular cargo to kinesin-1. Mol Biol Cell. 2006;17:3651–3663. doi: 10.1091/mbc.E06-02-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ludwig A, Blume J, Diep TM, Yuan J, Mateos JM, et al. Calsyntenins Mediate TGN Exit of APP in a Kinesin-1-Dependent Manner. Traffic. 2009;10:572–589. doi: 10.1111/j.1600-0854.2009.00886.x. [DOI] [PubMed] [Google Scholar]
- 52.Vogt L, Schrimpf SP, Meskenaite V, Frischknecht R, Kinter J, et al. Calsyntenin-1, a proteolytically processed postsynaptic membrane protein with a cytoplasmic calcium-binding domain. Mol Cell Neurosci. 2001;17:151–166. doi: 10.1006/mcne.2000.0937. [DOI] [PubMed] [Google Scholar]
- 53.Hintsch G, Zurlinden A, Meskenaite V, Steuble M, Fink-Widmer K, et al. The calsyntenins—a family of postsynaptic membrane proteins with distinct neuronal expression patterns. Mol Cell Neurosci. 2002;21:393–409. doi: 10.1006/mcne.2002.1181. [DOI] [PubMed] [Google Scholar]
- 54.Cho RW, Park JM, Wolff SB, Xu D, Hopf C, et al. mGluR1/5-dependent long-term depression requires the regulated ectodomain cleavage of neuronal pentraxin NPR by TACE. Neuron. 2008;57:858–871. doi: 10.1016/j.neuron.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Umezawa Y, Kuge S, Kikyo N, Shirai T, Watanabe J, et al. Identity of brain-associated small cell lung cancer antigen and the CD56 (NKH-1/Leu-19) leukocyte differentiation antigen and the neural cell adhesion molecule. Jpn J Clin Oncol. 1991;21:251–255. [PubMed] [Google Scholar]
- 56.Kakunaga S, Ikeda W, Itoh S, Deguchi-Tawarada M, Ohtsuka T, et al. Nectin-like molecule-1/TSLL1/SynCAM3: a neural tissue-specific immunoglobulin-like cell-cell adhesion molecule localizing at non-junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci. 2005;118:1267–1277. doi: 10.1242/jcs.01656. [DOI] [PubMed] [Google Scholar]
- 57.Gao J, Chen T, Hu G, Gong Y, Qiang B, et al. Nectin-like molecule 1 is a glycoprotein with a single N-glycosylation site at N290KS which influences its adhesion activity. Biochim Biophys Acta. 2008;1778:1429–1435. doi: 10.1016/j.bbamem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, et al. SynCAMs organize synapses through heterophilic adhesion. J Neurosci. 2007;27:12516–12530. doi: 10.1523/JNEUROSCI.2739-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hosaka M, Suda M, Sakai Y, Izumi T, Watanabe T, et al. Secretogranin III binds to cholesterol in the secretory granule membrane as an adapter for chromogranin A. J Biol Chem. 2004;279:3627–3634. doi: 10.1074/jbc.M310104200. [DOI] [PubMed] [Google Scholar]
- 60.Lechner T, Adlassnig C, Humpel C, Kaufmann WA, Maier H, et al. Chromogranin peptides in Alzheimer's disease. Exp Gerontol. 2004;39:101–113. doi: 10.1016/j.exger.2003.09.018. [DOI] [PubMed] [Google Scholar]
- 61.Lassmann H, Weiler R, Fischer P, Bancher C, Jellinger K, et al. Synaptic pathology in Alzheimer's disease: immunological data for markers of synaptic and large dense-core vesicles. Neuroscience. 1992;46:1–8. doi: 10.1016/0306-4522(92)90003-k. [DOI] [PubMed] [Google Scholar]
- 62.Eder U, Leitner B, Kirchmair R, Pohl P, Jobst KA, et al. Levels and proteolytic processing of chromogranin A and B and secretogranin II in cerebrospinal fluid in neurological diseases. J Neural Transm. 1998;105:39–51. doi: 10.1007/s007020050036. [DOI] [PubMed] [Google Scholar]
- 63.Kaufmann WA, Barnas U, Humpel C, Nowakowski K, DeCol C, et al. Synaptic loss reflected by secretoneurin-like immunoreactivity in the human hippocampus in Alzheimer's disease. Eur J Neurosci. 1998;10:1084–1094. doi: 10.1046/j.1460-9568.1998.00121.x. [DOI] [PubMed] [Google Scholar]
- 64.Paco S, Pozas E, Aguado F. Secretogranin III is an astrocyte granin that is overexpressed in reactive glia. Cereb Cortex. 2010;20:1386–1397. doi: 10.1093/cercor/bhp202. [DOI] [PubMed] [Google Scholar]
- 65.Hosaka M, Watanabe T. Secretogranin III: a bridge between core hormone aggregates and the secretory granule membrane. Endocr J. 2010;57:275–286. doi: 10.1507/endocrj.k10e-038. [DOI] [PubMed] [Google Scholar]
- 66.Bozdagi O, Rich E, Tronel S, Sadahiro M, Patterson K, et al. The neurotrophin-inducible gene Vgf regulates hippocampal function and behavior through a brain-derived neurotrophic factor-dependent mechanism. J Neurosci. 2008;28:9857–9869. doi: 10.1523/JNEUROSCI.3145-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rüetschi U, Zetterberg H, Podust VN, Gottfries J, Li S, et al. Identification of CSF biomarkers for frontotemporal dementia using SELDI-TOF. Exp Neurol. 2005;196:273–281. doi: 10.1016/j.expneurol.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 68.Levi A, Ferri GL, Watson E, Possenti R, Salton SR. Processing, distribution, and function of VGF, a neuronal and endocrine peptide precursor. Cell Mol Neurobiol. 2004;24:517–533. doi: 10.1023/B:CEMN.0000023627.79947.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Alder J, Thakker-Varia S, Bangasser DA, Kuroiwa M, Plummer MR, et al. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23:10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Steiner DF. The proprotein convertases. Curr Opin Chem Biol. 1998;2(1):31–39. doi: 10.1016/s1367-5931(98)80033-1. [DOI] [PubMed] [Google Scholar]
- 71.Zhu X, Wu K, Rife L, Cawley NX, Brown B, et al. Carboxypeptidase E is required for normal synaptic transmission from photoreceptors to the inner retina. J Neurochem. 2005;95:1351–1362. doi: 10.1111/j.1471-4159.2005.03460.x. [DOI] [PubMed] [Google Scholar]
- 72.Hosaka M, Watanabe T, Sakai Y, Kato T, Takeuchi T. Interaction between secretogranin III and carboxypeptidase E facilitates prohormone sorting within secretory granules. J Cell Sci. 2005;118:4785–4795. doi: 10.1242/jcs.02608. [DOI] [PubMed] [Google Scholar]
- 73.Park JJ, Koshimizu H, Loh YP. Biogenesis and transport of secretory granules to release site in neuroendocrine cells. J Mol Neurosci. 2009;37:151–159. doi: 10.1007/s12031-008-9098-y. [DOI] [PubMed] [Google Scholar]
