Abstract
Dcp1 plays a key role in the mRNA decay process in Saccharomyces cerevisiae, cleaving off the 5′ cap to leave an end susceptible to exonucleolytic degradation. The eukaryotic initiation factor complex eIF4F, which in yeast contains the core components eIF4E and eIF4G, uses the cap as a binding site, serving as an initial point of assembly for the translation apparatus, and also binds the poly(A) binding protein Pab1. We show that Dcp1 binds to eIF4G and Pab1 as free proteins, as well as to the complex eIF4E–eIF4G–Pab1. Dcp1 interacts with the N-terminal region of eIF4G but does not compete significantly with eIF4E or Pab1 for binding to eIF4G. Most importantly, eIF4G acts as a function-enhancing recruitment factor for Dcp1. However, eIF4E blocks this effect as a component of the high affinity cap-binding complex eIF4E–eIF4G. Indeed, cooperative enhancement of the eIF4E–cap interaction stabilizes yeast mRNAs in vivo. These data on interactions at the interface between translation and mRNA decay suggest how events at the 5′ cap and 3′ poly(A) tail might be coupled.
Keywords: cap-binding proteins/decapping enzyme/eukaryotic translation initiation factor complex 4F/mRNA stability/poly(A) binding protein
Introduction
mRNA molecules leaving the eukaryotic nucleus become incorporated into polysome complexes that use the mRNA as a template for protein synthesis. The intact, capped and polyadenylated mRNA is loaded with up to approximately two actively translating ribosomes per 100 nucleotides. However, at some point this translating polysome complex enters a process of disassembly that is associated with degradation of the mRNA. The associated half-life varies by up to a factor of 50 for different mRNA species. The current working model of this process in Saccharomyces cerevisiae envisages that progressive deadenylation eventually triggers decapping by virtue of an as yet unknown mechanism of coupling between events occurring at the 3′ and 5′ ends of the mRNA (Caponigro and Parker, 1996). This type of pathway may also apply to at least some mRNAs in higher eukaryotes (Ross, 1996; Couttet et al., 1997). Decapping is the process of removing the 5′ cap of the mRNA, which thus effectively eliminates the assembly point for the cap-binding complex eIF4F, a key factor involved in mediating translation initiation. A decapping enzyme, Dcp1, has been described in S.cerevisiae (Stevens, 1980, 1988; Beelman et al., 1996; LaGrandeur and Parker, 1998). Moreover, decapping also renders the 5′ end of the mRNA susceptible to 5′→3′ exonucleolytic degradation, which in yeast is catalysed by Xrn1 (Larimer and Stevens, 1990; Caponigro and Parker, 1996).
Given the dual roles of the 5′ cap in promoting translation initiation and protecting mRNA against 5′→3′ exonucleolytic degradation, control of the decapping event is of major significance as a switching point between active protein synthesis and mRNA decay. As a step that exerts strong rate control over the overall process of mRNA decay, decapping plays an important role as a determinant of mRNA half-lives in the cell (Beelman et al., 1996; Caponigro and Parker, 1996). Equally, removal of the cap eliminates the major anchor point for the translational components that mediate early events in the initiation pathway (McCarthy, 1998). It is therefore a central question for the field whether there are physical and/or functional interactions between the cap-binding complex eIF4F and the decapping activity.
The eukaryotic initiation factor complex eIF4F comprises the cap-binding protein eIF4E (∼25 kDa), a much larger factor called eIF4G and the DEAD box (helicase) protein eIF4A (Merrick and Hershey, 1996; Gingras et al., 1999). Saccharomyces cerevisae has two versions of eIF4G (Goyer et al., 1993), eIF4G1 (107 kDa) and eIF4G2 (104 kDa). The association between eIF4G and eIF4A appears to be much less stable in yeast, and the latter factor binds in greatly substoichiometric amounts to eIF4G (Dominguez et al., 1999; Neff and Sachs, 1999). eIF4G acts like a scaffolding protein, in that it has binding sites for other translation-related factors (Lamphear et al., 1995; Mader et al., 1995; Tarun and Sachs, 1996; Morley et al., 1997; Pyronnet et al., 1999), including eIF4A, eIF4E, eIF3 and poly(A) binding protein (Pab1).
It now seems likely that the structural and functional properties of the macromolecular complex bound to the mRNA cap are dynamic and responsive to intermolecular interactions (McCarthy, 1998). For example, experiments with yeast translation factors have shown that, at least in vitro, interactions between the eIF4E-binding domain of eIF4G and eIF4E can exert a positive modulatory effect on cap binding. This is mediated by a dorsal binding site on eIF4E that is bound by both eIF4G and the yeast eIF4E-binding protein (4E-BP) called p20 (Ptushkina et al., 1998). p20 can compete with eIF4G for binding to eIF4E (Altmann et al., 1997), although it does not seem to act as a strong regulator of translation in vivo (Altmann et al., 1997; de la Cruz et al., 1997). The fact that p20 is a phosphoprotein has raised the possibility that its function is regulatable (Zanchin and McCarthy, 1995). Other reports have indicated that binding of the poly(A) binding protein (PABP) to eIF–iso4F in wheat germ extracts enhances eIF–iso4F–cap interactions (Wei et al., 1998), that the binding of RNA to yeast Pab1 enhances this protein’s affinity for eIF4G (Tarun and Sachs, 1996), and that the cap-binding affinity of mammalian eIF4E is subject to modulation by protein ligand binding to this factor’s dorsal face (Ptushkina et al., 1999). All these observations paint a picture of eIF4F as potentially a key player in a network of modulatory interactions based on cooperativity effects. Previous to the present study, however, there was no direct evidence of interactions of this kind that act across the interface between translation and mRNA decay.
In this study we have discovered that both eIF4G and the poly(A) binding protein Pab1 bind to Dcp1, either independently, or when these proteins are in the 5′–3′ translation complex involving eIF4F and Pab1. Moreover, eIF4G acts as a potent modulator of Dcp1 activity, while eIF4E blocks this effect. These results provide new insight into the functional interactions that could underlie communication between events at the 5′ and 3′ ends of eukaryotic mRNA within the cell, and establish a basis for understanding the relationship between translation and mRNA decay.
Results
Dcp1 binds to eIF4G and Pab1
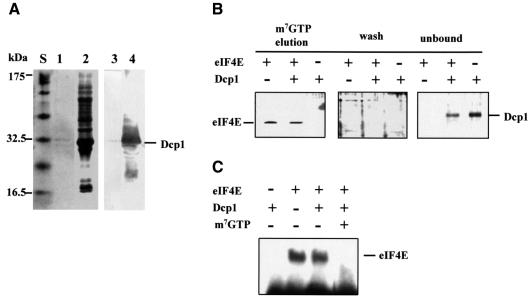
We asked whether a direct link exists between the macromolecular assemblies of translation initiation and mRNA degradation that act at the 5′ end of the mRNA. In particular, is Dcp1 a ligand of proteins associated with the eIF4F complex? We generated FLAG-tagged Dcp1 and poly(His)-tagged Pab1 using inducible expression plasmids in Escherichia coli, while producing poly(His)-tagged eIF4G in an insect cell line (see Materials and methods). The FLAG-tagged Dcp1 protein used in most of the experiments described in this study was purified from an E.coli strain transformed with a suitable expression construct (PET5AFLAG-Dcp1; Figure 1A and B). We observed that if induction times longer than 2 h were used for the expression phase, or an inappropriate purification procedure was followed, this protein was largely cleaved to yield a smaller product (see Materials and methods). This may explain why in a previous report by LaGrandeur and Parker (1998), the Dcp1 purified from E.coli was found to have a reduced decapping activity relative to the corresponding protein isolated from S.cerevisiae. In this study we have used the active Dcp1 purified according to our new procedure from E.coli (see Figure 7) because this protein could be obtained at high levels of purity and free from contamination by other yeast proteins.
