Abstract
Although widely used for the characterization of the transmission of intestinal Cryptosporidium spp., genotyping tools are not available for C. muris and C. andersoni, two of the most common gastric Cryptosporidium spp. infecting mammals. In this study, we screened the C. muris whole-genome sequencing data for microsatellite and minisatellite sequences. Among the 13 potential loci (6 microsatellite and 7 minisatellite loci) evaluated by PCR and DNA sequencing, 4 were eventually chosen. DNA sequence analyses of 27 C. muris and 17 C. andersoni DNA preparations showed the presence of 5 to 10 subtypes of C. muris and 1 to 4 subtypes of C. andersoni at each locus. Altogether, 11 C. muris and 7 C. andersoni multilocus sequence typing (MLST) subtypes were detected among the 16 C. muris and 12 C. andersoni specimens successfully sequenced at all four loci. In all analyses, the C. muris isolate (TS03) that originated from an East African mole rat differed significantly from other C. muris isolates, approaching the extent of genetic differences between C. muris and C. andersoni. Thus, an MLST technique was developed for the high-resolution typing of C. muris and C. andersoni. It should be useful for the characterization of the population genetics and transmission of gastric Cryptosporidium spp.
Cryptosporidium muris and Cryptosporidium andersoni are gastric Cryptosporidium species of various mammals. Cryptosporidium muris was first identified in the gastric glands of mice over a century ago, whereas C. andersoni was long considered C. muris because of the genetic and biological similarity between the two and was only recently established as a separate species (25, 42). Cryptosporidium muris is well known to have broad host specificity. In addition to various rodent species, natural C. muris infections have been documented for pigs, Bactrian camels, giraffes, dogs, cats, nonhuman primates, seals, bilbies, and tawny frogmouth (3, 16, 17, 20, 22, 26, 27, 29, 32, 34, 37, 38, 40, 44). In contrast, C. andersoni is mostly a parasite of cattle, having been found only occasionally in other animals such as Bactrian camels, sheep, and goats. In recent years, C. muris or C. andersoni infection has been reported in a few human cases (1, 8, 11, 13, 15, 19, 24, 30, 31, 33, 39). Thus, both C. muris and C. andersoni are considered zoonotic Cryptosporidium species. Because of its biological uniqueness and zoonotic potential, the complete genome of C. muris is being sequenced.
Recently, various molecular diagnostic tools have been used for the characterization of the transmission of human-pathogenic intestinal Cryptosporidium spp. such as C. hominis and C. parvum (35, 41). These tools have proven to be especially useful for comparisons of parasite population genetics among hosts or Cryptosporidium species, characterization of host specificity of Cryptosporidium spp., identification of infection sources in humans, tracking of the temporal and geographical spread of pathogens, and investigation of outbreaks and endemicity. One such high-resolution subtyping tool is multilocus sequence typing (MLST). For C. hominis and C. parvum, MLST tools have recently been developed using the polymorphic microsatellite and minisatellite markers identified in the recently published whole-genome sequencing data (9, 10, 12).
In this study, we screened the C. muris genome for microsatellite and minisatellite sequences and developed an MLST technique for the high-resolution typing of C. muris and C. andersoni isolated from humans and various animals.
MATERIALS AND METHODS
Cryptosporidium specimens.
A total of 27 DNA extractions from 25 C. muris specimens and 17 extractions from 17 C. andersoni specimens were used in the study (Table 1). The C. muris specimens were from humans, various rodents, Bactrian camels, and one mountain goat, snake, dog, cat, and tawny frogmouth each in the Czech Republic, Egypt, Kenya, and Peru. In contrast, all C. andersoni specimens were from cattle in the United States, Canada, the Czech Republic, and China. A few of the isolates were maintained in laboratory rodents (mice, SCID mice, and Mastomys coucha) (Table 1). Prior to experimental infection, these animals were maintained in breeding facilities designed for experimental animals and determined to be free of Cryptosporidium infection by microscopy of consecutive fecal samples and, in the case of SCID mice and Mastomys coucha, by PCR analysis. Three of the DNA preparations were from the same C. muris specimen from a child in Kenya, extracted at different storage times. These specimens were diagnosed as being positive for C. muris or C. andersoni by DNA sequence analysis of an ∼830-bp fragment of the small-subunit (SSU) rRNA gene (43). Seven C. muris DNA preparations (preparations 6853, 7379, 9412, 14260, 14272, 14713, and 14714) and two C. andersoni preparations (preparations 1808 and 11084) were used for initial primer evaluations and the selection of markers. These DNA extractions were chosen for the initial screening of PCR primers and targets because of their various efficiencies of amplification at the SSU rRNA locus, with DNAs 9412 and 14272 producing amplification in only 1 of 3 PCR runs. The remaining specimens were analyzed only at the selected microsatellite and minisatellite loci.
TABLE 1.