- 74.Woronowicz A, Koshimizu H, Chang SY, Cawley NX, Hill JM, et al. Absence of carboxypeptidase E leads to adult hippocampal neuronal degeneration and memory deficits. Hippocampus. 2008;18:1051–1063. doi: 10.1002/hipo.20462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Woronowicz A, Cawley NX, Chang SY, Koshimizu H, Phillips AW, et al. Carboxypeptidase E knockout mice exhibit abnormal dendritic arborization and spine morphology in central nervous system neurons. J Neurosci Res. 2010;88:64–72. doi: 10.1002/jnr.22174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arun P, Moffett JR, Namboodiri AM. Evidence for mitochondrial and cytoplasmic N-acetylaspartate synthesis in SH-SY5Y neuroblastoma cells. Neurochem Int. 2009;55:219–225. doi: 10.1016/j.neuint.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 77.Schmidbaur JM, Kugler P, Horvath E. Glutamate producing aspartate aminotransferase in glutamatergic perforant path terminals of the rat hippocampus. Cytochemical and lesion studies. Histochemistry. 1990;94:427–433. doi: 10.1007/BF00266451. [DOI] [PubMed] [Google Scholar]
- 78.Würdig S, Kugler P. Histochemistry of glutamate metabolizing enzymes in the rat cerebellar cortex. Neurosci Lett. 1991;130:165–168. doi: 10.1016/0304-3940(91)90388-a. [DOI] [PubMed] [Google Scholar]
- 79.Riemenschneider M, Buch K, Schmolke M, Kurz A, Guder WG. Diagnosis of Alzheimer's disease with cerebrospinal fluid tau protein and aspartate aminotransferase. Lancet. 1997;351:63–64. doi: 10.1016/s0140-6736(05)62568-8. [DOI] [PubMed] [Google Scholar]
- 80.D'Aniello A, Fisher G, Migliaccio N, Cammisa G, D'Aniello E, et al. Amino acids and transaminases activity in ventricular CSF and in brain of normal and Alzheimer patients. Neurosci Lett. 2005;388:49–53. doi: 10.1016/j.neulet.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 81.Jansen Steur E, Vermes I, de Vos RA. Cerebrospinal-fluid tau protein and aspartate aminotransferase in Parkinson's disease. Lancet. 1998;351:1105–1106. doi: 10.1016/s0140-6736(05)79387-9. [DOI] [PubMed] [Google Scholar]
- 82.Tapiola T, Lehtovirta M, Pirttilä T, Alafuzoff I, Riekkinen P, et al. Increased aspartate aminotransferase activity in cerebrospinal fluid and Alzheimer's disease. Lancet. 1998;352:287. doi: 10.1016/S0140-6736(05)60260-7. [DOI] [PubMed] [Google Scholar]
- 83.Wright NT, Cannon BR, Zimmer DB, Weber DJ. S100A1: Structure, Function, and Therapeutic Potential. Curr Chem Biol. 2009;3:138–145. doi: 10.2174/187231309788166460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Liang D, Nunes-Tavares N, Xie HQ, Carvalho S, Bon S, et al. Protein CutA undergoes an unusual transfer into the secretory pathway and affects the folding, oligomerization, and secretion of acetylcholinesterase. J Biol Chem. 2009;284:5195–5207. doi: 10.1074/jbc.M806260200. [DOI] [PubMed] [Google Scholar]
- 85.Perrier AL, Cousin X, Boschetti N, Haas R, Chatel JM, et al. Two distinct proteins are associated with tetrameric acetylcholinesterase on the cell surface. J Biol Chem. 2000;275:34260–34265. doi: 10.1074/jbc.M004289200. [DOI] [PubMed] [Google Scholar]
- 86.Ablonczy Z, Prakasam A, Fant J, Fauq A, Crosson C, et al. Pigment epithelium-derived factor maintains retinal pigment epithelium function by inhibiting vascular endothelial growth factor-R2 signaling through gamma-secretase. J Biol Chem. 2009;284:30177–30186. doi: 10.1074/jbc.M109.032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Castano E, Roher A, Esh C, Kokjohn T, Beach T. Comparative proteomics of cerebrospinal fluid in neuropathologically-confirmed Alzheimer's disease and non-demented elderly subjects. Neurol Res. 2006;28:155–163. doi: 10.1179/016164106X98035. [DOI] [PubMed] [Google Scholar]
- 88.Yamagishi S, Inagaki Y, Takeuchi M, Sasaki N. Is pigment epithelium-derived factor level in cerebrospinal fluid a promising biomarker for early diagnosis of Alzheimer's disease? Med Hypotheses. 2004;63:115–117. doi: 10.1016/j.mehy.2004.02.022. [DOI] [PubMed] [Google Scholar]
- 89.Takanohashi A, Yabe T, Schwartz JP. Pigment epithelium-derived factor induces the production of chemokines by rat microglia. Glia. 2005;51:266–278. doi: 10.1002/glia.20203. [DOI] [PubMed] [Google Scholar]
- 90.Bilak MM, Corse AM, Bilak SR, Lehar M, Tombran-Tink J, et al. Pigment epithelium-derived factor (PEDF) protects motor neurons from chronic glutamate-mediated neurodegeneration. J Neuropathol Exp Neurol. 1999;58:719–728. doi: 10.1097/00005072-199907000-00006. [DOI] [PubMed] [Google Scholar]
- 91.Davidsson P, Sjögren M, Andreasen N, Lindbjer M, Nilsson CL, et al. Studies of the pathophysiological mechanisms in frontotemporal dementia by proteome analysis of CSF proteins. Brain Res Mol Brain Res. 2002;109:128–133. doi: 10.1016/s0169-328x(02)00549-1. [DOI] [PubMed] [Google Scholar]
- 92.Kuncl RW, Bilak MM, Bilak SR, Corse AM, Royal W, et al. Pigment epithelium-derived factor is elevated in CSF of patients with amyotrophic lateral sclerosis. J Neurochem. 2002;81:178–184. doi: 10.1046/j.1471-4159.2002.00813.x. [DOI] [PubMed] [Google Scholar]
- 93.Yabe T, Sanagi T, Yamada H. The neuroprotective role of PEDF: implication for the therapy of neurological disorders. Curr Mol Med. 2010;10:259–266. doi: 10.2174/156652410791065354. [DOI] [PubMed] [Google Scholar]
- 94.Sanagi T, Yabe T, Yamada H. The regulation of pro-inflammatory gene expression induced by pigment epithelium-derived factor in rat cultured microglial cells. Neurosci Lett. 2005;380:105–110. doi: 10.1016/j.neulet.2005.01.035. [DOI] [PubMed] [Google Scholar]
- 95.Sanagi T, Yabe T, Yamada H. Gene transfer of PEDF attenuates ischemic brain damage in the rat middle cerebral artery occlusion model. J Neurochem. 2008;106:1841–1854. doi: 10.1111/j.1471-4159.2008.05529.x. [DOI] [PubMed] [Google Scholar]
- 96.Pang IH, Zeng H, Fleenor DL, Clark AF. Pigment epithelium-derived factor protects retinal ganglion cells. BMC Neurosci. 2007;8:11. doi: 10.1186/1471-2202-8-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Perretti M, D'Acquisto F. Annexin A1 and glucocorticoids as effectors of the resolution of inflammation. Nat Rev Immunol. 2009;9:62–70. doi: 10.1038/nri2470. [DOI] [PubMed] [Google Scholar]
