Abstract
Hereditary hemochromatosis (HH) is a common disorder of iron metabolism caused by mutation in HFE, a gene encoding an MHC class I-like protein. Clinical studies demonstrate that the severity of iron loading is highly variable among individuals with identical HFE genotypes. To determine whether genetic factors other than Hfe genotype influence the severity of iron loading in the murine model of HH, we bred the disrupted murine Hfe allele onto three different genetically defined mouse strains (AKR, C57BL/6, and C3H), which differ in basal iron status and sensitivity to dietary iron loading. Serum transferrin saturations (percent saturation of serum transferrin with iron), hepatic and splenic iron concentrations, and hepatocellular iron distribution patterns were compared for wild-type (Hfe +/+), heterozygote (Hfe +/−), and knockout (Hfe −/−) mice from each strain. Although the Hfe −/− mice from all three strains demonstrated increased transferrin saturations and liver iron concentrations compared with Hfe +/+ mice, strain differences in severity of iron accumulation were striking. Targeted disruption of the Hfe gene led to hepatic iron levels in Hfe −/− AKR mice that were 2.5 or 3.6 times higher than those of Hfe −/− C3H or Hfe −/− C57BL/6 mice, respectively. The Hfe −/− mice also demonstrated strain-dependent differences in transferrin saturation, with the highest values in AKR mice and the lowest values in C3H mice. These observations demonstrate that heritable factors markedly influence iron homeostasis in response to Hfe disruption. Analysis of mice from crosses between C57BL/6 and AKR mice should allow the mapping and subsequent identification of genes modifying the severity of iron loading in this murine model of HH.
Hereditary hemochromatosis (HH) is a common autosomal recessive disorder of iron metabolism characterized by excessive dietary iron absorption and progressive iron deposition in the parenchymal cells of many tissues (1, 2). Untreated HH leads to the failure of multiple organs and contributes to early death. The gene carrying the mutation responsible for HH encodes a nonclassical MHC class-I protein designated HFE. The most common HFE gene mutation leads to the substitution of tyrosine for cysteine at amino acid 282 (C282Y) (3). Homozygosity for the C282Y mutation accounts for about 85% or more of HH cases in populations of northern European extraction (1, 2, 4). The severity of iron loading, however, is variable in human patients with identical HFE genotypes, and some individuals homozygous for the C282Y mutation do not demonstrate iron overload (5–7). Some of the variation in phenotypic expression of HH may be attributed to nongenetic factors, such as diet, alcohol intake, and iron loss (menstruation or pregnancy). However, population studies suggest that the variation in severity of iron loading in HH may be caused by the additive or interactive effect of other genes (8, 9).
The generation of mice with targeted disruption of the orthologous murine gene Hfe (10–12) provides a means of examining the HH phenotype under controlled environmental conditions. The marked variability in hepatic iron loading observed in Hfe −/− progeny from crosses of mixed-strain heterozygous mice suggested the segregation of other genes modifying the phenotype (10). Crosses between Hfe −/− mice and mice carrying other mutations that impair normal iron homeostasis provided specific examples for genetic modification of the HH phenotype (13).
Inbred mouse strains exhibit considerable variability in several parameters of iron metabolism (14–16). Serum iron levels, serum transferrin saturations, and hepatic iron stores vary as much as 2-fold among inbred strains on a basal diet. Inbred strains also differ in severity of iron loading on an iron-supplemented diet. We hypothesized that the heritable factors determining differences in iron status among mouse strains would contribute to the phenotypic variability seen with Hfe disruption. To test this hypothesis, we examined the effect of Hfe disruption on the iron status of three inbred mouse strains that differ in basal iron status and sensitivity to iron loading.
Methods
Generation of Inbred Strains of Hfe −/− Mice.
The disrupted Hfe allele (10) on a mixed-strain mouse background (C57BL/6 × 129/SVJ) was bred by successive crosses for 6–10 generations onto C57BL/6, C3H, or AKR backgrounds. The transmission of the disrupted Hfe allele was determined by PCR analysis of tail DNA as described (17). Mice were provided ad libitum a standard chow (Purina 5001) that contains 0.02% iron. At 10 weeks of age, mice were fasted overnight and anesthetized before blood was collected by cardiac puncture and tissue samples were obtained. The studied AKR population consisted of four male and five female Hfe +/+ mice, two male and five female Hfe +/− mice, and three male and three female Hfe −/− mice. The studied C57BL/6 population consisted of four female and two male Hfe +/+ mice, four female and two male Hfe +/− mice, and three female and three male Hfe −/− mice. The studied C3H population consisted of three female and three male Hfe +/+ mice, three female and three male Hfe +/− mice, and two female and three male Hfe −/− mice.
