Abstract
The density of introns is both an important feature of genome architecture and a highly variable trait across eukaryotes. This heterogeneity has posed an evolutionary puzzle for the last 30 years. Recent evidence is consistent with novel introns being the outcome of the error-prone repair of DNA double-stranded breaks (DSBs) via non-homologous end joining (NHEJ). Here we suggest that deletion of pre-existing introns could occur via the same pathway. We propose a novel framework in which species-specific differences in the activity of NHEJ and homologous recombination (HR) during the repair of DSBs underlie changes in intron density.
Is intron density controlled by selection or mutation?
All eukaryotes hold in common a highly complex spliceosome devoted to the identification and removal of introns from the nascent mRNA. Although speculative, it is probable that both the very first introns and core components of the spliceosome arose from the mutational decay and cooption of self-splicing group II introns during early eukaryotic evolution [1–3]. Subsequent evolution has involved both extensive intron gain and loss, leading to the current distribution of intron density that varies by several orders of magnitude between species [4–6]. With as few as four introns in the entire Giardia intestinalis genome [7], and more than eight per gene in most mammals, intron density is a key determinant of genome architecture. Although this highly variable trait has important phenotypic consequences, both the mutational mechanisms and the evolutionary conditions that alter intron density have remained unclear.
The absence of group II introns from eukaryotic nuclear genomes, and the fact that novel spliceosomal introns do not share length or sequence characteristics with group II introns, have argued for a two-tier model of intron evolution in which different mechanisms underlie intron-density variation [8,9]. It now appears likely that intron gain is mediated by the capture of DNA fragments during NHEJ of DSBs [10]. The presence of short direct repeats overlapping the splice sites of a subset of novel introns in Daphnia [10], Drosophila [11], and Aspergillus [12] is consistent with the capture of an exogenous fragment at the overhanging ends of a staggered breakpoint (Box 1) [10–13]. Based on patterns of intron gain and loss in Drosophila, we propose that a major proportion of intron loss also occurs as an outcome of NHEJ (in addition to the previously established mechanism of HR-mediated intron loss).
Box 1. DNA double-strand break repair.
The cell utilises two major pathways to repair DNA double-strand breaks (DSBs) [19]. Non-homologous end joining (NHEJ) is the rapid (approx. 30 min) and error-prone religation of two free DNA ends (Figure I), whereas homologous recombination (HR) involves the accurate replacement of a broken segment by copying the homologous chromosome (or sister chromatid), a process that can take more than 7 h to complete [55]. Both pathways compete for the repair of a DSB and the ratio of NHEJ to HR activity is highly dependent on the type of damage, the stage of the cell cycle (HR is often restricted to S/G2-phase), the chromosomal location of the damage and the organism involved [43,55].
We suggest that a mechanism of intron turnover based on DSB repair could provide insight into the evolution of intron density. To date, much of this discussion has focused on the relative importance of selection and drift in shaping taxon-specific variation in intron density (e.g. [14,15]). An alternative explanation is that the dynamics of intron evolution depend largely on changes in the rates of mutations that generate or remove an intron [16]. A genome-wide change in intron density (as has occurred multiple times throughout eukaryotic evolution) requires non-randomness in either the introduction of variants (mutation bias) or in the transmission of variants (selection). As such, the equilibrium intron density can be expressed as:
| [1] |
where μ is the mutation rate and p is the probability of fixation [17]. The acceptance factor (p) is a function of the selection coefficient (s) and the population size (which is equal for gain and loss within a species). Thus, differential acceptance only occurs if there is a systematic difference in s between the presence and absence of an intron. Numerous studies have sought such a difference (reviewed in [8]), proposing that in general novel introns are deleterious (e.g. [10,15]), or that introns are beneficial and that intron density is therefore adaptive (e.g. [18]).
Here, we suggest that changes in mutation bias are a major factor underlying species-specific intron density. Given that both intron gain and loss might be outcomes of DSB repair, we propose that the relative importance of NHEJ and HR (a ratio that does vary between species) might alter the rate of mutations that generate novel introns or remove existing ones.
Does microhomology between the 5′ and 3′ splice sites promote NHEJ-mediated intron deletion?