- 98.Lim LH, Pervaiz S. Annexin 1: the new face of an old molecule. FASEB Journal. 2007;21:968–975. doi: 10.1096/fj.06-7464rev. [DOI] [PubMed] [Google Scholar]
- 99.Eberhard DA, Brown MD, VandenBerg SR. Alterations of annexin expression in pathological neuronal and glial reactions. Immunohistochemical localization of annexins I, II (p36 and p11 subunits), IV, and VI in the human hippocampus. Am J Pathol. 1994;145:640–649. [PMC free article] [PubMed] [Google Scholar]
- 100.Misasi R, Hozumi I, Inuzuka T, Capozzi A, Mattei V, et al. Biochemistry and neurobiology of prosaposin: a potential therapeutic neuro-effector. Cent Nerv Syst Agents Med Chem. 2009;9:119–131. doi: 10.2174/187152409788452045. [DOI] [PubMed] [Google Scholar]
- 101.Ochiai T, Takenaka Y, Kuramoto Y, Kasuya M, Fukuda K, et al. Molecular mechanism for neuro-protective effect of prosaposin against oxidative stress: its regulation of dimeric transcription factor formation. Biochim Biophys Acta. 2008;1780:1441–1447. doi: 10.1016/j.bbagen.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 102.Sikora J, Harzer K, Elleder M. Neurolysosomal pathology in human prosaposin deficiency suggests essential neurotrophic function of prosaposin. Acta Neuropathol. 2007;113:163–175. doi: 10.1007/s00401-006-0148-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.O'Brien JS, Carson GS, Seo HC, Hiraiwa M, Kishimoto Y. Identification of prosaposin as a neurotrophic factor. Proc Natl Acad Sci U S A. 1994;91:9593–9596. doi: 10.1073/pnas.91.20.9593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li L, Hung AC, Porter AG. Secretogranin II: a key AP-1-regulated protein that mediates neuronal differentiation and protection from nitric oxide-induced apoptosis of neuroblastoma cells. Cell Death Differ. 2008;15:879–888. doi: 10.1038/cdd.2008.8. [DOI] [PubMed] [Google Scholar]
- 105.Shyu WC, Lin SZ, Chiang MF, Chen DC, Su CY, et al. Secretoneurin promotes neuroprotection and neuronal plasticity via the Jak2/Stat3 pathway in murine models of stroke. J Clin Invest. 2008;118:133–148. doi: 10.1172/JCI32723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gasser MC, Berti I, Hauser KF, Fischer-Colbrie R, Saria A. Secretoneurin promotes pertussis toxin-sensitive neurite outgrowth in cerebellar granule cells. J Neurochem. 2003;85:662–669. doi: 10.1046/j.1471-4159.2003.01677.x. [DOI] [PubMed] [Google Scholar]
- 107.Hipkiss AR. Could carnosine or related structures suppress Alzheimer's disease? J Alzheimers Dis. 2007;11:229–240. doi: 10.3233/jad-2007-11210. [DOI] [PubMed] [Google Scholar]
- 108.Guiotto A, Calderan A, Ruzza P, Borin G. Carnosine and carnosine-related antioxidants: a review. Curr Med Chem. 2005;12:2293–2315. doi: 10.2174/0929867054864796. [DOI] [PubMed] [Google Scholar]
- 109.Yin GN, Lee HW, Cho JY, Suk K. Neuronal pentraxin receptor in cerebrospinal fluid as a potential biomarker for neurodegenerative diseases. Brain Res. 2009;1265:158–170. doi: 10.1016/j.brainres.2009.01.058. [DOI] [PubMed] [Google Scholar]
- 110.Teufel M, Saudek V, Ledig JP, Bernhardt A, Boularand S, et al. Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J Biol Chem. 2003;278:6521–6531. doi: 10.1074/jbc.M209764200. [DOI] [PubMed] [Google Scholar]
- 111.Balion CM, Benson C, Raina PS, Papaioannou A, Patterson C, et al. Brain type carnosinase in dementia: a pilot study. BMC Neurol. 2007;7:38. doi: 10.1186/1471-2377-7-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zelko IN, Mariani TJ, Folz RJ. Superoxide dismutase multigene family: a comparison of the CuZn-SOD (SOD1), Mn-SOD (SOD2), and EC-SOD (SOD3) gene structures, evolution, and expression. Free Radic Biol Med. 2002;33:337–349. doi: 10.1016/s0891-5849(02)00905-x. [DOI] [PubMed] [Google Scholar]
- 113.Folz RJ, Crapo JD. Extracellular superoxide dismutase (SOD3): tissue-specific expression, genomic characterization, and computer-assisted sequence analysis of the human EC SOD gene. Genomics. 1994;22:162–171. doi: 10.1006/geno.1994.1357. [DOI] [PubMed] [Google Scholar]
- 114.Levin ED. Extracellular superoxide dismutase (EC-SOD) quenches free radicals and attenuates age-related cognitive decline: opportunities for novel drug development in aging. Curr Alzheimer Res. 2005;2:191–196. doi: 10.2174/1567205053585710. [DOI] [PubMed] [Google Scholar]
- 115.Kothakota S, Azuma T, Reinhard C, Klippel A, Tang J, et al. Caspase-3-generated fragment of gelsolin: effector of morphological change in apoptosis. Science. 1997;278:294–298. doi: 10.1126/science.278.5336.294. [DOI] [PubMed] [Google Scholar]
- 116.Nag S, Ma Q, Wang H, Chumnarnsilpa S, Lee WL, et al. Ca2+ binding by domain 2 plays a critical role in the activation and stabilization of gelsolin. Proc Natl Acad Sci U S A. 2009;106:13713–13718. doi: 10.1073/pnas.0812374106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chauhan VP, Ray I, Chauhan A, Wisniewski HM. Binding of gelsolin, a secretory protein, to amyloid beta-protein. Biochem Biophys Res Commun. 1999;258:241–246. doi: 10.1006/bbrc.1999.0623. [DOI] [PubMed] [Google Scholar]
- 118.Ray I, Chauhan A, Wegiel J, Chauhan VP. Gelsolin inhibits the fibrillization of amyloid beta-protein, and also defibrillizes its preformed fibrils. Brain Res. 2000;853:344–351. doi: 10.1016/s0006-8993(99)02315-x. [DOI] [PubMed] [Google Scholar]
- 119.Ji L, Chauhan A, Wegiel J, Essa MM, Chauhan V. Gelsolin is proteolytically cleaved in the brains of individuals with Alzheimer's disease. J Alz Dis. 2009;18:105–111. doi: 10.3233/JAD-2009-1127. [DOI] [PubMed] [Google Scholar]
- 120.Slee EA, Adrain C, Martin SJ. Executioner caspase-3, -6, and -7 perform distinct, non-redundant roles during the demolition phase of apoptosis. J Biol Chem. 2001;276:7320–7326. doi: 10.1074/jbc.M008363200. [DOI] [PubMed] [Google Scholar]
