Abstract
The application of molecular diagnostic techniques along with nucleotide sequence determination to permit contemporary phylogenetic analysis of European field isolates of equine infectious anemia virus (EIAV) has not been widely reported. As a result, of extensive testing instigated following the 2006 outbreak of equine infectious anemia in Italy, 24 farms with a history of exposure to this disease were included in this study. New PCR-based methods were developed, which, especially in the case of DNA preparations from peripheral blood cells, showed excellent correlation with OIE-approved agar gel immunodiffusion (AGID) tests for identifying EIAV-infected animals. In contrast, the OIE-recommended oligonucleotide primers for EIAV failed to react with any of the Italian isolates. Similar results were also obtained with samples from four Romanian farms. In addition, for the first time complete characterization of gag genes from five Italian isolates and one Romanian isolate has been achieved, along with acquisition of extensive sequence information (86% of the total gag gene) from four additional EIAV isolates (one Italian and three Romanian). Furthermore, in another 23 cases we accomplished partial characterization of gag gene sequences in the region encoding the viral matrix protein. Analysis of this information suggested that most Italian isolates were geographically restricted, somewhat reminiscent of the “clades” described for human immunodeficiency virus type 1 (HIV-1). Collectively this represents the most comprehensive genetic study of European EIAV isolates conducted to date.
The host range of equine infectious anemia virus (EIAV), a lentivirus related to HIV-1, is restricted to members of the Equidae. Clinical signs following exposure to this virus can range from undetectable to life threatening, although pyrexia and thrombocytopenia are frequently reported in acute cases (6, 26). The course of the disease, equine infectious anemia (EIA), is also variable, although many infected animals will experience multiple recurrent febrile disease episodes lasting at least 12 months, followed by a prolonged period, the “inapparent carrier stage,” where they appear clinically normal but remain as active reservoirs for the virus (6, 26).
As there are no effective vaccines, control of EIA is currently dependent on serological diagnostic assays for the identification of infected animals prior to their removal from the general population, thereby preventing subsequent transmission. Although a number of EIA enzyme-linked immunosorbent assays (ELISAs) are available, the only method that has been shown to correlate with the presence of virus in horse inoculation tests is the Coggins or agar gel immunodiffusion (AGID) assay (3, 22). Therefore, in most countries, this assay has been adopted as the officially recognized test, and very often it is mandatory that positive reactions in any EIA ELISA be confirmed by AGID testing before regulatory actions are taken. Unfortunately, despite its excellent specificity, the AGID test has a number of deficiencies. These include a lack of sensitivity, because relatively large amounts of antibody are required to generate precipitin lines, and the fact that interpretation of results can be highly subjective. Furthermore, there can be significant delays between initial exposure and the first positive serological response. Under carefully controlled experimental conditions with known virus strains, most horses produce positive reactions in AGID tests within 45 days of infection with EIAV (9). However, the situation in the field is more variable, and time intervals of as long as 157 days between infection and the first seropositive reaction have been reported (7). Therefore, during the initial stages of infection, there is certainly a requirement for alternative forms of diagnostic testing involving the detection of virus or virus-derived products. Unfortunately, in vitro replication of wild-type strains of EIAV is restricted to equid leukocyte cultures, and these are too variable and insensitive for routine diagnostic use. While PCR-based methods represent a potentially highly sensitive alternative for the detection of viral nucleic acids, these techniques require extensive nucleotide sequence information. Recently, however, an increase in the number of viral sequences from EIAV field isolates submitted to the public databases from different parts of the world has facilitated the design of reverse transcription-PCR (RT-PCR) and PCR assays with the potential to be more broadly reactive.
The 2006 EIA outbreak in Italy, involving several high-profile cases in valuable racehorses, precipitated the introduction of mandatory AGID testing for all equids older than 6 months, with the exception of animals imported or bred for food production. This represented a significant departure from previous testing requirements, as since 1995 regulations to control EIA had been restricted to the most economically important areas of the equine industry and compulsory serological screening for EIA using the AGID test was required only for stallions at stud and horses destined for export. Consequently, during this period, the majority of the equid population went untested and so the actual prevalence of EIAV in Italy was an unknown quantity, although serologically positive individuals were occasionally identified in unofficial surveys. Furthermore, positive EIA AGID test results were regularly reported to occur in horses for human or animal consumption, either imported (mainly from Eastern Europe) or locally bred. It is not surprising, therefore, that the renewed interest in the disease along with a greatly expanded testing program resulted in the identification of many new cases of EIA unrelated to the 2006 outbreak. This has provided an unprecedented opportunity to systematically evaluate PCR- and RT-PCR-based methods for the detection of EIAV proviral DNA and viral RNA in the context of field isolates with unknown genotypes. Furthermore, sequence analysis of PCR-generated products has enabled us to study the molecular epidemiology of natural EIAV infections in Central Europe.
