Abstract
SecA insertion and deinsertion through SecYEG drive preprotein translocation at the Escherichia coli inner membrane. We present three assessments of the theory that oligomers of SecYEG might form functional translocation sites. (i) Formaldehyde cross- linking of translocase reveals cross-links between SecY, SecE and SecG, but not higher order oligomers. (ii) Cross-linking of membranes containing unmodified SecE and hemagglutinin-tagged SecE (SecEHA) reveals cross-links between SecY and SecE and between SecY and SecEHA. However, anti-HA immunoprecipitates contain neither untagged SecE nor SecY cross-linked to SecE. (iii) Membranes containing similar amounts of SecE and SecEHA were saturated with translocation intermediate (I29) and detergent solubilized. Anti-HA immunoprecipitation of I29 required SecYEHAG and SecA, yet untagged SecE was not present in this translocation complex. Likewise, anti-HA immunoprecipitates of membranes containing equal amounts of SecY and SecYHA were found to contain SecYHA but not SecY. Both immunoprecipitates contain more moles of I29 than of the untagged subunit, again suggesting that translocation intermediates are not engaged with multiple copies of SecYEG. These studies suggest that the active form of preprotein translocase is monomeric SecYEG.
Keywords: membrane proteins/preprotein translocase/SecYEG
Introduction
The export of most proteins across the Escherichia coli inner membrane is catalyzed by a multisubunit complex termed preprotein translocase. Proteins destined for transport by translocase are synthesized as preproteins containing a cleavable N-terminal leader (signal) sequence and associate with chaperones such as SecB. Translocase consists of the peripheral membrane ATPase SecA bound to an integral membrane core of SecY and SecE, and is aided by the accessory integral membrane subunits SecG and/or SecD, SecF and yajC (Duong and Wickner, 1997a). Biochemical studies have led to a working model for translocase activity. SecB–preprotein complexes are targeted to translocase by the binding affinity of SecB for SecA and by the affinities of SecA for leader sequences and preprotein mature domains (Lill et al., 1989; Hartl et al., 1990). SecA then binds ATP, resulting in a conformational change that inserts domains of SecA into and partially across the cytoplasmic membrane (Economou and Wickner, 1994; Kim et al., 1994; Eichler and Wickner, 1997). Approximately 20–30 N-terminal residues of preprotein are inserted across the membrane concomitant with the membrane insertion of SecA. SecA then hydrolyzes ATP, resulting in the release of preprotein from SecA. An additional round of ATP binding and hydrolysis results in the deinsertion of SecA from the membrane (Economou and Wickner, 1994). Deinserted SecA is then free to exchange with cytosolic SecA and undergo repeated cycles of translocation (Economou and Wickner, 1994). Each additional cycle of SecA insertion and deinsertion is accompanied by the translocation of 20–30 residues of preprotein. Once SecA has translocated the first segment of preprotein, translocation can also be driven by the proton-motive force during that part of the catalytic cycle when SecA is not bound with translocation intermediate at SecYEG (Schiebel et al., 1991).
Several lines of evidence suggest that SecYEG forms the pathway for translocating preproteins: (i) overexpres sion of SecYEG increases the number of translocation sites (Douville et al., 1995); (ii) preproteins in transit across the cytoplasmic membrane can be cross-linked to SecY and SecA (Joly and Wickner, 1993); and (iii) purified SecYEG reconstituted into proteoliposomes supports translocation (Brundage et al., 1990). These data have established that SecA and SecYEG form the core of the translocase. SecYEG forms a stable heterotrimeric complex. In vivo, the stability of overexpressed SecY is dependent upon the co-overexpression of SecE, suggesting a direct interaction between SecY and SecE (Matsuyama et al., 1990). A direct interaction between the three subunits of SecYEG has been demonstrated by co-chromatography and co-immunoprecipitation, and points of contact between the transmembrane domains of SecY and SecE have been mapped (Brundage et al., 1990; Nishiyama et al., 1994; Harris and Silhavy, 1999). Once assembled, the individual SecYE subunits do not readily exchange from one SecYE core complex to another (Joly et al., 1994). This was established by inducing the expression of hemagglutinin (HA) epitope-tagged SecE (SecEHA) in cells in which the chromosomally encoded SecYEG had been radioactively labeled and observing that 35S-labeled SecY or SecE did not associate with SecEHA in anti-HA immunoprecipitates.
The discovery of the cycle of SecA insertion and deinsertion, and the finding that this cycle is closely linked to preprotein movement raise questions as to the path whereby SecA inserts. SecA is a homodimer of an entirely polar 102 kDa polypeptide. Purified SecA is water soluble at concentrations of up to 50 mg/ml, yet the association of SecA with its many physiological ligands, which include preprotein, SecB, SecYEG, acidic lipids and ATP, causes a profound conformational change in which a 30 kDa C-terminal domain and a 65 kDa N-terminal domain of SecA insert so far into the membrane as to become inaccessible to protease from the cytoplasmic membrane surface and accessible to probes added to the periplasmic surface of the membrane (Ramamurthy and Oliver, 1997; Eichler and Wickner, 1998; van der Does et al., 1998). Since the membrane-spanning helices of SecYEG are apolar, and since membrane-spanning translocation intermediate and inserted SecA are apparently shielded from the lipid bilayer by SecYEG (Eichler et al., 1997), it will be important to determine the pathway of preprotein transit and of SecA insertion/deinsertion.
Although much is known about the interactions among the three polypeptide subunits of SecYEG, little is known about the potential interactions between SecYEG complexes in forming oligomers [n(SecYEG)], in E.coli or in other organisms. The eukaryotic endoplasmic reticulum (ER) has a protein translocation system with a heterotrimeric membrane-embedded domain that is homologous to SecYEG of E.coli (Rapoport et al., 1996). This complex (Sec61p in mammals) is made up of Sec61α, Sec61β and Sec61γ (Rapoport et al., 1996). Sec61α and Sec61γ are homologous to SecY and SecE, respectively, suggesting a potential conservation in function, while SecG is thought to represent a functional homolog of Sec61β. Electron microscopic examination of purified Sec61p complex revealed an oligomeric assembly consisting of 3–4 trimeric Sec61p complexes in a ring (Hanein et al., 1996). The lumen of this ring is proposed to be the conductive channel for translocating polypeptide chains. These studies recently have been extended to the bacterial translocases of Bacillus subtillus and E.coli (Meyer et al., 1999; Manting et al., 2000). In both cases, examination of purified translocase in the electron microscope revealed an oligomeric assembly of 3–4 SecYE(G) complexes similar to that seen for Sec61p. Although these studies suggest that translocase might function as an oligomer with a single translocation channel, neither the functional activity of oligomers for translocation nor the stoichiometry with which they might interact with preprotein are known.