- 121.Antequera D, Vargas T, Ugalde C, Spuch C, Molina JA, et al. Cytoplasmic gelsolin increases mitochondrial activity and reduces Abeta burden in a mouse model of Alzheimer's disease. Neurobiol Dis. 2009;36:42–50. doi: 10.1016/j.nbd.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 122.Owen D, Lowe PN, Nietlispach D, Brosnan CE, Chirgadze DY, et al. Molecular dissection of the interaction between the small G proteins Rac1 and RhoA and protein kinase C-related kinase 1 (PRK1). J Biol Chem. 2003;278:50578–50587. doi: 10.1074/jbc.M304313200. [DOI] [PubMed] [Google Scholar]
- 123.Palmer RH, Parker PJ. Expression, purification and characterization of the ubiquitous protein kinase C-related kinase 1. Biochem J. 1995;309:315–320. doi: 10.1042/bj3090315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Okii N, Amano T, Seki T, Matsubayashi H, Mukai H, et al. Fragmentation of protein kinase N (PKN) in the hydrocephalic rat brain. Acta Histochem Cytochem. 2007;40:113–121. doi: 10.1267/ahc.07011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takahashi M, Mukai H, Toshimori M, Miyamoto M, Ono Y. Proteolytic activation of PKN by caspase-3 or related protease during apoptosis. Proc Natl Acad Sci U S A. 1998;95:11566–11571. doi: 10.1073/pnas.95.20.11566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ueyama T, Ren Y, Sakai N, Takahashi M, Ono Y, et al. Generation of a constitutively active fragment of PKN in microglia/macrophages after middle cerebral artery occlusion in rats. J Neurochem. 2001;79:903–913. doi: 10.1046/j.1471-4159.2001.00624.x. [DOI] [PubMed] [Google Scholar]
- 127.Henion TR, Raitcheva D, Grosholz R, Biellmann F, Skarnes WC, et al. Beta1,3-N-acetylglucosaminyltransferase 1 glycosylation is required for axon pathfinding by olfactory sensory neurons. J Neurosci. 2005;25:1894–1903. doi: 10.1523/JNEUROSCI.4654-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Puche AC, Bartlett PF, Key B. Substrate-bound carbohydrates stimulate signal transduction and neurite outgrowth in an olfactory neuron cell line. Neuroreport. 1997;8:3183–3188. doi: 10.1097/00001756-199710200-00001. [DOI] [PubMed] [Google Scholar]
- 129.Svichar N, Esquenazi S, Waheed A, Sly WS, Chesler M. Functional demonstration of surface carbonic anhydrase IV activity on rat astrocytes. Glia. 2006;53:241–247. doi: 10.1002/glia.20277. [DOI] [PubMed] [Google Scholar]
- 130.Svichar N, Waheed A, Sly WS, Hennings JC, Hubner CA, et al. The Carbonic Anhydrases CA4 and CA14 Both Enhance AE3-Mediated Cl–HCO3 Exchange in Hippocampal Neurons. J Neurosci. 2009;29:3252–3258. doi: 10.1523/JNEUROSCI.0036-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shah GN, Ulmasov B, Waheed A, Becker T, Makani S, et al. Carbonic anhydrase IV and XIV knockout mice: roles of the respective carbonic anhydrases in buffering the extracellular space in brain. Proc Natl Acad Sci U S A. 2005;102:16771–16776. doi: 10.1073/pnas.0508449102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Benfenati F, Ferrari R, Onofri F, Arcuri C, Giambanco I, et al. S100A1 codistributes with synapsin I in discrete brain areas and inhibits the F-actin-bundling activity of synapsin I. J Neurochem. 2004;89:1260–1270. doi: 10.1111/j.1471-4159.2004.02419.x. [DOI] [PubMed] [Google Scholar]
- 133.Redondo RL, Okuno H, Spooner PA, Frenguelli BG, Bito H, et al. Synaptic tagging and capture: differential role of distinct calcium/calmodulin kinases in protein synthesis-dependent long-term potentiation. J Neurosci. 2010;30:4981–4989. doi: 10.1523/JNEUROSCI.3140-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhong L, Cherry T, Bies CE, Florence MA, Gerges NZ. Neurogranin enhances synaptic strength through its interaction with calmodulin. EMBO J. 2009;28:3027–3039. doi: 10.1038/emboj.2009.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Liu X, Yang PS, Yang W, Yue DT. Enzyme-inhibitor-like tuning of Ca(2+) channel connectivity with calmodulin. Nature. 2010;463:968–972. doi: 10.1038/nature08766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Supnet C, Bezprozvanny I. Neuronal calcium signaling, mitochondrial dysfunction, and Alzheimer's disease. J Alzheimers Dis. 2010;20(Suppl 2):S487–498. doi: 10.3233/JAD-2010-100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Craig-Schapiro R, Perrin RJ, Roe CM, Xiong C, Carter D, et al. YKL-40: A Novel Prognostic Fluid Biomarker for Preclinical Alzheimer's Disease. Biol Psychiatry. 2010;68:903–12. doi: 10.1016/j.biopsych.2010.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Colton CA, Mott RT, Sharpe H, Xu Q, Van Nostrand WE, et al. Expression profiles for macrophage alternative activation genes in AD and in mouse models of AD. J Neuroinflammation. 2006;3:27. doi: 10.1186/1742-2094-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kolson DL. YKL-40: a candidate biomarker for simian immunodeficiency virus and human immunodeficiency virus encephalitis? Am J Pathol. 2008;173:25–29. doi: 10.2353/ajpath.2008.080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bonneh-Barkay D, Bissel SJ, Wang G, Fish KN, Nicholl GC, et al. YKL-40, a marker of simian immunodeficiency virus encephalitis, modulates the biological activity of basic fibroblast growth factor. Am J Pathol. 2008;173:130–143. doi: 10.2353/ajpath.2008.080045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ostergaard C, Johansen JS, Benfield T, Price PA, Lundgren JD. YKL-40 is elevated in cerebrospinal fluid from patients with purulent meningitis. Clin Diagn Lab Immunol. 2002;9:598–604. doi: 10.1128/CDLI.9.3.598-604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kaynar MY, Tanriverdi T, Kafadar AM, Kacira T, Yurdakul F, et al. YKL-40 levels in the cerebrospinal fluid and serum of patients with aneurysmal subarachnoid hemorrhage: preliminary results. J Clin Neurosci. 2005;12:754–757. doi: 10.1016/j.jocn.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 143.Junker N, Johansen JS, Hansen LT, Lund EL, Kristjansen PE. Regulation of YKL-40 expression during genotoxic or microenvironmental stress in human glioblastoma cells. Cancer Sci. 2005;96:183–190. doi: 10.1111/j.1349-7006.2005.00026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hakala BE, White C, Recklies AD. Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J Biol Chem. 1993;268:25803–25810. [PubMed] [Google Scholar]
