Abstract
The purpose of this study was to define the effects of individual polymorphisms within the haplotypes of the TAS2R38 taste receptor gene on human bitter taste perception. A racially and ethnically diverse sample of children and adults (N = 980) was phenotyped for thresholds of 6-n-propylthiouracil (PROP) and genotyped for 3 polymorphisms of the TAS2R38 gene (A49P, V262A, I296V). Subjects were grouped according to their diplotype (i.e., specific combinations of haplotypes) and compared for PROP thresholds. By contrasting subjects with particular diplotypes, we found that in addition to A49P, V262A and I296V were related to the ability of the subjects to detect PROP. The V262A variant site affected the ability of subjects to detect mid-range concentrations of PROP, whereas the I296V variant site affected the ability of subjects to perceive PROP at the lowest concentration. These data agree with results from previous studies using cell-based assays for 2 variant sites (A49P and V262A) but not those for the I296V variant site. The reason for the discordant results is not known but it highlights the need for psychophysical as well as cell-based methods to understand the genotype–phenotype relationship for taste receptors. Human PROP sensitivity is determined by the combination of each of these 3 polymorphisms within the TAS2R38 gene.
Keywords: alleles, bitter, genetics
Introduction
Humans perceive bitterness when specific chemicals contact particular receptors on the apical surface of taste cell membranes. There are approximately 25 bitter receptors in the human genome, and some of their ligands have been identified through cell-based assay and transgenic methods (Chandrashekar et al. 2000; Bufe et al. 2002, 2005; Behrens et al. 2004, 2009; Kuhn et al. 2004; Pronin et al. 2004, 2007; Mueller et al. 2005; Soranzo et al. 2005; Conte et al. 2006; Sainz et al. 2007; Meyerhof et al. 2009). Genetic association studies have also provided clues about receptor–ligand pairs (Kim et al. 2003; Duffy et al. 2004; Prodi et al. 2004; Mennella et al. 2005; Pronin et al. 2007; Reed et al. 1999, 2010; Timpson et al. 2007; Tepper et al. 2008). Currently, not all bitter chemicals have a known receptor and not all receptors have a known ligand (Meyerhof et al. 2009).
One of the most intensively studied bitter receptor genes is TAS2R38, alleles of which are responsible in large part for the taste blindness to phenylthiocarbamide and other structurally similar chemicals such as propylthiouracil (PROP) and goitrin (Wooding et al. 2010). The observation that some people were insensitive to these chemicals, whereas others perceive them as intensely bitter was made in the early 1930s (Fox 1932) and was of interest to a range of researchers from different academic disciplines such as physical anthropology, medicine, genetics, and sensory biology (Guo and Reed 2001). Because the transmission of the trait followed a roughly Mendelian inheritance pattern, it was assumed a single polymorphism was involved, but this assumption was not quite correct: in fact, there are 3 variant sites within one gene that account for variations in human perception (Kim et al. 2003; Bufe et al. 2005). Investigators also assumed that because this taste trait was rare in some geographic regions and common in others that the allele frequencies would vary by race and ethnicity, and this assumption proved to be correct (Kim et al. 2003; Wooding et al. 2004). The nature of the collective and individual contributions to perceptual differences among people of different TAS2R38 genotype and haplotypes described below is the focus of the current study.
Recent research on the molecular characterization of the TAS2R38 gene revealed 3 variant sites (A49P, V262A, and I296V), and these haplotypes are found in 2 common (AVI and PAV), 2 less common (AAI and AAV), and 2 rare forms (PVI and PAI) (Kim et al. 2003; Wooding et al. 2004; Wang et al. 2007). Two haplotypes have not been reported in any subjects to date (AVV and PVV). Initially, it was not clear whether the main effect on taste perception was due to only one polymorphism, and thus the others within the haplotype were inconsequential (but found together because of linkage disequilibrium), or whether each polymorphism in the haplotype contributed to the increased or decreased bitter sensitivity. Threshold data from a few subjects with less common haplotypes suggested that they had intermediate phenotypes, but specific comparisons to assess the effects of each polymorphism could not be made with certainty (Kim et al. 2003). To try to surmount these difficulties, cell-based assays were developed to test the effects of individual variant sites (Bufe et al. 2005). From these in vitro studies, we learned that the A49P variant site had the greatest effect on taste transduction, the V262A variant site had weaker effects, and I296V variant site had no detectable effect. However, cell-based assays do not always recapitulate the natural signaling within cells, and so whether these observations would generalize to human taste was not known. To that end, we measured PROP thresholds from 980 human subjects and compared people with different diplotypes to determine the effect of each variant site on their taste response. (The term “diplotype” refers to a specific combination of haplotypes, e.g., someone homozygous for the nontaster haplotype would have an AVI/AVI diplotype.). The large sample size and its diversity of ancestry ensured that a number of rare haplotypes informative for this analysis would be represented.