Specimens used in the study and their subtype identity at the four selected microsatellite and minisatellite loci
| Specimen | Species | Host | Location and/or source | MLST subtypeb |
Reference | |||
|---|---|---|---|---|---|---|---|---|
| MS1 | MS2 | MS3 | MS16 | |||||
| Referencea | C. muris | RN66 | Japan | M5 | M4 | M1 | M5 | 18 |
| 6853 | C. muris | Human | Lima, Peru | M1 | M2 | M4 | M5 | 33 |
| 9412 | C. muris | Human | Nairobi, Kenya | Noisy | — | — | — | 13 |
| 14713 | C. muris | Human (same as 9412) | Nairobi, Kenya | M6 | M4 | — | M2 | 13 |
| 14714 | C. muris | Human (same as 9412) | Nairobi, Kenya | M6 | M4 | M1 | M2 | 13 |
| 13460 | C. muris | Camel via mice | Primary passage in Giza, Egypt | M5 | M4 | M2 | M3 | This study |
| 13461 | C. muris | Dog via mice | Primary passage in Giza, Egypt | M5 | M4 | M2 | M3 | This study |
| 7379 | C. muris | Domestic mouse | Mouse breeder in Vyškov, Czech Republic | M7 | M4 | M2 | M1 | This study |
| 1666 | C. muris | Siberian chipmunk (Eutamius sibiricus) | Imported into Czech Republic from Southeast Asia | M7 | M4 | M1 | M5 | 17 |
| 7380 | C. muris | Mara (Dolichotis patagonum) | Born in a zoo in Plzeň, Czech Republic | M9 | M4 | M1 | M1 | This study |
| 7511 | C. muris | Bactrian camel | Born in a zoo in Plzeň, Czech Republic | M1 | M1 | M4 | M5 | This study |
| 7512 | C. muris | Mountain goat (Oreamnos americanus) | Born in a zoo in Plzeň, Czech Republic | M8 | M4 | M2 | M4 | This study |
| 14242 | C. muris | Bactrian camel via Mastomys coucha | 14th passage of isolate 7511 in Czech Republic | M10 | M6 | M5 | M6 | 23 |
| 14243 | C. muris | RN66 via SCID mice | 10th passage in Czech Republic of oocyst from Waterborne, Inc. | M10 | M6 | M5 | M6 | 23 |
| 14260 | C. muris | Tachyoryctes splendens via Mastomys coucha | 14th passage in Czech Republic of oocysts from wild Tachyoryctes splendens in Kakamega, Kenya | M10 | M6 | M5 | M6 | 23 |
| 14272 | C. muris | Giraffe (Giraffa camelopardalis reticulate) | Born in a zoo in Dvůr Králové nad Labem, Czech Republic | — | — | — | — | 20 |
| 14905 | C. muris | Tawny frogmouth, Podargus strigoides (CZ-B2-2) | Zoo in Prague, Czech Republic | M1 | M3 | — | Noisy | 36 |
| 14906 | C. muris | Laboratory mouse (CZ-B2-11) | Prague, Czech Republic | M6 | M4 | M2 | M4 | 36 |
| 14907 | C. muris | Yellow rat snake, Elaphe obsolete quadrivittata (CZ-B2-17) | Zoo in Prague, Czech Republic | M2 | M2 | M3 | M5 | 36 |
| 14908 | C. muris | Laboratory mouse (CZ-B3-5) | Prague, Czech Republic | — | — | — | M5 | This study |
| 14910 | C. muris | Laboratory mouse (CZ-B3-8) | Prague, Czech Republic | M1 | M4 | — | Noisy | This study |
| 14911 | C. muris | Laboratory mouse (CZ-B4-11) | Prague, Czech Republic | M1 | M2 | M4 | M5 | This study |
| 14913 | C. muris | Cat (CZ-B6-2) | Prague, Czech Republic | M3 | M4 | — | M1 | 34 |
| 14914 | C. muris | Laboratory mouse (CZ-B6-6) | Prague, Czech Republic | M9 | M4 | M1 | M1 | This study |
| 14915 | C. muris | Rat (CZ-B6-18) | Prague, Czech Republic | Noisy | Noisy | M2 | M1 | This study |
| 14916 | C. muris | Laboratory mouse (CZ-B6-26) | Prague, Czech Republic | M4 | M4 | — | M5 | This study |
| 14917 | C. muris | Laboratory mouse (CZ-B6-27) | Prague, Czech Republic | Noisy | M4 | — | M5 | This study |
| 14919 | C. muris | Prevost's squirrel (Callosciurus prevostii; CZ-B6-86) | Zoo in Prague, Czech Republic | M4 | M4 | — | M5 | This study |
| 590 (M356) | C. andersoni | Cattle | Calgary, Canada | A2 | A3 | A4 | A1 | 29 |
| 14934 | C. andersoni | Cattle | Henan, China | A1 | A2 | A4 | A1 | This study |
| 14937 | C. andersoni | Cattle | Henan, China | A1 | A2 | A4 | A1 | This study |
| 14939 | C. andersoni | Cattle | Henan, China | A2 | A1 | A3 | A1 | This study |
| 14942 | C. andersoni | Cattle | Henan, China | A2 | A1 | A2 | A1 | This study |
| 14943 | C. andersoni | Cattle | Henan, China | A2 | A1 | A2 | A1 | This study |
| 14944 | C. andersoni | Cattle | Henan, China | A2 | Noisy | A2 | — | This study |
| 14945 | C. andersoni | Cattle | Henan, China | A1 | — | A4 | — | This study |
| 14948 | C. andersoni | Cattle | Henan, China | A1 | A2 | A4 | A1 | This study |
| 845 | C. andersoni | Cattle | Sumava Mountains, Czech Republic | A2 | A3 | A4 | A1 | 21 |
| 14237 | C. andersoni | Cattle | Ceske Budejovice, Czech Republic | A1 | A3 | A4 | A1 | This study |
| 1808 | C. andersoni | Cattle | Indiana | A2 | A3 | A1 | A1 | This study |
| 1809 | C. andersoni | Cattle | Indiana | A2 | A3 | A4 | A1 | This study |
| 11084 | C. andersoni | Cattle | Georgia | A2 | A3 | — | — | 7 |
| 14857 | C. andersoni | Cattle | North Carolina | A2 | A3 | A2 | A1 | 4 |
| 14860 | C. andersoni | Cattle | New York | A2 | — | — | — | 5 |
| 14862 | C. andersoni | Cattle | Pennsylvania | — | — | Noisy | A1 | 5 |
Reference sequences from RN66 were obtained from the C. muris whole-genome sequencing project.
MS1, coding for the hypothetical protein CMU_036310 (GenBank accession no. XM_002141771); MS2, coding for the 90-kDa heat shock protein (accession no. XM_002141007); MS3, coding for the hypothetical protein CMU_020660 (accession no. XM_002142635); MS16, coding for the leucine-rich repeat family protein CMU_035650 (accession no. XM_002141705). Dashes indicate no PCR amplification.
Microsatellite and minisatellite identification.
A search for microsatellite and minisatellite sequences in the C. muris genome was conducted on 8 March 2007. The first 2,500 sequences (traces 1649410657 to 1649413156) in the C. muris whole-genome sequencing database were retrieved from the website of the Institute for Genomic Research (TIGR, now the J. Craig Venter Institute [http://gsc.jcvi.org/projects/msc/Cryptosporidium_muris/). Microsatellite and minisatellite sequences in the retrieved sequences were identified by using the software Tandem Repeat Finder (http://tandem.bu.edu/trf/trf.html).