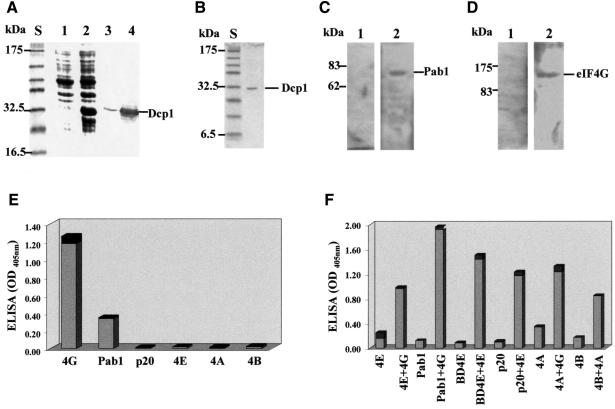
Fig. 1. Dcp1 binds eIF4G and Pab1 in vitro. (A) FLAG-Dcp1 in total cell extracts from E.coli BL21 before (lane 1) and after (lane 2) induction. Lanes 3 and 4 show the western blots (using anti-FLAG antibody) corresponding to lanes 1 and 2, respectively. (B) The purity of the final FLAG-Dcp1 preparation was assessed by means of SDS–PAGE and Coomassie Blue staining. Far-western analysis revealed binding of Dcp1 to Pab1 (C) and to eIF4G (D). Following SDS–PAGE (10 and 7.5% gels, respectively) and immobilization of purified recombinant Pab1 and eIF4G, FLAG-Dcp1 was overlaid, as described in Materials and methods, and binding was detected by western blot analysis using anti-FLAG antibody (lanes 2). The control lanes labelled 1 in (C) and (D) show the results of the western blot analysis performed before incubation with FLAG-Dcp1. (E) An ELISA gave positive results for the Dcp1–eIF4G and Dcp1–Pab1 interactions, whilst revealing no binding to eIF4E, eIF4A, eIF4B or p20. In this series of assays, the proteins indicated were immobilized on the plate and FLAG-Dcp1, followed by anti-FLAG and the secondary antibody, were overlaid. (F) A series of ELISA controls demonstrated that the proteins used in (E) could all react with other known ligands. BD4E is the eIF4E-binding domain of eIF4G (see Figure 4A). The binding experiments were performed in pairs, comparing the signal obtained for each protein with and without a known protein ligand. Since a different primary antibody was used for each binding partner, the results in (F) are not directly quantitatively comparable. The results shown are averages of at least three separate experiments, and the black sections of the columns represent the standard deviation values. The positions of molecular weight standards [S in (A) and (B)] on the respective gels are indicated (A–D).
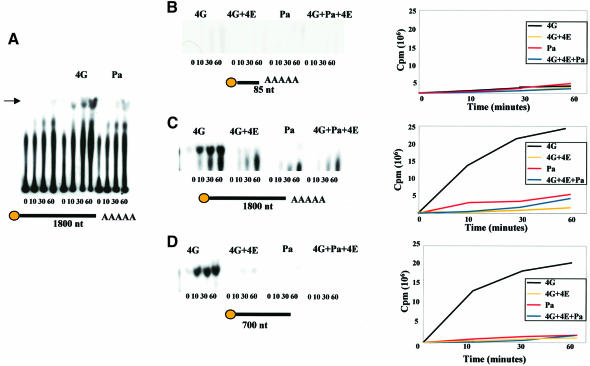
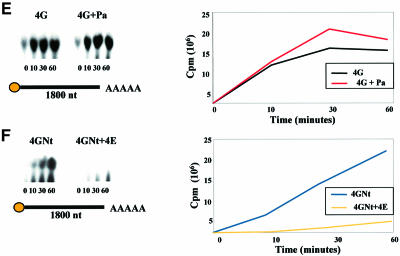
Fig. 7. eIF4G and eIF4E are modulators of Dcp1 function. Analysis of decapping activity by FLAG-Dcp1 in the presence of eIF4G, eIF4E and Pab1. Decapping by FLAG-Dcp1 was examined over a 1 h time course, with aliquots of the decapping reactions removed at the indicated time points. The products of the reaction were separated by PEI-cellulose TLC; equal amounts of material were loaded on each lane. Each mRNA used in the experiments is schematically represented under each plate, with the length (in nucleotides), and presence of a cap or poly(A) tail indicated. The arrow in (A) indicates the position of the 32P-labelled m7GDP generated by decapping. The band running below m7GDP on these plates corresponds to GDP that was probably released from capped mRNA whose cap was not fully methylated during substrate preparation, due to incomplete methylation by guanyltransferase. (A) Dcp1 activity in the presence of eIF4G. The enhancement of decapping induced by eIF4G is suppressed in the presence of eIF4E (C and D). Moreover, decapping is a function of substrate length, being hardly detectable with the 86 nucleotide mRNA (B), but does not depend on the presence of a poly(A) tail (D). Pab1 has a relatively minor positive effect on the decapping activity (A, C, D and E). The N-terminal fragment of eIF4G (4GNt) also enhances decapping activity, albeit rather less effectively than whole eIF4G (F). The data shown are typical for the respective experiments, which were all performed at least three times each.
Far-western analyses using the intact material purified from E.coli according to our procedure (Figure 1B) revealed that Dcp1 can form complexes with both Pab1 and eIF4G (Figure 1C and D). The same proteins were used to establish a sandwich ELISA procedure, which confirmed the Dcp1–eIF4G and Dcp1–Pab1 interactions (Figure 1E). In control experiments, no evidence of Dcp1 binding to other eIF4F-associated proteins (p20, eIF4E, eIF4A or eIF4B) was found (Figure 1E), while the various potential binding partners for Dcp1 were found to react positively with known protein ligands (Figure 1F).
Next we investigated whether Dcp1–eIF4G and Dcp1–Pab1 interactions could be detected in extracts prepared from yeast cells. The FLAG-tagged version of Dcp1 was expressed in the yeast strain W303. Co-immunoprecipitation experiments were performed using antibodies specific for Pab1 and the FLAG sequence tag on Dcp1, respectively (Figure 2). From the results we conclude that Dcp1 associates with both eIF4G and Pab1 in vivo.
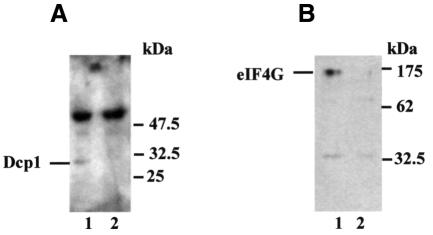
Fig. 2. Dcp1 binds eIF4G and Pab1 in vivo. Co-immunoprecipitation was performed using antibodies against Pab1 (A) or the FLAG sequence tag on Dcp1 (B), followed by loading on SDS–polyacrylamide gels. Western blot analysis then demonstrated the presence of Dcp1 (A) and eIF4G (B), respectively, in the immunoprecipitates. The experiments were performed using S.cerevisiae strain W303 as a control (lanes labelled 2), or W303 in which FLAG-Dcp1 is expressed from a 2µ plasmid (lanes labelled 1). The secondary peroxidase-coupled anti-rabbit IgG antibody reacted with the rabbit anti-Pab1 immunoglobulins in (A) (see strong higher molecular weight bands in both lanes 1 and 2).