Materials and methods
Subjects
Subjects who participated in research studies on taste and smell preferences during the years 2003–2007 were phenotyped for PROP threshold and genotyped for 3 variant sites within the TAS2R38 gene. Included in this sample of 980 individuals were 448 children (241 females, 207 males), 100 adolescents (55 females, 45 males), and 432 adults (425 females, 7 males). The majority of the adult subjects (N = 345) were the mothers of the children or adolescent participants. Children and adolescents ranged in age from 3 to 19 years (mean 7 ± 2) and adults from 20 to 55 years (mean 34 ± 7). Race/ethnicity was assigned by maternal (or adult) report according to standard US Census categories. We used the term race/ethnicity in describing our groups because it represents both the genetic and cultural components of this sample. These categories reflect the population of the urban setting (Philadelphia, PA) from which it was drawn: 56% Americans of African descent (Non-Hispanic; Black), 29% Americans of European descent (Non-Hispanic; White), and 15% other groups (Mixed ancestry, Asian, or Hispanic) (Anonymous 2006). All testing procedures were approved by the Office of Regulatory Affairs at the University of Pennsylvania. Informed consent was obtained from each adult, and assent was obtained from each child who was 7 years of age or older. Age-, sex-, and race-related effects have been reported elsewhere (Mennella et al. 2010).
Phenotyping for PROP perception
Following a 1-h fast, each subject was tested individually in a closed room designed for sensory studies. Most of the children younger than 7 years were tested with their mothers present. The mothers, who sat behind the children and out of view, refrained from talking during the test session and listened to music with headphones to prevent them from hearing their children's answers. All other subjects were tested individually.
To allow for comparisons, all procedures were identical for children and adults, and several steps were undertaken to make sure that the younger subjects understood the task before testing. The forced-choice procedures and concentrations of PROP used were based on previous research (Anliker et al. 1991; Mennella et al. 2005). PROP was chosen for the study because it is a medication used to treat thyroid disorders and thus more safety data are available regarding its use when compared with phenylthiocarbamide (PTC), about which comparably little is known (Wheatcroft and Thornburn 1972). Subjects were presented with a cup containing 5 mL of water and told to rinse the contents in their mouth and spit it out. If the solution tasted like water, they were told to give it to a stuffed toy of Big Bird (a likeable, well-known television character puppet), but if it tasted “yucky” or bitter, they should give it to Oscar the Grouch so that he can “throw it in his trash can” (Schmidt and Beauchamp 1988). The procedure was repeated and subjects tasted, in ascending order, 3 solutions of PROP (56, 180, and 560 μM) rinsing with water before and after each tasting. Subjects were classified into 4 groups based on the lowest concentration, if any, that they reported bitterness and, in turn, gave the sample to Oscar the Grouch. Those who gave all samples to Big Bird were classified as “None (of the samples) tasted bitter.”