PCR analysis.
Nested PCR was used for the amplification of microsatellite and minisatellite targets. Primary and secondary PCR primers were designed based on nucleotide sequences of the potential microsatellite and minisatellite loci. The potential targets were amplified by nested PCR, using 1 μl of DNA in the primary PCR and 2 μl of primary PCR products in the secondary PCR. For both the primary and secondary PCRs, the PCR mixture consisted of 200 mM (each) deoxynucleotide triphosphate, 1× PCR buffer (Perkin-Elmer, Foster City, CA), 3.0 mM MgCl2, 5.0 U of Taq polymerase (Promega, Madison, WI), and 100 nM primers in a total volume of 100 μl. The reactions were performed with a GeneAmp PCR 9700 thermocycler (Perkin-Elmer) for 35 cycles of 94°C for 45 s, the annealing temperatures specified in Table 2 for 45 s, and 72°C for 60 s, with an initial denaturation step (94°C for 5 min) and a final extension step (72°C for 10 min). To neutralize PCR inhibitors, 400 ng/μl of nonacetylated bovine serum albumin (Sigma-Aldrich, St. Louis, MO) was used in the primary PCR. The secondary PCR products were detected by agarose gel electrophoresis and ethidium bromide staining.
TABLE 2.
Primer sequences and amplification efficiencies of microsatellite and minisatellite loci selected for evaluations
| Locus | Trace | Targeted repeat(s)a | Primer sequence (5′-3′) | Annealing temp (°C) | Expected size (bp) | Amplification efficiency (no. of amplified samples/total no. of samples)b |
|---|---|---|---|---|---|---|
| CM-MS1 | 1649410765 | (GAACGAGATAGG)15 | ACCATCTAGAGATAACGAGCGA (F1) | 55 | 550 | 8/9 |
| GAATCAGAAGATGAGCGACAA (R1) | ||||||
| CGTGATAGTGGGTATGAATTGGACA (F2) | 55 | |||||
| CGACTGCGATACTCACGTCCT (R2) | ||||||
| CM-MS2 (HSP90) | 1649411347 | (CCATATCCC)6 | TTGCAACTGTACCTAAATTAGTA (F1) | 55 | 457 | 7/9 |
| GTGAGACTTCTGGGGTCCTGA (R1) | ||||||
| TCATGACGCGTCATACCAACA (F2) | 52 | |||||
| ACTTAGACAGTTCTATGCTGA (R2) | ||||||
| CM-MS3 | 1649411585 | (TGTTGG)9 and (GCTGCA)6 | AACCAAGTGAATCACGAACTT (F1) | 55 | 536 | 4/9 |
| TTGCTTTAAGTGTAGAGCATACAA (R1) | ||||||
| TCAAGTACAGCAGTCTATTGCTT (R1r) | ||||||
| GCAATATCTTCGACGATCCCA (F2) | 55 | |||||
| ATGGGAATAATTCTTCATCATCAA (R2) | ||||||
| CM-MS4 | 1649411639 | (ATC)73 | GTTGGCTGCATCTATGTT (F1) | 48 | 676 | 2/9 |
| TATACTTGATGATGTACTGGAT (R1) | ||||||
| TGTGGTGGAGTTGTAATTAGGA (F2) | 55 | |||||
| TAGATGGTGACTCAGATTCTT (R2) | ||||||
| CM-MS6 | 1649411735 | (AATGCAGATACAAGTATC)11 | TCTCTTTCCAACTCGATGCTT (F1) | 55 | 751 | 4/9 (weak bands) |
| ACCAAGAGTGATATTCATTCCTCT (R1) | ||||||
| GACTTACAATCAGATGGATACAGC (F2) | 58 | |||||
| TCTGTTCCTTGGTAAGTGGGCTGT (R2) | ||||||
| CM-MS8 | 1649411889 | (AT)44 | GCAACGTGACATGGATGGA (F1) | 52 | 307 | 0/7 |
| CGCTAGAGTCAAGAGTCAT (R1) | ||||||
| TTGTGGAGATAAATTAACAAACA (F2) | 52 | |||||
| CTCATCATCTGAAGTTGACAA (R2) | ||||||
| CM-MS11 | 1649412113 | (AT)21 | AGTCTTCATCATCTGGATATAGCA (F1) | 55 | 467 | 2/9 |
| GCTTTAGATTCTCGTACTATCA (R1) | ||||||
| ATCTAGAACAGTTCCCATTACCA (F2) | 52 | |||||
| AATGATGTCGAGTTAGCTAAA (R2) | ||||||
| CM-MS12 | 1649412180 | (TTTATTTT)6 | CTTGAAGGTTTAGAACCGGGA (F1) | 52 | 450 | 6/9c |
| (intergenic) | TTCGTGAAGGTACTTCATATG (R1) | |||||
| GGATCAATCCAAGTGAGTTCT (F2) | 55 | |||||
| CTCACATTGATATGGCTGGTT (R2) | ||||||
| CM-MS13 | 1649412266 | (TA)21 | GGACCACATGATAAAGAGCCA (F1) | 55 | 579 | 0/9 |
| TTGCGCTCTTGGGAAAGGTT (R1) | ||||||
| GACTTAGCAGATATAACTGGT (F2) | 52 | |||||
| CTCGCGCATGCATCAATGCAA (R2) | ||||||
| CM-MS14 | 1649412309 | (GTAGTAGTT)7 and (GTTGTAGTAGTA)11 | GTACCATCAGCTCCTATAGGA (F1) | 55 | 764 | 0/9 |
| GATCTGGCAGATCCAATTCAT (R1) | ||||||
| GGATCTTGCCAAATCACTTCT (F2) | 55 | |||||
| CTATCCTATAAGAGGTGATGTACT (R2) | ||||||
| CM-MS16 | 1649412803 | (CTTCTTCAT)9 | GAAGAGGTCGAAGTTAAGCTA (F1) | 50 | 597 | 7/9 |
| GACAATCATCTAAATCGTGTT (R1) | ||||||
| AAGTTTCATCTAGGTACACTAAGA (F2) | 55 | |||||
| CACTACCTAATCTCGTGTACTT (R2) | ||||||
| CM-MS17 | 1649413150 | (CTCGTTCCCTAT)15 | TCACTTGACTGCGATCTC (F1) | 50 | 532 | 2/9 |
| GTTCTTCAAGGTCCAGATCTCCTT (R1) | ||||||
| TACCCTAAGGAATAACGATCA (F2) | 52 | |||||
| ATGAGAAGTATCGCTCTTATGGCT (R2) | ||||||
| CM-MS18 | 1649412657 | (TA)15 | AGAGCCATCAGTACCACTGTT (F1) | 55 | 590 | 8/9c |
| TCGATTGGCCTTACCGGTGCA (R1) | ||||||
| CAGCTGTTCCAGAACCACTTGA (F2) | 55 | |||||
| CCAACAGACATAGACATGCTT (R2) |
Tandem repeat identified in the sequence of the C. muris whole-genome sequencing project.