Genetic variation is a hallmark of the lentiviruses, and EIAV is no exception. The viral genotype in part determines clinical outcome following initial exposure (4, 20), and the ability to undergo extensive genetic/antigenic variation without loss of infectivity, thereby avoiding elimination by the immune response, is one of the major strategies employed in the establishment of a persistent infections within the host (11-16, 19). At one time, it was believed that most of this variation occurred within sequences encoding the viral envelope glycoproteins plus the Rev regulatory protein (1, 13, 14, 21, 25, 28, 29) and that EIAV genes encoding the group-specific antigens (gag) and replicase proteins (pol) were relatively conserved both during the course of infection within an individual host and between geographically distinct isolates (25). However, recent studies have suggested that in addition to env/rev, there may be significant between-isolate variation in other major viral genes, such as gag (23), although more work is needed before the full extent of this variation is appreciated. Despite this considerable capacity to undergo change at the genetic level, some lentiviruses, such as HIV-1, can be classified into distinct genomic subtypes or clades based on their geographical location (reviewed in reference 27). It remains to be determined if geographically restricted genetic subtypes or clades exist in EIAV.
MATERIALS AND METHODS
Sample collection.
Samples were collected during a 4-year period from 2006 until 2009 (collection dates are indicated in the GenBank record). Spleen, liver, bone marrow, and lymph node tissues were harvested shortly after death or euthanasia from adult horses and foals on those farms (farms 1 to 3) at the epicenter of the 2006 outbreak, immediately frozen in liquid nitrogen, and maintained at −80°C until extraction of nucleic acids. As a result of heightened awareness and increased mandatory AGID testing, a number of additional EIA cases that were not connected with the 2006 outbreak on farms 1 to 3 (defined as being geographically distinct with no shared personnel and no evidence for the use of contaminated plasma products) were identified. Although 12 of these new cases had overt clinical signs, the majority of EIAV-infected equids were asymptomatic at the time of detection, suggesting that they had attained inapparent carrier status. In total, blood and/or tissue samples were collected from 400 horses maintained on 24 farms predominantly in northern and central regions of Italy along with 4 farms in the Transylvanian region of Romania. Included in this total were all potential “in-contact” equids on each farm where infection with EIAV was diagnosed (see Tables 2 and 3).
All blood samples were collected by licensed veterinary practitioners using jugular venipuncture into sterile glass evacuated tubes for serum or with EDTA for buffy coat preparations.
Nucleic acid extraction.
Total RNA was extracted from 100 mg of homogenized tissue sample or buffy coat cells derived from 5 ml of whole blood using commercially available reagents (Aurum Total RNA Fatty and Fibrous Tissue kit; Bio-Rad, Hercules, CA) according to the manufacturer's recommendations.
Genomic DNA was extracted from 200 μl of whole blood with a GenElute blood genomic DNA kit (Sigma, St. Louis, MO) following the manufacturer's instructions. Nucleic acid concentrations were determined using a spectrophotometer (DNA) or fluorometer (RNA), with the integrity of these samples monitored by gel electrophoresis.
Primer design.
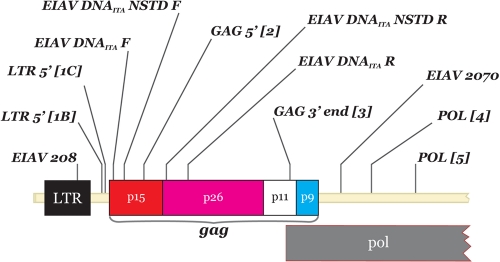
The primers used in this study (Table 1; Fig. 1) were designed using Primer3 (http://frodo.wi.mit.edu/primer3/) and based on consensus sequences derived from previously published studies (18, 23) and/or previously submitted to GenBank (accession numbers AB008196, AB008197, AF016316, AF028232, AF247394, AF327877, AF327878, AF033820, EF418582, EF418583, EF418584, EF418585, EU240733, EU375543, EU375544, EU741609, M16575, and NC001450).
TABLE 1.
Primers and reaction conditions for EIAV gag gene amplification and diagnostic PCR
| Assay | Primer name, sequence | PCR conditions (30 cycles)a | Amplicon length (bp) | Template | Reference or source |
|---|---|---|---|---|---|
| Full length, no. 1, first round | EIAV 208, CGGTCTGAGTCCCTTCTCTG | 94°C, 20 s; 62°C, 30 s; 68°C, 2 min | 1,862 | cDNA/DNA | 23 |
| EIAV 2070, TTCCACATTTCCTCCCACTC | |||||
| Full length, no. 1, second round | EIAV 450, ATGGGAGACCCAGTGACATGGAGCAAA | 94°C, 20 s; 68°C, 2 min | 1,429 | cDNA/DNA | 23 |
| EIAV 1879, CTCCCACAAACTGTTCAAATTGAGATCCT | |||||
| Full length, no. 2, first round | LTR 5′ [1B], GGACAGCAGAGGAGAACTTACAGA | 94°C, 20 s; 52°C, 20 s; 68°C, 2 min | 2,234 | cDNA/DNA | Consensus sequence derived |
| POL [5], GTGAAAGCTGTATATGGTCTAAACTCTGGA | |||||
| Full length, no. 2, 5′, second round | LTR 5′ [1C], GTCTTCTGGAGGTGTTCCTGGCCA | 94°C, 20 s; 57°C, 20 s; 68°C, 1 min 30 s | 1,325 | cDNA/DNA | Consensus sequence derived |
| GAG 3′ end [3], TGAGCCCCTTGYTTCCCGTTTTTTGG | |||||
| Full length, no. 2 3′, second round | GAG 5′ [2], AACATGGTGGGCAATTKYTGCTGT | 94°C, 20 s; 57°C, 20 s; 68°C, 1 min 30 s | 1,625 | cDNA/DNA | Consensus sequence derived |
| POL [4], GTGAGTGGCCATTGAGGAATTTTTGGCC | |||||
| Diagnostic PCR, first round | EIAV DNAITA F, GACATGGAGCAAAGCGCTCA | 94°C, 20 s; 66°C, 30 s; 68°C, 30 s | 547 | DNA | Consensus sequence derived |
| EIAV DNAITA R, CTGCCCAGGCACCACATCTA | |||||
| Diagnostic PCR, second round | EIAV DNAITA NSTD F, TGTGGGCGCTAAGTTTGGTG | 94°C, 20 s; 64°C, 30 s; 68°C, 30 s | 313 | DNA | Consensus sequence derived |
| EIAV DNAITA NSTD R, TTTCTGTTTCCAGCCCCATC | |||||
| cDNA check | βACTINE R, GAGCAAGAGGGGCATCCTGA | 94°C, 20 s; 64°C, 30 s; 68°C, 30 s | 184 | cDNA | 24 |
| βACTINE R, GGTCATCTTCTCGCGGTTGG |
All PCR protocols also had an initial denaturation step (94° for 2 min) and a final extension (72°C for 7 min).