Measurement of Serum Transferrin Saturation.
Blood was obtained by cardiac puncture. Serum iron and total iron binding capacity were measured as described by Fielding (18). Transferrin saturation was calculated as follows: (serum iron/total iron binding capacity) × 100%.
Measurement of Tissue Iron Content.
Nonheme iron concentration in liver and spleen tissue was measured by the bathophenanthroline method as described by Torrance and Bothwell (19) and the values were expressed as μg of iron per g of dry tissue.
Histology of Liver Iron Deposition.
Liver tissue samples were fixed in neutral-buffered 10% formalin for 18 h and subjected to routine histological processing. The sections were stained with Perls' Prussian blue and counterstained with hematoxylin. Iron distribution was determined by light microscopy.
Statistical Analysis.
Mean values for transferrin saturation, liver nonheme iron concentration, and splenic nonheme iron concentration were compared separately across mice with the same Hfe genotype but from different strains and across mice from the same strain but with different Hfe genotypes, by one-way ANOVA and Newman–Keuls posttest analysis. The relative contributions of genotype, strain, and the interaction between genotype and strain on mean transferrin saturation, liver nonheme iron concentration, and splenic nonheme iron concentration were determined by two-way ANOVA. P < 0.05 was considered statistically significant.
Results
Strain Differences Influence Serum Transferrin Saturation in Response to Hfe Knockout.
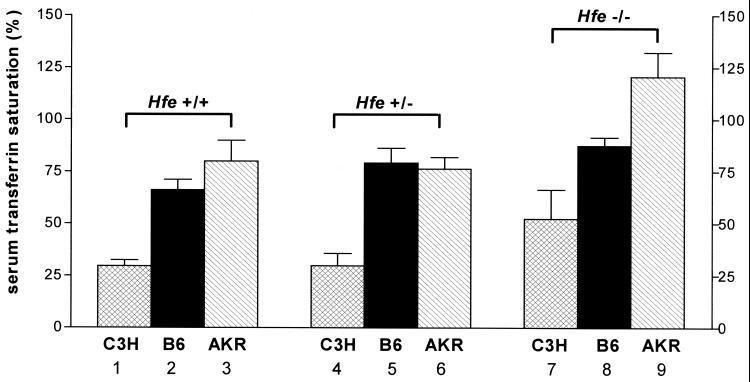
Serum transferrin saturations were measured in Hfe +/+, Hfe +/−, and Hfe −/− mice from each of the three strains, C3H, C57BL/6, and AKR (Fig. 1). On basal diets, the mean transferrin saturations in the Hfe +/+ mice differed across strains. The C3H Hfe +/+ strain had the lowest transferrin saturations (mean = 27%) and AKR the highest (mean = 77%). As seen in previous studies on mice from a mixed-strain genetic background (10), the Hfe −/− mice within each strain had higher transferrin saturations than the Hfe +/+ mice. Mean transferrin saturations of the Hfe +/− mice did not differ significantly from the Hfe +/+ mice within each strain. Hfe disruption led to the highest degree of transferrin saturation in the AKR strain (in excess of 100%). Two-way ANOVA showed that both strain and Hfe genotype contribute to the observed variance in serum transferrin saturation across the strain-genotype combinations. These studies demonstrate that strain differences strongly influence the degree of serum transferrin saturation in the presence of Hfe disruption.
Figure 1.
Effect of strain differences and Hfe genotype on serum transferrin saturation. Serum transferrin saturation was measured in wild-type (Hfe +/+), heterozygote knockout (Hfe +/−), and knockout (Hfe −/−) mice from three inbred mouse strains: C3H (hatched bars), C57BL/6 (B6, solid bars), and AKR (slashed bars). Data are presented as the mean ± SEM. Differences across strains within each genotype and across genotypes within each strain were determined separately by using a one-way ANOVA. P < 0.05 across strains within genotype: bars 1 vs. 2, bars 1 vs. 3, bars 4 vs. 5, bars 5 vs. 6, bars 7 vs. 8, bars 7 vs. 9, and bars 8 vs. 9. P < 0.05 across genotypes within strain: bars 1 vs. 7, bars 2 vs. 8, bars 3 vs. 9, and bars 6 vs. 9.