Intron loss is thought to be mediated by either HR or genomic deletion [8]. HR typically utilises the homologous chromosome or sister chromatid as a template during the repair of a DSB [19]. However, in rare cases a reverse-transcriptase-generated cDNA copy of the same gene [20–22] or an intronless retrogene elsewhere in the genome could serve as a template. Both scenarios result in the precise deletion of the genomic intron because the repair template has previously undergone splicing (Figure I in Box 1).
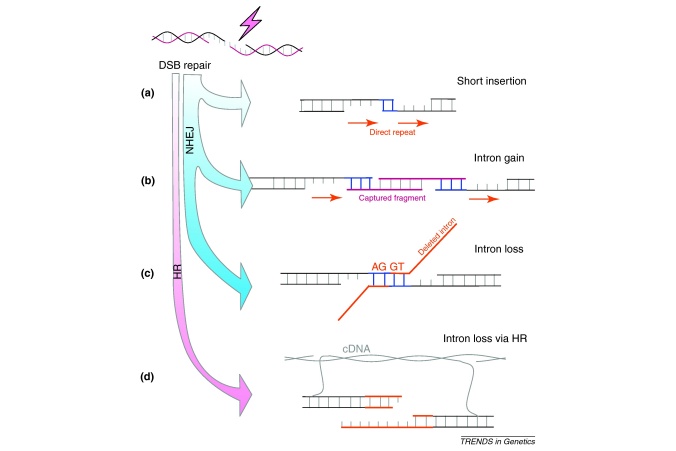
Figure I.
Intron gain and loss as outcomes of DSB repair. (a) NHEJ repair is stabilised by short ‘microhomology’ (blue bases) after 5′ to 3′ resection to generate single-stranded overhangs. Repair could be clean, or lead to a deletion or insertion (as shown). Microhomology pairing within the overhangs results in the insertion of a short direct repeat (red arrows). (b) Microhomology between the overhangs and an exogenous/free DNA fragment can result in a large insertion [68,69] that might or might not be flanked by short direct repeats (as seen for novel introns [10,11]). (c) Microhomology pairing between the 5′ and 3′ splice sites flanking an intronic DSB will cause the precise deletion of the intron, leaving only the original AGGT motif. (d) DNA repair also occurs via homologous recombination. If the template for repair is an intronless cDNA then the genomic intron is lost.
Interestingly, the two ends of a full-length cDNA will resemble DSBs and can thus trigger HR [20]. This would generate a double-crossover event at each end of a gene leading to a long tract of gene conversion potentially spanning (and thus deleting) several introns at once. This model has a powerful advantage in that it offers an explanation for the simultaneous and precise deletion of multiple adjacent introns [12,22–24]. However, despite strong evidence for precise HR-mediated intron loss, several species show a large number of imprecise intron deletions (20% in Drosophila [11,25] and ∼28% in Caenorhabditis [26] for example) that are inconsistent with HR, indicating that more than one mechanism is causing intron loss.
Although it is generally accepted that genomic deletions are an alternative intron-loss pathway, mechanistic details are lacking. Does the error-prone repair of DSBs via NHEJ, which often generates deletions, offer an explanation [26]? The rejoining of single-stranded overhangs by NHEJ is stabilised by the pairing of short (1–6 bp) identical motifs (often referred to as microhomology) on either side of a breakpoint [19]. In many cases the first such similarity encountered on either side of an intronic DSB will be the consensus motif AG|GT of the 5′ and 3′ splice sites (where | indicates the splice site). If pairing occurs between the splice sites, the subsequent repair event will cause the precise deletion of the intronic sequence (Figure I in Box 1) [26]. If an alternative microhomology in the proximity of the DSB is used, this would generate an imprecise deletion. Therefore, NHEJ-mediated deletion is consistent with both the precise and imprecise deletion of one intron at a time.
Experimental support for NHEJ-mediated deletion comes from Caenorhabditis and Drosophila where lost introns show an overly strong adherence to the consensus motif AG|GT at both their 5′ and 3′ splice sites (based on the sequence in the closest neighbouring species) [25,26]. Furthermore, lineages with very few introns tend to have 5′ and 3′ splice sites with high sequence similarity, whereas intron-rich species show highly degenerate splicing motifs [27–29]. Within longer introns one might expect to encounter short identical motifs before reaching the splice sites, hence precise deletion should favour shorter introns, and this is in fact the case in mammals [30], Drosophila [11] and yeast [12].