FIG. 1.
Primer positions in the EIAV genome.
Reverse transcription of EIAV gag gene sequences.
Five micrograms of total RNA was reverse transcribed with random hexamer primers at 52°C for 60 min using the Superscript III cDNA synthesis kit (Invitrogen, Carlsbad, CA) according to the manufacturer's specifications. Successful reverse transcription was verified by PCR amplification of Equus caballus β-actin using the βACTINE F and βACTINE R primer pair (24) (Table 1) with a template consisting of 1 μl of undiluted cDNA from the Superscript III cDNA synthesis reaction.
Diagnostic PCR Assay.
For detection of EIAV gag sequences in diagnostic specimens, a nested PCR assay was developed (detailed in Table 1) based on consensus information from Asian, North American, and European viral isolates. For the first-stage reaction, 8 μl of undiluted cDNA or 50 to 250 ng of genomic DNA was added to 1 U Platinum Taq high-fidelity DNA polymerase (Invitrogen) and 0.2 μM of each primer in a buffer containing 2 mM MgSO4 and 50 μM of each deoxynucleoside triphosphate (dNTP). In the second-stage reaction, 5 μl of the first PCR product was combined with 1 U Platinum Taq high-fidelity DNA polymerase (Invitrogen) and 0.2 μM of each primer in a buffer containing 2 mM MgSO4 plus 50 mM of each dNTP. The thermal cycle conditions are listed in Table 1. For each assay, sterile, nuclease-free water was employed instead of cDNA or genomic DNA as negative controls, and PCR products were sequenced to ensure their identity.
PCR amplification of full-length EIAV gag gene sequences.
Complete EIAV gag gene sequences for comparative genetic analysis were amplified from cDNA or proviral DNA, again using a nested-PCR strategy (detailed in Table 1). In the first stage, 8 μl of undiluted cDNA or 50 to 250 ng of genomic DNA was mixed with 1 U Platinum Taq high-fidelity DNA polymerase (Invitrogen) and 0.2 μM of primers (Table 1) in a buffer containing 2 mM MgSO4 and 50 μM of each dNTP. In the second-stage reaction, 5 μl of the first PCR product was combined with 1 U Platinum Taq high-fidelity DNA polymerase (Invitrogen) and 0.2 μM of primers (Table 1) in buffer containing 2 mM MgSO4 and 50 mM of each dNTP. The thermal cycle conditions are listed in Table 1.
Cloning and sequencing.
Where available, full-length gag PCR products were molecularly cloned using the pCR4-TOPO vector in conjunction with the TOPO TA cloning system (Invitrogen) and subjected to nucleotide sequencing (http://www.primm.it). The DNA product fragments derived from diagnostic PCR assays were directed purified from agarose gels using the Wizard SV gel and PCR clean-up system (Promega, Madison, WI) and sequenced.
Sequence assembly.
Raw sequences were assembled with ContigExpress (VectorNTI Suite 8.0; Invitrogen) and aligned along with other EIAV reference sequences deposited in GenBank using AlignX (VectorNTI Suite 8.0; Invitrogen).
2.9. Phylogenetic analysis.
gag gene sequences from different Italian and Romanian EIAV isolates were imported into a sequence editor (BioEdit; http://www.mbio.ncsu.edu/bioedit/bioedit.html), along with equivalent information from previously published reference virus strains, and translated into their corresponding amino acids prior to alignment using the built-in ClustalX (2.0.9) algorithm. These amino acid alignments were back translated to ensure that all the nucleotide alignments generated were based on the correct open reading frame, with the final data set comprising 1,521 aligned nucleotides.