Strain Differences Influence Severity of Hepatic Iron Loading in Response to Hfe Knockout.
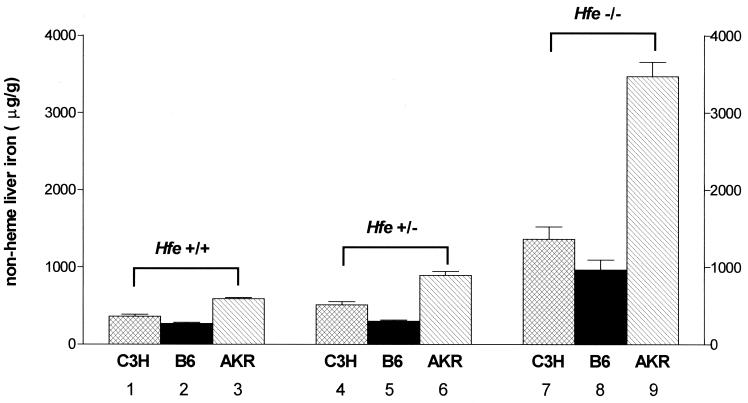
Hepatic nonheme iron concentrations were measured in Hfe +/+, Hfe +/−, and Hfe −/− mice in the three strains (Fig. 2). As observed with serum transferrin saturation, liver iron concentration differed across strains in the Hfe +/+ mice. Within each strain, the Hfe −/− mice had higher liver iron concentrations than the Hfe +/+ mice. The relative order of liver iron concentration across strains was similar (AKR > C3H > C57BL/6) in the Hfe +/+ and Hfe −/− mice. The AKR Hfe −/− mice not only had the highest absolute level of liver iron but also had the greatest relative increase in liver iron (6-fold) with Hfe disruption (C57BL/6, 3.6-fold; C3H, 3.8-fold). These observations indicate that the AKR strain has a much greater propensity for liver iron loading in response to Hfe disruption than the other two strains. In fact, the Hfe +/− AKR mice demonstrated significantly higher liver iron concentrations than the Hfe +/+ AKR mice, whereas the liver iron concentrations of the Hfe +/− C3H and C57BL/6 mice did not differ significantly from the Hfe +/+ mice of the same strain. Two-way ANOVA demonstrated that strain, Hfe genotype, and the interaction between strain and Hfe genotype each contribute to the variance in hepatic iron concentration across the strain-genotype combinations. These studies thus show that strain differences strongly influence hepatic iron accumulation resulting from Hfe disruption.
Figure 2.
Effect of strain differences and Hfe genotype on hepatic iron concentration. Hepatic nonheme iron concentrations were measured in wild-type (Hfe +/+), heterozygote knockout (Hfe +/−), and knockout (Hfe −/−) mice from three inbred mouse strains: C3H (hatched bars), C57BL/6 (B6, solid bars), and AKR (slashed bars). Data are presented as the mean ± SEM. Differences across strains within each genotype and across genotypes within each strain were determined separately by a one-way ANOVA. P < 0.05 across strains within genotype: bars 1 vs. 2, bars 1 vs. 3, bars 2 vs. 3, bars 4 vs. 5, bars 4 vs. 6, bars 5 vs. 6, bars 7 vs. 8, bars 7 vs. 9, and bars 8 vs. 9. P < 0.05 across genotypes within strain: bars 1 vs. 7, bars 2 vs. 8, bars 3 vs. 6, bars 3 vs. 9, bars 4 vs. 7, bars 5 vs. 8, bars 6 vs. 9.
The relationship between transferrin saturation and liver iron concentration was analyzed across strains and Hfe genotype. Although the Hfe +/+ C3H strain had the lowest mean transferrin saturation, this strain had an intermediate level of hepatic iron deposition. Likewise, the Hfe −/− C3H mice, despite a mean transferrin saturation of only 53%, had a hepatic iron concentration similar to the Hfe −/− C57BL/6 mice, with a mean transferrin saturation of 86%. Furthermore, there was no correlation between serum transferrin saturation and liver iron concentration among the animals within each strain. These observations suggest that different genetic factors determine serum transferrin concentration and hepatic iron concentration. LeBoeuf et al. (14) reached a similar conclusion from studies of strain differences in sensitivity to dietary iron loading.
