Abstract
The reverse tetracycline-dependent transactivator system was employed in insulinoma INS-1 cells to achieve controlled inducible expression of hepatocyte nuclear factor-1α (HNF1α)-P291fsinsC, the most common mutation associated with subtype 3 of maturity-onset diabetes of the young (MODY3). Nuclear localized HNF1α-P291fsinsC protein exerts its dominant-negative effects by competing with endogenous HNF1α for the cognate DNA-binding site. HNF1α controls multiple genes implicated in pancreatic β-cell function and notably in metabolism– secretion coupling. In addition to reduced expression of the genes encoding insulin, glucose transporter-2, l-pyruvate kinase, aldolase B and 3-hydroxy-3-methylglutaryl coenzyme A reductase, induction of HNF1αP291fsinsC also significantly inhibits expression of mitochondrial 2-oxoglutarate dehydrogenase (OGDH) E1 subunit mRNA and protein. OGDH enzyme activity and [14C]pyruvate oxidation were also reduced. In contrast, the mRNA and protein levels of mitochondrial uncoupling protein-2 were dramatically increased by HNF1α-P291fsinsC induction. As predicted from this altered gene expression profile, HNF1α-P291fsinsC also inhibits insulin secretory responses to glucose and leucine, correlated with impaired nutrient-evoked mitochondrial ATP production and mitochondrial membrane hyperpolarization. These unprecedented results suggest the molecular mechanism of HNF1αP291fsinsC causing β-cell dysfunction.
Keywords: β-cell gene expression/HNF1α/insulin secretion/mitochondrial function/MODY3
Introduction
Mutations in the gene encoding hepatocyte nuclear factor-1α (HNF1α) are the cause of maturity-onset diabetes of the young type 3 (MODY3). MODY3 is characterized by autosomal dominant inheritance, early onset (generally before the age of 25 years) and severe impairment of glucose-stimulated insulin secretion (Fajans, 1990; Byrne et al., 1996; Lehto et al., 1997). Most MODY3 subjects under the age of 10 years have a normal fasting blood glucose and normal glucose tolerance (Hattersley, 1998). With a progressive impairment in insulin secretion, they develop marked hyperglycemia often associated with microvascular complications, but no insulin resistance (Lehto et al., 1997; Hattersley, 1998). In ∼30% of cases, the patients require insulin therapy, although no signs of autoimmune destruction of the β-cells have been observed (Lehto et al., 1997; Hattersley, 1998; Iwasaki et al., 1998). It has been reported that 5–10% of type I diabetes in Japanese and Danish populations are actually caused by mutations in HNF1α (Yamagata et al., 1997; Møller et al., 1998).
HNF1α is expressed in liver, kidney, intestine and pancreatic islets (Blumenfeld et al., 1991). By now, >100 genes regarded as liver specific are regulated potentially by HNF1α (Tronche et al., 1997). The human HNF1α gene is comprised of 10 exons encoding a transcription factor of 631 amino acids composed of a short N-terminal dimerization domain (amino acids 1–32), a POU-homeobox DNA-binding domain (amino acids 150–280) and a C-terminal transactivation domain (amino acids 281–631) (Mendel and Crabtree, 1991; Kaisaki et al., 1997). Numerous mutations linked to MODY3 have been found scattered throughout the HNF1α gene and consist of frameshift, missense, nonsense and splice site mutations (Glucksmann et al., 1997; Kaisaki et al., 1997). The majority of mutations located in the DNA-binding domain or transactivation domain of HNF1α would function in a dominant-negative manner. Other mutations associated with the MODY3 phenotype that have been identified in the promoter region (Gragnoli et al., 1997) and dimerization domain (Vaxillaire et al., 1997) of HNF1α would result in reduced HNF1α gene dosage. MODY3 mutants that form proteins with reduced stability or decreased transactivation would also elicit haploinsufficiency. HNF1α-P291fsinsC, the most common HNF1α mutation, is generated by a mutational hotspot in exon 4 occurring in unrelated families (Hattersley, 1998). This frameshift mutation, resulting from insertion of a C in a poly(C) tract centered around codon 290, encodes a truncated protein of 315 amino acids. It contains intact dimerization and DNA-binding domains but lacks the transactivation domain (Glucksmann et al., 1997; Kaisaki et al., 1997).
Transient expression of HNF1α-P291fsinsC in mouse insulinoma MIN6 cells has been shown to inhibit the transactivation of the l-type pyruvate kinase (l-PK) promoter by wild-type HNF1α (HNF1α-WT) (Yamagata et al., 1998). In addition, in vitro translated HNF1α-P291fsinsC and HNF1α-WT proteins were demonstrated to form heterodimers (Yamagata et al., 1998). This suggests that HNF1α-P291fsinsC has a dominant-negative effect on the transactivation of HNF1α-WT (Yamagata et al., 1998).
We have reported that the inducible expression of an artificial, DNA-binding-deficient dominant-negative mutant of HNF1α (DNHNF1α) in insulinoma INS-1 cells results in impaired metabolism–secretion coupling (Wang et al., 1998). To elucidate the HNF1α target genes and the molecular mechanism implicated in impaired metabolism–secretion coupling, we investigated the consequences of inducible expression of HNF1α-P291fsinsC on various β-cell mRNA species. The present study provides the following novel findings: (i) it demonstrates in a cellular system that the nuclear localized HNF1α-P291fsinsC functions as a dominant-negative mutant; (ii) the glycolytic enzyme, aldolase B, is also a target of HNF1α, in addition to l-PK and glucose transporter-2 (GLUT2); (iii) induction of HNF1α-P291fsinsC results in diminished expression of 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase that controls the mevalonate pathway important for cholesterol synthesis (Goldstein and Brown, 1990); (iv) HNF1α-P291fsinsC reduces expression of the E1 subunit of 2-oxoglutarate dehydrogenase (α-ketoglutarate dehydrogenase, OGDH), a rate-limiting enzyme of the tricarboxylic acid (TCA) cycle; and (v) HNF1α-P291fsinsC increases expression of the mitochondrial uncoupling protein-2 (UCP-2), which may also impair glucose-stimulated insulin secretion by uncoupling oxidative phosphorylation (Chan et al., 1999).