Interestingly, recently lost introns in Drosophila are more likely to contain motifs imparting a higher twist angle on the DNA backbone than do stable introns (see supplementary material online). Such motifs have been associated with a high propensity to suffer DSBs [31,32] due to the formation of a non-canonical DNA secondary structure [33] and replication stress and instability [34]. Although indirect, this might suggest that some introns have a higher than average chance of undergoing intron loss via NHEJ or HR.
DSB repair: a common mechanism for intron gain and loss?
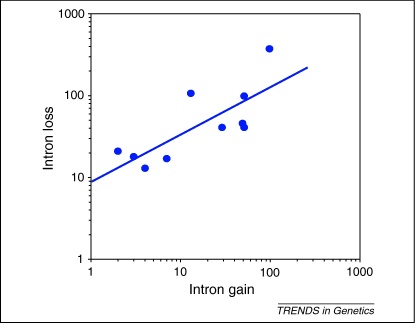
If repair of DSBs is the mechanistic basis of both intron gain (via NHEJ) and intron loss (via a combination of NHEJ and HR) then one might expect a positive correlation between the rates of intron gain and loss across species and over time. A survey of these rates across the ∼40 million years of Drosophila evolution shows just such a strong positive correlation (Spearman correlation coefficient = 0.89, P < 0.0001) (Figure 1) [11]. Likewise, the same positive relationship is observed over much deeper branches of eukaryotic evolution (Spearman correlation coefficient = 0.69, P < 0.003 [4]). Although it could be possible that other factors influence this positive correlation, we suggest that, in general, the rate of intron gain is linked to the rate of intron loss – because both processes are at least partly an outcome of DSB repair.
Figure 1.
A highly significant positive correlation between the rate of intron gain and intron loss is consistent with commonality in the underlying mutational mechanism. The number of intron gain and loss events that have occurred along each branch of the Drosophila clade was taken as previously published [11]. Each datapoint represents one branch of Drosophila evolution, allowing the level of intron gain and loss over the same time period to be compared (Figure S2 in the supplementary material online).
Several points in eukaryotic evolution have been marked by either a dramatic increase in intron density (leading to Metazoa and Deuterostomia for example) or the overwhelming loss of introns [4,35,36]. One common hypothesis is that selection drives changes in intron density, for example through selection for genome reduction or an increase in alternative splicing [5,6,37]. However, if selection is the dominant factor determining intron density then the rates of intron gain and loss should be negatively correlated because the same evolutionary process would drive both rates in opposite directions [5]. Likewise, a negative correlation is expected if changes in intron density are purely the result of changes in population size [38]. Although these expectations could be considered overly simplistic (by ignoring any number of more complex evolutionary scenarios), the observation of a positive correlation across several timescales and many species argues against a general role for selection in determining intron density. Furthermore, despite a great deal of effort attempting to link intron density to either adaptation (via selection for or against introns) or genetic drift (and population size), no strong connection has been established [4–6,35,36,39,40].
Changing intron density: the relative usage of NHEJ and HR differs between species
The large change in intron density that has taken place at several points in eukaryotic evolution requires a shift in the ratio of intron gain to loss. We suggest that this change could be modulated by differences in the activity of NHEJ and HR during DSB repair. Both pathways are largely separate and compete for the repair of DSBs [41,42] and, significantly, the relative contribution of the two pathways is highly species-specific (Table 1) [19]. NHEJ is predominantly used in mammals [43,44] and Drosophila [45,46], whereas HR is the major pathway in Saccharomyces cerevisiae [47], a species having undergone almost complete intron loss. By contrast, NHEJ is the major repair pathway in two comparatively intron-rich fungal species Schizosaccharomyces pombe [48] [∼1 intron/gene (Wellcome Trust Sanger Institute, http://www.sanger.ac.uk/Projects/S_pombe/genome_stats.shtml), and in Cryptococcus neoformans [49] (5.3 introns/gene [50]) suggesting a correspondence between NHEJ usage and intron density.