- 145.Chupp GL, Lee CG, Jarjour N, Shim YM, Holm CT, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 146.Ling H, Recklies AD. The chitinase 3-like protein human cartilage glycoprotein 39 inhibits cellular responses to the inflammatory cytokines interleukin-1 and tumour necrosis factor-alpha. Biochem J. 2004;380:651–659. doi: 10.1042/BJ20040099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Létuvé S, Kozhich A, Arouche N, Grandsaigne M, Reed J, et al. YKL-40 is elevated in patients with chronic obstructive pulmonary disease and activates alveolar macrophages. J Immunol. 2008;181:5167–5173. doi: 10.4049/jimmunol.181.7.5167. [DOI] [PubMed] [Google Scholar]
- 148.Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, et al. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162:2041–2057. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Choi-Miura NH, Takahashi K, Yoda M, Saito K, Hori M, et al. The novel acute phase protein, IHRP, inhibits actin polymerization and phagocytosis of polymorphonuclear cells. Inflamm Res. 2000;49:305–310. doi: 10.1007/PL00000211. [DOI] [PubMed] [Google Scholar]
- 150.Choi-Miura NH. Novel human plasma proteins, IHRP (acute phase protein) and PHBP (serine protease), which bind to glycosaminoglycans. Curr Med Chem Cardiovasc Hematol Agents. 2004;2:239–248. doi: 10.2174/1568016043356327. [DOI] [PubMed] [Google Scholar]
- 151.Akiyama H, Kawamata T, Dedhar S, McGeer PL. Immunohistochemical localization of vitronectin, its receptor and beta-3 integrin in Alzheimer brain tissue. J Neuroimmunol. 1991;32:19–28. doi: 10.1016/0165-5728(91)90067-h. [DOI] [PubMed] [Google Scholar]
- 152.McGeer PL, Kawamata T, Walker DG. Distribution of clusterin in Alzheimer brain tissue. Brain Res. 1992;579:337–341. doi: 10.1016/0006-8993(92)90071-g. [DOI] [PubMed] [Google Scholar]
- 153.Shin TM, Isas JM, Hsieh CL, Kayed R, Glabe CG, et al. Formation of soluble amyloid oligomers and amyloid fibrils by the multifunctional protein vitronectin. Mol Neurodegener. 2008;3:16. doi: 10.1186/1750-1326-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Milner R, Campbell IL. The extracellular matrix and cytokines regulate microglial integrin expression and activation. J Immunol. 2003;170:3850–3858. doi: 10.4049/jimmunol.170.7.3850. [DOI] [PubMed] [Google Scholar]
- 155.Milner R, Crocker SJ, Hung S, Wang X, Frausto RF, et al. Fibronectin- and vitronectin-induced microglial activation and matrix metalloproteinase-9 expression is mediated by integrins alpha5beta1 and alphavbeta5. J Immunol. 2007;178:8158–8167. doi: 10.4049/jimmunol.178.12.8158. [DOI] [PubMed] [Google Scholar]
- 156.Lambert JC, Heath S, Even G, Campion D, Sleegers K, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- 157.Zanjani H, Finch CE, Kemper C, Atkinson J, McKeel D, et al. Complement activation in very early Alzheimer disease. Alzheimer Dis Assoc Disord. 2005;19:55–66. doi: 10.1097/01.wad.0000165506.60370.94. [DOI] [PubMed] [Google Scholar]
- 158.Stoltzner SE, Grenfell TJ, Mori C, Wisniewski KE, Wisniewski TM, et al. Temporal accrual of complement proteins in amyloid plaques in Down's syndrome with Alzheimer's disease. Am J Pathol. 2000;156:489–499. doi: 10.1016/S0002-9440(10)64753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Loeffler DA, Camp DM, Schonberger MB, Singer DJ, LeWitt PA. Early complement activation increases in the brain in some aged normal subjects. Neurobiol Aging. 2004;25:1001–1007. doi: 10.1016/j.neurobiolaging.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 160.Finehout EJ, Franck Z, Lee KH. Complement protein isoforms in CSF as possible biomarkers for neurogenerative disease. Dis Markers. 2005;21:93–101. doi: 10.1155/2005/806573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Masaki T, Matsumoto M, Nakanishi I, Yasuda R, Seya T. Factor I-dependent inactivation of human complement C4b of the classical pathway by C3b/C4b receptor (CR1, CD35) and membrane cofactor protein (MCP, CD46). J Biochem. 1992;111:573–578. doi: 10.1093/oxfordjournals.jbchem.a123799. [DOI] [PubMed] [Google Scholar]
- 162.Puchades M, Hansson S, Nilsson C, Andreasen N, Blennow K, et al. Proteomic studies of potential cerebrospinal fluid protein markers for Alzheimer's disease. Mol Brain Res. 2003;118:140–146. doi: 10.1016/j.molbrainres.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 163.Bergamaschini L, Donarini C, Gobbo G, Parnetti L, Gallai V. Activation of complement and contact system in Alzheimer's disease. Mech Ageing Dev. 2001;122:1971–1983. doi: 10.1016/s0047-6374(01)00311-6. [DOI] [PubMed] [Google Scholar]
- 164.Murphy BF, Saunders JR, O'Bryan MK, Kirszbaum L, Walker ID, et al. SP-40 is an inhibitor of C5b-6-initiated haemolysis. Int Immunol. 1989;1:551–554. doi: 10.1093/intimm/1.5.551. [DOI] [PubMed] [Google Scholar]
- 165.Koch S, Donarski N, Goetze K, Kreckel M, Stuerenburg HJ, et al. Characterization of four lipoprotein classes in human cerebrospinal fluid. J Lipid Res. 2001;42:1143–1151. [PubMed] [Google Scholar]
- 166.Harr SD, Uint L, Hollister R, Hyman BT, Mendez AJ. Brain expression of apolipoproteins E, J, and A-I in Alzheimer's disease. J Neurochem. 1996;66:2429–2435. doi: 10.1046/j.1471-4159.1996.66062429.x. [DOI] [PubMed] [Google Scholar]
- 167.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet. 2009;41:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Perarnau B, Siegrist CA, Gillet A, Vincent C, Kimura S, et al. Beta 2-microglobulin restriction of antigen presentation. Nature. 1990;346:751–754. doi: 10.1038/346751a0. [DOI] [PubMed] [Google Scholar]
- 169.Vitiello A, Potter TA, Sherman LA. The role of beta 2-microglobulin in peptide binding by class I molecules. Science. 1990;250:1423–1426. doi: 10.1126/science.2124002. [DOI] [PubMed] [Google Scholar]
- 170.Berggard I, Bearn AG. Isolation and properties of a low molecular weight beta-2-globulin occurring in human biological fluids. J Biol Chem. 1968;243:4095–4103. [PubMed] [Google Scholar]
