Abstract
Glycosylphosphatidylinositols (GPIs) are attached to the C-termini of many proteins, thereby acting as membrane anchors. Biosynthesis of GPI is initiated by GPI-N-acetylglucosaminyltransferase (GPI-GnT), which transfers N-acetylglucosamine from UDP- N-acetylglucosamine to phosphatidylinositol. GPI-GnT is a uniquely complex glycosyltransferase, consisting of at least four proteins, PIG-A, PIG-H, PIG-C and GPI1. Here, we report that GPI-GnT requires another component, termed PIG-P, and that DPM2, which regulates dolichol-phosphate-mannose synthase, also regulates GPI-GnT. PIG-P, a 134-amino acid protein having two hydrophobic domains, associates with PIG-A and GPI1. PIG-P is essential for GPI-GnT since a cell lacking PIG-P is GPI-anchor negative. DPM2, but not two other components of dolichol-phosphate-mannose synthase, associates with GPI-GnT through interactions with PIG-A, PIG-C and GPI1. Lec15 cell, a null mutant of DPM2, synthesizes early GPI intermediates, indicating that DPM2 is not essential for GPI-GnT; however, the enzyme activity is enhanced 3-fold in the presence of DPM2. These results reveal new essential and regulatory components of GPI-GnT and imply co-regulation of GPI-GnT and the dolichol-phosphate-mannose synthase that generates a mannosyl donor for GPI.
Keywords: endoplasmic reticulum/glycosyltransferase/N-glycan/posttranslational modification
Introduction
Glycosylphosphatidylinositol (GPI), a complex glycolipid, acts as a membrane anchor of many cell surface proteins. It is synthesized in the endoplasmic reticulum (ER) and transferred en bloc to the C-termini of proteins that have a GPI attachment signal peptide. GPI-anchoring is a ubiquitous mode of posttranslational modification in eukaryotes (Herscovics and Orlean, 1993; Udenfriend and Kodukula, 1995; Schultz et al., 1998; Ferguson, 1999). In cells of protozoa, such as Plasmodium falciparum and Trypanosoma brucei, GPI-anchor is the predominant membrane attachment of cell surface proteins (Gerold et al., 1996; Ferguson, 1999). In both budding and fission yeasts, GPI-anchors are essential for growth (Leidich et al., 1994). Saccharomyces cerevisiae has >60 different GPI-anchored proteins. Many of them are incorporated into the cell wall by means of glycosidic linkages between the glycan portion of GPI and the cell wall glucans, generating firm structures (Kapteyn et al., 1999).
In mammalian cells, >100 different proteins are GPI-anchored (Kinoshita et al., 1995), including cell surface enzymes, receptors, adhesion molecules or immunologically important proteins. GPI-anchoring is not essential at the cell level in mammalian systems. In fact, a number of mutant cell lines defective in various steps of GPI-anchor biosynthesis have been established (Hyman, 1988). In contrast, GPI-anchoring is essential for embryogenesis (Nozaki et al., 1999) and development of skin (Tarutani et al., 1997) as shown by studies with gene knockout mice, indicating critical roles of GPI-anchored proteins in cell to cell and/or to environment interactions. In humans, somatic mutation of PIG-A, an X-linked gene involved in biosynthesis of GPI, in hematopoietic stem cells causes a hematologic disease, paroxysmal nocturnal hemoglobinuria (Takeda et al., 1993).
Biosynthesis of GPI is initiated by a transfer of N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to phosphatidylinositol (PI) to generate N-acetylglucos aminyl-PI (GlcNAc-PI) (Masterson et al., 1989). This reaction is catalyzed by GPI-N-acetylglucosaminyl transferase (GPI-GnT), which consists of at least four proteins, PIG-A, PIG-H, PIG-C and GPI1 (Watanabe et al., 1998). This complex structure is unusual for glycosyltransferases. PIG-A is essential and most likely a catalytic component because it has homology to a bacterial GnT involved in lipopolysaccharide biosynthesis and to many other glycosyltransferases (Kinoshita et al., 1997). The functions of PIG-H, PIG-C and GPI1 cannot be predicted from their primary sequences (Kamitani et al., 1993; Inoue et al., 1996; Watanabe et al., 1998). It is clear that PIG-H and PIG-C are essential for GPI-GnT because cells with mutations in these genes are completely deficient in the surface expression of GPI-anchored proteins (Stevens and Raetz, 1991). GPI1 is important for the formation of the enzyme complex because, although PIG-A and PIG-H associate with each other in GPI1-knockout cells, PIG-C does not stably associate with the complex of PIG-A and PIG-H (Hong et al., 1999). The GPI1-knockout cell expresses a small amount of GPI-anchored proteins but its GPI-GnT activity was below the detectable level (Hong et al., 1999).
In order to determine whether PIG-A, PIG-H, PIG-C and GPI1 are the only components of the GPI-GnT, and in light of recent genetic evidence of a fifth gene necessary for this step (see below), we isolated GPI-GnT for further characterization. Here, we report that GPI-GnT requires another component, termed PIG-P, and that DPM2, which is a regulatory component of dolichol-phosphate-mannose (Dol-P-Man) synthase, also regulates GPI-GnT activity.
Results
Isolation and characterization of GPI-GnT
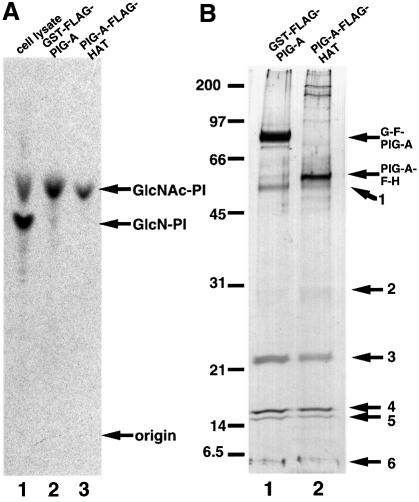
In order to isolate active human GPI-GnT in an amount sufficient for analysis of its subunit composition, we transfected JY5 cells with cDNA encoding PIG-A tandemly tagged with FLAG and glutathione S-trans ferase (GST), or FLAG and HAT. We isolated GPI-GnT complexes containing either of the two tagged PIG-A from these transfected cells by means of two-step affinity purification. We incubated the isolated complexes with radiolabeled UDP-GlcNAc and bovine PI to confirm the enzymatic activity of both complexes (Figure 1A).
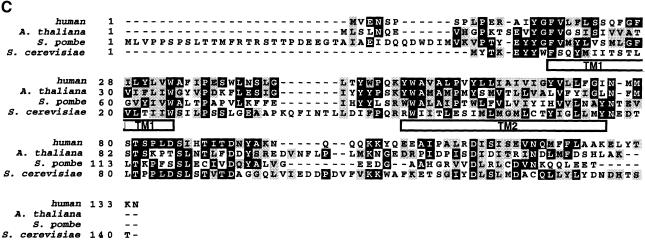
Fig. 1. Purification of GPI-GnT complexes. GST–FLAG-PIG-A and FLAG-HAT-PIG-A were isolated by two-step affinity purification from the digitonin lysates of 1.4 × 108 cells of JY5 transfectants. Samples of the purified proteins (equivalent to 6 × 107 cells) were used for in vitro GPI-GnT assay (A) and the rest (equivalent to 8 × 107 cells) were used for SDS–PAGE and silver staining (B). (A) Lysates derived from 107 cells of wild-type JY25 as a positive control (lane 1) and purified complexes containing GST–FLAG-PIG-A (lane 2) or FLAG-HAT-PIG-A (lane 3) were incubated with radiolabeled UDP-GlcNAc and bovine PI. Lipids were analyzed by TLC. Identities of spots are indicated on the right. (B) Silver-stained SDS–PAGE profiles of purified complexes containing GST–FLAG-PIG-A (lane 1) and FLAG-HAT-PIG-A (lane 2). Band numbers are shown on the right. The positions of the molecular size markers are indicated on the left (kDa).
Analysis by SDS–PAGE and silver staining of the complexes demonstrated six specific bands in addition to the tagged PIG-A (Figure 1B). Bands seen just above the 45 kDa marker are likely to be non-specific because they were greatly decreased after the second purification step and were seen in samples of similarly tagged non-relevant proteins prepared in parallel (data not shown). The bands numbered 1–3 correspond to GPI1, PIG-C and PIG-H, respectively, based on their molecular sizes. The three bands, numbered 4–6, may be other proteins associated with GPI-GnT.