Determining the monomeric or oligomeric character of SecYEG (Figure 1) as it functions with SecA and preprotein during translocation will be of fundamental importance to understanding the preprotein translocation pathway and the SecA cycle of insertion and deinsertion. There are daunting technical challenges that are inherent to this study. The normal catalytic cycle of translocase includes the release of SecA, and thus it is by no means clear that SecA will remain stoichiometrically bound with preprotein and SecYEG during translocation. The preprotein itself is capable of spontaneously ‘sliding’ both forward and backward in SecYEG when it is not bound by SecA, and thus may also be released from SecA and SecYEG (Schiebel et al., 1991; Duong and Wickner, 1997b). Finally, like most membrane-embedded integral membrane proteins, SecYEG has strong tendencies to denature in detergent solution through both disassembly (Brundage et al., 1992; Duong and Wickner, 1999) and aggregation. Thus the observation of SecYEG, whether biochemically or through electron microscopy, as either a simple heterotrimer or as an oligomer of heterotrimers is not sufficient to indicate its normal mode of physiological function. Solubilized translocation complexes often denature and are difficult to assay for function while in detergent micellar solution and may disassemble, aggregate and assume varied conformations during reconstitution into proteoliposomes. Nevertheless, the central attraction to the study of translocation for the last 30 years has been the belief that translocase will have a catalytic mechanism that is unusual among transport systems, and thus the determination of its functional subunit structure is essential.
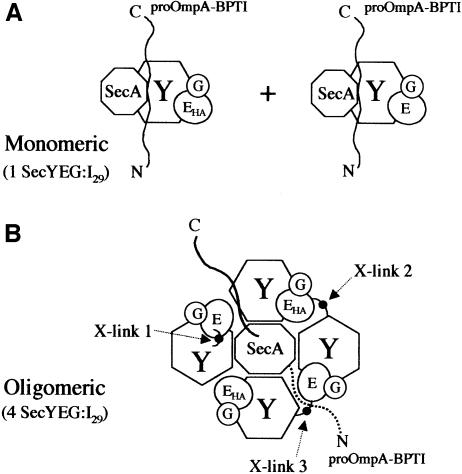
Fig. 1. Models of the proposed monomeric (A) and oligomeric (B) forms of SecYEG. Under the monomeric model for translocase, SecYEG and SecYEHAG complexes associate with one SecY molecule for each I29 translocation intermediate. In the oligomeric model, SecYEG and SecYEHAG assemble into an oligomeric complex with both the SecE and SecHA equally represented. Each oligomeric complex of four SecY molecules contains one I29 translocation intermediate. Under cross-linking conditions, cross-link 1 (X-link 1) represents an intra-SecYEG cross-link between SecY and SecE. Cross-links 2 and 3 represent inter-SecYEG cross-links. Cross-link 2 results in SecY cross-linked to SecEHA and would allow anti-HA immunoprecipitation of SecE. Cross-link 3 results in a SecY–SecE cross-link and is predicted to result in the anti-HA immunoprecipitation of SecY–SecE.
We now present studies that address this question of subunit structure. Cross-linking studies of intact membranes bearing saturating levels of preprotein translocation intermediate fail to reveal oligomers of SecYEG. We therefore solubilized membranes bearing an arrested preprotein translocation intermediate with detergent under conditions where the preprotein and the translocase remain associated. We find that this translocase, in association with preprotein chains, does not have multiple copies of SecE or SecY. These studies suggest that the active form of preprotein translocase functions as monomeric SecYEG.
Results
Studies of the Sec61 complex of yeast and SecYEG of E.coli suggest that an assembly step is required for the formation of oligomeric translocase. In E.coli, dimers of SecYEG are proposed to form tetramers in the presence of SecA or SecA–preprotein (Manting et al., 2000). Studies of the potential oligomeric nature of translocase must allow for such an assembly step. Our experimental approach is to examine the oligomeric state of SecYEG under conditions in which the translocase is saturated with translocation intermediate that would presumably satisfy this assembly requirement. To generate translocation intermediate on inner membrane vesicles (IMVs), we took advantage of a translocase substrate (proOmpA-BPTI) that previously has been shown to arrest during translocation (Schiebel et al., 1991). Using the thiol-reducible cross-linker N-succinimidyl 3-[2-pyridyldithio] proprionate, bovine pancreas trypsin inhibitor (BPTI) was conjugated to one of two unique cysteinyl residues (290 or 302) at the C-terminus of proOmpA, yielding proOmpA-BPTI. In standard translocation reactions using proOmpA as a substrate, proOmpA is completely translocated to the lumen of IMVs and therefore protected from added protease. In contrast, proOmpA-BPTI becomes arrested during its translocation, yielding a protease-protected domain of 29 kDa (I29) corresponding to the portion of proOmpA that has completed translocation.
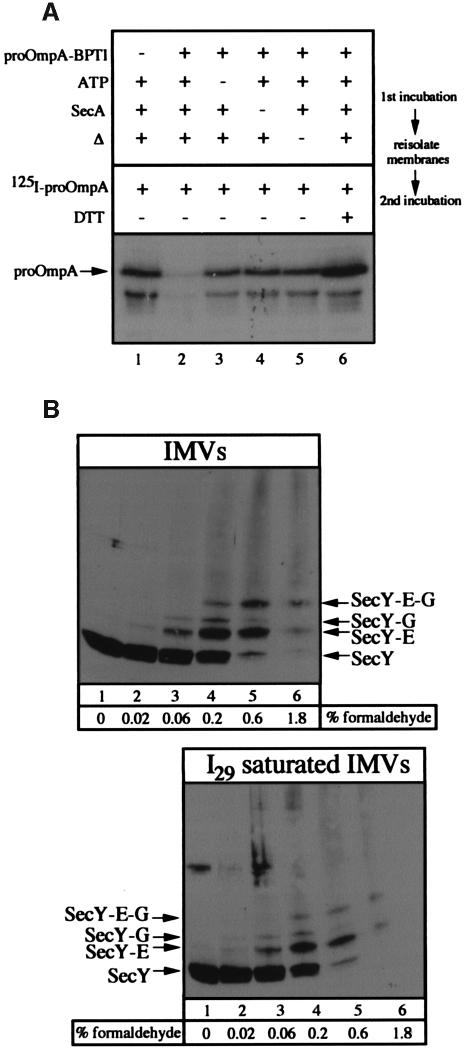
To confirm that all of the translocation sites are saturated with I29 arrested translocation intermediates, two-stage translocation reactions were performed. In the first incubation, complete reactions or reactions lacking proOmpA-BPTI, ATP or SecA were performed for 20 min at 37°C or on ice. Membranes were re-isolated through a sucrose cushion and incubated in a second translocation reaction with SecA, SecB, ATP, NADH and radiolabeled proOmpA. The omission of proOmpA-BPTI (Figure 2A, lane 1), ATP (lane 3), SecA (lane 4) or heat (lane 5) during the first incubation prevents the formation of I29 and allows the translocation of proOmpA during the second reaction. In contrast, a complete reaction (Figure 2A, lane 2) produces I29 that in turn blocks the translocation of proOmpA during the second incubation. The inclusion of dithiothreitol (DTT) during the second incubation reduces the cross-link between proOmpA and BPTI, allowing the I29 to complete its translocation and freeing the translocase for further proOmpA translocation (Figure 2A, lane 6). Quantitation of these data demonstrates a 7-fold reduction in the rate of translocation following saturation of translocase with I29, suggesting that the active translocation sites are largely occupied by I29 translocation intermediates under these reaction conditions. In theory, a logical approach to the question of oligomeric versus monomeric SecYEG would involve saturating IMVs with translocation intermediate and comparing the amount of SecY with the amount of I29 arrested translocation intermediate. In practice, however, we have found that this approach yields quite variable results. The ratio of SecY to I29 in IMVs saturated with translocation intermediate ranged from 2 to 6 in SecYEG-overexpressing membranes, indicating that at least a substantial fraction of the SecYEG on isolated IMVs is active for translocation.