Results
Inducible expression and cellular localization of HNF1α-P291fsinsC in INS-1 cells
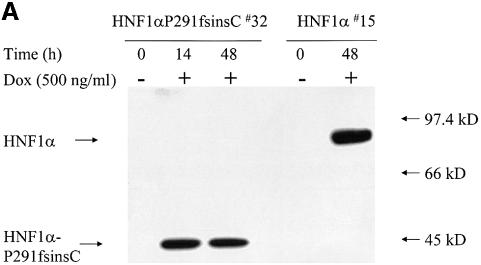
Four out of 40 hygromycin-resistant clones showed high level expression of HNF1α-P291fsinsC mRNA in initial screening by northern blotting (data not shown). The clone, termed HNF1α-P291fsinsC#32, was selected randomly for the present study. The MODY3 mutant protein was expressed efficiently after 14 h culture with 500 ng/ml doxycycline. The expression was similar at 48 h (Figure 1A). Nuclear extracts from HNF1α#15 cells, in which the overexpression of HNF1α-WT was induced by doxycycline (Wang et al., 1998), were used as control. As predicted from their sequences, the truncated HNF1α-P291fsinsC protein is half the size of the wild-type protein (44 and 88 kDa, respectively) (Figure 1A).
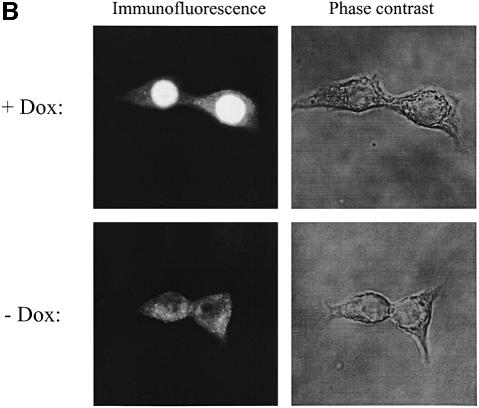
Fig. 1. (A) Rapid induction of HNF1α-P291fsinsC protein by doxycycline in INS-1 cells. Nuclear extracts from HNF1α-P291fsinsC#32 cells cultured with or without 500 ng/ml doxycycline for 14 or 48 h were resolved in 9% SDS–PAGE, transferred to nitrocellulose and immunoblotted with an antibody against the HNF1α N-terminus. Nuclear extract proteins from HNF1α#15 cells, which expressed the wild-type HNF1α under the control of doxycycline, were analyzed on the same gel. All lanes were loaded with 10 µg of nuclear extract proteins. (B) Inducible expression and cellular localization of HNF1α-P291fsinsC protein in INS-1 cells. Immunofluorescence using an antibody specific for the N-terminus of HNF1α was performed in HNF1α-P291fsinsC#32 cells cultured in the presence (upper left) or absence (lower left) of 500 ng/ml doxycycline for 24 h. The microscopic phase contrast images are shown in the right panel.
The intracellular distribution of MODY3 mutant protein was examined in HNF1α-P291fsinsC#32 cells cultured with 500 ng/ml doxycycline for 24 h. Immunostaining with antibody specific for HNF1α N-terminal sequences clearly showed that HNF1α-P291fsinsC protein was localized almost exclusively in the nucleus (Figure 1B).
Induction of HNF1α-P291fsinsC reduces insulin gene expression by direct suppression of insulin promoter activity
HNF1α-P291fsinsC is a truncated protein lacking the C-terminal transactivation domain but possessing intact dimerization and DNA-binding domains. Consequently, overexpression of HNF1α-P291fsinsC should prevent the binding of endogenous HNF1α to DNA by forming HNF1α-P291fsinsC homodimers and HNF1α-P291fsinsC–HNF1α-WT heterodimers. To test this hypothesis, we have performed electrophoretic mobility shift assay (EMSA) experiments with a probe corresponding to the rat insulin I FLAT element (InsFLAT), which contains the HNF1α-binding site (Emens et al., 1992). In accordance with our previous results using a substitution mutation of rat HNF1α (Wang et al., 1998), the induction of HNF1α-P291fsinsC protein abolished the binding of endogenous HNF1α-WT homodimers to the InsFLAT (data not shown). We found that the formation of the HNF1α-P291fsinsC homodimers was predominant in nuclear extracts from HNF1α-P291fsinsC#32 cells cultured with doxycycline for 48 h (data not shown). HNF1α-P291fsinsC–HNF1α-WT heterodimers were also present (data not shown). Cold probe competition and antibody supershift experiments were also performed to define these DNA-binding complexes (data not shown).
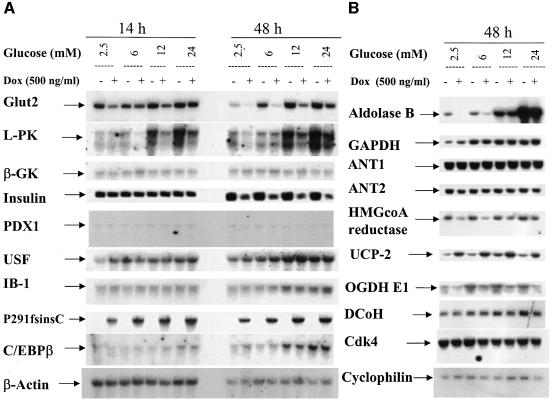
In agreement with its dominant-negative function on HNF1α DNA binding, HNF1α-P291fsinsC also directly suppressed insulin promoter activity and decreased insulin mRNA expression. As shown by northern analysis (Figure 2A), insulin mRNA expression was significantly inhibited after 48 h induction of HNF1α-P291fsinsC. In contrast, the expression of two transcription factors, PDX1 and CCAAT/enhancer-binding protein β (C/EBPβ), which are involved in insulin gene transcription (Seufert et al., 1998), was not altered by induction of HNF1α-P291fsinsC (Figure 2A). The expression of dimerization cofactor for HNF1 (DcoH) was also unchanged (Figure 2B).
Fig. 2. Effects of HNF1α-P291fsinsC on the expression of HNF1α target genes. (A and B) Northern blotting was employed to examine the gene expression in HNF1α-P291fsinsC#32 cells induced with 500 ng/ml doxycycline at 2.5 mM glucose for 14 or 48 h. Culture was continued for 8 h at 2.5 mM glucose or at the indicated glucose concentrations before cells were extracted for total RNA. RNA samples were analyzed by hybridizing with the indicated cDNA probes.