In order to identify these proteins, we determined the N-terminal sequences using samples derived from ∼4 × 109 cells expressing GST–FLAG-PIG-A. We obtained the sequence VLKAF from band 1 that corresponds to residues 2–6 of GPI1, confirming the prediction based on molecular size. We did not obtain any sequence information from bands 2 and 3, most likely because their N-termini were blocked. We obtained a new sequence, VENSPSPL, from band 4. From band 5, we obtained the sequence ATGTD, which corresponds to the sequence of DPM2 starting from the second residue. DPM2 is one of the components of Dol-P-Man synthase (Maeda et al., 1998) and its N-terminal methionine is known to be eliminated (Maeda et al., 2000). The amount of band 6 protein was so small that we did not obtain sequence information. The amounts of PTH amino acids detected were 0.65, 0.6, 0.5, 0.29 and 0.4 pmol for V, E, N, S and P of band 4, respectively, and 0.75, 0.46, 0.7, 0.41 and 0.64 pmol for A, T, G, T and D of band 5/DPM2, respectively.
Cloning a new gene, PIG-P, involved in GPI-GnT
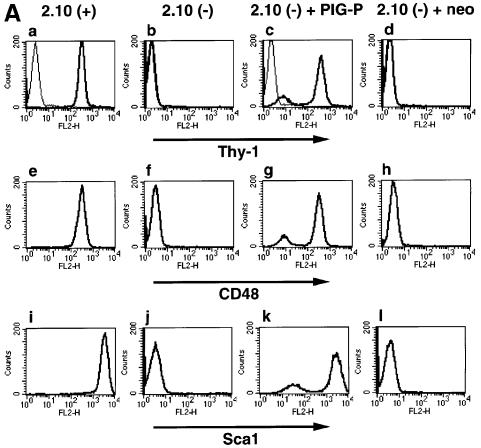
We cloned a cDNA that encodes a 134-amino acid protein whose sequence from amino acids 2–9 exactly matched with that of band 4, and named the gene PIG-P (phosphatidylinositol-glycan-class P) (Figure 2A). The predicted human PIG-P protein consists of two N-terminal hydrophobic regions and one C-terminal hydrophilic region (Figure 2B). PIG-P had no significant homology with other proteins of known functions. Human PIG-P is the same as a gene termed DSCR5a1 for Down syndrome critical region 5a1 (DDBJ/EMBL/GenBank accession No. AB035742) that has been mapped to chromosome 21q22.2 (DDBJ/EMBL/GenBank accession No. AP000150).
Fig. 2. (A) Amino acid sequence of human PIG-P. The sequence determined by protein sequencing is underlined. The DDBJ/EMBL/GenBank accession No. of human PIG-P cDNA is AB039659. (B) Hydropathy profile of human PIG-P (Kyte and Doolittle, 1982). (C) Alignment of amino acid sequences of human PIG-P and its homologs of A.thaliana, S.pombe and S.cerevisiae. Black and gray boxes indicate identical and similar amino acids, respectively. Amino acid numbers are indicated on the left. Two putative transmembrane regions of human PIG-P, TM1 and TM2 are indicated.
We found in databases the PIG-P homologs of mouse (DDBJ/EMBL/GenBank accession No. AAF32294), Arabidopsis thaliana (AAC13913), Schizosaccharomyces pombe (CAB16583) and S.cerevisiae (YDR437W) (Figure 2C), which had 90, 25, 30 and 22% amino acid identity with human PIG-P, respectively. PIG-P has two putative transmembrane regions (indicated as TM1 and TM2 in Figure 2C) predicted by the TMpred program provided by the BCM Search Launcher (Smith et al., 1996).
PIG-P complements a new mutant cell defective in GPI-GnT
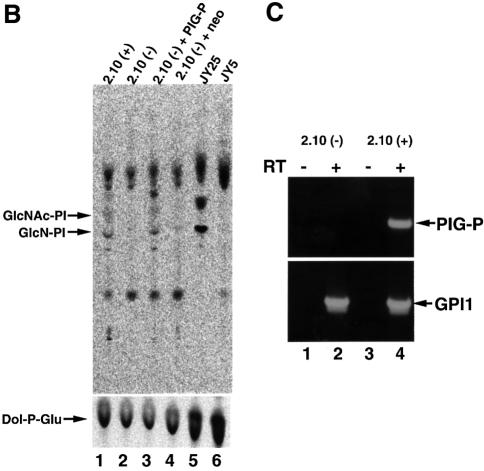
Recently, we found a mutant mouse T cell clone, 2.10 GPI (–), which is defective in biosynthesis of GPI-anchors. 2.10 GPI (–) lacked the surface expression of various GPI-anchored proteins, such as Thy-1, CD48 and Sca1 (Figure 3A, b, f and j), whereas its parental cell, 2.10 GPI (+), expressed them (a, e and i).
Fig. 3. (A) FACS analysis of 2.10 GPI (+), 2.10 GPI (–) and 2.10 GPI (–) cells transfected with pME-neo-PIG-P or pME-neo. Cells were stained by anti-Thy-1 (thick lines in a–d), isotype-matched control (thin lines in a–d), anti-CD48 (e–h) and anti-Sca1 (i–l) antibodies. (B) In vitro assay for the early steps of GPI biosynthesis. Cell lysates were incubated with radiolabeled UDP-GlcNAc (upper panel) and with radiolabeled UDP-glucose to assay Dol-P-Glu synthase as a measure of the amount of lysate (lower panel). The lipids were extracted and analyzed by TLC. Identities of spots are indicated on the left. Lane 1, 2.10 GPI (+); lane 2, 2.10 GPI (–); lane 3, 2.10 GPI (–) transfected with pME-neo-PIG-P; lane 4, 2.10 GPI (–) transfected with pME-neo; lane 5, wild-type JY25 cells; and lane 6, mutant JY5 cells. The radiolabeled products above GlcNAc-PI seen in all lanes are non-GPI products because they are seen with cell lysates from JY5 (lane 6), which is known to be deficient in the first step of GPI biosynthesis. (C) RT–PCR analysis of PIG-P in 2.10 GPI (+) and 2.10 GPI (–) cells. Samples of RNA were incubated in the presence (lanes 2 and 4) and absence (lanes 1 and 3) of reverse transcriptase. PCRs were done for the amplification of mouse PIG-P (upper panel) and GPI1 (lower panel).
To determine the defective biosynthetic step in this mutant cell, we measured the activities of the early GPI-anchor biosynthesis enzymes (Figure 3B). Lysates of parental 2.10 GPI (+) and wild-type B-lymphoblastoid cells, JY25, generated the first and second intermediates, GlcNAc-PI and GlcN-PI (Figure 3B, lanes 1 and 5). In contrast, 2.10 GPI (–) cells were defective in the synthesis of GlcNAc-PI (Figure 3B, lane 2), like the JY5 cells (lane 6) in which the first step is disrupted due to a defect in the PIG-A gene (Miyata et al., 1993).
We next performed somatic cell fusion analysis (Hyman, 1988) to determine whether 2.10 GPI (–) represents a new gene involved in the first step of GPI-anchor biosynthesis. We fused 2.10 GPI (–) with four mutant cells, JY5 (Miyata et al., 1993), S49Thy-1–h (Kamitani et al., 1993), T1M1Thy-1–c (Inoue et al., 1996) and GPI1-knockout F9 (Hong et al., 1999), which are defective in PIG-A, PIG-H, PIG-C and GPI1, respectively. In all combinations of cell fusions, the surface expression of GPI-anchored proteins was restored (data not shown), indicating that 2.10 GPI (–) is defective in a new gene that is involved in GPI-GnT.
To see whether PIG-P cDNA complements 2.10 GPI (–) mutant cells, we transfected it and assessed the surface expression of GPI-anchored proteins (Figure 3A) and GPI-GnT activity in vitro (Figure 3B). PIG-P cDNA (Figure 3A, c, g and k), but not an empty vector (d, h and l), restored the surface expression of GPI-anchored proteins on 2.10 GPI (–) mutant cells to the levels observed in wild-type 2.10 GPI (+) cells (a, e and i). GPI-GnT activity was also restored to the wild-type level (Figure 3B, lane 1) by transfection with PIG-P cDNA, but not an empty vector (Figure 3B, lanes 3 and 4).