Fig. 2. (A) Saturation of translocase by proOmpA-BPTI. Complete translocation reactions (lanes 2 and 6) or reactions lacking proOmpA-BPTI (lane 1), ATP (lane 3) or SecA (lane 4) were performed at 37°C or on ice (lane 5) as described in Materials and methods. Following a 15 min incubation at 37°C, reactions were layered over 100 µl of TL buffer containing 0.2 M sucrose and centrifuged at 200 000 g for 10 min. Membrane fractions were suspended in TL buffer and supplemented with BSA (200 µg/ml), SecA (50 µg/ml), SecB (66 µg/ml), ATP (2 mM), NADH (5 mM), [125I]proOmpA (50 000 c.p.m.) and DTT (2 mM) (lane 6 only). Following 15 min at 37°C, reactions were transferred to ice and treated with proteinase K (1 mg/ml) for 15 min. Proteins were precipitated with TCA, washed with acetone and analyzed by SDS–PAGE and autoradiography. The position of fully translocated proOmpA is indicated. The stimulation of proOmpA translocation seen in the presence of DTT (lane 6) results from reduction of the disulfide bridge between cysteine residues 290 and 302. (B) Formaldehyde cross-linking of SecYEG. IMVs (160 µg) in 800 µl of HEPES TL buffer or IMVs (160 µg) saturated with I29 translocation intermediate were layered over 200 µl of 0.2 M sucrose in HEPES TL buffer. Membranes were collected by centrifugation (TLA-120.2, 73 000 r.p.m., 10 min) and suspended in 300 µl of HEPES TL buffer. Aliquots of 47.5 µl were removed and treated with formaldehyde to the indicated concentrations for 40 min at 23°C. Following the addition of SDS–PAGE sample buffer, samples were heated for 10 min at 37°C and analyzed by 12% SDS–PAGE and anti-SecY immunoblot analysis. The positions of SecY and SecY cross-linked to SecG and SecE are indicated.
Nearest-neighbor analysis of SecYEG complexes
Formaldehyde cross-linking was employed to explore whether SecYEG forms oligomeric complexes. IMVs (Figure 2B, lanes 1–6) or IMVs saturated with I29 translocation intermediate (lanes 7–12) were incubated with increasing concentrations of formaldehyde and analyzed by SDS–PAGE and immunoblot to SecY (Figure 2B), or to SecE or SecG (data not shown). SecY was cross-linked efficiently to SecE, to SecG or to both, but higher oligomers, indicative of n(SecYEG) structures (Figure 1B), were not observed. At higher cross-linker concentrations (Figure 2B, lanes 5 and 6), these cross-linked species were lost to a smear of higher molecular weight species that presumably represent cross-linking to the other proteins of the IMV. Saturating the translocase in the IMVs with I29 prior to cross-linking did not change the pattern (Figure 2B). These data suggest that the nearest neighbors of SecYEG complexes are not additional SecYEG complexes; while it is possible that formaldehyde cross-links within a SecYEG complex are favored and cross-links between SecYEG complexes are less likely to occur, the simplest interpretation is that SecYEG does not form higher order oligomers in native membranes.
Analysis of the complex of SecYEG and preprotein translocation intermediate
As a complement to the formaldehyde cross-linking approach, we set out to establish a biochemical assay for the detection of oligomeric SecYEG. The two key features that the assay requires are (i) identifying solubilization conditions that maintain the associations between SecYEG and the proOmpA-BPTI translocation intermediate I29, and (ii) the ability to detect multiple copies of SecE or SecY in immunoprecipitates of SecYEG–proOmpA-BPTI complexes. We reasoned that if membranes contained similar amounts of untagged SecE and HA-tagged SecE (SecEHA) or untagged SecY and HA-tagged SecY (SecYHA), and if SecYEG formed higher order oligomers (Figure 1B), both the tagged and untagged forms would associate equally into the oligomeric complex (Figure 1B). If an oligomeric complex were present, immunoprecipitations using anti-HA would result in the cross-immunoprecipitation of untagged SecE or SecY.
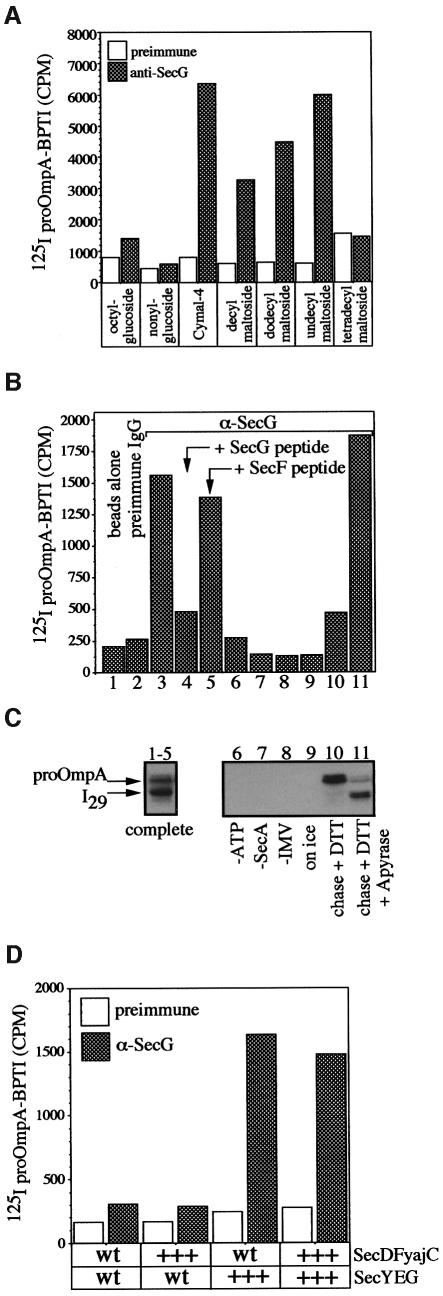
The first requirement for the oligomeric assay was the identification of conditions for the solubilization of SecYEG–proOmpA-BPTI. Since this complex is not stable under the detergent extraction conditions (β-octylglucoside) traditionally used to purify SecYEG (Brundage et al., 1990), we screened a larger set of detergents for their ability to solubilize the SecYEG–proOmpA-BPTI complex. Using radiolabeled proOmpA-BPTI as substrate, translocation intermediates were generated with IMVs derived from a SecYEG-overexpressing strain, solubilized with various detergents and immunoprecipitated with anti-SecG covalently bound to protein A–Sepharose beads. Immunoprecipitates were monitored by scintillation counting for the cross-immunoprecipitation of proOmpA-BPTI by anti-SecG. As previously observed, β-octylglucoside, though capable of solubilizing SecYEG, did not allow the cross-immunoprecipitation of proOmpA-BPTI by anti-SecG (Figure 3A), nor did digitonin (data not shown). However, several detergents can support the solubilization and cross-immunoprecipitation of proOmpA-BPTI by anti-SecG (Figure 3A). One of the more efficient detergents, cymal-4, was chosen for subsequent experiments.