To confirm that HNF1α directly affects the insulin promoter, we mutated the HNF1α-binding site in –410INSLuc that encodes the rat insulin I promoter luciferase reporter construct (German et al., 1992a,b). This indeed abolished HNF1α binding activity in EMSA experiments (data not shown). HNF1α-P291fsinsC#32 cells were transiently transfected with this HNF1α site-mutated plasmid, termed –410MFINSLuc, to compare its luciferase reporter activity with that of wild-type –410INSLuc. The mutation of the HNF1α-binding site resulted in 39.7 ± 4.3% (n = 6) loss of intrinsic promoter activity. The induction of HNF1α-P291fsinsC for 48 h caused 63.4 ± 6.1% (n = 6) reduction of the rat insulin I promoter activity. As expected, this inhibitory effect was no longer observed in parallel transfection experiments with –410MFINSLuc. The direct inhibitory effect of HNF1α-P291fsinsC on insulin promoter activity was also substantiated by transient transfection of HNF1α-P291fsinsC#32 cells with three luciferase constructs containing multimerized segments of the rat insulin I minienhancer: wild-type (Far-FLATwt), HNF1α-binding site-mutated (Far-FLATMF) and E element-mutated (Far-FLATME) (German et al., 1992a,b). Induction of HNF1α-P291fsinsC resulted in 65.2 ± 5.4% and 80.3 ± 7.8% reduction of luciferase activity in HNF1α-P291fsinsC#32 cells transfected respectively with Far-FLATwt and Far-FLATME, but had no further suppressive effect on the activity of Far-FLATMF.
GLUT-2 and glycolytic enzymes are suppressed by HNF1α-P291fsinsC
As previously reported (Marie et al., 1993; Waeber et al., 1994; Roche et al., 1997), the transcripts of l-PK, GLUT-2 and glyceraldehyde-3 phosphate dehydrogenase (GAPDH) were induced in response to an increased extracellular glucose concentration in insulinoma INS-1 cells (Figure 2A and B). The expression of aldolase B mRNA was also induced by glucose in a concentration-dependent manner (Figure 2B). The promoters of l-PK and GLUT-2 have the HNF1α-binding site in common. Induction of HNF1α-P291fsinsC inhibited endogenous HNF1α binding to its recognition site in the promoters of l-PK and GLUT-2 (data not shown) and consequently suppressed the basal expression of their transcripts (Figure 2A). Notably, HNF1α-P291fsinsC did not diminish the glucose responsiveness of these two genes (Figure 2A). HNF1α regulates aldolase B mRNA in a similar manner but the underlying mechanism remains to be established. In contrast to l-PK, GLUT-2 and aldolase B, the expression of two other glucose-responsive genes, GAPDH (Figure 2B) and acetyl-CoA carboxylase (data not shown), remained unchanged after induction of HNF1α-P291fsinsC. HNF1α-P291fsinsC did not affect the expression of upstream stimulatory factors (USFs), which have been proposed to mediate the glucose response of the l-PK promoter (Miquerol et al., 1994) (Figure 2A). Likewise, HNF1α-P291fsinsC had no effect on expression of islet brain-1 (IB1) and PDX1 (Figure 2A), which are implicated in regulation of GLUT-2 promoter activity (Waeber et al., 1996; Bonny et al., 1998).
HMGCoA reductase controls the mevalonate pathway important for diverse cellular functions, ranging from cholesterol synthesis to cell proliferation (Goldstein and Brown, 1990). As shown by northern analysis (Figure 2B), induction of HNF1α-P291fsinsC caused a significant decrease in HMGCoA reductase mRNA. The expression of cyclin-dependent kinase 4 (Cdk4), which regulates pancreatic β-cell proliferation (Rane et al., 1999), was not changed after induction of HNF1α-P291fsinsC.
HNF1α-P291fsinsC controls genes important for mitochondrial metabolism
RT–PCR screening of 21 genes encoding mitochondrial proteins after induction of HNF1α-P291fsinsC revealed significant and consistent down-regulation of OGDH E1 subunit and up-regulation of UCP-2 (data not shown). Using cDNA probes specific for OGDH E1 and UCP-2, respectively, we performed northern hybridization. As demonstrated in Figure 2B, the expression of OGDH E1 subunit was significantly suppressed while the expression of UCP-2 was dramatically increased. As predicted from RT–PCR results (data not shown), the expression of mitochondrial adenine nucleotide translocator 1 and 2 (ANT1 and ANT2) mRNAs remained constant (Figure 2B).
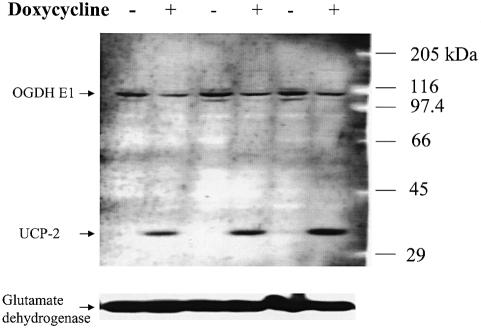
E1 is the catalytic subunit of OGDH and a rate-limiting enzyme in the mitochondrial TCA cycle. UCP-2 mediates proton transport and uncouples respiration from oxidative phosphorylation, which may dissipate the mitochondrial membrane potential and inhibit the efficiency of ATP synthesis (Boss et al., 2000; Ricquier and Bouillaud, 2000). In agreement with changes observed in OGDH E1 and UCP-2 mRNAs, induction of HNF1α-P291fsinsC also caused a significant reduction of OGDH E1 protein and a marked increase of UCP-2 protein (Figure 3). In contrast, another mitochondrial protein, glutamate dehydrogenase, remained constant irrespective of doxycycline induction (Figure 3). As observed in three independent western blots, OGDH E1 protein expression was decreased consistently by 63.2 ± 7.4% (n = 3). Accordingly, the enzyme activity of OGDH was also inhibited by 36% after induction of HNF1α-P291fsinsC (6.1 ± 0.5 versus 3.9 ± 0.7 nmol NADH/min/mg protein), without or with doxycycline induction, respectively (P <0.02, n = 5).
Fig. 3. A representative of three independent western blots performed on mitochondrial proteins (100 µg/lane) isolated from HNF1α-P291fsinsC cells treated or not with 500 ng/ml doxycycline for 48 h in normal culture medium. Samples were collected from 15 cm dishes in triplicate. Similar results were obtained when the membrane was blotted separately with individual antibody (data not shown).