We used RT–PCR to confirm that PIG-P is the gene responsible for the lack of GPI expression in 2.10 GPI (–) mutant (Figure 3C). The PIG-P transcript was detected in 2.10 GPI (+) cells (Figure 3C, upper panel, lane 4) but not in the 2.10 GPI (–) clone (upper panel, lane 2). Transcripts of the GPI1 gene were detected at similar levels in both cells (Figure 3C, lower panel, lanes 2 and 4). We concluded that PIG-P is the gene defective in 2.10 GPI (–) cells, hence it encodes an essential component of GPI-GnT.
PIG-P binds directly to PIG-A and GPI1
In order to determine the component(s) of GPI-GnT to which PIG-P binds, we constructed a cDNA encoding FLAG-PIG-P. FLAG-PIG-P was functional because it restored the surface expression of Thy-1 on 2.10 GPI (–) cells (data not shown). Using JY5 cells, we co-transfected FLAG-PIG-P with each of the four other components of GPI-GnT that are GST-tagged, or with GST-tagged microsomal aldehyde dehydrogenase (ALDH) as a control ER protein (Masaki et al., 1994). FLAG-PIG-P (Figure 4, top panel) and all GST-tagged proteins (bottom panel) were expressed well. Among five GST-tagged proteins, GST–PIG-A and –GPI1 were specifically co-precipitated with FLAG-PIG-P (Figure 4, middle panel, lanes 1 and 4). Since these experiments were performed using PIG-A-deficient cells, the interaction of PIG-P with GPI1 must be direct. We did not see specific co-precipitation of GST–PIG-H, –PIG-C and –ALDH with FLAG-PIG-P (Figure 4, middle panel, lanes 2, 3 and 5), suggesting that PIG-P directly associates with PIG-A and GPI1, but not with PIG-H and PIG-C.
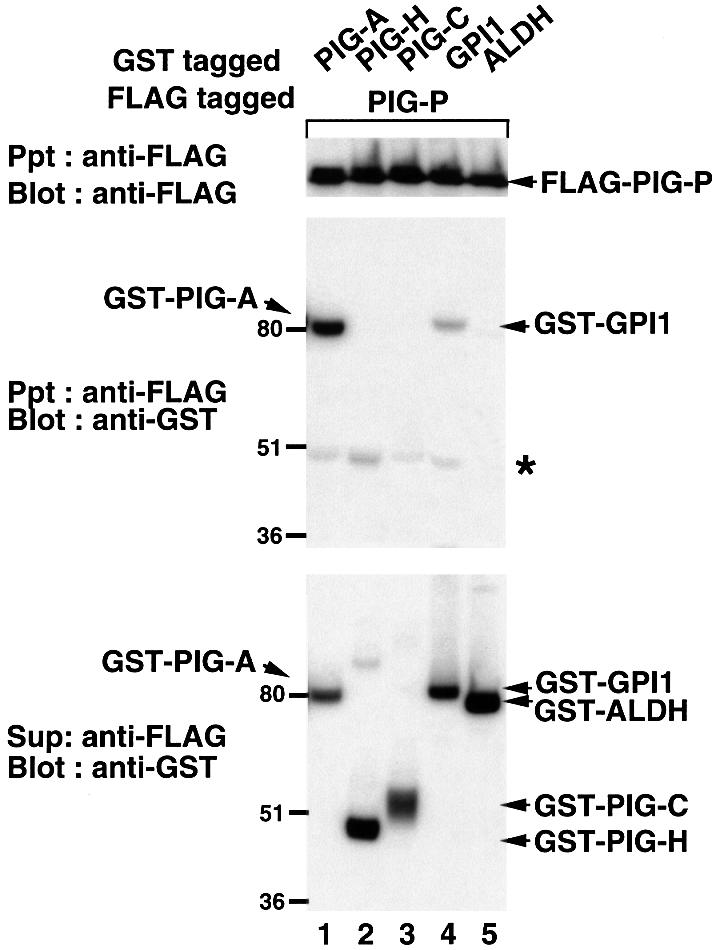
Fig. 4. Interaction of PIG-P with other components of GPI-GnT. FLAG-PIG-P was co-expressed in JY5 cells with GST–PIG-A (lane 1), –PIG-H (lane 2), –PIG-C (lane 3), –GPI1 (lane 4) or –ALDH (lane 5), and their associations were analyzed by co-precipitation. The lysates (in 1% digitonin) were precipitated with anti-FLAG beads and immunoprecipitates were analyzed by western blotting against anti-FLAG (top panel) and anti-GST (middle) antibodies. GST-tagged proteins were collected from the supernatant obtained after the immunoprecipitation by glutathione beads and analyzed by western blotting against anti-GST antibody (bottom). An asterisk in the middle panel indicates the heavy chain of mouse anti-FLAG antibody that was recognized by protein G. Positions of molecular size markers are indicated on the left (kDa).
DPM2 specifically binds to GPI-GnT complex via PIG-A, PIG-C and GPI1
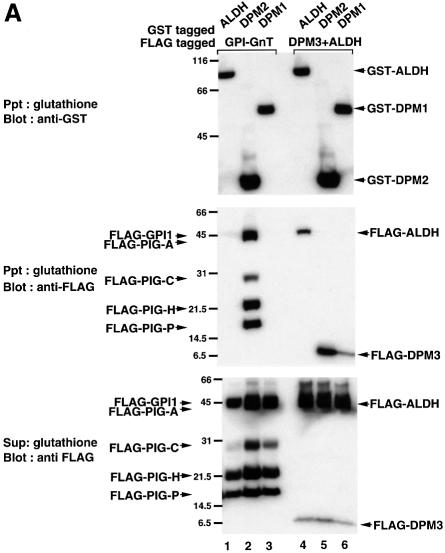
As shown in Figure 1B, the isolated GPI-GnT contained DPM2, a component of Dol-P-Man synthase. Mammalian Dol-P-Man synthase consists of three components, DPM1, DPM2 and DPM3 (Colussi et al., 1997; Maeda et al., 1998, 2000; Tomita et al., 1998). We analyzed the isolated GPI-GnT by western blotting with anti-human DPM3 antibody (Maeda et al., 2000) and found that DPM3 was not included in GPI-GnT (data not shown). To confirm the specificity of the association of DPM2 and to determine whether DPM1 also associates with GPI-GnT, we transfected JY5 cells with GST–ALDH, –DPM2 or –DPM1, and then with a mixture of five FLAG-tagged components of GPI-GnT (Figure 5A, lanes 1–3). When GST-tagged proteins were precipitated with glutathione beads from the cell extract in 1% digitonin (Figure 5A, top panel), all FLAG-tagged components of GPI-GnT were co-precipitated with GST–DPM2 (middle panel, lane 2) but not with GST–ALDH (lane 1), indicating a specific association of DPM2 with GPI-GnT. Components of GPI-GnT were not co-precipitated with GST–DPM1 (Figure 5A, lane 3). Therefore, of the three components of Dol-P-Man synthase, only DPM2 associated with GPI-GnT. It is also noted that the amounts of GPI-GnT components were higher in the DPM2-transfectants (Figure 5A, lane 2) than in the ALDH- and DPM1-transfectants (lanes 1 and 3).
Fig. 5. (A) Interaction of DPM2 with GPI-GnT complexes. GST–ALDH (lanes 1 and 4), –DPM2 (lanes 2 and 5) and –DPM1 (lanes 3 and 6) were co-expressed in JY5 cells with the five FLAG-tagged GPI-GnT components (lanes 1–3), or FLAG-DPM3 plus -ALDH (lanes 4–6). Physical associations between GST- and FLAG-tagged proteins were analyzed by co-precipitation. The lysates in 1% digitonin were precipitated with glutathione beads, and precipitates were analyzed by western blotting against anti-GST (top panel) and anti-FLAG (middle) antibodies. FLAG-tagged proteins remaining in the supernatant after precipitation with glutathione beads were immunoprecipitated and analyzed by western blotting against anti-FLAG antibody (bottom). Positions of molecular size markers are indicated on the left (kDa). (B) Association of DPM2 with components of GPI-GnT. GST–DPM2 was co-expressed in JY5 cells with FLAG-ALDH (lane 1), -PIG-A (lane 2), -PIG-H (lane 3), -PIG-C (lane 4), -GPI1 (lane 5) -PIG-P (lane 6) or -DPM3 (lane 7), and their associations were analyzed by co-precipitation. The lysates were precipitated with anti-FLAG and the immunoprecipitates were analyzed by western blotting against anti-FLAG (top panel) and anti-GST (middle) antibodies. GST–DPM2 remaining in the supernatants after immunoprecipitation was assessed by precipitation with glutathione beads and western blotting (bottom). An asterisk in the middle panel indicates a faint band seen in lane 1 showing a background level of GST–DPM2 present in the precipitates.