Fig. 3. (A) Solubilization and immunoprecipitation of SecYEG/proOmpA-BPTI. Translocation reactions (100 µl) containing [125I]proOmpA-BPTI (1 × 105 c.p.m.) were performed as described in Materials and methods using IMVs from SecYEG-overexpressing cells. Reactions were solubilized in 50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1.5 mg/ml E.coli phospholipids, 40% glycerol and 1% of the indicated detergents and processed for immunoprecipitation with pre-immune and anti-SecG protein A–Sepharose beads. After a 60 min incubation, beads were washed twice with 1 ml of solubilization buffer and bound proOmpA-BPTI was monitored by scintillation counting. (B and C) Assays of translocation intermediate formation. Complete translocation reactions (lanes 1–5 and 9–11) or reactions lacking either ATP (lane 6), SecA (lane 7) or IMVs (lane 8) were performed at 37°C or on ice (lane 9). After 15 min, potato apyrase (5 U) was added to reaction 11 for 3 min, followed by the addition of DTT (2 mM) and unlabeled proOmpA-BPTI (25 µg/ml) for an additional 10 min at 37°C. Samples were transferred to ice for 5 min, and ATP, SecA or IMV was added to reactions 5, 6 and 7, respectively. An aliquot of each reaction was transferred to ice, treated with proteinase K (10 mg/ml) for 15 min, TCA precipitated and analyzed by SDS–PAGE and autoradiography (C). The remaining portion of each reaction was layered over 0.2 M sucrose in TL buffer and centrifuged (10 min, 200 000 g) to recover membranes. Membrane fractions were solubilized in 500 µl of cymal buffer (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 1.5 mg/ml E.coli phospholipids, 25% glycerol, 1% cymal-4) on ice for 15 min. Samples were centrifuged (100 000 g, 10 min) to remove insoluble material. Soluble extracts were transferred to fresh tubes and incubated with 100 µl of protein A–Sepharose beads alone (reaction 1), beads coupled to pre-immune sera (reaction 2) or affinity-purified anti-SecG IgG (reactions 3–11) for 60 min at 4°C. The beads were washed three times with 1 ml of cymal buffer and proteins were eluted by the addition of 500 µl of 50 mM Tris–HCl pH 8.0, 1% SDS. Bound proOmpA-BPTI was monitored by scintillation counting (B). (D) The effects of membrane composition on the yield of intermediate. Translocation reactions (100 µl) were performed as described above using [125I]proOmpA-BPTI (1 × 105 c.p.m.) mixed with unradiolabeled proOmpA-BPTI (16 µg/ml) and 100 µg/ml of each of the indicated IMVs. Reactions were solubilized in cymal IP buffer and processed for immunoprecipitation with pre-immune and anti-SecG protein A–Sepharose beads. Immunoprecipitated proOmpA-BPTI was monitored by scinitillation counting. wt indicates wild-type levels, while +++ indicates overexpressed levels of the indicated proteins.
To demonstrate that the anti-SecG immunoprecipitates represent authentic translocation intermediates, complete translocation reactions or reactions lacking ATP, SecA, IMVs or incubation at physiological temperature were performed. An aliquot of each reaction was removed and treated with protease (Figure 3C). The remaining portion of each reaction was solubilized and immunoprecipitated with protein A–Sepharose beads, pre-immune IgG beads or anti-SecG beads. In complete reactions where the formation of I29 is observed in protease protection assays (Figure 3B and C, lanes 1–5), a specific immunoprecipitation of proOmpA-BPTI is seen with anti-SecG beads (Figure 3B, lane 3) or with anti-SecG beads pre-mixed with an irrelevant peptide (Figure 3B, lane 5). Beads alone (Figure 3B, lane 1), beads with pre-immune IgG (lane 2) or anti-SecG beads pre-mixed with the peptide used to raise the antisera (lane 4) did not immunoprecipitate the complex. Reactions lacking ATP (Figure 3B and C, lane 6), SecA (lane 7), IMVs (lane 8) or incubation at physiological temperature (lane 9) supported neither the formation of I29 as assayed by proteolysis of intact IMVs (Figure 3C) nor the cross-immunoprecipitation of proOmpA-BPTI from cymal extracts by anti-SecG (Figure 3B). Most strikingly, the addition of DTT allows the completion of I29 translocation to fully translocated proOmpA and results in a large reduction in the amount of immunoprecipitated proOmpA-BPTI (Figure 3B and C, lane 10). The addition of apyrase with the DTT prevents I29 from completing translocation and allows the recovery of I29 by anti-SecG immunoprecipitation (Figure 3B, lane 11). As a final test of this complex, I29 was generated in wild-type membranes or membranes from strains overexpressing SecYEG and/or SecDFyajC. Membranes from cells that overexpressed SecYEG allowed far more formation of I29 (data not shown) and immunoprecipitation of the proOmpA-BPTI–SecYEG intermediate complex (Figure 3D). These data establish that anti-SecG can immunoprecipitate an authentic SecYEG–preprotein translocation intermediate complex from detergent micellar solution.