Genotyping and haplotyping for the TAS2R38 gene
Cells from the cheek were obtained using swabs, and genomic DNA was extracted following the directions of the manufacturer (Epicenter). Three polymorphisms of the TAS2R38 gene (accession no. NM_176817) were genotyped using real-time polymerase chain reaction (PCR) single nucleotide polymorphism genotyping assays (rs713598, rs1726866, and rs10246939) with the Prism 7000, manufactured by Applied Biosystems. Other polymorphisms within the gene (Wooding et al. 2004) were not typed because their rarity precluded inclusion in the statistical analysis. Genotypes were checked for appropriate segregation to detect genotyping or family history errors. DNA samples from 6 subjects failed to amplify despite appropriate concentration and purity and 5 other samples amplified but failed to separate into genotype groups, even after several assay attempts. These unusual results could be due to other variant sites that prevented primer binding (and thus resulted in failed amplification), or to copy number variants, which might result in PCR products that fail to cluster into genotypic groups. Regardless of the explanation, the individuals who contributed these DNA samples were not included in the current analysis. Haplotypes were unequivocally identified, for example, by tracing the parental origin of the variant sites (61% of subjects) or otherwise they were inferred by expectation–maximization methods using an algorithm implemented by the computer program fastPHASE (39% of subjects) (Scheet and Stephens 2006).
Data analyses
The goal of data analysis was to describe the genotypes and haplotypes observed and select and compare people with particular haplotypes to determine how each variant site affected taste perception. All analyses followed the same method, which was to stratify subjects into groups by the variables of interest, and when appropriate, to detect frequency differences between the groups with an omnibus χ2 analyses for k independent samples, followed by a partitioned χ2 to determine where the difference occurred (Siegel and Castellan 1988). Because this partitioning analysis cannot be undertaken if there are no observations in a particular cell, or when the number of observations in 20% of the cells is less than 5, in some cases, we conducted analysis only on groups with sufficient sample size or we combined related groups to increase the sample size per cell.
The sample contained males and females, people of different ages (adults and children) and subjects from several racial/ethnic groups. The effects of sex, age, and race are reported elsewhere (Mennella et al. 2010). There is no difference between males and females or between children and adults in TAS2R38 diplotype frequency. There are racial differences in polymorphism frequencies for TAS2R38, but there is no additional effect of race. In other words, regardless of race/ethnicity, the relationship between diplotype and its effects on tasting ability is the same. Thus, age, sex, and race groups were combined for analysis.
To isolate the effect of each polymorphism on bitter taste perception, subjects were grouped by a diplotype of interest and the percentage of people from each PROP taste threshold group was assessed. For example, we compared PROP thresholds of people homozygous for the AAI versus AVI haplotype (underlined polymorphisms emphasize the specific comparison) to evaluate the effect of the V262A polymorphism. Descriptive analyses and the proportion test were conducted with procedures in STATISTICA (StatSoft). Criterion for statistical significance for all analyses was P ≤ 0.05.
Results
Haplotypes and diplotypes
From all combinations of the 3 polymorphisms, 6 of the 8 possible haplotypes and 13 of the 36 possible diplotypes were observed. Two haplotypes accounted for over 84% of all haplotypes (AVI, 41.2%, nontaster and PAV, 43.1%, taster), whereas the remaining 4 haplotypes were less common: rare (AAI, 12.2%; AAV, 3.3%), or extremely rare (PAI, <1% and PVI, <1%). The 2 remaining possible combinations, AVV and PVV, were not observed in this sample. We refer to the AVI as “nontaster” haplotype and PAV as the “taster” haplotype for simplicity, recognizing that there is a range of tasting ability within each haplotype. Likewise, subjects with 2 copies of the taster or nontaster haplotype are referred to as having the taster or nontaster diplotype, respectively. The most frequent diplotype was the combination of the 2 most frequent haplotypes and these AVI/PAV heterozygous subjects accounted for 37% of all subjects, followed in frequency by the homozygous subjects: 18% (nontaster) and 17% (taster). The remaining diplotypes were combinations of one rare and one common haplotype, except for 3 subjects, who each had a different combination of rare haplotypes.
Individual TAS2R38 polymorphisms and PROP perception
To gauge the contribution of each of the 3 TAS2R38 polymorphisms to taste perception, we identified groups of subjects that differed for a particular polymorphism and compared them for taste sensitivity. For the A49P polymorphism, we attempted to compare subjects who were homozygous for the PAV haplotype versus those that were homozygous for the AAV haplotype but because only one subject had an AAV/AAV diplotype, no statistical comparisons could be performed. As an alternative strategy, we compared subjects with one versus 2 P genotypes at the first position, that is, PAV/PAV versus PAV/AAV and observed no difference in the proportion of subjects with particular PROP thresholds (P > 0.05). (To increase sample size in this analysis, subjects who detected a taste in one of the 3 PROP concentrations were combined into one group and compared with subjects who reported no taste at all [all solutions were given to Big Bird].).