Table 1.
The relative contributions of NHEJ and HR differ between species
| Species | Introns per gene |
Estimated contributiona |
Approximate ratio | DSB typeb | Refs | |
|---|---|---|---|---|---|---|
| NHEJ | HR | NHEJ:HR | ||||
| S. cerevisiae | 0.07 | <1% | >99% | 1:100 | Complex | [58] |
| S. cerevisiae | 2–18% | 82–98% | 1:9 | HO | [19] | |
| S. cerevisiae | Minimal | Major | Complex (bleomycin) | [59] | ||
| S. pombec | 0.8 | ∼14% | 66% | 1:5 | HO | [60] |
| D. melanogaster | ∼4 | 19% | 11% | 3:2 | I-SceI | [45,61] |
| C. elegans | 4.7 | ∼0 | Dominantd | Ionising radiation | [62] | |
| C. neoformans | 4.7 | Major | 0–47% | >1:1 | Transgene integration | [49] |
| Mus musculuse | ∼8 | 57% | 19% | 3:1 | I-SceI | [63] |
| Homo sapiens | ∼9 | 86% | 14% | 9:1 | I-SceI | [55] |
| Homo sapiens | 75% | 25% | 3:1 | Incompatible ends | [55] | |
Several studies also differentiate repair via single-strand annealing (SSA) which generates large deletions. There is no mechanistic basis for intron gain or loss via SSA [19], therefore the contribution of this pathway is not included here.
I-SceI and HO endonucleases generate 4 bp complementary overhangs.
NHEJ becomes the dominant pathway during the G1 phase of the haploid cell cycle because sister chromatids are not available as templates for HR [64].
Germline DSBs in C. elegans are exclusively repaired via HR. Interestingly, intron loss is 400-fold higher in nematodes than in mammals [4,65], and most novel introns arise via the unusual process of intronisation [66].
Genetic differences in the efficiency of HR segregate within natural populations of mouse [67].
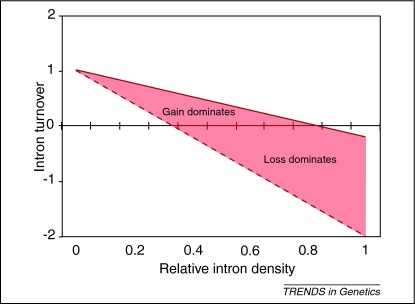
We propose a simple model in which the relative rate of these two DSB repair pathways (in combination with intron density) can produce either an increase or decrease in intron number (Figure 2). Given that the relative contribution of NHEJ and HR to DSB repair varies over several orders of magnitude between intron-rich and -poor species, it is difficult to know the relevant values for each parameter that existed within a species over evolutionary time. However, using a conservative range of values clearly demonstrates that intron turnover can shift from net gain to net loss. Although this simple model does not consider variables such as the rate of exogenous DNA capture during NHEJ, or variation in splicing efficiency and intron size between clades, it does illustrate how adjusting a single parameter (the relative activity of NHEJ and HR) could be sufficient to explain the heterogeneity in intron density among eukaryotic species, an observation which has puzzled researchers for three decades.
Figure 2.
Small changes to the relative roles of NHEJ and HR are sufficient to increase or decrease intron numbers depending on intron density. This simple model highlights the intuitive finding that intron density presents a limiting factor to intron proliferation, consistent with the excess of intron loss observed in intron-dense genomes [30,56,57]. Although the parameters used are largely arbitrary (Table S2), with the solid line representing a fivefold excess of NHEJ over HR and the broken line a twofold excess of HR, these values are reasonable considering the >100-fold preference of HR in budding yeast [47] and the general preference for NHEJ in metazoa [19,45] (Table 1) (see also Table S2 in the supplementary material online).
This model might partly explain the curious observation that, whereas almost all intron-poor species have a strong 5′ bias in intron position, intron-rich species do not [51,52]. The dominant action of cDNA-mediated HR in these species might lead to the preferential loss of 3′ [53] and internal introns [54] due to the directionality of reverse transcriptase. However, a more dominant role of NHEJ-mediated intron loss in intron-rich species would not generate this 3′ bias.