HNF1α-P291fsinsC affects mitochondrial membrane potential, ATP generation and pyruvate oxidation
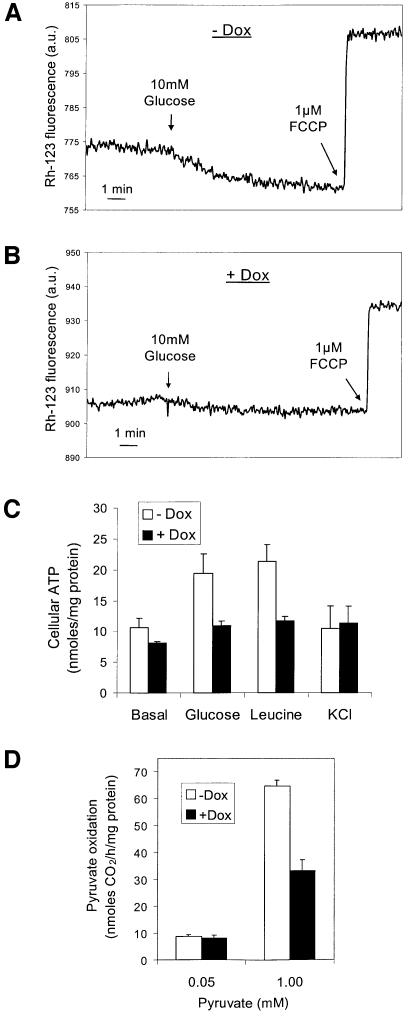
HNF1α-P291fsinsC thus controls multiple genes involved in regulation of glucose metabolism for both glycolysis and mitochondrial oxidation. The effects of HNF1α-P291fsinsC on OGDH E1 and UCP-2 expression should impair mitochondrial function. We therefore examined the consequences of induction of HNF1α-P291fsinsC on mitochondrial membrane potential (ΔΨm), ATP generation and pyruvate oxidation. The ΔΨm was measured in suspensions of HNF1α-P291fsinsC#32 cells by monitoring rhodamine-123 fluorescence. As shown in Figure 4A, the addition of 10 mM glucose (12.5 mM final) hyperpolarized ΔΨm, and 1 µM of the protonophore FCCP depolar ized ΔΨm. After induction of HNF1α-P291fsinsC, glucose-induced mitochondrial hyperpolarization was inhibited by 66% (Figure 4B).
Fig. 4. Effects of HNF1α-P291fsinsC on mitochondrial membrane potential (ΔΨm), ATP generation and [14C]pyruvate oxidation. HNF1α-P291fsinsC#32 cells were treated or not with 500 ng/ml doxycycline at 2.5 mM glucose for 48 h prior to experiments. The ΔΨm was measured using rhodamine-123 in cells treated without (A) or with 500 ng/ml doxycycline (B). Delta of fluorescence (hyperpolarization) 10 min after 10 mM glucose addition: –Dox (A) 9.27 ± 0.96 versus +Dox (B) 3.12 ± 0.13, respectively, n = 3 independent experiments, P <0.005. (C) Cellular ATP production was measured during 8 min incubation with 24 mM glucose, 20 mM leucine or 20 mM KCl in KRBH containing 2.5 mM glucose. Data represent the mean ± SEM of five independent experiments. (D) [14C]Pyruvate oxidation was measured during 1 h incubation in KRBH containing 0.05 or 1 mM pyruvate. Data represent the mean ± SE performed in quadruplicate from one of four similar experiments.
Defective mitochondrial function was substantiated further by the results of ATP measurement. ATP production is a key element in nutrient-stimulated insulin secretion, which for glucose occurs predominantly (98%) and for leucine exclusively in the mitochondria (Matschinsky, 1996). In non-induced cells, both glucose and leucine caused 2-fold increases in cellular ATP after 8 min incubation. K+, which directly stimulates insulin secretion by raising cytosolic Ca2+ (Wang et al., 1998), did not alter cellular ATP levels (Figure 4C). Induction of HNF1α-P291fsinsC completely abolished the ATP generation by glucose and leucine (Figure 4C).
As demonstrated in Figure 4D, induction of HNF1α-P291fsinsC resulted in 47% reduction in [14C]pyruvate oxidation. This also indicates defective mitochondrial function.
HNF1α-P291fsinsC impairs insulin secretion
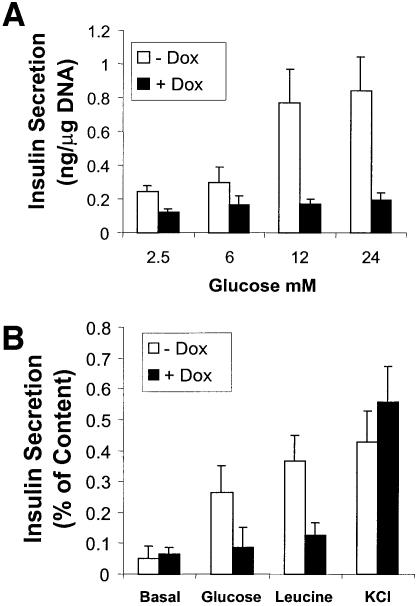
The progressively developing hyperglycemia in MODY3 patients is due primarily to defective glucose-stimulated insulin secretion from pancreatic β-cells (Byrne et al., 1996; Lehto et al., 1997; Hattersley, 1998). The impact of forced expression of MODY3 mutant protein on insulin secretory responses to glucose, leucine and K+ was investigated in the experiments shown in Figure 5. The induction of HNF1α-P291fsinsC for 48 h led to a drastic decrease in the glucose-stimulated insulin secretion. As predicted from the diminished insulin transcription, insulin content in HNF1α-P291fsinsC#32 cells cultured with doxycycline for 48 h was reduced by 55.4 ± 8.9% (n = 5). The reduced insulin content was reflected in diminished basal insulin secretion (Figure 5A). When secretion is related to insulin content, it is apparent that HNF1α-P291fsinsC selectively inhibited the insulin secretory responses to glucose and leucine but not to K+ depolarization (Figure 5B).
Fig. 5. Effects of HNF1α-P291fsinsC on insulin secretion. Cells were cultured at 2.5 mM glucose with or without 500 ng/ml doxycycline for 48 h. For insulin secretion experiments, cells were incubated in KRBH with specified stimulators for 30 min. (A) Glucose-stimulated insulin secretion from HNF1α-P291fsinsC#32 cells was quantified by radioimmunoassay and normalized by cellular DNA content. (B) Insulin secretion from HNF1α-P291fsinsC#32 cells measured during 30 min incubation with 24 mM glucose, 20 mM leucine or 20 mM KCl in KRBH containing 2.5 mM glucose was expressed as a percentage of cellular insulin content.
In negative control experiments, neither the glucose-evoked insulin release nor the β-cell gene expression was altered by similar induction of a MODY1 nonsense mutant, HNF4α-Q268X (Stoffel and Duncan, 1997) (data not shown).
Discussion
MODY is a genetically heterogeneous group of disorders, with at least six subtypes caused by mutations in the genes coding for HNF4α (MODY1) (Yamagata et al., 1996b), glucokinase (MODY2) (Froguel et al., 1993), HNF1α (MODY3) (Yamagata et al., 1996a), PDX1 (MODY4) (Stoffers et al., 1997), HNF1β (MODY5) (Lindner et al., 1998) and NeuroD/β2 (MODY6) (Malecki et al., 1999). MODY3, accounting for 65% of MODY cases in the UK, is the most common form of MODY (Hattersley, 1998). The severe hyperglycemia developing progressively in MODY3 patients is due primarily to severely impaired insulin secretory responses to glucose in pancreatic β-cells (Byrne et al., 1996; Lehto et al., 1997; Hattersley, 1998). To characterize the mechanism underlying the impairment of insulin secretion, we generated a β-cell model by inducible expression of HNF1α-P291fsinsC, the most common mutation identified in MODY3 pedigrees.