- 171.Nissen MH, Roepstorff P, Thim L, Dunbar B, Fothergill JE. Limited proteolysis of beta 2-microglobulin at Lys-58 by complement component C1s. Eur J Biochem. 1990;189:423–429. doi: 10.1111/j.1432-1033.1990.tb15505.x. [DOI] [PubMed] [Google Scholar]
- 172.Lob HE, Marvar PJ, Guzik TJ, Sharma S, McCann LA, et al. Induction of hypertension and peripheral inflammation by reduction of extracellular superoxide dismutase in the central nervous system. Hypertension. 55:277–283, 276p following 283. doi: 10.1161/HYPERTENSIONAHA.109.142646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hansson SF, Andréasson U, Wall M, Skoog I, Andreasen N, et al. Reduced levels of amyloid-beta-binding proteins in cerebrospinal fluid from Alzheimer's disease patients. J Alzheimers Dis. 2009;16:389–397. doi: 10.3233/JAD-2009-0966. [DOI] [PubMed] [Google Scholar]
- 174.Kanekiyo T, Ban T, Aritake K, Huang ZL, Qu WM, et al. Lipocalin-type prostaglandin D synthase/beta-trace is a major amyloid beta-chaperone in human cerebrospinal fluid. Proc Natl Acad Sci U S A. 2007;104:6412–6417. doi: 10.1073/pnas.0701585104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Lovell MA, Lynn BC, Xiong S, Quinn JF, Kaye J, et al. An aberrant protein complex in CSF as a biomarker of Alzheimer disease. Neurology. 2008;70:2212–2218. doi: 10.1212/01.wnl.0000312383.39973.88. [DOI] [PubMed] [Google Scholar]
- 176.Paula-Lima AC, Tricerri MA, Brito-Moreira J, Bomfim TR, Oliveira FF, et al. Human apolipoprotein A-I binds amyloid-beta and prevents Abeta-induced neurotoxicity. Int J Biochem Cell Biol. 2009;41:1361–1370. doi: 10.1016/j.biocel.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 177.Wisniewski T, Golabek AA, Kida E, Wisniewski KE, Frangione B. Conformational mimicry in Alzheimer's disease. Role of apolipoproteins in amyloidogenesis. Am J Pathol. 1995;147:238–244. [PMC free article] [PubMed] [Google Scholar]
- 178.Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Biroccio A, Del Boccio P, Panella M, Bernardini S, Di Ilio C, et al. Differential post-translational modifications of transthyretin in Alzheimer's disease: a study of the cerebral spinal fluid. Proteomics. 2006;6:2305–2313. doi: 10.1002/pmic.200500285. [DOI] [PubMed] [Google Scholar]
- 180.Wati H, Kawarabayashi T, Matsubara E, Kasai A, Hirasawa T, et al. Transthyretin accelerates vascular Abeta deposition in a mouse model of Alzheimer's disease. Brain Pathol. 2009;19:48–57. doi: 10.1111/j.1750-3639.2008.00166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Buxbaum JN, Ye Z, Reixach N, Friske L, Levy C, et al. Transthyretin protects Alzheimer's mice from the behavioral and biochemical effects of Abeta toxicity. Proc Natl Acad Sci U S A. 2008;105:2681–2686. doi: 10.1073/pnas.0712197105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Buxbaum JN, Reixach N. Transthyretin: the servant of many masters. Cell Mol Life Sci. 2009;66:3095–3101. doi: 10.1007/s00018-009-0109-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Costa R, Ferreira-da-Silva F, Saraiva MJ, Cardoso I. Transthyretin protects against A-beta peptide toxicity by proteolytic cleavage of the peptide: a mechanism sensitive to the Kunitz protease inhibitor. PLoS One. 2008;3:e2899. doi: 10.1371/journal.pone.0002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Choi SH, Leight SN, Lee VM, Li T, Wong PC, et al. Accelerated Abeta deposition in APPswe/PS1deltaE9 mice with hemizygous deletions of TTR (transthyretin). J Neurosci. 2007;27:7006–7010. doi: 10.1523/JNEUROSCI.1919-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Kaeser SA, Herzig MC, Coomaraswamy J, Kilger E, Selenica ML, et al. Cystatin C modulates cerebral beta-amyloidosis. Nat Genet. 2007;39:1437–1439. doi: 10.1038/ng.2007.23. [DOI] [PubMed] [Google Scholar]
- 186.Sun B, Zhou Y, Halabisky B, Lo I, Cho SH, et al. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer's disease. Neuron. 2008;60:247–257. doi: 10.1016/j.neuron.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mi W, Pawlik M, Sastre M, Jung SS, Radvinsky DS, et al. Cystatin C inhibits amyloid-beta deposition in Alzheimer's disease mouse models. Nat Genet. 2007;39:1440–1442. doi: 10.1038/ng.2007.29. [DOI] [PubMed] [Google Scholar]
- 188.Selenica ML, Wang X, Ostergaard-Pedersen L, Westlind-Danielsson A, Grubb A. Cystatin C reduces the in vitro formation of soluble Abeta1-42 oligomers and protofibrils. Scand J Clin Lab Invest. 2007;67:179–190. doi: 10.1080/00365510601009738. [DOI] [PubMed] [Google Scholar]
- 189.Wood JA, Wood PL, Ryan R, Graff-Radford NR, Pilapil C, et al. Cytokine indices in Alzheimer's temporal cortex: no changes in mature IL-1 beta or IL-1RA but increases in the associated acute phase proteins IL-6, alpha 2-macroglobulin and C-reactive protein. Brain Res. 1993;629:245–252. doi: 10.1016/0006-8993(93)91327-o. [DOI] [PubMed] [Google Scholar]
- 190.Narita M, Holtzman DM, Schwartz AL, Bu G. α2-Macroglobulin complexes with and mediates the endocytosis of β-amyloid peptide via cell surface low-density lipoprotein receptor-related protein. J Neurochem. 1997;69:1904–1911. doi: 10.1046/j.1471-4159.1997.69051904.x. [DOI] [PubMed] [Google Scholar]
- 191.Hye A, Lynham S, Thambisetty M, Causevic M, Campbell J, et al. Proteome-based plasma biomarkers for Alzheimer's disease. Brain. 2006;129:3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 192.Kovacs DM. alpha2-macroglobulin in late-onset Alzheimer's disease. Exp Gerontol. 2000;35:473–479. doi: 10.1016/s0531-5565(00)00113-3. [DOI] [PubMed] [Google Scholar]
- 193.French K, Yerbury JJ, Wilson MR. Protease activation of alpha2-macroglobulin modulates a chaperone-like action with broad specificity. Biochemistry. 2008;47:1176–1185. doi: 10.1021/bi701976f. [DOI] [PubMed] [Google Scholar]