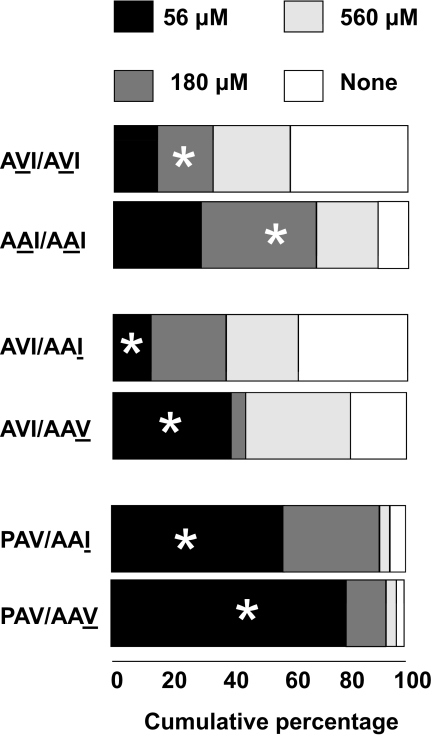
For the second position (V262A), we compared subjects who were homozygous for the AVI haplotype with those who were homozygous for the AAI haplotype. When the second variant was an “A” rather than a “V,” the proportion of people who could perceive the bitterness of PROP increased (omnibus, = 12.83, P = 0.005; partition between people who could perceive a bitter taste at 56 and 180 μM vs. 560 μM, = 4.42, P = 0.04; people who reported that “no solution tasted bitter” vs. people who could taste it at 56, 180, or 560 μM, = 8.39, P = 0.004). When people with the AVI/AAI diplotype are compared with those with the AVI/AVI diplotype, there is no difference in PROP tasting ability = 2.31, P = 0.51). It was not possible to compare the same polymorphism substitution on the alternative haplotype background because a haplotype needed for this comparison was not observed (PVV). One effect of the V262A polymorphism is to decrease the threshold of people at the middle-range concentrations of PROP (180 μM; Figure 1).
Figure 1.
Comparisons of PROP thresholds for paired groups of subjects with specific diplotypes. Shown here is the cumulative percentage of subjects for each diplotype who first reported a bitter taste when sampling 56, 180, or 560 μM PROP or who never reported a bitter taste when sampling each of these PROP solutions (None = none of the solutions offered tasted bitter). Asterisks denote a significant difference by χ2 partition. The number of subjects per diplotype group is shown in Table 1.
Table 1.
Number of subjects stratified by TAS2R38 diplotype and PROP taste threshold
| Diplotypes | All subjects |
Threshold concentration |
||||
| Total | Percent | 56 μM | 180 μM | 560 μM | None tasted bitter | |
| AVI/AVI | 172 | 18 | 26 | 31 | 47 | 68 |
| AAI/AVI | 84 | 9 | 11 | 22 | 21 | 30 |
| AAV/AVI | 22 | 2 | 9 | 1 | 8 | 4 |
| AAI/AAI | 23 | 2 | 7 | 9 | 5 | 2 |
| AAV/AAI | 3 | <1 | 2 | 1 | 0 | 0 |
| AAV/AAV | 1 | <1 | 1 | 0 | 0 | 0 |
| AVI/PAV | 358 | 37 | 222 | 109 | 18 | 9 |
| AAI/PAV | 106 | 11 | 61 | 36 | 4 | 5 |
| AAV/PAV | 37 | 4 | 30 | 5 | 1 | 1 |
| AAI/PAI | 1 | <1 | 1 | 0 | 0 | 0 |
| PAV/PVI | 2 | <1 | 1 | 1 | 0 | 0 |
| PAV/PAI | 1 | <1 | 1 | 0 | 0 | 0 |
| PAV/PAV | 170 | 17 | 147 | 19 | 1 | 3 |
| Total | 980 | 519 | 234 | 105 | 122 | |
The panel labeled “All Subjects” (left) contains the number and percent of subjects grouped by TAS2R38 diplotype, as defined by 3 polymorphisms (A49P, V262A, and I296V). The panel labeled “Threshold concentration” (right) contains the PROP thresholds for each of the diplotypes. Subjects are grouped by the concentration of PROP they first reported to taste bitter (56, 180, and 560 uM) or if None tasted bitter = None of the solutions tasted bitter to the subject.