MrModeltest (http://www.abc.se/∼nylander/) and PAUP* (http://paup.csit.fsu.edu/) were used to perform a hierarchical likelihood ratio test to determine the best evolutionary model and parameter values among those supported by MrBayes, and the model HKY+I was selected based on the AIC information criterion, which ranks the competing models with negative log likelihood. The model was incorporated into MrBayes, with parameter values free to vary. The run consisted of two times four chains (one hot and three cold) of 5 million generations each, with sampling of trees and parameters every 100th generation. At the end of the search, the two runs converged appropriately to an average standard deviation of split frequencies of 0.008. Sampled parameters were imported into Tracer (1.4.1) and plotted to ensure stability. Both runs converged, and 500,000 generations were set as a burn-in of the analysis. Trees of both runs (stationary phase only) were collated and summed using MrBayes to calculate the posterior probabilities (frequency of each node in the sample trees).
Serological testing.
All the serological tests were performed with the DiaSystems EIA AGID test (Idexx, Westbrook, MA) following the manufacturer's instructions.
Nucleotide sequence accession numbers.
All assembled sequences were submitted to GenBank (see Table 3 for accession numbers).
RESULTS
Comparison between EIAV antibody testing and detection of viral nucleic acids by modified diagnostic PCR.
With the OIE-recommended PCR primers and conditions for amplification of EIAV genetic material, no products were detected in nucleic acids derived from plasma and tissue samples harvested during febrile episodes from confirmed clinical cases of EIA (data not shown) on farms 1 and 3. As these can be considered “optimal” samples in terms of high plasma and tissue viral burdens (8) for inclusion in PCR-based testing, the most likely explanation for this result is the presence of mismatches between OIE-recommended primers and European isolates of the virus. Indeed, subsequent nucleotide sequence analysis demonstrated that only OIE primer 3 (17) was homologous with sequences from the Italian EIAV isolates, with five, two, and six mismatches present in OIE primers 1, 2, and 4, respectively. In contrast, inclusion of primers designed in this laboratory in nested PCR assays with these same samples from farms 1 and 3 produced a 547-bp fragment after the first round of amplification and a 313-bp fragment following the second stage. Furthermore, these fragments were confirmed as EIAV specific by nucleotide sequencing.
This new diagnostic PCR assay was then tested on samples from EIA cases that were asymptomatic and therefore predicted to have significantly lower tissue- and plasma-associated viral burdens (8) than those for the clinical cases on farms 1 and 3. The efficacy of the new diagnostic PCR assay for detection of EIAV-infected equids was compared to that of conventional OIE-approved AGID test kits.
Of the 400 samples, 13.25% produced positive reactions in the AGID test, compared with 17.5% in diagnostic PCR, with 82.5% of the samples being negative in both test formats. Although some samples were PCR positive and AGID negative, there were none that were negative in PCR but positive in the AGID assay (Table 2).
TABLE 2.
Comparison of diagnostic PCR and AGID test for detection of EIAV-infected equids
| Farm | Breed | No. of samples |
|||
|---|---|---|---|---|---|
| Total | From symptomatic cases | AGID+ | Diagnostic PCR+ | ||
| F-1 | Standard breed | 66 | 12 | 19 | 21 |
| F-2 | Thoroughbred | 18 | 3 | 4 | 4 |
| F-3 | Thoroughbred | 23 | 1 | 1 | 1 |
| F-4 | Heavy horse | 72 | 0 | 2 | 2 |
| F-5 | Warm blood | 3 | 1 | 0 | 1 |
| F-6 | Arabian horse | 1 | 0 | 1 | 1 |
| F-7 | Mule | 48 | 0 | 7 | 7 |
| F-8 | Warm blood | 1 | 0 | 1 | 1 |
| F-9 | Warm blood | 1 | 0 | 1 | 1 |
| F-10 | Warm blood | 8 | 2 | 0 | 1 |
| F-11 | Warm blood | 34 | 3 | 0 | 3 |
| F-12 | Warm blood | 62 | 0 | 5 | 6 |
| F-13 | Warm blood | 4 | 2 | 0 | 2 |
| F-14 | Warm blood | 12 | 1 | 0 | 1 |
| F-15 | Standard breed | 6 | 1 | 0 | 1 |
| F-16 | Warm blood | 1 | 0 | 1 | 1 |
| F-17 | Warm blood | 3 | 0 | 1 | 1 |
| F-18 | Warm blood | 2 | 1 | 0 | 1 |
| F-19 | Warm blood | 5 | 0 | 0 | 1 |
| F-20 | Donkey | 4 | 0 | 1 | 1 |
| F-21 | Warm blood | 10 | 0 | 2 | 2 |
| F-22 | Warm blood | 1 | 0 | 1 | 1 |
| F-23 | Warm blood | 1 | 1 | 0 | 1 |
| F-24 | Arabian horse | 8 | 0 | 0 | 2 |
| FROM-1 | Heavy horse | 1 | 0 | 1 | 1 |
| FROM-2 | Heavy horse | 2 | 0 | 2 | 2 |
| FROM-3 | Heavy horse | 1 | 0 | 1 | 1 |
| FROM-4 | Heavy horse | 2 | 0 | 2 | 2 |
| Total | 400 | 28 | 53 | 70 | |
In Italy, seropositive reactions in AGID tests conducted in government-approved laboratories are required before any regulatory actions can be taken for cases of EIA. With the exception of some animals, such as a 14-year-old mare on farm 1 (Table 3) that had particularly severe clinical signs and died, all equids that were PCR positive and AGID negative in this laboratory (Tables 2 and 3) eventually became AGID test seropositive in official test procedures. Moreover, preliminary side-by-side comparisons had demonstrated that especially in asymptomatic cases, amplification of proviral DNA from blood cell chromatin samples using nested PCR was a more sensitive indicator of EIAV infection than attempts to detect viral RNA in plasma with RT-PCR-based techniques. Therefore, the results shown in Table 2 were obtained with the diagnostic PCR assay using DNA isolated from blood buffy coat cells.