We previously reported that DPM1, DPM2 and DPM3 bind to each other, and that the Dol-P-Man synthase complex is maintained in 1% digitonin (Maeda et al., 2000). To verify that GST–DPM2 and –DPM1 associate with DPM3 under similar conditions, we transfected JY5 cells with GST–ALDH, –DPM2 or –DPM1, and then with a mixture of FLAG-DPM3 and -ALDH (Figure 5A, lanes 4–6). FLAG-DPM3 was co-precipitated with GST–DPM2 (Figure 5A, middle panel, lane 5) and –DPM1 (lane 6) but not with –ALDH (lane 4). FLAG-ALDH was not co-precipitated with GST–DPM2 (Figure 5A, lane 5) and –DPM1 (lane 6), showing the specificity of co-precipitation of FLAG-DPM3. FLAG-ALDH was co-precipitated with GST–ALDH (Figure 5A, lane 4), suggesting a homophilic association. Taken together, it seems unlikely that DPM1 and DPM3 had associated with GPI-GnT but dissociated during preparation. We therefore conclude that DPM2, but not the two other components of Dol-P-Man synthase, specifically associates with GPI-GnT. These results also indicate that there are two fractions of DPM2, one acting as a regulatory component of Dol-P-Man synthase and the other associating with GPI-GnT.
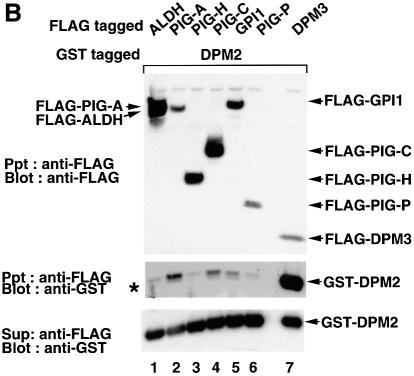
We next determined to which component(s) of GPI-GnT DPM2 binds. Using JY5 cells, we transfected GST–DPM2 with each of the FLAG-tagged components of GPI-GnT, FLAG-ALDH (as a negative control) or -DPM3 (as a positive control), and then examined the co-precipitation of GST–DPM2 with FLAG-tagged proteins (Figure 5B). GST–DPM2 was efficiently co-precipitated with FLAG-DPM3 (Figure 5B, middle panel, lane 7) as expected. GST–DPM2 was significantly co-precipitated with FLAG-PIG-A (Figure 5B, lane 2) and less efficiently with FLAG-PIG-C and -GPI1 (lanes 4 and 5) but not with -PIG-H and -PIG-P (lanes 3 and 6). These results suggest that DPM2 binds to GPI-GnT via PIG-A, PIG-C and GPI1, and that each of the three interactions alone is not strong but together they support the stable association of DPM2 with the GPI-GnT complex.
Enhancement of GPI-GnT by DPM2
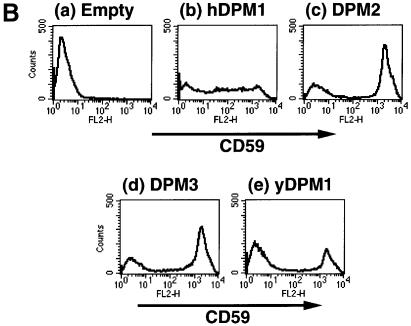
In order to determine whether DPM2 modulates GPI-GnT activity, we used Lec15 cells defective in the DPM2 (Maeda et al., 1998). We transfected DPM2 cDNA and an empty vector into Lec15/B5 (Lec15 stably transfected with cDNAs encoding CD59 and DAF as marker GPI-anchored proteins) (Maeda et al., 1998), and compared their GPI-GnT activities using microsomes (Figure 6A). Both microsomes had GPI-GnT activity, generating GlcNAc-PI and GlcN-PI; however, microsomes from the DPM2-transfectants (Figure 6A, lane 3) had a significantly higher activity than those from the vector-transfectants (lane 1). All the microsomes had similar levels of Dol-P-Glu synthase activity, a control ER enzyme (Figure 6A, bottom panel). To estimate transfection efficiencies, we analyzed the surface expression of GPI-anchored proteins by flow cytometry (Figure 6B). DPM2 (Figure 6B, c), but not an empty vector (Figure 6B, a), restored the surface expression of GPI-anchored proteins on 68% of Lec15/B5 cells. Taking this transfection efficiency into consideration, we evaluated that the DPM2-transfected cells had 3.0-fold higher GPI-GnT activity than the vector-transfected cells. Similar levels of enhancement of GPI-GnT by DPM2 were seen in two subsequent repeated experiments. These results indicate that DPM2 enhances GPI-GnT activity, although it is not an essential component of GPI-GnT.
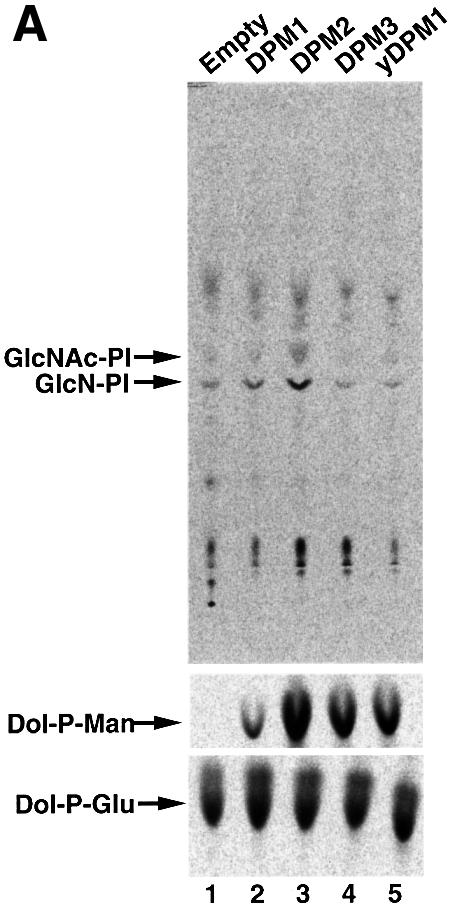
Fig. 6. GPI-GnT activity of Lec15/B5 cells transfected with various vectors. (A) GPI-GnT (top panel), Dol-P-Man synthase (middle) and Dol-P-Glu synthase (bottom) activities of Lec15/B5 cells transfected with an empty vector (lane 1), DPM1 (lane 2), DPM2 (lane 3), DPM3 (lane 4) or yeast DPM1 (lane 5). Cell lysates were incubated with radiolabeled UDP-GlcNAc (top), GDP-mannose (middle) or UDP-glucose (bottom). The lipids were analyzed by TLC. The identities of spots are indicated on the left. (B) Restoration of the surface expression of GPI-anchored proteins on Lec15/B5 cells transfected with an empty vector (a), DPM1 (b), DPM2 (c), DPM3 (d) or yeast DPM1 (e).
Because the transfection of DPM2 restores synthesis of Dol-P-Man, we tested whether Dol-P-Man alone enhances GPI-GnT. It is known that human DPM1, DPM3 and yeast DPM1 can restore synthesis of Dol-P-Man in Lec15 cells in the absence of DPM2 (Beck et al., 1990; Maeda et al., 1998, 2000). We transfected these three genes into Lec15/B5 cells. Human DPM3 and yeast DPM1 restored the surface expression of GPI-anchored proteins to the level in DPM2-transfectants on 68 and 35% of cells, respectively (Figure 6B, d and e), and human DPM1 partially restored expression (panel b) as described previously (Maeda et al., 1998). Microsomes from human DPM1-, human DPM3- and yeast DPM1-transfectants had 5.5, 40 and 27% of Dol-P-Man synthase activity compared with those from DPM2-transfectants (Figure 6A, middle panel, lanes 1–5). We saw no enhancement of GPI-GnT activity in these cells (Figure 6A, upper panel, lanes 2, 4 and 5), indicating that in the absence of DPM2, Dol-P-Man does not enhance GPI-GnT activity.