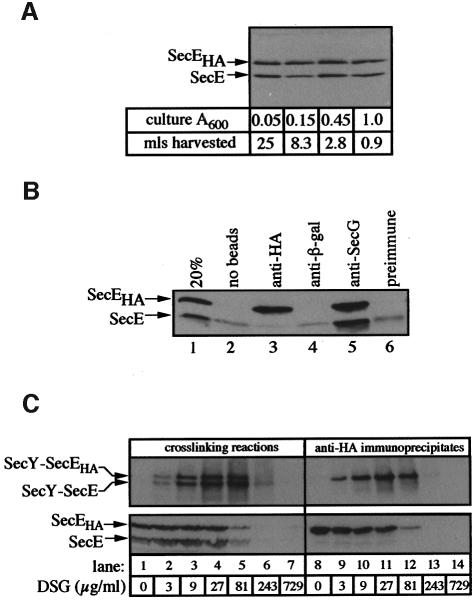
The second requirement for the assay of oligomeric translocase was the generation of membranes with comparable amounts of tagged and untagged translocase components. To prepare membranes with similar amounts of SecEHA and SecE or of SecYHA and SecY, untagged proteins were expressed at basal levels from the endogenous chromosomal copy of the respective genes. Expression of plasmid-encoded SecYEHAG or SecYHA EG was driven from an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter. By titrating the amount of IPTG used for induction, conditions were identified that resulted in approximately equal expression of both SecE and SecEHA (Figure 4A) or SecY and SecYHA (data not shown) throughout the growth of the cells. The ratio of epitope-tagged to untagged proteins in these membranes varied between one and two. Both the HA-tagged SecE and untagged SecE, present in the IMVs (Figure 4B, lane 1), were immunoprecipitated efficiently by anti-SecG (lane 5) as part of SecYEG. However, there was almost no untagged SecE associated with HA-tagged SecE (lane 3), and even the minor amounts of untagged SecE recovered corresponded to non-specific binding to the beads (lanes 2 and 4). These data suggest that SecYEG and SecYEHAG are separate complexes (Figure 1A) rather than associated (Figure 1B). However, in case the associations between trimeric SecYEG complexes were labile under the solubilization conditions or required an assembly step (i.e. generation of I29), IMVs with similar amounts of SecE and SecEHA were saturated with proOmpA-BPTI translocation intermediate and exposed to a range of disuccinimidyl glutarate (DSG) cross-linker concentrations. Reactions were then solubilized in detergent and immunoprecipitated with anti-HA coupled to protein A–Sepharose beads (Figure 4C). Lanes 1–7 represent 15% of the material used in each anti-HA immunoprecipitation (Figure 4C, lanes 8–14). The addition of cross-linker resulted in the appearance of two higher molecular weight species (Figure 4C, lanes 2–6). Immunoblotting with anti-SecE (Figure 4C, lanes 2–6), or anti-SecY and anti-HA (data not shown) identified these species as SecY cross-linked to SecE or SecEHA (Figure 4C, lanes 2–6). At the highest concentration of cross-linker (lanes 7 and 14), all of the SecE has been cross-linked into high molecular weight species that do not enter the gel. Immuno precipitations with anti-HA resulted in the specific immunoprecipitation of SecEHA and SecY–SecEHA (Figure 4C, lanes 8–12) with only very small amounts of SecE, which correspond to non-specific adsorption to the beads (Figure 4B). The specific anti-HA immunoprecipitation of SecY–SecE or SecE, which would have been expected for a cross-linked oligomeric complex (Figure 1B, cross-links 2 and 3), was not detected under any of the cross-linker concentrations (Figure 4C) or with other cross-linkers (data not shown).
Fig. 4. Nearest-neighbor analysis using cross-linking reagents. (A) SecE and SecEHA are expressed at similar levels. Cultures of BL21 ptrc99YEHAG were diluted to A600 0.01 into 100 ml of LB containing 0.025 mM IPTG. At the indicated culture absorbance, samples of the indicated volumes were removed and cells were collected by centrifugation. Cell pellets were suspended in SDS–PAGE sample buffer, sonicated briefly and analyzed by SDS–PAGE and anti-SecE immunoblot analyses. (B) Specificity of SecE immunoprecipitation. IMVs prepared from cells expressing similar amounts of SecE and SecEHA were solubilized and immunoprecipitated without beads (lane 2), or with anti-HA (lane 3), anti-β-galactosidase (lane 4), anti-SecG (lane 5) or pre-immune IgG beads (lane 6). Lane 1 is 20% of the extract used in each immunoprecipitation. The positions of SecE and SecEHA are indicated. (C) Anti-HA does not immunoprecipitate SecY–SecE or SecE following cross-linking. Translocation reactions (800 µl for each cross-linker concentration) were performed as described in Materials and methods using IMVs (200 µg/ml, with similar amounts of HA epitope-tagged and untagged SecE) and proOmpA-BPTI (20 µg/ml). After 20 min at 37°C, reactions were layered over 200 µl of HEPES TL buffer (50 mM HEPES–KOH pH 8.0, 50 mM KCl, 5 mM MgCl2) containing 0.2 M sucrose. Membranes were re-isolated by centrifugation (TLA-120.2, 73 000 r.p.m., 10 min), washed with 1 ml of HEPES TL buffer and suspended in 100 µl of HEPES TL buffer containing 5% (v/v) dimethylsulfoxide (DMSO). Cross-linkers were dissolved in 95% DMSO and added in four separate 2.5 µl aliquots at 10 min intervals to the indicated final concentrations. Cross-linking reactions were performed at 23°C. After the 40 min incubation, cross-linker was quenched by the addition of 10 µl of 1 M Tris–HCl pH 8.0. Samples were solubilized by the addition of 400 µl of 1.25× cymal IP buffer [62.5 mM HEPES pH 8.0, 188 mM NaCl, 50% glycerol (w/v), 1.875 mg/ml E.coli phospholipids, 1.25% cymal-4], nutated at 4°C for 15 min and centrifuged at 100 000 g (TLA-45) for 10 min to remove insoluble material. Detergent extracts were transferred to fresh tubes and mixed with 500 µl of cymal solubilization buffer containing 2% (w/v) BSA and 50 µl of packed protein G–Sepharose beads coupled to 128 µg of anti-HA monoclonal antibody. After 60 min at 4°C, beads were washed three times with 1 ml of cymal IP buffer and eluted with 120 µl of 50 mM Tris–HCl pH 8.0, 1% SDS at 37°C for 10 min. After a brief centrifugation, supernatants were collected and mixed with 20 µl of 5× SDS–PAGE loading buffer, heated at 37°C for 10 min and analyzed by High-Tris (lower panel) (Brundage et al., 1990) and 12% (upper panel) SDS–PAGE, and anti-SecE immunoblot analyses.