For the I296V polymorphism, we could not study subjects who were homozygous for the key haplotypes (AAI vs. AAV or PAV vs. PAI) because of their rarity, but we could compare subjects who differed in the last polymorphism (I or V) on only one chromosome, for example, (AVI/AAI vs. AVI/AAV). In this analysis, we pooled the 2 groups of subjects who were most insensitive to PROP to meet cell size requirements. Individuals with a “V” in the last position were more likely to detect bitterness at the lowest concentration compared with subjects with the same diplotype but with an “I” in the last position (omnibus, = 11.02, P = 0.004; people who could perceive a bitter taste at 56 μM vs. 180 μM, = 10.75, P = 0.001). Likewise, similar results were observed when studying the same substitution among subjects who had a PAV haplotype on the opposite chromosome, that is, PAV/AVI versus PAV/AAV, (omnibus, = 6.73, P = 0.03; people who could perceive a bitter taste at 56 μM vs. 180 μM, = 6.36, P = 0.01, Figure 1). The I296V polymorphism appeared to shift the threshold downward, making sensitive people even more so.
Discussion
The study objective was to take a psychophysical approach to examine the effects of the individual TAS2R38 polymorphisms on bitter perception. Previous studies suggested that different amino acids at position 49 account for the greatest fraction of differences in phenotype, followed by V262A, and that I296V has subtle or undetectable effects (Kim et al. 2003; Bufe et al. 2005). Because of the large and genetically diverse sample recruited herein, we were able to find sufficient numbers of subjects with rare diplotypes to more precisely define the roles of the last 2 polymorphisms. The results for V262A agreed with past research that demonstrates that an alanine in this position increases sensitivity to moderate concentrations of PROP. However, we found that a valine, rather than isoleucine, in the last position (I296V) was associated with increased sensitivity at the lowest concentration of PROP, and this result was apparent regardless of the upstream haplotype. The main point is that genotyping only the A49P polymorphism, while convenient for statistical analysis, does not fully capture the genotype–phenotype association (Mennella et al. 2005).
The observation that the I296V polymorphism has an effect on the perception of PROP does not agree with data obtained from cell-based assay experiments of receptor function (Bufe et al. 2005). This discrepancy might potentially be resolved by the results of structural modeling studies of the TAS2R38 protein, which suggested that the I296V polymorphism is within a motif that interacts with G-proteins (i.e., intracellular signaling molecules) (Floriano et al. 2006; Miguet et al. 2006). Therefore, one explanation is that the I296V interacts with the native G-protein and results in greater or lesser PROP sensitivity, but in the cell-based assay system, it does not have the same effect because it interacts with different nonnative G-proteins. In keeping with this explanation, failures of heterologous systems to fully capture the action of the receptor in vivo have been reported for olfactory receptors (Grosmaitre et al. 2009). Overall, these data highlight the limitations of heterologous cell-based assay methods to study human taste systems.
Race and ethnicity are important variables in this study because the uncommon haplotypes used to draw conclusions about the effects of particular polymorphisms are more common in some racial groups than others. For instance, the AAI haplotype is more common in Americans of African descent, whereas the AAV haplotype is more common in Americans of European descent. However, although polymorphism frequencies differ among these groups, race had no effect on the bitter tasting ability of people with the same diplotype. As an example, people with the AVI/AVI diplotype were equally insensitive to PROP, regardless of race (Mennella et al. 2010). This result suggests that there are no residual effects of race-specific genetic background on this taste trait and also that environmental or cultural differences which have been reported for some aspects of taste (Moskowitz et al. 1975) do not influence the genotype–phenotype relationship here.