Dcp1 is not a major competitor for ligands of eIF4E
Since Dcp1, like eIF4E, is a cap-interacting factor, we sought to determine whether these two proteins compete for binding to the same ligands. The FLAG-Dcp1 prepared according to our procedure bound m7GTP–Sepharose only weakly compared with eIF4E (Figure 3A; compare Figures 3B, 4B and 5C). Indeed, we found that at least 1000 times more Dcp1 needs to be added to a given volume of m7GTP–Sepharose in order for its retention to be detectable by the procedure used for eIF4E. Moreover, Dcp1 competed at most very poorly with eIF4E binding to the same affinity resin (Figure 3B). In a further experimental approach, we found that Dcp1 exerted at most a minimal effect on the cross-linking of eIF4E to radiolabelled, capped mRNA at equimolar levels (Figure 3C).
Fig. 3. Dcp1 shows very weak cap-binding and does not compete with eIF4E for binding to the cap structure. (A) A cell extract from the E.coli BL21 strain producing Dcp1 was incubated with m7GTP–Sepharose and the eluted fractions (lane 1) were collected for SDS–PAGE and Coomassie Blue staining. Lane 2 shows the material that flowed through the column without binding. More than 1 mg of FLAG-Dcp1 was present in the cell extract passed through the column. The identity of Dcp1 in these lanes was confirmed via western blotting using an anti-Dcp1 antibody (lanes 3 and 4; compare Figure 1A). (B) Purified recombinant eIF4E and Dcp1, individually or combined, were allowed to bind to m7GTP–Sepharose. The amount of each protein added was identical (5 µg). After elution with m7GTP, the fractions were rendered visible by silver-staining. The staining results obtained with the wash and unbound fractions (see Materials and methods) are shown for comparison. (C) The ability of Dcp1 to bind to the mRNA was further assessed by means of UV crosslinking analysis. An 86 nucleotide capped mRNA was radioactively labelled using [α-32P]ATP (see Materials and methods) and incubated together with either Dcp1 or eIF4E, or with both proteins simultaneously. RNase treatment was used to remove non-crosslinked RNA after UV irradiation, thus generating a labelled eIF4E–RNA complex visible in lanes 2 and 3. As a control, the cap analogue m7GTP was added, as indicated in the figure, which competed with the capped mRNA for binding to eIF4E. These results are typical for the experiments shown, which were performed at least three times each.
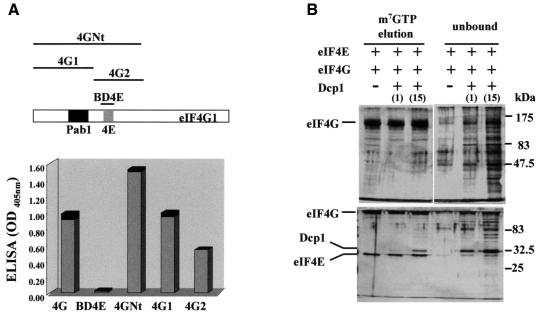
Fig. 4. Dcp1 does not bind the eIF4E-binding site in eIF4G. (A) Schematic representation of eIF4G and the fragments generated from this protein that were used in this study. ELISA analysis results for interactions between Dcp1 and the eIF4E-binding domain of eIF4G (BD4E), the N-terminal region of this protein (4GNt, amino acids 1–513) and the eIF4G fragments corresponding to amino acids 1–329 (4G1) and 327–539 (4G2), are shown. The proteins indicated on the x-axis were immobilized on the microtitre plate and allowed to react with FLAG-Dcp1, which was detected using an anti-FLAG antibody. The black sections of the columns represent the respective standard deviations. (B) m7GTP–Sepharose chromatography was used to assess whether Dcp1 competes effectively with eIF4G for binding to eIF4E. The proteins indicated were incubated with the affinity resin and, after elution, analysed using SDS–PAGE [7.5% (upper) or 12.5% (lower) gels). Dcp1 was added to either the same concentration as eIF4E (1), or to 15 times the concentration of eIF4E (15). Silver staining was allowed to develop until the weakest bands were visible. Unbound fractions of the material that ran through the column were also loaded on the gels. Under these conditions, no Dcp1 binding to the affinity resin could be detected (data not shown; compare Figure 3B). The identities of eIF4G, eIF4E and Dcp1 were confirmed independently by means of western blotting using the appropriate specific antibodies (data not shown). The results shown are typical for the experiments shown, which were performed at least three times each.
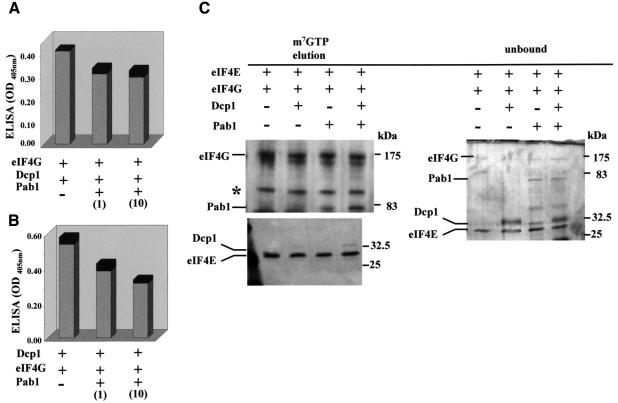
Fig. 5. Pab1 and Dcp1 can bind simultaneously to eIF4G. ELISA sandwich analysis revealed minimal competition between Pab1 and Dcp1 for binding to eIF4G (A), or between Pab1 and eIF4G for binding to Dcp1 (B), even in the presence of a 10-fold excess of Pab1 relative to the other free ligand (see figures in parentheses for relative concentrations). In (A), eIF4G was immobilized to the plate and the proteins indicated were overlaid; in (B), Dcp1 was the immobilized binding partner. (C) eIF4E, eIF4G, Pab1 and Dcp1 were allowed to bind to m7GTP–Sepharose, as indicated in the figure. After elution with m7GTP, the proteins were subjected to SDS–PAGE and silver-staining. The Dcp1 band in the experiment involving incubation with eIF4E and eIF4G was very weak (and is hardly visible in this figure), but became significantly stronger when Pab1 was added (see text for details). Most of the Dcp1 remained unbound. In further control experiments (see e.g. Figure 3B), we were unable to observe any binding of Dcp1 to the affinity resin under the stated conditions. One of the eIF4G degradation fragments visible in the unbound fragments showed a similar mobility to Dcp1, but is still clearly distinguishable as a separate band. These results are typical for the experiments shown, which were performed at least three times each. The band marked with an asterisk is a further breakdown product of eIF4G.
We next determined whether Dcp1 binds to the same region of eIF4G as eIF4E. ELISA experiments (Figure 4A) demonstrated that Dcp1 binds to complete eIF4G and the N-terminal region of eIF4G (4GNt), but not to the eIF4E-binding domain of eIF4G (BD4E). Further far-western analyses confirmed these results (data not shown). It also binds the region of eIF4G comprising amino acids 1–319 (4G1) and, to a lesser degree, the region comprising amino acids 317–539.
Moreover, Dcp1 competes poorly with eIF4G for binding to eIF4E on m7GTP–Sepharose (Figure 4B); it seems capable of binding simultaneously with eIF4G, albeit with a lower affinity. Given the poor affinity of Dcp1 for the m7GTP–Sepharose matrix (Figure 3), the increased amount of Dcp1 in eluted fractions in the presence of eIF4G and eIF4E (Figure 4B) indicates that Dcp1 binds to the eIF4E–eIF4G complex. A superstoichiometric ratio of Dcp1 to eIF4E and eIF4G was required in order to obtain comparable levels of retention on the affinity matrix, presumably because of the relatively low affinity of Dcp1 for eIF4G. In a further experiment, we found that neither eIF4G nor 4GNt enhances the binding of Dcp1 to m7GTP–Sepharose (data not shown).