Seven C. muris specimens (specimens 6853, 7379, 9412, 14260, 14272, 14713, and 14714) and two C. andersoni specimens (specimens 1808 and 11084) were used in initial primer evaluations. Specimens 9412 (except for MS1 and MM16) and 14272 (except for MS18) were not amplified at most loci.
Noisy at sequencing.
Sequence analysis.
The secondary PCR products were sequenced in both directions with an ABI 3130 genetic analyzer (Applied Biosystems, Foster City, CA) using the secondary primers and the BigDye1 Terminator V3.1 cycle sequencing kit (Applied Biosystems). The sequences obtained were aligned with each other and reference sequences downloaded from the C. muris whole-genome database using ClustalX (http://www.clustal.org/). To assess the genetic relatedness of various C. muris and C. andersoni subtypes, neighbor-joining trees were constructed by using the program TreeconW (http://bioinformatics.psb.ugent.be/software/details/3), based on the evolutionary distances calculated by the Kimura two-parameter model.
Nucleotide sequence accession numbers.
Unique sequences generated in this study have been deposited in the GenBank database under accession numbers HM565066 to HM565101.
RESULTS
Identification of microsatellite and minisatellite sequences in the C. muris genome.
The search for tandem repeats in the 2,500 short sequences retrieved from the C. muris whole-genome sequencing project identified 101 sequences with microsatellites and minisatellites. Based on the nature (largely an absence of imperfect repeats) and length (>6 copies for minisatellite targets and >10 copies for microsatellite targets) of the repeats and the availability of suitable sequences for primer design (excluding those with short or AT-rich 5′- or 3′-flanking nucleotide sequences), 13 loci were chosen from the 101 potential targets, including 6 microsatellite loci and 7 minisatellite loci. The location of the loci was not considered, as the C. muris genome was not assembled and annotated at the execution of the study. Primers for nested PCR were designed for each locus (Table 2). Because DNA sequencing rather than fragment length measurement was used for the determination of polymorphism, the final PCR product was larger than that normally used for microsatellite and minisatellite analysis, with expected PCR products ranging from 307 bp to 751 bp.
Initial evaluation of the amplification efficiency.
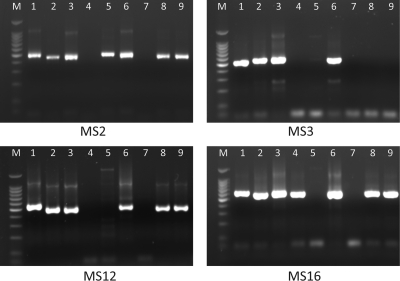
The amplification efficiency of the 13 sets of nested PCR primers was initially evaluated by using seven C. muris DNA preparations (preparations 6853, 7379, 9412, 14260, 14272, 14713, and 14714) and two C. andersoni DNA preparations (preparations 1808 and 11084) (Fig. 1). The primers of three loci (CM-MS8, CM-MS13, and CM-MS14) did not produce the expected PCR products. Primers of the remaining loci produced the expected PCR products for 2 to 8 of the DNA preparations used in the analysis, with one locus (CM-MS6) generating only light bands in gel electrophoresis analyses of the PCR products (Fig. 1, Table 2). DNA preparations 9412 (except for MS1 and MS16) and 14272 (except for MS18) were not amplified at most loci. Positive PCR products of the amplified loci were sequenced mostly successfully, with the exception of two loci, CM-MS12 and CM-MS18, which produced unreadable sequences with numerous underlying signals in the electropherogram.
FIG. 1.
PCR amplification of C. muris and C. andersoni DNA at four of the microsatellite and minisatellite loci (MS2, MS3, MS12, and MS16). M, 100-bp molecular markers. Lanes 1 to 9, DNAs 1808 (C. andersoni), 6853 (C. muris), 7379 (C. muris), 9412 (C. muris), 11084 (C. andersoni), 14260 (C. muris variant), 14272 (C. muris), 14713 (C. muris), and 14714 (C. muris), respectively.
Amplification efficiency of the selected genetic loci.
Four loci, CM-MS1, CM-MS2, CM-MS3, and CM-MS16, were chosen for sequence polymorphism evaluations using a total of 44 DNA extractions from C. muris and C. andersoni specimens. Of these, 41, 39, 24, and 38 DNA preparations were amplified at the CM-MS1, CM-MS2, CM-MS3, and CM-MS16 loci, respectively (Table 1). These PCR products were sequenced successfully, except for 2 to 3 DNA preparations at each locus, which produced mixed signals in electropherograms. The nucleotide sequences generated at each locus were homologous to those downloaded from the database of the C. muris whole-genome sequencing project. A BLAST analysis of the CM-MS3 sequences against the GenBank sequence database identified three sequencing errors in the primary reverse primer (CM-MS3-R1) designed based on initial sequences obtained from TIGR project database. A replacement primer (CM-MS3-R1r) was then designed, which led to the PCR amplification of nine additional DNA preparations at the CM-MS3 locus.
Genetic relatedness among C. muris and C. andersoni subtypes.