HA-tagged and untagged SecE in translocation complexes
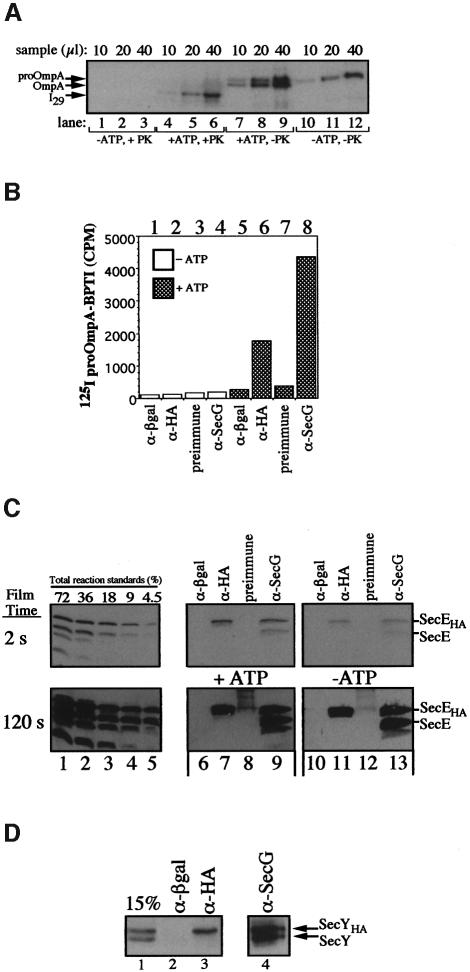
In a further attempt to detect an oligomeric complex, IMVs with similar amounts of SecE and SecEHA were saturated with translocation intermediate, a step that has been proposed to induce oligomerization of SecYEG (Manting et al., 2000). I29 proOmpA-BPTI-arrested translocation intermediate was found to have blocked >80% of the active translocation sites (Figure 2A and data not shown). Membranes bearing translocation intermediate were sedimented through a sucrose cushion and suspended in TL buffer. Aliquots were removed and a portion of each was incubated with or without proteinase K to determine the percentage of the total proOmpA-BPTI that was I29 (Figure 5A). ProOmpA-BPTI was largely recovered with the membranes due to ATP-dependent translocation (Figure 5A, lanes 10–12 versus 7–9) and translocation was accompanied by some leader sequence cleavage (lanes 7–9). Proteinase K treatment (Figure 5A, lanes 4–6) showed that bound proOmpA had been converted to I29. The remaining portion of the IMVs bearing I29 was solubilized and processed for immunoprecipitation. Bound material was eluted from beads and aliquots were monitored for immunoprecipitated proOmpA-BPTI by scintillation counting (Figure 5B), and SecEHA and SecE were analyzed by immunoblot analysis with antibodies to SecE (Figure 5C). In reactions lacking ATP, generation of I29 was not observed, as judged by protease protection (Figure 5A) and immunoprecipitation (Figure 5B) assays. In parallel reactions containing ATP, formation of I29 was detected (Figure 5A and B). Scanning and densitometric analysis of samples incubated with (+ATP, +PK) or without (+ATP, –PK) protease revealed that 44% of the total proOmpA-BPTI bound to IMVs was I29 while the remaining 56% resulted from non-specific association (Figure 5A). Immunoprecipitations using anti-HA and anti-SecG resulted in the specific recovery of I29 (Figure 5B, lanes 6 and 8). The recovery of I29 in the immunoprecipitates was 10 and 24% of the total I29 input for anti-HA and anti-SecG, respectively; the lower yield with anti-HA is consistent with SecEHA and SecE association with distinct populations of I29, both of which are in association with SecG. These data also demonstrate that complexes containing I29, SecG and SecE or SecEHA are fully competent for translocation. Immunoprecipitations with anti-SecG also resulted in the specific immunoprecipitation of SecE and SecEHA in ratios comparable to the starting membranes (Figure 5C, lane 5 versus 9). In contrast, while anti-HA and anti-SecG immunoprecipitated comparable amounts of SecEHA (Figure 5C, lanes 7 versus 9), the anti-HA immunoprecipitation of untagged SecE was >60-fold less than with anti-SecG (Figure 5C, lane 7 versus 9, see 120 s exposure). The immunoprecipitates in reactions lacking ATP, and thus translocation intermediate, were nearly identical (Figure 5C, lanes 10–13). Quantitation of the I29, SecE and SecEHA immunoprecipitation data from the reaction performed in the presence of ATP is summarized in Table I. Less than 1 mol of SecE is recovered with anti-HA-immunoprecipitated complexes of translocase for each 50 mol of associated I29. If there is undetected oligomeric n(SecYEG), these data indicate that the immunoprecipitation of I29 is not dependent upon the presence of oligomeric translocase. Likewise, using membranes with similar amounts of SecY and SecYHA (Figure 5D, lane 1), translocase was saturated with I29 intermediate and subjected to anti-HA immunoprecipitation. While both anti-SecG and anti-HA immunoprecipitated SecYHA (Figure 5D, lanes 3 and 4) and I29, the immunoprecipitation of untagged SecY was not detected in anti-HA immunoprecipitates (Figure 5D, lane 3) and more moles of I29 were immunoprecipitated along with SecYHA than moles of untagged SecY (Table II).
Fig. 5. (A–C) Immunoisolation of proOmpA-BPTI with translocase. Translocation reactions (1.5 ml) were performed as described above using IMVs with similar amounts of SecE and SecEHA in the presence or absence of ATP. Following a 20 min incubation at 37°C, membranes were re-isolated over a sucrose cushion and suspended in 100 µl of TL buffer. Aliquots (5 µl) were removed, mixed with 95 µl of TL buffer containing BSA (200 µg/ml), incubated with or without proteinase K (10 mg/ml) and analyzed by SDS–PAGE and autoradiography (A). The remaining portion of each sample was solubilized with 1 ml of cymal IP buffer, divided into four aliquots and processed for immunoprecipitation with anti-β-galactosidase, anti-HA, pre-immune and anti-SecG beads. Following 60 min on ice, the beads were washed twice with 1 ml of cymal IP buffer and eluted with 120 µl of 50 mM Tris–HCl pH 8.0, 1% SDS. Portions of the eluate were monitored for proOmpA-BPTI by scintillation counting (B) and SecE–SecEHA by SDS–PAGE and anti-SecE immunoblot analyses (C). (D) Immunoisolation of preprotein–translocase complex from cells expressing SecY and SecYHA. A complete translocation reaction (1.5 ml) was performed as described above using IMVs with similar amounts of SecY and SecYHA. The reaction was solubilized with 1 ml of cymal IP buffer, divided into four aliquots and processed for immunoprecipitation with anti-β-galactosidase, anti-HA, anti-preimmune and anti-SecG beads.
Table I. Quantitation of translocation components following immunoprecipitation.
| Antibody | SecEHA |
SecE |
I29 |
|---|---|---|---|
| fmol immunoprecipitated | |||
| Anti-HA | 2120 | <5 | 270 |
| Anti-SecG | 3320 | 3180 | 810 |
| Total input into IP (fmol) | 5470 | 5130 | 4890 |
Table II. Quantitation of translocation components following immunoprecipitation.
| Antibody | SecYHA |
SecY |
I29 |
|---|---|---|---|
| fmol immunoprecipitated | |||
| Anti-HA | 950 | <7 | 60 |
| Anti-SecG | 1630 | 1520 | 330 |
| Total input into IP (fmol) | 3200 | 2950 | 2490 |
Discussion
As the components of translocases have largely been identified in bacteria, mitochondria, chloroplasts and ER, the next logical question is their stoichiometry at functional translocation sites. This a difficult question for several reasons. SecA binds to, and is released from, translocase as part of the normal catalytic cycle (Schiebel et al., 1991). The preprotein chain itself can slide forward and back in the translocase when not bound by SecA and, at sites in the preprotein of even quite limited hydrophobicity, can be released laterally from translocase (Duong and Wickner, 1998). Like many other membrane-embedded integral membrane proteins, SecYEG can aggregate and denature upon solubilization. Conversely, the fundamental association of the three polypeptides in SecYEG is labile, and studies of their association in prl mutants show that this lability is important for translocase function (Brundage et al., 1992; Duong and Wickner, 1999). For each of these reasons, simply measuring a stoichiometry or observing a SecYEG oligomer may not resolve the issue of functional translocase structure.