Liver Histopathology of Hfe Knockout Mice from Each Strain.
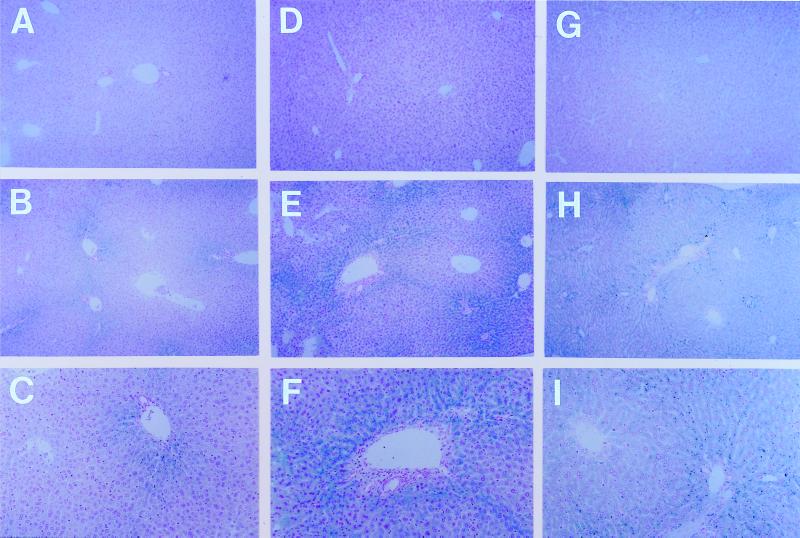
Perls' Prussian blue staining and histological examination were performed on liver specimens from the Hfe +/+, Hfe +/−, and Hfe −/− mice from the three strains tested. By this method, no iron was detected in liver sections from Hfe +/+ mice regardless of strain (Fig. 3 A, D, and G). However, abundant iron was detected in liver sections from the Hfe −/− mice of all three strains (Fig. 3 B, C, E, F, H, and I). The hepatocellular reactivity in the Hfe −/− mice from the C3H (Fig. 3 B and C) and C57BL/6 (Fig. 3 E and F) strains was predominantly periportal, with a gradient decreasing from zone 1 to zone 3. Periportal iron staining was seen in the original Hfe −/− mice on a mixed genetic background (10) and is typical of that observed for patients with early HH. The Hfe −/−AKR mice, which had the highest liver iron concentrations (Fig. 2), had a more diffuse distribution of hepatocellular iron staining (Fig. 3 H and I) as observed in more advanced HH patients. Storage iron was also detected in some sinusoidal lining cells in the Hfe −/− mice with a similar distribution from zone 1 to zone 3. Stainable hepatic iron was not detected in Hfe +/− mice from the C3H and C57BL/6 strains (data not shown). However, occasional areas of periportal granular hepatocellular staining were observed in some Hfe +/− AKR mice (data not shown).
Figure 3.
Hepatocellular iron distribution in Hfe +/+ and Hfe −/− C3H (A–C), C57BL/6 (D–F), and AKR (G–I) mice. Representative Perls' Prussian blue-stained liver sections from Hfe +/+ (A, D, and G) and Hfe −/− mice of the C3H (B and C), C57BL/6 (E and F), and AKR (H and I) strains are shown, with two magnifications for the Hfe −/− specimens (×100, B, E, and H; ×200, C, F, and I; original magnifications). Dark blue granules represent storage iron. Note predominant distribution of iron in periportal (zone 1) hepatocytes in Hfe −/− mice from the C3H (B and C) and C57BL/6 (E and F) strains, and the more panlobular staining of hepatocytes in AKR Hfe −/− (H and I) mice.
Spleen Resists Iron Loading in Hfe −/− Mice Regardless of Strain.