Concluding remarks
Based on the suggestion that intron gain and loss are outcomes of DSB repair, we propose that intron density is consistent with the long-term efficiency of the two repair pathways, NHEJ and HR. Although this hypothesis is consistent with much of the available data, we hope our proposal could serve as a null hypothesis against which new genomic data can be tested. This points future work in two directions: experimental evolution and population genetic surveys to establish the molecular outcomes of NHEJ and HR, and estimation of the relative activity of these two pathways in species that have undergone recent changes in intron density. Whereas a full account of intron evolution will include examples where intron density is influenced by selection and drift, here we suggest a dominant role for genetic changes to the activity of NHEJ and HR in generating species-specific differential intron gain/loss mutation rates.
Acknowledgements
We are grateful A. McGregor for raising our awareness of the different rates of NHEJ and HR in Drosophila, and thank A. Tucker and J. Heraud for feedback on this manuscript. Many thanks to other members of the Institute of Population Genetics for helpful discussion. This work was supported by grants of the FWF Austrian Science Fund (P19832).
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.tig.2010.10.004.
Appendix A. Supplementary data
References
- 1.Koonin E.V. The origin of introns and their role in eukaryogenesis: a compromise solution to the introns-early versus introns-late debate? Biol. Direct. 2006;1:22. doi: 10.1186/1745-6150-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Toor N. Crystal structure of a self-spliced group II intron. Science. 2008;320:77–82. doi: 10.1126/science.1153803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chalamcharla V.R. Nuclear expression of a group II intron is consistent with spliceosomal intron ancestry. Genes Dev. 2010;24:827–836. doi: 10.1101/gad.1905010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmel L. Three distinct modes of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roy S.W. Intron-rich ancestors. Trends Genet. 2006;22:468–471. doi: 10.1016/j.tig.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Stajich J.E. Comparative genomic analysis of fungal genomes reveals intron-rich ancestors. Genome Biol. 2007;8:R223. doi: 10.1186/gb-2007-8-10-r223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franzen O. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5:e1000560. doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S.W., Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat. Rev. Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- 9.Poole A.M. Did group II intron proliferation in an endosymbiont-bearing archaeon create eukaryotes? Biol. Direct. 2006;1:36. doi: 10.1186/1745-6150-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farlow A. Nonsense-mediated decay enables intron gain in Drosophila. PLoS Genet. 2010;6:e1000819. doi: 10.1371/journal.pgen.1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L.Y. Evaluation of models of the mechanisms underlying intron loss and gain in Aspergillus Fungi. J. Mol. Evol. 2010 doi: 10.1007/s00239-010-9391-6. [DOI] [PubMed] [Google Scholar]
- 13.Ragg H. Intron creation and DNA repair. Cell Mol. Life Sci. 2010 doi: 10.1007/s00018-010-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeffares D. The biology of intron gain and loss. Trends Genet. 2006;22:16–22. doi: 10.1016/j.tig.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 15.Lynch M., Richardson A.O. The evolution of spliceosomal introns. Curr. Opin. Genet. Dev. 2002;12:701–710. doi: 10.1016/s0959-437x(02)00360-x. [DOI] [PubMed] [Google Scholar]
- 16.Roy S.W., Hartl D.L. Very little intron loss/gain in Plasmodium: intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–756. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stoltzfus A. Mutationism and the dual causation of evolutionary change. Evol. Dev. 2006;8:304–317. doi: 10.1111/j.1525-142X.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 18.Niu D.K. Protecting exons from deleterious R-loops: a potential advantage of having introns. Biol. Direct. 2007;2:11. doi: 10.1186/1745-6150-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber J.E. Partners and pathways repairing a double-strand break. Trends Genet. 2000;16:259–264. doi: 10.1016/s0168-9525(00)02022-9. [DOI] [PubMed] [Google Scholar]