Pab1 and Dcp1 are non-competitive ligands of eIF4G
We examined the competitiveness of Pab1 and Dcp1 binding to eIF4G. An ELISA procedure revealed at most minimal competition for binding of Dcp1 to eIF4G, even in the presence of a 10-fold excess of Pab1 (Figure 5A and B). Moreover, all four proteins, eIF4E, eIF4G, Pab1 and Dcp1, were able to bind simultaneously to m7GTP–Sepharose (Figure 5C). Indeed, the addition of Pab1 and Dcp1 mutually enhanced the amounts of these respective proteins retained on the cap-analogue resin compared with experiments in which one or the other was omitted. It therefore appears that a relatively stable eIF4E–eIF4G–Pab1–Dcp1 complex can be formed. In a control experiment, we observed that the presence of another eIF4G-binding protein, eIF4A, did not stabilize association of either Pab1 or Dcp1 with the complex (data not shown).
Dcp1 can destabilize the eIF4F complex on mRNA
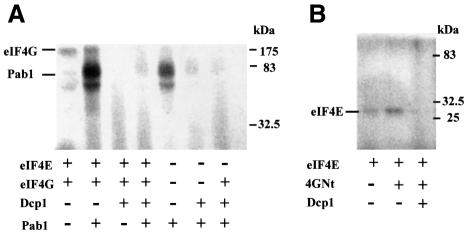
We asked the question whether Dcp1 binding is a neutral event in terms of the conformation of the eIF4F–Pab1 complex, or possibly influences the way that this complex interacts with the mRNA. Cross-linking experiments were performed using a relatively short (86 nucleotides) radioactively labelled mRNA (Figure 6). The eIF4E–eIF4G complex was readily cross-linked to the mRNA, yielding a major band plus smear after RNase treatment, most likely because eIF4G can bind to different stretches of the mRNA, thus allowing cleavage to different lengths in the RNase treatment phase of the procedure (Figure 6A). The addition of Pab1 leads to a refocusing of the cross-link bands, presumably because of the shifting of the complex into a different range of interactions with the mRNA. Dcp1 acts apparently to reverse the effect of Pab1 binding. It is noteworthy that strongly focused bands were also evident in the presence of Pab1 alone, and that the addition of Dcp1 again acted to eliminate them almost completely.
Fig. 6. Dcp1 can impose conformational changes on the eIF4F complex. A radiolabelled, capped and polyadenylated 86 nucleotide mRNA (see Materials and methods) was incubated with the proteins indicated in the figure. Either whole eIF4G (A) or 4GNt (B) was used in these experiments. After UV crosslinking, each fraction was analysed by SDS–PAGE. The positions of the eIF4G, Pab1 and eIF4E proteins, which were all confirmed by means of separate western blotting experiments (not shown here), are indicated. These results are typical for the experiments shown, which were performed at least three times each.
In a further set of experiments, 4GNt (Figure 4A) was used instead of whole eIF4G (Figure 6B). In this case, the defined band obtained with eIF4E alone was retained in the presence of 4GNt. This is likely to be related to the fact that 4GNt lacks the RNA-binding domains in the C-terminal region of eIF4G, so that the association with the mRNA is more dependent on binding to eIF4E. As with whole eIF4G, however, the addition of Dcp1 resulted in the loss of the main cross-linking band.
The defined cross-link bands observed in the above experiments constitute evidence of close association between the eIF4F-associated proteins and the mRNA. The apparently disruptive effect of Dcp1 could theoretically be explained in either of two ways. First, Dcp1 remodels the complex between eIF4E–eIF4G–Pab1 and capped mRNA so that its mobility in SDS–PAGE is affected; secondly, Dcp1 decaps the mRNA, thus preventing stable maintenance of the complex. The major question raised by these observations could be resolved by the subsequent examination of the effects of protein ligands on Dcp1 decapping behaviour.
eIF4G, eIF4E and Pab1 are modulators of Dcp1 function
The observation that Dcp1 can bind to protein factors involved in translation prompted us to explore whether these interactions modulate the decapping activity. Measurements of decapping rate in vitro revealed that eIF4G and, to a much lesser extent, Pab1, enhance the decapping activity of Dcp1 (Figure 7A). However, this only applies to the two longer mRNAs used in this study (Figure 7C and D); the shorter mRNA of 86 nucleotides is not decapped at a measurable rate, even in the presence of eIF4G or Pab1 (Figure 7B). Selectivity for longer mRNAs was already shown for Dcp1 alone by LaGrandeur and Parker (1998), which means that eIF4G does not change this property of the protein. eIF4G and Pab1 apparently act independently on Dcp1, in that the Dcp1–eIF4G–Pab1 complex shows a comparable decapping rate to that of Dcp1–eIF4G (Figure 7E). A particularly striking result is the observation that the presence of eIF4E inhibits decapping by the Dcp1–eIF4G complex (Figure 7C and D). The N-terminal region of eIF4G (4GNt) also shows a substantial enhancing effect on Dcp1 activity (Figure 7F), but this is clearly reduced compared with the modulatory influence of whole eIF4G. Finally, it should be emphasized that we observed the same enhancing effect of eIF4G using Dcp1 purified directly from S.cerevisiae (data not shown), meaning that the dependence of Dcp1 activity on eIF4G is not a property peculiar to the recombinant protein from E.coli.
From the above results, we conclude that the very significant changes in the cross-linking patterns for eIF4E–eIF4G–Pab1 and eIF4E–eIF4G with the short mRNA (85 nucleotides) caused by the addition of Dcp1 (Figure 6) must be attributable to conformational changes in the complex, and not to decapping by Dcp1.
Stabilization of mRNA via eIF4E
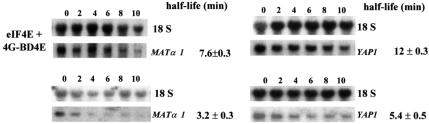
If the rate of decapping is regulated at the molecular level via the accessibility of the cap to Dcp1, it would be predicted that changes in the level of protection of the cap by eIF4E should affect mRNA stability. Since stronger binding of eIF4E to the cap is promoted by interaction with the eIF4E-binding domain of eIF4G (Ptushkina et al., 1998), we tested this prediction by inducing synthesis of 4G-BD4E in yeast. We chose to examine endogenous mRNAs with short- to mid-range half-lives (Caponigro and Parker, 1996; Vilela et al., 1999) since changes in their half-lives can be detected via measurements over a relatively short fraction of the cell division cycle. This minimizes the potential influence of longer-term general effects on translation. Moreover, the response of the stability of one of these mRNAs (MATα1) to changes in decapping rate has been studied before (Caponigro and Parker, 1996), and these previously reported results provide good reference values. Expression of the corresponding region of eIF4G (4G-BD4E; see Figure 4A) was found to stabilize the MATα1 and YAP1 mRNAs in vivo, especially under conditions of co-expression together with CDC33 (the yeast gene that encodes eIF4E; Figure 8). Expression of 4G-BD4E alone (MATα1: 5.5 ± 0.6 min; YAP1: 6.0 ± 0.2 min) or overexpression of only CDC33 (MATα1: 3.4 ± 0.1 min; YAP1: 5.4 ± 0.4 min) had more limited effects, or no effect, respectively, on the half-lives. The most likely explanation of this result is that modulation of the accessibility of the cap to Dcp1 via control of the eIF4G–eIF4E interaction is of critical importance to the control of decapping, and thereby of mRNA stability.