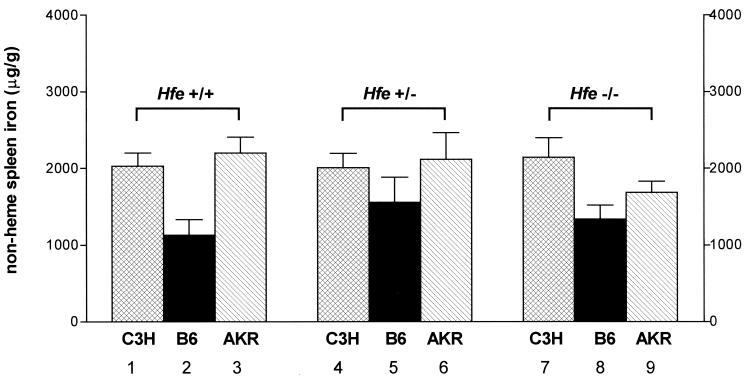
In contrast to secondary iron loading, where iron deposition in reticuloendothelial cells predominates, the reticuloendothelial cells in HH patients are relatively spared from iron loading until late in the disease (20, 21). Two studies have shown (10, 11) that Hfe −/− mice are also relatively resistant to splenic iron loading. To determine whether this relative sparing of spleen in Hfe −/− mice was strain-dependent, we measured splenic nonheme iron concentrations in Hfe +/+, Hfe +/−, and Hfe −/− mice, whose hepatic iron levels are presented in Fig. 2. Fig. 4 shows that none of the strains had a statistically significant increase in splenic iron concentration with Hfe disruption. The sparing effect on splenic iron loading was most evident in the AKR strain, which showed the greatest loading of hepatic iron (Fig. 2).
Figure 4.
Effect of strain differences and Hfe genotype on splenic iron concentration. Splenic nonheme iron concentrations were measured in wild-type (Hfe +/+), heterozygote knockout (Hfe +/−), and knockout (Hfe −/−) mice from three inbred mouse strains: C3H (hatched bars), C57BL/6 (B6, solid bars), and AKR (slashed bars). Data are presented as the mean ± SEM. Differences across strains within each genotype and across genotypes within each strain were determined separately by a one-way ANOVA. P < 0.05 across strains within genotype: bars 1 vs. 2, bars 2 vs. 3, bars 7 vs. 8. P < 0.05 across genotypes within strain: none.
Discussion
HH is a highly prevalent genetic disorder, affecting 3 to 5 individuals per 1000 individuals of northern European descent (1, 2, 4). In this condition, progressive accumulation of iron in tissues leads to liver cirrhosis and increased risk for hepatocellular carcinoma, diabetes mellitus, cardiomyopathy, impotence, and arthritis (1, 2, 4). Identification of the HFE gene mutations responsible for HH led to the hope of early diagnosis by genetic screening. Most patients with HH are homozygous for a missense mutation in the HFE gene that results in the C282Y substitution in the protein (3). Now that precise identification of HFE genotype is possible, it has become clear that some individuals homozygous for the C282Y mutation do not have evidence of an iron overload (5–7, 22–24). Furthermore, among HH patients with iron loading, wide variation is observed in measurements of body iron status (serum ferritin, transferrin saturation, and hepatic iron concentration) and in the severity of clinical disease (5–7, 25). The phenotypic variability among individuals with identical HFE genotypes is partially attributable to age and environmental factors, including diet, blood donation, and alcohol intake. Additionally, the expression of HH in premenopausal women may be delayed by the physiological loss of iron during pregnancy and/or menstruation (26, 27). Nonetheless, environmental factors do not completely account for the phenotypic variability in HH.
Several observations indicate that heritable factors other than HFE genotype also influence the severity of iron loading in HH. Greater concordance in biochemical measurements and clinical manifestations of an iron overload is observed within families than between families (28–30). A twin study concluded that 33%–47% of the variance in transferrin saturation and 47% of the variance in serum ferritin could be attributed to genetic factors other than HFE genotype (8). Linkage studies suggest that one or more genes other than HFE located near the HLA complex on human chromosome 6 may act to modify the HH phenotype (9, 31). Given the frequency of the disease and the severity when fully expressed, identification of the genes that modify the phenotype is of great practical importance.