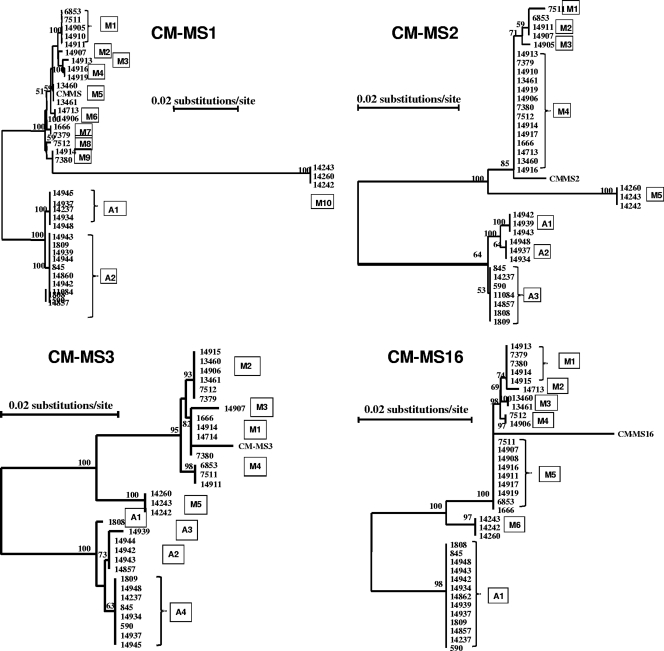
Multiple-sequence alignment analysis of the acquired sequences grouped the parasites into three groups at each of the four loci under analysis. One major group was formed by most C. muris specimens, including the C. muris reference sequence; one group (referred to hereafter as the C. muris variant) was formed by three Czech C. muris specimens, 14242, 14243, and 14260; and one was formed by C. andersoni. The formation of the three sequence groups was supported by results of phylogenetic analyses, as three distinct groups were seen in neighbor-joining trees constructed with the sequences (Fig. 2).
FIG. 2.
Phylogenetic relationships among subtypes of C. muris, the C. muris variant, and C. andersoni at four microsatellite and minisatellite loci (MS1, MS2, MS3, and MS16) as assessed by a neighbor-joining analysis of the nucleotide sequences, using distance calculated by the Kimura two-parameter model. CM-MS1, CM-MS2, CM-MS3, and CM-MS4 are reference sequences from the whole-genome sequencing project.
Nature of polymorphism in microsatellite and minisatellite sequences.
The three groups of parasites identified differed from each other by having numerous nucleotide substitutions in the nonrepeat region. Within each group, sequences differed from each other only in the number of microsatellite and minisatellite repeats. As the microsatellite and minisatellite repeats occurred in the coding region of the genes, the insertions and deletions were always in trinucleotides. The only exceptions were reference sequences of C. muris from the genome sequencing project, which differed in the nonrepeat region (by single-nucleotide deletions/insertions and substitutions) from the C. muris sequences acquired in this project at three of the four loci: CM-MS2, CM-MS3, and CM-MS16. These nucleotide deletions, insertions, and substitutions, however, were likely due to sequencing errors, as indicated by a comparison of the original reference sequences retrieved from the C. muris genome sequence project website and their respective sequences (GenBank accession no. XM_002141771 for CM-MS1, XM_002141007 for CM-MS2, XM_002142635 for CM-MS3, and XM_002141705 for CM-MS16) recently downloaded from the GenBank database.
The three groups of parasites further differed from each other in the nature of microsatellite and minisatellite repeats at each locus. With the exception of CM-MS16, the C. muris variant had repeat sequences different from those of C. andersoni and C. muris. Cryptosporidium andersoni also had repeat sequences different from those of C. muris at the CM-MS2 and CM-MS3 loci. Sometimes, the difference was just a slight modification of the repeat, such as one of the two minisatellite regions in the C. muris variant at the CM-MS1 locus, whereas sometimes the repeat sequences were totally different, such as those at the CM-MS3 locus (Table 3).
TABLE 3.
Nature of microsatellite and minisatellite repeats in four genetic loci
| Locus | Repeat(s) |
||
|---|---|---|---|
| C. muris | C. muris variant | C. andersoni | |
| CM-MS1 | (TAAAGGGAGAGA)3 and (GAACGAGATAGG)14-18 | (TAAAGAGAGAGA)6 and (GAACGAGATAGG)14 | (TAAAGGGCGAGA)3 and (GAACGAGATAGG)13-14 |
| CM-MS2 | (CCATATCCC)3-4 and (CCATACCTC)3 | (CCCATTCCT)4 | (CCATACCTC)10-11 |
| CM-MS3 | (TGTTGG)8-9 and (GCTGCA)6 | (TGTTGG)7 and (TGC)10 | (TGTTGGTGTTGCTGT)2 and (TGCTGCAGCTGC)2-3 |
| CM-MS16 | (CTTCTTCAT)9-12 | (CTTCTTCAT)8 | (CTTCTTCAT)14 |
Multilocus sequence subtypes.
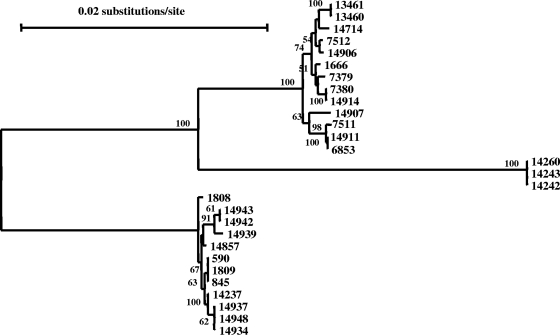
Altogether, there were 10, 5, 5, and 6 subtypes for C. muris and 2, 3, 4, and 1 subtypes for C. andersoni at the CM-MS1, CM-MS2, CM-MS3, and CM-MS16 loci, respectively (Table 1). A total of 16 C. muris specimens and 12 C. andersoni specimens were subtyped successfully at all four genetic loci, forming 11 C. muris and 7 C. andersoni multilocus sequence typing (MLST) subtypes. Most of the MLST subtypes had only one specimen, with the exception of four C. muris MLST subtypes and three C. andersoni MLST subtypes, which had two or three specimens (Table 1). A neighbor-joining tree was constructed with concatenated sequences from the four genes. The tree topology obtained was identical to that obtained with sequences from individual loci, with the formation of three distinct groups (Fig. 3).
FIG. 3.