Fig. 8. Modulation of mRNA stability in vivo via eIF4E. Northern blots show the results of hybridization using RNA preparations from the S.cerevisiae strain JDS+S taken at various time-points during half-life determination experiments. The wild-type endogenous 18S rRNA was used as an internal reference for correction of mRNA values for variations in the amount of total RNA isolated. Typical northern blot data are displayed for non-transformed JDS+S, and for JDS+S transformed with plasmids directing the synthesis of eIF4E and 4G-BD4E (as indicated to the left of the figure). The estimated half-life values represent averages of measurements performed using at least three independent sets of RNA preparations (± SD).
Discussion
In this study we have described functional and/or physical interactions between an enzyme that participates in mRNA degradation, Dcp1, and proteins that act to promote translational initiation, eIF4G, eIF4E and Pab1. The results indicate that Dcp1 alone shows only a basal level of decapping activity, and that it needs to be recruited to the mRNA and activated by eIF4G in order to be fully functional. This effect is only partially lost when the C-terminal part containing the RNA-binding domains is deleted from eIF4G. This is consistent with a model in which the RNA-binding capacity of eIF4G contributes to, but is not essential for, this protein’s function as a Dcp1 recruitment and activation factor. Since eIF4G promotes neither decapping of the 86 nucleotide mRNA nor tighter binding of Dcp1 to m7GTP–Sepharose, we conclude that eIF4G-enhanced decapping is not simply attributable to an increased affinity of Dcp1 for the cap structure. Instead, a combination of the properties of eIF4G, including RNA binding and the molecular consequences of the eIF4G–Dcp1 interaction, is responsible for its stimulatory function.
The binding of eIF4G to eIF4E localizes eIF4G near the cap. Indeed, the eIF4G–eIF4E interaction stabilizes association of eIF4F with the cap at least partly by virtue of positive cooperativity between eIF4G binding at the eIF4E dorsal binding site and the cap interaction (Ptushkina et al., 1998). While this anchoring of eIF4G to the 5′ end of the mRNA will provide a docking site for Dcp1 that is close to its natural place of action, the presence of eIF4E blocks access for Dcp1 to the cap. The further observation that Dcp1 also binds Pab1 independently of eIF4G suggests that there can be direct communication between the decapping process and the 3′ end/poly(A)–Pab1 complex, although further investigation of this interaction is not within the scope of the present work.
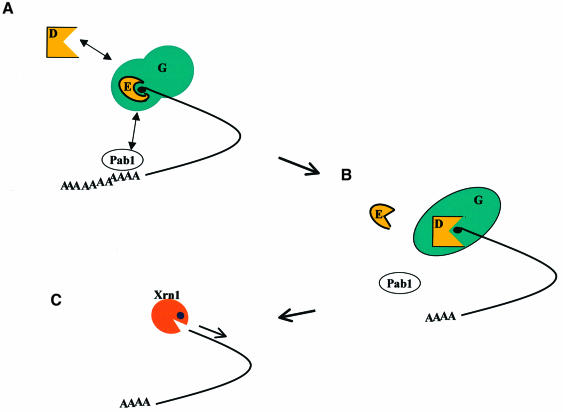
Overall, our results indicate that the eIF4E–eIF4G complex suppresses Dcp1 activity. Thus, access of Dcp1 to the cap may require remodelling of the eIF4E–eIF4G complex and/or release of eIF4E (Figure 9). It is noteworthy that there is a marked change in the conformation of eIF4E–eIF4G or of eIF4E–eIF4G–Pab1 when Dcp1 binds. This remodelling process is not, in itself, sufficient to allow Dcp1 activation. However, combined with an as yet undefined event that allows release of eIF4E from the eIF4F complex (Figure 9), it may be an important step towards activation of Dcp1. Also taking into account the positive effect on mRNA stability of stabilizing the eIF4E–cap interaction (Figure 8), our working hypothesis predicts that the switching of eIF4G-mediated stabilization of the eIF4E–cap interaction to eIF4G-mediated activation of Dcp1 governs a transition from translation to mRNA decay. As a subsequent project following on from this study, it will be important to define in quantitative terms the kinetic and regulatory characteristics of the molecular interactions between eIF4F, Pab1 and Dcp1, and how these relate to the shutdown of translation and the triggering of decapping.
Fig. 9. A working hypothesis for modulatory interactions between eIF4F–Pab1 and Dcp1. (A) The eIF4F core complex, eIF4E–eIF4G, binds tightly to capped mRNA. Dcp1 can bind eIF4G, but has no access to the cap. Pab1 molecules associated with the poly(A) tail can bind to eIF4G, forming a 5′–3′ translation initiation complex. The presence of eIF4E in the stable initiation complex prevents Dcp1 from decapping the mRNA. (B) Pab1 may be involved in signalling to the 5′ end that deadenylation has occurred, thus triggering rearrangement of the 5′ complex. eIF4E is released from eIF4F, probably in its lower affinity form, thus allowing Dcp1 access to the cap. (C) Decapping generates an mRNA that is susceptible to exonucleolytic degradation by Xrn1. This model makes predictions that can be readily tested experimentally.
In previous work, we showed that changes in the cellular abundance of free eIF4E do not modulate mRNA stability (Linz et al., 1997). This indicated that free eIF4E does not compete effectively with Dcp1 for access to the cap. We can now understand this in the context of our interactive model, which predicts that the cellular level of eIF4E bound to eIF4G, rather than of free eIF4E, determines the accessibility of the cap to Dcp1. Another important conclusion arising from the present study is that eIF4G is the only member of the eIF4 group of factors that enhances Dcp1 decapping activity, thus demonstrating that this is a specific function of the largest eIF4 factor.
There have been reports indicating that proteins other than eIF4G and Pab1 interact, directly or indirectly, with Dcp1. The DCP2 gene has been identified as a multicopy suppressor of both a nuclear petite mutant (when it was named PSU1) and the growth defect of a dcp1Δski8Δ strain (Dunckley and Parker, 1999). Dcp2 may be multifunctional, but one of its properties is to promote decapping via an as yet undefined, indirect mechanism. The efficiency of decapping is also affected by mutations in a number of other genes, including MRT1, MRT3 (Hatfield et al., 1996), SPB8 (Boeck et al., 1998) and VPS16 (Zhang et al., 1999), although the mode of action of the encoded proteins is unknown. Two-hybrid screening has identified other potentially significant interactions with Dcp1, including some that are involved in RNA splicing (Uetz et al., 2000). Indeed, the so-called Sm-like proteins (Lsm1-7) co-immunoprecipitate with Dcp1, and lsm mutations accumulate oligoadenylated, full-length mRNAs (Bouveret et al., 2000; Tharun et al., 2000).
None of the above proteins has been shown to modulate the specific activity of Dcp1, and it is likely that in most cases the relationship is indirect. The existence of so many functional interactions is by no means peculiar to Dcp1, and is consistent with the idea that interaction networks are likely to represent a general theme in the cellular environment (McCarthy, 1998). The functional significance of at least some of these interactions may not be evident when studied in purified systems, perhaps only being amenable to meaningful analysis in the context of the overall network. In a wider context, the apparent ‘networking’ of molecular interactions is becoming increasingly evident as various groups discover complexes between components of the cellular machineries that perform processes that were previously believed to occur independently.
The reason why Dcp1–eIF4G and Dcp1–Pab1 binding has only now been identified by biochemical procedures is likely to be related to the fact that these interactions, like eIF4A–eIF4G binding (Dominguez et al., 1999; Neff and Sachs, 1999), are effectively substoichiometric and are, at least under certain conditions, relatively weak. However, as with eIF4A–eIF4G, this apparently low affinity does not mean that the interaction is functionally insignificant. Indeed, quite the opposite seems to be true in the case of eIF4G–Dcp1. Moreover, the observation that these interactions are not of a very high affinity is by no means anomalous, since tight binding that is not subject to modulation would impose a permanent change in the behaviour of Dcp1, rather than allowing for a controlled mode of action. Genetically, of course, recognition of the functional interaction between eIF4G and Dcp1 would have been complicated by the presence of two eIF4G-encoding genes in S.cerevisiae.