We have observed (10) that Hfe −/− progeny from crosses of F1 129/Sv × C57BL/6 Hfe +/− mice had variable hepatic iron loading and suggested that segregating genes may influence the susceptibility to iron loading in the mouse. Studies on several genetically defined mouse strains have demonstrated considerable differences in basal iron status and in sensitivity to dietary iron loading (14–16). Furthermore, strain-specific differences have been observed in measurements of duodenal mucosal iron uptake and in transfer and clearance of iron from the circulation to the liver (16). We hypothesized that factors leading to strain differences in iron homeostasis in wild-type mice might also determine the sensitivity to iron loading consequent to Hfe disruption. To test this hypothesis, we used cross-breeding to introduce the disrupted Hfe allele into three mouse strains that differ in parameters of iron homeostasis. The results presented herein show marked differences between the strains resulting from disruption of the Hfe allele. The most striking difference was in the propensity for hepatic iron loading on a basal diet. The AKR strain was particularly susceptible to hepatic iron loading, and the C57BL/6 strain was particularly resistant.
The present studies show that two well-characterized strains, C57BL/6 and AKR, differ strikingly in the severity of hepatic iron loading in response to Hfe disruption. Studies of crosses between these strains should allow a straightforward genetic analysis of the loci that account for differences in iron loading, therefore providing a basis for the genetic dissection of iron homeostasis. Our observations suggest the existence of polymorphic genes that modify the severity of iron loading in this murine Hfe knockout model of HH. The identification of genes modifying the murine HH phenotype will certainly add to our understanding of regulation of iron homeostasis in the mouse. Whether the murine gene(s) responsible are related to those involved in susceptibility to iron loading in humans with HFE mutations is a question of great interest. If they were, they could suggest screening strategies to identify individuals likely to develop more severe clinical manifestations of HH.
Acknowledgments
We thank Mary Migas, Tammi Holmes, and Rosemary O'Neill for technical assistance and Elizabeth Torno for editorial help. This research was funded by National Institutes of Health Grants DK53405, GM34182, and DK40163 to W.S.S.; HL66225 to R.E.F.; DK41816 to B.R.B.; and DK56597, AI28802, and HL65749 to D.C.R.
Abbreviation
- HH
hereditary hemochromatosis
Footnotes
To whom reprint requests should be addressed at: Edward A. Doisy Department of Biochemistry and Molecular Biology, Saint Louis University School of Medicine, 1402 South Grand Boulevard, St. Louis, MO 63104. E-mail slyws@slu.edu.
References
- 1.Bacon B R, Powell L W, Adams P C, Kresina T F, Hoofnagle J H. Gastroenterology. 1999;116:193–207. doi: 10.1016/s0016-5085(99)70244-1. [DOI] [PubMed] [Google Scholar]
- 2.Powell L W. Pathology. 2000;32:24–36. doi: 10.1080/003130200104529. [DOI] [PubMed] [Google Scholar]
- 3.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 4.Adams P, Brissot P, Powell L W. J Hepatol. 2000;33:485–504. doi: 10.1016/s0168-8278(01)80874-6. [DOI] [PubMed] [Google Scholar]
- 5.Adams P C, Chakrabarti S. Gastroenterology. 1998;114:319–323. doi: 10.1016/s0016-5085(98)70483-4. [DOI] [PubMed] [Google Scholar]
- 6.Crawford D H G, Jazwinska E C, Cullen L M, Powell L W. Gastroenterology. 1998;114:1003–1008. doi: 10.1016/s0016-5085(98)70320-8. [DOI] [PubMed] [Google Scholar]
- 7.McDonnell S M, Hover A, Gloe D, Ou C Y, Cogswell M E, Grummer-Strawn L. Am J Med. 1999;107:30–37. doi: 10.1016/s0002-9343(99)00163-1. [DOI] [PubMed] [Google Scholar]