Phylogenetic relationships among subtypes of C. muris, the C. muris variant, and C. andersoni as assessed by a neighbor-joining analysis of the concatenated nucleotide sequences of four microsatellite and minisatellite loci (MS1, MS2, MS3, and MS16), using distance calculated by the Kimura two-parameter model.
DISCUSSION
In this study, an MLST tool targeting microsatellite and minisatellite sequences was developed for C. muris and C. andersoni. This tool allowed the identification of at least 11 MLST subtypes of C. muris and 7 MLST subtypes of C. andersoni. Unlike what was previously observed for C. hominis and C. parvum (9, 10, 12), the sequence polymorphism in C. muris and C. andersoni was largely in the form of differences in the copy numbers of the microsatellite and minisatellite repeats. In contrast, both C. hominis and C. parvum have extensive single-nucleotide substitutions in the nonrepeat regions of most microsatellite and minisatellite targets. The coding nature of the targets was probably not responsible for the differences observed between the gastric and intestinal Cryptosporidium spp., as most microsatellites and minisatellites in Cryptosporidium occur in coding regions of protein genes because of the presence of few introns and short intergenic regions as the result of a compact genome. This difference might be a reflection of intrinsic biological and genetic differences between gastric and intestinal Cryptosporidium species, as indicated by previous data on the phylogenetic relationships and G/C contents of the SSU rRNA genes between the two groups of Cryptosporidium spp. (42).
Two groups of C. muris were identified in this study. Most C. muris specimens had sequences similar to each other at the four genetic loci examined, with differences only in the copy number of microsatellite and minisatellite repeats. They formed a cluster in the phylogenetic analysis. In contrast, three C. muris isolates from the Czech Republic, including an isolate (TS03) that originated from an East African mole rat (Tachyoryctes splendens) and was maintained in Mastomys coucha, had sequences very different from those of most C. muris specimens. This isolate was previously shown to have different infectivity and/or host specificity in experimental animal models (23). It was also shown previously that the East African mole rat C. muris isolate had sequence differences in the SSU rRNA gene that were comparable to those between C. andersoni and most C. muris isolate. Surprisingly, two other C. muris isolates maintained in laboratory animals by the same research group, RN66 and Bactrian camel isolate CB03, also had sequences identical to those of the East African mole rat C. muris isolate. The genetic similarity of the three C. muris isolates was also confirmed by the sequencing of the SSU rRNA gene, which produced sequences identical to the one from the East African mole rat isolate. A contamination of C. muris isolates could have happened during the animal passage of C. muris isolates RN66 and CB03. The substantial sequence differences between the C. muris East African mole rat isolate and other C. muris isolates in the SSU rRNA gene and four loci in this study indicate that the East African mole rat isolate could present a different species. A parasite identical to the C. muris East African mole rat isolate was previously identified in an eastern gray squirrel in New York (6), suggesting that this parasite is probably widespread. A C. muris isolate from Japanese field mice was also shown to have minor differences in mouse infectivity and the sequence of the SSU rRNA gene from other C. muris isolates (16). Thus, genetic and biological diversities exist in C. muris, and with the inclusion of more genetic loci and samples from a wide range of hosts and geographical areas, the MLST approach developed in this study should be useful in elucidating the genetic basis for the difference in host specificity among C. muris isolates and in examining the spread of the parasite in geographically isolated areas such as the continent of Australia.
The genetic diversity of C. andersoni appears to be much lower than that of C. muris. Only three of the four loci examined in this study were polymorphic in C. andersoni, and only 2 to 4 subtypes of C. andersoni were seen at each polymorphic locus. The low genetic diversity of C. andersoni in comparison with that of C. muris is expected, as the domestication of cattle is a recent event. Thus, modern cattle are thought to have originated from a few places in the Near East and Europe and introduced to the rest of the world during the last 15,000 years (2, 14). The narrow host specificity of C. andersoni has probably further reduced its genetic diversity. The recent introduction of C. andersoni into many areas is supported by the finding of one MLST subtype of C. andersoni in the United States, Canada, and the Czech Republic (Table 2 and Fig. 3). In contrast, rodents are abundant and widespread in distribution and consist of numerous species living in diverse ecological niches. The broad host specificity of C. muris and the geographical isolation of some rodent species have probably led to the emergence of host-adapted subtypes, as seen for the better-known C. parvum (41). Nevertheless, biological differences are known to exist in C. andersoni. Isolates of C. andersoni in Japan, the so-called strain Kawatabi, differ from C. andersoni isolates in other areas in its ability to infect SCID mice (28). The genetic difference between the two biological types of C. andersoni is not yet clear. Again, the inclusion of more genetic loci and a large number of samples from different geographical areas are needed before we can have a better understanding of the geographical spread of C. andersoni and genetic determinants of host specificity.
Among the 18 MLST subtypes identified, 7 were found in multiple specimens. The number of specimens with complete data for all four loci was limited by the fact that some DNA preparations were not amplified at all loci, especially in the initial PCR analysis of the CM-MS3 locus. The reason for the poor initial PCR amplification (only 24 of 44 produced the expected PCR product) of CM-MS3 was due largely to sequencing errors in the primary reverse primer region in the original C. muris sequence downloaded from the C. muris genome sequencing project. The primer sequence used was 5′-TTGCTTTAAGTGTAGAGCATAGAA-3′. A comparison with the corrected sequence (GenBank accession no. XM_002142635) recently downloaded from the GenBank database indicated that the primers had three nucleotide errors, and the corrected primer sequence should be 5′-TTGCTTTAAGTGTAAATAATACAA-3′. Because the correct primer sequence had a much lower annealing temperature, a new primary reverse primer was designed using a sequence 17 nucleotides downstream of the original location: 5′-TCAAGTACAGCAGTCTATTGCTT-3′. It is not clear whether the poor amplification efficiency of other loci was also caused by the sequencing errors in the genome sequencing project. More microsatellite and minisatellite targets may also be needed to increase the differentiation power of the MLST tool.