Rapoport and colleagues have shown that the bacterial SecYE complex can form ring structures like their mammalian counterparts (Hanein et al., 1996; Meyer et al., 1999). These pioneering studies are the foundation of the postulate that a ring of SecYEG or (eukaryotic) Sec61 complexes forms a translocation pore. However, neither the translocation-dependent formation of these structures nor the stoichiometry of leader sequence-dependent targeting of preprotein to these rings has been demonstrated, and it is not known whether the path of preprotein translocation is actually through the center of the ring. Structural studies of the yeast Sec61p complex bound to ribosomes indicated an alignment of complex with the site of exiting polypeptides from the ribosome, suggesting that the co-translational secretion of preproteins into the ER was directly through the center of the Sec61p ring, yet these electron microscopic pictures could not reveal the path of the preprotein (Beckmann et al., 1997), i.e. whether truly through the lumen of the ring rather than through a single Sec61 complex of the ring. Driessen and colleagues have shown that there are two SecYEG complexes in association with each nascent chain for each SecA (Manting et al., 2000). In light of the fact that SecA functions as a dimer, this suggests that SecYEG may function as a tetramer, and indeed larger structures are seen in their electron microscopic images of detergent-solubilized translocase with proOmpA than for SecYEG alone. However, the distribution of particle sizes in their study is still quite heterogeneous and the subunit composition of the various particles has not been determined. It is also not known whether SecA remains stoichiometrically associated with translocase during this detergent extraction.
We now report three independent approaches to the question of the structure of functioning translocase. Two of these approaches rest on the co-expression of comparable amounts of HA epitope-tagged and unlabeled SecE in the same cells. Since SecEHA associates normally with SecY and SecG (Figure 5C) and the resulting complexes are fully functional for translocation (Duong and Wickner, 1997a), the ring model of oligomeric SecYEG would suggest that almost all oligomers would include both SecE and SecEHA. Nevertheless, we do not see cross-immunoprecipitation of SecE and SecY–E by anti-HA or cross-linking between SecYEG and SecYEHAG. Furthermore, immunoprecipitation of translocase bearing the I29 translocation intermediate by anti-HA shows less than one-fiftieth of a mole of associated SecE per mole of translocation intermediate. These studies suggest that SecYEG may not function as an oligomer. An alternative explanation for these data is that SecYEG functions as a tetramer and, upon solubilization in detergent, dissociates into monomeric SecYEG that retains its associations with translocation intermediate. Although this explanation would rationalize our data with the oligomeric model, it would imply that the preprotein that can slide through translocase (Schiebel et al., 1991) is tightly bound to one of the SecYEGs that line the channel, and we view this as unlikely.
A recent report suggests that a significant fraction of SecYEG from dodecyl-maltoside detergent extracts of IMVs is dimeric (Manting et al., 2000). Upon the addition of SecA, they report that SecYEG is shifted from a dimeric to a tetrameric conformation (Manting et al., 2000). In our assay for oligomeric SecYEG, this model would predict that 50% of the dimeric SecYEG would be heterodimers containing equal amounts of tagged and untagged SecE. However, anti-HA immunoprecipitations from cymal-4 (Figure 5C) or dodecyl-maltoside (data not shown) detergent extracts failed to detect this interaction. The major difference between our results and those of previous studies is the source of SecYEG. While our experiments used SecYEG expressed at essentially wild-type levels in IMVs, previous studies (Meyer et al., 1999; Manting et al., 2000) used purified SecYEG or SecYE from overexpressing strains and examined either purified SecYEG in micellar solution or SecYEG reconstituted into proteoliposomes (Meyer et al., 1999; Manting et al., 2000). Overexpression or purification of SecYEG might lead to aggregation and the oligomerization seen in electron micrographs. To establish definitively that either monomeric or oligomeric SecYEG functions during translocation, it will be necessary to isolate homogeneous I29–SecA–SecYEG, to determine the I29 to SecYEG stoichiometry and to establish that all of this isolated complex is functional, perhaps as measured by release of the preprotein due to completion of translocation from the complex in micellar solution upon addition of both ATP and reducing agent. Despite an extensive search of experimental conditions, this goal remains elusive.
Materials and methods
Proteins were quantitated using Bradford (Bio-Rad) and BCA (Pierce) protein assay reagents. Escherichia coli phospholipids were from Avanti Polar Lipids. Detergents were from Anatrace. Peptide antisera against SecY, SecE and SecG have been described previously (Joly et al., 1994; Douville et al., 1995). Antibodies were affinity purified using Sulfo-link resin (Pierce) as previously described (Duong and Wickner, 1997). Anti-HA and anti-β-galactosidase were from Roche. For immunoprecipitations, antibodies were covalently coupled to protein A–Sepharose (Roche Molecular Biochemicals) or protein G–agarose (Roche) using dimethylpimelimidate. Anti-mouse and anti-rabbit horseradish peroxidase conjugates and ECL reagent were from Amersham Pharmacia Biotech. For densitometry, autoradiograms were scanned (Lacie, Silverscanner III) and analyzed using IPLab gel software (Signal Analytics).
Bacterial strains and plasmids
Escherichia coli strains UT5600 (Elish et al., 1988), KM9 (Klionsky et al., 1984) and BL21 were used for all IMV preparations. The pBAD22 SecYEHAG, pBAD22 SecYHAEG, pBAD33 SecDFyajC and pTrc99A YEHAG overexpression plasmids have been described previously (Douville et al., 1995; Duong and Wickner, 1997a). For preparation of IMVs with equal amounts of SecY and SecYHA, secYHAEG was PCR amplified from pBAD22 SecYHAEG (Douville et al., 1995) and cloned into the XbaI and SstI sites of pTrc99A (Amersham Pharmacia Biotech). Transformants were grown and induced as described in Figure 4A.
Translocation reagents
SecA (Cunningham et al., 1989), proOmpA (Crooke et al., 1988), proOmpA-BPTI (Schiebel et al., 1991), SecB (Weiss et al., 1988) and IMVs (Douville et al., 1995) were prepared as previously described. proOmpA, proOmpA-BPTI and SecA were radio-iodinated as previously described (Economou and Wickner, 1994). For quantitative immunoblots, SecY was purified from IMVs as a hexahistidine-tagged fusion protein (van der Does et al., 1998). IMVs were solubilized in β-octylglucoside and chromatographed on DE52 resin as previously described (Douville et al., 1995). ProOmpA was purified from inclusion bodies as previously described (Crooke et al., 1988) and solubilized in 50 mM Tris–HCl pH 8.0, 8 M urea, 2 mM DTT. Solubilized proOmpA was bound to a 3 ml bed of Fast Flow Q resin (Pharmacia Biotech) in 50 mM Tris–HCl pH 8.0, 8 M urea, 2 mM DTT and eluted with a linear gradient of NaCl (0–300 mM). Following chromatography, peak fractions of SecY and proOmpA were subjected to SDS–PAGE, stained with Coomassie blue and electroeluted. Electroeluted SecY and proOmpA were desalted on a bed of Sephacryl S200 resin equilibrated in 0.1% SDS and subjected to quantitative amino acid analysis. The results from two independent sources were in close agreement (3.7 and 3.1 µM for SecY and 17 and 20 µM for proOmpA).
Translocation reactions
For generation and quantitation of proOmpA-BPTI translocation intermediate, 100 µl reactions contained 10 µl of 10× TL buffer [500 mM Tris–HCl pH 8.0, 500 mM KCl, 50 mM MgCl2], SecA (50 µg/ml), SecB (66 µg/ml), bovine serum albumin (BSA; 200 µg/ml), ATP (2 mM), NADH (5 mM) and IMVs (100 µg/ml). Reactions were initated by the addition of a mixture of [125I]proOmpA-BPTI (5 × 105 or 1 × 106 c.p.m./ml) and proOmpA-BPTI (16 or 32 µg/ml) in 50 mM Tris–HCl pH 8.0, 6 M urea and incubated for 20 min at 37°C. Samples were transferred to ice and digested with proteinase K (1 mg/ml) for 15 min. Protein was precipitated by the addition of 165 µl of 25% trichloroacetic acid (TCA), acetone washed, suspended in 40 µl of SDS–PAGE sample buffer and heated for 5 min at 100°C. Samples were analyzed by 12% SDS–PAGE followed by autoradiography.