- 8.Whitfield J B, Cullen L M, Jazwinska E C, Powell L W, Heath A C, Zhu G, Duffy D L, Martin N G. Am J Hum Genet. 2000;66:1246–1258. doi: 10.1086/302862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pratiwi R, Fletcher L M, Pyper W R, Do K A, Crawford D H G, Powell L W, Jazwinska E C. J Hepatol. 1999;31:39–46. doi: 10.1016/s0168-8278(99)80161-5. [DOI] [PubMed] [Google Scholar]
- 10.Zhou X Y, Tomatsu S, Fleming R E, Parkkila S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, et al. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levy J E, Montross L K, Cohen D E, Fleming M D, Andrews N C. Blood. 1999;94:9–11. [PubMed] [Google Scholar]
- 12.Bahram S, Gilfillan S, Kuhn L C, Moret R, Schulze J B, Lebeau A, Schumann K. Proc Natl Acad Sci USA. 1999;96:13312–13317. doi: 10.1073/pnas.96.23.13312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy J E, Montross L K, Andrews N C. J Clin Invest. 2000;105:1209–1216. doi: 10.1172/JCI9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leboeuf R C, Tolson D, Heinecke J W. J Lab Clin Med. 1995;126:128–136. [PubMed] [Google Scholar]
- 15.Morse A C, Beard J L, Jones B C. Proc Soc Exp Biol Med. 1999;220:147–152. doi: 10.1046/j.1525-1373.1999.d01-22.x. [DOI] [PubMed] [Google Scholar]
- 16.Clothier B, Robinson S, Akhtar R A, Francis J E, Peters T J, Raja K, Smith A G. Biochem Pharmacol. 2000;59:115–122. doi: 10.1016/s0006-2952(99)00306-8. [DOI] [PubMed] [Google Scholar]
- 17.Fleming R E, Migas M C, Zhou X, Jiang J, Britton R S, Brunt E M, Tomatsu S, Waheed A, Bacon B R, Sly W S. Proc Natl Acad Sci USA. 1999;96:3143–3148. doi: 10.1073/pnas.96.6.3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fielding J. Methods Hematol. 1980;1:15–43. [Google Scholar]
- 19.Torrance J D, Bothwell T H. Methods Hematol. 1980;1:90–115. [Google Scholar]
- 20.McLaren G D. J Lab Clin Med. 1989;113:137–138. [PubMed] [Google Scholar]
- 21.Brunt E M, Olynyk J K, Britton R S, Janney C G, Di Bisceglie A M, Bacon B R. Am J Gastroenterol. 2000;95:1788–1793. doi: 10.1111/j.1572-0241.2000.02175.x. [DOI] [PubMed] [Google Scholar]
- 22.Olynyk J K, Cullen D J, Aquilia S, Rossi E, Summerville L, Powell L W. N Engl J Med. 1999;341:718–724. doi: 10.1056/NEJM199909023411002. [DOI] [PubMed] [Google Scholar]
- 23.Adams P C, Kertesz A E, McLaren C E, Barr R, Bamford A, Chakrabarti S. Hepatology. 2000;31:1160–1164. doi: 10.1053/he.2000.6984. [DOI] [PubMed] [Google Scholar]
- 24.Beutler E, Felitti V, Gelbart T, Ho N. Ann Intern Med. 2000;133:329–337. doi: 10.7326/0003-4819-133-5-200009050-00008. [DOI] [PubMed] [Google Scholar]
- 25.Bacon B R, Olynyk J K, Brunt E M, Britton R S, Wolff R K. Ann Intern Med. 1999;130:953–962. doi: 10.7326/0003-4819-130-12-199906150-00002. [DOI] [PubMed] [Google Scholar]
- 26.Edwards C Q, Dadone M M, Skolnick M H, Kushner J P. Clin Haematol. 1982;11:411–435. [PubMed] [Google Scholar]
- 27.Barton J C, Harmon L, Rivers C, Acton R T. Blood Cells Mol Dis. 1996;22:195–204. doi: 10.1006/bcmd.1996.0100. [DOI] [PubMed] [Google Scholar]
- 28.Crawford D H G, Halliday J W, Summers K M, Bourke M J, Powell L W. Hepatology. 1993;17:833–837. [PubMed] [Google Scholar]
- 29.Muir W A, McLaren G D, Braun W, Askari A. Am J Med. 1984;76:806–814. doi: 10.1016/0002-9343(84)90991-4. [DOI] [PubMed] [Google Scholar]
- 30.Bulaj Z J, Ajioka R S, Phillips J D, LaSalle B A, Jorde L B, Griffen L M, Edwards C Q, Kushner J P. N Engl J Med. 2000;343:1529–1535. doi: 10.1056/NEJM200011233432104. [DOI] [PubMed] [Google Scholar]
- 31.Barton J C, Shih W W, Sawada-Hirai R, Acton R T, Harmon L, Rivers C, Rothenberg B E. Blood Cells Mol Dis. 1997;23:135–145. doi: 10.1006/bcmd.1997.0129. [DOI] [PubMed] [Google Scholar]