As we have shown previously for eIF4E–eIF4G (Ptushkina et al., 1998), conformational changes may underlie dynamic alterations in binding affinity. This seems to be a common theme in the mode of function of the eIF4F complex. In this case, it is possible that the binding affinity of Dcp1 for eIF4G or for eIF4F:Pab1 might be variable, perhaps in response to the series of events: translation of nascent mRNA→deadenylation/loss of translatability→mRNA degradation (Figure 9). Further work will be needed to test this possibility. Moreover, molecular channelling plays an undeniably important role in guiding and kinetically controlling the complex steps of post-transcriptional control (Negrutskii et al., 1994; McCarthy, 1998; Suppmann et al., 1999). The channelling effects are imposed by macromolecular supercomplexes that create a microenvironment in which the progress of sequential steps of a pathway is subject to altered kinetic and thermodynamic parameters relative to ideal solution conditions. Future work will therefore also have to examine how the subcellular organization of the tightly packed components of the translation and mRNA decay machineries influences the kinetics and control of these two processes.
Finally, the identification of the modulatory interactions between Dcp1, eIF4G, eIF4E and Pab1 may open the door to significant advances towards understanding the molecular mechanisms defining the interface between translation and mRNA decay in eukaryotes. Recently, another group (Dehlin et al., 2000) has suggested that in mammalian cells, another cap-binding protein [poly(A)-specific exoribonuclease, or PARN] might couple translational events at the 5′ end with the mRNA decay process. Future work will reveal whether in yeast the system we have described is the only form of molecular interface between translation and degradation.
Materials and methods
Plasmids and yeast strains
An NdeI–EcoRI fragment encoding the His6-tagged version of 4G-BD4E was cut from the E.coli expression plasmid used previously (Belev et al., 1991; Ptushkina et al., 1998) and inserted into the inducible yeast expression vector (YCpSUPEX1; Oliveira et al., 1993). YCpSUPEX1-4G-BD4E was restricted with HindIII and the fragment containing the PGPF promoter plus the His6-4G-BD4E subsequently inserted into pFL39 (Brachman et al., 1998). The YCpSUPEX1-eIF4E plasmid (Ptushkina et al., 1998) was also used in this study. The plasmid pG-1 Flag-Dcp1 (kindly provided by Dr Stuart Peltz) was used as the template for PCR to add NdeI and XbaI restriction sites. The oligonucleotides 5′-GGGAAT TCCATATGGACTACAAGGACGACGATGAC-3′ and 5′-CTAGTC TAGATCAAGCAAAAGAATCTTTTGGCT-3′ were used in the amplification procedure. The resulting Flag-Dcp1 fragment was ultimately inserted into the yeast expression vector pRS402 (Brachman et al., 1998). For Dcp1 production in E.coli the fragment NdeI–BamHI was inserted into the PET5A vector (Novagen).
The following yeast strains were used in this study: JDS+S, MATα ura3-52 trp1-Δ1 his4-38 leu2-1 rbp1-1 (kindly provided by Dr Audrey Atkin) and W303, MAT ura3 ade2 his3 leu2 trp1 (kindly provided by Dr Patrick Linder). Yeast transformation was performed according to standard procedures (Schiestl and Gietz, 1989) using single- stranded nucleic acids as carrier. The E.coli strain BL21 hsdS gal (λcIts857 ind1 Sam7 nin5 lacUV5-T7 gene 1) was used for protein production.
Protein production and purification
For the preparation of recombinant Dcp1, a 500 ml culture of the E.coli BL21 strain harbouring PET5A FLAG-Dcp1 was induced for 1 h in TB medium containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and harvested by centrifugation. Cells were washed in buffer A [20 mM HEPES pH 7.4, 100 mM KCl, 2 mM MgCl2 and 1 mM dithiothreitol (DTT)], suspended in 15 ml of buffer A and sonicated for 3 min. After centrifugation, the supernatant was precipitated in 90% (NH4)2SO4 and dialysed against buffer B (20 mM HEPES pH 7.4, 150 mM NaCl, 2 mM MgCl2 and 1 mM DTT). The resulting fraction was then loaded on a 1 ml anti-FLAGM2 monoclonal antibody immunoaffinity column (Kodak) and allowed to equilibrate with buffer B at 4°C. The unbound material was collected and reapplied to the column. The column was washed with 15 ml of buffer B and the protein was eluted with 6 ml of 0.1 M glycine. The protein was subsequently dialysed against buffer A, separated into aliquots and stored at –80°C. The correct identity of the recombinant Dcp1 was confirmed by means of western blotting and N-terminal protein sequencing. Yeast FLAG-Dcp1 was purified as described in LaGrandeur and Parker (1998), except that the host strain was W303 and the expression plasmid was the pRS402 derivative described above.
For the preparation of recombinant Pab1, 2 l of BL21 carrying pPET5A-His6-tagged-PAB1 were centrifuged and sonicated. The supernatant was precipitated with 90% (NH4)2SO4 and the fractions were loaded onto a Ni-affinity chromatography column and eluted with 400 mM imidazole in buffer containing 20 mM HEPES pH 7.5 and 0.2 M KCl. Fractions containing Pab1 were applied to a Heparin column using a BioCAD 700E (PerSeptive Biosystems Inc., USA). Elution was performed with 20 mM HEPES pH 7.5 and 1 M KCl and followed by multiple dialysis with buffer containing 20 mM HEPES, 0.2 M KCl, 5% glycerol, 2 mM MgCl2 and 1 mM DTT.
Recombinant eIF4G was generated in Sf9-insect cells transformed with BacPac8-His6-tagged-eIF4G1. After centrifugation, the pellet was resuspended in lysis buffer (20 mM HEPES, 100 mM NaCl, 5 mM EDTA, 5 mM EGTA, 5 mM DTT, 1 mM PFSM, 10 mM E-64). Tween 20 was added to a final concentration of 1%, and after mixing and further centrifugation the supernatant was precipitated with 50% (NH4)2SO4. The resulting pellet was resuspended in buffer containing 20 mM HEPES, 30 mM KCl, 1 mM DTT, 1 mM MgCl2 and applied on a Heparin column. Buffer containing 20 mM HEPES, 1 M KCl, 1 mM DTT, 1 mM MgCl2 was used for elution. Fractions with a high concentration of eIF4G1 were further purified using cation exchange chromatography on the BioCAD 700E.
Analytical m7GTP–Sepharose chromatography
m7GTP–Sepharose chromatography was performed as described previously (Ptushkina et al., 1999) with minor modifications. Briefly, each binding partner was incubated together in 20 mM HEPES pH 7.4, 100 mM KCl, 2 mM MgCl2 and 1 mM DTT at 4°C for 5 min, then 50 µl of m7GTP–Sepharose were added and the total volume was made up to 300 µl with further buffer. Incubation was continued at 4°C for 2 h with moderate shaking. The resin was subsequently washed twice with 1 ml of buffer A and bound proteins were eluted with 50 µl of 0.1 mM m7GTP in the same buffer. Eluted fractions (18 µl) were analysed on both 7.5 and 12.5% SDS–polyacrylamide gels and the proteins rendered visible by silver staining. The identities of the bands appearing in the analytical gels were confirmed by means of western blotting using specific antibodies raised against eIF4E, eIF4G, Pab1 and FLAG-Dcp1, respectively.