In conclusion, an MLST tool for subtyping C. muris and C. andersoni was developed. With further refinement, especially the inclusion of more loci, the tool should be useful for the characterization of the population genetics and the dispersal of the two parasites and especially the potential role of either host species or geography in genetic structuring. It should also be useful for the epidemiological investigation of cryptosporidiosis outbreaks caused by C. muris in some animals (26) and the public health significance of parasites of animal origin. These studies should analyze a larger number of specimens from more diverse regions and assess the relationship among MLST subtypes, host specificity, virulence or clinical presentations, and risk factors.
Acknowledgments
We thank Wangeci Gatei, Mosaad Hilali, and Carol Palmer for providing specimens.
This work was supported in part by the Shanghai Science and Technology Committee (grant no. 09540704400), the National Natural Science Foundation of China (grant no. 30771881 and 30928019), and Fundamental Research Funds for the Central Universities, China (grant no. WB0914044).
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Footnotes
Published ahead of print on 27 October 2010.
REFERENCES
- 1.Al-Brikan, F. A., H. S. Salem, N. Beeching, and N. Hilal. 2008. Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J. Egypt. Soc. Parasitol. 38:645-658. [PubMed] [Google Scholar]
- 2.Beja-Pereira, A., D. Caramelli, C. Lalueza-Fox, C. Vernesi, N. Ferrand, A. Casoli, F. Goyache, L. J. Royo, S. Conti, M. Lari, A. Martini, L. Ouragh, A. Magid, A. Atash, A. Zsolnai, P. Boscato, C. Triantaphylidis, K. Ploumi, L. Sineo, F. Mallegni, P. Taberlet, G. Erhardt, L. Sampietro, J. Bertranpetit, G. Barbujani, G. Luikart, and G. Bertorelle. 2006. The origin of European cattle: evidence from modern and ancient DNA. Proc. Natl. Acad. Sci. U. S. A. 103:8113-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubey, J. P., J. E. Markovits, and K. A. Killary. 2002. Cryptosporidium muris-like infection in stomach of cynomolgus monkeys (Macaca fascicularis). Vet. Pathol. 39:363-371. [DOI] [PubMed] [Google Scholar]
- 4.Fayer, R., M. Santin, and J. M. Trout. 2007. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet. Parasitol. 145:260-266. [DOI] [PubMed] [Google Scholar]
- 5.Fayer, R., M. Santin, J. M. Trout, and E. Greiner. 2006. Prevalence of species and genotypes of Cryptosporidium found in 1-2-year-old dairy cattle in the eastern United States. Vet. Parasitol. 135:105-112. [DOI] [PubMed] [Google Scholar]
- 6.Feng, Y., K. A. Alderisio, W. Yang, L. A. Blancero, W. G. Kuhne, C. A. Nadareski, M. Reid, and L. Xiao. 2007. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 73:6475-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, Y., Y. Ortega, G. He, P. Das, M. Xu, X. Zhang, R. Fayer, W. Gatei, V. Cama, and L. Xiao. 2007. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 144:1-9. [DOI] [PubMed] [Google Scholar]
- 8.Gatei, W., R. W. Ashford, N. J. Beeching, S. K. Kamwati, J. Greensill, and C. A. Hart. 2002. Cryptosporidium muris infection in an HIV-infected adult, Kenya. Emerg. Infect. Dis. 8:204-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gatei, W., D. Barrett, J. F. Lindo, D. Eldemire-Shearer, V. Cama, and L. Xiao. 2008. Unique Cryptosporidium population in HIV-infected persons, Jamaica. Emerg. Infect. Dis. 14:841-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gatei, W., P. Das, P. Dutta, A. Sen, V. Cama, A. A. Lal, and L. Xiao. 2007. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect. Genet. Evol. 7:197-205. [DOI] [PubMed] [Google Scholar]
- 11.Gatei, W., J. Greensill, R. W. Ashford, L. E. Cuevas, C. M. Parry, N. A. Cunliffe, N. J. Beeching, and C. A. Hart. 2003. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J. Clin. Microbiol. 41:1458-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gatei, W., C. A. Hart, R. H. Gilman, P. Das, V. Cama, and L. Xiao. 2006. Development of a multilocus sequence typing tool for Cryptosporidium hominis. J. Eukaryot. Microbiol. 53:S43-S48. [DOI] [PubMed] [Google Scholar]
- 13.Gatei, W., C. N. Wamae, C. Mbae, A. Waruru, E. Mulinge, T. Waithera, S. M. Gatika, S. K. Kamwati, G. Revathi, and C. A. Hart. 2006. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am. J. Trop. Med. Hyg. 75:78-82. [PubMed] [Google Scholar]
- 14.Gotherstrom, A., C. Anderung, L. Hellborg, R. Elburg, C. Smith, D. G. Bradley, and H. Ellegren. 2005. Cattle domestication in the Near East was followed by hybridization with aurochs bulls in Europe. Proc. Biol. Sci. 272:2345-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hikosaka, K., and Y. Nakai. 2005. A novel genotype of Cryptosporidium muris from large Japanese field mice, Apodemus speciosus. Parasitol. Res. 97:373-379. [DOI] [PubMed] [Google Scholar]
- 17.Hurkova, L., O. Hajdusek, and D. Modry. 2003. Natural infection of Cryptosporidium muris (Apicomplexa: Cryptosporiidae) in Siberian chipmunks. J. Wildl. Dis. 39:441-444. [DOI] [PubMed] [Google Scholar]
- 18.Iseki, M., T. Maekawa, K. Moriya, S. Uni, and S. Takada. 1989. Infectivity of Cryptosporidium muris (strain RN 66) in various laboratory animals. Parasitol. Res. 75:218-222. [DOI] [PubMed] [Google Scholar]
- 19.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohno. 2000. Possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 20.Kodadkova, A., M. Kvac, O. Ditrich, B. Sak, and L. Xiao. 2010. Cryptosporidium muris in a reticulated giraffe (Giraffa camelopardalis reticulata). J. Parasitol. 96:211-212. [DOI] [PubMed] [Google Scholar]