Acknowledgments
Acknowledgements
We thank Dr Arnold Driessen for generously providing the SecY(His6)EG overexpression strain. We also thank Dr Tom Rapoport and members of the Wickner laboratory for insightful discussions concerning this work. This study was supported by a grant from the National Institute of General Medical Sciences (to W.T.W.) and a National Research Service Award (to T.L.Y.).
References
- Beckmann R., Bubeck,D., Grassucci,R., Penczek,P., Verschoor,A., Blobel,G. and Frank,J. (1997) Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science, 278, 2123–2126. [DOI] [PubMed] [Google Scholar]
- Brundage L., Hendrick,J.P., Schiebel,E., Driessen,A.J.M. and Wickner,W. (1990) The purified E.coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell, 62, 649–657. [DOI] [PubMed] [Google Scholar]
- Brundage L., Fimmel,C.J., Mizushima,S. and Wickner,W. (1992) SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J. Biol. Chem., 267, 4166–4170. [PubMed] [Google Scholar]
- Crooke E., Brundage,L., Rice,M. and Wickner,W. (1988) ProOmpA spontaneously folds in a membrane assembly competent state which trigger factor stabilizes. EMBO J., 7, 1831–1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham K., Lill,R., Crooke,E., Rice,M., Moore,K., Wickner,W. and Oliver,D. (1989) SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J., 8, 955–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douville K., Price,A., Eichler,J., Economou,A. and Wickner,W. (1995) SecYEG and SecA are the stoichiometric components of preprotein translocase. J. Biol. Chem., 270, 20106–20111. [DOI] [PubMed] [Google Scholar]
- Duong F. and Wickner,W.T. (1997a) Distinct catalytic roles of the SecYE, SecG, and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J., 16, 2756–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W.T. (1997b) The SecDFyajC domain of preprotein translocase controls preprotein movement by regulating SecA membrane cycling. EMBO J., 16, 4871–4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1998) Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity, and translocation kinetics control the stop-transfer function. EMBO J., 17, 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong F. and Wickner,W. (1999) The PrlA and PrlG phenotypes are caused by a loosened association among the translocase SecYEG subunits. EMBO J., 18, 3263–3270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A. and Wickner,W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell, 78, 835–843. [DOI] [PubMed] [Google Scholar]
- Eichler J. and Wickner,W. (1997) Both an N-terminal 65-kDa domain and a C-terminal 30-kDa domain of SecA cycle into the membrane at SecYEG during translocation. Proc. Natl Acad. Sci. USA, 94, 5574–5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J. and Wickner,W. (1998) The SecA subunit of Esherichia coli preprotein translocase is exposed to the periplasm. J. Bacteriol., 180, 5776–5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J., Brunner,J. and Wickner,W. (1997) The protease-protected 30 kDa domain of SecA is largely inaccessible to the membrane lipid phase. EMBO J., 16, 2188–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elish M.E., Pierce,J.R. and Earhart,C.F. (1988) Biochemical analysis of spontaneous fepA mutants of Escherichia coli.J. Gen. Microbiol., 134, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Hanein D., Matlack,K.E.S., Jungnickel,B., Plath,K., Kalies,K., Miller,K.R., Rapoport,T.A. and Akey,C.W. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Harris C.R. and Silhavy,T.J. (1999) Mapping an interface of SecY (PrlA) and SecE (PrlG) using synthetic phenotypes and in vivo cross-linking. J. Bacteriol., 181, 3438–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.-H., Lecker,S., Schiebel,E., Hendrick,J.P. and Wickner,W. (1990) The binding of SecB to SecA to SecY/E mediates preprotein targeting to the E.coli membrane. Cell, 63, 269–279. [DOI] [PubMed] [Google Scholar]
- Joly J.C. and Wickner,W. (1993) The SecA and SecY subunits of translocase are the nearest neighbors of a translocating preprotein, shielding it from phospholipids. EMBO J., 12, 255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly J.C., Leonard,M.R. and Wickner,W.T. (1994) Subunit dynamics in Escherichia coli preprotein translocase. Proc. Natl Acad. Sci. USA, 91, 4703–4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.J., Rajapandi,T. and Oliver,D. (1994) SecA protein is exposed to the periplasmic surface of the E.coli inner membrane in its active state. Cell, 78, 845–853. [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Brusilow,W.M. and Simoni,R.D. (1984) In vivo evidence for the role of the ε subunit as an inhibitor of the proton translocating ATPase of Escherichia coli. J. Bacteriol., 160, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R., Cunningham,K., Brundage,L., Ito,K., Oliver,D. and Wickner,W. (1989) The SecA protein hydrolyzes ATP and is an essential component of the preprotein translocation ATPase of E.coli. EMBO J., 8, 961–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manting E.H., van der Does,C., Remigy,H., Engel,A. and Driessen,A.J.M. (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J., 19, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Akimanu,J. and Mizushima,S. (1990) SecE-dependent overproduction of SecY in Escherichia coli.FEBS Lett., 269, 96–100. [DOI] [PubMed] [Google Scholar]
- Meyer T.H., Menetret,J., Breitling,R., Miller,K.R., Akey,C.W. and Rapoport,T.A. (1999) The bacterial SecY/E translocation complex forms oligomeric channel-like structures similar to those of the eukaryotic Sec61p complex. J. Mol. Biol., 285, 1789–1800. [DOI] [PubMed] [Google Scholar]
- Nishiyama K.-I., Hanada,M. and Tokuda,H. (1994) Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J., 13, 3272–3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy V. and Oliver,D. (1997) Topology of the integral membrane form of Escherichia coli SecA protein reveals multiple periplasmically exposed regions and modulation by ATP binding. J. Biol. Chem., 272, 23239–23246. [DOI] [PubMed] [Google Scholar]
- Rapoport T.A., Jungnickel,B. and Kutay,U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Schiebel E., Driessen,A.J.M., Hartl,F.-U. and Wickner,W. (1991) ΔµH+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell, 64, 927–939. [DOI] [PubMed] [Google Scholar]
- van der Does C., Manting,E.H., Kaufmann,A., Lutz,M. and Driessen,A.J. (1998) Interaction between SecA and SecYEG in micellar solution and formation of the membrane-inserted state. Biochemistry, 37, 201–210. [DOI] [PubMed] [Google Scholar]
- Weiss J.B., Ray,P.H. and Bassford,P.J.,Jr (1988) Purified SecB protein of Escherichia coli retards folding and promotes membrane translocation of the maltose-binding protein in vitro. Proc. Natl Acad. Sci. USA, 85, 8978–8982. [DOI] [PMC free article] [PubMed] [Google Scholar]