Preparation of mRNA templates
Uncapped mRNAs were synthesized in vitro using T7 RNA polymerase. The DNA template for the 86 nt RNA was created by annealing the oligonucleotides: GAATTGTAATACGACTCACTATAGTTTCACCA CCTCCACCACC TC C ACCACCTCAAAAAAAAAAAAAAAAAA AAAAAAAAAAAA (RNA53) and TTTTTTTTTTTTTTTTTTTTT TTTTTTTTTGAGGTGGTGGAGGTGGTGGAGGTGGTGAAACTAT AGTGAGTCGTATTACAATTC (RNA35). The 700 nucleotide mRNA is a truncated version of the LUC gene generated from a T7 promoter plasmid cleaved with EcoRI within the reading frame (Lang et al., 1994). Full-length LUC mRNA was transcribed from a similar plasmid digested with NsiI (Oliveira et al., 1993; Lang et al., 1994). T7 transcriptions and capping reactions were performed as described previously (Knapp, 1989; Lang et al., 1994).
Decapping assays
Decapping assays used in this study followed the protocol described in LaGrandeur and Parker (1998). The reactions generally contained 66 ng of Dcp1, equimolar amounts of eIF4G, 4GNt, Pab1 or eIF4E, 0.2 pmol of each m7G [32P]mRNA, 20 mM HEPES pH 7.5, 1 mM MgCl2, 1 U RNasin (Promega) and 1 mM DTT in a volume of 15–20 µl. The products of the reaction were separated using PEI-cellulose thin layer chromatography developed in 0.45 M (NH4)2SO4 and detected using a Typhoon 8600 Imager (Molecular Dynamics). The RNA substrates with 32P-labelled caps were prepared as described previously (Knapp, 1989).
Cross-linking assays
Cross-linking reactions generally contained 0.1 µg of Dcp1 and equimolar amounts of eIF4F proteins in 20 µl of buffer containing 40 U of RNasin and 1 µg/µl wheat germ tRNA (Sigma). After incubation at 25°C for 5 min, 10–20 fmol of the 32P-labelled 86 nucleotide mRNA were added and the reaction continued for another 25 min. After incubation, reaction mixtures were transferred to ice and UV-irradiated for 10 min in a Stratalinker 1800. Samples were digested with 40 µg of RNase A for 30 min at 37°C and analysed by SDS–PAGE.
ELISAs
The ELISAs (see Harlow and Lane, 1988) were performed by immobilizing 0.1 µg of each protein antigen in 50 mM NaHCO3 pH 9.6 per microtitre plate well overnight at 4°C. After blocking with 1% gelatine for 2 h at 37°C, each protein ligand was incubated within the appropriate wells for 1 h at room temperature in Tris-buffered saline pH 7.5 with 0.2% gelatine and subsequently analysed for binding using antibodies specific for each ligand. Incubation with each antibody was carried out in the same solution overnight at 4°C. All samples were assayed in triplicate, and the mean of each triplex was determined. Each mean value was first normalized by subtracting a set of control values based on the background signal due to binding of the antibody to the immobilized protein antigen in the absence of protein ligand. A further set of control values, representing samples in which no protein antigens were immobilized, were then subtracted from the the first normalized values to yield the data presented in the respective figures.
mRNA half-lives
Half-life analysis of endogenous mRNAs was performed using yeast transformants harbouring a temperature-sensitive allele of RNA polymerase II (rbp1-1) grown in selective media. RNA extractions (Schmitt et al., 1990) and mRNA decay rates (Linz et al., 1997) were performed as described previously, and the results of these experiments were quantified using a Typhoon 8600 Imager. The mRNA abundance at each time point was normalized using 18S rRNA as a reference for the amount of total RNA isolated.
Immunoprecipitation
The immunoprecipitation experiments were carried out using standard methodology (Harlow and Lane, 1988). Total protein extracts from yeast strain W303 transformed with the plasmid pRS402-FLAG Dcp1 were used for these assays. After a preclearing of the extracts with protein A–Sepharose (Sigma) for 1 h at 4°C, the appropriate antibodies were added and the incubation continued for a further 1 h. The Sepharose beads were sedimented and washed with buffer containing 20 mM HEPES pH 7.4, 100 mM KCl, 2 mM MgCl2 and 1 mM DTT. The proteins were loaded on 10% SDS gels. Western blotting was performed also using conventional protocols (Harlow and Lane, 1988).
Far-western analysis
Far-western blots were performed following the protocol of Krieger et al. (1999). Pab1 and whole eIF4G were resolved on a 10% SDS gel and transferred to Immobilon P (Sigma). After incubation with blocking solution containing bovine serum albumin (BSA) for 30 min, the membrane was washed with buffer containing phosphate-buffered saline (PBS) pH 7.5 and 0.1% Tween 20. Recombinant FLAG-Dcp1 was added in the same buffer used for the blocking step. The proteins were incubated for 1 h at room temperature, then FLAG-tagged Dcp1 that had bound to Pab1 or eIF4G was detected by western blotting using an anti-FLAG antibody. Secondary detection of the bound antibodies in western blots was achieved using peroxidase-coupled anti rabbit IgG.
Acknowledgments
Acknowledgements
We are grateful to Zivar Salehi, Dr Tobias von der Haar and Anthea Scothern of the McCarthy laboratory for gifts of p20, Pab1, 4G-BD4E, 4GNt, eIF4A, eIF4B and eIF4G, and to Dr Tobias von der Haar for helpful discussions. Dr Maria Nardelli and Sabina Picchinini kindly provided assistance with some of the experimental work. We thank Dr Audrey Atkins (Lincoln, NE) for the S.cerevisiae strain JDS+S, Dr Patrick Linder (Genève, Switzerland) for strain W303, Dr Roy Parker (Tucson, AZ) for the pGEM-3Z-MATα1 plasmid, Dr Stuart Peltz (Piscataway, NJ) for a FLAG-DCP1 construct, and Dr Alan Sachs (Berkeley, CA) for an anti-eIF4G antibody preparation. The Biotechnology and Biological Sciences Research Council (BBSRC) provided grants for C.Velasco and M.P., while C.Vilela was supported by the Portuguese Fundação para a Ciência e Tecnologia.