In the present study, we demonstrate that HNF1α-P291fsinsC is expressed efficiently and localized almost exclusively in nuclei of INS-1 cells. HNF1α-P291fsinsC inhibits the DNA-binding activity of endogenous HNF1α by forming HNF1α-P291fsinsC homodimers and HNF1α-P291fsinsC–HNF1α-WT heterodimers (data not shown). We have provided extensive evidence that HNF1α-P291fsinsC directly and specifically suppresses the rat insulin I promoter activity. This observation is in full agreement with our previous report on INS-1 cell lines overexpressing the rat hnf1α-WT and an artificial dominant-negative mutant (Wang et al., 1998). We therefore conclude that normal HNF1α function is necessary for insulin gene transcription. This contrasts with results obtained in one of the hnf1α knock-out mouse models. Pontoglio et al. (1998) have reported that the insulin mRNA levels in hnf1α-null mice were unchanged. Studies on another knock-out mouse model (Lee et al., 1998), which demonstrated reduced pancreatic insulin protein levels, support our observation. The results of Lee et al. (1998) and our findings explain the phenotype of MODY3, which exhibits diminished plasma insulin concentration not only under glucose-stimulated but also under basal conditions (Byrne et al., 1996; Lehto et al., 1997). The latter should reflect the reduced pancreatic insulin content and/or decreased β-cell mass. Indeed, this reduction may underlie the requirement for insulin replacement therapy in at least 30% of MODY3 patients (Hattersley, 1998; Iwasaki et al., 1998).
Metabolism of glucose including glycolysis and mitochondrial substrate oxidation in pancreatic β-cells generates critical signals for insulin secretion (Matschinsky, 1996). Mitochondrial ATP generation results in closure of ATP-sensitive K+ channels, depolarization of the plasma membrane, opening of voltage-dependent Ca2+ channels, an increase in [Ca2+]i and, consequently, insulin exocytosis (Matschinsky, 1996; Wollheim, 2000). We found that forced expression of HNF1α-P291fsinsC selectively inhibits the insulin secretory responses to glucose and leucine, but not to K+ depolarization (Figure 5). This is linked to the impaired ATP generation by glucose and leucine (Figure 4C). The studies on a transgenic mouse model (Dukes et al., 1998) also demonstrate reduced glucose-stimulated insulin secretion associated with decreased glucose-promoted ATP production in the hnf1α-deficient islets. However, Dukes et al. (1998) suggested that the defect in ATP production is secondary to reduced formation of NADH from glycolytic substrates. Indeed, we previously have observed a significant decrease in glycolytic rate in INS-1 cells overexpressing a dominant-negative mutant of the rat hnf1α (Wang et al., 1998). The inhibition of GLUT2 expression may not be the major factor underlying the defective glycolysis, since induction of a PDX1 dominant-negative mutant resulted in a similar inhibitory effect on the GLUT2 mRNA level but did not cause impaired metabolism–secretion coupling (H.Wang and C.B.Wollheim, unpublished results). Therefore, the suppression of aldolase B and l-PK could explain reduced glycolysis after induction of HNF1α-P291fsinsC (Figure 2). However, reduced glycolysis cannot be the mechanism underlying the diminished leucine-stimulated insulin secretion, since leucine is metabolized directly in the mitochondria (Patterson et al., 2000). We hypothesized that in addition to diminished glycolysis, loss of HNF1α function also leads to impaired mitochondrial oxidation and consequently defective ATP production. In the present study, the molecular basis of the impaired mitochondrial function was determined. We demonstrate that the leucine-evoked ATP production, which is due exclusively to mitochondrial oxidation, was completely inhibited by induction of HNF1α-P291fsinsC (Figure 4C). Furthermore, induction of HNF1α-P291fsinsC resulted in severe impairment of mitochondrial oxidation, as indicated by the pronounced reduction in 14CO2 formation from the mitochondrial substrate [14C]pyruvate (Figure 4D). Decreased oxidative metabolism was also reflected by diminished glucose-evoked mitochondrial membrane hyperpolarization (Figure 4A and B).
All of these results can be explained by the marked down-regulation of the catalytic E1 subunit of OGDH. The promoter of the OGDH E1 subunit indeed contains a putative HNF1α-binding site (Koike, 1998). OGDH E1, which is the catalytic subunit of OGDH and constitutes the rate-limiting enzyme in the mitochondrial TCA cycle, was reduced at both transcript and protein levels. The diminished OGDH E1 activity may be partially responsible for the HNF1α-P291fsinsC-induced defective mitochondrial function including oxidation, ATP generation, membrane potential and, consequently, impaired insulin secretory responses to nutrients (Figures 4 and 5). The observed up-regulation of UCP-2, which favors the uncoupling of respiration from mitochondrial oxidative phosphorylation and increases the rate of proton leak (Boss et al., 2000; Ricquier and Bouillaud, 2000), would reduce the efficiency of mitochondrial ATP generation and partially dissipate the mitochondrial membrane potential. Overexpression of UCP-2 has been demonstrated to inhibit glucose-stimulated insulin secretion from rat islets (Chan et al., 1999).
In conclusion, we have established a cell model to investigate the impact of human HNF1α mutations on β-cell function. The most common MODY3 mutation, HNF1α-P291fsinsC, has dominant-negative effects on endogenous HNF1α function. HNF1α-P291fsinsC regulates multiple genes implicated in pancreatic β-cell function. Induction of HNF1α-P291fsinsC inhibits insulin gene transcription and consequently reduces the cellular insulin content. HNF1α-P291fsinsC also suppresses the expression of genes involved in glucose transport and glycolysis. The similarly impaired glucose- and leucine-induced insulin secretion strongly suggests a common mitochondrial defect substantiated by diminished ATP generation and substrate oxidation. Down-regulation of OGDH E1 subunit and up-regulation of UCP-2 constitute the molecular targets underlying the impaired metabolism–secretion coupling. It remains to be established whether the same targets are implicated in the defective insulin secretion in animal models of MODY3.