Although the TAS2R38 gene and its variants account for a large fraction of the heritable variation in the perception of PTC and PROP bitterness, other genetic modifiers may exist. Evidence for this assertion comes from the observation that within groups of people with only the nontaster TAS2R38 diplotype, the ability to taste PTC is heritable (Kim et al. 2003). The genetic contributors that restore the ability to taste these bitter stimuli may include other members in the bitter receptor family (Reed et al. 1999, 2010) and other nonbitter receptor genes (Drayna et al. 2003; Prodi et al. 2004; Reed et al. 2010). Other modifiers of taste ability are receptor cell number and density (Hayes et al. 2008) or influences like age and disease (Bartoshuk et al. 1996; Hayes et al. 2008; Mennella et al. 2010; Ventura et al., forthcoming).
The history of PTC taste genetics illustrates 2 viewpoints about how to characterize human traits, either as a dichotomization, for example, taster versus nontaster, or as a continuum. The most common haplotypes for TAS2R38, AVI, and PAV, are associated with the most extreme phenotypes, and the differences among individuals are so marked that the use of the terms taster and nontaster, although often adopted for convenience as we have done here, is probably warranted. However, there are haplotypes that are associated with intermediate phenotypes and it is also accurate to say that taste perception of PTC (and PROP) is on a continuum. Studies conducted by anthropologists on populations around the world prior to the discovery of the molecular basis of PTC genetics suggest that some people can detect it at very low concentrations (Ibraimov and Mirrakhimov 1979) and given what we now know, a supersensitive haplotype may exist. Overall, the choice of whether to consider this taste trait as a qualitative or quantitative one is probably a practical issue and depends on why the trait is measured and the scientific questions being addressed.
Funding
This work was supported by the National Institutes of Health [grants HD37119 and AA09523 to J.A.M. and DC004698 to D.R.R]. The work was also supported by a grant from the Pennsylvania Department of Health. Dr Pepino is currently a fellow of NIDA T32 DA07313 at the School of Medicine, Washington University in St. Louis.
Acknowledgments
We acknowledge the assistance of Kirsten J. Mascioli, Amanda H. McDaniel, Janice Kennedy, Greg Shaffer, Allison Steinmeyer, Dr Catherine Forestell, Minna Bak, Sarah Obenrader, Macrina Cooper-White, Abigail Bosk, and other technicians and students for expert assistance. The advice of Dr Wely Floriano regarding the interpretation of the modeling studies is acknowledged. Drs Michael G. Tordoff and Gary K. Beauchamp commented on the manuscript prior to publication. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the National Institutes of Health, or the Pennsylvania Department of Health.
References
- Anliker JA, Bartoshuk L, Ferris AM, Hooks LD. Children's food preferences and genetic sensitivity to the bitter taste of 6-n-propylthiouracil (PROP) Am J Clin Nutr. 1991;54:316–320. doi: 10.1093/ajcn/54.2.316. [DOI] [PubMed] [Google Scholar]
- Anonymous. 2006. Pennsylvania Vital Statistics; Pennsylvania Department of Health. Pennsylvania: Bureau of Health Statistics and Research. [Google Scholar]
- Bartoshuk LM, Duffy VB, Reed D, Williams A. Supertasting, earaches and head injury: genetics and pathology alter our taste worlds. Neurosci Biobehav Rev. 1996;20:79–87. doi: 10.1016/0149-7634(95)00042-d. [DOI] [PubMed] [Google Scholar]
- Behrens M, Brockhoff A, Batram C, Kuhn C, Appendino G, Meyerhof W. The human bitter taste receptor hTAS2R50 is activated by the two natural bitter terpenoids andrographolide and amarogentin. J Agric Food Chem. 2009;57:9860–9866. doi: 10.1021/jf9014334. [DOI] [PubMed] [Google Scholar]