Discussion
Isolated active GPI-GnT consists of seven proteins
By a two-step affinity purification, we isolated active GPI-GnT from human cells expressing tandem-tagged PIG-A (Figure 1A). Two GPI-GnT preparations isolated using different combinations of tags contained six common proteins (bands 1–6) in addition to tagged PIG-A (Figure 1B). A non-relevant protein isolated in a similar manner did not contain these proteins, indicating the specific association of these six proteins with PIG-A. We concluded that bands 1–3 correspond to GPI1, PIG-C and PIG-H, respectively, based on molecular size and N-terminal sequence. Therefore, GPI-GnT contained three additional proteins. Among them, band 4 was a new protein that we called PIG-P. Band 5 was DPM2, a protein known to be a positive regulator of Dol-P-Man synthase. Band 6, a 5 kDa protein, has yet to be molecularly cloned.
PIG-P, the fifth component of GPI-GnT
We demonstrated that PIG-P associates with PIG-A efficiently and with GPI1 less efficiently (Figure 4). PIG-P consists of a short hydrophilic N-terminal region (15 residues), TM1, a short hydrophilic region (19 residues), TM2 and a major hydrophilic region (∼60 residues) (Figure 2). According to the membrane topology prediction program (Smith et al., 1996), the N-terminus faces the cytoplasmic side, suggesting that the major hydrophilic region resides on the cytoplasmic side of the ER. The major portion of PIG-A also resides on the cytoplasmic side, followed by a transmembrane domain and a short lumenal region (Watanabe et al., 1996). It is likely that PIG-P associates with PIG-A through the major hydrophilic region on the cytoplasmic side, although another possibility is that the association is between their transmembrane domains.
We showed that PIG-P is essential for GPI-GnT activity. A T cell clone 2.10 (–) completely deficient in the surface expression of multiple GPI-anchored proteins (Figure 3A) and in GlcNAc-PI biosynthesis (Figure 3B) lacks the PIG-P transcript (Figure 3C). Biosynthesis of GlcNAc-PI and the surface expression of GPI-anchored proteins were restored by transfection of PIG-P cDNA (Figure 3A and B).
Therefore, at least five proteins, PIG-A, PIG-H, PIG-C, GPI1 and PIG-P, are important for GPI-GnT activity. We transfected cDNAs encoding these five components into CHO and JY cells, and found that both transfectants had two to three times the GPI-GnT activity of the control transfectants (data not shown). The 5 kDa band 6 protein found in GPI-GnT (Figure 1B) should be cloned to examine whether the GPI-GnT activity is intensified when this component is also overexpressed.
PIG-P is conserved among various eukaryotes (Figure 2C). It is not, however, possible to predict the function of PIG-P from its sequence because it has no significant homology with other proteins. According to the membrane topology prediction, only a less conserved hydrophilic region of 19 residues between TM1 and TM2 would be on the lumenal side. This orientation suggests that the functional sites of PIG-P reside either within the membrane or on the cytopasmic side of the ER, consistent with the idea that transfer of GlcNAc to PI occurs on the cytoplasmic surface (Vidugiriene and Menon, 1993; Watanabe et al., 1996).
We reported that mammalian GPI-GnT uses bovine PI more efficiently than soybean PI, suggesting that GPI-GnT recognizes alkyl and/or acyl chains of PI (Watanabe et al., 1998). There is a report that a minor pool of PI is used for protein GPI-anchor biosynthesis in Leishmania mexicana (Ralton and McConville, 1998). These substrate specificities for PI could be determined by a component bearing hydrophobic domains with a conserved sequence. PIG-P and PIG-C have such characteristics.
Association of DPM2 with GPI-GnT
GPI-GnT contained DPM2, which is one of the three subunits of Dol-P-Man synthase (Maeda et al., 1998). The two other subunits of Dol-P-Man synthase, DPM1 and DPM3, were not found in the isolated GPI-GnT (Figure 1). Within Dol-P-Man synthase, the catalytic DPM1 is stabilized by its association with DPM3, and DPM3 is stabilized by its association with DPM2 (Maeda et al., 2000). It is unlikely that DPM1 and DPM3 had associated with GPI-GnT in the ER membrane and dissociated during the isolation procedure, because the associations of DPM1 with DPM3 and of DPM3 with DPM2 were stable in a buffer containing 1% digitonin, which was used for the extraction and purification of GPI-GnT (Figure 5). Moreover, tagged DPM1 co-expressed with tagged PIG-A, PIG-C, PIG-H, GPI1 and PIG-P did not show a significant association with the complex of the five other proteins (Figure 5A). In contrast, tagged DPM2 associated efficiently with the complex of tagged PIG-A, PIG-C, PIG-H, GPI1 and PIG-P (Figure 5A). Therefore, GPI-GnT specifically contains DPM2 and not DPM1 and DPM3.
The associations of tagged DPM2 with each of the five other proteins were either very weak (with PIG-A, PIG-C and GPI1) or not significant (PIG-H and PIG-P) (Figure 5B), indicating that DPM2 associates with GPI-GnT via multiple interactions that together support the stable association. DPM2 is a hydrophobic protein of 84 amino acids consisting of two major hydrophobic domains and three short hydrophilic regions (Maeda et al., 1998). It is likely that DPM2 associates with PIG-A, PIG-C and GPI1 within the membrane.
The molar ratio of DPM2 to PIG-P in GPI-GnT was about one to one as determined from the amounts of PTH amino acids detected by sequencing. The relative intensity of silver-stained bands of DPM2 and PIG-P varied to some extent among samples of GPI-GnT (our unpublished result) but DPM2 is clearly a major rather than a minor component of GPI-GnT. These results indicate that there are two populations of DPM2, one contained in Dol-P-Man synthase and the other in GPI-GnT.
Enhancement of GPI-GnT activity by DPM2
DPM2 is not an essential component of GPI-GnT because Lec15 cells that lack DPM2 (Maeda et al., 1998) have GPI-GnT activity (Figure 6A, lane 1) and synthesize the third intermediate of GPI-anchor biosynthesis, glucosaminyl acyl-PI (GlcN-acyl-PI) (Camp et al., 1993). When DPM2 cDNA was transfected into Lec15 cells, GPI-GnT activity was enhanced 3-fold compared with the level in vector-transfected Lec15 cells (Figure 6A, lane 3), suggesting that association of DPM2 enhances GPI-GnT activity. Transfection of DPM2 cDNA also caused biosynthesis of Dol-P-Man in Lec15 cells; however, restoration of Dol-P-Man synthesis without DPM2 (achieved by transfection of DPM3 cDNA or yeast DPM1) did not enhance GPI-GnT (Figure 6A, lanes 4 and 5). Therefore, DPM2 itself is required for the enhancement of GPI-GnT, i.e. DPM2 is a non-essential, regulatory component of GPI-GnT. It is not known at the moment whether Dol-P-Man is also involved in the enhancement of GPI-GnT.
There was a possibility that GlcN-acyl-PI that accumulated in Lec15 cells might cause accumulations of earlier products, such as GlcN-PI and GlcNAc-PI, which might in turn inhibit further synthesis of GlcNAc-PI and GlcN-PI. If this feedback inhibition does occur, elimination of accumulated GlcN-acyl-PI following restoration of Dol-P-Man biosynthesis should result in increased synthesis of GlcNAc-PI. The demonstration that restoration of Dol-P-Man biosynthesis in the absence of DPM2 had no effect on GPI-GnT (Figure 6A) eliminated this possibility.
Further work is necessary to clarify the mechanism of enhancement of GPI-GnT by DPM2. DPM2 positively regulates Dol-P-Man synthase by increasing both the specific activity and the expression level of the enzyme. The complex of DPM1, DPM3 and DPM2 has ten times the specific activity to generate Dol-P-Man that the complex of DPM1 and DPM3 has (Maeda et al., 2000). DPM2 increases the level of catalytic DPM1 4- to 5-fold by stabilizing DPM3, which in turn stabilizes DPM1 (Maeda et al., 2000). Therefore, in the absence of DPM2, the activity of Dol-P-Man synthase is very weak or undetectable (Figure 6A, lane 1). The enhancement of GPI-GnT by DPM2 is ∼3-fold. It seems likely that increased levels of the components of GPI-GnT in the presence of DPM2 (Figure 5A) contribute to the enhancement of the activity.
UDP-GlcNAc, acyl-CoA, Dol-P-Man and phosphatidylethanolamine act as donors of the components of GPI. Among them, Dol-P-Man is of limited use whereas the others are of more general use. Whatever the mechanism of regulation of GPI-GnT by DPM2, it seems reasonable that the initial GPI biosynthesis enzyme is upregulated when biosynthesis of Dol-P-Man is upregulated by DPM2.