TABLE 3.
Background information for EIAV gag gene sequences
| Farm | GenBank accession no. for sequence | Age of animala | Symptomsb |
AGID result | Tissue | Test used | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| F | T | L | A | E | P | ||||||
| F-1 | EU240733 | 14 | + | + | + | − | Spleen | RT-PCR | |||
| F-1 | GQ927489 | 55 days | + | + | + | + | + | + | Blood | PCR | |
| F-2 | EU375544 | 16 | + | + | + | + | + | + | + | Bone marrow | RT-PCR |
| F-3 | EU375543 | 5 mo | + | + | + | + | + | + | + | Liver | RT-PCR |
| F-4 | EU741609 | 15 | + | Buffy coat | RT-PCR | ||||||
| F-5 | GQ927486 | 8 | + | + | + | + | + | + | − | Blood | PCR |
| F-6 | GQ265785 | 16 | + | Buffy coat | RT-PCR | ||||||
| F-7 | GQ927501 | 10 | + | Blood | PCR | ||||||
| F-8 | GQ927505 | 22 | + | Blood | PCR | ||||||
| F-9 | GQ927483 | 10 | + | Blood | PCR | ||||||
| F-10 | GQ927506 | 11 | + | + | + | − | Blood | PCR | |||
| F-11a | GQ927502 | 10 | + | + | + | + | + | − | Blood | PCR | |
| F-11b | GQ927484 | 5 | + | + | + | + | + | + | − | Blood | PCR |
| F-12a | GQ927500 | 15 | + | Blood | PCR | ||||||
| F-12b | GQ927492 | 14 | + | Blood | PCR | ||||||
| F-13 | GQ927482 | 10 | + | + | + | + | + | − | Blood | PCR | |
| F-14 | GQ927503 | 17 | + | + | + | − | Blood | PCR | |||
| F-15 | GQ927485 | 3 | + | + | + | + | + | + | − | Blood | PCR |
| F-16 | GU060664 | 30 | + | Blood | PCR | ||||||
| F-17 | GQ927497 | 14 | + | Blood | PCR | ||||||
| F-18 | GQ927495 | 18 | + | + | + | + | − | Blood | PCR | ||
| F-19 | GQ927487 | 5 | + | + | + | + | − | Blood | PCR | ||
| F-20 | GQ927504 | 9 | + | Blood | PCR | ||||||
| F-21a | GQ927498 | 8 | + | Blood | PCR | ||||||
| F-21b | GQ927496 | 12 | + | Blood | PCR | ||||||
| F-22a | GQ927494 | 8 mo | + | + | + | + | − | Blood | PCR | ||
| F-22b | GQ927493 | 19 | + | Blood | PCR | ||||||
| F-23 | GQ927490 | 8 | + | + | + | − | Blood | PCR | |||
| F-24 | GQ927488 | 3 | + | + | + | + | − | Blood | PCR | ||
| FROM-3 | GQ927499 | 6 | + | Blood | PCR | ||||||
| FROM-1 | GQ229581 | 16 | + | Blood | PCR | ||||||
| FROM-2 | GQ923952 | 12 | + | Blood | PCR | ||||||
| FROM-4 | GU060662 | 10 | + | Blood | PCR | ||||||
| FROM-4 | GU060663 | 8 | + | Blood | PCR | ||||||
Age is in years unless otherwise indicated.
F, fever (>39°C); T, thrombocytopenia (16 × 103/μl < T < 40 × 103/μl); L, lethargy; A, anemia (2 × 1012/μl < A < 5 × 1012/μl); E, edema; P: petechial hemorrhage.
These results demonstrate that PCR-based methods are effective for the diagnosis of EIAV infections.
Sequence analysis of EIAV strains.
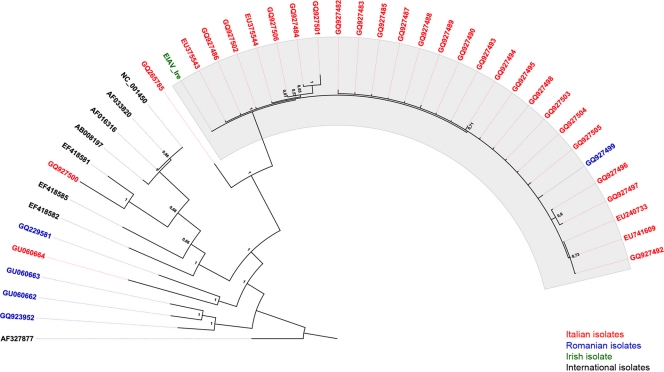
All PCR-generated fragments, including those spanning the entire gag gene, were sequenced both to ensure their authenticity and for subsequent phylogenetic analysis (Fig. 2).
FIG. 2.