Materials and methods
Generation of stable cell lines
The rat insulinoma INS-1 cell line-derived stable clones were cultured in RPMI 1640 in 11 mM glucose, unless indicated otherwise (Asfari et al., 1992). The establishment of the first-step stable clone INS-r3, which expresses the reverse tetracycline-dependent transactivator, was reported previously (Wang and Iynedjian, 1997). Plasmids used in the secondary stable transfection were constructed by subcloning the cDNAs encoding the human HNF1α-P291fsinsC (kindly supplied by Dr M.A.Glucksmann, Millenium Pharmaceuticals, Boston, MA) into the expression vector PUHD10-3 (a kind gift from Dr H.Bujard, Universität Heidelberg, Germany). Transfection, clone selection and screening procedures were described previously (Wang and Iynedjian, 1997).
Immunoblot and immunofluorescence
Immunoblotting procedures were performed as described previously using enhanced chemiluminescence (Pierce, Illinois) for detection (Wang et al., 1998). Dilutions for antibodies against HNF1α (a kind gift from Dr R.Cortese), OGDH E1 (kindly supplied by Dr J.G.Lindsay), UCP-2 (Research Diagnostics Inc., Flanders, NJ) and glutamate dehydrogenase (Rockland, Gilbertsville, PA) were 1:4000, 1:2000, 1:500 and 1:5000, respectively.
For immunofluorescence, cells were grown on polyornithine-treated glass coverslips for 3 days prior to 24 h treatment with 500 ng/ml doxycycline to induce HNF1α-P291fsinsC expression. The cells were then washed, fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline containing 1% bovine serum albumin (PBS-BSA). The preparation was then blocked with PBS-BSA before staining with rabbit antibody specific for HNF1α N-terminal sequences followed by rhodamine-labeled goat anti-rabbit IgG as the second antibody. The resultant immunofluorescence was viewed using a Zeiss laserscan confocal 410 microscope (Zurich, Switzerland).
Total RNA isolation and northern blotting
Cells in 10 cm dishes were cultured in 2.5 mM glucose medium with or without 500 ng/ml doxycycline for 14 or 48 h, followed by an additional 8 h in culture medium with 2.5, 6, 12 and 24 mM glucose. Total RNA was extracted and blotted to nylon membranes as described previously (Wang et al., 1998). The membrane was pre-hybridized and then hybridized to 32P-labeled random primer cDNA probes by the technique of Sambrook et al. (1989). To ensure equal RNA loading and even transfer, all membranes were stripped and re-hybridized with ‘house-keeping gene’ probes such as β-actin or cyclophilin. cDNA fragments used as probes for l-PK, GLUT-2, glucokinase, insulin, PDX1, HNF1α-P291fsinsC, USF, IB-1 and C/EBPβ mRNA detection were digested from corresponding expression vectors kindly provided by Drs A.Kahn, B.Thorens, P.B.Iynedjian, J.Philippe, T.Edlund, M.A.Glucksmann, M.Sawadogo, G.Waeber and U.Schibler. cDNA probes for rat aldolase B, GAPDH, HMGCoA reductase, DcoH, ANT1 and ANT2, UCP-2, OGDC E1 subunit and cyclophilin were prepared by RT–PCR and confirmed by sequencing.
Site-directed mutagenesis, transient transfection and luciferase assay
The following oligonucleotides corresponding to the rat FLAT I element with a mutated HNF1α-binding site were used for generation of –410MFINSluc by PCR-mediated site-directed mutagenesis according to the manufacturer’s protocol (Stratagene, La Jolla, CA): 5′-CCATCTGGCCCCTTGGTCCGAATCTAATTACCCTAG-3′. The mutated nucleotides are underlined. Plasmids –410INSLuc, Far-FLATwt, Far-FLATMF and Far-FLATME were kindly supplied by Dr M.S.German (University of California, San Francisco, CA) (German et al., 1992a,b).
Transient transfection experiments and luciferase reporter enzyme assays were carried out as previously reported (Wang et al., 1998).
Insulin secretion and cellular insulin content
Cells in 24-well dishes were cultured in 2.5 mM glucose medium with or without 500 ng/ml doxycycline for 48 h. Insulin secretion was measured over a period of 30 min, in Krebs–Ringer–bicarbonate–HEPES buffer (KRBH; 140 mM NaCl, 3.6 mM KCl, 0.5 mM NaH2PO4, 0.5 mM MgSO4, 1.5 mM CaCl2, 2 mM NaHCO3, 10 mM HEPES, 0.1% BSA) containing the indicated stimulators. Insulin content was determined after extraction with acid ethanol following the procedures of Asfari et al. (1992). Insulin was detected by radioimmunoassay using rat insulin as standard (Wang et al., 1998).
Intracellular ATP
Cells in 6-well dishes were cultured in 2.5 mM glucose medium with or without 500 ng/ml doxycycline for 48 h. The production of ATP was measured during 8 min stimulation in KRBH. ATP was assayed as previously reported (Wang et al., 1998).
Mitochondrial membrane potential (ΔΨm)
The ΔΨm was measured in a cell suspension previously loaded with 10 µg/ml rhodamine-123. The fluorescence was excited at 490 nm and measured at 530 nm in a fluorimeter at 37°C (Maechler et al., 1997).
[14C]pyruvate oxidation
The production of 14CO2 from [14C]pyruvate was measured over 1 h in KRBH containing either 0.05 or 1.0 mM pyruvate as previously described (Noel, 1997; Antinozzi et al., 1998).
OGDH activity assay
Mitochondria were prepared as described in Rustin et al. (1994). Mitochondria suspensions containing 25 µg of total protein were resuspended in 2 ml of assay buffer (50 mM HEPES, 1 mM MgCl2, 1 mM CaCl2, 1 mM EGTA, 0.3% Triton X-100). The reaction was initiated with the sequential addition to final concentrations of 60 µM CoA, 1.25 mM NAD+ and 1 mM 2-oxoglutarate. The generation of NADH was measured by fluorescence spectroscopy (340 nm excitation, 460 nm emission).
Acknowledgments
Acknowledgements
We are grateful to D.Harry, C.Bartley, G.Chaffard and E.-J.Sarret for expert technical assistance. We are indebted to Drs J.G.Lindsay (OGDH E1 antibody), M.S.German (–410INSLuc and three insulin minienhancer constructs), R.Cortese (HNF1α antibodies), M.A.Glucksmann (HNF1α-P291fsinsC and HNF4α-Q268X cDNAs), P.B.Iynedjian (INS-r3 cell line and glucokinase cDNA), U.Schibler (C/EBPβ cDNA), T.Edlund (PDX1 cDNA), G.Waeber (IB-1 cDNA), M.Sawadogo (USF cDNA), A.Kahn (l-PK cDNA), B.Thorens (GLUT2 cDNA), J.Philippe (insulin I cDNA), H.Bujard (PUHD 10-3 plasmid) and N.Quintrell (pTKhygro plasmid). We thank Dr B.Ritz-Laser for helpful advice on EMSA. This work was supported by the Swiss National Science Foundation (grant no. 32-49755.96) and by a European Union Network grant (through the Swiss Federal Office for Education and Science).