- 20.Hu K. Intron exclusion and the mystery of intron loss. FEBS Lett. 2006;580:6361–6365. doi: 10.1016/j.febslet.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 21.Derr L.K. RNA-mediated recombination in S. cerevisiae. Cell. 1991;67:355–364. doi: 10.1016/0092-8674(91)90187-4. [DOI] [PubMed] [Google Scholar]
- 22.Hu K.J., Leung P.C. Complete, precise, and innocuous loss of multiple introns in the currently intronless, active cathepsin L-like genes, and inference from this event. Mol. Phylogenet. Evol. 2006;38:685–696. doi: 10.1016/j.ympev.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 23.Stajich J.E., Dietrich F.S. Evidence of mRNA-mediated intron loss in the human-pathogenic fungus Cryptococcus neoformans. Eukaryot. Cell. 2006;5:789–793. doi: 10.1128/EC.5.5.789-793.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharpton T. Mechanisms of intron gain and loss in Cryptococcus. Genome Biol. 2008;9:R24. doi: 10.1186/gb-2008-9-1-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coulombe-Huntington J., Majewski J. Intron loss and gain in Drosophila. Mol. Biol. Evol. 2007;24:2842–2850. doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- 26.Kent W.J., Zahler A.M. Conservation, regulation, synteny, and introns in a large-scale C. briggsae–C. elegans genomic alignment. Genome Res. 2000;10:1115–1125. doi: 10.1101/gr.10.8.1115. [DOI] [PubMed] [Google Scholar]
- 27.Irimia M. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 28.Irimia M., Roy S.W. Evolutionary convergence on highly-conserved 3′ intron structures in intron-poor eukaryotes and insights into the ancestral eukaryotic genome. PLoS Genet. 2008;4:e1000148. doi: 10.1371/journal.pgen.1000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Irimia M. Complex selection on 5′ splice sites in intron-rich organisms. Genome Res. 2009;19:2021–2027. doi: 10.1101/gr.089276.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coulombe-Huntington J., Majewski J. Characterization of intron loss events in mammals. Genome Res. 2006;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zlotorynski E. Molecular basis for expression of common and rare fragile sites. Mol. Cell Biol. 2003;23:7143–7151. doi: 10.1128/MCB.23.20.7143-7151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan Y.F. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Travers A.A., Thompson J.M. An introduction to the mechanics of DNA. Philos. Trans. R. Soc. Lond. A Math. Phys. Sci. 2004;362:1265–1279. doi: 10.1098/rsta.2004.1392. [DOI] [PubMed] [Google Scholar]
- 34.Durkin S.G. Replication stress induces tumor-like microdeletions in FHIT/FRA3B. Proc. Natl. Acad. Sci. U. S. A. 2008;105:246–251. doi: 10.1073/pnas.0708097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nguyen H. New maximum likelihood estimators for eukaryotic intron evolution. PLoS Comput. Biol. 2005;1:e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Csuros M. Malin: maximum likelihood analysis of intron evolution in eukaryotes. Bioinformatics. 2008;24:1538–1539. doi: 10.1093/bioinformatics/btn226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carmel L. Evolutionarily conserved genes preferentially accumulate introns. Genome Res. 2007;17:1045–1050. doi: 10.1101/gr.5978207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lynch M. Intron evolution as a population-genetic process. Proc. Natl. Acad. Sci. U. S. A. 2002;99:6118–6123. doi: 10.1073/pnas.092595699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy S.W., Gilbert W. Rates of intron loss and gain: implications for early eukaryotic evolution. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5773–5778. doi: 10.1073/pnas.0500383102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roy S.W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mills K.D. Rad54 and DNA Ligase IV cooperate to maintain mammalian chromatid stability. Genes Dev. 2004;18:1283–1292. doi: 10.1101/gad.1204304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Preston C.R. Efficient repair of DNA breaks in Drosophila: evidence for single-strand annealing and competition with other repair pathways. Genetics. 2002;161:711–720. doi: 10.1093/genetics/161.2.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takata M. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rebuzzini P. New mammalian cellular systems to study mutations introduced at the break site by non-homologous end-joining. DNA Repair (Amst.) 2005;4:546–555. doi: 10.1016/j.dnarep.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 45.Preston C.R. Differential usage of alternative pathways of double-strand break repair in Drosophila. Genetics. 2006;172:1055–1068. doi: 10.1534/genetics.105.050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beumer K.J. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. U. S. A. 2008;105:19821–19826. doi: 10.1073/pnas.0810475105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jeggo P.A. DNA breakage and repair. Adv. Genet. 1998;38:185–218. doi: 10.1016/s0065-2660(08)60144-3. [DOI] [PubMed] [Google Scholar]