References
- Altmann M., Schmitz,N., Berset,C. and Trachsel,H. (1997) A novel inhibitor of cap-dependent translation initiation in yeast: p20 competes with eIF4G for binding to eIF4E. EMBO J., 16, 1114–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman C.A., Stevens,A., Caponigro,G., LaGrandeur,T.E., Hatfield,L., Fortner,D.M. and Parker,R. (1996) An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature, 382, 642–646. [DOI] [PubMed] [Google Scholar]
- Belev T.N., Singh,M. and McCarthy,J.E.G. (1991) A fully modular vector system for the optimisation of gene expression in Escherichia coli. Plasmid, 26, 147–150. [DOI] [PubMed] [Google Scholar]
- Boeck R., Lapeyre,B., Brown,C.E. and Sachs,A.B. (1998) Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell. Biol., 18, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret E., Rigaut,G., Shevchenko,A., Wilm,M. and Séraphin,B. (2000) A Sm-like protein complex that participates in mRNA degradation. EMBO J., 19, 1661–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachman C.B., Davies,A., Cost,G.J., Caputo,E., Li,J., Hieter,P. and Boeke,J. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast, 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Caponigro G. and Parker,R. (1996) Mechanisms and control of mRNA turnover in Saccharomyces cerevisiae. Microbiol. Rev., 60, 233–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P., Fromont-Racine,M., Steel,D., Pictet,R. and Grange,T. (1997) Messenger RNA deadenylation precedes decapping in mammalian cells. Proc. Natl Acad. Sci. USA, 94, 5628–5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehlin E., Wormington,M., Körner,C.G. and Wahle,E. (2000) Cap-dependent deadenylation of mRNA. EMBO J., 19, 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J., Iost,I., Kressler,D. and Linder,P. (1997) The p20 and DED1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 94, 5201–5206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D., Altmann,M., Benz,J., Baumann,U. and Trachsel,H. (1999) Interaction of translation initiation factor eIF4G with eIF4A in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 274, 26720–26726. [DOI] [PubMed] [Google Scholar]
- Dunckley T. and Parker,R. (1999) The Dcp2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J., 18, 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught,B. and Sonenberg,N. (1999) eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem., 68, 913–963. [DOI] [PubMed] [Google Scholar]
- Goyer C., Altmann,M., Lee,H.S., Blanc,A., Deshmukh,M., Woolford,J.L., Trachsel,H. and Sonenberg,N. (1993) TIF4631 and TIF4632: two yeast genes encoding the high-molecular-weight subunits of the cap-binding protein complex (eukaryotic initiation factor 4F) contain an RNA recognition motif-like sequence and carry out an essential function. Mol. Cell. Biol., 13, 4860–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E. and Lane,D. (1988) Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Hatfield L., Beelman,C., Stevens,A. and Parker,R. (1996) Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol., 16, 5830–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp G. (1989) Preparation of yeast transfer RNA precursors in vitro. Methods Enzymol., 180, 192–212. [DOI] [PubMed] [Google Scholar]
- Krieger I.V., Revina,L.P., Kostina.,L.I., Buzdin,A.A., Zalunin,I.A., Chestukhina,G.G. and Stepanov,V.M. (1999) Membrane proteins of Aedes aegypti larvae bind toxins Cry4B and Cry11A of Bacillus thuringiensis ssp. Israelensis. Biochemistry (Moscow), 64, 1163–1168. [PubMed] [Google Scholar]
- LaGrandeur T.E. and Parker,R. (1998) Isolation and characterization of Dcp1, the yeast mRNA decapping enzyme. EMBO J., 17, 1487–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamphear B.J., Kirchweger,R., Skern,T. and Rhoads,R.E. (1995) Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. J. Biol. Chem., 270, 21975–21983. [DOI] [PubMed] [Google Scholar]
- Lang V., Zanchin,N., Lünsdorf,H., Tuite,M.F. and McCarthy,J.E.G. (1994) Initiation factor eIF-4E of Saccharomyces cerevisiae: distribution within the cell, binding to mRNA and consequences of its overproduction. J. Biol. Chem., 269, 6117–6123. [PubMed] [Google Scholar]
- Larimer F.W. and Stevens,A. (1990) Disruption of the gene XRN1, encoding a 5′–3′ exoribonuclease, restricts yeast cell growth. Gene, 59, 85–90. [DOI] [PubMed] [Google Scholar]
- Linz B., Koloteva,N., Vasilescu,S. and McCarthy,J.E.G. (1997) Disruption of ribosomal scanning on the 5′-untranslated region and not restriction of translational initiation per se, modulates the stability of nonaberant mRNAs in the yeast Saccharomyces cerevisiae. J. Biol. Chem., 272, 9131–9140. [DOI] [PubMed] [Google Scholar]
- Mader S., Lee,H., Pause,A. and Sonenberg,N. (1995) The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4γ and the translational repressors 4E-binding proteins. Mol. Cell. Biol., 15, 4990–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J.E.G. (1998) Posttranscriptional control of gene expression in yeast. Microbiol. Mol. Biol. Rev., 62, 1492–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick W.C. and Hershey,J.W.B. (1996) The pathway and mechanism of eukaryotic protein synthesis. In Hershey,J.W.B., Matthews,M.B. and Sonenberg,N. (eds), Translational Control. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 31–70. [Google Scholar]
- Morley S.J., Curtis,P.S. and Pain,V.M. (1997) eIF4G: translation’s mystery factor begins to yield its secrets. RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- Neff C.L. and Sachs,A.B. (1999) Eukaryotic translation initiation factors 4G and 4A from Saccharomyces cerevisiae interact physically and functionally. Mol. Cell. Biol., 19, 5557–5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutskii B.S., Stapulionis,R. and Deutscher,M.P. (1994). Supra molecular organisation of the mammalian translation system. Proc. Natl Acad. Sci. USA, 91, 964–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira C.C., Van den Heuvel,J.J. and McCarthy,J.E.G. (1993). Inhibition of translational initiation in Saccharomyces cerevisiae by secondary structure: the role of the stability and position of stem loops in a mRNA leader. Mol. Microbiol., 9, 521–532. [DOI] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Vasilescu,S., Frank,R., Birkenhäger,R. and McCarthy,J.E.G. (1998) Cooperative modulation by eIF4G of eIF4E binding to the mRNA 5′ cap in yeast involves a site partially shared by p20. EMBO J., 17, 4798–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptushkina M., von der Haar,T., Karim,M.M., Hughes,J.M. and McCarthy,J.E. (1999) Repressor binding to a dorsal regulatory site traps human eIF4E in a high cap-affinity state. EMBO J., 18, 4068–4075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyronnet S., Imataka,H., Gingras,A.-C., Fukunaga,R., Hunter,T. and Sonenberg,N. (1999) Human eukaryotic translation initiation factor 4G (eIF4G) recruits Mnk1 to phosphorylate eIF4E. EMBO J., 18, 270–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J. (1996) Control of messenger mRNA stability in higher eukaryotes. Trends Genet., 12, 171–175. [DOI] [PubMed] [Google Scholar]
- Schiestl R.H. and Gietz,R.D. (1989) High efficiency transformation of intact yeast cells using single stranded nucleic acids as carrier. Curr. Genet., 16, 339–346. [DOI] [PubMed] [Google Scholar]
- Schmitt M.E., Brown,T.A. and Trumpower,B.L. (1990) A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res., 18, 3091–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens A. (1980) An mRNA-decapping enzyme from ribosomes of Saccharomyces cerevisiae.Biochim. Biophys. Res. Commun., 81, 656–661. [DOI] [PubMed] [Google Scholar]
- Stevens A. (1988) mRNA-decapping enzyme from Saccharomyces cerevisiae: purification and unique specificity for long RNA chains. Mol. Cell. Biol., 8, 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppmann S., Persson,B.C. and Böck,A. (1999) Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion on the ribosome. EMBO J., 18, 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun S.Z. and Sachs,A.B. (1996) Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- Tharun S., Weihai,H., Mayes,A.E., Lennertz,P., Beggs,J. and Parker,R. (2000) Yeast Sm-like proteins function in mRNA decapping and decay. Nature, 404, 515–518. [DOI] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Vilela C., Linz,B., Velasco Ramirez,C., Rodrigues-Pousada,C. and McCarthy,J.E.G. (1999) Posttermination ribosome interactions with the 5′UTR modulate yeast mRNA stability. EMBO J., 18, 3139–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C.C., Balasta,M.L., Ren,J.H. and Goss,D.J. (1998) Wheat germ poly(A)binding protein enhances the binding affinity of eukaryotic initiation factor 4F and (iso)4F for cap analogues. Biochemistry, 37, 1910–1916. [DOI] [PubMed] [Google Scholar]
- Zanchin N.I.T. and McCarthy,J.E.G. (1995) Characterisation of the in vivo phosphorylation sites of the mRNA-cap-binding complex proteins eukaryotic initiation factor-4E and p20 in Saccharomyces cerevisiae. J. Biol. Chem., 270, 26505–26510. [DOI] [PubMed] [Google Scholar]
- Zhang S., Williams,C.J., Hagan,K. and Peltz,S.W. (1999) Mutations in VPS16 and MRT1 stabilize mRNAs by activating an inhibitor of the decapping enzyme. Mol. Cell. Biol., 19, 7568–7576. [DOI] [PMC free article] [PubMed] [Google Scholar]