- 194.Hughes SR, Khorkova O, Goyal S, Knaeblein J, Heroux J, et al. Alpha2-macroglobulin associates with beta-amyloid peptide and prevents fibril formation. Proc Natl Acad Sci U S A. 1998;95:3275–3280. doi: 10.1073/pnas.95.6.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Abraham C, Selkoe D, Potter H. Immunochemical identification of the serine protease inhibitor alpha 1-antichymotrypsin in the brain amyloid deposits of Alzheimer's disease. Cell. 1988;52:487–501. doi: 10.1016/0092-8674(88)90462-x. [DOI] [PubMed] [Google Scholar]
- 196.Harigaya Y, Shoji M, Nakamura T, Matsubara E, Hosoda K, et al. Alpha 1-antichymotrypsin level in cerebrospinal fluid is closely associated with late onset Alzheimer's disease. Intern Med. 1995;34:481–484. doi: 10.2169/internalmedicine.34.481. [DOI] [PubMed] [Google Scholar]
- 197.Abraham CR, McGraw WT, Slot F, Yamin R. Alpha 1-antichymotrypsin inhibits A beta degradation in vitro and in vivo. Ann N Y Acad Sci. 2000;920:245–248. [PubMed] [Google Scholar]
- 198.DeKosky S, Ikonomovic MD, Wang X, Farlow M, Wisniewski S, et al. Plasma and cerebrospinal fluid 1-Antichymotrypsin levels in Alzheimer's disease: correlation with cognitive impairnent. Ann Neurol. 2003;53:81–90. doi: 10.1002/ana.10414. [DOI] [PubMed] [Google Scholar]
- 199.Nielsen HM, Minthon L, Londos E, Blennow K, Miranda E, et al. Plasma and CSF serpins in Alzheimer disease and dementia with Lewy bodies. Neurology. 2007;69:1569–1579. doi: 10.1212/01.wnl.0000271077.82508.a0. [DOI] [PubMed] [Google Scholar]
- 200.Vukovic J, Marmorstein LY, McLaughlin PJ, Sasaki T, Plant GW, et al. Lack of fibulin-3 alters regenerative tissue responses in the primary olfactory pathway. Matrix Biol. 2009;28:406–415. doi: 10.1016/j.matbio.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 201.Hu B, Thirtamara-Rajamani KK, Sim H, Viapiano MS. Fibulin-3 is uniquely upregulated in malignant gliomas and promotes tumor cell motility and invasion. Mol Cancer Res. 2009;7:1756–1770. doi: 10.1158/1541-7786.MCR-09-0207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Klenotic PA, Munier FL, Marmorstein LY, Anand-Apte B. Tissue inhibitor of metalloproteinases-3 (TIMP-3) is a binding partner of epithelial growth factor-containing fibulin-like extracellular matrix protein 1 (EFEMP1). Implications for macular degenerations. J Biol Chem. 2004;279:30469–30473. doi: 10.1074/jbc.M403026200. [DOI] [PubMed] [Google Scholar]
- 203.Vollbach H, Heun R, Morris CM, Edwardson JA, McKeith IG, et al. APOA1 polymorphism influences risk for early-onset nonfamiliar AD. Ann Neurol. 2005;58:436–441. doi: 10.1002/ana.20593. [DOI] [PubMed] [Google Scholar]
- 204.Montine TJ, Montine KS, Swift LL. Central nervous system lipoproteins in Alzheimer's disease. Am J Path. 1997;151:1571–1575. [PMC free article] [PubMed] [Google Scholar]
- 205.Gunzburg MJ, Perugini MA, Howlett GJ. Structural basis for the recognition and cross-linking of amyloid fibrils by human apolipoprotein E. J Biol Chem. 2007;282:35831–35841. doi: 10.1074/jbc.M706425200. [DOI] [PubMed] [Google Scholar]
- 206.Sun Y, Shi J, Zhang S, Tang M, Han H, et al. The APOC3 SstI polymorphism is weakly associated with sporadic Alzheimer's disease in a Chinese population. Neurosci Lett. 2005;380:219–222. doi: 10.1016/j.neulet.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 207.Houlden H, Crook R, Duff K, Hutton M, Collinge J, et al. Apolipoprotein E alleles but neither apolipoprotein B nor apolipoprotein AI/CIII alleles are associated with late onset, familial Alzheimer's disease. Neurosci Lett. 1995;188:202–204. doi: 10.1016/0304-3940(95)11422-s. [DOI] [PubMed] [Google Scholar]
- 208.Pan S, Rush J, Peskind ER, Galasko D, Chung K, et al. Application of targeted quantitative proteomics analysis in human cerebrospinal fluid using a liquid chromatography matrix-assisted laser desorption/ionization time-of-flight tandem mass spectrometer (LC MALDI TOF/TOF) platform. J Proteome Res. 2008;7:720–730. doi: 10.1021/pr700630x. [DOI] [PubMed] [Google Scholar]
- 209.Katzav A, Faust-Socher A, Kvapil F, Michaelson DM, Blank M, et al. Neurobiol Aging; 2009. Antiphospholipid syndrome induction exacerbates a transgenic Alzheimer disease model on a female background. [DOI] [PubMed] [Google Scholar]
- 210.Zimmer DB, Chaplin J, Baldwin A, Rast M. S100-mediated signal transduction in the nervous system and neurological diseases. Cell Mol Biol (Noisy-le-grand) 2005;51:201–214. [PubMed] [Google Scholar]
- 211.Loeffler DA, LeWitt PA, Juneau PL, Sima AA, Nguyen HU, et al. Increased regional brain concentrations of ceruloplasmin in neurodegenerative disorders. Brain Res. 1996;738:265–274. doi: 10.1016/s0006-8993(96)00782-2. [DOI] [PubMed] [Google Scholar]
- 212.Castellani RJ, Smith MA, Nunomura A, Harris PL, Perry G. Is increased redox-active iron in Alzheimer disease a failure of the copper-binding protein ceruloplasmin? Free Radic Biol Med. 1999;26:1508–1512. doi: 10.1016/s0891-5849(99)00016-7. [DOI] [PubMed] [Google Scholar]
- 213.Kaneko K, Yoshida K, Arima K, Ohara S, Miyajima H, et al. Astrocytic deformity and globular structures are characteristic of the brains of patients with aceruloplasminemia. J Neuropathol Exp Neurol. 2002;61:1069–1077. doi: 10.1093/jnen/61.12.1069. [DOI] [PubMed] [Google Scholar]
- 214.Kessler H, Pajonk FG, Meisser P, Schneider-Axmann T, Hoffmann KH, et al. Cerebrospinal fluid diagnostic markers correlate with lower plasma copper and ceruloplasmin in patients with Alzheimer's disease. J Neural Transm. 2006;113:1763–1769. doi: 10.1007/s00702-006-0485-7. [DOI] [PubMed] [Google Scholar]
- 215.Capo CR, Arciello M, Squitti R, Cassetta E, Rossini PM, et al. Features of ceruloplasmin in the cerebrospinal fluid of Alzheimer's disease patients. Biometals. 2008;21:367–372. doi: 10.1007/s10534-007-9125-4. [DOI] [PubMed] [Google Scholar]
- 216.Squitti R, Quattrocchi CC, Salustri C, Rossini PM. Ceruloplasmin fragmentation is implicated in ‘free’ copper deregulation of Alzheimer's disease. Prion. 2008;2:23–27. doi: 10.4161/pri.2.1.6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Squitti R, Bressi F, Pasqualetti P, Bonomini C, Ghidoni R, et al. Longitudinal prognostic value of serum “free” copper in patients with Alzheimer disease. Neurology. 2009;72:50–55. doi: 10.1212/01.wnl.0000338568.28960.3f. [DOI] [PubMed] [Google Scholar]