- 48.Wilson S. The role of Schizosaccharomyces pombe Rad32, the Mre11 homologue, and other DNA damage response proteins in non-homologous end joining and telomere length maintenance. Nucleic Acids Res. 1999;27:2655–2661. doi: 10.1093/nar/27.13.2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimizu K. Deletion of CnLIG4 DNA ligase gene in the fungal pathogen Cryptococcus neoformans elevates homologous recombination efficiency. Mycoscience. 2010;51:28–33. [Google Scholar]
- 50.Loftus B.J. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mourier T., Jeffares D.C. Eukaryotic intron loss. Science. 2003;300:1393. doi: 10.1126/science.1080559. [DOI] [PubMed] [Google Scholar]
- 52.Lin K., Zhang D.Y. The excess of 5′ introns in eukaryotic genomes. Nucleic Acids Res. 2005;33:6522–6527. doi: 10.1093/nar/gki970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sverdlov A.V. Preferential loss and gain of introns in 3′ portions of genes suggests a reverse-transcription mechanism of intron insertion. Gene. 2004;338:85–91. doi: 10.1016/j.gene.2004.05.027. [DOI] [PubMed] [Google Scholar]
- 54.Nielsen C. Patterns of intron gain and loss in fungi. PloS Biol. 2004;2:e422. doi: 10.1371/journal.pbio.0020422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mao Z. Comparison of nonhomologous end joining and homologous recombination in human cells. DNA Repair (Amst.) 2008;7:1765–1771. doi: 10.1016/j.dnarep.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh Y.H. Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon) Mol. Biol. Evol. 2008;25:526–535. doi: 10.1093/molbev/msm278. [DOI] [PubMed] [Google Scholar]
- 57.Putnam N. Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science. 2007;317:86–94. doi: 10.1126/science.1139158. [DOI] [PubMed] [Google Scholar]
- 58.Lee S.E. Role of yeast SIR genes and mating type in directing DNA double-strand breaks to homologous and non-homologous repair paths. Curr. Biol. 1999;9:767–770. doi: 10.1016/s0960-9822(99)80339-x. [DOI] [PubMed] [Google Scholar]
- 59.Letavayova L. Relative contribution of homologous recombination and non-homologous end-joining to DNA double-strand break repair after oxidative stress in Saccharomyces cerevisiae. DNA Repair (Amst.) 2006;5:602–610. doi: 10.1016/j.dnarep.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 60.Prudden J. Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J. 2003;22:1419–1430. doi: 10.1093/emboj/cdg119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Johnson-Schlitz D.M. Multiple-pathway analysis of double-strand break repair mutations in Drosophila. PLoS Genet. 2007;3:e50. doi: 10.1371/journal.pgen.0030050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clejan I. Developmental modulation of nonhomologous end joining in Caenorhabditis elegans. Genetics. 2006;173:1301–1317. doi: 10.1534/genetics.106.058628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Johnson R.D., Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000;19:3398–3407. doi: 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raji H., Hartsuiker E. Double-strand break repair and homologous recombination in Schizosaccharomyces pombe. Yeast. 2006;23:963–976. doi: 10.1002/yea.1414. [DOI] [PubMed] [Google Scholar]
- 65.Cho S. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Irimia M. Origin of introns by ‘intronization’ of exonic sequences. Trends Genet. 2008;24:378–381. doi: 10.1016/j.tig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 67.Kujjo L.L. Enhancing survival of mouse oocytes following chemotherapy or aging by targeting bax and rad51. PLoS One. 2010;5:e9204. doi: 10.1371/journal.pone.0009204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin Y., Waldman A.S. Capture of DNA sequences at double-strand breaks in mammalian chromosomes. Genetics. 2001;158:1665–1674. doi: 10.1093/genetics/158.4.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hazkani-Covo E., Covo S. Numt-mediated double-strand break repair mitigates deletions during primate genome evolution. PLoS Genet. 2008;4:e1000237. doi: 10.1371/journal.pgen.1000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.