References
- Antinozzi P.A., Segall,L., Prentki,M., McGarry,J.D. and Newgard,C.B. (1998) Molecular or pharmacologic perturbation of the link between glucose and lipid metabolism is without effect on glucose-stimulated insulin secretion: a re-evaluation of the long-chain acyl CoA hypothesis. J. Biol. Chem., 273, 16146–16154. [DOI] [PubMed] [Google Scholar]
- Asfari M., Janjic,D., Meda,P., Li,G., Halban,P.A. and Wollheim,C.B. (1992) Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology, 130, 167–178. [DOI] [PubMed] [Google Scholar]
- Blumenfeld M., Maury,M., Chouard,T., Yaniv,M. and Condamine,H. (1991) Hepatic nuclear factor 1 (HNF1) shows a wider distribution than products of its known target genes in the developing mouse. Development, 113, 589–599. [DOI] [PubMed] [Google Scholar]
- Bonny C., Pascal,N. and Waeber,G. (1998) IB1, a JIP-1-related nuclear protein present in insulin-secreting cells. J. Biol. Chem., 273, 1843–1846. [DOI] [PubMed] [Google Scholar]
- Boss O., Hagen,T. and Lowell,B.B. (2000) Uncoupling proteins 2 and 3: potential regulators of mitochondrial energy metabolism. Diabetes, 49, 143–156. [DOI] [PubMed] [Google Scholar]
- Byrne M.M. et al. (1996) Altered insulin secretory responses to glucose in diabetic and nondiabetic subjects with mutations in the diabetes susceptibility gene MODY3 on chromosome 12. Diabetes, 45, 1503–1510. [DOI] [PubMed] [Google Scholar]
- Chan C.B., MacDonald,P.E., Saleh,M.C., Johns,D.C., Marbàn,E. and Wheeler,M.B. (1999) Overexpression of uncoupling protein 2 inhibits glucose-stimulated insulin secretion from rat islets. Diabetes, 48, 1482–1486. [DOI] [PubMed] [Google Scholar]
- Dukes I.D. et al. (1998) Defective pancreatic β-cell glycolytic signaling in hepatocyte nuclear factor-1α-deficient mice. J. Biol. Chem., 273, 24457–24464. [DOI] [PubMed] [Google Scholar]
- Emens L.A., Landers,D.W. and Moss,L.G. (1992) Hepatocyte nuclear factor 1α is expressed in a hamster insulinoma line and transactivates the rat insulin I gene. Proc. Natl Acad. Sci. USA, 89, 7300–7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajans S.S. (1990) Scope and heterogeneous nature of MODY. Diabetes Care, 13, 49–64. [DOI] [PubMed] [Google Scholar]
- Froguel P. et al. (1993) Familial hyperglycemia due to mutations in glucokinase: definition of a subtype of diabetes mellitus. N. Engl. J. Med., 328, 697–702. [DOI] [PubMed] [Google Scholar]
- German M.S., Moss,L.G., Wang,J. and Rutter,W.J. (1992a) The insulin and islet amyloid polypeptide genes contain similar cell-specific promoter elements that bind identical β-cell nuclear complexes. Mol. Cell. Biol., 12, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German M.S., Wang,J., Chadwick,R.B. and Rutter,W.J. (1992b) Synergistic activation of the insulin gene by a LIM-homeo domain protein and a basic helix–loop–helix protein: building a functional insulin minienhancer complex. Genes Dev., 6, 2165–2176. [DOI] [PubMed] [Google Scholar]
- Glucksmann M.A. et al. (1997) Novel mutations and a mutational hotspot in the MODY3 gene. Diabetes, 46, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Goldstein J.L. and Brown,M.S. (1990) Regulation of the mevalonate pathway. Nature, 343, 425–430. [DOI] [PubMed] [Google Scholar]
- Gragnoli C., Lindner,T., Cockburn,B.N., Kaisaki,P., Gragnoli,F., Marozzi,G. and Bell,G.I. (1997) Maturity-onset diabetes of the young due to a mutation in the hepatocyte nuclear factor-4α binding site in the promoter of the hepatocyte nuclear factor-1α gene. Diabetes, 46, 1648–1651. [DOI] [PubMed] [Google Scholar]
- Hattersley A.T. (1998) Maturity-onset diabetes of the young: clinical heterogeneity explained by genetic heterogeneity. Diabetic Med., 15, 15–24. [DOI] [PubMed] [Google Scholar]
- Iwasaki N., Ogata,M., Tomonaga,O., Kuroki,H., Kasahara,T., Yano,N. and Iwamoto,Y. (1998) Liver and kidney function in Japanese patients with maturity-onset diabetes of the young. Diabetes Care, 21, 2144–2148. [DOI] [PubMed] [Google Scholar]
- Kaisaki P.J. et al. (1997) Mutations in the hepatocyte nuclear factor-1α gene in MODY and early-onset NIDDM. Evidence for a mutational hotspot in exon 4. Diabetes, 46, 528–535. [DOI] [PubMed] [Google Scholar]
- Koike K. (1998) Cloning, structure, chromosomal localization and promoter analysis of human 2-oxoglutarate dehydrogenase gene. Biochim. Biophys. Acta, 1385, 373–384. [DOI] [PubMed] [Google Scholar]
- Lee Y.-H., Sauer,B. and Gonzalez,F.J. (1998) Laron dwarfism and non-insulin-dependent diabetes mellitus in the hnf-1α knock out mouse. Mol. Cell. Biol., 18, 3059–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehto M. et al. (1997) Characterization of the MODY3 phenotype. Early-onset diabetes caused by an insulin secretion defect. J. Clin. Invest., 99, 582–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner T., Yamagata,K., Ogata,M.M., Tomonaga,O., Kuroki,H., Kasahara,T., Iwamoto,Y. and Bell,G.I. (1998) Mutation in hepatocyte nuclear factor-1β gene (TCF2) associated with MODY. Nature Genet., 17, 384–385. [DOI] [PubMed] [Google Scholar]
- Maechler P., Kennedy,E.D., Pozzan,T. and Wollheim,C.B. (1997) Mitochondrial activation directly triggers the exocytosis of insulin in permeabilized pancreatic β-cells. EMBO J. 16, 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M.T. et al. (1999) Mutations in neuroD1 are associated with the development of type 2 diabetes mellitus. Nature Genet., 23, 323–328. [DOI] [PubMed] [Google Scholar]
- Marie S., Diaz-Guerra,M.-J., Miquerol,L., Kahn,A. and Iynedjian,P.B. (1993) The pyruvate kinase gene as a model for studies of glucose-dependent regulation of gene expression in the endocrine pancreatic β-cell type. J. Biol. Chem., 268, 23881–23890. [PubMed] [Google Scholar]