- 218.Okada M, Hatakeyama T, Itoh H, Tokuta N, Tokumitsu H, et al. S100A1 is a novel molecular chaperone and a member of the Hsp70/Hsp90 multichaperone complex. J Biol Chem. 2004;279:4221–4233. doi: 10.1074/jbc.M309014200. [DOI] [PubMed] [Google Scholar]
- 219.Lein ES, Callaway EM, Albright TD, Gage FH. Redefining the boundaries of the hippocampal CA2 subfield in the mouse using gene expression and 3-dimensional reconstruction. J Comp Neurol. 2005;485:1–10. doi: 10.1002/cne.20426. [DOI] [PubMed] [Google Scholar]
- 220.Hosokawa N, You Z, Tremblay LO, Nagata K, Herscovics A. Stimulation of ERAD of misfolded null Hong Kong alpha1-antitrypsin by Golgi alpha1,2-mannosidases. Biochem Biophys Res Commun. 2007;362:626–632. doi: 10.1016/j.bbrc.2007.08.057. [DOI] [PubMed] [Google Scholar]
- 221.Schweden J, Bause E. Characterization of trimming Man9-mannosidase from pig liver. Purification of a catalytically active fragment and evidence for the transmembrane nature of the intact 65 kDa enzyme. Biochem J. 1989;264:347–355. doi: 10.1042/bj2640347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Kehoe PG, Miners S, Love S. Angiotensins in Alzheimer's disease - friend or foe? Trends Neurosci. 2009;32:619–628. doi: 10.1016/j.tins.2009.07.006. [DOI] [PubMed] [Google Scholar]
- 223.Blennow K, Davidsson P, Wallin A, Ekman R. Chromogranin A in cerebrospinal fluid: a biochemical marker for synaptic degeneration in Alzheimer's disease? Dementia. 2004;6:306–11. doi: 10.1159/000106963. [DOI] [PubMed] [Google Scholar]
- 224.Massaro AR, De Pascalis D, Carnevale A, Carbone G. The neural cell adhesion molecule (NCAM) present in the cerebrospinal fluid of multiple sclerosis patients is unsialylated. Eur Rev Med Pharmacol Sci. 2009;13:397–9. [PubMed] [Google Scholar]
- 225.Todaro L, Puricelli L, Gioseffi H, Pallotta MG, Lastiri J, et al. Neural cell adhesion molecule in human serum. Increased levels in dementia of the Alzheimer type. Neurobiol Dis. 2004;15:387–393. doi: 10.1016/j.nbd.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 226.Kapaki E, Paraskevas GP, Papageorgiou SG, Bonakis A, Kalfakis N, et al. Diagnostic value of CSF biomarker profile in frontotemporal lobar degeneration. Alzheimer Dis Assoc Disord. 2008;22:47–53. doi: 10.1097/WAD.0b013e3181610fea. [DOI] [PubMed] [Google Scholar]
- 227.Bian H, Van Swieten JC, Leight S, Massimo L, Wood E, et al. CSF biomarkers in frontotemporal lobar degeneration with known pathology. Neurology. 2008;70:1827–1835. doi: 10.1212/01.wnl.0000311445.21321.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mollenhauer B, Cullen V, Kahn I, Krastins B, Outeiro TF, et al. Direct quantification of CSF alpha-synuclein by ELISA and first cross-sectional study in patients with neurodegeneration. Exp Neurol. 2008;213:315–325. doi: 10.1016/j.expneurol.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 229.Bateman RJ, Siemers ER, Mawuenyega KG, Wen G, Browning KR, et al. A gamma-secretase inhibitor decreases amyloid-beta production in the central nervous system. Ann Neurol. 2009;66:48–54. doi: 10.1002/ana.21623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Hansson O, Zetterberg H, Buchhave P, Londos E, Blennow K, et al. Association between CSF biomarkers and incipient Alzheimer's disease in patients with mild cognitive impairment: A follow-up study. Lancet Neurol. 2006;5:228–234. doi: 10.1016/S1474-4422(06)70355-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ApoE protein isoforms appear in different gel features on 2D-DIGE. Overlays of fluorescent 2D-DIGE images from gels representing CSF from two individuals with homozygosity for APOE-ε2 (green) or APOE-ε3 (red) (panel A) and for APOE-ε3 (green) or APOE-ε4 (red) (panel B) illustrate the heterogeneity of signal distribution by isoelectric point and molecular weight among apoE protein isoforms derived from different alleles. In panels C, D, E, F, G, H, signal intensities of individual CSF samples, grouped by genotype (2/2, 3/3 and 4/4 represent homozygotes; 2/3, 3/4 represent heterozygotes) are indicated for six apoE gel features (labeled C, D, E, F, G, H in panels A and B), illustrating that gel features C and D represent apoE2; gel feature E represents multiple forms; gel feature F represents apoE3; and gel features G and H, apoE4.
(TIF)
Mass spectrometry and protein identification data for 2D-DIGE gel features that differ in AD CSF. Results are ordered sequentially by “heat map #” [column A], corresponding to the ‘heat map’ row numbers in Figure 2. “Spot” [column B] refers to BVA number (see Methods). “(Accession) primary protein name” [column C] provides the gi number and protein name from the NCBI database. “Protein molecular weight” [column D] is the gene product molecular weight in Daltons. “Protein score” [column E] is the MASCOT-generated protein score. “Protein ID probability” [column F] indicates Scaffold's percent probability that the protein identification is correct. “Spectral count” [column G] is the number of spectra assigned to the protein by Scaffold. “Unique proteins” [column H] refers to the number of recognized tryptic peptides attributed to the protein by MASCOT. “Peptide sequence” [column I] indicates the amino acid sequence of the tryptic peptide predicted by MASCOT. “MASCOT ion score” [column J] is the MASCOT quality assessment of the peptide sequence assignment. “M/Z (observed)” [column K] is mass/charge ratio. “Mass (observed)” [column L] of peptide is indicated in Daltons. “Mass (theoretical)” [column M] is idealized tryptic peptide mass as predicted by NCBI. “Mass error (ppm)” [column N] is the error in parts per million determined through comparison of theoretical peptide mass to data generated by mass spectrometry. “MS source” [column O] reflects the mass spectrometer that produced the observed data (Q-STAR or LTQ-FT). “Modifications” [column P] lists variable post-translational modifications identified by mass spectrometry peptide sequence analysis.
(XLS)