Phylogenetic tree demonstrating the relationships between Central European, American, and Asian EIAV strains. Posterior probabilities are indicated at each node. Topology was made with FigTree 1.2.3. The highlighted zone indicates the main Italian gag gene subtype. International sequences are as follows: NC_001450, reference genome; AF033820, Wyoming; AF016316, United Kingdom; AF327877, Liaoning; AB008197, Japan; EF418582, Canada-1; EF418585, Canada-10; and EF418581, Argentina 1. The 2006 Irish isolate of EIAV is also included (23).
Complete gag gene sequences encoding the p15, p26, p11, and p9 antigens were obtained from animals located at all three farms (F-1 to F-3 in Tables 2 and 3) associated with the 2006 EIA outbreak (Ita-1, EU240733; Ita-2, EU375543; and Ita-3, EU375544). In addition, complete and partially (86%) complete gag gene sequences were determined for three Italian EIAV isolates that were geographically and/or temporally unrelated to the 2006 outbreak (Ita-4, EU741609; Ita-5, GQ265785; and Ita-6, GU060664). Finally, we determined, the complete (Rom-1, GQ229581) or partially (86%) complete (Rom-2, GQ923952; Rom-4, GU060662; and Rom-5, GU060663) gag gene sequences for four Romanian EIAV isolates.
The analysis of sequence variability using a panel of bioinformatics algorithms demonstrated very close phylogenetic relationships between the gag gene sequences of viruses obtained from each of the three farms (farms 1 to 3) associated with the 2006 Italian outbreak. When these sequences were translated into amino acid sequences, the overall homology between Ita-1, Ita-2, and Ita-3 at the predicted coding level was 97.7%. An alignment between the predicted amino acid sequences encoded by the gag genes of Ita-1 to Ita-3 with the published predicted amino acid sequence of the gag gene from a virus associated with the 2006 outbreak of EIA in Ireland (EIAVIRE) also demonstrated a high degree of similarity. For example, comparison between Ita-1 and EIAVIRE revealed rates of homology in p15, p26, p11, and p9 of 98.4%, 99.6%, 98.7%, and 100%, respectively, strongly supporting the hypothesis (23) that these viruses were derived from a common or related source. Not surprisingly, the Ita-5 and Ita-6 (GQ265785 and GU060664), isolates from asymptomatic horses, collected in 2007 and 2008, respectively, and not connected with the four farms involved in the 2006 outbreak, had similarity levels of 88.5% with Ita-1 and 90% with EIAVIRE. The Romanian isolates proved to be even more distantly related to the viruses responsible for the 2006 outbreak, with the Rom-1, Rom-2, Rom-4, and Rom-5 isolates (GQ229581, GQ923952, GU060662, and GU060663, respectively) sharing only 84.6, 76.4, 76.8, and 77.2% identity, respectively, with Ita-1.
The ability of the diagnostic PCR to detect EIAV sequences in blood cell DNA has enabled the analysis of sequence information even from inapparent carrier horses, where viral burdens are usually very low (2, 8). Therefore, sequences with a minimal length of 313 bp, encompassing regions of the gag gene encoding the p15 and p26 antigens from 23 individual, clinically apparently normal Italian horses and 1 Romanian animal (Table 2), were subjected to a comprehensive phylogenetic analysis in combination with the longer gag sequences described above and previously published gag gene information from North American, Asian and Irish EIAV strains (Fig. 2). The phylogenetic tree in Fig. 2 clearly shows the presence of at least three gag gene subtypes among strains of EIAV currently circulating in Italy. The predominant subtype includes viruses associated with the 2006 EIA outbreaks in both Italy and Ireland. A second gag subtype, consisting of Ita-5 and Ita-6, grouped between the bulk of the Italian and the Romanian isolates, while the third subtype, represented by DNA-Ita-12A, clustered with Japanese and North American strains.
DISCUSSION
The 2006 EIA outbreaks in Italy and Ireland that precipitated this wider study have been attributed to the transfusion of closely related batches of contaminated plasma into foals. This type of plasma administration is a widespread practice within the equine industry and is claimed to provide clinical protection against certain bacterial diseases in neonatal foals (7). To our knowledge, the results presented in this report are the first to provide direct support, based on molecular characterization techniques, for the contaminated-plasma hypothesis to explain the simultaneous occurrence of EIA in Italy and Ireland during 2006. Complete EIAV gag gene sequences were obtained from foals on farms 1 and 3 (GQ927489 and EU375543, respectively) that were infused with batches of this plasma. In addition to iatrogenic transmission, insect-mediated infections were an important factor in the 2006 Italian outbreak, as evidenced by the fact similar gag gene sequences (EU375544) were isolated from an adult horse housed on premises (farm 2) adjacent to farm 3 that had never been exposed to the contaminated plasma.
The 2006 EIA outbreak in Italy was managed according to current European Union regulations. However, eight new clinical cases were observed on farm 1 (five mares and three foals) 5 months after the end of compulsory 90-day quarantine period. Extensive gag gene homology between later (EU240733) and earlier farm 1 isolates suggested that these viruses were very closely related and therefore probably derived from the same source. Consequently, it is likely that at least one of the eight new cases had been infected but did not produce a seropositive reaction in AGID tests conducted before the horses were released from quarantine. Although most equids become seropositive in standard AGID assays within 45 days of an initial exposure, this process can take considerably longer in some cases. Interestingly, this was reported during the 2006 EIA outbreak in Ireland, where in one case there was a delay of 157 days between the probable time of exposure and the first positive serological test result (7). The fact that all PCR-positive, AGID-negative equids shown in Tables 2 and 3 that survived long enough eventually became seropositive in AGID assays performed in government-approved laboratories demonstrates that PCR-based assays have the power to detect animals recently infected with EIAV.