- Matschinsky F.M. (1996) Banting Lecture 1995. A lesson in metabolic regulation inspired by the glucokinase glucose sensor paradigm. Diabetes, 45, 223–241. [DOI] [PubMed] [Google Scholar]
- Mendel D.B. and Crabtree,G.R. (1991) A member of a novel class of dimerizing homeodomain proteins. J. Biol. Chem., 266, 677–680. [PubMed] [Google Scholar]
- Miquerol L., Lopez,S., Cartier,N., Tulliez,M., Raymondjean,M. and Kahn,A. (1994) Expression of the l-type pyruvate kinase gene and the hepatocyte nuclear factor 4 transcription factor in exocrine and endocrine pancreas. J. Biol. Chem., 269, 8944–8955. [PubMed] [Google Scholar]
- Møller A.M., Dalgaard,L.T., Pociot,F., Nerup,J., Hansen,T. and Pedersen,O. (1998) Mutations in the hepatocyte nuclear factor-1α gene in Caucasian families originally classified as having type 1 diabetes. Diabetologia, 41, 1528–1531. [DOI] [PubMed] [Google Scholar]
- Noel R.J., Antinozzi,P.A., McGarry,J.D. and Newgard,C.B. (1997) Engineering of glycerol-stimulated insulin secretion in islet β cells. J. Biol. Chem., 272, 18621–18627. [DOI] [PubMed] [Google Scholar]
- Patterson G.H., Knobel,S.M., Arkhammar,P., Thastrup,O. and Piston,D.W. (2000) Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet β cells. Proc. Natl Acad. Sci. USA, 97, 5203–5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontoglio M. et al. (1999) Defective insulin secretion in hepatocyte nuclear factor 1α-deficient mice. J. Clin. Invest., 101, 2215–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane G.R., Dubus,P., Mettus,R.V., Galbreath,E.J., Boden,G., Reddy,E.P. and Barbacid,M. (1999) Loss of Cdk4 expression causes insulin-deficient diabetes and Cdk4 activation results in β-islet cell hyperplasia. Nature Genet., 22, 44–52. [DOI] [PubMed] [Google Scholar]
- Ricquier D. and Bouillaud,F. (2000) The uncoupling protein homologues: UCP1, UCP2, UCP3, StUCP and AtUCP. Biochem. J., 345, 161–179. [PMC free article] [PubMed] [Google Scholar]
- Roche E., Assimacopoulos-Jeannet,F., Witters,L.A., Perruchoud,B., Yaney,G., Corkey,B., Asfari,M. and Prentki,M. (1997) Induction by glucose of genes coding for glycolytic enzymes in a pancreatic β-cell line (INS-1). J. Biol. Chem., 272, 3091–3098. [DOI] [PubMed] [Google Scholar]
- Rustin P., Chretien,D., Bourgeron,B., Gerard,B., Rotig,A., Saudubray,J.M. and Munnich,A. (1994) Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta, 228, 35–51. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Seufert J., Weir,G.C. and Habener,J.F. (1998) Differential expression of the insulin gene transcriptional repressor CCAAT/enhancer-binding protein β and transactivator islet Duodenum Homeobox-1 in rat pancreatic β cells during the development of diabetes mellitus. J. Clin. Invest., 101, 2528–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel M. and Duncan,S.A. (1997) The maturity-onset diabetes of the young (MODY1) transcription factor HNF4α regulates expression of genes required for glucose transport and metabolism. Proc. Natl Acad. Sci. USA, 94, 13209–13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffers D.A., Ferrer,J., Clarke,W.L. and Habener,J.F. (1997) Early-onset type-II diabetes mellitus (MODY 4) linked to IPF-1. Nature Genet., 17, 138–139. [DOI] [PubMed] [Google Scholar]
- Tronche F., Ringeisen,F., Blumenfeld,M., Yaniv,M. and Pontoglio,M. (1997) Analysis of the distribution of binding sites for a tissue-specific transcription factor in the vertebrate genome. J. Mol. Biol., 266, 231–245. [DOI] [PubMed] [Google Scholar]
- Vaxillaire M. et al. (1997) Identification of nine novel mutations in the hepatocyte nuclear factor 1α gene associated with maturity-onset diabetes of the young (MODY3). Hum. Mol. Genet., 6, 583–586. [DOI] [PubMed] [Google Scholar]
- Waeber G., Thompson,N., Haefliger,J.-A. and Nicod,P. (1994) Characterization of the murine high Km glucose transporter GLUT2 gene and its transcriptional regulation by glucose in a differentiated insulin-secreting cell line. J. Biol. Chem., 269, 26912–26919. [PubMed] [Google Scholar]
- Waeber G., Thompson,N., Nicord,P. and Bonny,C. (1996) Transcriptional activation of the GLUT2 gene by the IPF-1/STF-1/IDX-1 homeobox factor. Mol. Endocrinol., 10, 1327–1334. [DOI] [PubMed] [Google Scholar]
- Wang H. and Iynedjian,P.B. (1997) Modulation of glucose responsiveness of insulinoma β-cells by graded overexpression of glucokinase. Proc. Natl Acad. Sci. USA, 99, 4372–4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Maechler,P., Hagenfeldt,K.A. and Wollheim,C.B. (1998) Dominant-negative suppression of HNF1α function results in defective insulin gene transcription and impaired metabolism–secretion coupling in a pancreatic β-cell line. EMBO J., 17, 6701–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollheim C.B. (2000) β-Cell mitochondria in the regulation of insulin: a new culprit in type II diabetes. Diabetologia, 43, 265–278. [DOI] [PubMed] [Google Scholar]
- Yamagata K. et al. (1996a) Mutations in the hepatocyte nuclear factor-1α gene in maturity-onset diabetes of the young (MODY3). Nature, 384, 455–458. [DOI] [PubMed] [Google Scholar]
- Yamagata K. et al. (1996b) Mutations in the hepatocyte nuclear factor-4α gene in maturity-onset diabetes of the young (MODY1). Nature, 384, 458–460. [DOI] [PubMed] [Google Scholar]
- Yamagata K. et al. (1998) Mutation P291fsinsC in the transcription factor hepatocyte nuclear factor-1α is dominant negative. Diabetes, 47, 1231–1235. [DOI] [PubMed] [Google Scholar]
- Yamagata S. et al. (1997) Identification of mutations in the hepatocyte nuclear factor (HNF)-1α gene in Japanese subjects with IDDM. Diabetes, 46, 1643–1647. [DOI] [PubMed] [Google Scholar]