Stimulation of the initial enzyme of N-glycan biosynthesis by Dol-P-Man
It was reported that Dol-P-Man stimulates UDP-GlcNAc: Dol-P GlcNAc-1-phosphate transferase (GlcNAc-P transferase) involved in the first step of biosynthesis of lipid-linked oligosaccharide precursors of N-glycans (Kean, 1985, 1996). Exogenous Dol-P-Man added to microsomes from various cell types stimulated GlcNAc-P transferase seven to eight times (Kean, 1985). It was also reported that microsomes derived from BW5147 class E Thy-1-negative cells that lack Dol-P-Man synthase had GlcNAc-P transferase activity that was stimulated by exogenously added Dol-P-Man (Kean, 1986). This indicates that DPM1 is not required for the stimulation because class E cells are defective in DPM1 (Tomita et al., 1998). Therefore, DPM1 is not involved in the enhancement of either GlcNAc-P transferase or GPI-GnT. It is unknown whether DPM2 is involved in enhancement of GlcNAc-P transferase. It would be interesting to examine whether exogenously added Dol-P-Man stimulates GlcNAc-P transferase in microsomes from Lec15 cells.
There are reports that isoproterenol and estrogen treatment that enhanced protein N-glycosylation also enhanced Dol-P-Man synthase activity in mammalian cells (Banerjee et al., 1987; Carson et al., 1990). These results together with the present finding that DPM2 enhances GPI-GnT suggest that cells may have a system that coordinates protein N-glycosylation, GPI-anchoring and Dol-P-Man synthesis.
Materials and methods
Cells, culture conditions and transfection
The mouse T cell clone 2.10 GPI (+), its GPI (–) variant clone and their transfectants were cultured as described (Haughn et al., 1992; Marmor et al., 1999). Human B-lymphoblastoid JY25 and JY5 cells (Hollander et al., 1988) were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum. Lec15/B5 cells (Maeda et al., 1998) and their transfectants were cultured in Ham’s F-12 medium supplemented with 10% fetal bovine serum. 2.10 cells were transfected by electroporation as described previously using 10 µg of plasmid DNA (Marmor et al., 1999). The transformants were selected and maintained in culture medium supplemented with 800 µg/ml G-418. JY5 cells were electroporated at 250 V and 960 µF in a Gene Pulser (Bio-Rad) (Miyata et al., 1993). Transfectants were selected and maintained in culture medium supplemented with 200 µg/ml hygromycin B.
Plasmids
Expression plasmids pMEEB-GST–PIG-A, pMEEB-GST–PIG-H, pMEEB-GST–PIG-C, pMEEB-GPI1–GST, pMEEB-GST–ALDH, pMEEB-FLAG-PIG-A, pMEEB-FLAG-PIG-H, pMEEB-FLAG-PIG-C, pMEEB-FLAG-GPI1, pMEEB-FLAG-ALDH and pME-neo (Watanabe et al., 1996, 1998), PME-Pyori18sf-, pME-Py-DPM1, pME-Py-yeastDPM1 and pME-neo-FLAG-DPM1 (Maeda et al., 1998), and pMEEB-GST–FLAG-DPM1, pME-Py-DPM2 and pME-Py-DPM3 (Maeda et al., 2000) were described previously. pMEEB-GST–FLAG-PIG-A was generated from pMEEB-GST–FLAG-DPM1 by replacing its SalI–XbaI fragment bearing DPM1 with a SalI–XbaI fragment of pMEEB-GST–PIG-A bearing PIG-A. pMEEB-PIG-A-FLAG-HAT, pMEEB-GST–DPM1, pMEEB-GST–DPM2, pMEEB-FLAG-DPM3 and pMEEB-FLAG-PIG-P were generated from pMEEB-GPI1 by replacing its XhoI–XbaI fragment bearing GPI1 with XhoI–XbaI fragments from pBS-PIG-A-FLAG-HAT, pME-Py-GST–DPM1, pME-Py-GST–DPM2, pME-Py-FLAG-DPM3 (Maeda et al., 2000) and pME-Py-FLAG-PIG-P, respectively. pBS-PIG-A-FLAG-HAT was generated by inserting the PIG-A fragment derived from pMEEB-PIG-A-GST into pBS-FLAG-HAT cut with XhoI and MluI. pBS-FLAG-HAT was generated by quadruple ligation of pBS cut with PstI and BamHI and three chemically synthesized oligo DNA fragments. One oligo DNA fragment, containing PstI and MluI sites and FLAG-tag, was generated by annealing two single stranded DNAs, 5′-GACGCGTGACTACAA GGACGACGATGACAAGG and 5′-TCGACCTTGTCATCGTCGTC CTTGTAGTCACGCGTCTGCA. Two other oligo DNA fragments each containing the first and second half of the HAT-tag were generated by annealing oligonucleotides 5′-TCGACAAGGATCATCTCATCCAC AATGTC and 5′-TTTGTGGACATTGTGGATGAGATGATCCTTG, and 5′-CACAAAGAGGAGCACGCTCATGCCCACAACAAGTAGG and 5′-GATCCCTACTTGTTGTGGGCATGAGCGTGCTCCTC, respectively. pME-neo-PIG-P was generated by ligation of an SfiI fragment containing the neo resistance gene derived from pME-neo-GST–PIG-H and an SfiI fragment of pMEEB-PIG-P containing pME-PIG-P.
Purification of GPI-GnT complexes
Purification of proteins double-tagged by GST and FLAG was as described previously (Maeda et al., 2000). Briefly, JY5 cells transfected with pMEEB-GST–FLAG-PIG-A (1.1 × 1010 cells) were solubilized in 1 l of lysis buffer A [1% digitonin, 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.1 mM tosyl-lysine chloromethyl ketone (TLCK) and 2 µg/ml leupeptin] at 4°C for 2 h. After the removal of insoluble materials by centrifugation at 100 000 g and 4°C for 1 h, the supernatant was mixed with 0.4 ml of anti-FLAG-M2 beads (Sigma) and agitated overnight. The beads were collected and washed with lysis buffer A. The bound proteins were eluted with 1.5 ml of 1 mg/ml FLAG-peptide in the same buffer. The eluate was incubated with 40 µl of gluthathione beads (Pharmacia) for 30 min at room temperature. The beads were collected, washed with lysis buffer A and eluted with 40 µl of 2× concentrated SDS–PAGE sample buffer. The eluted sample was subjected to 10–20% gradient SDS–PAGE and transferred to PVDF membrane (ProBlott, Applied Biosystems Inc.). The N-terminal sequences of proteins revealed by staining with Coomassie Brilliant Blue R250 were determined with a 494 sequencer (Applied Biosystems Inc.).
For purification of FLAG-HAT-tagged PIG-A, we diluted the eluate from anti-FLAG beads ten times with lysis buffer A containing 50 mM Tris–HCl pH 8 instead of 20 mM Tris–HCl pH 7.5, and purified with TALON Superflow Metal Affinity Resin (Clontech) instead of glutathione beads. When we isolated double-tagged PIG-A proteins for silver staining and in vitro GPI-GnT activity, we used 1.4 × 108 cells and eluted the proteins from glutathione beads and Metal Affinity Resin with elution buffer A (20 mM reduced glutathione, 1% digitonin, 50 mM Tris–HCl pH 9.5 and 150 mM NaCl) and elution buffer B (100 mM imidazole, 1% digitonin, 50 mM Tris–HCl pH 8 and 150 mM NaCl), respectively.
Assay for GPI-GnT
The GPI-GnT complexes eluted from glutathione beads were collected with anti-FLAG M2 beads and used for GPI-GnT assay because GPI-GnT is inhibited by glutathione. Anti-FLAG beads bearing protein complexes and eluates from Metal Affinity Resin were incubated with 100 µl of a GPI-GnT reaction mixture [2 µCi of UDP-6[3H]GlcNAc (American Radiolabeled Chemicals, MO), 100 µM bovine PI (Sigma), 50 mM HEPES–NaOH pH 7.4, 25 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 1 mM ATP, 0.5 mM dithiothreitol, 0.2 µg/ml tunicamycin, 0.1 mM TLCK and 1 µg/ml leupeptin] for 4 h at 37°C. For anti-FLAG beads, we added 1 mg/ml FLAG peptide to liberate GPI-GnT complexes. The reactions were terminated by adding 1 ml of chloroform:methanol (1:1) and the solutions were dried. The lipids were extracted by 1-butanol partition, separated by thin layer chromatography (TLC) and analyzed by an Image Analyzer BAS 1500 (Fuji Film Co., Tokyo) (Watanabe et al., 1998).