- Behrens M, Brockhoff A, Kuhn C, Bufe B, Winnig M, Meyerhof W. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 2004;319:479–485. doi: 10.1016/j.bbrc.2004.05.019. [DOI] [PubMed] [Google Scholar]
- Bufe B, Breslin PA, Kuhn C, Reed DR, Tharp CD, Slack JP, Kim UK, Drayna D, Meyerhof W. The molecular basis of individual differences in phenylthiocarbamide and propylthiouracil bitterness perception. Curr Biol. 2005;15:322–327. doi: 10.1016/j.cub.2005.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 2002;32:397–401. doi: 10.1038/ng1014. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba N. T2Rs function as bitter taste receptors. Cell. 2000;100:703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Conte C, Guarin E, Marcuz A, Andres-Barquin PJ. Functional expression of mammalian bitter taste receptors in Caenorhabditis elegans. Biochimie. 2006;88:801–806. doi: 10.1016/j.biochi.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Drayna D, Coon H, Kim UK, Elsner T, Cromer K, Otterud B, Baird L, Peiffer AP, Leppert M. Genetic analysis of a complex trait in the Utah Genetic Reference Project: a major locus for PTC taste ability on chromosome 7q and a secondary locus on chromosome 16p. Hum Genet. 2003;112:567–572. doi: 10.1007/s00439-003-0911-y. [DOI] [PubMed] [Google Scholar]
- Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28:1629–1637. doi: 10.1097/01.ALC.0000145789.55183.D4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floriano WB, Hall S, Vaidehi N, Kim U, Drayna D, Goddard WA., 3rd Modeling the human PTC bitter-taste receptor interactions with bitter tastants. J Mol Model. 2006;12:931–941. doi: 10.1007/s00894-006-0102-6. [DOI] [PubMed] [Google Scholar]
- Fox AL. The relationship between chemical constitution and taste. Proc Natl Acad Sci U S A. 1932;18:115–120. doi: 10.1073/pnas.18.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosmaitre X, Fuss SH, Lee AC, Adipietro KA, Matsunami H, Mombaerts P, Ma M. SR1, a mouse odorant receptor with an unusually broad response profile. J Neurosci. 2009;29:14545–14552. doi: 10.1523/JNEUROSCI.2752-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 2001;28:111–142. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JE, Bartoshuk LM, Kidd JR, Duffy VB. Supertasting and PROP bitterness depends on more than the TAS2R38 gene. Chem Senses. 2008;33:255–265. doi: 10.1093/chemse/bjm084. [DOI] [PubMed] [Google Scholar]
- Ibraimov A, Mirrakhimov MM. PTC-tasting ability in populations living in Kirghizia with special reference to hypersensitivity: its relation to sex and age. Hum Genet. 1979;46:97–105. doi: 10.1007/BF00278907. [DOI] [PubMed] [Google Scholar]
- Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science. 2003;299:1221–1225. doi: 10.1126/science.1080190. [DOI] [PubMed] [Google Scholar]
- Kuhn C, Bufe B, Winnig M, Hofmann T, Frank O, Behrens M, Lewtschenko T, Slack JP, Ward CD, Meyerhof W. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 2004;24:10260–10265. doi: 10.1523/JNEUROSCI.1225-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella JA, Duke F, Pepino MY, Reed DR. Age modifies the genotype-phenotype relationship for the bitter receptor TAS2R38. BMC Genetics. 2010 doi: 10.1186/1471-2156-11-60. 11:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella JA, Pepino MY, Reed DR. Genetic and environmental determinants of bitter perception and sweet preferences. Pediatrics. 2005;115:e216–e222. doi: 10.1542/peds.2004-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 2009;35:157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Miguet L, Zhang Z, Grigorov MG. Computational studies of ligand-receptor interactions in bitter taste receptors. J Recept Signal Transduct Res. 2006;26:611–630. doi: 10.1080/10799890600928210. [DOI] [PubMed] [Google Scholar]
- Moskowitz HW, Kumaraiah V, Sharma KN, Jacobs HL, Sharma SD. Cross-cultural differences in simple taste preferences. Science. 1975;190:1217–1218. doi: 10.1126/science.1198109. [DOI] [PubMed] [Google Scholar]
- Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJ. The receptors and coding logic for bitter taste. Nature. 2005;434:225–229. doi: 10.1038/nature03352. [DOI] [PubMed] [Google Scholar]
- Prodi DA, Drayna D, Forabosco P, Palmas MA, Maestrale GB, Piras D, Pirastu M, Angius A. Bitter taste study in a Sardinian genetic isolate supports the association of phenylthiocarbamide sensitivity to the TAS2R38 bitter receptor gene. Chem Senses. 2004;29:697–702. doi: 10.1093/chemse/bjh074. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Tang H, Connor J, Keung W. Identification of ligands for two human bitter T2R receptors. Chem Senses. 2004;29:583–593. doi: 10.1093/chemse/bjh064. [DOI] [PubMed] [Google Scholar]
- Pronin AN, Xu H, Tang H, Zhang L, Li Q, Li X. Specific alleles of bitter receptor genes influence human sensitivity to the bitterness of aloin and saccharin. Curr Biol. 2007;17:1403–1408. doi: 10.1016/j.cub.2007.07.046. [DOI] [PubMed] [Google Scholar]
- Reed DR, Nanthakumar E, North M, Bell C, Bartoshuk LM, Price RA. Localization of a gene for bitter-taste perception to human chromosome 5p15. Am J Hum Genet. 1999;64:1478–1480. doi: 10.1086/302367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DR, Zhu G, Breslin PA, Duke FF, Henders AK, Campbell MJ, Montgomery GW, Medland SE, Martin NG, Wright MJ. The perception of quinine taste intensity is associated with common genetic variants in a bitter receptor cluster on chromosome 12. Hum Mol Genet. 2010;19(21):427842–427885. doi: 10.1093/hmg/ddq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainz E, Cavenagh MM, Gutierrez J, Battey JF, Northup JK, Sullivan SL. Functional characterization of human bitter taste receptors. Biochem J. 2007;403:537–543. doi: 10.1042/BJ20061744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HJ, Beauchamp GK. Adult-like odor preferences and aversions in three-year-old children. Child Dev. 1988;59:1136–1143. [PubMed] [Google Scholar]
- Siegel S, Castellan NJ. Nonparametric statistics for the behavioral sciences. Boston (MA): McGraw Hill; 1988. [Google Scholar]
- Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15:1257–1265. doi: 10.1016/j.cub.2005.06.042. [DOI] [PubMed] [Google Scholar]
- Tepper BJ, Koelliker Y, Zhao L, Ullrich NV, Lanzara C, d'Adamo P, Ferrara A, Ulivi S, Esposito L, Gasparini P. Variation in the bitter-taste receptor gene TAS2R38, and adiposity in a genetically isolated population in Southern Italy. Obesity (Silver Spring) 2008;16:2289–2295. doi: 10.1038/oby.2008.357. [DOI] [PubMed] [Google Scholar]
- Timpson NJ, Heron J, Day IN, Ring SM, Bartoshuk LM, Horwood J, Emmett P, Davey-Smith G. Refining associations between TAS2R38 diplotypes and the 6-n-propylthiouracil (PROP) taste test: findings from the Avon Longitudinal Study of Parents and Children. BMC Genet. 2007;8:51. doi: 10.1186/1471-2156-8-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura AK, Reed DR, Mennella JA Forthcoming. Ear infections, bitter taste sensitivity, and weight status in children. Physiol Behav [Google Scholar]
- Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, et al. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcohol Clin Exp Res. 2007;31:209–215. doi: 10.1111/j.1530-0277.2006.00297.x. [DOI] [PubMed] [Google Scholar]
- Wheatcroft PE, Thornburn CC. Toxicity of the taste testing compound phenylthiocarbamide. Nat New Biol. 1972;235:93–94. doi: 10.1038/newbio235093a0. [DOI] [PubMed] [Google Scholar]
- Wooding S, Gunn H, Ramos P, Thalmann S, Xing C, Meyerhof W. Genetics and bitter taste responses to goitrin, a plant toxin found in vegetables. Chem Senses. 2010;35:685–692. doi: 10.1093/chemse/bjq061. [DOI] [PubMed] [Google Scholar]
- Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–646. doi: 10.1086/383092. [DOI] [PMC free article] [PubMed] [Google Scholar]