In contrast to the success obtained with the primers based on consensus sequences described here, the PCR primers based exclusively on North American isolates (5, 17), as recommended by the OIE for the detection of EIAV, proved to be completely unsuitable for the reliable amplification of genetic material from the EIAV strains currently circulating in Italy. These results demonstrate that once appropriate primers are identified, PCR-based methods can serve as a valuable adjunct to conventional serological testing for the routine diagnosis of EIA.
In addition to highlighting the efficacy of nucleic acid detection systems for EIA diagnosis, this work contributes significant amounts of new information expanding our understanding about the extent of naturally occurring variation within EIAV gag gene sequences. For the first time, the complete characterization of gag genes from five Italian isolates and one Romanian isolate has been achieved, along with extensive sequence information (86% of the total gag gene) from four (one Italian and three Romanian) additional EIAV isolates. Furthermore, in another 23 cases we accomplished partial characterization of gag gene sequences in the region encoding the viral matrix protein. Collectively, this represents the most comprehensive analysis of European EIAV isolates conducted to date.
A comparison of gag gene sequences from viruses isolated during the 2006 Irish EIA outbreak with limited numbers of Asian and North American strains suggested that variation within this gene was much more extensive than previously thought (23). In addition, it appeared that the nucleotide substitutions were not evenly distributed throughout this gene and that while sequences encoding p26 shared 90% homology, there was only 50% identity in the region encoding p9 (23). Our more exhaustive analysis supports these preliminary conclusions and demonstrates that in those cases where complete gag gene sequences are available, predicted amino acid identity is 63.8% for p26, 56.4% for p11, 40.5% for p15, and only 28.3% for p9. Therefore, as suggested previously (23), conservation of the amino acid sequences encoded by the EIAV gag gene conforms to a hierarchy in which p26 > p11 > p15 > p9, a fact that presumably reflects the different functional and structural constraints that are imposed on each gag antigen. These observations have important implications for the diagnosis and epidemiological investigation of EIAV outbreaks. As p26 is the major antigen employed in the AGID (Coggins) test as well as in most ELISA formats (9, 10) for the serological detection of EIAV infections, it is somewhat reassuring to observe that it is relatively well conserved between viral isolates from very different geographical locations. On the other hand, the significant divergence in p9 detected between viruses from different countries suggest that analysis of variation within this antigen might be particularly useful as an epidemiological marker to investigate the origin and progression of EIA outbreaks. The fact that the predicted p9 amino acid sequence of Ita-1 is identical to that of the 2006 Irish strain while retaining only 59.3% and 51.9% homology, respectively, to North American isolates such as EIAVWYO and EIAVCAN3 strongly supports this inference.
The phylogenic analysis (Fig. 2) suggests there at least three gag gene subtypes currently circulating in Italy, two of which are also present in Romania. Furthermore, the major subtype appears to be geographically restricted. Although geographical restriction among EIAV genotypes is not altogether surprising, considering the measures in force to prevent the export of infected animals and the mechanisms of transmission associated with this virus, further studies are required before the term “clade” can be applied to the epidemiology of EIAV in the same context as it is currently used for HIV-1 (27).
Acknowledgments
We are grateful to Francesco Nardi for his advice on the phylogenetic analysis, to Gianluca Alunni for his valuable technical assistance, and to Kathy Shuck for critical reading of the manuscript.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Belshan, M., M. E. Harris, A. E. Shoemaker, T. J. Hope, and S. Carpenter. 1998. Biological characterization of Rev variation in equine infectious anemia virus. J. Virol. 72:4421-4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheevers, W. P., and T. C. McGuire. 1985. Equine infectious anemia virus: immunopathogenesis and persistence. Rev. Infect. Dis. 7:83-88. [DOI] [PubMed] [Google Scholar]
- 3.Coggins, L., N. L. Norcross, and S. R. Nusbaum. 1972. Diagnosis of equine infectious anemia by immunodiffusion test. Am. J. Vet. Res. 33:11-18. [PubMed] [Google Scholar]
- 4.Cook, R. F., et al. 2003. Enhancement of equine infectious anemia virus virulence by identification and removal of suboptimal nucleotides. Virology 313:588-603. [DOI] [PubMed] [Google Scholar]
- 5.Cook, R. F., S. J. Cook, F. L. Li, R. C. Montelaro, and C. J. Issel. 2002. Development of a multiplex real-time reverse transcriptase-polymerase chain reaction for equine infectious anemia virus (EIAV). J. Virol. Methods 105:171-179. [DOI] [PubMed] [Google Scholar]
- 6.Cook, R. F., C. J. Issel, and R. C. Montelaro. 1996. Equine infectious anemia, vol. 6. Elsevier, Amsterdam, Netherlands.