Cloning human PIG-P cDNA
We searched the Expressed Sequence Tag (EST) database (National Center for Biotechnology Information, Bethesda, MD) using a tBLASTn program (Altschul et al., 1990) for sequences corresponding to the amino acid sequence of band 4 and found several human PIG-P ESTs. Based on their sequences, we synthesized the primers PIG-PU1 (5′-GCT CGGCTCGAGGTCTAAAGCCCCAGGAAAAATGGT) and PIG-PL1 (5′-GCTCGGTCTAGAGTGTTACTATGGTTACACACAGTTCA) and amplified the coding region by PCR from a HeLa cell cDNA library (Miyata et al., 1993). We subcloned the PCR product into the EcoRV site of pBluescript and confirmed the sequence. To make pMEEB-PIG-P for expression in mammalian cells, the XhoI–XbaI fragment including PIG-P derived from the pBluescript bearing PIG-P was ligated into the pMEEB vector cut with XhoI and XbaI. To fuse a FLAG tag to the N-terminus of PIG-P, we designed a primer (PIG-P N-SalI, 5′-GCTCGGGTCGAC GTGGAAAATTCACCGTCGCCATTG) to replace the initiation methionine codon with the sequence of a SalI site. Using primers PIG-P N-SalI and PIG-P L1, we amplified the coding region of PIG-P by PCR, digested it with SalI and XbaI and ligated it with the SalI–XbaI fragment of pME-Py-FLAG derived from pME-Py-FLAG-hGPI8 (Ohishi et al., 2000).
FACS analysis
Cells were stained for Thy-1 (2.5 µg/ml biotinylated G7), CD59 (10 µg/ml biotinylated 5H8), CD48 [1:25 dilution of phycoerythrin (PE)-conjugated anti-CD48] and Sca1 (1:100 dilution of rat anti-Sca1 IgG). The secondary reagents used were Phycoprobe PE Streptavidin (Biomeda) and PE-conjugated anti-Rat IgG. Stained cells were analyzed in a FACS caliber (Becton-Dickinson).
Complementation analysis with somatic cell fusion and immunofluorescence staining
Somatic cell fusion and immunofluorescence staining using anti-Thy-1 antibody were done as described previously (Takahashi et al., 1993).
RT–PCR
Total RNA was extracted from 5 × 106 cells of wild-type 2.10 GPI (+) and mutant 2.10 GPI (–) cells using TRIzol reagent (Gibco-BRL), treated with RT-grade deoxyribonuclease (Wako, Japan) and reverse transcribed using random primers and SuperscriptII (Gibco-BRL). PCRs were hot started and followed by cycles of a reaction consisting of 95°C for 30 s, 63°C for 30 s and 68°C for 2 min using two sets of primers for mouse PIG-P and GPI1. The primers used for the amplification of the full coding region of PIG-P were 5′-GTGGAAAATTCACCGTCGCCATTG and 5′-GTTTTAGGTATTGAGTTCTTTGGCTC. The primers used for the amplification of mouse GPI1 were 5′-TATACTCCCTTGGCCTTCG ACTCTG and 5′-TTTGGGACGGTTGAGAACCACTGTGC.
Analyses of protein interactions
To analyze the interactions of PIG-P with other components of GPI-GnT, we co-transfected 15 µg of pMEEB-FLAG-PIG-P together with 15 µg of pMEEB-GST–PIG-A, –PIG-H, –PIG-C, –GPI1 or –ALDH into JY5 cells. Cells (6–10 × 107) were solubilized in 6 ml of lysis buffer A at 4°C for 2 h. After removal of insoluble materials by centrifugation at 100 000 g and 4°C for 1 h, the supernatants were mixed with 20 µl of anti-FLAG-M2 beads and agitated overnight at 4°C. The beads were collected and the supernatants were mixed with 20 µl of glutathione beads and agitated for 3 h at 4°C. Both beads were washed with lysis buffer A, eluted with 2× sample buffer and analyzed by SDS–PAGE and western blotting using biotinylated anti-FLAG-M2 antibody plus horseradish peroxidase-conjugated streptavidin (Amersham Life Science) or goat anti-GST antibody plus horseradish peroxidase-conjugated protein G (Bio-Rad) (Maeda et al., 1998). To see the interaction of DPM2 with GPI-GnT complexes, we first transfected JY5 cells with pMEEB-GST–DPM2, pMEEB-GST–DPM1 or pMEEB-GST–ALDH. We further transfected these cells with a mixture of pMEEB-FLAG-PIG-A, -PIG-H, -PIG-C, -GPI1 and -PIG-P or of pMEEB-FLAG-ALDH and -DPM3. We first precipitated GST fusion proteins with glutathione beads, and then the unbound FLAG-tagged proteins with anti-FLAG-M2 beads. Both precipitates were analyzed by SDS–PAGE/western blotting as described above. To reveal the interaction of DPM2 with each of the GPI-GnT components, we transfected 30 µg of pMEEB-FLAG-ALDH, -PIG-A, -PIG-H, -PIG-C, -GPI1, -PIG-P or -DPM3 into GST–DPM2-transfected cells and analyzed the interactions between proteins as described above.
In vitro assays of GPI-GnT, Dol-P-Man synthase and Dol-P-Glu synthase
Lec 15/B5 (1.5 × 107) cells were transfected with 30 µg each of pME-Py, pME-Py-DPM1, pME-Py-DPM2, pME-Py-DPM3 or pME-Py-yeast DPM1, cultured for 2 days and incubated in medium containing 5 µg/ml tunicamycin (Sigma) for 2 h. After collection, cell lysates (1 ml) were prepared (Watanabe et al., 1998) and stored at –80°C until use. Samples of 0.8 ml of cell lysates were used for the GPI-GnT assay, and 60 µl each of the cell lysates were used for Dol-P-Man and Dol-P-Glu synthase assays (Maeda et al., 1998). GPI-GnT activity was measured by incubation with 150 µl of GPI-GnT reaction mixture for 30 min at 37°C.
Acknowledgments
Acknowledgements
We thank Kazuhito Ohishi and Yeongjin Hong for discussion and Keiko Kinoshita for technical assistance. This work was supported by grants from the Ministry of Education, Science, Sports and Culture of Japan.