- 21.Koudela, B., D. Modry, and J. Vitovec. 1998. Infectivity of Cryptosporidium muris isolated from cattle. Vet. Parasitol. 76:181-188. [DOI] [PubMed] [Google Scholar]
- 22.Kvac, M., D. Hanzlikova, B. Sak, and D. Kvetonova. 2009. Prevalence and age-related infection of Cryptosporidium suis, C. muris and Cryptosporidium pig genotype II in pigs on a farm complex in the Czech Republic. Vet. Parasitol. 160:319-322. [DOI] [PubMed] [Google Scholar]
- 23.Kvac, M., B. Sak, D. Kvetonova, O. Ditrich, L. Hofmannova, D. Modry, J. Vitovec, and L. Xiao. 2008. Infectivity, pathogenicity, and genetic characteristics of mammalian gastric Cryptosporidium spp. in domestic ruminants. Vet. Parasitol. 153:363-367. [DOI] [PubMed] [Google Scholar]
- 24.Leoni, F., C. Amar, G. Nichols, S. Pedraza-Diaz, and J. McLauchlin. 2006. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J. Med. Microbiol. 55:703-707. [DOI] [PubMed] [Google Scholar]
- 25.Lindsay, D. S., S. J. Upton, D. S. Owens, U. M. Morgan, J. R. Mead, and B. L. Blagburn. 2000. Cryptosporidium andersoni n. sp. (Apicomplexa: Cryptosporiidae) from cattle, Bos taurus. J. Eukaryot. Microbiol. 47:91-95. [DOI] [PubMed] [Google Scholar]
- 26.Lupo, P. J., R. C. Langer-Curry, M. Robinson, P. C. Okhuysen, and C. L. Chappell. 2008. Cryptosporidium muris in a Texas canine population. Am. J. Trop. Med. Hyg. 78:917-921. [PubMed] [Google Scholar]
- 27.Lv, C., L. Zhang, R. Wang, F. Jian, S. Zhang, C. Ning, H. Wang, C. Feng, X. Wang, X. Ren, M. Qi, and L. Xiao. 2009. Prevalence and molecular characterization of Cryptosporidium spp. in wild, laboratory, and pet rodents in China. Appl. Environ. Microbiol. 75:7692-7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsubayashi, M., I. Kimata, M. Iseki, T. Hajiri, H. Tani, K. Sasai, and E. Baba. 2005. Infectivity of a novel type of Cryptosporidium andersoni to laboratory mice. Vet. Parasitol. 129:165-168. [DOI] [PubMed] [Google Scholar]
- 29.Morgan, U. M., L. Xiao, P. Monis, I. Sulaiman, I. Pavlasek, B. Blagburn, M. Olson, S. J. Upton, N. V. Khramtsov, A. Lal, A. Elliot, and R. C. Thompson. 2000. Molecular and phylogenetic analysis of Cryptosporidium muris from various hosts. Parasitology 120:457-464. [DOI] [PubMed] [Google Scholar]
- 30.Morse, T. D., R. A. Nichols, A. M. Grimason, B. M. Campbell, K. C. Tembo, and H. V. Smith. 2007. Incidence of cryptosporidiosis species in paediatric patients in Malawi. Epidemiol. Infect. 135:1307-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muthusamy, D., S. S. Rao, S. Ramani, B. Monica, I. Banerjee, O. C. Abraham, D. C. Mathai, B. Primrose, J. Muliyil, C. A. Wanke, H. D. Ward, and G. Kang. 2006. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J. Clin. Microbiol. 44:632-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ng, J., I. Pavlasek, and U. Ryan. 2006. Identification of novel Cryptosporidium genotypes from avian hosts. Appl. Environ. Microbiol. 72:7548-7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palmer, C. J., L. Xiao, A. Terashima, H. Guerra, E. Gotuzzo, G. Saldias, J. A. Bonilla, L. Zhou, A. Lindquist, and S. J. Upton. 2003. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg. Infect. Dis. 9:1174-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavlasek, I., and U. Ryan. 2007. The first finding of a natural infection of Cryptosporidium muris in a cat. Vet. Parasitol. 144:349-352. [DOI] [PubMed] [Google Scholar]
- 35.Plutzer, J., and P. Karanis. 2009. Genetic polymorphism in Cryptosporidium species: an update. Vet. Parasitol. 165:187-199. [DOI] [PubMed] [Google Scholar]
- 36.Ryan, U., L. Xiao, C. Read, L. Zhou, A. A. Lal, and I. Pavlasek. 2003. Identification of novel Cryptosporidium genotypes from the Czech Republic. Appl. Environ. Microbiol. 69:4302-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santin, M., B. R. Dixon, and R. Fayert. 2005. Genetic characterization of Cryptosporidium isolates from ringed seals (Phoca hispida) in Northern Quebec, Canada. J. Parasitol. 91:712-716. [DOI] [PubMed] [Google Scholar]
- 38.Santin, M., J. M. Trout, J. A. Vecino, J. P. Dubey, and R. Fayer. 2006. Cryptosporidium, Giardia and Enterocytozoon bieneusi in cats from Bogota (Colombia) and genotyping of isolates. Vet. Parasitol. 141:334-339. [DOI] [PubMed] [Google Scholar]
- 39.Tiangtip, R., and S. Jongwutiwes. 2002. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop. Med. Int. Health 7:357-364. [DOI] [PubMed] [Google Scholar]
- 40.Warren, K. S., R. A. Swan, U. M. Morgan-Ryan, J. A. Friend, and A. Elliot. 2003. Cryptosporidium muris infection in bilbies (Macrotis lagotis). Aust. Vet. J. 81:739-741. [DOI] [PubMed] [Google Scholar]
- 41.Xiao, L. 2010. Molecular epidemiology of cryptosporidiosis: an update. Exp. Parasitol. 124:80-89. [DOI] [PubMed] [Google Scholar]
- 42.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, L., I. M. Sulaiman, U. M. Ryan, L. Zhou, E. R. Atwill, M. L. Tischler, X. Zhang, R. Fayer, and A. A. Lal. 2002. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int. J. Parasitol. 32:1773-1785. [DOI] [PubMed] [Google Scholar]
- 44.Zintl, A., D. Neville, D. Maguire, S. Fanning, G. Mulcahy, H. V. Smith, and T. De Waal. 2007. Prevalence of Cryptosporidium species in intensively farmed pigs in Ireland. Parasitology 134:1575-1582. [DOI] [PubMed] [Google Scholar]