- 7.Cullinane, A., et al. 2007. Diagnosis of equine infectious anaemia during the 2006 outbreak in Ireland. Vet. Rec. 161:647-652. [DOI] [PubMed] [Google Scholar]
- 8.Harrold, S. M., et al. 2000. Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J. Virol. 74:3112-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Issel, C. J., and R. F. Cook. 1993. A review of techniques for the serologic diagnosis of equine infectious anemia. J. Vet. Diagn. Invest. 5:137-141. [DOI] [PubMed] [Google Scholar]
- 10.Issel, C. J., S. J. Cook, R. F. Cook, and T. R. Cordes. 1999. Optimal paradigms to detect reservoirs of equine infectious anemia virus (EIAV). J. Equine Vet. Sci. 19:728-732. [Google Scholar]
- 11.Kono, Y. 1969. Viremia and immunological responses in horses infected with equine infectious anemia virus. Natl. Inst. Anim. Health Q. (Tokyo) 9:1-9. [PubMed] [Google Scholar]
- 12.Kono, Y., K. Kobayashi, and Y. Fukunaga. 1973. Antigenic drift of equine infectious anemia virus in chronically infected horses. Arch. Gesamte Virusforsch. 41:1-10. [DOI] [PubMed] [Google Scholar]
- 13.Leroux, C., J. K. Craigo, C. J. Issel, and R. C. Montelaro. 2001. Equine infectious anemia virus genomic evolution in progressor and nonprogressor ponies. J. Virol. 75:4570-4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leroux, C., C. J. Issel, and R. C. Montelaro. 1997. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J. Virol. 71:9627-9639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mealey, R. H., B. Zhang, S. R. Leib, M. H. Littke, and T. C. McGuire. 2003. Epitope specificity is critical for high and moderate avidity cytotoxic T lymphocytes associated with control of viral load and clinical disease in horses with equine infectious anemia virus. Virology 313:537-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montelaro, R. C., B. Parekh, A. Orrego, and C. J. Issel. 1984. Antigenic variation during persistent infection by equine infectious anemia virus, a retrovirus. J. Biol. Chem. 259:10539-10544. [PubMed] [Google Scholar]
- 17.Nagarajan, M. M., and C. Simard. 2001. Detection of horses infected naturally with equine infectious anemia virus by nested polymerase chain reaction. J. Virol. Methods 94:97-109. [DOI] [PubMed] [Google Scholar]
- 18.Nagarajan, M. M., and C. Simard. 2007. Gag genetic heterogeneity of equine infectious anemia virus (EIAV) in naturally infected horses in Canada. Virus Res. 129:228-235. [DOI] [PubMed] [Google Scholar]
- 19.O'Rourke, K. I., L. E. Perryman, and T. C. McGuire. 1989. Cross-neutralizing and subclass characteristics of antibody from horses with equine infectious anemia virus. Vet. Immunol. Immunopathol. 23:41-49. [DOI] [PubMed] [Google Scholar]
- 20.Payne, S. L., X. M. Qi, H. Shao, A. Dwyer, and F. J. Fuller. 1998. Disease induction by virus derived from molecular clones of equine infectious anemia virus. J. Virol. 72:483-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Payne, S. L., K. Rushlow, B. R. Dhruva, C. J. Issel, and R. C. Montelaro. 1989. Localization of conserved and variable antigenic domains of equine infectious anemia virus envelope glycoproteins using recombinant env-encoded protein fragments produced in Escherichia coli. Virology 172:609-615. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, J. E., C. S. Becvar, and L. O. Mott. 1971. Evaluation of the immunodiffusion tests for the diagnosis of equine infectious anemia. Proc. U. S. Anim. Health Assoc. 74:259-267. [Google Scholar]
- 23.Quinlivan, M., R. F. Cook, and A. Cullinane. 2007. Real-time quantitative RT-PCR and PCR assays for a novel European field isolate of equine infectious anaemia virus based on sequence determination of the gag gene. Vet. Rec. 160:611-618. [DOI] [PubMed] [Google Scholar]
- 24.Rieder, S., S. Taourit, D. Mariat, B. Langlois, and G. Guerin. 2001. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm. Genome 12:450-455. [DOI] [PubMed] [Google Scholar]
- 25.Salinovich, O., et al. 1986. Rapid emergence of novel antigenic and genetic variants of equine infectious anemia virus during persistent infection. J. Virol. 57:71-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sellon, D. C. 1993. Equine infectious anemia. Vet. Clin. North Am. Equine Pract. 9:321-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spira, S., M. A. Wainberg, H. Loemba, D. Turner, and B. G. Brenner. 2003. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. J. Antimicrob. Chemother. 51:229-240. [DOI] [PubMed] [Google Scholar]
- 28.Zheng, Y. H., et al. 1997. Insertions, duplications and substitutions in restricted gp90 regions of equine infectious anaemia virus during febrile episodes in an experimentally infected horse. J. Gen. Virol. 78:807-820. [DOI] [PubMed] [Google Scholar]
- 29.Zheng, Y. H., H. Sentsui, T. Nakaya, Y. Kono, and K. Ikuta. 1997. In vivo dynamics of equine infectious anemia viruses emerging during febrile episodes: insertions/duplications at the principal neutralizing domain. J. Virol. 71:5031-5039. [DOI] [PMC free article] [PubMed] [Google Scholar]