References
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Banerjee D.K., Kousvelari,E.E. and Baum,B.J. (1987) cAMP-mediated protein phosphorylation of microsomal membranes increases mannosylphosphodolichol synthase activity. Proc. Natl Acad. Sci. USA, 84, 6389–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck P.J., Orlean,P., Albright,C., Robbins,P.W., Gething,M.J. and Sambrook,J.F. (1990) The Saccharomyces cerevisiae DPM1 gene encoding dolichol-phosphate-mannose synthase is able to complement a glycosylation-defective mammalian cell line. Mol. Cell. Biol., 10, 4612–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp L.A., Chauhan,P., Farrar,J.D. and Lehrman,M.A. (1993) Defective mannosylation of glycosylphosphatidylinositol in Lec35 Chinese hamster ovary cells. J. Biol. Chem., 268, 6721–6728. [PubMed] [Google Scholar]
- Carson D.D., Farrar,J.D., Laidlaw,J. and Wright,D.A. (1990) Selective activation of the N-glycosylation apparatus in uteri by estrogen. J. Biol. Chem., 265, 2947–2955. [PubMed] [Google Scholar]
- Colussi P.A., Taron,C.H., Mack,J.C. and Orlean,P. (1997) Human and Saccharomyces cerevisiae dolichol phosphate mannose synthases represent two classes of the enzyme, but both function in Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA, 94, 7873–7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M.A. (1999) The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors and the contributions of trypanosome research. J. Cell Sci., 112, 2799–2809. [DOI] [PubMed] [Google Scholar]
- Gerold P., Schofield,L., Blackman,M.J., Holder,A.A. and Schwarz,R.T. (1996) Structural analysis of the glycosyl-phosphatidylinositol membrane anchor of the merozoite surface protein-1 and -2 of Plasmodium falciparum. Mol. Biochem. Parasitol., 75, 131–143. [DOI] [PubMed] [Google Scholar]
- Haughn L., Gratton,S., Caron,L., Sekaly,R.-P., Veillette,A. and Julius,M. (1992) Association of tyrosine kinase p56lck with CD4 inhibits the induction of growth through the αβ T-cell receptor. Nature, 358, 328–331. [DOI] [PubMed] [Google Scholar]
- Herscovics A. and Orlean,P. (1993) Glycoprotein biosynthesis in yeast. FASEB J., 7, 540–550. [DOI] [PubMed] [Google Scholar]
- Hollander N., Selvaraj,P. and Springer,T.A. (1988) Biosynthesis and function of LFA-3 in human mutant cells deficient in phosphatidylinositol-anchored proteins. J. Immunol., 141, 4283–4290. [PubMed] [Google Scholar]
- Hong Y., Ohishi,K., Watanabe,R., Endo,Y., Maeda,Y. and Kinoshita,T. (1999) GPI1 stabilizes an enzyme essential in the first step of glycosylphosphatidylinositol biosynthesis. J. Biol. Chem., 274, 18582–18588. [DOI] [PubMed] [Google Scholar]
- Hyman R. (1988) Somatic genetic analysis of the expression of cell surface molecules. Trends Genet., 4, 5–8. [DOI] [PubMed] [Google Scholar]
- Inoue N., Watanabe,R., Takeda,J. and Kinoshita,T. (1996) PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis is a homologue of Saccharomyces cerevisiae GPI2. Biochem. Biophys. Res. Commun., 226, 193–199. [DOI] [PubMed] [Google Scholar]
- Kamitani T., Chang,H.M., Rollins,C., Waneck,G.L. and Yeh,E.T.H. (1993) Correction of the class H defect in glycosyl phosphatidylinositol anchor biosynthesis in Ltk– cells by a human cDNA clone. J. Biol. Chem., 268, 20733–20736. [PubMed] [Google Scholar]
- Kapteyn J.C., van den Ende,H. and Klis,F.M. (1999) The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta, 1426, 373–383. [DOI] [PubMed] [Google Scholar]
- Kean E.L. (1985) Stimulation by dolichol phosphate-mannose and phospholipids of the biosynthesis of N-acetylglucosaminyl pyrophosphoryl dolichol. J. Biol. Chem., 260, 12561–12571. [PubMed] [Google Scholar]
- Kean E.L. (1986) Stimulation by dolichol phosphate-mannose of N-acetylglucosaminyl-lipid biosynthesis by membranes from class E Thy-1-negative mutant mouse lymphoma cells which are defective in dolichol phosphate-mannose biosynthesis. Arch. Biochem. Biophys., 250, 146–152. [DOI] [PubMed] [Google Scholar]
- Kean E.L. (1996) Site of stimulation by mannosyl-P-dolichol of GlcNAc-lipid formation by microsomes of embryonic chick retina. Glycoconj. J., 13, 675–680. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Inoue,N. and Takeda,J. (1995) Defective glycosyl phosphatidylinositol anchor synthesis and paroxysmal nocturnal hemoglobinuria. Adv. Immunol., 60, 57–103. [DOI] [PubMed] [Google Scholar]
- Kinoshita T., Ohishi,K. and Takeda,J. (1997) GPI-anchor synthesis in mammalian cells: genes, their products and a deficiency. J. Biochem., 122, 251–257. [DOI] [PubMed] [Google Scholar]
- Kyte J. and Doolittle,R.F. (1982) A simple method for displaying the hydropathic character of a protein. J. Mol. Biol., 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Leidich S.D., Drapp,D.A. and Orlean,P. (1994) A conditionally lethal yeast mutant blocked at the first step in glycosyl phosphatidylinositol anchor synthesis. J. Biol. Chem., 269, 10193–10196. [PubMed] [Google Scholar]
- Maeda Y., Tomita,S., Watanabe,R., Ohishi,K. and Kinoshita,T. (1998) DPM2 regulates biosynthesis of dolichol phosphate-mannose in mammalian cells: correct subcellular localization and stabilization of DPM1 and binding of dolichol phosphate. EMBO J., 17, 4920–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda Y., Tanaka,S., Hino,J., Kangawa,K. and Kinoshita,T. (2000) Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2 and DPM3. EMBO J., 19, 2475–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor M.D., Bachmann,M.F., Ohashi,P.S., Malek,T.R. and Julius,M. (1999) Immobilization of glycosylphosphatidylinositol-anchored proteins inhibits T cell growth but not function. Int. Immunol., 11, 1381–1393. [DOI] [PubMed] [Google Scholar]
- Masaki R., Yamamoto,A. and Tashiro,Y. (1994) Microsomal aldehyde dehydrogenase is localized to the endoplasmic reticulum via its carboxyl-terminal 35 amino acids. J. Cell Biol., 126, 1407–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masterson W.J., Doering,T.L., Hart,G.W. and Englund,P.T. (1989) A novel pathway for glycan assembly: biosynthesis of the glycosyl-phosphatidylinositol anchor of the trypanosome variant surface glycoprotein. Cell, 56, 793–800. [DOI] [PubMed] [Google Scholar]
- Miyata T., Takeda,J., Iida,Y., Yamada,N., Inoue,N., Takahashi,M., Maeda,K., Kitani,T. and Kinoshita,T. (1993) Cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science, 259, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Nozaki M., Ohishi,K., Yamada,N., Kinoshita,T., Nagy,A. and Takeda,J. (1999) Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab. Invest., 79, 293–299. [PubMed] [Google Scholar]
- Ohishi K., Inoue,N., Maeda,Y., Takeda,J., Riezman,H. and Kinoshita,T. (2000) Gaa1p and gpi8p are components of a glycosyl phosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell, 11, 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralton J.E. and McConville,M.J. (1998) Delineation of three pathways of glycosylphosphatidylinositol biosynthesis in Leishmania mexicana. Precursors from different pathways are assembled on distinct pools of phosphatidylinositol and undergo fatty acid remodeling. J. Biol. Chem., 273, 4245–4257. [DOI] [PubMed] [Google Scholar]
- Schultz C., Gilson,P., Oxley,D., Youl,J. and Bacic,A. (1998) GPI-anchors on arabinogalactan-proteins: implication for signalling in plants. Trends Plant Sci., 3, 426–431. [Google Scholar]
- Smith R.F., Wiese,B.A., Wojzynski,M.K., Davison,D.B. and Worley,K.C. (1996) BCM Search Launcher—an integrated interface to molecular biology data base search and analysis services available on the World Wide Web. Genome Res., 6, 454–462. [DOI] [PubMed] [Google Scholar]
- Stevens V.L. and Raetz,C.R. (1991) Defective glycosyl phosphatidyl inositol biosynthesis in extracts of three Thy-1 negative lymphoma cell mutants. J. Biol. Chem., 266, 10039–10042. [PubMed] [Google Scholar]
- Takahashi M. et al. (1993) Deficient biosynthesis of N-acetylglucos aminyl phosphatidylinositol, the first intermediate of glycosyl phosphatidylinositol anchor biosynthesis, in cell lines established from patients with paroxysmal nocturnal hemoglobinuria. J. Exp. Med., 177, 517–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda J., Miyata,T., Kawagoe,K., Iida,Y., Endo,Y., Fujita,T., Takahashi,M., Kitani,T. and Kinoshita,T. (1993) Deficiency of the GPI anchor caused by a somatic mutation of the PIG-A gene in paroxysmal nocturnal hemoglobinuria. Cell, 73, 703–711. [DOI] [PubMed] [Google Scholar]
- Tarutani M., Itami,S., Okabe,M., Ikawa,M., Tezuka,T., Yoshikawa,K., Kinoshita,T. and Takeda,J. (1997) Tissue specific knock-out of the mouse Pig-a gene reveals important roles for GPI-anchored proteins in skin development. Proc. Natl Acad. Sci. USA, 94, 7400–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita S., Inoue,N., Maeda,Y., Ohishi,K., Takeda,J. and Kinoshita,T. (1998) A homologue of Saccharomyces cerevisiae Dpm1p is not sufficient for synthesis of dolichol-phosphate-mannose in mammalian cells. J. Biol. Chem., 273, 9249–9254. [DOI] [PubMed] [Google Scholar]
- Udenfriend S. and Kodukula,K. (1995) How glycosylphosphatidyl inositol-anchored membrane proteins are made. Annu. Rev. Biochem., 64, 563–591. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J. and Menon,A.K. (1993) Early lipid intermediates in glycosyl-phosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol., 121, 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Kinoshita,T., Masaki,R., Yamamoto,A., Takeda,J. and Inoue,N. (1996) PIG-A and PIG-H, which participate in glycosylphosphatidylinositol anchor biosynthesis, form a protein complex in the endoplasmic reticulum. J. Biol. Chem., 271, 26868–26875. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue,N., Westfall,B., Taron,C.H., Orlean,P., Takeda,J. and Kinoshita,T. (1998) The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J., 17, 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]